Summary
The gene transfer agent produced by Rhodobacter capsulatus (RcGTA) resembles a small tailed bacteriophage that packages almost random genomic DNA segments that may be transferred to other R. capsulatus cells. Gene transfer agents are produced by a number of prokaryotes; however, no receptors have been identified. We investigated the RcGTA recipient capability of wild-type R. capsulatus cells at different culture growth phases, and found that the frequency of RcGTA-dependent acquisition of an allele increases as cultures enter the stationary phase. We also found that RcGTA adsorption to cells follows a similar trend. RcGTA recipient capability and adsorption were found to be dependent on the GtaR/I quorum-sensing (QS) system. Production of an extracellular polysaccharide was found to be regulated by GtaR/I QS, as was production of the cell capsule. A number of QS-regulated putative polysaccharide biosynthesis genes were identified, and mutagenesis of two of these genes, rcc01081 and rcc01932, yielded strains that lack a capsule. Furthermore, these mutants were impaired in RcGTA recipient capability and adsorption, as was a non-encapsulated wild-type isolate of R. capsulatus. Overall, our results indicate that capsular polysaccharide is a receptor for the gene transfer agent of R. capsulatus, RcGTA.
Introduction
The term gene transfer agent (GTA) is used for genetic exchange elements that resemble bacteriophages, but unlike bacteriophages, exclusively package almost random fragments of the host cell genome into a transducing particle (Humphrey et al., 1997; Eiserling et al., 1999; Hynes et al., 2012; Lang et al., 2012). The size of DNA in the particles is smaller than the DNA encoding the GTA structural proteins, thus a single particle is unable to transfer the genes required for its own synthesis to a new host (Lang et al., 2012). This is in contrast to generalized transducing bacteriophage, where the DNA molecule packaged is equivalent in size to the phage genome, and the frequency of packaging of host cell DNA is very low (Ikeda and Tomizawa, 1965). Numerous phylogenetically distinct bacteria produce functional GTA particles, including the Δ-proteobacterium Desulfovibrio desulfuricans (the GTA Dd1) (Rapp and Wall, 1987), the spirochaete Brachyspira hydrosenteriae (VSH-1) (Matson et al., 2005), the archaeon Methanococcus voltae (VTA) (Eiserling et al., 1999), and the α-proteobacterium Rhodobacter capsulatus (RcGTA) (Marrs, 1974; Lang et al., 2012).
A model system for studying GTAs is R. capsulatus RcGTA. This GTA may have evolved from a bacteriophage soon after the α-proteobacteria branched from the other proteobacteria, because RcGTA gene homologues appear to have been inherited vertically as part of the cell genome in a wide variety of species (Lang and Beatty, 2007). The RcGTA resembles a small tailed bacteriophage that packages ~ 4.3 kb of donor cell genomic DNA (Yen et al., 1979) that can be transduced in to an R. capsulatus recipient cell and integrated into genome, most likely through homologous recombination (Genthner and Wall, 1984). The RcGTA structural genes are encoded by a ~ 15 kb gene cluster, ruling out the possibility of self-propagation by a single RcGTA particle (Lang and Beatty, 2000). In addition to the RcGTA main cluster, genes required for regulation, release and infectivity have been found in distant genome locations (Chen et al., 2009; Lang et al., 2012).
Production and release of RcGTA is dependent on a number of factors, including growth phase (Florizone, 2006), phosphate concentration (Westbye et al., manuscript in preparation), and a putative phosphorelay system involving homologues of the Caulobacter crescentus cell cycle regulatory proteins CtrA, ChpT and CckA (Lang and Beatty, 2000; Mercer et al., 2010). An additional regulatory system depends on the GtaR and GtaI quorum-sensing (QS) proteins (Schaefer et al., 2002; Leung et al., 2012). GtaR and GtaI are homologous to LuxR- and LuxI-like proteins (Lazdunski et al., 2004) respectively, where LuxI-like proteins synthesize N-acyl-homoserine lactone (acyl-HSL) signalling molecules, and LuxR-like proteins function as cognate receptors that bind these signals and regulate transcription. LuxI/R-type QS systems regulate a variety of typically group-oriented bacterial behaviours, such as the pathogenicity of bacteria infecting plants (Piper et al., 1993) or humans (Winson et al., 1995; Williams et al., 2000; Bottomley et al., 2007), stem-nodule symbiosis in plants (Daniels et al., 2002; Tun-Garrido et al., 2003), horizontal gene transfer (Piper et al., 1993; Leung et al., 2012), extracellular polysaccharide (EPS) production and biofilm formation (Davies et al., 1998; Gonzalez and Keshavan, 2006; Nadell et al., 2008). Depending on the system, transcription of target genes may be induced or repressed when a threshold concentration of acyl-HSL is reached (Lazdunski et al., 2004). The GtaI protein synthesizes two long-chain acyl-HSLs, C14 and C16-acyl-HSL, either of which causes the induction of RcGTA production (Schaefer et al., 2002), and the production of RcGTA is stimulated by several acyl-HSLs that are not produced by R. capsulatus itself (Leung et al., 2012). The GtaR protein functions as a negative regulator of the gtaRI operon in the absence of acyl-HSLs synthesized by GtaI, and GtaR modulates the production of RcGTA by indirectly repressing RcGTA transcription in the absence of acyl-HSL (Leung, 2010; Leung et al., 2012).
Although the structure, regulation and genetics of RcGTA have been studied for decades, the identity of the bacterial receptor used for RcGTA attachment has been a mystery. Many surface-exposed structures of Gram-negative bacteria, including lipopolysaccharide (LPS), outer membrane proteins, and EPS such as a cell capsule function as phage receptors (Choy et al., 1975; Rakhuba et al., 2010). For example, it was reported that a phage-resistant R. capsulatus strain lacked a capsule, indicating a role of the bacterial capsule in the adsorption of that phage (Flammann and Weckesser, 1984).
In this study, we investigated factors affecting the ability of R. capsulatus to act as a recipient for RcGTA-mediated gene transduction. Through a variety of analyses, we found that that capsule synthesis in R. capsulatus is quorum-sensing regulated, and that the capsule polysaccharide functions in RcGTA adsorption during gene transduction, and so is the first example of a GTA receptor.
Results
RcGTA recipient capability and attachment of RcGTA to R. capsulatus cells are growth phase-dependent
We investigated the effect of growth phase on RcGTA recipient capability of R. capsulatus cells by quantifying the number of cells that received and expressed a rifampicin-resistance marker from a stock solution of RcGTA of known titre, using cells harvested at different time points during culture growth (Fig. 1A). The recipient capability of R. capsulatus cells at early growth phases was much less than those at high cell densities, showing a positive correlation between the optical density (OD) of cultures at time of harvest and recipient capability (Fig. 1B). The recipient capability does not address whether cells in early growth phases lack an RcGTA receptor molecule, the required recombination machinery, or other possible factors required for RcGTA-mediated gene transduction. We investigated the possibility of a receptor deficiency by employing an RcGTA adsorption assay with the same culture samples used as RcGTA recipients. The ability of RcGTA to adsorb to cells was measured by quantifying the transduction efficiency of cell-free (non-adsorbed) RcGTA after incubation with cells. It was found that the ability of cells to adsorb RcGTA increased in tandem with the culture density of the culture at time of harvest, indicating that cells increased their ability to adsorb RcGTA particles in later growth phases (Fig. 1C).
Fig. 1. Effect of growth phase on RcGTA recipient capability and adsorption.
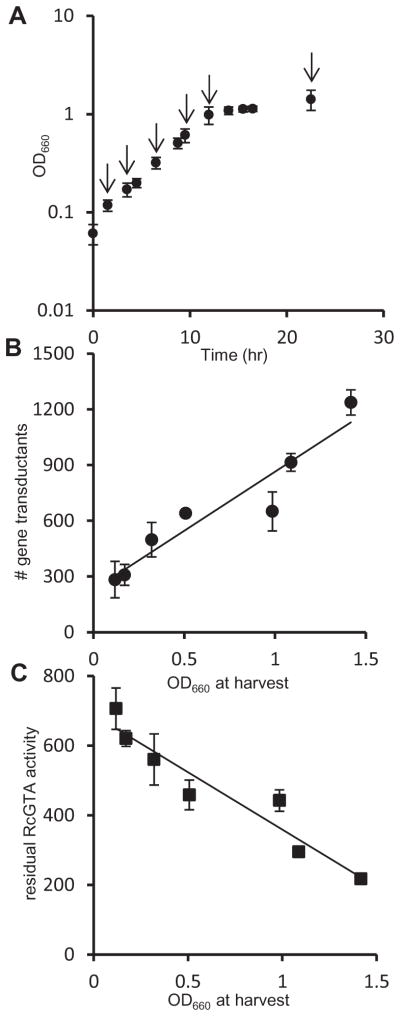
A. Growth curve of R. capsulatus B10 culture used for recipient capability and adsorption experiments. Arrows indicate time points where samples were taken for recipient capability and adsorption experiments. Error bars represent the variation in OD660 between two duplicate cultures.
B. Number of rifampicin-resistant colonies obtained in a transduction assay using equal numbers of cells at different phases of growth as recipients for RcGTA. Error bars represent the variation between two samples (one sample from each culture).
C. RcGTA adsorption to cells at different growth phases. The vertical axis shows the number of rifampicin-resistant colonies derived from an adsorption assay (see Experimental procedures), and the error bars represent the variation between two samples (one sample from each culture). A statistical correlation analysis of the data in Fig. 1B and C is given in Table S2.
These two types of experiment show that the ability of cells to bind RcGTA particles and acquire a new allele is culture growth phase-dependent, and maximal in the stationary phase. By analogy with the attachment of bacteriophage to cell surface-exposed receptors, we hypothesized that a receptor required for RcGTA binding is produced, or becomes available for attachment, as cultures increase in cell concentration.
