Abstract
It is known that cyanobacteria negatively affect herbivores due to their production of toxins such as protease inhibitors. In the present study we investigated potential interspecific differences between two major herbivores, Daphnia magna and Daphnia pulex, in terms of their tolerance to cyanobacteria with protease inhibitors. Seven clones each of D. magna and of D. pulex were isolated from different habitats in Europe and North America. To test for interspecific differences in the daphnids’ tolerance to cyanobacteria, their somatic and population growth rates were determined for each D. magna and D. pulex clone after exposure to varying concentrations of two Microcystis aeruginosa strains. The M. aeruginosa strains NIVA and PCC− contained either chymotrypsin or trypsin inhibitors, but no microcystins. Mean somatic and population growth rates on a diet with 20% NIVA were significantly more reduced in D. pulex than in D. magna. On a diet with 10% PCC−, the population growth of D. pulex was significantly more reduced than that of D. magna. This indicates that D. magna is more tolerant to cyanobacteria with protease inhibitors than D. pulex. The reduction of growth rates was possibly caused by an interference of cyanobacterial inhibitors with proteases in the gut of Daphnia, as many other conceivable factors, which might have been able to explain the reduced growth, could be excluded as causal factors. Protease assays revealed that the sensitivities of chymotrypsins and trypsins to cyanobacterial protease inhibitors did not differ between D. magna and D. pulex. However, D. magna exhibited a 2.3-fold higher specific chymotrypsin activity than D. pulex, which explains the observed higher tolerance to cyanobacterial protease inhibitors of D. magna. The present study suggests that D. magna may control the development of cyanobacterial blooms more efficiently than D. pulex due to differences in their tolerance to cyanobacteria with protease inhibitors.
Introduction
The frequency of cyanobacterial blooms in many marine and freshwater environments has increased world wide during the last century, partly due to increasing temperatures as a consequence of global warming and partly due to the eutrophication of lakes [1]. Blooms of cyanobacteria and their toxins may sometimes be associated with harmful effects on human health and livestock [2], [3]. When the temperature of the epilimnion reaches its maximum in late summer and early fall [4], the phytoplankton of many eutrophic lakes and ponds is often dominated by bloom-forming cyanobacterial species of the genera Microcystis, Anabaena and/or Oscillatoria [5]. During this time cyanobacteria are often an important food source for herbivorous zooplankton in freshwater ecosystems, such as for Daphnia, which often provides an important link for the transfer from primary production, e.g. from cyanobacteria to higher trophic levels. In cases when growth of Daphnia is mainly restricted by food quantity, non-toxic cyanobacteria can act as a complementary food source for Daphnia [6], [7]. However, since in eutrophic lakes growth of Daphnia is rather constrained by food quality than by food quantity, bloom-forming cyanobacteria in those habitats have been claimed to be a major factor for a constrained mass and energy transfer from primary producers to organisms of higher trophic levels [8], [9].
Negative relationships between bloom-forming cyanobacteria and the abundance of Daphnia have been discussed extensively over the years, and three major quality constraints of cyanobacteria as a food source have been revealed so far: (1) The occurrence of cyanobacterial filaments and the formation of colonies hinder ingestion by interfering with the filtering apparatus of Daphnia [10]. (2) Compared to most green algae, cyanobacteria contain low levels of essential lipids such as highly unsaturated fatty acids and sterols, which leads to reduced somatic and population growth of Daphnia due to constrained carbon assimilation [11]–[14]. (3) Many cyanobacteria produce a variety of bioactive secondary metabolites such as hepatotoxins like microcystins [15] and/or protease inhibitors [16]–[18]. These compounds reduce the fitness of Daphnia in terms of survival, growth and reproduction [19], [20]. In addition to microcystins (which are the most extensively investigated class of cyanobacterial toxins), the role of protease inhibitors in herbivore/cyanobacteria interaction has recently also become a focus of attention. More than twenty depsipeptides, which specifically inhibit the serine proteases chymotrypsin and trypsins, have been found in different genera of marine and freshwater cyanobacteria [16]. These two classes of proteases are the most important digestive enzymes in the gut of D. magna and are responsible for more than 80% of the proteolytic activity [21].
It is known that the edible size fraction of natural phytoplankton can contain compounds that inhibit Daphnia’s trypsins and chymotrypsins [22]. This inhibitory potential of seston can be in the same order of magnitude as of pure cyanobacterial cultures [23]. Hence, it is reasonable to assume that an interference of cyanobacterial protease inhibitors with Daphnia’s digestive proteases occurs in nature and is ecologically relevant.
However, several studies have also demonstrated that Daphnia may develop tolerances against cyanobacterial toxins at the population level [24]–[27]: D. magna populations that were pre-exposed to toxic cyanobacteria exhibited a higher tolerance to microcystin producing M. aeruginosa than populations that were not pre-exposed [25]. Furthermore, Sarnelle & Wilson [24] suggested that D. pulicaria populations, exposed to high cyanobacterial levels over long periods of time, can adapt in terms of being more tolerant to dietary toxic cyanobacteria. With regard to protease inhibitors Blom et al. [27] have shown that Daphnia sp. coexisting with Planktothrix rubescens (a cyanobacterium that contains the trypsin inhibitor oscillapeptin-J) was significantly more tolerant to oscillapeptin-J than Daphnia sp. from a lake free of this cyanobacterium. Considering the finding that almost 60% of 17 cyanobacterial blooms isolated from 14 distinct water-bodies in India contained protease inhibitors [28], it is reasonable to assume that increased tolerance to cyanobacteria in Daphnia populations may be caused by an enhanced tolerance to the cyanobacterial protease inhibitors. It has been suggested that at least two fundamental mechanisms underlie the increased tolerance to these dietary inhibitors: (1) Colbourne et al. [29] have hypothesized that the ability of Daphnia to cope with different environmental conditions is a consequence of an elevated rate of gene duplications resulting in tandem gene clusters. And indeed, a surprisingly high number of genes of digestive serine proteases have been found in the recently published genome of D. pulex [29]. (2) Von Elert et al. [18] have shown that a physiological plasticity at the protein level in Daphnia in terms of expressing different isoforms of digestive enzymes leads to increased tolerance against cyanobacterial protease inhibitors.
