Abstract
The enzymatic addition of a single β-D-N-acetylglucosamine sugar molecule on serine and/or threonine residues of protein chains is referred to as O-GlcNAcylation. This novel form of post-translational modification, first reported in 1984, is extremely abundant on nuclear and cytoplasmic proteins and has site specific cycling dynamics comparable to that of protein-phosphorylation. A nutrient and stress sensor, O-GlcNAc abnormalities underlie insulin resistance and glucose toxicity in diabetes, neurodegenerative disorders and dysregulation of tumor suppressors and oncogenic proteins in cancer. Recent advances have helped understand the biochemical mechanisms of GlcNAc addition and removal and have opened the door to developing key inhibitors towards this type of protein modification. Advanced methods in detecting and measuring O-GlcNAcylation have assisted in delineating its biological roles in a variety of cellular processes and diseased states. Availability of facile glycomic techniques are allowing for the exponential growth in the study of protein O-GlcNAacylation and are helping to elucidate key biological roles of this novel PTM.
1. Introduction
Generation of diversity within the proteome of a cell can occur either at a transcriptional level via RNA splicing, or at the post-translational level via covalent modifications of protein amino acid backbones. These protein modifications can take place sometimes by proteolysis but most often by the enzymatic addition of a chemical group to an amino acid side chain.1 Of the different forms of post-translational modifications (PTMs), glycosylation is the most abundant and structurally diverse, and is predicted to occur in as many as 80–90% of all extracellular and nucleocytoplasmic proteins.2 Prior to the early 1980’s, glycosylation generally referred to the N-linked and O-linked glycosylation present predominantly on cell surface and secreted proteins. These cell surface or extracellular glycans are often large complex sugar structures added onto protein chains maturing through the ER and Golgi apparatus that mediate protein quality control, cell-cell adhesion, antibody recognition, extracellular signaling and myriad biological processes. In the early 1980s, a previously undetected form of protein glycosylation, O-linked N-acetylglucosamine (O-GlcNAc) was found to occur on proteins within the nucleus and cytoplasm.3 This form of protein glycosylation is distinctly different from the canonical multimeric long chain glycan structures added onto extracellular proteins. O-GlcNAc is a single N-acetylglucosamine monomer covalently attached to serine or threonine residues via a β-C2 linkage. O-GlcNAc occurs nearly exclusively on nuclear, cytosolic or mytochondrial proteins without being further elongated with complex sugar structures. The cycling dynamics of O-GlcNAc (the addition and removal of N-acetylglucosamine at a particular site, which may occur multiple times within a protein’s lifetime) bears semblance to protein phosphorylation. And over the past three decades a wealth of information regarding the functional and regulatory aspects pertaining to this post-translational modification have been discovered.
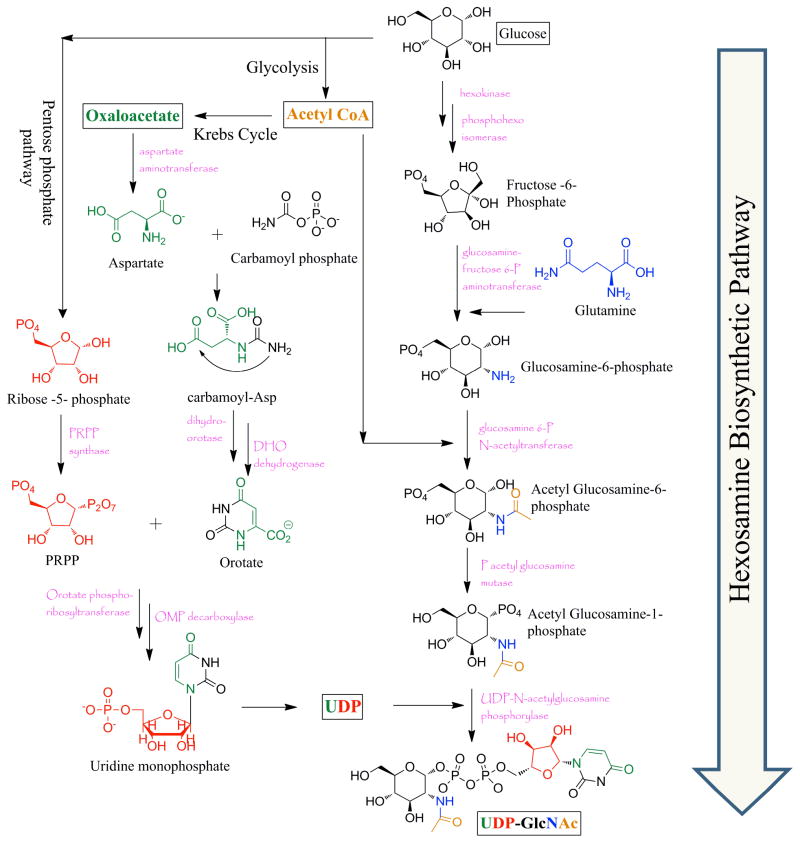
O-GlcNAcylation has been documented in some bacteria, filamentous fungi, all metazoans examined to date, including, insects, plants and animals. Within cells the highest density of O-GlcNAc is found on nuclear pore proteins, as well as cytoskeletal proteins. However, overall abundance of O-GlcNAcylation is highest within the nucleus. Like phosphorylation, O-GlcNAcylation is sub-stoichiometric at any single modification site, contributing to difficulties in detection by standard proteomic techniques. The extent of O-GlcNAcylation on polypeptides is also highly dependent on the cellular concentrations of its donor substrate (UDP-N-acetylglucosamine, UDP-GlcNAc), sub-cellular compartmentalization of the protein, and differentiation state of the cell. Flux through glucose, amino acid, fatty acid and nucleotide metabolic pathways feed the flux of the hexosamine biosynthetic pathway (HBP), which generates UDP-GlcNAc (Figure 1). Approximately 2 to 5% of all cellular glucose is estimated to enter the HBP where Glucosamine-6-phosphate is generated via an amino-transfer reaction between Fructose-6-phosphate and the amino acid glutamine by the action of the rate limiting enzyme, glutamine:fructose-6-phosphate amidotransferase (GFAT). This pathway also intersects with production of acetyl-CoA, the product of glycolysis, to give rise to N-acetylglucosamine. Ribose sugar moieties processed via the pentose phosphate pathway feed into pyrimidine biosynthesis (oxaloacetate from kreb’s cycle forms aspartate; that along with carbamoyl phosphate and phosphoribosyl pyrophosphate (PRPP) fluxes nucleotide biosynthesis) that ultimately attaches the uridine -di-phosphate moiety in the synthesis of UDP-GlcNAc. GFAT is the rate limiting enzyme that controls the HBP flux and is regulated by feedback mechanisms. Exquisite control of the donor sugar generation, thus points towards functional aspects of O-GlcNAcylation as being regulatory to cellular signaling, while also being dependent upon nutrient availability.4, 5
Figure 1.
The hexosamine biosynthetic pathway showing the overall biosynthesis of UDP-GlcNAc. Flux from glycolysis, Kreb’s cycle, the pentose phosphate pathway, pyrimidine biosynthesis and amino acid metabolism feeds in to the production of the UDP donor sugar.
The incorporation of O-linked N-acetylglucosamine, is enzymatically accomplished by a single protein called the O-GlcNAc transferase (OGT). Unlike other PTMs this one enzyme is capable of modifying the whole gamut of proteins currently known to be O-GlcNAcylated. Also no consensus sequences have been identified in substrate proteins. Thus a prevailing hypothesis suggests that this PTM is not only directed to specific sites by peptide sequence, but also is mainly directed via protein-protein interaction networks through OGT targeting proteins. The hydrolysis of O-GlcNAc from a protein is also carried out by a single enzyme O-GlcNAc hydrolase, known as O-GlcNAcase (OGA). Akin to OGT, the function of OGA is also predicted to be targeted via protein interaction domains, with both proteins sometimes occurring within the same complex. These enzymes and their modes of action will be discussed in greater detail below.