Effects of mutations in gtaR and gtaI on RcGTA recipient capability and attachment to cells
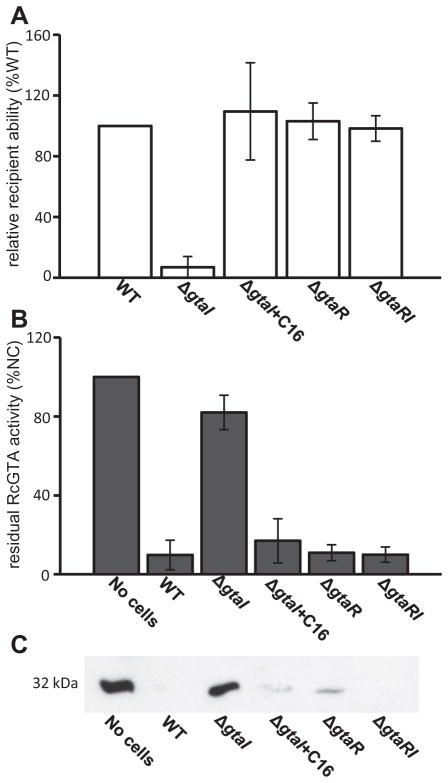
RcGTA production is regulated by the gtaR and gtaI QS genes (Schaefer et al., 2002; Leung et al., 2012). The correlations of the increases in recipient capability and RcGTA cell-adsorption to culture density indicated a possible link to the GtaR/I QS system. To evaluate this possible link, we determined the recipient capability of stationary phase WT, ΔgtaI, ΔgtaR and ΔgtaRI cells, as well as the ΔgtaI mutant supplemented with C16-acyl-HSL. It was found that the ΔgtaI mutant yielded on average 7% as many gene recipients as the WT strain, and recipient capability was restored to 109% of the WT level by the addition of exogenous C16-acyl-HSL (Fig. 2A). Additionally, we observed that the ΔgtaR and ΔgtaRI mutants yielded average recipient frequencies of 103% and 98% of WT respectively (Fig. 2A).
Fig. 2. Comparison of RcGTA recipient capability of QS mutant strains with their ability to adsorb RcGTA particles.
A. Ability of WT, ΔgtaI, ΔgtaR, ΔgtaRI, and ΔgtaI supplemented with C16-acyl-HSL strains to be transduced by RcGTA containing a Rifampicin resistance marker.
B. RcGTA adsorption ability of strains (values are presented as a percentage of the number of RifR transductants obtained in the unabsorbed control where no cells were added to the reaction mixture, indicated by %NC).
C. Western blot of adsorption assay filtrate probed with RcGTA capsid protein antiserum. Error bars represent the standard deviation between samples (n ≥ 3). Statistical analysis was done by One-Way ANOVA, given in Tables S3 and S4.
To determine whether the decrease in the ΔgtaI mutant recipient capability was a result of decreased binding of RcGTA to cells, we performed an adsorption assay on all the cultures evaluated in the recipient capability experiments described above. The ΔgtaI mutant was greatly impaired in the adsorption of RcGTA particles, and adsorption was restored to WT levels by the addition of C16-acyl-HSL (Fig. 2B). Furthermore, WT amounts of RcGTA adsorption were observed for the ΔgtaR and ΔgtaRI mutants, with the amounts of cell-free RcGTA inversely proportional to the recipient capability of these mutants (Fig. 2B). In Western blots probed for the RcGTA capsid protein remaining in the filtrate of the adsorption assay, the amounts of capsid paralleled the residual RcGTA activity values, confirming that the amount of RcGTA had decreased (Fig. 2C). These results indicate that GtaR/I QS regulates the expression of genes encoding a cell surface factor required for RcGTA adsorption to cells.
Analysis of EPS production in gtaR and gtaI mutant strains
Rhodobacter capsulatus
WT strain B10 cells form a loose cell pellet when centrifuged and this pellet is easily dissociated with a single inversion of the tube (Fig. 3A). The ΔgtaI mutant, under the same conditions, produces a tight pellet that requires considerable mixing force to re-suspend. The ΔgtaR mutant has a loose pellet phenotype, as does the ΔgtaRI double mutant (Fig. 3A). These results indicate that one or more genes required for the loose pellet phenotype is negatively regulated by the GtaR protein, because disruption of the gtaR gene does not change this phenotype, whereas mutation of the gtaR gene suppresses the effects of the gtaI mutation.
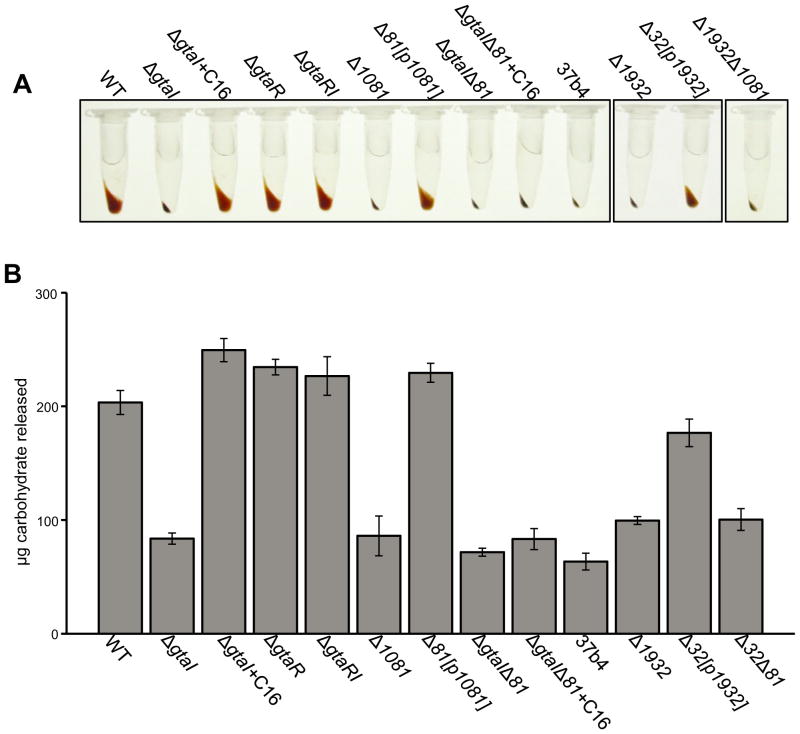
Fig. 3. Evaluation of EPS production in R. capsulatus QS and EPS biosynthesis mutants.
A. Pellet formation of equal numbers of cells of phototrophically grown B10 (WT), ΔgtaI, ΔgtaI + C16-acyl-HSL, ΔgtaR, ΔgtaRI, Δ1081, Δ1081[p1081], ΔgtaIΔ1081, ΔgtaI/Δ1081 + C16-acyl-HSL, Δ1932, Δ1932[p1932], Δ1932/Δ1081 and WT 37b4 R. capsulatus strains after centrifugation for 1 min.
B. Levels of cell-associated EPS production in an extract of each strain as measured by a phenol-sulfuric acid assay. Error bars represent the standard deviation between samples (n = 3), values were calculated based on standard samples with a known amount of a 1:1 mixture of glucose and fructose. Statistical analysis was done by One-Way ANOVA, given in Table S5.
The loose cell pellet persisted after several washes in 50 mM NaCl, indicating that the looseness of the cell pellet is a property due to a substance that is tightly associated with cells (data not shown). To evaluate the causative agent of the loose cell pellet phenotype, WT cells were treated with a selection of enzymes or EDTA. We found that treatment with EDTA, which is known to disrupt the outer membrane of Gram-negative bacteria (Hancock, 1984), or proteinase K, resulted in a tight cell pellet whereas other chemicals and enzymes tested did not have a similar effect (Fig. S1). These results indicate that the loose pellet phenotype is due to a cellular structure attached to the outer membrane of R. capsulatus B10 cells, and it may involve a protein component. In other bacteria, similar cell pellet phenotypes are caused by EPS such as capsular polysaccharide (Ionescu and Belkin, 2009). Because R. capsulatus B10 has a capsule (Weaver et al., 1975), we hypothesized that the loose pellet was caused by the production of an EPS that is part of the capsule.
We roughly quantified the total amount of EPS and LPS in EDTA extracts of cells using a phenol-sulfuric acid method and found that the levels of cell-associated EPS/LPS correlate with the loose pellet phenotype (Fig. 3B). The ΔgtaI strain produced much less EPS/LPS than WT cells, and this production was restored by addition of exogenous C16-acyl-HSL. Furthermore, both the ΔgtaR and ΔgtaRI mutants produced WT levels of EPS/LPS (Fig. 3B). These results indicate that the GtaR/GtaI QS system regulates EPS/LPS production, but do not determine whether this EPS/LPS is the R. capsulatus capsule, or whether the loose cell pellet phenotype relates to the binding of RcGTA.
There was a consistently high background signal when quantifying surface polysaccharides with the phenol-sulfuric acid method. EDTA extracts contain many molecules associated with the outer membrane. Because the phenol-sulfuric acid assay measures total carbohydrate levels, the consistently high background measurements are likely a result of non-capsular polysaccharide (such as LPS) being present in extracts of strains lacking the capsule.
Microarray and bioinformatic identification of putative EPS biosynthesis genes
To identify genes potentially involved in the production of EPS/LPS, we utilized microarrays to evaluate the relative gene expression in the ΔgtaI mutant versus WT cells [NCBI Gene Expression Omnibus (GEO) database Accession No. GSE41014]. A number of potential polysaccharide biosynthesis genes were downregulated in the ΔgtaI mutant, between 2.1- and 3.1-fold in the stationary phase (Table 1). We selected a putative polysaccharide biosynthesis operon, containing open reading frames (ORFs) rcc01081-rcc01086 as candidates for the production of an EPS. The rcc01081-rcc01086 ORFs (Fig. 4) are annotated in the R. capsulatus strain SB1003 genome as two group 1 glycosyltransferases (GTase), an epimerase, two putative membrane proteins, and a group 1 GTase respectively (summarized in Table 2). To more accurately annotate these ORFs, we performed BLASTp searches and hydropathicity analyses (Whitfield, 2006; Moreno-Hagelsieb and Latimer, 2008). The top BLASTp hits to Rcc01081-Rcc01086 include components present in both LPS O-antigen biosynthesis systems and capsular polysaccharide biosynthesis operons (Cuthbertson et al., 2009; Raetz and Whitfield, 2002; Whitfield, 2006). These include three GTases, an epimerase, and genes predicted to encode a polysaccharide flippase (wzx) and a polymerase (wzy) (Table 2). Hydropathicity analysis of the Rcc01081-Rcc01086 proteins lends support to these annotations, notably the Wzx (encoded by rcc01084) and Wzy (rcc01085) protein homologues. Although the amino acid sequence conservation of Wzx and Wzy proteins is often low, a hydropathy profile of multiple transmembrane segments is a strongly conserved characteristic (Whitfield, 2006). The genes we suggest to encode Wzx- and Wzy-like proteins have a hydropathy profile indicating eight to nine transmembrane segments (Table 2 and Fig. S2).
