Abstract
Purpose
Lung cancer in never-smokers (LCINS) is increasingly recognized as a distinct disease from that in ever-smokers owing to substantial differences in etiology, clinical characteristics, and prognosis. Therefore, we aimed to identify prognostic markers specific for LCINS.
Experimental Design
First, 11,930 single-nucleotide polymorphisms (SNP) in 904 inflammation-related genes were genotyped, and their associations with overall survival in 411 patients with LCINS at MD Anderson Cancer Center were analyzed. Next, validation of the top 27 SNPs in 311 patients with LCINS at Mayo Clinic was conducted.
Results
Three SNPs (IL17RA:rs879576, BMP8A:rs698141, and STY:rs290229) were validated (P < 0.05), and two SNPs (CD74:rs1056400 and CD38:rs10805347) reached borderline significance (P = 0.08) in the Mayo Clinic population. We validated a survival-tree created in the MD Anderson population exploring gene–gene interactions in the Mayo Clinic population. This survival-tree stratified patients into subsets with significantly different risks of death: patients with the rs1056400_GG/rs698141_GA+AA genotype had significantly higher risk of death in both MD Anderson (HR:2.32, 95%CI: 1.58–3.41) and Mayo (HR:1.97, 95%CI: 1.11–3.50) populations compared with those with the rs1056400_GG/rs698141_GG or rs1056400_GA + AA genotype. We evaluated these five SNPs in 996 ever-smokers from MD Anderson and found no significant associations.
Conclusions
Our study provides strong evidence that inflammation-related genetic variations can affect clinical outcomes in LCINS, which may lead to significant biologic insight into these outcomes.
Introduction
Smoking remains the number one established risk factor for lung cancer. However, 15% of male and 53% of female patients with lung cancer are never smokers worldwide (1, 2). Over the past few decades, the proportion of never-smokers with lung cancer has increased strikingly (2). Previous studies have reported differing tumor etiology and clinicopathologic presentation according to smoking status in patients with lung cancer with never-smokers more likely to be women, having adenocarcinomas, and less-differentiated tumors (1–4). Genetic and epigenetic alterations also differ, with fewer changes overall. Tumors from never-smokers also have a unique and predominant profile compared with those from smokers, such as chromosomal gains at 16p, promoter hypermethylation of hMLH1 and hMLH2, and distinct mutations of major oncogenes and tumor suppressor genes (1–3). These findings suggest different paths of carcinogenesis in ever- and never-smokers with lung cancer. Moreover, studies have repeatedly identified differences in prognosis for lung cancer in ever- and never-smokers (2, 3, 5, 6). Given all of these differences, the identification of specific prognostic and predictive markers for lung cancer in never-smokers (LCINS) beyond the general markers for lung cancer is warranted.
Inflammation is an important cellular process that is activated in response to tissue damage, infections and other cellular stress. There is a well-established relationship between inflammation and cancer with many cancers initiated at the site of inflammation (7, 8). A growing body of evidence supported a relationship between inflammation and cancer. Products of the inflammatory response, such as free oxygen radicals, may induce harmful DNA alterations resulting in carcinogenesis and formation of invasive and/or metastatic phenotypes (9–14). Inflammatory cells and related signaling molecules could also be used by tumor to facilitate its progression and metastasis by generating a favorable microenvironment as well as promoting genetic instability and angiogenesis (10). Moreover, poorer survival was found in patients with cancer with elevated inflammatory markers (15).
The lung is a frequent site of infection and occasional site of chronic inflammation owing to environmental exposures. Furthermore, accumulating evidence shows that inflammation is associated with clinical outcomes of various cancers, including lung cancer (16–19). Previous studies have explored associations between selected inflammation gene polymorphisms and lung cancer prognosis, with inconsistent results potentially due to small sample sizes (20, 21). In the present study, we genotyped a comprehensive panel of inflammation-related single-nucleotide polymorphisms (SNP) in a large number of never-smokers with lung cancer at The University of Texas MD Anderson Cancer Center with comprehensive epidemiological, clinical, and follow-up information to determine the effect of inherited genetic variations on clinical outcome of this cancer. To validate our findings, we studied these SNPs in an independent patient population from Mayo Clinic. To our knowledge, this is the first study to systematically evaluate the associations of genetic variations in major inflammation-related genes with overall survival of LCINS that uses a 2-stage study design.
Materials and Methods
Study population and data collection
Written informed consent to participate in the study was obtained from all participants. The study protocol was approved by the Institutional Review Boards at MD Anderson (Houston, TX) and Mayo Clinic (Rochester, MN). Patients at MD Anderson had newly diagnosed, histologically confirmed non–small cell lung cancer (NSCLC) enrolled in an ongoing lung cancer study initiated in 1995. Demographic information, smoking status and environmental exposures were collected from the patients by trained staff using a structured questionnaire in-person interviews. Patients’ clinical and follow-up data were abstracted from medical records. The primary analysis focused on never-smokers; a group of ever-smokers with lung cancer was also studied to compare the effect of SNPs by smoking status. Patients at Mayo Clinic had newly diagnosed, histopathologically confirmed primary NSCLC who are never smokers. A structured questionnaire was used to collect detailed epidemiologic data on the patients. These patients participated in a long-term follow-up study from 1997 to 2008 described in detail previously (22, 23). All analyses were restricted to Caucasian patients to minimize effects of population structure.