In the present study we tested for interspecific differences between two Daphnia species (D. magna and D. pulex) in their tolerance to cyanobacteria which produce protease inhibitors. D. pulex and D. magna are both large-bodied species and are frequently encountered in fishless ponds [30]. Due to the availability of full-genome data (D. pulex, [29]) or EST libraries (D. magna; [31]), both Daphnia species are ideal for ecological investigations and were therefore chosen for use in the present study. To determine potential differences between D. pulex and D. magna in their tolerance to cyanobacteria containing protease inhibitors, we performed single-clone somatic and population growth experiments in which the clones were fed with various cyanobacterial mixtures containing trypsin or chymotrypsin inhibitors. Both M. aeruginosa strains used in the present study (NIVA Cya 43 and PCC7806−) produce exclusively either the chemically known chymotrypsin inhibitors cyanopeptolin 954 and nostopeptin 920 (NIVA, [32]) or specific cyanopeptolins (A-D) which are known to inhibit trypsins (PCC−, [33]). Possible differences in tolerance to cyanobacteria with protease inhibitors might have several causes and are therefore tested in the present study: (1) We determined the specific trypsin and chymotrypsin activity of each of the investigated D. magna and D. pulex clones and hypothesized that high growth rates on cyanobacterial diets might result from high specific protease activities. (2) For each Daphnia clone, we determined the sensitivity of gut chymotrypsins and trypsins to the respective cyanobacterial protease inhibitors. We assumed that higher sensitivity values of Daphnia’s gut proteases might cause reduced somatic and population growth rates for diets with cyanobacterial protease inhibitors.
Materials and Methods
Origin and Cultivation of Organisms
Two cyanobacterial strains and one green alga were used in the single-clone growth experiments: The cyanobacterium Microcystis aeruginosa NIVA Cya 43 (Culture Collection of Algae, Norwegian Institute for Water Research), subsequently labeled as ‘NIVA’, is known to contain the chymotrypsin inhibitors cyanopeptolin 954 and nostopeptin 920 [32]. NIVA was cultured in 2 l chemostates in sterile cyano medium [34] at a dilution rate of 0.1 d−1 (20°C; illumination: 40 µmol m−2 s−1). M. aeruginosa PCC 7806 Mut contains the trypsin inhibitors cyanopeptolin A–D [33] and was grown in 0.75 l chemostates under otherwise identical conditions as for NIVA. M. aeruginosa PCC 7806 Mut is a genetically engineered microcystin synthetase knock-out mutant of M. aeruginosa PCC 7806 [35] and is subsequently labeled as ‘PCC−’. Neither M. aeruginosa strain contains microcystins. The green algae Chlamydomonas sp. (strain 56, culture collection of the Limnological Institute at the University of Constance) was grown in 5 l semi-continuous batch cultures (20°C; illumination: 120 µmol m−2 s−1) by replacing 20% of the culture with sterile cyano medium every Monday, Wednesday and Friday in the late exponential phase of the culture. Chlamydomonas sp. contains neither chymotrypsin/trypsin inhibitors nor microcystins.
Seven D. magna and seven D. pulex clones originating from different habitats (Table 1) were used in the somatic and population growth experiments. All clones were cultured separately in aged, membrane-filtered tap water and fed with saturating concentrations of Chlamydomonas sp. for at least three generations prior to the experiment.
Table 1. Geographic origin of the Daphnia clones used in the experiments.
| Daphnia spp. | Clone | Location | Latitude | Longitude | Reference |
| D. pulex | Gerstel | Germany | N/A | N/A. | [51] |
| D. pulex | NFL3 | USA | N39°54′ | W84°55′ | [52] |
| D. pulex | Gräf | Germany | N50°49′04′′ | E10°42′02′′ | [53] |
| D. pulex | Disp14 | Canada | N42°13′ | W83°02′ | [54] |
| D. pulex | Povi113 | USA | N42°45′ | W85°21′ | [52] |
| D. pulex | Giev08 | Germany | N51°57′48′′ | E7°34′38′′ | Y. Reydelet1 |
| D. pulex | TCO | USA | N43°49′48′′ | W124°08′53′′ | [29] |
| D. magna | F10 | Germany | N50°56′02′′ | E6°55′41′′ | [22] |
| D. magna | G38 | Belgium | N51°04′04′′ | E3°46′25′′ | This study |
| D. magna | S15 | Sweden | N55°40′31′′ | E13°32′42′′ | [55] |
| D. magna | P6 | Poland | N52°19′21′′ | E20°43′49′′ | [55] |
| D. magna | P | The Netherlands | N51°44′01′′ | E5°08′17′′ | [56] |
| D. magna | B | Germany | N54°19′39′′ | E10°37′45′′ | [57] |
| D. magna | W | Poland | N/A | N/A | [58] |
Y. Reydelet, personal communication, 2011.
Somatic and Population Growth Assays
Each of the 14 clones was assayed in single-clone experiments for somatic and population growth. Five (D. magna) to seven (D. pulex) juveniles from the same cohort of the third clutch and no older than 24 h were kept in 0.25 l aged and filtered tap water (membrane filter of 0.45 µm pore size) under constant dim light at 20°C. The animals were fed with non-limiting food concentrations (2 mg C/l) either of 100% Chlamydomonas sp. or of various mixtures of Chlamydomonas sp. and the two M. aeruginosa strains: In two treatments the animals were fed either with a mixture of 80% C. sp and 20% NIVA or with a mixture of 50% Chlamydomonas sp. and 50% NIVA. In one further treatment the animals were fed with a mixture of 90% Chlamydomonas sp. and 10% PCC−. Each treatment was triplicated, and animals were transferred daily into fresh water with saturating food concentrations. Somatic growth rates were calculated on day six as according to Wacker & Von Elert [36] as
for which (G) is the body dry-weight of a subsample of the animals at the beginning (G 0) and end (G t) of the experiment. Mean individual dry weights were mean values of two individuals. As according to Brzezinski & Von Elert [37] population growth rates (r) were calculated from daily survival and fecundity of the first clutch by using Euler’s equation,
in which lx is the survival rate and mx the size of the first clutch on day x. Population and somatic growth rates were calculated for each replicate and subsequently averaged to give the mean of the treatment. Population growth rates, calculated on the basis of fecundity at first reproduction, may be a better predictor of fitness in the presence of size-selective mortality than the calculation based on the first three clutches as used in Lampert & Trubetskova [38], since larger individuals are more vulnerable to visually orientated predators. We performed an equal variance test (Levene’s test) for each treatment to ensure a comparable intraspecific variability of both Daphnia species.