Table 1.
Relative transcription of putative EPS/capsular polysaccharide gene cluster and additional required genes as analysed by microarray analysis.
| Locus | Genome annotation | WT/ΔgtaI loga | WT/ΔgtaI statb | stat/logc |
|---|---|---|---|---|
| rcc01081 | Glycosyl transferase, group 1 | 4.53 | 3.06 | 2.02 |
| rcc01082 | Glycosyl transferase, group 1 | 4.02 | 2.83 | 1.49 |
| rcc01083 | UDP-glucuronate 5′-epimerase (UDP-glucuronic acid epimerase) | 2.59 | 2.16 | 1.51 |
| rcc01084 | Membrane protein, putative | 2.94 | 2.12 | 1.44 |
| rcc01085 | Hypothetical protein | 3.73 | 2.36 | 1.48 |
| rcc01086 | Glycosyl transferase, group 1 | 2.65 | 2.92 | 1.5 |
| rcc01932 | Glycosyl transferase, family 4 | 8.93 | 5.36 | 3.45 |
| rcc01958 | Tyrosine-protein kinase etk | 1.66 | 1.39 | 1.57 |
| rcc01959 | Low molecular weight protein-tyrosine-phosphatase etp | 2.08 | 1.27 | 1.8 |
| rcc01960 | Polysaccharide export protein | 2.67 | 1.75 | 1.01 |
Ratio of WT to ΔgtaI microarray values for cells grown to the exponential phase of growth.
Ratio of WT to ΔgtaI microarray values for cells grown to the stationary phase of growth.
Ratio of microarray values of cells in the stationary phase to cells in the exponential phase.
Values are expressed as the fold change in a gtaI mutant relative to wild type, and log versus stationary phase in the wild type. All cultures grown in RCV defined medium.
Fig. 4.
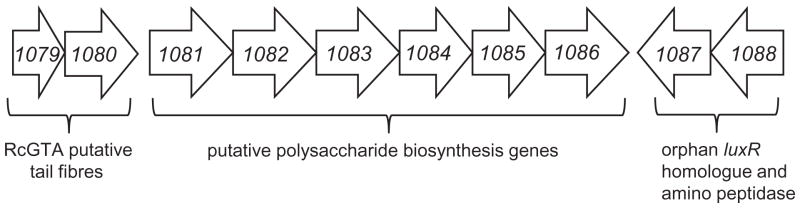
Genomic organization of the rcc01081-1086 gene cluster, and surrounding genes (not to scale). Arrows indicate the direction of transcription. Immediately upstream of rcc01081-1086 are two predicted bacteriophage tail fibre genes, rcc01079 and 1080, and downstream is a putative amino peptidase, rcc01087, and a LuxR homologue, rcc01088.
Table 2.
Bioinformatic analysis of putative EPS/capsular polysaccharide biosynthesis gene cluster and additional related genes.
| Locus | Genome annotation | Best experimentally verified BLASTp hit | % ID | # Predicted TMS | Predicted function |
|---|---|---|---|---|---|
| rcc01081 | Glycosyl transferase, group 1 | PglA; Helicobacter pullorum | 28.7 | 0 | Repeat unit assembly |
| rcc01082 | Glycosyl transferase, group 1 | WffV; Shigella dysenteriae | 39 | 1 | Repeat unit assembly |
| rcc01083 | UDP-glucuronate 5′-epimerase | Gla; Cronobacter malonaticus | 50.4 | 0 | Repeat unit monosaccharide biosynthesis |
| rcc01084 | Membrane protein, putative | Wzx; Cronobacter malonaticus | 21.3 | 8 | Repeat unit flippase |
| rcc01085 | Hypothetical protein | Wzy; Escherichia coli | 22.8 | 9 | Polysaccharide polymerase |
| rcc01086 | Glycosyl transferase, group 1 | WbpR; Pseudomonas aeurginosa | 17.5 | 0 | Repeat unit assembly |
| rcc01932 | Glycosyl transferase, family 4 | WecA; Ralstonia solnacearum | 33.7 | 9 | Repeat unit chain initiation |
| rcc01958 | Tyrosine-protein kinase etk | Wzc; Vibrio vulnificus | 38 | 2 | Polysaccharide export |
| rcc01959 | Low molecular weight protein-tyrosine-phosphatase etp | Wzb; Vibrio vulnificus | 41.1 | 1 | Polysaccharide export |
| rcc01960 | Polysaccharide export protein | Wza; Vibrio vulnificus | 33.4 | 0 | Polysaccharide export |
Data shown include best BLASTp hits, pairwise alignment identities, number of predicted transmembrane segments (TMS) as indicated by a hydropathicity plot (window size = 19), and predicted function in EPS/CPS biosynthesis.
To verify that that these genes are QS-regulated, we measured the promoter activity of an rcc01081::lacZ in-frame fusion plasmid, which contains approximately 1.5 kb of sequences 5′ of the rcc01081 start codon. The WT, ΔgtaI, ΔgtaR, ΔgtaRI strains containing this plasmid were grown phototrophically in RCV defined medium to the stationary phase, and the β-galactosidase specific activity measured. We found that rcc01081::lacZ expression in the ΔgtaI strain was reduced to 25% of the WT activity, and that expression was restored to WT levels upon addition of C16-acyl-HSL to cultures (Fig. S3). Furthermore, both the ΔgtaR and ΔgtaRI strains yielded WT levels of rcc01081::lacZ expression (Fig. S3), verifying that rcc01081 expression is GtaR/GtaI-regulated.
The foregoing analysis prompted us to search the R. capsulatus genome for homologues of the initiating GTases WbaP and WecA, as well as Wzb, Wzc and Wza homologues, which function as machinery for polymer export to the cell surface (Whitfield, 2006; Cuthbertson et al., 2009). A BLASTp search identified ORFs rcc01932, encoding a WecA homologue (34% amino acid identity), and rcc01958-1960, which encode homologues of: Wzc (Rcc01958); Wzb (Rcc01959); and Wza (Rcc01960); (38%, 41% and 33% amino acid identity respectively) (Table 2). The hydropathy profile of Rcc01932 matched the profile expected for this protein (Fig. S2). The rcc01932 gene (encoding the putative initiating GTase, WecA) is of particular interest because it is downregulated under the same conditions as the rcc01081-1086 cluster, and could encode an enzyme catalysing a key step in polysaccharide chain biosynthesis. On the basis of these analyses, we hypothesized that the rcc01081-rcc01086 and rcc01932 genes are involved in capsule production in R. capsulatus, and that this capsular polysaccharide is important for RcGTA adsorption to cells.
Analysis of capsule production and mutagenesis of rcc01081 and rcc01932
Capsule production in the ΔgtaI, ΔgtaR, ΔgtaRI mutants, and ΔgtaI supplemented with C16-acyl-HSL, was evaluated using a capsule stain (Anthony, 1931) and microscopic examination of cells. We found that WT R. capsulatus B10 cells have a clear halo surrounding the cells (Fig. 5C), indicative of the presence of a capsule. The ΔgtaI mutant lacks this halo, indicating the lack of a capsule. Capsule production was restored in the ΔgtaI mutant by addition of C16-acyl-HSL; furthermore, both the ΔgtaR and ΔgtaRI strains produced capsules similar to those of WT cells. These results indicate that the GtaR protein negatively regulates production of the capsule, analogous to the GtaR-regulation of RcGTA production (Leung et al., 2012) and recipient capability.
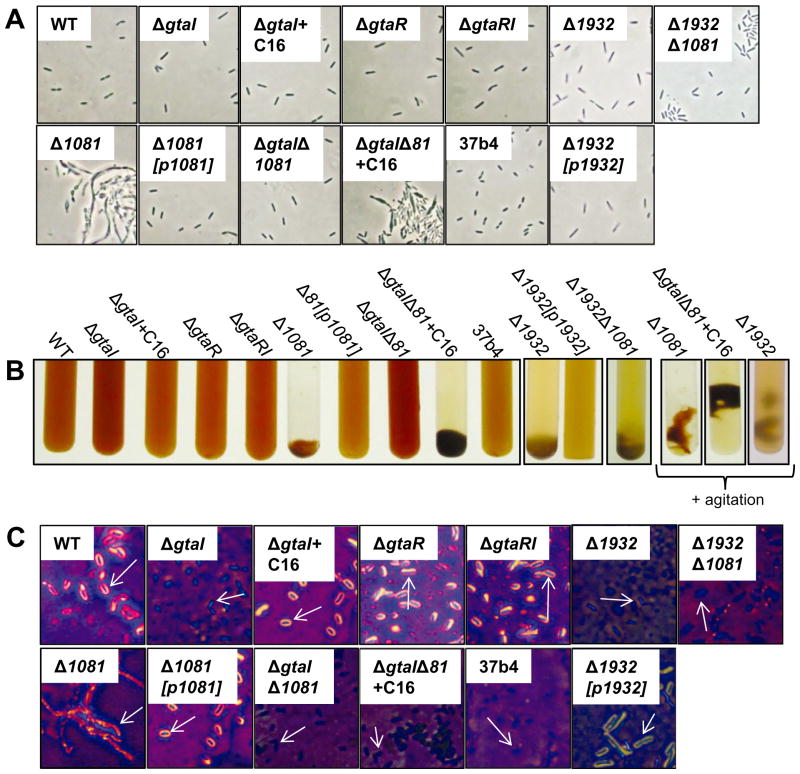
Fig. 5.
Phase contrast microscopy, capsule stain images, and culture macroscopic morphology of WT B10 (WT), ΔgtaI, ΔgtaI + C16-acyl-HSL, ΔgtaR, ΔgtaRI, Δ1081, Δ1081[p1081], ΔgtaI/Δ1081, ΔgtaI/Δ1081 + C16-acyl-HSL, Δ1932, Δ1932[p1932], Δ1932/Δ1081 and WT 37b4 R. capsulatus strains.
A. Phase contrast microscopy images of the strains listed above, at 100× magnification.
B. Images of tubes of phototrophically grown cultures. On the far right, images of agitated cultures of Δ1081, ΔgtaI/Δ1081, and Δ1932 are shown.