SNP selection
Compilation of the genes involved in the inflammatory response was conducted on the basis of a published panel of inflammation-associated genes (24) and a database of diabetes and inflammation genes [T1DBase (http://www.t1dbase.org); University of Cambridge, Cambridge, UK]. Tagging SNPs for candidate genes based on data from an European population were identified using data from the International HapMap Project, based on National Center for Biotechnology Information B36 assembly and dbSNP b126. For each gene, sequences 10 kb before the transcription start site and 10 kb after the transcription end site were included in the tag SNP selection using the Tagger pairwise method (Broad Institute, Cambridge, MA) with an r2 threshold of 0.8 and minor allele frequency of at least 0.05 (25). The compiled SNP list was sent to Illumina (San Diego, CA) for designability analysis using their array design tool, which uses a proprietary algorithm to predict successful design of the genotyping assay for each SNP. Each SNP is scored from 0 to 1 with higher scores having a higher likelihood of assay success. A score of 0.4 was considered as the cut-off value. Only SNPs that exceeded the threshold score (>0.4) were considered designable. In total, 11,930 SNPs (Supplementary Table S1) were included for construction of an Infinium II iSelect Custom Genotyping BeadChip (Illumina).
Genotyping and quality control
Genomic DNA was extracted from peripheral blood samples obtained from the study patients using a QIAamp DNA Mini Kit (QIAGEN, Valencia, CA). iSelect Custom Genotyping BeadChips were processed according to the Infinium II assay protocol (Illumina). Standard quality control procedures were conducted including only SNPs with genotyping data in more than 95% of all samples and samples with genotyping data for more than 95% of all SNPs. SNPs selected for validation were genotyped at Mayo Clinic’s Genotyping Core Facility using a Fluidigm Dynamic Array (Fluidigm, South San Francisco, CA) and a Human-Hap317 BeadChip (Illumina) according to a standard protocol and using quality control measures.
Statistical analysis
Overall survival, defined as the duration from diagnosis to death of any cause or the last follow-up, was the primary endpoint of our analysis. Never-smokers were defined as patients who had smoked fewer than 100 cigarettes over their lifetimes, ever-smokers are those who had smoked more than 100 cigarettes over their lifetimes. The effect of each SNP on survival was assessed using multivariate Cox proportional hazards regression analysis adjusted for age, sex, clinical stage, and treatment regimen. Kaplan–Meier survival curves and log-rank tests were used to evaluate the effect of variants on the time to death. Multiple testing adjustment was conducted to calculate P-value after adjusting for 11,930 tests in the discovery phase setting a false discovery rate (FDR) of 10% using the “q-value” package in R (26). Between-study heterogeneity for each SNP was examined using the χ2-based Q test (27). A fixed effects model was used to estimate the summarized effect on survival with absence of heterogeneity (P > 0.05). Survival tree analysis was conducted to identify higher-order gene–gene interactions affecting survival using the STREE software (http://c2s2.yale.edu/software/stree/). STREE uses a log-rank statistical method to select optimal and subsequent splits of datasets.
Results
Patient characteristics
In the MD Anderson study, we identified 411 never-smokers with NSCLC (Table 1). Sixty-seven percent of them were women, and adenocarcinoma was the most common histology (77%). The mean age at diagnosis was 61.5 years. The median survival time (MST) was 23.2 months, and the median follow-up time (MFT) was 54.2 months. Most of the cases (77%) were diagnosed at a late stage (stage III/IV). Fifty-three percent of the patients received only chemotherapy, 33% underwent surgery, and 24% received radiotherapy. At the time of the current study, 276 (67%) of the patients had died. In the Mayo Clinic study, 311 never-smokers with NSCLC were identified and included as the validation population (Table 1). The mean age was 61.7 years, with the majority being female (73%). Sixty-one percent of the patients had late-stage disease at diagnosis. Fifty-nine percent of the patients received chemotherapy, 53% underwent surgery, and 25% received radiotherapy. At the time of this study, 59% of the patients had died. Because of the greater proportions of patients with early-stage disease and who had undergone surgery in the Mayo Clinic population than in the MD Anderson population, the MST (44.6 months) and MFT (73.6 months) were longer in the former population than in the latter.
Table 1.
Characteristics of the never-smokers with lung cancer
| Characteristic | Number of patients (%)
|
|
|---|---|---|
| MD Anderson (discovery) | Mayo Clinic (validation) | |
| MST, mo | 23.2 | 44.6 |
| MFT, mo | 54.2 | 73.6 |
| Mean age, y (SD) | 61.5 (13.0) | 61.7 (13.1) |
| Sex | ||
| Male | 135 (33) | 84 (27) |
| Female | 276 (67) | 227 (73) |
| Stage | ||
| I | 93 (23) | 105 (34) |
| II | 15 (4) | 15 (5) |
| III | 91 (22) | 90 (29) |
| IV | 212 (52) | 101 (32) |
| Histology | ||
| Adenocarcinoma | 316 (77) | 213 (68) |
| Squamous cell carcinoma | 28 (7) | 14 (5) |
| Non–small cell carcinoma | 41 (10) | 18 (6) |
| Bronchoalveolar carcinoma | 20 (5) | 11 (4) |
| Other | 6 (1) | 55 (18) |
| Treatment | ||
| Surgery | 135 (33) | 165 (53) |
| Radiotherapy | 100 (24) | 77 (25) |
| Chemotherapy | 218 (53) | 182 (59) |
| Concurrent chemoradiation | 38 (9) | 36 (12) |
| Vital status | ||
| Dead | 276 (67) | 182 (59) |
| Alive | 135 (33) | 129 (41) |
| Total | 411 | 311 |
Main effect of individual SNP on survival in the discovery, replication, and combined analysis
In the discovery phase, after carrying out quality control measures, 11,689 SNPs were included in our analysis. Of these SNPs, 1,538 were significantly associated with overall survival (P < 0.05), with 14 of these variants being significant at the P < 10−4 level. These associations remained significant after removal of patients who reported passive smoking exposures from the analysis.