Functionally, post-translational addition/removal of O-GlcNAc have been shown to be extremely important in cell cycle progression, transcription, intracellular signaling, nutrient sensing, and neuronal plasticity. Knockout of the OGT gene in mice has been shown to be embryonically lethal and OGT mutation in somatic cells causes T-cell death, neuronal tau hyperphosphorylation, and growth arrest in fibroblasts with altered expression profiles in a number of transcription factors – thus highlighting the extreme importance of O-GlcNAcylation in development.6 OGT deletion in mouse fibroblasts is associated with reduced growth, increased p27 – the cyclin inhibitor, as well as increased cell death. Analyses of conditional knockout phenotypes are complicated by tissue targeted lethality. Conversely OGA overexpression has been shown to induce a mitotic exit phenotype, delineating the significant role of O-GlcNAc in cell survival and growth.
One of the critical ways that O-GlcNAc regulates signaling pathways is via direct crosstalk with protein phosphorylation. An O-GlcNAc sensitive fluorescence resonance energy transfer (FRET) probe found that changes in O-GlcNAcylation is spatiotemporally controlled in response to signal inducing stimuli.7, 8 Dynamics of O-GlcNAcylation was affected downstream of the PI3 kinase pathway – pointing to a complex interplay between phosphorylation and GlcNAcylation. Overexpression of OGT has been shown to cause dramatic reduction in cyclin dependent kinase 1 (CDK1) while changing the phosphorylation profiles for aurora and polo kinases.9 Reduction in OGA activity via treatment with highly specific cell permeable inhibitors, raised overall protein O-GlcNAc levels three fold with concomitant changes at 700 different phosphorylation sites.10 Such data underscores the depth of crosstalk between these two important intracellular signaling networks. RNA polymerase II O-GlcNAcylation though known to occur since 1993, remained functionally obscure, till recent studies showed that O-GlcNAc cycling is important for the formation of the preinitiation complex.11 OGT was also shown to form a component of this complex, with both OGT and OGA being enriched at the start sites of transcription, thereby regulating the transcription cycle. O-GlcNAc was also recently shown to block CREB dependent transcription and thus has a role in cellular plasticity and axonal and dendritic growth.12 Global glycoproteomic studies identified a number of O-GlcNAc modified ribosomal proteins. Subsequently, this modification was found enriched within active polysomes and some core ribosomal proteins with the modifying enzymes associating with active ribosomes.13 Nucleolar fractions were shown to actively exclude OGT though not OGA, pointing to the importance of this PTM in translation and ribosomal biogenesis. Overexpression of OGT results in polyploidy and affects cytokinesis by associating with the spindle and cellular mid-body during M phase.14 Inhibitory phosphorylation of CDK1 is increased during OGT overexpression thus resulting in decreased activity of proteins necessary for cell cycle progression – Map4, kinesin and others.9 During DNA damage and concomitant increased cell death, p53 gets activated and stabilized.15 This stabilization has been shown to act via O-GlcNAcylation of p53 that ultimately lowers the degradative phosphorylation signal – highlighting the importance of protein O-GlcNAcylation during stress conditions, which may be important in cancers. Altered O-GlcNAcylation has long been implicated in cancer cell survival by overcoming hypoxia and hypoglycemia.16 Recently Linda Hsieh-Wilson’s lab showed that O-GlcNAc on phosphofructokinase 1(PFK1) lowered its activity thereby increasing glucose flux through the pentose phosphate pathway and aiding in the growth of cancer cells. These examples point to the critical importance of protein-O-GlcNAcylation in cell growth, survival as well as in disease progression.
In this review we primarily focus on the chemistry of O-GlcNAc, the cycling enzymes and their mode of actions and also take a general overview of some of the methods developed to study protein O-GlcNAc modification. Each of these specific fields is broad and has been extensively reviewed by us and others elsewhere.10, 17–20 The examples presented are by no means exhaustive but are given to provide a taste of the recently published work done in this area.
2. O-GlcNAc as a chemical modification – chemistry of the catalyzing enzymes
2.1. O-GlcNAc Transferase
β-N-acetylglucosaminyltransferase, OGT, is a unique glycosyltransferase in that it is capable of GlcNAcylating myriad different protein substrates with no well-defined consensus sequence among them (though roughly 50% of OGT substrates share a proline-valine-serine motif). The OGT gene resides on the X chromosome at locus Xq13 occurring near the centromere – a region implicated in Parkinson’s disease.21 It is expressed ubiquitously throughout all tissues, but is most abundant in the brain, pancreas, thymus, uterus, heart and kidneys. Structurally OGT consists of N-terminal tetratricopeptide repeat (TPR) domains and a multiunit catalytic C-terminal. The TPRs are predicted to be engaging in protein-protein interaction networks and identifying substrate peptides. Three different isoforms of OGT are reported to exist, differing mainly in the length of their TPRs. The nuclear and cytoplasmic OGT (ncOGT) consists of 13 TPRs while the mitochondrial isoform contains 9.5 TPRs. The predicted mitochondrial isoform also has a distinct 50 amino acid N-terminal sequence for mitochondrial targeting and presumed inner-membrane association. Another isoform of OGT known as the short form contains only 3.5 TPRs. This isoform has been found to be active against peptides, but not very active on proteins.21 Very little is known about the mitochondrial and soluble isoforms and almost nothing is known about their activities and substrate specificities. Native OGT occurs as a trimeric holoenzyme with the TPRs acting as trimerization clusters. Sometimes full length ncOGT is also associated with the soluble isoform, especially within neuronal post-synaptic densities. The crystal structure of OGT shows the multiunit catalytic C-terminal to be composed of 3 different domains – an N-Cat, a C-Cat and an intervening Int-D region. Both the N-Cat and C-Cat domains are characterized by the presence of Rossman like folds typical of GT-B family of glycosyltransferases.22–24 The TPR and the catalytic domain are joined by an inter-domain helical insertion that acts like a hinge and appear to be controlling the substrate binding cleft.23, 25 Interestingly the intervening region is reportedly absent from plant and bacterial homologs of OGT.22, 25
OGT catalyzes the addition of GlcNAc moieties onto protein serine and threonine residues utilizing the activated sugar donor UDP-GlcNAc. Falling in the family of inverting glycosyltransferases, OGT converts the α-donor sugar anomer to a peptide β-linked residue – the reaction thus following an inversion of configuration around the C-1 sugar carbon. This likely occurs via a single displacement mechanism where nucleophilic attack by Ser/Thr hydroxyl groups from the β face of the C-1 sugar results in a loss of the α-linked UDP moiety.25 Structural analyses showed that OGT probably operates via an ordered sequential mechanism in which the UDP-donor sugar binds to the catalyzing protein first. Subsequent binding of the target peptide by the amino acids surrounding the UDP-binding cleft closes the entire structure and facilitates nucleophilic displacement. The crystal structure also indicates that the substrate peptide lies over the nucleotide-sugar binding pocket. Histidine 498 in the active site acts as the general base and activates the incoming ser/thr nucleophile, which displaces the UDP moiety in a concerted SN2-like reaction.23 Mutational analysis of this histidine residue resulted in greater than 90% loss of OGT activity. Structural proximity hints at lysine 842 as being the general acid assisting in the release of the uridine leaving group – though this has not been confirmed. The activity of OGT is highly dependent on the concentration of both UDP-GlcNAc as well as the substrate polypeptide. The Km values of UDP-GlcNAc for different proteins reportedly vary between 1 to 20 μM, while Km for proteins occur within 2 to 7 μM.26 This means that different proteins get O-GlcNAcylated at varying rates in specific concentrations of glucose or UDP-GlcNAc, pointing towards differential cellular activity and effects of OGT in response to glucose/nutrient level alterations. The activity of OGT is also controlled by various post-translational modifications on OGT. Tyrosine phosphorylation of OGT due to insulin receptor and CaMKIV signaling increases its activity. O-GlcNAcylation of OGT gets reduced during M phase and increases during G1 phase, thus affecting cell cycle progression. Finally multimeric structures of OGT can potentially impact its activity though no studies of this issue have been reported.