Preparation of Cyanobacterial Extracts and Daphnia Homogenates
Freeze-dried NIVA or PCC− were thoroughly homogenized and mixed separately. 50 mg of the resulting powder was suspended in 500 µl of 60% methanol and sonicated for 15 min followed by centrifugation (3 min at 104×g). The supernatant was subsequently separated from the residue and used as an extract in the protease assays. Controls with 60% methanol had no effects on protease activities.
Seven-day-old individuals of each of the seven D. magna and D. pulex clones grown on non-limiting food concentrations of Chlamydomonas sp. were homogenized with a Teflon pestle. Subsequently the homogenate was centrifuged (3 min at 1.4×104×g). In order to minimize protease inactivation due to autolytic degradation, the resulting supernatant was kept permanently on ice until it was used in the enzyme assays.
Protease Activity and Protease Inhibition Assays
The activity of the proteases chymotrypsin and trypsin in homogenates of all Daphnia clones was measured as according to Von Elert et al. [21]. The protein concentration of the supernatant was analyzed using a Qubit fluorometer and the appropriate Quant-iT™ Protein Assay Kit (Invitrogen, Carlsbad, USA) as according to the manufacturer’s standard protocol. For the protease activity and protease inhibition assays, SuccpNA (N-succinyl-L-alanyl-L-alanyl-L-prolyl-Lphenylalanine 4-nitroanilide, Sigma, 125 µM in DMSO) was used as a substrate for chymotrypsins, while BapNA (N-R-benzoyl-DL-arginine 4-nitroanilide hydrochloride, Sigma, 1.8 mM in DMSO) served as a substrate for trypsins. Trypsin and chymotrypsin assays were performed in a potassium phosphate buffer (0.1 M, pH 7.5). The absorption change was measured continuously for 10 min at 30°C at 390 nm with a Cary 50 photometer (Varian). Specific proteolytic activity was determined as nmol para-nitroanilide liberated per minute and µg protein for synthetic substrates. With regard to the chymotrypsin inhibition assays, homogenates of each of the D. magna and D. pulex clones were assayed after addition of 10–15 different concentrations of the NIVA extract. The resulting chymotrypsin activities were plotted as a function of extracted NIVA biomass per ml assay volume. By fitting a sigmoidal dose response curve, the concentration of extracted NIVA biomass which resulted in a 50% inhibition of Daphnia chymotrypsin activity (IC50) was calculated. The higher the IC50 values for the analyzed D. magna and D. pulex homogenates, the more tolerant their respective chymotrypsins were to chymotrypsin inhibitors from NIVA. Trypsin inhibition of the D. magna and D. pulex clones was assayed as above, except that different concentrations of the PCC− extract were used.
Data Analysis
For the inhibition assays of each single clone of D. magna and D. pulex, the protease activities were plotted as a function of extracted NIVA or PCC− biomass. Resulting IC50 values were calculated by fitting a sigmoid dose-response curve using the software Graph Pad Prism (GraphPad Software, Inc.). A Mann-Whitney U-test was used to determine whether one of the two Daphnia species exhibits stronger growth reductions on mixtures of M. aeruginosa, while a Student’s t-test was used to determine interspecific differences in protease inhibition and specific protease activity assays between the two species. Population growth rates were calculated for each individual per replicate independently using an R-script and subsequently averaged for each replicate. Mean Population growth rate of a Daphnia clone on a diet derived from the average of three replicates. All other statistical tests were performed using SigmaPlot 11 (Systat Software, Inc.). A significance level of p = 0.05 was applied to all statistical analyses.
Results
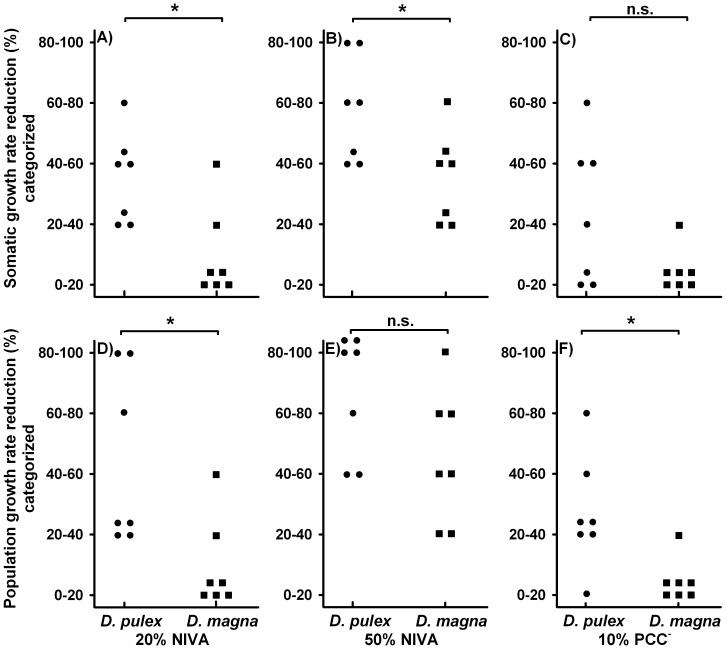
In order to test for interspecific differences between D. pulex and D. magna with regard to their tolerance to cyanobacteria, we performed population and somatic growth experiments on four different diets. Seven clones of each species, each originating from different habitats, were fed either with pure Chlamydomonas sp. or with mixtures of Chlamydomonas sp. and either of two M. aeruginosa strains, NIVA and PCC−. For each treatment (20% NIVA, 50% NIVA and 10% PCC−) as well as for both growth rate calculations (somatic growth and population growth) all D. magna and D. pulex clones were categorized into five groups according to their respective relative growth rate reduction (Figure 1a–f), whereas values ≤0 were regarded as no growth rate reduction on respective diet and were consequently grouped in the category 0–20%. Raw data underlying this classification are presented in Table 2. Equal variance tests revealed that the variability for both Daphnia species were not different for each treatment (Levene’s test, p>0.05). On a mixture of 80% Chlamydomonas sp. and 20% NIVA the categorized growth rates of the D. pulex clones were significantly more reduced than those of the D. magna clones (Figure 1a, d). This applies equally for the reduction of population growth rates (U-test: U = 6, p<0.05) and of somatic growth rates (U-test: U = 6, p<0.05): While five of seven D. magna clones were grouped into the first category (0%–20%) of relative somatic and population growth rate reduction, no D. pulex clones were grouped in this category when grown on a mixture with 20% NIVA. With increasing NIVA concentration from 20% to 50%, the categorized somatic and population growth rates decreased for both Daphnia species (Figure 1b, e). On 50% NIVA, the somatic growth rates for D. pulex were significantly more reduced than for D. magna (Figure 1b, U-test: U = 8.5, p<0.05), while this was not the case for the population growth rates (Figure 1e, U-test: U = 12, p = 0.128). However, with regard to the population growth rate reduction on 50% NIVA, four of seven D. pulex clones were grouped in the category of strongest growth reduction, whereas only one clone of D. magna was grouped in this category (Figure 1e). On 10% PCC−, the majority of the D. magna clones (six of seven) were grouped in the category of weakest somatic- and population growth rate reduction, representing a reduction of 0%–20% compared to the control treatment on 100% Chlamydomonas sp. (Figure 1c, f). Population growth rates of the D. pulex clones exhibited a more variable classification when grown on 10% PCC− and were significantly more reduced than those of the D. magna clones (Figure 1f, U-test: U = 6, p = 0.017). With regard to the somatic growth rate reduction on 10% PCC− no statistical differences were found between D. pulex and D. magna (Figure 1c, U-test: U = 12.5, p = 0.128).