We selected 27 top SNPs for validation in the Mayo Clinic population. The entire 27 SNPs remained significant after multiple comparisons adjustment in the MD Anderson discovery population (q < 0.1). Eighteen SNPs had a consistent direction of the effect (HR same direction) in both populations (Table 2). Of these 18, 3 SNPs [interleukin 17 receptor A (IL17RA):rs879576, bone morphogenetic protein 8A (BMP8A):rs698141, and spleen tyrosine kinase (SYK):rs290229] in the Mayo population were significant (P < 0.05) with an additional 2 (CD74:rs1056400 and CD38:rs10805347) reaching borderline significance (P < 0.1)
Table 2.
SNPs with the same trend in both the MD Anderson and Mayo Clinic populations
| Position | Gene | SNP | Model | MD Anderson (discovery)
|
Mayo Clinic (validation)
|
Combined analysisb
|
||||
|---|---|---|---|---|---|---|---|---|---|---|
| HR (95% CI)a | P | HR (95% CI)a | P | HR (95% CI)b | P | P-het | ||||
| Chr22:15969246 | IL17RA | rs879576 | DOM | 0.57 (0.41–0.78) | 5.49 × 10−4 | 0.65 (0.44–0.94) | 0.023 | 0.60 (0.47–0.77) | 4.13 × 10−5 | 0.610 |
| Chr9:92674234 | SYK | rs290229 | DOM | 1.58 (1.23–2.03) | 3.03 × 10−4 | 1.43 (1.01–2.02) | 0.046 | 1.53 (1.25–1.87) | 4.15 × 10−5 | 0.635 |
| Chr1:39738348 | BMP8A | rs698141 | DOM | 1.89 (1.33–2.68) | 4.04 × 10−4 | 1.73 (1.03–2.91) | 0.038 | 1.84 (1.37–2.46) | 4.29 × 10−5 | 0.789 |
| Chr9:21397604 | IFNA8 | rs4978115 | REC | 1.79 (1.35–2.36) | 4.39 × 10−5 | 1.24 (0.86–1.79) | 0.240 | 1.56 (1.25–1.95) | 7.43 × 10−5 | 0.123 |
| Chr5:149761820 | CD74 | rs1056400 | DOM | 0.58 (0.43–0.78) | 3.54 × 10−4 | 0.71 (0.48–1.04) | 0.080 | 0.62 (0.49–0.79) | 1.00 × 10−4 | 0.403 |
| Chr9:21403703 | IFNA8 | rs13296822 | REC | 1.83 (1.36–2.47) | 7.76 × 10−5 | 1.23 (0.75–2.04) | 0.412 | 1.65 (1.28–2.13) | 1.36 × 10−4 | 0.188 |
| Chr4:15449937 | CD38 | rs10805347 | REC | 0.45 (0.28–0.72) | 7.93 × 10−4 | 0.55 (0.28–1.07) | 0.080 | 0.48 (0.33–0.70) | 1.75 × 10−4 | 0.616 |
| Chr20:36392996 | BPI | rs5743539 | DOM | 2.77 (1.61–4.77) | 2.45 × 10−4 | 1.59 (0.87–2.92) | 0.134 | 2.16 (1.44–3.25) | 1.90 × 10−4 | 0.184 |
| Chr13:101691503 | FGF14 | rs1336726 | ADD | 0.71 (0.58–0.86) | 5.34 × 10−4 | 0.80 (0.60–1.07) | 0.133 | 0.74 (0.63–0.87) | 2.10 × 10−4 | 0.474 |
| Chr16:86446704 | SLC7A5 | rs4240803 | DOM | 0.66 (0.52–0.84) | 8.39 × 10−4 | 0.82 (0.60–1.12) | 0.219 | 0.72 (0.59–0.87) | 6.78 × 10−4 | 0.290 |
| Chr9:92684769 | SYK | rs1755938 | DOM | 1.63 (1.25–2.14) | 3.68 × 10−4 | 1.17 (0.80–1.73) | 0.417 | 1.47 (1.17–1.83) | 7.17 × 10−4 | 0.168 |
| Chr7:2744970 | GNA12 | rs11971014 | DOM | 0.59 (0.44–0.80) | 4.77 × 10−4 | 0.87 (0.58–1.30) | 0.488 | 0.67 (0.53–0.86) | 1.18 × 10−3 | 0.141 |
| Chr21:33599261 | IL10RB | rs2834178 | DOM | 0.66 (0.51–0.84) | 6.96 × 10−4 | 0.86 (0.63–1.18) | 0.363 | 0.73 (0.60–0.88) | 1.19 × 10−3 | 0.178 |
| Chr6:152461219 | ESR1 | rs9341066 | DOM | 1.96 (1.37–2.79) | 2.01 × 10−4 | 1.13 (0.74–1.75) | 0.568 | 1.57 (1.20–2.07) | 1.20 × 10−3 | 0.056 |
| Chr9:92665566 | SYK | rs1572104 | DOM | 0.63 (0.49–0.81) | 3.12 × 10−4 | 0.92 (0.65–1.31) | 0.660 | 0.72 (0.59–0.88) | 1.43 × 10−3 | 0.082 |
| Chr12:6766579 | LAG3 | rs11064386 | DOM | 1.68 (1.26–2.23) | 3.74 × 10−4 | 1.10 (0.76–1.57) | 0.618 | 1.42 (1.14–1.78) | 1.92 × 10−3 | 0.070 |
| Chr5:172137235 | DUSP1 | rs4868204 | DOM | 0.69 (0.53–0.91) | 9.40 × 10−3 | 0.84 (0.58–1.23) | 0.373 | 0.74 (0.59–0.93) | 8.67 × 10−3 | 0.421 |
| Chr7:41713523 | INHBA | rs12532252 | REC | 1.34 (1.00–1.79) | 0.050 | 1.17 (0.79–1.72) | 0.442 | 1.27 (1.01–1.61) | 0.042 | 0.580 |
Abbreviations: ADD, additive model; DOM, dominant model; p-het, P for heterogeneity test; REC, recessive model.