C. Capsule stain images of WT, QS and EPS biosynthesis mutant strains.
To test the hypothesis that the ORFs rcc01081-1086 are involved in capsular polysaccharide production, we generated a marker-less, in-frame knockout mutation of the first ORF in the cluster, rcc01081, which is annotated as a group 1 glycosyltransferase. A capsule stain of Δ1081 mutant cells showed that they do not produce an extracellular capsule and the cells are weakly stained with crystal violet/copper sulphate (Fig. 5C). The Δ1081 mutation was observed to cause gross cell defects: cultures form a large aggregate of cells when grown phototrophically in RCV medium, which associate as a floating mass when agitated (Fig. 5B). After centrifugation, the Δ1081 cells form a tight cell pellet similar to that of ΔgtaI cells, and produce a low amount of EPS as measured in the phenol-sulfuric acid assay (Fig. 3A and B). Phase contrast microscopy revealed that Δ1081 cells are irregular in shape, filamentous, and appear as large aggregates, in contrast to the WT morphology (Fig. 5A). The Δ1081 mutant was complemented in trans, and the complemented strain, Δ1081[p1081], was phenotypically similar to the WT in liquid culture growth, cell morphology, formation of a cell pellet, levels of EPS, and production of a capsule (Figs 3 and 5).
The microarray and promoter::lacZ analysis indicated that the rcc01081-1086 gene cluster is downregulated in the ΔgtaI mutant (Table 1 and Fig. S3). If the phenotype of the Δ1081 mutant strain is due to a block in a polysaccharide biosynthesis pathway that is downregulated in the ΔgtaI strain, a ΔgtaI/Δ1081 double mutant would be expected to lack the gross morphological defects of the Δ1081 mutant. We therefore generated a marker-less double mutant ΔgtaI/Δ1081. The cell morphology, growth, pellet formation, levels of EPS and capsule of the double mutant were similar to those of the ΔgtaI mutant in liquid culture (Figs 3 and 5). Furthermore, supplementation of ΔgtaI/Δ1081 cultures with C16-acyl-HSL resulted in a phenotype that was similar to that of a Δ1081 single mutant: cells aggregated, yielded tight pellets, produced a low amount of EPS, and lacked a capsule (Figs 3 and 5).
To evaluate whether rcc01932, encoding the predicted WecA homologue, is involved in capsule production, we generated a marker-less, in-frame knockout of this gene. Capsule stains of Δ1932 mutant cells show that they lack a capsule (Fig. 5C). Additionally, the Δ1932 mutant produces less EPS than the WT strain as measured by the phenol-sulfuric acid method, and forms a tight pellet after centrifugation (Fig. 3). Unlike the Δ1081 mutant, however, Δ1932 cells are morphologically normal when viewed in a microscope (Fig. 5A). When grown phototrophically in RCV defined medium, Δ1932 cells clump to a lesser extent than Δ1081 mutant cells, and the Δ1932 cell mass disintegrates readily with agitation (Fig. 5B). The Δ1932 mutation was complemented in trans in an equivalent fashion to the Δ1081 mutation, and the complemented strain Δ1932[p1932] was phenotypically similar to WT cells (Figs 3 and 5). To provide additional evidence that rcc01932 and rcc01081 function in the same capsule biosynthesis system, a double mutant of rcc01081 and rcc01932 was created. The Δ1932/Δ1081 strain showed a similar phenotype to the Δ1932 single mutant (Figs 3 and 5), indicating that mutation of rcc01932 suppresses the morphological defect of the Δ1081 mutant, and provides evidence that these two genes function in the same capsule biosynthesis system.
To confirm that the foregoing results were consistent with a capsule biosynthesis defect, we compared our mutant strains with R. capsulatus strain 37b4, a non-encapsulated WT isolate (Omar et al., 1983). The 37b4 strain pelleted tightly, produced less EPS than WT B10 cells, and did not have a capsule halo (Figs 3 and 5). The properties of strain 37b4 were essentially identical to the ΔgtaI strain phenotype, supporting the idea that the EPS described here is indeed a capsule polysaccharide.
Recipient capability of and RcGTA attachment to strains Δ1081, Δ1932 and 37b4
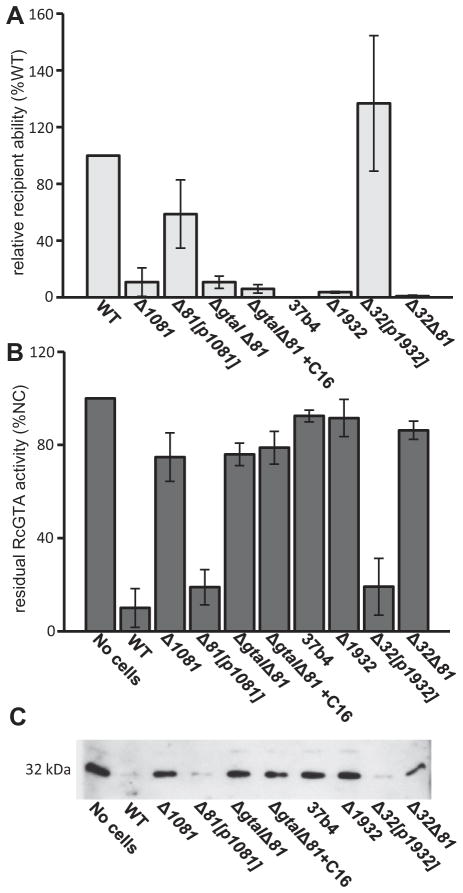
We tested the RcGTA recipient capability and adsorption ability of the strains Δ1081, Δ1081[p1081], ΔgtaI/Δ1081, ΔgtaI/Δ1081 supplemented with C16-acyl-HSL, Δ1932, Δ1932[p1932], Δ1932/Δ1081, and 37b4. It was found that the Δ1081, ΔgtaI/Δ1081, Δ1932, and Δ1932/Δ1081 mutants were greatly impaired in recipient capability, yielding on average 10%, 6%, 4% and 1% respectively, as many transductants as the WT strain B10. This defect was somewhat restored in the Δ1081[p1081] complemented strain, which yielded 58% of the WT level (Fig. 6A), and fully restored in the Δ1932[p1932] strain, which yielded 136% of the WT level. Unlike the ΔgtaI mutant, however, recipient capability was not restored to the ΔgtaI/Δ1081 mutant by addition of C16-acyl-HSL (Fig. 6A), supporting the interpretation that the defect in recipient capability and attachment in the ΔgtaI strain involves downregulation of the rcc01081-1086 genes. Additionally, the considerable reduction in recipient capability of the Δ1932 strain indicates that this gene is also involved in the production of the capsule. This interpretation was confirmed by RcGTA adsorption assays which showed that the Δ1081 and Δ1932, and Δ1932/Δ1081 mutants were impaired in RcGTA attachment. As in the recipient capability experiments, RcGTA-adsorption was not restored in the ΔgtaI/Δ1081 double mutant by the addition of C16-acyl-HSL (Fig. 6B and C). To confirm the interpretation that the capsule is required for RcGTA attachment, we tested the 37b4 strain for recipient capability. Attempts at transduction into 37b4 were unsuccessful, and this strain was effectively unable to adsorb RcGTA (Fig. 6). These observations indicate that production of a capsule polysaccharide requires the rcc01081-1086 and rcc01932 genes, and that the capsule is needed for maximal adsorption of RcGTA – and hence, acquisition of alleles originating in another cell.
Fig. 6. Comparison of WT B10 (WT), Δ1081, Δ1081[p1081], ΔgtaIΔ1081, ΔgtaI/Δ1081 + C16-acyl-HSL, Δ1932, Δ1932[p1932], Δ1932/Δ1081 and 37b4 WT strains recipient capability and ability to adsorb RcGTA particles.
A. Ability of strains to be transduced by RcGTA as measured by the number of rifampicin-resistant colonies arising in a transduction assay.
B. Adsorption ability of strains as measured by residual RcGTA activity in culture filtrate (values are presented as a percentage of the number of RifR transductants obtained in the unabsorbed control where no cells were added to the reaction mixture, indicated by %NC).
C. Western blot adsorption assay of a residual RcGTA sample, probed with RcGTA capsid protein antiserum. Error bars represent the standard deviation between samples (n ≥ 3). Statistical analysis was done by One-Way ANOVA, given in Tables S3 and S4.
Inhibition of RcGTA adsorption by surface polysaccharide extracts
Although our results indicate that the rcc01081 and rcc01932 are involved in capsule polysaccharide biosynthesis, mutagenesis of such genes can also cause pleotropic effects (as is seen in the Δ1081 mutant), and may interfere in other biosynthetic pathways such as LPS O-antigen biosynthesis. Because LPS is often used as a bacteriophage receptor, we investigated the possibility that the reduced RcGTA adsorption in our mutants was due to an altered LPS structure. We visualized the LPS of all our strains by silver staining (data not shown), and did not observe differences in the banding pattern in any of our B10-derived mutant strains, indicating that these strains produce a WT LPS. However, there was a difference in LPS banding of the 37b4 strain. As a further step, we evaluated whether surface extracts from the WT B10 encapsulated strain, but not from non-encapsulated mutants, inhibit RcGTA adsorption to cells. It was found that addition of exogenous WT B10 extracts, but not extracts from the ΔgtaI or Δ1932 strains (both of which lack a capsule), significantly blocked RcGTA adsorption to cells (Fig. 7A and B), supporting the conclusion that capsular polysaccharide is involved in RcGTA adsorption.
Fig. 7. Inhibition of RcGTA adsorption by addition of cell surface extracts.