2.2. O-GlcNAc Hydrolase
O-GlcNAc hydrolase (OGA) catalyzes the loss of O-linked N-acetylglucosamine from polypeptide chains. A member of G84 family of glyosyl hydrolases, it is highly conserved, with pancreas, brain and thymus showing the highest levels of OGA expression. The chromosome 10q24.1 locus, which is associated with Alzheimer’s disease (AD), is where the OGA gene has been mapped. Two other genes HexA and HexB encode similar glycolytic hydrolases, which are located within the lysosome. But the cytoplasmic OGA differs from these lysosomal analogs in being optimal at neutral pH and having no activity towards O-GalNAc residues. Though no crystal structure of the human enzyme has been solved, sequence analysis and structure determination of two bacterial homologs indicate the presence of 2 distinct domains27 – consisting of an N-terminal catalytic domain and a C-terminal putative histone acetyltransferase (HAT) domain.28 However, there is controversy as to whether OGA actually possesses any HAT activity. The linker region between these domains is cleaved by caspase-3 without loss of substantial activity or overall structure. OGA exists in two distinct isoforms, the full length and a smaller isoform, which has a portion of its C-terminal tail cut off.
All glycosyl hydrolases catalyze their hydrolytic reactions either by retaining or inverting the configuration at the anomeric sugar carbon. OGA falls within the family of retaining type glycosidases, indicating that the β linkage between the C1 of N-acetylglucosamine and the Ser/Thr hydroxyl of the substrate peptide is broken to generate a free β-O-GlcNAc residue. Mechanistic analyses of glycosidases present two alternative routes for retention of configuration both of which proceed via the formation of an oxocarbenium ion transition state. In one case, an enzyme assisted SN1 type catalytic reaction occurs in which a nucleophilic residue within the hydrolyzing enzyme first attacks the α face of C-1 GlcNAc to form a covalent intermediate by inversion of configuration. This intermediate then gets hydrolyzed by water molecules, followed by another inversion at C1 to ultimately retain the β stereochemistry of free O-GlcNAc. A second possibility, especially for sugars containing an N-acetyl linkage is dubbed substrate assisted or anchimeric assisted catalysis. In such a scenario, the carboxylic oxygen of the 2-acetamido group within the sugar moiety acts as a nucleophile to attack the C-1 carbon generating an oxazoline intermediate. An aspartic or glutamic acid residue in the enzyme active site polarizes the N-acetyl side group to increase nucleophilicity of the amide oxygen promoting intramolecular attack at the anomeric carbon. An adjacent acidic amino acid acts as a general acid to facilitate breakdown of the peptide-sugar linkage. For ring opening of the cyclic oxazoline intermediate, the amino acid acting as the general acid now acts as a general base activating a water molecule to attack the anomeric carbon, this time from the β face, to expel the acetamido oxygen. That the OGA reaction mechanism proceeds via substrate assisted catalysis was shown by Macauley et al. by reducing the nucleophilicity of the acetamido side chain. Substrate analogs containing electron withdrawing fluorine atoms on the acetyl carbon resulted in a concomitant reduction in the rate of hydrolytic reaction with human OGA (hOGA).29 Using a bacterial homolog of OGA, BtGH84, He et al. were successful in obtaining a crystal structure of the cyclized oxazoline structure associated with the enzyme active site.30 These studies as well as other analyses with OGA homologs from Clostridium perfringens have now demonstrated the substrate participation of N-acetyl group in O-GlcNAc hydrolysis.27 Mutational analysis of OGA identified two adjacent acidic aspartic acid residues, Asp 174 and Asp 175 within the active site as being responsible for the aforementioned polarization and acid/base catalysis that assist in the overall hydrolytic reaction.31 The presence of TPR regions in OGT may explain its ability for substrate recognition. No such structure has been identified for OGA. Though active site residues may be involved in recognition of O-GlcNAcylated polypeptides, kinetic data seems to suggest that OGA activity may depend on the presence of O-GlcNAc and not on the peptide sequence.26, 32
2.3. Structural consequence of O-GlcNAc
An important consequence of protein O-GlcNAc modification may rest on the secondary structure of the protein – ultimately affecting its activity or interactions with other potential binding partners. Unlike a phosphate addition, N-acetylglucosamine does not confer any charge on the resultant peptide. Yet the increased stokes radius probably has conformational effects on the peptide secondary structure. Liang et al. showed that addition of O-GlcNAc induces a β turn formation of a helix33 while Chen and co-workers reported O-linked glycosylations to have impact on the kinetics of amyloid formation in the prion peptide.34 O-phosphate and O-GlcNAc were also shown to induce different conformations to the murine estrogen receptor.35 Tau is a key protein whose oligomerization contributes to neuronal death in Alzheimer’s disease. Recently it has also been shown that via its reciprocity with phosphorylation, O-GlcNAc reduces tau aggregation thus lowering chances of Alzheimer’s disease. Changing O-GlcNAcylation on substrate proteins may thus prove to be an attractive target for therapeutic purposes. Apart from a few papers, no comprehensive work has been carried out to look at these structural effects of O-GlcNAc, but its tremendous impact in neurons and neurodegenerative disorders calls for a more in-depth analysis in protein structural implications as a result of O-GlcNAcylation.
2.4. Small molecule inhibitors of OGT and OGA
Development of cell permeable, small molecule inhibitors of the enzymes OGT and OGA are extremely important for detailed understanding of the biochemistry of these proteins, elucidating their functional impacts on the biology of the cell and in generating therapeutics for diseases caused due to aberrant O-GlcNAcylation. Such molecules can also be used to parse out overlapping signaling networks, especially phosphorylation. They can also be used to generate animal models of particular diseases e.g. diabetes, cardiovascular diseases and neurodegenerative disorders. As with other glycosyltransferases, inhibitors of OGT have been difficult to synthesize, especially ones that work in living cells. This is probably because substrate analogs are rarely ably to bind the OGT active site competitively, which has a nanomolar affinity for its primary substrate UDP-GlcNAc. Other chemical structures have been plagued by solubility issues and by their inability to be effectively transported into the cell. Also, absence of a high throughput assay has hindered screening of large compound libraries. Finally, the prolonged absence of structural data on the transferase, also arrested efforts for designing chemically relevant molecules for OGT inhibition via rational design or by computational analysis.
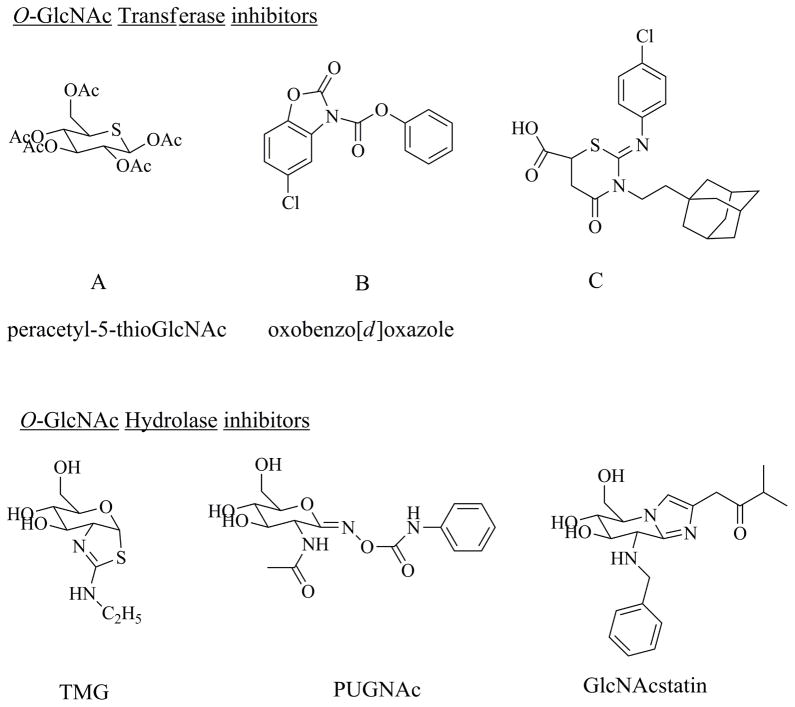
In late 2005 the Walker lab at Harvard Medical School developed a ligand displacement assay to interrogate OGT binding, which they had previously used for high throughput analysis of MurG, an important glycosyltransferase in bacterial peptidoglycan biosynthesis. By monitoring the fluorescence polarization of an UDP-GlcNAc analog containing a fluorescent probe attached to the N-acetyl group, they screened around sixty thousand compounds and validated 3 molecules as having OGT inhibitory activity with IC50 values ranging from 10 to 100 μM.36 The compound with the strongest inhibitory activity came from a family of molecules having an oxobenzo[d]oxazole core structure (Figure 2, compounds B and C). These compounds are irreversible inhibitors of OGT modifying the active site catalytic base. Analyses of the mechanistic details of the inhibitor action showed the formation of a covalent carbonyl adduct between the active site lysine 842, thought to be the reactive base, and an adjacent cystine residue.37 This reduces UDP-GlcNAc binding to OGT and thus inhibits O-GlcNAcylation of substrate proteins. The oxazole group of compounds has low water solubility and has not been shown to work within cell culture systems – thus limiting their use in vivo applications. Also the covalently bound carbonyl at the reaction site of the transferase effectively kills the enzyme and may result in metabolic instability due to buildup of substrate or accumulation of an intermediate in the HBP.