Adjusted according to age, sex, clinical stage, and treatment regimen.
Combined (meta) analysis based on fixed effects model. Boldface: P < 0.1.
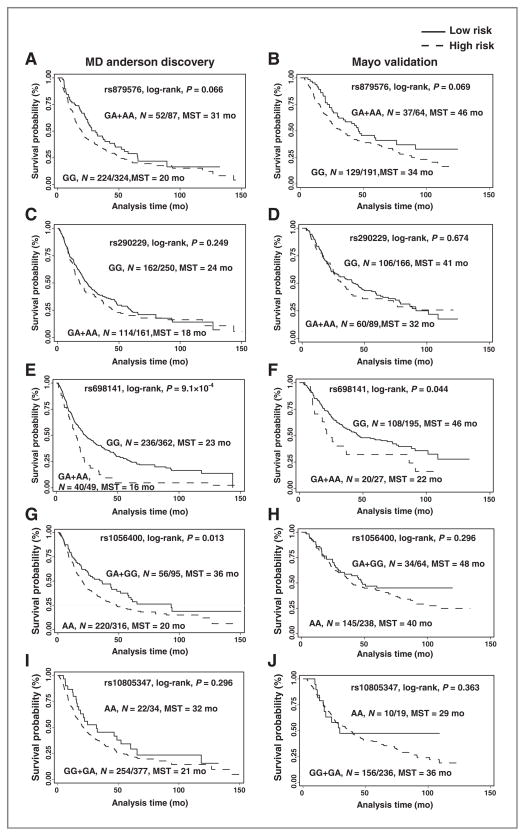
The most significant SNP was rs879576, a synonymous variant in the last exon of the proinflammatory cytokine IL17RA. Rs879576 was associated with a significantly decreased risk of death in the discovery phase [hazard ratio (HR): 0.57; 95% confidence interval (CI) 0.41–0.78; P = 5.49 × 10−4], validation phase (HR: 0.65; 95%CI: 0.44–0.94; P = 0.023) and combined population (HR: 0.60; 95%CI: 0.47–0.77; P = 4.13 × 10−5; Table 2). This decreased risk of dying resulted in enhanced survival duration. Compared with patients with variant genotypes, a prolonged MST was observed in patients with the common homozygous genotype in both discovery (31 vs. 20 months, P =0.066, log-rank test) and validation (46 vs. 34 months, P = 0.069, log-rank test) phases. (Fig. 1A and B)
Figure 1.
Kaplan–Meier estimates of the effect of selected SNPs on survival probability in never-smokers with lung cancer. A, IL17RA: rs879576 in the MD Anderson population (discovery phase). B, IL17RA:rs879576 in the Mayo Clinic population (validation phase). C, SYK:rs290229 in the MD Anderson population (discovery phase). D, SYK:rs290229 in the Mayo Clinic population (validation phase). E, BMP8A:rs698141 in the MD Anderson population (discovery phase). F, BMP8A:rs698141 in the Mayo Clinic (validation phase). G, CD74:rs1056400in the MD Anderson population (discovery phase). H, CD74:rs1056400 in the Mayo Clinic population (validation phase). I, CD38:rs10805347 in the MD Anderson population (discovery phase). J, CD38:rs10805347 in the Mayo Clinic population (validation phase). N = A/B, A: number of patients with event, B: total number of patients.
Rs290229 is an intronic SNP in SYK, a gene that encodes for a nonreceptor type Tyr protein kinase. This SNP was associated with a significantly increased risk of death in both the MD Anderson (HR: 1.58; 95%CI: 1.23–2.03; P = 3.03 × 10−4), Mayo Clinic (HR: 1.43; 95%CI: 1.01–2.02; P = 0.046), and combined (HR: 1.53; 95%CI: 1.25–1.87; P = 4.15 × 10−5) populations. Although not significant, both study populations had the same trend of decreased MST (Fig. 1C and D).
Rs698141 is located in intron of BMP8A, a gene involved in cytokine signaling transduction. Patients who had at least one variant allele had a nearly 2-fold increase in risk of death in both the MD Anderson (HR: 1.89; 95%CI: 1.33–2.68; P = 4.04 × 10−4) and Mayo Clinic (HR: 1.73; 95%CI: 1.03–2.91; P = 0.038) and combined (HR: 1.84; 95%CI: 1.37–2.46; P = 4·29 × 10−5) populations (Table 2). The MST was 23 months in patients with the common homozygous genotype and 16 months in patients with the heterozygous or homozygous variant genotypes in the MD Anderson population (P = 9.1 × 10−4, log-rank test; Fig. 1E). We also observed a similar longer MST (24 months) in the Mayo population (P = 0.044, log-rank test; Fig. 1F).
CD74:rs1056400 (3′-untranslated region) and CD38: rs10805347 (intronic) were significantly associated with an increased risk of death in the MD Anderson population but were borderline significant in the Mayo Clinic population (Table 2). Although not statistically significant, the trend of differing survival times by genotype was observed (Figs. 1G–J).
Stratified analysis of individual SNPs on survival
The majority of never-smokers with lung cancer are female with adenocarcinoma. We conducted a subgroup analysis of survival in female patients with adenocarcinoma. The results were similar to those of the overall analysis of all study patients (Supplementary Table S2). We further conducted a stratified analysis of the 5 top SNPs according to disease stage. Specifically, we combined the MD Anderson and Mayo Clinic patients and stratified them according to early-stage (I and II) and late-stage (III and IV) lung cancer. The results showed that all 5 SNPs were significantly associated with survival in the patients with late-stage, an association that was comparable with or even stronger than that in the overall population. Because of the limited sample size and number of deaths in the patients with early-stage, this association was not as robust. However, the same trend of effect for all 5 SNPs was observed in the patients with early-stage (Supplementary Table S3). In addition, stratified analysis was also conducted in treatment subgroups (chemotherapy only, surgery only, surgery plus chemotherapy, radiation ± chemotherapy) and similar effects were observed (data not shown). To identify potential differences by gender, we conducted a stratified analysis. The results in the female population were similar to the combined population. In male cases, 3 SNPs were not significant in Mayo Clinic population, which might be due to the limited number of male cases (Supplementary Table S4).