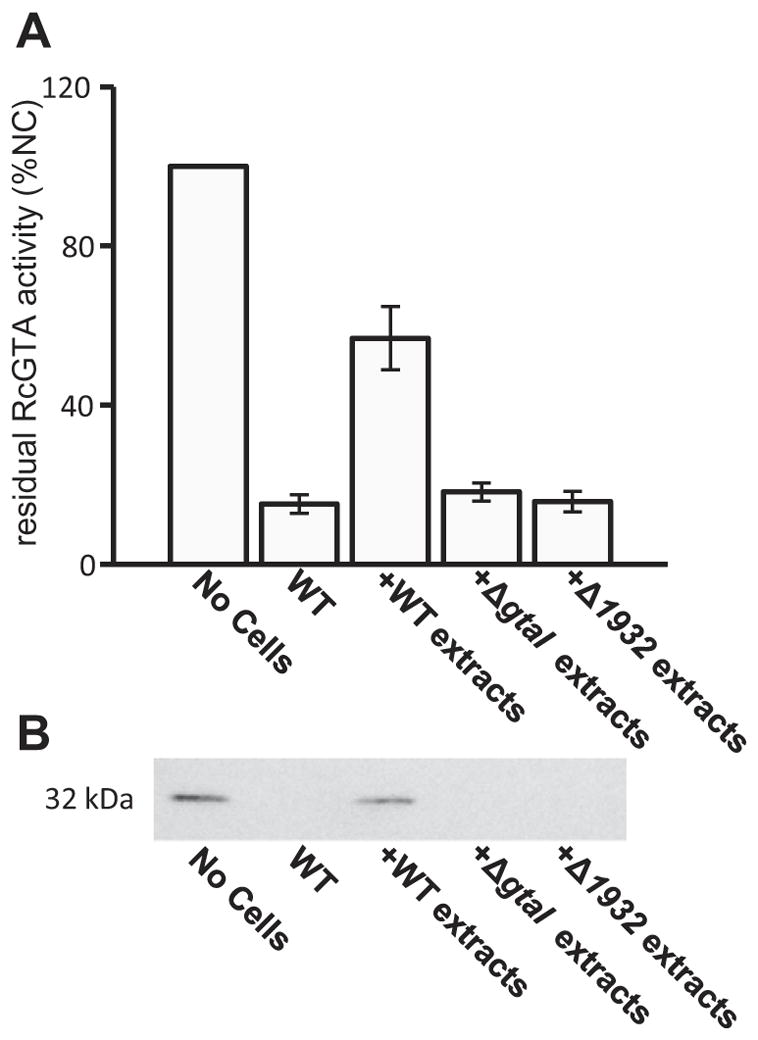
A. Relative amount of transduction-proficient RcGTA present after an adsorption assay, with cell surface extracts added to the reaction mixture where indicated; No Cells denotes a control, in which RcGTA was added directly to recipient cells; WT indicates the addition of WT B10 cells to RcGTA before removal of cells by filtration and use of the RcGTA-containing filtrate for gene transduction; +WT extracts denotes the addition of cell surface extracts of WT B10 cells to RcGTA before removal of cells by filtration and use of the RcGTA-containing filtrate for transduction; +ΔgtaI extracts, and +Δ1932 extracts denote adsorption assays where cell surface extracts from each indicated strain were added to the transduction mixture (containing WT B10 cells). Adsorption values were measured by the amount of residual RcGTA in the filtrate (values are presented as a percentage of the number of RifR transductants obtained in the control where no cells were added to the reaction mixture).
B. Western blot of cells treated as described in (A), and probed with capsid antiserum. Statistical analysis was done by One-Way ANOVA, given in Table S7.
Discussion
We report the discovery of a receptor involved in the adsorption of RcGTA to R. capsulatus cells. The receptor is an EPS that we ascribe to the capsule layer surrounding most strains of R. capsulatus (Weaver et al., 1975). This finding has interesting implications in terms of horizontal gene transfer in laboratory and environmental contexts, as well as possibly in biotechnological applications by extending the host range of GTAs.
The initial observation of changes in RcGTA recipient capability and attachment of R. capsulatus B10 cells in different growth phases prompted us to evaluate whether the GtaR/I QS system is involved in the process. As hypothesized, we found that RcGTA recipient ability and adsorption are negatively regulated by the GtaR protein, as was shown for RcGTA production (Leung et al., 2012). The regulation by the GtaR/I system of not only the production of RcGTA particles in donor cells, but also the display of an RcGTA receptor on the surface of recipient cells indicates a long and complex evolutionary history. We speculate that part of the natural selection for QS regulation of this RcGTA receptor stems from an advantage gained by restraining the energetically expensive production of the receptor polysaccharide unless potential RcGTA donors are present in the environment, as indicated by the presence of the acyl-HSL signal. Similarly, the ultimate cost associated with the production of RcGTA particles (cell death) would provide no benefit to kin unless there were nearby cells potentially capable of taking up RcGTA.
The rcc01081-1086 gene cluster has similarities to both group 1 LPS O-antigen biosynthesis genes and group 1 or 4 capsule biosynthesis genes. When sequence analysis alone cannot reliably distinguish between O-antigen, capsular polysaccharide or other EPS biosynthesis genes, genomic location may provide some insight (Whitfield, 2006). LPS O-antigen biosynthesis clusters are typically located in close proximity to other biosynthesis genes, such as those that synthesize the core oligosaccharide (Raetz and Whitfield, 2002); however, no such genes were found in the proximity of rcc01081-1086. For group 4 capsules, the genes required to synthesize a polysaccharide repeat unit usually are separate from the other required genes. For example, in Escherichia coli, functions additional to the synthesis of a repeat unit, such as polysaccharide chain initiation and export, are carried out by functional homologues in other gene clusters, such as the ‘22 minute’ locus (Whitfield, 2006). Our search for the required chain initiation and export machinery yielded clear homologues of such proteins encoded in the R. capsulatus genome, namely Rcc01932 (WecA; chain initiator), and Rcc01958-1960 (Wzc, Wzb and Wza; export). Furthermore, the pattern of regulation of these genes is similar to that of the rcc01081-1086 genes (Table 1). Based on this information, and the fact that most R. capsulatus strains have a capsule, we hypothesized that the rcc01081-1086 gene cluster is involved in capsule biosynthesis.
Initially, we were surprised by the phenotype of the Δ1081 strain, as it forms a massive aggregate when grown in RCV defined medium and large clumps of swollen, filamentous cells when viewed in the microscope. We also observed that the rcc01081 mutant strain pellets well, and produces a level of cell-associated EPS similar to the ΔgtaI and 37b4 strains, consistent with a defect in the same EPS biosynthetic pathway. We ascribe the aggregation of strain Δ1081 misshapen cells to the build-up of an intracellular polymer (Clarke et al., 2004). It is possible that this polymer is assembled on an polyisoprenoid lipid undecaprenyl diphosphate (und-PP) carrier that is also used in peptidoglycan biosynthesis (Xayarath and Yother, 2007), and so this phenotype may be the result of a defective peptidoglycan layer due to the continual unproductive occupancy of an und-PP carrier. The weak staining of Δ1081 cells with crystal violet (Fig. 5C) is consistent with a defect in peptidoglycan production. Future experiments aimed at identifying the precise function of the Rcc01081 protein may include a comparative analysis of EPS from WT and Δ1081 strains, as was done by others to propose the sugar composition of the R. capsulatus capsule (Omar et al., 1983).
Loss of function mutagenesis of rcc01932 indicated that this gene is involved in the same pathway as the rcc01081-1086 genes, because cells neither produce a capsule nor adsorb RcGTA effectively. Unlike the Δ1081 strain, however, Δ1932 mutant cells have a near-normal morphology (Fig. 5). Because the Rcc01932 protein is most likely involved in polysaccharide chain initiation, we predicted that mutation of the rcc01932 gene would result in a strain lacking a particular EPS. Indeed, the phenotype of the Δ1932 mutant strain strengthens the suggestion of a group 4-like capsule biosynthesis system, because mutagenesis of a gene in a distant genomic location from the rcc01081-1086 cluster generated a phenotype similar to that of the Δ1081 mutant. Additionally, the Δ1081/Δ1932 double mutant has a phenotype similar to that of the Δ1932 single mutant, providing strong evidence that these two distantly linked genes function in the same capsule biosynthesis system (i.e. mutagenesis of rcc01932 suppresses the cell shape defect caused by the rcc01081 mutation).
A previous analysis of the active RcGTA recipient population in a culture found that nearly all cells were active recipients (Solioz et al., 1975). Our capsule stains indicated that all cells in a population produce a capsule under the conditions tested, and so our results are in good agreement that the entire population can act as RcGTA recipients, whereas RcGTA is produced by a sub-population of cells (Fogg et al., 2012; Hynes et al., 2012).
It is interesting that the genes rcc01079 and rcc01080, adjacent to the rcc01081-1086 cluster (Fig. 4), encode putative tail fibre proteins that co-purified with RcGTA particles in a proteomic study (Chen et al., 2009). Other experiments indicated that rcc01079 and rcc01080 are required for maximal RcGTA-mediated transduction frequencies (Lang et al., 2012). Thus it appears that RcGTA receptor and receptor-binding genes are adjacent on the chromosome, and separate from the structural gene cluster (rcc01682-01698).
Many bacteriophages that target capsular polysaccharide as a receptor, such as coliphage K5a, K1-5, and the Vi phages of Salmonella enterica, possess enzymes that degrade capsular polysaccharide upon binding (Gupta et al., 1983; Scholl et al., 2001; Pickard et al., 2010). Such a mechanism allows the phage to traverse the capsule layer, which can span a relatively large distance (~ 0.5 μm). One mechanistic example is the KflA protein of coliphage K5a, which was shown to function as a polysaccharide lyase that degrades K5 capsular polysaccharide, and is a tail-spike protein (Clarke et al., 2000). It is possible that RcGTA uses a similar mechanism to traverse the capsule layer and deliver DNA into cells. It was previously noted that RcGTA orf g15 has weak similarity to a putative rhamnosyl transferase and that rhamnose is a component of the R. capsulatus capsule (Lang and Beatty, 2000).
In a previous study, it was reported that the capsular polysaccharide of R. capsulatus strain St Louis was initially obtained as part of an SDS–solution–insoluble complex. This complex was found to contain peptidoglycan, protein and capsular polysaccharide, and the capsular polysaccharide was liberated by addition of a protease (Bräutigam et al., 1988). Furthermore, the same complex purified from the non-encapsulated strain 37b4 was found to lack the polysaccharide component attributed to the capsule, although the protein–peptidoglycan complex remained; the same complex purified from a phage-resistant strain of St Louis, RC1-, was also found to lack attached capsular polysaccharide (Bräutigam et al., 1988). These findings are of note for two reasons. First, the fact that capsular polysaccharide attachment involved a protein component may explain our observation that proteinase K abolishes the loose pellet phenotype. Second, it is an interesting parallel that an R. capsulatus genuine phage also appears to utilize the capsule as a receptor in a fashion analogous to RcGTA.
The discovery of a receptor involved in RcGTA adsorption is an important step forward in terms of understanding how and when RcGTA-mediated genetic exchange functions. It may also provide insight into how R. capsulatus interacts with its own species and in mixed species communities. Now that a receptor involved in adsorption has been discovered, the groundwork for further characterization of exactly how the RcGTA attaches to and transfers DNA into cells has been established, and may allow for future application in gene transduction between species.