Figure 2.
Structure of some of the often used O-GlcNAc Transferase and O-GlcNAc hydrolase inhibitors
Recently Vocadlo and co-workers reported the development of a substrate analog of O-GlcNAc, 5-thio glucosamine that acts as a competitive inhibitor of OGT (Figure 2).38 The peracetylated form of 5SGlcNAc, after easily crossing the cell membrane due to its hydrophobic nature, gets rapidly de-acetylated via cellular esterases and is subsequently converted to the corresponding UDP-5SGlcNAc, probably via the GlcNAc salvage pathway. UDP-5SGlcNAc binds the active site of OGT with markedly reduced turnover. Preliminary experiments in cell culture have revealed reduction of overall protein O-GlcNAcylation without any observable effects to cellular viability for up to day five in a variety of cell types. It remains to be seen as to its effects on intracellular pools of UDP-GlcNAc – variation of which can have impacts in cellular signaling. In addition, many controls are still needed to validate the inhibitor’s off-target affects.
Unlike for OGT, there have existed a number of efficient and highly selective in vivo OGA inhibitors that have been readily used by researchers to study functional roles of O-GlcNAc. This is probably due to the existence of fluorometric assays for OGA that are highly amenable for large scale screening. One of the earliest known compounds, Streptozotocin (STZ) is widely used for studying hyperglycemia models – as it elicits a type I diabetic phenotype especially in rodents. Though initial hypotheses suggested STZ interacting with the active site of OGA by acting as a transition state analog, recent structural data on bacterial OGA homologs have suggested that this may not be the case. While STZ is a GlcNAc analog, it is likely selectively taken up by the beta cells of the pancreas, where it acts as a general chemical poison rather that a specific OGA inhibitor. Within β cells of the pancreas, its primary mode of action is through DNA alkylation and strand breakage, production of NO and activation of ADP-ribose polymerase. In spite of such off target effects, the clinical symptoms of hyperglycemia are highly correlated in STZ treated animals. Another widely used inhibitor of OGA is O-(2-acetamido-2-deoxy-d-glucopyranosylidene)amino-N-phenylcarbamate (PUGNAc), shown by Dong et al. to have Ki values of about 50 nM against purified rat spleen OGA (Figure 2). Subsequent biochemical analyses and molecular modeling experiments have shown PUGNAc to behave as a transition state analog for OGA.39, 40 Though it possesses a sp2 hybridized carbon at C1 – giving it partial geometric requirements of the oxocarbenium ion transition state of OGA, it lacks the relative positive charge development at O5-C1 centers and is less conformationally constrained. Structural data on PUGNAc binding to CpNagJ (homolog of OGA) shows that the GlcNAc moiety forms ten hydrogen bonds within the catalytic site and the phenyl ring of the inhibitor stacks up against a tryptophan residue of the protein that projects from the C-terminal domain.27 These interactions make PUGNAc a more general inhibitor of glycosyl hydrolases. Thus it can also inhibit the lysosomal hexosaminidases with Ki of 30 to 40 nM.29 Nonetheless PUGNAc was shown to be a cell permeable OGA inhibitor, which has led to it being widely used as a modulator of cellular O-GlcNAcylation levels, and PUGNAc has assisted in understanding of the dynamic nature of this ubiquitous post-translational modification.
Since neither the structure nor the mechanistic details of OGA was completely well understood for quite some time, most preliminary development of inhibitory molecules followed up on core structures previously reported to be active for other glycosidases, especially those belonging to the same family. The development and usage of STZ and PUGNAc as O-GlcNAc hydrolase inhibitors followed such reasoning. With the understanding of the substrate assisted catalytic reaction of OGA and the crystallization of the bacterial OGA homolog, different groups started to develop a number of molecules starting from preexisting core structures that were already known to have activity against similar acting glycosyl hydrolases. Thus, upon identifying the two step catalytic mechanism for OGA, Vocadlo et al. tested the molecule 1,2-dideoxy-2′-methyl-α-d-glucopyranoso[2,1-d]-Δ2′-thiazoline (NAG-thiazoline) for activity against OGA.29 NAG-thiazoline was synthesized as a mimetic of the bicyclic oxazoline intermediate by the Withers lab as inhibitors for the family 20 and 84 hexosaminidases. This compound showed a modest Ki of 180 nM against OGA but showed even stronger binding for the lysosomal β-hexosaminidases. To increase selectivity for the hOGA and maintain reasonable potency, NAG-thiazoline was used as a core molecule for further improving its inhibitory activity. Increasing bulky groups on the thiazoline ring lead to the development of 1,2-dideoxy-2′-propyl-α-D-glucopyranoso-[2,1-D]-Δ2′-thiazoline (NButGT) with an increased selectivity for OGA albeit with loss of potency.39 In the hydrolytic mechanism, the OGA active site contains an aspartic acid residue interacting with the amide proton acting as a catalytic base and increasing the nucleophilicity of the amide carbonyl. The bicyclic oxazoline intermediate thus has an inherent charged interaction with the enzyme. Mimicking such association within the inhibitor, an aminothiazoline molecule – thiamet-G (Figure 2), was reported which showed a higher Ki for OGA (21 nM) and increased selectivity compared to Hex A/B.41 TMG has subsequently been shown to be a stable bioactive inhibitor of OGA, increasing tau O-GlcNAcylation in mice brain thus reducing the aggregation of phosphorylated tau and is being pursued as a therapeutic for AD.42
Given that PUGNAc was a less selective inhibitor of the hOGA compared to the lysosomal hexosaminidases, rational design was also applied to the PUGNAc core structure to generate a potent and highly selective drug for hOGA. Based on the crystal structure of bacterial GlcNAcase, the binding interactions of PUGNAc with it and structure activity relationship analyses of nagstatin analogs as glycosidase inhibitors, Dorfmuller and co-workers reported the development of GlcNAc configured nagstatin moieties.43 These series of molecules called GlcNAcstatins (Figure 2) showed a high degree of selectivity towards the bacterial hydrolase compared to the lysosomal enzymes. They were also plagued by low solubility in aqueous solutions. Subsequent queries into the N-acetyl chemical space helped generate a highly specific hOGA inhibitor designated GlcNAcstatin G. This particular derivative showed a 9 × 105 fold increased selectivity towards the human isoform compared to Hex A/B with Ki of about 5 nM.44 These derivatives were also shown to be cell-penetrant and increased overall cellular O-GlcNAcylation by 2 to 3-fold when treated for 6 hours.