Main effects of individual SNPs on survival in ever-smokers
We next analyzed overall survival in the 996 ever-smokers at MD Anderson to assess the effects of the 5 SNPs described earlier on survival according to smoking status. The ever-smokers were slightly older than the never-smokers (mean age, 64.8 years vs. 61.5 years) and had a smaller proportion of women (42% vs. 67%) and adenocarcinoma cases (52% vs. 77%). The treatment regimens in the 2 groups were similar. None of the SNPs validated in the never-smokers were significantly associated with survival in the ever-smokers (Table 3). We further stratified the ever–smoker patients into former and current smokers and did not observe any significant associations within these subgroups (data not shown).
Table 3.
Effect of selected SNPs on survival according to smoking status in the MD Anderson population
| Gene | SNP | Model | Never-smokers
|
Ever-smokers
|
||
|---|---|---|---|---|---|---|
| HR (95% CI)a | P | HR (95% CI)a | P | |||
| SYK | rs290229 | DOM | 1.58 (1.23–2.03) | 3.03 × 10−4 | 1.12 (0.94–1.33) | 0.214 |
| CD74 | rs1056400 | DOM | 0.58 (0.43–0.78) | 3.54 × 10−4 | 0.93 (0.75–1.15) | 0.487 |
| BMP8A | rs698141 | DOM | 1.89 (1.33–2.68) | 4.04 × 10−4 | 1.06 (0.81–1.40) | 0.655 |
| IL17RA | rs879576 | DOM | 0.57 (0.41–0.78) | 5.49 × 10−4 | 0.89 (0.72–1.10) | 0.266 |
| CD38 | rs10805347 | REC | 0.45 (0.28–0.72) | 7.93 × 10−4 | 0.91 (0.67–1.24) | 0.557 |
Abbreviations: DOM, dominant model; REC, recessive model.
Adjusted according to age, sex, clinical stage, and treatment regimen. Boldface: P < 0.1.
Survival tree analysis
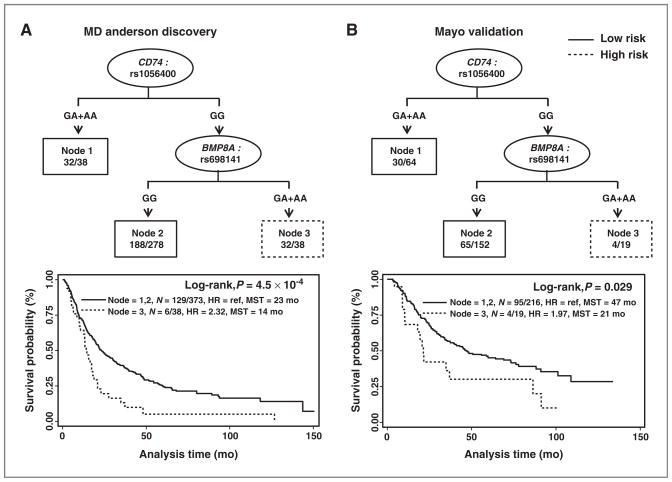
Survival tree analysis was used to identify higher order gene–gene interactions among these 5 SNPs in modulating risk of death. Using the MD Anderson never-smoker population as a training set, we identified 2 SNPs (CD74: rs1056400 and BMP8A:rs698141) potentially having gene–gene interactions. Patients with the rs1056400_GG/rs698141_GA+AA genotype (node 3) had a 2.32-fold greater risk of death (HR: 2.32; 95%CI: 1.58–3.41; P = 1.72 × 10−5) and significantly shorter MST (14 months vs. 23 months; P = 4.5 × 10−4, log-rank test) than did patients with the rs1056400_GG/rs698141_GG or rs1056400_ GA + AA genotype (nodes 1 and 2). This tree model was validated in the Mayo Clinic population: patients with the rs1056400_GG/rs698141_GA+AA genotype (node 3) had a nearly 2-fold greater risk of death (HR: 1.97; 95%CI: 1.11–3.50; P = 0.02) and a strikingly shorter MST (by 26 months) than did patients with the rs1056400_GG/rs698141_GG or rs1056400_GA+AA genotype (nodes 1 and 2; P = 0.029, log-rank test; Fig. 2).
Figure 2.
Potential gene–gene interactions among SNPs validated in the survival tree analysis. A, survival tree analysis results and Kaplan–Meier estimates in the MD Anderson population (discovery phase). B, survival tree analysis results and Kaplan–Meier estimates in the Mayo Clinic population (validation phase). MST, median survival time in months. N = A/B, A: number of patients with event, B: total number of patients.
Discussion
NSCLC in never-smokers is unique from that in ever-smokers due to distinct clinical, histological, and genetic characteristics. These attributes warrant specific investigation of never-smokers. Although we are in the era of the genome-wide association study (GWAS), the coverage of certain genetic region on commercial available GWAS chips is not sufficient for detailed genetic analysis; this limits the power of GWAS to identify all genetic determinants. Thus study design based on prior knowledge focusing on known cancer relations is indispensable. In this context, we conducted a 2-stage, discovery-validation study to identify genetic predictors of overall survival in never-smokers with lung cancer using a pathway-based approach. By systematically evaluating SNPs in major inflammatory pathways, we found 5 SNPs in CD74, CD38, SYK, BMP8A, and IL17RA that were significantly associated with overall survival in these patients. Furthermore, we analyzed and validated a survival tree model in predicting survival that takes gene–gene interactions into consideration. In comparing the associations of SNPs with survival in ever- and never-smokers, we provided evidence of distinct roles for inflammatory genetic determinants of prognosis in never-smokers with lung cancer. Moreover, we conducted a stratified analysis by clinical stage and treatments. The similar results observed in the subgroup analysis indicated the global prognostic role of these top markers regardless of treatments received.