In summary, we propose that the capsule polysaccharide of R. capsulatus is a receptor involved in RcGTA adsorption to cells, and that production of the capsule is dependent on culture growth phase and the GtaR/I QS system. We identified genes that are required for capsule production and demonstrated that their expression is dependent on QS. Our findings show that the regulation of production of an RcGTA receptor as well as the production of RcGTA itself are controlled by the same QS system. These results indicate that RcGTA, an RcGTA receptor, and QS genes have evolved in concert. We anticipate that these findings will lead to additional development of RcGTA as a tool and a better understanding of the function of RcGTA and other GTAs in natural settings.
Experimental procedures
Bacterial strains and growth of cultures
The E. coli strains DH5α and S17-1 (Simon et al., 1983) were used for cloning and conjugation of plasmids into R. capsulatus respectively. E. coli strains were grown at 37°C in Luria–Bertani (LB) medium (Sambrook et al., 1989) supplemented with the appropriate antibiotics at the following concentrations (μg ml−1): ampicillin, 150; gentamicin sulphate, 10; kanamycin sulphate, 50.
The R. capsulatus strains B10 (WT) (Marrs, 1974), ΔgtaI, ΔgtaR, ΔgtaRI, Δ1081, Δ1081[p1081], ΔgtaI/Δ1081, Δ1932, Δ1932[p1932], Δ1932/Δ1081 and 37b4 (WT) (Weckesser et al., 1972) were grown at 30°C in defined RCV medium (Beatty and Gest, 1981) phototrophically for EPS measurements, culture images, capsule-staining, and β-galactosidase assays. For RcGTA adsorption assays, cells were grown aerobically in RCV medium with shaking (flask filled to 50% of nominal capacity) at 200 r.p.m. With the exception of the growth curve experiment, cells were harvested in the stationary phase for use in assays. When appropriate, media were supplemented with (μg ml−1): gentamicin sulphate, 3; kanamycin sulphate, 10; rifampicin, 80, and tetracycline, 1.
Culture turbidity was used as a measure of the number of cells per ml, and monitored in a Klett-Summerson photometer (red filter #66); 100 Klett units represents approximately 4 × 108 R. capsulatus colony-forming units per ml (cfu ml−1). For RcGTA recipient capability and adsorption assays, cell turbidity was measured at 660 nm in a spectrophotometer; an OD660 of 1 = ~ 4.5 × 108 cfu ml−1.
Recombinant DNA techniques and plasmids
Standard methods of DNA purification, restriction enzyme digestion, and other modification techniques were used (Sambrook et al., 1989). The plasmid pUC19 (Invitrogen) was used for subcloning, pIND4 (Ind et al., 2009) as an expression vector for the Δrcc01081 and Δrcc01932 complementation experiments, and pXCA601(Adams et al., 1989) for the rcc01081 promoter lacZ fusion experiments.
The plasmids used in this work are listed in Table S1. The rcc01081::lacZ promoter fusion plasmid containing ~ 1.5 kb of sequence 5′ of the rcc01081 start codon was used to evaluate transcription and translation of the rcc01081 gene. The insert was made by PCR amplification using WT R. capsulatus genomic DNA as a template, and the primer set 1081_lacZ_for and 1081_lacZ_rev. The resultant amplicon was digested with PstI and BglII, and ligated into pXCA601. The ligation resulted in a translationally in-frame fusion between the rcc01081 promoter sequences and the lacZ gene.
Creation of the suicide vector pZDJ
Generating silent knockouts in R. capsulatus using the suicide plasmid pZJD29a (J. Jiang and C.E. Bauer, unpublished) is relatively inefficient due to recombination of the puc promoter region into the R. capsulatus chromosome. Additionally, counter-selection efficiency on sucrose is low, as the puc promoter does not effectively induce the expression of the sacB gene under aerobic conditions. We thus created a new suicide vector, pZDJ, created by replacing the puc promoter with the tetA promoter, allowing for constitutive expression of the sacB gene, and to allow for use of this vector in other Gram-negative bacteria, such as Rhodobacter sphaeroides.
Briefly, pZJD29a was linearized with ApaI and inverse PCR amplified using the primer set sacB:accC1_for and sacB:accC1_rev. This reaction yielded an ApaLI to MfeI fragment, with the R. capsulatus puc promoter removed. The tetA promoter from the broad-host range vector pRK415 was amplified using the primers tetAp_for and tetAp_rev with modifications from (Larsen et al., 2002), generating an MfeI to ApaLI fragment. These two PCR fragments were digested with ApaLI and MfeI, ligated together, and transformed into E. coli DH5α λpir, resulting in the 4.8 kb suicide vector pZDJ (Fig. S4).
Construction of chromosomal deletion mutant strains
All gene disruptions made in this study were markerless in-frame deletions of the majority of the gene. All mutant strains were constructed from the R. capsulatus B10 strain. The ΔgtaI, ΔgtaR, and ΔgtaRI strains were constructed as described previously (Leung et al., 2012). The Δ1081 mutant was constructed by amplifying ~ 700 bp flanking regions of the rcc01081 gene using the primer pairs 1081_up_for and 1081_up_rev, and 1081_down_for and 1081_down_rev. The PCR products were inserted into the vector pUC19 as a SalI-HindIII fragment (upstream) and a HindIII-SacI fragment (downstream). The resultant vectors were then digested with the appropriate enzymes, ligated together to generate a SalI-SacI fragment, and inserted into the suicide plasmid pZDJ. This vector was conjugated into R. capsulatus B10 and allowed to recombine into the chromosome, selected for by gentamicin resistance. Cells were then selected for a second recombination event involving the loss of the plasmid by aerobic growth on medium containing 7% sucrose. The resultant colonies were screened by PCR for a change in the size of the rcc01081 locus amplicon and selected mutants were confirmed by sequencing using the primers 1081_seq_for and 1081_seq_rev. The Δ1932 mutant was generated in a similar fashion, using the primers 1932_up_for, 1932 up_rev, 1932_down_for, and 1932_down_rev. The Δ1932/Δ1081 double mutant was generating by conjugating pZDJΔ1081 into the Δ1932 strain and screening for mutants as described above (see Table S1 for primer sequences).
Complementation of the Δrcc01081 and Δrcc01932 mutations
The primers 1081_comp_up and 1081_comp_down, and 1932_comp_up and 1932_comp_down were used to amplify the entire rcc01081 and rcc01932 genes respectively. Amplicons were ligated into the multiple cloning site of the plasmid pIND4 (Ind et al., 2009) as NcoI to HindIII fragments to produce the plasmids pIND1081, and pIND1932. These constructs allow for inducible expression of the rcc01081 and rcc01932 genes under control of an isopropyl β-D-1-thiogalactopyranoside (IPTG)-inducible lac promoter. The plasmid pIND1081 was then conjugated into R. capsulatus mutant strain Δ1081, and the plasmid pIND1932 was conjugated into the strain Δ1932, generating the strains Δ1081[p1081] and Δ1932[p1932]. For experiments using these complemented strains, the cultures were grown with 1 mM IPTG during growth to induce expression of each gene.
RcGTA recipient capability assay
A rifampicin-resistant RcGTA overproducer strain (Yen et al., 1979), DE442, was used as a source of RcGTA for recipient capability and adsorption experiments. A sample of stationary phase culture grown phototrophically in YPS complex medium (Wall et al., 1975), passed through a 0.2 μm filter, was titred for RcGTA activity, and a diluted stock solution that produced ~ 800 rifampicin-resistant transductants per 100 μl of donor filtrate (with stationary phase B10 cells as a recipient) was used for all experiments. All strains used as RcGTA recipients, other than 37b4, were derived from WT B10. Cells were grown aerobically in RCV defined medium with shaking at 200 r.p.m. to the stationary phase, harvested, and re-suspended in an equal volume of G-buffer [10 mM Tris-HCL (pH 7.8), 1 mM MgCl2, 1 mM CaCl2, 1 mM NaCl, 500 μg ml−1 BSA] (Solioz et al., 1975). A transduction assay was then performed, where 100 μl of RcGTA stock, 100 μl of recipient cells, and 400 μl of G-buffer are mixed together. This mixture was incubated for 90 min at 30°C with gentle agitation, after which 900 μl of RCV medium were added, followed by incubation under the same conditions for 3 h. Cells were spread on RCV plates containing rifampicin and incubated aerobically for 3 days. Rifampicin-resistant colonies were counted and the average was corrected by subtracting the number of spontaneous rifampicin-resistant colonies (no addition of RcGTA, usually no more than 3%). Because of variability in total numbers of transductants between individual experiments, RcGTA recipient capability efficiencies are expressed as % WT (B10), with each experimental strain being compared with the WT control within the same experiment. In the case of the growth phase recipient capability experiment, total numbers of rifampicin-resistant colonies from a single representative experiment are shown. The statistical significance of results was evaluated by One-Way ANOVA. A P-value of 0.05 (95% confidence interval) was set as a cut-off for significance.
RcGTA adsorption assay
To quantitatively measure the ability of different strains to bind to RcGTA, cells were grown aerobically in RCV defined medium with shaking at 200 r.p.m. to the stationary phase, harvested, and re-suspended at the same density in G-buffer. The first step of a transduction assay was then performed, where 100 μl of RcGTA solution, 100 μl of recipient cells, and 400 μl of G-buffer are mixed together. This mixture was incubated for 90 min at 30°C with gentle agitation, after which this mixture was passed through a 0.2 μm filter. The residual levels of RcGTA in the filtrate (RcGTA that did not adsorb to cells) was quantified by performing another standard transduction assay using 100 μl of this filtrate as the RcGTA donor, and WT R. capsulatus B10 cells as the recipient. As two separate controls, an assay that included no recipient cells for adsorption (no cells), and an assay that contained no RcGTA were also performed. For cell surface extract competitive inhibition assays, WT B10 cells were used as a RcGTA recipient strain in all cases. Because of variability in total numbers between experiments, adsorption efficiencies are expressed as a % of the value obtained with no cells, with each experimental strain being compared with the un-adsorbed control (with the exception of the growth curve experiment). The statistical significance of results was evaluated by One-Way ANOVA. A P-value of 0.05 (95% confidence interval) was set as a cut-off for significance.