Figure 1. Reduction of growth rates of each Daphnia clone fed on NIVA and PCC−.
Reduction of relative somatic (a, b, c) and population (c, d, e) growth rates of clones of D. pulex (circles) and D. magna (squares) in response to different mixtures of Chlamydomonas sp. and either of two Microcystis aeruginosa strains, NIVA or PCC−. Animals were fed either with 80% Chlamydomonas sp. and 20% NIVA (a, d), with 50% Chlamydomonas sp. and 50% NIVA (b, e) or with 90% Chlamydomonas sp. and 10% PCC− (c, f). Daphnia clones were classified into growth reduction categories according to the relative growth rate reduction on respective treatments compared to the control treatment on 100% Chlamydomonas sp. Significant differences (Mann-Whitney U-test, p<0.05) between species are indicated by an asterisk, while no differences are labeled with “n.s.”.
Table 2. Relative reduction of somatic and population growth rate of all D. magna and D. pulex clones on mixtures containing 20% NIVA, 50% NIVA and 10% PCC− as well as specific activity and IC50 values of Daphnia’s chymotrypsins (CT) and trypsins (T).
| Somatic growth rate reduction (%) | Population growth rate reduction (%) | Specific protease activity | IC50 values | ||||||||
| on | on | (nmol/min*µg prot) | (ng/ml) | ||||||||
| Daphnia spp. | Clone | 20% NIVA | 50% NIVA | 10% PCC− | 20% NIVA | 50% NIVA | 10% PCC− | CT | T | CT | T |
| D. pulex | Gerstel | 27.7 | 48.0 | 6.6 | 89.2 | 98.7 | 25.1 | 306.43 | 39.7 | 230.7 | 540.8 |
| D. pulex | NFL3 | 42.6 | 84.6 | 64.9 | 39.4 | 59.4 | 37.6 | 95.18 | 30.2 | 417.5 | 577.6 |
| D. pulex | Gräf | 59.1 | 57.6 | 7.7 | 92.6 | 100.0 | 30.1 | 228.88 | 67.56 | 311 | 1052 |
| D. pulex | Disp14 | 38.6 | 64.0 | 41.2 | 23.7 | 68.9 | 63.4 | 23.45 | 25.4 | 281.1 | 678.6 |
| D. pulex | Povi113 | 42.4 | 86.8 | 45.5 | 36.1 | 92.9 | 45.7 | 38.6 | 21.81 | 228.4 | 803.3 |
| D. pulex | Giev08 | 73.4 | 59.4 | 0.4 | 79.4 | 100.0 | 17.5 | 116.98 | 66.72 | 296.1 | 944.9 |
| D. pulex | TCO | 27.5 | 63.6 | 20.8 | 32.0 | 57.7 | 25.0 | 222.55 | 64.99 | 304.6 | 994 |
| D. magna | F10 | 52.4 | 76.5 | 17.2 | 46.7 | 80.8 | 19.1 | 353.63 | 51.7 | 195 | 388.5 |
| D. magna | G38 | 7.2 | 50.2 | −12.9 | 15.4 | 64.3 | 5.2 | 226.48 | 41.57 | 216.9 | 990.4 |
| D. magna | S15 | 25.2 | 52.2 | 12.1 | 34.2 | 73.8 | 14.6 | 545.09 | 92.53 | 269.3 | 893.3 |
| D. magna | P6 | 17.2 | 43.3 | 18.4 | 17.4 | 26.4 | 15.0 | 385.33 | 50.11 | 251.1 | 587.8 |
| D. magna | P | −1.4 | 39.9 | −9.8 | −0.1 | 53.6 | −10.9 | 358.1 | 43.24 | 244.2 | 1135 |
| D. magna | B | 10.9 | 22.4 | 28.9 | 9.8 | 24.3 | 36.6 | 215.75 | 45.23 | 271.1 | 920.3 |
| D. magna | W | 1.7 | 35.4 | 5.3 | −43.6 | 53.2 | −2.9 | 238.46 | 56.34 | 230.7 | 1222 |
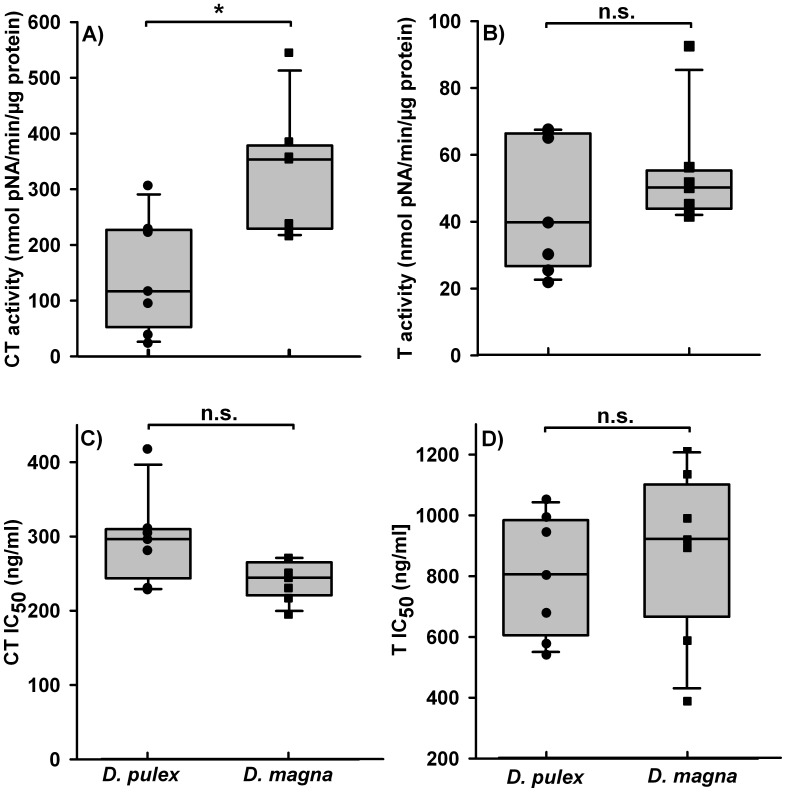
We furthermore quantified the specific activity of trypsins and chymotrypsins for each of the D. magna and D. pulex clones as possible causes for the observed differences in tolerance of the two species to cyanobacterial protease inhibitors. Mean specific chymotrypsin activity of D. magna (331.8±117.5 nmol/min/mg protein standard deviation (SD)) was dramatically higher (+225%) than the activity of D. pulex (147.4±106.8 nmol/min/mg protein SD; Figure 2a, t -test: t12 = −3.073, p<0.05). Mean specific trypsin activity for D. pulex was 45.2±20.6 nmol/min/mg protein SD, whereas it was 54.4±17.6 nmol/min/mg protein SD for D. magna (Figure 2b); however, these values were not different (t-test: t12 = −0.897, p = 0.387).