Two SNPs—IL17RA:rs879576 and BMP8A:rs698141—are related to cytokine signaling. IL17RA is an isoform of the interleukin (IL)-17 receptors. In the presence of IL-17 ligands, these receptors can activate various downstream signaling pathways to induce macrophage recruitment, angiogenesis, and inflammatory lung diseases (28, 29). In our study, IL17RA:rs879576 was associated with a consistent protective effect against death and corresponding prolonged MSTs in both the MD Anderson and Mayo Clinic populations. This is a synonymous SNP located in the last exon of IL17RA that may influence the structure and/or regulation of its host gene. BMP8A is a member of the transforming growth factor β superfamily (30). BMP proteins play important roles in cell differentiation, proliferation, survival, and apoptosis and are implicated in tumor cell migration, metastasis, and angiogenesis in various cancers (31–34). Rs698141 is located in the first intron of BMP8A, and not in any obvious functional elements. Therefore, it is most likely linked with other functional SNPs that result in BMP8A altered function. Authors have reported that tobacco smoking can lead to immunosuppression and downregulation of proinflammatory cytokines specifically in the lung tissues, suggesting important roles for cytokines in lung pathology (35). Cytokine signaling pathway variants were predominant in our validated SNPs highlighted the potential roles of cytokines in determining prognosis for LCINS.
SYK belongs to the Syk family of tyrosine kinases and plays an oncogenic role in different cancers (36). In lung cancer cells, SYK is silenced owing to hypermethylation in its promoter region (37). SYK:rs290229 was associated with an increased risk of death and reduced survival in our populations. This SNP is located in an intron; it is possible that this SNP tagged another causal variant that affects the function of SYK. Further deep sequencing would be warranted to identify the potential casual locus responsible for this finding.
Two other SNPs—CD38:rs10805347 and CD74: rs1056400—were borderline significant in our validation. CD74 is a member of a class of polypeptides involved in antigen presentation that is a potential therapeutic target and prognostic factor for cancer (38–41) with involvement in lung adenocarcinoma. Our results suggested the potential prognostic role of CD74:rs1056400 about overall survival in patients with lung cancer. CD38 is a multifunctional single-chain type II transmembrane glycoprotein, related to the development of viral infections, diabetes, and cancer (42). Studies have shown a prognostic role for CD38 in patients with leukemia (43). We observed a consistent protective effect for CD38:rs10805347 against death, which indicated a potential role for this gene in solid cancers in addition to leukemia.
In the current study, we aimed at identifying specific prognostic markers for never smokers. Although incidence is increasing, LCINS represents only approximately 10% of all lung cancer cases. Thus, to identify a homogeneous never-smoking patient cohort with adequate demographic/clinical variables is a challenge. In this study, we were able to identify relatively large and well-characterized study populations from 2 study sites with complete collection of clinical and epidemiologic data that enabled us to recruit a sufficient study population. This provided an important resource contributing to the understanding of this disease, which has emerged as a major public health problem tracking smoking and smoking cessation rate. Interestingly, none of the 5 SNPs were significantly associated with overall survival in ever-smokers, providing additional evidence of LCINSasa distinct disease and requires identifying specific prognostic markers.
Moreover, to control for potential false discoveries, we have adopted both a FDR-based multiple testing adjustment in the discovery phase and an independent external validation, which largely reduced the likelihood of false-positive results. Another significant finding in our study was the identification and validation of a survival tree, which has proven to be a powerful analytic tool about survival in patients with cancer based on higher-order gene–gene interactions (37–39). The survival tree analysis stratified the Mayo Clinic patients into significantly different risk subgroups in a manner similar to that in the MD Anderson patients. Beyond the effect of a single SNP on survival, the survival tree takes into account the complicated interactions of genes, which are yet not discovered and has high predictive power about patients’ prognosis that may be clinically applicable.
In conclusion, this is the first large-scale study to examine the association of SNPs in 800 inflammation-related genes with survival in never-smokers with lung cancer. The identified individual SNPs and the survival tree may be applicable to future modeling of clinical outcome for prediction of survival following validation in other independent populations of never-smokers with lung cancer.
Supplementary Material
Translational Relevance.
Lung cancer in never-smokers (LCINS) is increasingly recognized as a distinct disease from that in ever-smokers owing to substantial differences in etiology, clinical characteristics, and prognosis. Identification of specific prognostic and predictive markers for LCINS beyond the general markers for lung cancer is warranted. Inflammation plays an important role in cancer initiation and progression, as well as influence patients’ clinical outcomes. In this study, we conducted a large-scale two-phase study to identify and validate inflammation-related genetic variations as prognostic markers specific for LCINS using two independent lung cancer patient cohorts. Our study provides strong evidence that inflammation-related genetic variations can affect clinical outcomes in LCINS, which may lead to significant biologic insight into these outcomes and guide personalized treatment and follow-up care regimens for LCINS.
Acknowledgments
Grant Support
This research is supported by National Cancer Institute grants [R01 CA111646, P50 CA070907, and R01 CA055769] and in part by the NIH through MD Anderson’s Cancer Center Support Grant [CA016672].