Western blot
Filtrate from RcGTA adsorption assays was concentrated 10-fold in a SpeedVac concentrator (Thermo Electron Corp) and samples were boiled for 10 min in sample loading buffer (50 mM Tris-HCl pH 6.8, 2% SDS, 10% glycerol, 0.1% bromophenol blue, 1% β-mercaptoethanol) prior to gel loading. Samples were separated on a 12% SDS-PAGE gel and blotted on to a nitrocellulose membrane (Perkin-Elmer). The blotting was done using a Mini Trans-Blot apparatus (Bio-Rad) according to the manufacturer’s specifications in electroblot buffer (27.5 mM Tris-Base, 192 mM glycine, 20% methanol) at 100 V constant voltage for ~ 1.5 h.
The primary antibody (rabbit) was raised against the R. capsulatus RcGTA capsid protein (Taylor, 2004). Primary antibody binding was detected using a peroxidase-linked anti-rabbit Ig secondary antibody (from donkey; Amersham) as part of the enhanced chemiluminescence (ECL) kit according to the manufacturer’s instructions (Amersham).
Densitometry measurements of relative band intensity were performed using the program ImageJ (http://rsbweb.nih.gov/ij/).
Phenol-sulfuric acid carbohydrate quantification
Estimation of cell-associated carbohydrate (EPS) relative amounts was done using a modified phenol-sulfuric acid method (Liu et al., 1973). R. capsulatus cells were grown phototrophically to the stationary phase in RCV defined medium, and the cell concentration normalized to 450 Klett units by the addition of medium. The cells from 1 ml of each normalized culture were harvested in a microcentrifuge at 16 500 g for 1 min (same centrifugation for all steps), and washed three times in 50 mM NaCl. Cells were re-suspended in 1 ml of 50 mM EDTA and incubated at 37°C for 30 min. Cells were then pelleted, and the resultant supernatant containing LPS and any tightly associated EPS, such as capsule, was subjected to a phenol-sulfuric acid colourimetric carbohydrate quantification in the following manner: 200 μl of the sample and 200 μl of 5% phenol were mixed together in a glass test tube; 1 ml of 93% sulfuric acid was added, and the tube agitated to ensure mixing. Colour was allowed to develop for 10 min. Colour intensity was then measured at 490 nm in a spectrophotometer, and compared with standards of known carbohydrate concentration. The sugar standards were a 50:50 mixture of sucrose and fructose diluted from a stock of known concentration. The statistical significance of results was evaluated by One-Way ANOVA. A P-value of 0.05 (95% confidence interval) was set as a cut-off for significance.
Capsule stain
Capsules were negatively stained using Anthony’s capsule stain (Anthony, 1931). In brief, 1 ml of stationary phase R. capsulatus cultures was harvested by centrifugation, and re-suspended in 1 ml of carnation skim milk (skim milk powder was re-suspended in H2O at 1% w/v). An inoculating loop of each re-suspended culture was spread onto a clean microscope slide and allowed to dry in the air. Smears were then stained with 1% crystal violet for 5 min, and washed gently and thoroughly with 20% copper sulphate. Slides were then allowed to dry in the air, and subsequently examined using oil-immersion phase contrast microscopy at 100× magnification.
Bioinformatic analyses
The predicted amino acid sequences for Rcc01081-1086, Rcc01932, and Rcc01958-1960 were used to perform a BLASTp search against the nr database (Altschul et al., 1990). Selected sequences were globally aligned using the EMBOSS Needle program for pairwise alignments.
β-Galactosidase assays
The β-galactosidase specific activity of cells containing the rcc01081::lacZ promoter fusion was assayed as previously described (Leung et al., 2012). Sonication was used to break cells, and total protein was measured using the Lowry method (Peterson, 1983), with bovine serum albumin as the standard. β-Galactosidase specific activities are presented as units per milligram of total protein.
Expression analysis of wild-type and gtaI mutant strains of R. capsulatus
Photoheterotrophic cultures for microarray analysis were grown in RCV defined medium at 30°C. Cells were harvested at the logarithmic and stationary phases of growth as determined by culture turbidity. RNA extraction, cDNA synthesis, target labelling and hybridization were all performed as previously described (Mercer et al., 2010). Microarrays for a single sample of each strain in both phases of growth were performed by The Centre for Applied Genomics, The Hospital for Sick Children, Toronto, Canada. The raw data from the scanned arrays were normalized by Robust Multi-Array Analysis (RMA) (Irizarry et al., 2003) in Affymetrix Expression Console v1.1 (Santa Clara, CA) and exported for analyses. The relationships between the gene features on the custom array and the sequenced genome (NCBI Accession Number NC_014034) have been previously described (Mercer et al., 2010).
LPS silver stains
The LPS from R. capsulatus was visualized as previously described (Tsai and Frasch, 1982). LPS was purified by harvesting 1 ml of culture by centrifugation, followed by three washes in 1 ml of 50 mM NaCl. Cells were then re-suspended in 500 μl of 20 mM EDTA and incubated for 30 min. Cells were then pelleted by centrifugation (14 500 r.p.m. for 1 min) and the supernatant (containing LPS) was removed, treated with proteinase K for 4 h at 50°C, and concentrated in a SpeedVac concentrator (Thermo Electron Corp). For LPS gels, material was loaded and electrophoresed as described in the Western blot section.
Surface polysaccharide extraction for inhibition of RcGTA binding
Surface polysaccharides (EPS/LPS) were purified in the following manner: 50 ml of each strain was grown aerobically in RCV medium with shaking at 200 r.p.m. until cultures reached the stationary phase. Cells were then harvested by centrifugation at 12 000 g for 30 min, followed by two consecutive washes in 50 mM NaCl. Cell pellets were then re-suspended in 10 ml of 50 mM EDTA, and incubated overnight at room temperature. Cells were then removed by centrifugation, and the resultant supernatant (containing EPS/LPS) was subsequently treated with proteinase K (1 mg ml−1) for 4 h, followed by heating at 80°C for 1 h to deactivate the protease. Samples were then dialysed against dH2O for 2 days with several buffer changes to remove EDTA, followed by quantification using the phenol-sulfuric acid method. Extracts were added to RcGTA adsorption reactions at a final concentration of 100 μg ml−1.
Supplementary Material
Acknowledgments
We thank J. Armitage for providing the plasmid pIND4, the Canadian Institutes for Health Research (CIHR) for an Operating Grant (JTB), and the University of British Columbia for a postgraduate scholarship (CB). RM was supported by fellowships from Memorial University School of Graduate Studies and the Natural Sciences and Engineering Research Council (NSERC). Research in ASL’s lab was supported by funding from NSERC and the Canada Foundation for Innovation.
Footnotes
Additional supporting information may be found in the online version of this article.
References
- Adams CW, Forrest ME, Cohen SN, Beatty JT. Structural and functional analysis of transcriptional control of the Rhodobacter capsulatus puf operon. J Bacteriol. 1989;171:473–482. doi: 10.1128/jb.171.1.473-482.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anthony EE., Jr A note on capsule staining. Science. 1931;73:319–320. doi: 10.1126/science.73.1890.319. [DOI] [PubMed] [Google Scholar]
- Beatty JT, Gest H. Generation of succinyl-coenzyme A in photosynthetic bacteria. Arch Microbiol. 1981;129:335–340. [Google Scholar]
- Bottomley MJ, Muraglia E, Bazzo R, Carfi A. Molecular insights into quorum sensing in the human pathogen Pseudomonas aeruginosa from the structure of the virulence regulator LasR bound to its autoinducer. J Biol Chem. 2007;282:13592–13600. doi: 10.1074/jbc.M700556200. [DOI] [PubMed] [Google Scholar]
- Bräutigam E, Fiedler F, Woitzik D, Flammann HT, Weckesser J. Capsule polysaccharide-protein-peptidoglycan complex in the cell envelope of Rhodobacter capsulatus. Arch Microbiol. 1988;150:567–573. [Google Scholar]
- Chen F, Spano A, Goodman BE, Blasier KR, Sabat A, Jeffery E, et al. Proteomic analysis and identification of the structural and regulatory proteins of the Rhodobacter capsulatus gene transfer agent. J Proteome Res. 2009;8:967–973. doi: 10.1021/pr8006045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy YM, Fehmel F, Frank N, Stirm S. Escherichia coli capsule bacteriophages. IV. Primary structure of the bacteriophage 29 receptor, the E. coli serotype 29 capsular polysaccharide. J Virol. 1975;16:581–590. doi: 10.1128/jvi.16.3.581-590.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke BR, Esumeh F, Roberts IS. Cloning, expression, and purification of the K5 capsular polysaccharide lyase (KflA) from coliphage K5A: evidence for two distinct K5 lyase enzymes. J Bacteriol. 2000;182:3761–3766. doi: 10.1128/jb.182.13.3761-3766.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke BR, Cuthbertson L, Whitfield C. Non-reducing terminal modifications determine the chain length of polymannose O antigens of Escherichia coli and couple chain termination to polymer export via an ATP-binding cassette transporter. J Biol Chem. 2004;279:35709–35718. doi: 10.1074/jbc.M404738200. [DOI] [PubMed] [Google Scholar]
- Cuthbertson L, Mainprize IL, Naismith JH, Whitfield C. Pivotal roles of the outer membrane polysaccharide export and polysaccharide copolymerase protein families in export of extracellular polysaccharides in gram-negative bacteria. Microbiol Mol Biol Rev. 2009;73:155–177. doi: 10.1128/MMBR.00024-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels R, De Vos DE, Desair J, Raedschelders G, Luyten E, Rosemeyer V, et al. The cin quorum sensing locus of Rhizobium etli CNPAF512 affects growth and symbiotic nitrogen fixation. J Biol Chem. 2002;277:462–468. doi: 10.1074/jbc.M106655200. [DOI] [PubMed] [Google Scholar]
- Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- Eiserling F, Pushkin A, Gingery M, Bertani G. Bacteriophage-like particles associated with the gene transfer agent of Methanococcus voltae PS. J Gen Virol. 1999;80:3305–3308. doi: 10.1099/0022-1317-80-12-3305. [DOI] [PubMed] [Google Scholar]