3. Methods for study and detection of O-GlcNAc
Akin to other post translational modifications, O-GlcNAcylation at a particular protein can be highly dynamic, sub-stoichiometric, compartment specific, cell type and cell state dependent. Additionally this single sugar addition is highly chemically and enzymatically labile. Lacking in formal charge and being of comparative negligible size, addition or deletion of N-acetylglucosamine rarely changed migration patterns for the protein in gel electrophoresis techniques. The O-sugar linkage being of low energy – O-GlcNAc easily fell off on most mass spectrometric analysis. Also ion suppression occurs for O-GlcNAc modified peptides compared to unmodified ones, even when both occur at similar concentrations, making it difficult to observe. Finally, hexosaminidases occurring within the cells readily hydrolyze this PTM during cell damage or protein isolation unless suitable inhibitors are added. Thus, in spite being of critical signaling importance for a variety of proteins, it remained undetected for a long while. Unlike phosphorylation or some of the other PTMs, O-GlcNAcylation lacks any consensus sequence within proteins. Often serine and/or threonine rich regions may contain a site of modification, but the presence of a large number of such residue obfuscates site identification. It is becoming increasingly clear that understanding site specificity of O-GlcNAc dynamics lies at the heart of its regulatory properties. This poses another layer of complexity in the detection and understanding of O-GlcNAc.
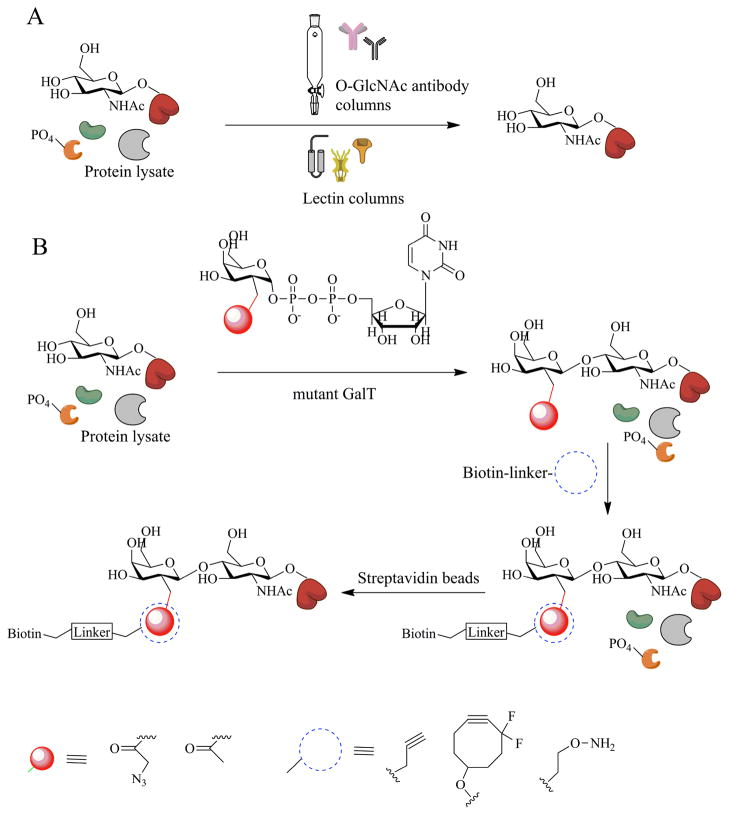
One of the earliest and often used methods to detect the presence of O-GlcNAc was its binding interactions with lectins. Lectins are proteins that are known to bind sugars with varying specificities. Succinylated wheat germ agglutinin, sWGA, is a common lectin used in the study β-O-GlcNAc. Columns packed with sWGA have been used to enrich protein fragments rich in O-GlcNAc – especially useful on low copy number regulatory proteins e.g. transcription factors. In contrast to WGA, which can bind to both sialic acid and O-GlcNAc, sWGA is negatively charged at physiological pH. Therefore sWGA cannot bind sialylated glycoproteins but can mainly bind to O-GlcNAcylated proteins. Another method traditionally used to study proteome wide O-GlcNAc changes is via the use of pan specific antibodies. RL2 and CTD 110.1 are a couple of the most widely used antibodies to detect O-GlcNAc by Western blotting. A number of other commercially available antibodies are also available. Some of these antibodies have preference towards specific peptide sequences or secondary structures that mimic GlcNAc residues, a few may also detect other types of cell surface glycosylation. Unlike phosphorylation, very few site-specific O-GlcNAc antibodies exist, to-date. Enrichment columns using antibodies have also been used in a number of studies (Figure 3). Site-mapping using lectin or antibody based enrichment columns involve HPLC purification of glycopeptides and Edman degradation methods for modification site determination. However these methods are complicated and time consuming and are compounded by low stoichiometries of O-GlcNAc modified peptide. Here we discuss some of the more recent methods developed to specifically study overall cellular protein O-GlcNAcylation as well as determination of site specific modification.
Figure 3.
Enrichment strategies used to separate O-GlcNAc modified proteins from nuclear and cytosolic fragments. A) Use of O-GlcNAc specific antibodies and sWGA helps to bind a number of glycosylated proteins. B) Chemoenzymatic method using a mutant form of GalT that transfers a galactose analog bearing a chemically reactive group onto O-GlcNAc residues. Subsequent chemistry with an orthogonal probe allows for tagging and enrichment via biotin based labels
3.1. β-elimination followed by Michael addition (BEMAD)
Mapping sites of O-GlcNAcylation is important for the understanding of functional roles of site specific O-GlcNAc in a given biological context. But usage of mass spectrometric collision induced dissociation result in loss of the highly labile O-GlcNAc moiety from the peptide backbone. To circumvent these issues a previously utilized technique for interrogation of protein phosphorylation sites was modified for use with O-glycosylations. This method involved the β-elimination of a hydrogen atom from the glycosylated serine backbone carbon to generate an alkene intermediate. This was followed by a Michael addition step in which a dithiothreitol (DTT) or biotin pentamine monomer was added onto the labeled serine to generate a unique mass tag. Following the ‘tag’ in mass spectrometry not only informed of the presence of O-GlcNAc but also facilitated site identification.45 This technique was further modified using light and heavy DTT molecules under specific conditions to discriminate between phosphate, GlcNAc and nonspecific residues.46
3.2. Enzymatic labeling by Galactosyltransferases
The enzyme β-1, 4-galactosyltransferase (GalT) catalyzes the addition of a galactose residue from a UDP-Galactose donor to sugar structures containing terminal GlcNAc residues. This method was originally used by Hart and co-workers to label nuclear/cytoplasmic terminal glucosamine with tritiated galactose residues. Radioactive labeling of O-GlcNAcylated proteins is an excellent way to confirm presence of this PTM, but often, due to the sub-stoichiometric nature of GlcNAc and the low abundance of regulatory proteins, sensitivity maybe an issue. The βGal-1, 4-GlcNAc disaccharide can also be preferentially bound to the lectin Ricin – thus providing another level of enrichment/detection strategy that can be coupled with the enzymatic reaction to determine site level occupancy of O-GlcNAc.
In 2003 Khidekel et al. developed a mutant GalT that had increased substrate tolerance, and could utilize a keto-derivative of UDP-Galactose to label O-GlcNAc residues.47 The incorporated keto group could be used as an inert tag to chemically label modified disaccharides with an amino-biotin probe. That can subsequently be detected by its interaction with streptavidin. Biotinylated proteins can also be immunoprecipitated with streptavidin beads and enriched for glycosylated proteins. Using this method on nucleocytoplasmic lysates, Tai and co-workers identified a number of cellular proteins that are O-GlcNAcylated like CREB, ATF-1 and some AP-1 family of transcription factors.48 This method was later modified to use an azido analog of galactose to incorporate the biologically inert azide on glycosylated proteins. Subsequent “click” reaction with alkyne bearing fluorescent tags was used by Clark et al. to label proteins within live cells.49 This method could also be used with alkynylated biotin tags to carry out pull down analysis of entire cellular proteomes (Figure 3). A final application of the GalT labeling method was reported by Reach et al. when they modified the amino tagging of keto-galactose derivatized sugars using polyethylene glycol molecules. Attachment of long chain PEG groups (~20,000 daltons) allowed on gel separation of multiply glycosylated protein. The facile method allows for estimation of both amounts as well as determination of the stoichiometries of O-GlcNAcylated protein at any given cellular state or under specific stimuli.50 Such analyses are instrumental in understanding the dynamics and regulation of protein O-GlcNAc modification and have been used to study the interplay between O-GlcNAcylation and phosphorylation on transcriptional repressors within the rat brain cortex. This GalT labeling method has been coupled with mass spectrometry based proteomics analyses to look into protein glycosylations in the brain, within diabetic tissues and for cancer biomarker analyses, as is discussed in greater detail below.