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors’ Contributions
Conception and design: X. Pu, W.K. Hong, J.D. Minna, P. Yang, X. Wu
Development of methodology: X. Pu, J. Gu, W.K. Hong, P. Yang, X. Wu
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): X. Pu, M.R. Spitz, L. Wang, J.A. Roth, P. Yang, X. Wu
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): X. Pu, Y. Ye, M.R. Spitz, L. Wang, M.A.T. Hildebrandt, P. Yang, X. Wu
Writing, review, and/or revision of the manuscript: X. Pu, Y. Ye, M.R. Spitz, L. Wang, J. Gu, S.M. Lippman, M.A.T. Hildebrandt, W.K. Hong, J.D. Minna, J.A. Roth, P. Yang, X. Wu
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): M.R. Spitz, S.M. Lippman, J.A. Roth, P. Yang, X. Wu
Study supervision: P. Yang, X. Wu
Principal investigator of large program project NCI Special Program of Research Excellence grant that provided part of the support for this research: J.D. Minna
Provided funding: P. Yang, X. Wu
References
- 1.Subramanian J, Govindan R. Lung cancer in never smokers: a review. J Clin Oncol. 2007;25:561–70. doi: 10.1200/JCO.2006.06.8015. [DOI] [PubMed] [Google Scholar]
- 2.Yano T, Miura N, Takenaka T, Haro A, Okazaki H, Ohba T, et al. Never-smoking nonsmall cell lung cancer as a separate entity: clinicopathologic features and survival. Cancer. 2008;113:1012–8. doi: 10.1002/cncr.23679. [DOI] [PubMed] [Google Scholar]
- 3.Bryant A, Cerfolio RJ. Differences in epidemiology, histology, and survival between cigarette smokers and never-smokers who develop non–small cell lung cancer. Chest. 2007;132:185–92. doi: 10.1378/chest.07-0442. [DOI] [PubMed] [Google Scholar]
- 4.Meguid RA, Hooker CM, Harris J, Xu L, Westra WH, Sherwood JT, et al. Long-term survival outcomes by smoking status in surgical and nonsurgical patients with non–small cell lung cancer: comparing never smokers and current smokers. Chest. 2010;138:500–9. doi: 10.1378/chest.08-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 6.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 7.Leibovici D, Grossman HB, Dinney CP, Millikan RE, Lerner S, Wang Y, et al. Polymorphisms in inflammation genes and bladder cancer: from initiation to recurrence, progression, and survival. J Clin Oncol. 2005;23:5746–56. doi: 10.1200/JCO.2005.01.598. [DOI] [PubMed] [Google Scholar]
- 8.Lu H, Ouyang W, Huang C. Inflammation, a key event in cancer development. Mol Cancer Res. 2006;4:221–33. doi: 10.1158/1541-7786.MCR-05-0261. [DOI] [PubMed] [Google Scholar]
- 9.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–99. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–7. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khatami M. Inflammation, aging, and cancer: tumoricidal versus tumorigenesis of immunity: a common denominator mapping chronic diseases. Cell Biochem Biophys. 2009;55:55–79. doi: 10.1007/s12013-009-9059-2. [DOI] [PubMed] [Google Scholar]
- 12.Sorrentino C, Di Carlo E. Expression of IL-32 in human lung cancer is related to the histotype and metastatic phenotype. Am J Respir Crit Care Med. 2009;180:769–79. doi: 10.1164/rccm.200903-0400OC. [DOI] [PubMed] [Google Scholar]
- 13.Gocheva V, Wang HW, Gadea BB, Shree T, Hunter KE, Garfall AL, et al. IL-4 induces cathepsin protease activity in tumor-associated macrophages to promote cancer growth and invasion. Genes Dev. 2010;24:241–55. doi: 10.1101/gad.1874010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeng L, O’Connor C, Zhang J, Kaplan AM, Cohen DA. IL-10 promotes resistance to apoptosis and metastatic potential in lung tumor cell lines. Cytokine. 2010;49:294–302. doi: 10.1016/j.cyto.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 15.Pierce BL, Ballard-Barbash R, Bernstein L, Baumgartner RN, Neuhouser ML, Wener MH, et al. Elevated biomarkers of inflammation are associated with reduced survival among breast cancer patients. J Clin Oncol. 2009;27:3437–44. doi: 10.1200/JCO.2008.18.9068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Proctor MJ, Morrison DS, Talwar D, Balmer SM, O’Reilly DS, Foulis AK, et al. An inflammation-based prognostic score (mGPS) predicts cancer survival independent of tumour site: a Glasgow Inflammation Outcome Study. Br J Cancer. 2011;104:726–34. doi: 10.1038/sj.bjc.6606087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishizuka M, Nagata H, Takagi K, Horie T, Kubota K. Inflammation-based prognostic score is a novel predictor of postoperative outcome in patients with colorectal cancer. Ann Surg. 2007;246:1047–51. doi: 10.1097/SLA.0b013e3181454171. [DOI] [PubMed] [Google Scholar]
- 18.Hara M, Matsuzaki Y, Shimuzu T, Tomita M, Ayabe T, Enomoto Y, et al. Preoperative serum C-reactive protein level in non–small cell lung cancer. Anticancer Res. 2007;27:3001–4. [PubMed] [Google Scholar]
- 19.O’Dowd C, McRae LA, McMillan DC, Kirk A, Milroy R. Elevated preoperative C-reactive protein predicts poor cancer specific survival in patients undergoing resection for non–small cell lung cancer. J Thorac Oncol. 2010;5:988–92. doi: 10.1097/JTO.0b013e3181da78f9. [DOI] [PubMed] [Google Scholar]