Figure 2. Sensitivity and specific activity of trypsins and chymotrypsins of each Daphnia clone.
Box plots showing two possible causes for the observed differences in the tolerance of D. magna and D. pulex to cyanobacteria with protease inhibitors were tested: (a) specific chymotrypsin (CT) and (b) trypsin (T) activity of the D. pulex (circles) and D. magna clones (squares). Inhibition of digestive proteases from homogenates of clones of D. pulex (circles) and D. magna (squares): (c) effects of extracts of M. aeruginosa strain NIVA on chymotrypsins, and (d) effects of extracts of M. aeruginosa strain PCC− on trypsins. Depicted are IC50 values, which represent the concentration of extracted dry weight of cyanobacterial biomass that is required to cause a 50% inhibition of respective proteases. Low IC50 values indicate a high sensitivity of proteases from the respective Daphnia clones. Medians are denoted by solid black lines, while the top and bottom box edges denote the first and third quartile. Whiskers denote the largest and smallest data within 1.5 times the interquartile range. Significant differences (Student’s t-test, p<0.05) among species are indicated by an asterisk; no differences are labeled with “n.s.”.
In order to test for possible additional causes of the interspecific differences in their tolerance to cyanobacterial protease inhibitors, the sensitivity of Daphnia’s trypsins and chymotrypsins to cyanobacterial protease inhibitors was determined. This was achieved by specifying the concentration of extracted cyanobacterial biomass that was needed to inhibit 50% (IC50) of either Daphnia’s chymotrypsins or their trypsins (Fig. 2c, d).Mean IC50 values of the effects of NIVA extracts on Daphnia’s chymotrypsins were 295.6±63.4 ng/ml SD for D. pulex and 239.8±27.7 ng/mL SD for D. magna, although these effects were not different (Figure 2c, t -test: t12 = 2.136, p = 0.054). When specifying the IC50 values of effects of PCC− extracts on Daphnia’s trypsins, no differences between D. pulex (798.7±205.6 ng/ml SD) and D. magna (876.8 ng/ml ±295.2 ng/ml SD) were detected (Figure 2d, t -test: t12 = −0.574, p = 0.577).
Discussion
Cyanobacterial blooms are harmful to many freshwater herbivorous grazers such as Daphnia. Owing to the dominance of cyanobacteria in eutrophic lakes and ponds in late summer [4], Daphnia genotypes from these habitats have to cope more frequently with the poor food quality of cyanobacterial carbon than genotypes from habitats free of cyanobacterial blooms. The causes for the poor assimilation of cyanobacterial carbon by Daphnia have been studied extensively in past decades, leading to the following observations: Firstly, colonial or filamentous cyanobacteria can mechanically interfere with Daphnia’s filtering apparatus [10]. Secondly, cyanobacteria lack essential sterols [12] and sufficient amounts of polyunsaturated fatty acids [14]. Thirdly, the production of hepaptotoxins such as microcystins can significantly reduce the fitness of Daphnia [19]. In the present study, all D. pulex clones and five out of seven D. magna clones exhibited a reduction in somatic and population growth rates at a concentration of 20% NIVA. The negative effects mentioned above can be ruled out as causal factors for the observed growth reductions, since the food mixtures used here consisted of saturating concentrations of the widely used, high quality reference food Chlamydomonas sp. Furthermore, both cyanobacterial strains as well as Chlamydomonas sp. were cultured as single cells and did not contain any microcystins. However, with two D. magna clones reductions were observed only with 50% NIVA (D. magna clone P and W, Table 2). This supports data of Lürling [39], who used the same strain of M. aeruginosa (NIVA) and reported a reduction in growth of D. magna clones at a concentration of ≥25% NIVA. On the mixture with PCC−, some D. magna clones and the majority of the D. pulex clones exhibited a reduction of population and somatic growth rate already at a concentration of 10%. Higher concentrations would probably have resulted in a significant growth rate reduction in all D. magna and D. pulex clones, since several other studies [40], [41] have reported a clear reduction in growth of daphnids at a concentration of 20% PCC−.
One possible explanation for the observed somatic and population growth rate reduction of the D. magna and D. pulex clones in response to cyanobacteria could be the result of dietary inhibition of either Daphnia’s digestive chymotrypsins or trypsins. In this study somatic and population growth rates of D. magna and D. pulex served as a measure of tolerance to microcystin-free cyanobacteria and as an approach to test for interspecific differences. In several cases it has been demonstrated that coexistence of Daphnia with microcystin-producing cyanobacteria leads to local adaptation of the Daphnia populations, as was evidenced by increased tolerance to cyanobacteria [24], [42]. These findings support the notion that the presence of cyanobacteria positively selects for more tolerant Daphnia genotypes.