3.3. Metabolic labeling of O-GlcNAc residues using non-canonical substrates
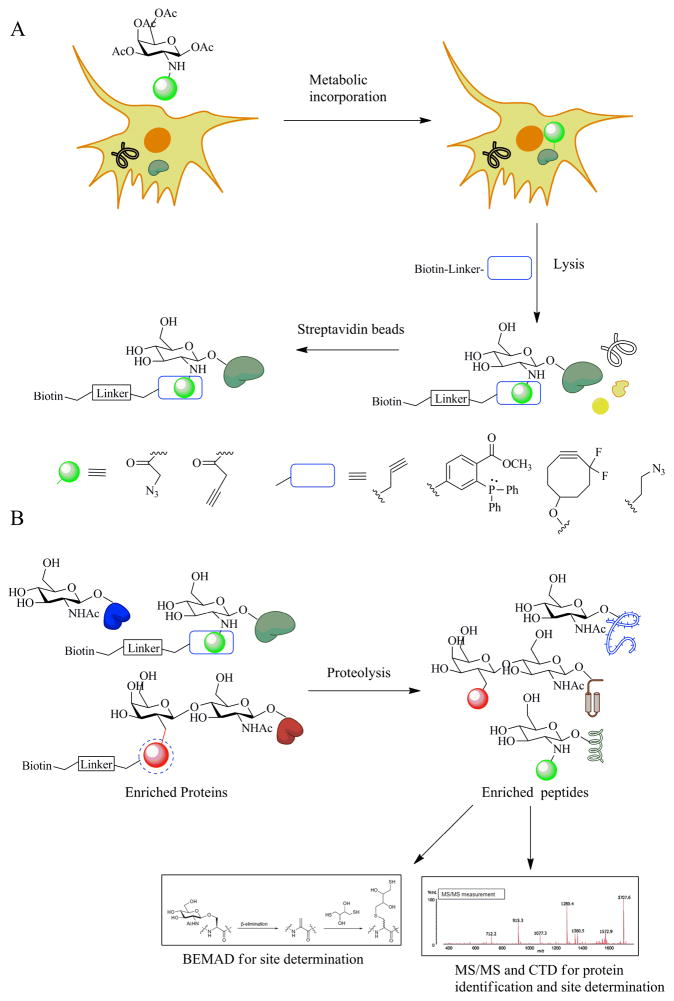
One of the more recent developments in chemical biology is the usage of substrate analog monomers of biological polymers to selectively tag a biopolymer like proteins, oligosaccharides, lipids or any bioactive molecules like nucleotide bases, ATP molecules. The researcher exploits the promiscuity of the cellular pathways or biosynthetic machinery to achieve such incorporations. Incorporation of selenomethionine molecules for X-ray crystal structure determination or radioactive S-35 methionine labeling are better known traditional methods using the same approach. Almost always the incorporated molecule contains a chemical handle that can subsequent be modified with a bio-orthogonal reaction. These reactions do not have biological counterparts and hence do not produce off target effects or false positives during further analysis. Apart from the de novo routes of sugar biosynthesis, there exists within cells salvage pathways through which such unnatural sugar incorporations have been achieved. Such a technique was initially reported by Vocadlo et al. in 2003 when it was shown that by feeding cells a peracetylated form of azidoacetylglucosamine (Ac4GlcNAz), O-GlcNAc residues within cellular proteins could be labeled by O-GlcNAz.51 Subsequently this azide group – the chemical handle, could be modified by either Staudinger ligation reactions or alkyne based “click” reactions (Figure 4A). This proved a non-invasive method to efficiently label O-GlcNAc residues in proteins and subsequently analyze them via immunoprecipitations techniques, proteomic methods or fluorescent tagging approaches. Experiments have shown that galactosamine analogs are more efficiently incorporated within cells compared to glucosamine derivatives and that the presence of an alkyne handle on the incoming sugar is more readily tolerated by the cellular machinery.52, 53 Modifications to the labeling chemistry recently generated the development of metal free click reactions, which take advantage of ring strain within cyclooctyne based probes to achieve efficient reactivity with the incorporated azide handle.
Figure 4.
Metabolic incorporation strategies and proteomic analyses of enriched O-GlcNAcylated samples. A) Metabolic incorporation of a variety of peracetylated analogs result in labeling of intracellular proteins with O-GlcNAc substrate analogs bearing a chemical handle. Bioorthogonal reactions assist in tagging of labeled proteins and their purifications. B) Tagged proteins analyzed via proteomic tools for peptide identification - ms/ms analysis and BEMAD technique for site determination.
3.4. Proteomic methods for the detection and analysis of O-GlcNAc
Proteomic based techniques are the most readily utilized for detection and site specific analysis of O-GlcNAcylation. Almost all such methods utilized one of the aforementioned enrichment strategies. Use of O-GlcNAc specific antibodies for enrichment suffers from sensitivity issues while lectin based approaches pick up false positives in the form of N-linked glycosylations. Thus, the chemo-enzymatic and metabolic tagging techniques are the most widely reported ways for O-GlcNAc protein and/or peptide enrichment. In proteomic analyzes, it is often easier to proteolyze the chemoenzymatically or metabolically labeled sample before performing streptavidin or other bead based purifications. This reduces the overall complexity of the protein sample and further enriches only the O-GlcNAc containing peptides from different proteins. A subsequent MS/MS analysis is used to identify the O-GlcNAc modified proteins (Figure 4B). They are often coupled with BEMAD technique for detection and confirmation of site specificity and have been more recently updated to facilitate mass spectrometry based analyzes. In a number of cases SILAC or ICAT based techniques are multiplexed with the O-glyco proteomic analysis to get a more quantitative understanding of peptide modification stoichiometries.
The GalT based chemoenzymatic tagging method with a keto-galactose tag and an amino-biotin probe was utilized by Khidekel and co-workers to look at O-GlcNAc dynamics within cortical neurons.54 Post enrichment with streptavidin beads, proteolysis was carried out to reduce the complexity of the analyzed proteomic sample. Capture was combined with mass spectrometry based CAD and ETD techniques for identification of glycosylation sites. Wang et al. used a similar tagging method to study erythrocyte proteins obtained from diabetic and normal patients for biomarker discovery.55 Here the keto-galactose tag was replaced by an azido-galactose and an alkyne-biotin probe utilized for enrichment. In this case the BEMAD technique was utilized for site-specific analysis of identified glyco-peptides. The researchers later updated this technique with a UV cleavable alkyne-biotin probe and used ETD based mass spec proteomic analysis for identification of GlcNAc sites.56 The specificity of this strategy was underscored when over 120 O-GlcNAc specific proteins were identified utilizing the same technique while studying cell cytokinesis.9 The metabolic labeling strategy has also been used to study O-GlcNAc proteomics by a number of groups. Nandi et al. used the azido glucosamine incorporation technique to label O-GlcNAc sites which were subsequently enriched using an alkyne-biotin reactive tag.57, 58 In a more recent study, Zaro et al. compared four different metabolic labels, azido glucosamine, azido galactosamine, alkyne galactosamine and alkynyl glucosamine to analyze whole cell O-GlcNAc proteomics.52 This study identified more than 250 new O-GlcNAcylated proteins while also suggesting that the alkynyl glucosamine (GlcNAlk) surrogate was the most efficient chemical reporter for O-linked glucosamine.
A couple of other groups have tried to develop titanium oxide labeled and periodate oxidation followed by hydrazide resin capture based enrichment strategies for O-GlcNAc proteomics.59, 60 But these methods are plagued by low sensitivity issues. Also cross reactivity with phosphates and other glycosylations containing vicinal diols generate false positive that need to be addressed with better experimental controls.