- 20.Minamiya Y, Miura M, Hinai Y, Saito H, Ito M, Imai K, et al. The CRP 1846T/T genotype is associated with a poor prognosis in patients with non–small cell lung cancer. Tumour Biol. 2010;31:673–9. doi: 10.1007/s13277-010-0086-9. [DOI] [PubMed] [Google Scholar]
- 21.Pine SR, Mechanic LE, Ambs S, Bowman ED, Chanock SJ, Loffredo C, et al. Lung cancer survival and functional polymorphisms in MBL2, an innate-immunity gene. J Natl Cancer Inst. 2007;99:1401–9. doi: 10.1093/jnci/djm128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang P, Allen MS, Aubry MC, Wampfler JA, Marks RS, Edell ES, et al. Clinical features of 5,628 primary lung cancer patients: experience at Mayo Clinic from 1997 to 2003. Chest. 2005;128:452–62. doi: 10.1378/chest.128.1.452. [DOI] [PubMed] [Google Scholar]
- 23.Li Y, Sheu CC, Ye Y, de Andrade M, Wang L, Chang SC, et al. Genetic variants and risk of lung cancer in never smokers: a genome-wide association study. Lancet Oncol. 2010;11:321–30. doi: 10.1016/S1470-2045(10)70042-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loza MJ, McCall CE, Li L, Isaacs WB, Xu J, Chang BL. Assembly of inflammation-related genes for pathway-focused genetic analysis. PLoS One. 2007;2:e1035. doi: 10.1371/journal.pone.0001035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–23. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 26.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–58. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 28.Kolls JK, Linden A. Interleukin-17 family members and inflammation. Immunity. 2004;21:467–76. doi: 10.1016/j.immuni.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 29.Zhou Y, Toh ML, Zrioual S, Miossec P. IL-17A versus IL-17F induced intracellular signal transduction pathways and modulation by IL-17RA and IL-17RC RNA interference in AGS gastric adenocarcinoma cells. Cytokine. 2007;38:157–64. doi: 10.1016/j.cyto.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 30.Katoh Y, Katoh M. Comparative integromics on BMP/GDF family. Int J Mol Med. 2006;17:951–5. [PubMed] [Google Scholar]
- 31.Ye L, Bokobza SM, Jiang WG. Bone morphogenetic proteins in development and progression of breast cancer and therapeutic potential (review) Int J Mol Med. 2009;24:591–7. doi: 10.3892/ijmm_00000269. [DOI] [PubMed] [Google Scholar]
- 32.Singh A, Morris RJ. The Yin and Yang of bone morphogenetic proteins in cancer. Cytokine Growth Factor Rev. 2010;21:299–313. doi: 10.1016/j.cytogfr.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Corey E, Vessella RL. Bone morphogenetic proteins and prostate cancer: evolving complexities. J Urol. 2007;178:750–1. doi: 10.1016/j.juro.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 34.Ye L, Lewis-Russell JM, Kyanaston HG, Jiang WG. Bone morphogenetic proteins and their receptor signaling in prostate cancer. Histol Histopathol. 2007;22:1129–47. doi: 10.14670/HH-22.1129. [DOI] [PubMed] [Google Scholar]
- 35.Chen H, Cowan MJ, Hasday JD, Vogel SN, Medvedev AE. Tobacco smoking inhibits expression of proinflammatory cytokines and activation of IL-1R-associated kinase, p38, and NF-kappaB in alveolar macrophages stimulated with TLR2 and TLR4 agonists. J Immunol. 2007;179:6097–106. doi: 10.4049/jimmunol.179.9.6097. [DOI] [PubMed] [Google Scholar]
- 36.Medves S, Demoulin JB. Tyrosine kinase gene fusions in cancer: translating mechanisms into targeted therapies. J Cell Mol Med. 2012;16:237–48. doi: 10.1111/j.1582-4934.2011.01415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma L, Dong S, Zhang P, Xu N, Yan H, Liu H, et al. The relationship between methylation of the Syk gene in the promoter region and the genesis of lung cancer. Clin Lab. 2010;56:407–16. [PubMed] [Google Scholar]
- 38.Nagata S, Jin YF, Yoshizato K, Tomoeda M, Song M, Iizuka N, et al. CD74 is a novel prognostic factor for patients with pancreatic cancer receiving multimodal therapy. Ann Surg Oncol. 2009;16:2531–8. doi: 10.1245/s10434-009-0532-3. [DOI] [PubMed] [Google Scholar]
- 39.Borghese F, Clanchy FI. CD74: an emerging opportunity as a therapeutic target in cancer and autoimmune disease. Expert Opin Ther Targets. 2011;15:237–51. doi: 10.1517/14728222.2011.550879. [DOI] [PubMed] [Google Scholar]
- 40.Stein R, Mattes MJ, Cardillo TM, Hansen HJ, Chang CH, Burton J, et al. CD74: a new candidate target for the immunotherapy of B-cell neoplasms. Clin Cancer Res. 2007;13:5556s–63s. doi: 10.1158/1078-0432.CCR-07-1167. [DOI] [PubMed] [Google Scholar]
- 41.Burton JD, Ely S, Reddy PK, Stein R, Gold DV, Cardillo TM, et al. CD74 is expressed by multiple myeloma and is a promising target for therapy. Clin Cancer Res. 2004;10:6606–11. doi: 10.1158/1078-0432.CCR-04-0182. [DOI] [PubMed] [Google Scholar]
- 42.Malavasi F, Deaglio S, Funaro A, Ferrero E, Horenstein AL, Ortolan E, et al. Evolution and function of the ADP ribosyl cyclase/CD38 gene family in physiology and pathology. Physiol Rev. 2008;88:841–86. doi: 10.1152/physrev.00035.2007. [DOI] [PubMed] [Google Scholar]
- 43.Mougalian SS, O’Brien S. Adverse prognostic features in chronic lymphocytic leukemia. Oncology (Williston Park) 2011;25:692–6. 9. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.