Several studies have already demonstrated high intraspecific variability of Daphnia’s tolerance to cyanobacteria [24], [25]. In the present study we also found intraspecific variability of D. magna and D. pulex in the tolerance to cyanobacteria with protease inhibitors. However, an equal variance test revealed that the variability for both Daphnia species were not different. With regard to interspecific comparisons we have shown for the first time that D. magna exhibits a higher mean tolerance to cyanobacteria with protease inhibitors than D. pulex. Microevolutionary adaptation of Daphnia populations to cyanobacteria has been experimentally confirmed: Exposure of a mixed population of several Daphnia clones to a microcystin-producing strain of M. aeruginosa resulted in an enhanced tolerance in subsequent generations [25]. Such a maternally transferred increase in tolerance of Daphnia’s offspring generation has also been demonstrated for the microcystin-free M. aeruginosa strain PCC− [43] which was also used in the present study.
Currently it is not clear to which extent the observed interspecific differences between D. magna and D. pulex in tolerance to cyanobacteria with protease inhibitors are affected by physiological plasticity of the clones investigated here. Using a single clone of D. magna, Von Elert et al. [18] have shown that a physiological plasticity at the protein level in Daphnia in terms of expressing different isoforms of digestive enzymes leads to increased tolerance to cyanobacterial protease inhibitors. In the present study we could not find a correlation between less sensitive proteases and higher tolerance to the cyanobacterial strains. The protease inhibition assays revealed that the tolerance of chymotrypsins and trypsins to cyanobacterial protease inhibitors did not differ between D. magna and D. pulex. However, the mean specific chymotrypsin activity of D. magna was significantly higher than that of D. pulex. This elevated specific chymotrypsin activity of D. magna coincides with less reduced somatic and population growth rates in the presence of chymotrypsin inhibitors. Schwarzenberger et al. [40] have demonstrated that the same D. magna clone as used by Von Elert et al. [18] responded to dietary protease inhibitors by increased expression of trypsin and chymotrypsin genes. However, it remains to be tested, if differences in plasticity of protease expression cause the observed higher tolerance to cyanobacteria with protease inhibitors in D. magna than in D. pulex. With regard to the significantly higher population growth rates of D. magna on PCC− than those of D. pulex, we couldn’t find differences in species-specific trypsin characteristics, neither with respect to the specific activity nor to the respective sensitivity.
Besides protease inhibitors, the observed differences in growth on cyanobacteria between D. magna and D. pulex could also result from differences in the tolerance to other toxic compounds such as lipopolysaccharides [44] or unidentified toxins. Additionally, we cannot rule out the possibility that potentially synergistic interactions between bioactive compounds may have important negative effects on the fitness of Daphnia. Such synergistic effects have been reported for the toxicity of artificial nano-particles and arsenic on the fitness of daphnids [45]. However, we are not aware of any cyanobacterial metabolites, for which synergistic effects on herbivores have been demonstrated. Furthermore, when looking at possible defense mechanisms of Daphnia against cyanobacterial toxins, more efficient detoxification mechanisms in D. magna than in D. pulex could also be a reason for the higher tolerance of D. magna to the cyanobacterial diets. One possibility of reducing toxic effects in Daphnia is to biotransform toxins by conjugation to glutathione, which has already been demonstrated for the cyanobacterial hepatotoxin microcystin-LR [46]. However, even for this conjugation of microcystins by Daphnia no intra- or interspecific comparisons are available. Although similar detoxification mechanisms of protease inhibitors in Daphnia have not been reported so far, they cannot be excluded as a possible causal factor for the higher tolerance of D. magna to diets containing cyanobacteria with protease inhibitors. Additionally, Rohrlack et al. [47] reported a reduction of the ingestion rate in D. galeata after a short time exposure (15 min.) with 100% PCC−, which could also result in a reduction of somatic and population growth. Here, experimental individuals were fed for at least six days a PCC− proportion of 10%, while 90% of the carbon resulted from the good reference food Chlamydomonas sp. With regard to NIVA there was no detectable inhibition of the ingestion process of one D. magna clone even at a proportion of 100% NIVA in the diet (Kuster, unpublished). Thus, it seems unlikely that inhibition effects of the ingestion process contributed to the observed growth reduction on cyanobacterial diets. However, since we haven’t measured ingestion rates for all D. magna and D. pulex clones used here, we could not clearly rule out this possibility.
As a result of continuing global warming, negative effects of toxic cyanobacteria on human health, livestock and the whole ecosystem will become stronger in the near future [1]. Much effort is therefore invested in preventing or controlling cyanobacterial blooms. Besides reducing nutrient input into lakes and ponds, the control of cyanobacterial blooms through biological agents such as bacteria, viruses and unicellular grazers has been discussed extensively in the last decades [48], [49]. Depending on initial conditions, Daphnia may also control the development of bloom-forming cyanobacteria and may even suppress established cyanobacterial blooms [50]. The present study suggests that D. magna may control the development of cyanobacterial blooms more efficiently than D. pulex due to significantly higher tolerance to cyanobacteria with protease inhibitors. In light of the increasing temperatures as a consequence of global warming, our results suggest that toxic cyanobacterial blooms and coinciding harmful effects to the ecosystem will occur more frequently in lakes with D. pulex than in lakes with D. magna.
Acknowledgments
We thank L. Schäfer for her invaluable help in conducting the experiments, Y. Reydelet for providing the D. pulex clone Giev08, P. Fink, T. Sadler and C. Effertz for their helpful comments and Hanne Krisch for technical assistance.