3.5. Methods for the study of O-GlcNAc in living cells systems and some recent advances
Given the involvement of O-GlcNAc in the plethora of biological signaling pathways ranging from cell state to external stimuli and the rapid dynamics of this modification with and in response to other PTMs make it imperative to study its dynamics inside live cells. Such studies are fraught with challenges involving optimum readouts and reversibility of designed system. One of the ways to tackle this problem is to take snapshot pictures of the cellular signaling cascade at specific time points. The chemoenzymatic tagging method has been used to label fixed cells with an azido galactose derivative via the GalT reaction. Probing with fluorescent probes showed robust labeling of GlcNAcylated proteins within cells. Another way to study signaling dynamics is designing of sensors preferably with reversible fluorescence based read outs that can be expressed within cells and their dynamics followed. Such in vivo probes have been designed for a wide variety of signaling events.61 Carrilo and co-workers followed the FRET based sensor design for live cell imaging of O-GlcNAc dynamics.8 They designed an effective probe that could be used to study changes in O-GlcNAcylation patterns in response to various stimuli.7 Tethering of this probe to specific cellular compartments gave an overall dynamic view of O-GlcNAc changes in response to various types of stimulation patterns and revealed an intricate relationship with protein phosphorylation.
One of the most important regulatory effects of protein O-GlcNAcylation is defined by its site specificity. Certain site modifications can cause changes in protein function via phosphorylation, while others have not yet revealed any effect on activity. For example on the protein CaMKIV, O-GlcNAcylation inhibits phosphorylation and hence regulation of its Ca2+ dependent gene expression. Site directed studies indicate that though CaMKIV contains five O-GlcNAclation sites, only Ser-189 modification has the highest observable effect on its activation.62 The other reported sites show little effect on its overall GlcNacylation and phosphorylation levels. Similar effects occur in the insulin receptor substrate protein where only O-GlcNAc sites close to SH2 domains effect its interaction with PI3 kinase, while the functions of other glycosylation residues have not yet been revealed.63 Thus understanding functional impacts of residue specific O-GlcNAc changes are important in determining its overall role in signaling, transcription or protein-protein interaction. Recently Tarrant et al. used intein based protein semi-synthesis to incorporate site specific glycosylation and phosphorylation mimetics within Casein Kinase 2, and showed differential substrate specificity for the same kinase based on its post translational modification.64 This study underscores the importance of cross talk between O-GlcNAc and O-phosphate. Yu et al. reported the development of a selective cross linking strategy taking advantage of the metabolic incorporation methodology to attach azidirine tagged analogs of N-acetylglucosamine within O-GlcNAc residues.65 This strategy can help interrogate functional consequences of protein -protein interaction effects due to site specific O-GlcNAcylation changes.
4. Conclusion and Outlook
Here we have summarized the various research findings in O-GlcNAc research field that have been using the chemical research approach. Although much research on O-GlcNAc has been conducted, further research is needed in order to answer many questions in biological and chemical aspects. Especially in the case of ncOGT, while it is assumed that it has nuclear localization signals (NLS), the simultaneous existence in the nucleus and cytoplasm despite putative NLS still needs to be explained. Also, the fact that mOGT exists in mitochondria should be supported by the existence of mechanisms that either synthesize UDP-GlcNAc within the mitochondria or transport UDP-GlcNAc into the mitochondria. Also, in a regulation perspective, OGA should certainly exist in mitochondria and this should be verified. Similar to the simultaneous existence of ncOGT in the nucleus and cytoplasm, the reason for the simultaneous existence of OGA in both nucleus and cytoplasm also needs to be revealed. Although the OGA inhibitor is somewhat developed and is being used in biological research as well as medical development for AD, OGT inhibitor is still in its developing stages. If a convenient OGT assay system is built then new OGT inhibitors can be created in a much faster fashion. There are various methods of finding O-GlcNAc modification sites, but a simple way is yet to be found. Therefore, a mass spectrometry technique that can easily find O-GlcNAc modification sites should be developed. The report stating that treatment of OGA inhibitors increases O-GlcNAc modification in cells which will lead to alterations in 700 different phosphorylation sites probably signifies the fact that the changes of the nutrition condition of cells may affect different signal transductions within cells. Thus through the artificial regulation of O-GlcNAc modification, we may develop new treatments for diseases caused by abnormal O-GlcNAc modification such as diabetic complications, AD, and occurrence and metastasis of cancer.
Acknowledgments
This work was supported by NIH R01CA42486, R01DK61671; N01-HV-00240; P01HL107153, R24DK084949 and the Patrick C. Walsh Prostate Cancer Research Fund to G.W.H and by the National Research Foundation (NRF) funded by the Korean Government (2010-0018923 to J.W.C.) and World Class University Program (R31-2008-000-10086-0 to J.W.C.).
Footnotes
Part of the carbohydrate chemistry themed issue
References and notes
- 1.Walsh CT, Garneau-Tsodikova S, Gatto GJ. Angewandte Chemie International Edition. 2005;44:7342–7372. doi: 10.1002/anie.200501023. [DOI] [PubMed] [Google Scholar]
- 2.Hart GW, Copeland RJ. Cell. 2010;143:672–676. doi: 10.1016/j.cell.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Torres CR, Hart GW. Journal of Biological Chemistry. 1984;259:3308–3317. [PubMed] [Google Scholar]
- 4.Zachara NE, Hart GW. Trends in cell biology. 2004;14:218–221. doi: 10.1016/j.tcb.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Zachara NE, O’Donnell N, Cheung WD, Mercer JJ, Marth JD, Hart GW. Journal of Biological Chemistry. 2004;279:30133–30142. doi: 10.1074/jbc.M403773200. [DOI] [PubMed] [Google Scholar]
- 6.O’Donnell N, Zachara NE, Hart GW, Marth JD. Molecular and Cellular Biology. 2004;24:1680–1690. doi: 10.1128/MCB.24.4.1680-1690.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrillo LD, Froemming JA, Mahal LK. Journal of Biological Chemistry. 2011;286:6650–6658. doi: 10.1074/jbc.M110.191627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrillo LD, Krishnamoorthy L, Mahal LK. Journal of the American Chemical Society. 2006;128:14768–14769. doi: 10.1021/ja065835+. [DOI] [PubMed] [Google Scholar]