- Flammann HT, Weckesser J. Composition of the cell wall of the phage resistant mutant Rhodopseudomonas capsulata St. Louis RC1. Arch Microbiol. 1984;139:33–37. [Google Scholar]
- Florizone SM. MSc Thesis. Vancouver: Department of Microbiology & Immunology, University of British Columbia; 2006. Studies on the regulation of the Gene Transfer Agent (GTA) of Rhodobacter capsulatus; p. 78. [Google Scholar]
- Fogg PCM, Westbye AB, Beatty JT. One for all or all for one: heterogeneous expression and host cell lysis are key to gene transfer agent activity in Rhodobacter capsulatus. PLoS ONE. 2012;7:e43772. doi: 10.1371/journal.pone.0043772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genthner FJ, Wall JD. Isolation of a recombination-deficient mutant of Rhodopseudomonas capsulata. J Bacteriol. 1984;160:971–975. doi: 10.1128/jb.160.3.971-975.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez JE, Keshavan ND. Messing with bacterial quorum sensing. Microbiol Mol Biol Rev. 2006;70:859–875. doi: 10.1128/MMBR.00002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta DS, Jann B, Jann K. Enzymatic degradation of the capsular K5-antigen of E. coli by coliphage K5. FEMS Microbiol Lett. 1983;16:13–17. [Google Scholar]
- Hancock RE. Alterations in outer membrane permeability. Annu Rev Microbiol. 1984;38:237–264. doi: 10.1146/annurev.mi.38.100184.001321. [DOI] [PubMed] [Google Scholar]
- Humphrey SB, Stanton TB, Jensen NS, Zuerner RL. Purification and characterization of VSH-1, a generalized transducing bacteriophage of Serpulina hyodysenteriae. J Bacteriol. 1997;179:323–329. doi: 10.1128/jb.179.2.323-329.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes AP, Mercer RG, Watton DE, Buckley CB, Lang AS. DNA packaging bias and differential expression of gene transfer agent genes within a population during production and release of the Rhodobacter capsulatus gene transfer agent, RcGTA. Mol Microbiol. 2012;85:314–325. doi: 10.1111/j.1365-2958.2012.08113.x. [DOI] [PubMed] [Google Scholar]
- Ikeda H, Tomizawa JI. Transducing fragments in generalized transduction by phage P1. I. Molecular origin of the fragments. J Mol Biol. 1965;14:85–109. doi: 10.1016/s0022-2836(65)80232-7. [DOI] [PubMed] [Google Scholar]
- Ind AC, Porter SL, Brown MT, Byles ED, de Beyer JA, Godfrey SA, Armitage J. Inducible-expression plasmid for Rhodobacter sphaeroides and Paracoccus denitrificans. Appl Environ Microbiol. 2009;75:6613–6615. doi: 10.1128/AEM.01587-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionescu M, Belkin S. Simple quantification of bacterial envelope-associated extracellular materials. J Microbiol Methods. 2009;78:302–306. doi: 10.1016/j.mimet.2009.06.020. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Lang AS, Beatty JT. Genetic analysis of a bacterial genetic exchange element: the gene transfer agent of Rhodobacter capsulatus. Proc Natl Acad Sci USA. 2000;97:859–864. doi: 10.1073/pnas.97.2.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang AS, Beatty JT. Importance of widespread gene transfer agent genes in alpha-proteobacteria. Trends Microbiol. 2007;15:54–62. doi: 10.1016/j.tim.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Lang AS, Zhaxybayeva O, Beatty JT. Gene transfer agents: phage-like elements of genetic exchange. Nat Rev Microbiol. 2012;10:472–482. doi: 10.1038/nrmicro2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen RA, Wilson MM, Guss AM, Metcalf WW. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch Microbiol. 2002;178:193–201. doi: 10.1007/s00203-002-0442-2. [DOI] [PubMed] [Google Scholar]
- Lazdunski AM, Ventre I, Sturgis JN. Regulatory circuits and communication in Gram-negative bacteria. Nat Rev Microbiol. 2004;2:581–592. doi: 10.1038/nrmicro924. [DOI] [PubMed] [Google Scholar]
- Leung MM, Brimacombe CA, Spiegelman GB, Beatty JT. The GtaR protein negatively regulates transcription of the gtaRI operon and modulates gene transfer agent (RcGTA) expression in Rhodobacter capsulatus. Mol Microbiol. 2012;83:759–774. doi: 10.1111/j.1365-2958.2011.07963.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung MM-L. PhD Thesis. Vancouver: Department of Microbiology & Immunology, University of British Columbia; 2010. CtrA and GtaR: two systems that regulate the Gene Transfer Agent of Rhodobacter capsulatus; p. 168. [Google Scholar]
- Liu D, Wong PTS, Dutka BJ. Determination of carbohydrate in lake sediment by a modified phenol-sulfuric acid method. Water Res. 1973;7:741–746. [Google Scholar]
- Marrs B. Genetic recombination in Rhodopseudomonas capsulata. Proc Natl Acad Sci USA. 1974;71:971–973. doi: 10.1073/pnas.71.3.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson EG, Thompson MG, Humphrey SB, Zuerner RL, Stanton TB. Identification of genes of VSH-1, a prophage-like gene transfer agent of Brachyspira hyodysenteriae. J Bacteriol. 2005;187:5885–5892. doi: 10.1128/JB.187.17.5885-5892.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer RG, Callister SJ, Lipton MS, Pasa-Tolic L, Strnad H, Paces V, et al. Loss of the response regulator CtrA causes pleiotropic effects on gene expression but does not affect growth phase regulation in Rhodobacter capsulatus. J Bacteriol. 2010;192:2701–2710. doi: 10.1128/JB.00160-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Hagelsieb G, Latimer K. Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics. 2008;24:319–324. doi: 10.1093/bioinformatics/btm585. [DOI] [PubMed] [Google Scholar]
- Nadell CD, Xavier JB, Levin SA, Foster KR. The evolution of quorum sensing in bacterial biofilms. PLoS Biol. 2008;6:e14. doi: 10.1371/journal.pbio.0060014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omar AS, Flammann HT, Golecki JR, Weckesser J. Detection of capsule and slime polysaccharide layers in two strains of Rhodopseudomonas capsulata. Arch Microbiol. 1983;134:114–117. [Google Scholar]
- Peterson G. Determination of total protein. Methods Enzymol. 1983;91:95–119. doi: 10.1016/s0076-6879(83)91014-5. [DOI] [PubMed] [Google Scholar]
- Pickard D, Toribio AL, Petty NK, van Tonder A, Yu L, Goulding D, et al. A conserved acetyl esterase domain targets diverse bacteriophages to the Vi capsular receptor of Salmonella enterica serovar Typhi. J Bacteriol. 2010;192:5746–5754. doi: 10.1128/JB.00659-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper KR, Beck S, Farrand SK. Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature. 1993;362:448–450. doi: 10.1038/362448a0. [DOI] [PubMed] [Google Scholar]
- Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakhuba DV, Kolomiets EI, Dey ES, Novik GI. Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Pol J Microbiol. 2010;59:145–155. [PubMed] [Google Scholar]
- Rapp BJ, Wall JD. Genetic transfer in Desulfovibrio desulfuricans. Proc Natl Acad Sci USA. 1987;84:9128–9130. doi: 10.1073/pnas.84.24.9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning : A Laboratory Manual. Plainview, New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schaefer AL, Taylor TA, Beatty JT, Greenberg E. Long-chain acyl-homoserine lactone quorum-sensing regulation of Rhodobacter capsulatus gene transfer agent production. J Bacteriol. 2002;184:6515–6521. doi: 10.1128/JB.184.23.6515-6521.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl D, Rogers S, Adhya S, Merril CR. Bacteriophage K1-5 encodes two different tail fiber proteins, allowing it to infect and replicate on both K1 and K5 strains of Escherichia coli. J Virol. 2001;75:2509–2515. doi: 10.1128/JVI.75.6.2509-2515.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat Biotechnol. 1983;1:784–791. [Google Scholar]
- Solioz M, Yen HC, Marris B. Release and uptake of gene transfer agent by Rhodopseudomonas capsulata. J Bacteriol. 1975;123:651–657. doi: 10.1128/jb.123.2.651-657.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor TA. MSc Thesis. Vancouver: Department of Microbiology & Immunology, University of British Columbia; 2004. Evolution and regulation of the Gene Transfer Agent (GTA) of Rhodobacter capsulatus; p. 66. [Google Scholar]
- Tsai CM, Frasch CE. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 1982;119:115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- Tun-Garrido C, Bustos P, Gonzalez V, Brom S. Conjugative transfer of p42a from Rhizobium etli CFN42, which is required for mobilization of the symbiotic plasmid, is regulated by quorum sensing. J Bacteriol. 2003;185:1681–1692. doi: 10.1128/JB.185.5.1681-1692.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JD, Weaver PF, Gest H. Gene transfer agents, bacteriophages, and bacteriocins of Rhodopseudomonas capsulata. Arch Microbiol. 1975;105:217–224. doi: 10.1007/BF00447140. [DOI] [PubMed] [Google Scholar]
- Weaver PF, Wall JD, Gest H. Characterization of Rhodopseudomonas capsulata. Arch Microbiol. 1975;105:207–216. doi: 10.1007/BF00447139. [DOI] [PubMed] [Google Scholar]
- Weckesser J, Drews G, Fromme I. Chemical analysis of and degradation studies on the cell wall lipopolysaccharide of Rhodopseudomonas capsulata. J Bacteriol. 1972;109:1106–1113. doi: 10.1128/jb.109.3.1106-1113.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem. 2006;75:39–68. doi: 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- Williams P, Camara M, Hardman A, Swift S, Milton D, Hope VJ, et al. Quorum sensing and the population-dependent control of virulence. Philos Trans R Soc Lond B Biol Sci. 2000;355:667–680. doi: 10.1098/rstb.2000.0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winson MK, Camara M, Latifi A, Foglino M, Chhabra SR, Daykin M, et al. Multiple N-acyl-L-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeuginosa. Proc Natl Acad Sci USA. 1995;92:9427–9431. doi: 10.1073/pnas.92.20.9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xayarath B, Yother J. Mutations blocking side chain assembly, polymerization, or transport of a Wzy-dependent Streptococcus pneumoniae capsule are lethal in the absence of suppressor mutations and can affect polymer transfer to the cell wall. J Bacteriol. 2007;189:3369–3381. doi: 10.1128/JB.01938-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen HC, Hu NT, Marrs BL. Characterization of the gene transfer agent made by an overproducer mutant of Rhodopseudomonas capsulata. J Mol Biol. 1979;131:157–168. doi: 10.1016/0022-2836(79)90071-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.