Funding Statement
This study was supported by two grants to EvE from the German Research Foundation (DFG): grant number El 179/6-1 and by a grant within the Collaborative Research Centre 680 Molecular Basis of Evolutionary Innovations. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Paerl HW, Huisman J (2008) Climate - Blooms like it hot. Science 320: 57–58. [DOI] [PubMed] [Google Scholar]
- 2. Carmichael WW (1994) Toxins of cyanobacteria. Scientific American 270: 78–86. [DOI] [PubMed] [Google Scholar]
- 3. Codd GA, Morrison LF, Metcalf JS (2005) Cyanobacterial toxins: risk management for health protection. Toxicology and Applied Pharmacology 203: 264–272. [DOI] [PubMed] [Google Scholar]
- 4. Johnk KD, Huisman J, Sharples J, Sommeijer B, Visser PM, et al. (2008) Summer heatwaves promote blooms of harmful cyanobacteria. Global Change Biology 14: 495–512. [Google Scholar]
- 5. Dokulil MT, Teubner K (2000) Cyanobacterial dominance in lakes. Hydrobiologia 438: 1–12. [Google Scholar]
- 6. DeMott WR, Tessier AJ (2002) Stoichiometric constraints vs. algal defenses: Testing mechanisms of zooplankton food limitation. Ecology 83: 3426–3433. [Google Scholar]
- 7. Demott WR, Müller-Navarra DC (1997) The importance of highly unsaturated fatty acids in zooplankton nutrition: evidence from experiments with Daphnia, a cyanobacterium and lipid emulsions. Freshwater Biology 38: 649–664. [Google Scholar]
- 8. Threlkeld ST (1979) Midsummer dynamics of 2 Daphnia species in Wintergreen Lake, Michigan. Ecology 60: 165–179. [Google Scholar]
- 9. Hansson LA, Gustafsson S, Rengefors K, Bomark L (2007) Cyanobacterial chemical warfare affects zooplankton community composition. Freshwater Biology 52: 1290–1301. [Google Scholar]
- 10. Porter KG, Mcdonough R (1984) The energetic cost of response to blue-green-algal filaments by cladocerans. Limnology and Oceanography 29: 365–369. [Google Scholar]
- 11. Von Elert E (2002) Determination of limiting polyunsaturated fatty acids in Daphnia galeata using a new method to enrich food algae with single fatty acids. Limnology and Oceanography 47: 1764–1773. [Google Scholar]
- 12. Von Elert E, Martin-Creuzburg D, Le CozJR (2003) Absence of sterols constrains carbon transfer between cyanobacteria and a freshwater herbivore (Daphnia galeata). Proceedings of the Royal Society of London Series B-Biological Sciences 270: 1209–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Martin-Creuzburg D, Von Elert E (2004) Impact of 10 dietary sterols on growth and reproduction of Daphnia galeata . Journal of Chemical Ecology 30: 483–500. [DOI] [PubMed] [Google Scholar]
- 14. Martin-Creuzburg D, Von Elert E, Hoffmann KH (2008) Nutritional constraints at the cyanobacteria-Daphnia magna interface: The role of sterols. Limnology and Oceanography 53: 456–468. [Google Scholar]
- 15.Sivonen K, Jones G (1999) Cyanobacterial toxins. Toxic cyanobacteria in water: A guide to their public health consequences. Monitoring and Management: 41–111.
- 16. Gademann K, Portmann C (2008) Secondary metabolites from cyanobacteria: Complex structures and powerful bioactivities. Current Organic Chemistry 12: 326–341. [Google Scholar]
- 17. Agrawal MK, Zitt A, Bagchi D, Weckesser J, Bagchi SN, et al. (2005) Characterization of proteases in guts of Daphnia magna and their inhibition by Microcystis aeruginosa PCC 7806. Environmental Toxicology 20: 314–322. [DOI] [PubMed] [Google Scholar]
- 18. von Elert E, Zitt A, Schwarzenberger A (2012) Inducible tolerance to dietary protease inhibitors in Daphnia magna . Journal of Experimental Biology 215: 2051–2059. [DOI] [PubMed] [Google Scholar]
- 19. Lürling M, van der Grinten E (2003) Life-history characteristics of Daphnia exposed to dissolved microcystin-LR and to the cyanobacterium Microcystis aeruginosa with and without microcystins. Environmental Toxicology and Chemistry 22: 1281–1287. [PubMed] [Google Scholar]
- 20. Rohrlack T, Dittmann E, Börner T, Christoffersen K (2001) Effects of cell-bound microcystins on survival and feeding of Daphnia spp. Applied & Environmental Microbiology 67: 3523–3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Von Elert E, Agrawal MK, Gebauer C, Jaensch H, Bauer U, et al. (2004) Protease activity in gut Daphnia magna: evidence for trypsin and chymotrypsin enzymes. Comparative Biochemistry and Physiology B-Biochemistry & Molecular Biology 137: 287–296. [DOI] [PubMed] [Google Scholar]
- 22.Kuster C, Schwarzenberger A, von Elert E (2012) Seasonal dynamics of sestonic protease inhibition: impact on Daphnia populations. Hydrobiologia 1–14.
- 23. Czarnecki O, Henning M, Lippert I, Welker M (2006) Identification of peptide metabolites of Microcystis (Cyanobacteria) that inhibit trypsin-like activity in planktonic herbivorous Daphnia (Cladocera). Environmental Microbiology 8: 77–87. [DOI] [PubMed] [Google Scholar]
- 24. Sarnelle O, Wilson AE (2005) Local adaptation of Daphnia pulicaria to toxic cyanobacteria. Limnology and Oceanography 50: 1565–1570. [Google Scholar]
- 25. Gustafsson S, Hansson LA (2004) Development of tolerance against toxic cyanobacteria in Daphnia . Aquatic Ecology 38: 37–44. [Google Scholar]
- 26. Hairston NG, Holtmeier CL, Lampert W, Weider LJ, Post DM, et al. (2001) Natural selection for grazer resistance to toxic cyanobacteria: Evolution of phenotypic plasticity? Evolution 55: 2203–2214. [DOI] [PubMed] [Google Scholar]
- 27. Blom JF, Baumann HI, Codd GA, Juttner F (2006) Sensitivity and adaptation of aquatic organisms to oscillapeptin J and [D-Asp(3),(E)-Dhb(7)]microcystin-RR. Archiv für Hydrobiologie 167: 547–559. [Google Scholar]
- 28. Agrawal MK, Bagchi D, Bagchi SN (2001) Acute inhibition of protease and suppression of growth in zooplankter, Moina macrocopa, by Microcystis blooms collected in Central India. Hydrobiologia 464: 37–44. [Google Scholar]
- 29. Colbourne JK, Pfrender ME, Gilbert D, Thomas W, Tucker A, et al. (2011) The ecoresponsive genome of Daphnia pulex . Science 331: 555–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. DeMott WR, Pape BJ (2005) Stoichiometry in an ecological context: testing for links between Daphnia P-content, growth rate and habitat preference. Oecologia 142: 20–27. [DOI] [PubMed] [Google Scholar]
- 31. Watanabe H, Tatarazako N, Oda S, Nishide H, Uchiyama I, et al. (2005) Analysis of expressed sequence tags of the water flea Daphnia magna . Genome 48: 606–609. [DOI] [PubMed] [Google Scholar]
- 32. Von Elert E, Oberer L, Merkel P, Huhn T, Blom JF (2005) Cyanopeptolin 954, a chlorine-containing chymotrypsin inhibitor of Microcystis aeruginosa NIVA Cya 43. Journal of Natural Products 68: 1324–1327. [DOI] [PubMed] [Google Scholar]
- 33. Weckesser J, Martin C, Jakobi C (1996) Cyanopeptolins, depsipeptides from cyanobacteria. Systematic and Applied Microbiology 19: 133–138. [Google Scholar]
- 34. Von Elert E, Jüttner F (1997) Phosphorus limitation and not light controls the extracellular release of allelopathic compounds by Trichormus doliolum (cyanobacteria). Limnology and Oceanography 42: 1796–1802. [Google Scholar]
- 35. Dittmann E, Neilan BA, Erhard M, vonDohren H, Borner T (1997) Insertional mutagenesis of a peptide synthetase gene that is responsible for hepatotoxin production in the cyanobacterium Microcystis aeruginosa PCC 7806. Molecular Microbiology 26: 779–787. [DOI] [PubMed] [Google Scholar]
- 36. Wacker A, von Elert E (2001) Polyunsaturated fatty acids: Evidence for non-substitutable biochemical resources in Daphnia galeata . Ecology 82: 2507–2520. [Google Scholar]
- 37. Brzezinski T, von Elert E (2007) Biochemical food quality effects on a Daphnia hybrid complex. Limnology and Oceanography 52: 2350–2357. [Google Scholar]
- 38. Lampert W, Trubetskova I (1996) Juvenile growth rate as a measure of fitness in Daphnia. Functional Ecology 10: 631–635. [Google Scholar]
- 39. Lürling M (2003) Daphnia growth on microcystin-producing and microcystin-free Microcystis aeruginosa in different mixtures with the green alga Scenedesmus obliquus . Limnology and Oceanography 48: 2214–2220. [Google Scholar]
- 40. Schwarzenberger A, Zitt A, Kroth P, Mueller S, von Elert E (2010) Gene expression and activity of digestive proteases in Daphnia: effects of cyanobacterial protease inhibitors. BMC physiology 10: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schwarzenberger A, Kuster CJ, von Elert E (2012) Molecular mechanisms of tolerance to cyanobacterial protease inhibitors revealed by clonal differences in Daphnia magna . Mol Ecol 21: 4898–4911 10.1111/j.1365-294X.2012.05753.x. [DOI] [PubMed] [Google Scholar]
- 42. Hairston NG, Lampert W, Caceres CE, Holtmeier CL, Weider LJ, et al. (1999) Lake ecosystems - Rapid evolution revealed by dormant eggs. Nature 401: 446. [Google Scholar]
- 43.Schwarzenberger A, von Elert E (2012) Cyanobacterial protease inhibitors lead to maternal transfer of increased protease gene expression in Daphnia. Oecologia. 10.1007/s00442-012-2479-5. [DOI] [PubMed]
- 44. Pires LMD, Sarpe D, Brehm M, Ibelings BW (2011) Potential synergistic effects of microcystins and bacterial lipopolysaccharides on life history traits of Daphnia galeata raised on low and high food levels. Aquatic Toxicology 104: 230–242. [DOI] [PubMed] [Google Scholar]
- 45. Wang D, Hu J, Irons DR, Wang J (2011) Synergistic toxic effect of nano-TiO2 and As(V) on Ceriodaphnia dubia. Science of the Total Environment 409: 1351–1356. [DOI] [PubMed] [Google Scholar]
- 46. Pflugmacher S, Wiegand C, Oberemm A, Beattie KA, Krause E, et al. (1998) Identification of an enzymatically formed glutathione conjugate of the cyanobacterial hepatotoxin microcystin-LR: the first step of detoxication. Biochimica et Biophysica Acta-General Subjects 1425: 527–533. [DOI] [PubMed] [Google Scholar]
- 47. Rohrlack T, Dittmann E, Henning M, Borner T, Kohl JG (1999) Role of microcystins in poisoning and food ingestion inhibition of Daphnia galeata caused by the cyanobacterium Microcystis aeruginosa. Applied and Environmental Microbiology 65: 737–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sigee DC, Glenn R, Andrews MJ, Bellinger EG, Butler RD, et al. (1999) Biological control of cyanobacteria: principles and possibilities. Hydrobiologia 395: 161–172. [Google Scholar]
- 49. Tucker S, Pollard P (2005) Identification of cyanophage Ma-LBP and infection of the cyanobacterium Microcystis aeruginosa from an Australian subtropical lake by the virus. Applied and Environmental Microbiology 71: 629–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sarnelle O (2007) Initial conditions mediate the interaction between Daphnia and bloom-forming cyanobacteria. Limnology and Oceanography 52: 2120–2127. [Google Scholar]
- 51. Koch U, von Elert E, Straile D (2009) Food quality triggers the reproductive mode in the cyclical parthenogen Daphnia (Cladocera). Oecologia 159: 317–324. [DOI] [PubMed] [Google Scholar]
- 52. Schaack S, Pritham EJ, Wolf A, Lynch M (2010) DNA transposon dynamicsin populations of Daphnia pulex with and without sex. Proceedings of the Royal Society of London Series B-Biological Sciences 277: 2381–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Matthes M (2004) Low genotypic diversity in a Daphnia pulex population in a biomanipulated lake: the lack of vertical and seasonal variability. Hydrobiologia 526: 33–42. [Google Scholar]
- 54. Haag CR, McTaggart SJ, Didier A, Little TJ, Charlesworth D (2009) Nucleotide polymorphism and within-gene recombination in Daphnia magna and D. pulex, two cyclical parthenogens. Genetics 182: 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kuster CJ, von Elert E (2012) High-resolution melting analysis: a genotyping tool for population studies on Daphnia . Molecular Ecology Resources 12: 1048–1057. [DOI] [PubMed] [Google Scholar]
- 56. De Meester L (1994) Life-Histories and habitat selection in Daphnia - Divergent life-histories of Daphnia magna clones differing in phototactic behavior. Oecologia 97: 333–341. [DOI] [PubMed] [Google Scholar]
- 57. Lampert W, Rothhaupt KO (1991) Alternating dynamics of rotifers and Daphnia magna in a shallow lake. Archiv für Hydrobiologie 120: 447–456. [Google Scholar]
- 58. Pijanowska J, Weider LJ, Lampert W (1993) Predator-mediated genotypic shifts in a prey population - Experimental evidence. Oecologia 96: 40–42. [DOI] [PubMed] [Google Scholar]