- 9.Wang Z, Udeshi ND, Slawson C, Compton PD, Sakabe K, Cheung WD, Shabanowitz J, Hunt DF, Hart GW. Sci Signal. 2010;3:ra2. doi: 10.1126/scisignal.2000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Annual Review of Biochemistry. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ranuncolo SM, Ghosh S, Hanover JA, Hart GW, Lewis BA. Journal of Biological Chemistry. 2012 doi: 10.1074/jbc.M111.330910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rexach JE, Clark PM, Mason DE, Neve RL, Peters EC, Hsieh-Wilson LC. Nat Chem Biol. 2012;8:253–261. doi: 10.1038/nchembio.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeidan Q, Wang Z, De Maio A, Hart GW. Molecular Biology of the Cell. 2010;21:1922–1936. doi: 10.1091/mbc.E09-11-0941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Slawson C, Zachara NE, Vosseller K, Cheung WD, Lane MD, Hart GW. Journal of Biological Chemistry. 2005;280:32944–32956. doi: 10.1074/jbc.M503396200. [DOI] [PubMed] [Google Scholar]
- 15.Yang WH, Kim JE, Nam HW, Ju JW, Kim HS, Kim YS, Cho JW. Nat Cell Biol. 2006;8:1074–1083. doi: 10.1038/ncb1470. [DOI] [PubMed] [Google Scholar]
- 16.Kang JG, Park SY, Ji S, Jang I, Park S, Kim HS, Kim S-M, Yook JI, Park Y-I, Roth J, Cho JW. Journal of Biological Chemistry. 2009;284:34777–34784. doi: 10.1074/jbc.M109.026351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whelan SA, Hart GW. Circulation Research. 2003;93:1047–1058. doi: 10.1161/01.RES.0000103190.20260.37. [DOI] [PubMed] [Google Scholar]
- 18.Rexach JE, Clark PM, Hsieh-Wilson LC. Nat Chem Biol. 2008;4:97–106. doi: 10.1038/nchembio.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macauley MS, Vocadlo DJ. Biochimica et Biophysica Acta (BBA) - General Subjects. 2010;1800:107–121. doi: 10.1016/j.bbagen.2009.07.028. [DOI] [PubMed] [Google Scholar]
- 20.Kim EJ. Molecules. 2011;16:1987–2022. doi: 10.3390/molecules16031987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iyer SPN, Hart GW. Journal of Biological Chemistry. 2003;278:24608–24616. doi: 10.1074/jbc.M300036200. [DOI] [PubMed] [Google Scholar]
- 22.Martinez-Fleites C, Macauley MS, He Y, Shen DL, Vocadlo DJ, Davies GJ. Nat Struct Mol Biol. 2008;15:764–765. doi: 10.1038/nsmb.1443. [DOI] [PubMed] [Google Scholar]
- 23.Lazarus MB, Nam Y, Jiang J, Sliz P, Walker S. Nature. 2011;469:564–567. doi: 10.1038/nature09638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clarke AJ, Hurtado-Guerrero R, Pathak S, Schuttelkopf AW, Borodkin V, Shepherd SM, Ibrahim AFM, van Aalten DMF. EMBO J. 2008;27:2780–2788. doi: 10.1038/emboj.2008.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez-Fleites C, He Y, Davies GJ. Biochimica et Biophysica Acta (BBA) - General Subjects. 2010;1800:122–133. doi: 10.1016/j.bbagen.2009.07.019. [DOI] [PubMed] [Google Scholar]
- 26.Shen DL, Gloster TM, Yuzwa SA, Vocadlo DJ. Journal of Biological Chemistry. 2012;287:15395–15408. doi: 10.1074/jbc.M111.310664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao FV, Dorfmueller HC, Villa F, Allwood M, Eggleston IM, van Aalten DMF. EMBO J. 2006;25:1569–1578. doi: 10.1038/sj.emboj.7601026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toleman C, Paterson AJ, Whisenhunt TR, Kudlow JE. Journal of Biological Chemistry. 2004;279:53665–53673. doi: 10.1074/jbc.M410406200. [DOI] [PubMed] [Google Scholar]
- 29.Macauley MS, Whitworth GE, Debowski AW, Chin D, Vocadlo DJ. Journal of Biological Chemistry. 2005;280:25313–25322. doi: 10.1074/jbc.M413819200. [DOI] [PubMed] [Google Scholar]
- 30.He Y, Macauley MS, Stubbs KA, Vocadlo DJ, Davies GJ. Journal of the American Chemical Society. 2010;132:1807–1809. doi: 10.1021/ja9086769. [DOI] [PubMed] [Google Scholar]
- 31.Çetinbaş N, Macauley MS, Stubbs KA, Drapala R, Vocadlo DJ. Biochemistry. 2006;45:3835–3844. doi: 10.1021/bi052370b. [DOI] [PubMed] [Google Scholar]
- 32.Schimpl M, Borodkin Vladimir S, Gray Lindsey J, van Aalten Daan MF. Chemistry & Biology. 2012;19:173–178. doi: 10.1016/j.chembiol.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang F-C, Chen RPY, Lin C-C, Huang K-T, Chan SI. Biochemical and Biophysical Research Communications. 2006;342:482–488. doi: 10.1016/j.bbrc.2006.01.168. [DOI] [PubMed] [Google Scholar]
- 34.Chen P-Y, Lin C-C, Chang Y-T, Lin S-C, Chan SI. Proceedings of the National Academy of Sciences. 2002;99:12633–12638. doi: 10.1073/pnas.192137799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y-X, Du J-T, Zhou L-X, Liu X-H, Zhao Y-F, Nakanishi H, Li Y-M. Chemistry & Biology. 2006;13:937–944. doi: 10.1016/j.chembiol.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 36.Gross BJ, Kraybill BC, Walker S. Journal of the American Chemical Society. 2005;127:14588–14589. doi: 10.1021/ja0555217. [DOI] [PubMed] [Google Scholar]
- 37.Jiang J, Lazarus MB, Pasquina L, Sliz P, Walker S. Nat Chem Biol. 2012;8:72–77. doi: 10.1038/nchembio.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gloster TM, Zandberg WF, Heinonen JE, Shen DL, Deng L, Vocadlo DJ. Nat Chem Biol. 2011;7:174–181. doi: 10.1038/nchembio.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitworth GE, Macauley MS, Stubbs KA, Dennis RJ, Taylor EJ, Davies GJ, Greig IR, Vocadlo DJ. Journal of the American Chemical Society. 2006;129:635–644. doi: 10.1021/ja065697o. [DOI] [PubMed] [Google Scholar]
- 40.Lameira J, Alves CuN, Moliner V, Martí S, Castillo R, Tuñón Ia. The Journal of Physical Chemistry B. 2010;114:7029–7036. doi: 10.1021/jp9115673. [DOI] [PubMed] [Google Scholar]
- 41.Yuzwa SA, Macauley MS, Heinonen JE, Shan X, Dennis RJ, He Y, Whitworth GE, Stubbs KA, McEachern EJ, Davies GJ, Vocadlo DJ. Nat Chem Biol. 2008;4:483–490. doi: 10.1038/nchembio.96. [DOI] [PubMed] [Google Scholar]
- 42.Yuzwa SA, Shan X, Macauley MS, Clark T, Skorobogatko Y, Vosseller K, Vocadlo DJ. Nat Chem Biol. 2012;8:393–399. doi: 10.1038/nchembio.797. [DOI] [PubMed] [Google Scholar]
- 43.Dorfmueller HC, Borodkin VS, Schimpl M, Shepherd SM, Shpiro NA, van Aalten DMF. Journal of the American Chemical Society. 2006;128:16484–16485. doi: 10.1021/ja066743n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dorfmueller HC, Borodkin VS, Schimpl M, Zheng X, Kime R, Read KD, van Aalten DMF. Chemistry & Biology. 2010;17:1250–1255. doi: 10.1016/j.chembiol.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wells L, Vosseller K, Cole RN, Cronshaw JM, Matunis MJ, Hart GW. Molecular & Cellular Proteomics. 2002;1:791–804. doi: 10.1074/mcp.m200048-mcp200. [DOI] [PubMed] [Google Scholar]
- 46.Vosseller K, Hansen KC, Chalkley RJ, Trinidad JC, Wells L, Hart GW, Burlingame AL. PROTEOMICS. 2005;5:388–398. doi: 10.1002/pmic.200401066. [DOI] [PubMed] [Google Scholar]
- 47.Khidekel N, Arndt S, Lamarre-Vincent N, Lippert A, Poulin-Kerstien KG, Ramakrishnan B, Qasba PK, Hsieh-Wilson LC. Journal of the American Chemical Society. 2003;125:16162–16163. doi: 10.1021/ja038545r. [DOI] [PubMed] [Google Scholar]
- 48.Tai H-C, Khidekel N, Ficarro SB, Peters EC, Hsieh-Wilson LC. Journal of the American Chemical Society. 2004;126:10500–10501. doi: 10.1021/ja047872b. [DOI] [PubMed] [Google Scholar]
- 49.Clark PM, Dweck JF, Mason DE, Hart CR, Buck SB, Peters EC, Agnew BJ, Hsieh-Wilson LC. Journal of the American Chemical Society. 2008;130:11576–11577. doi: 10.1021/ja8030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rexach JE, Rogers CJ, Yu S-H, Tao J, Sun YE, Hsieh-Wilson LC. Nat Chem Biol. 2010;6:645–651. doi: 10.1038/nchembio.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vocadlo DJ, Hang HC, Kim E-J, Hanover JA, Bertozzi CR. Proceedings of the National Academy of Sciences. 2003;100:9116–9121. doi: 10.1073/pnas.1632821100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zaro BW, Yang Y-Y, Hang HC, Pratt MR. Proceedings of the National Academy of Sciences. 2011;108:8146–8151. doi: 10.1073/pnas.1102458108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boyce M, Carrico IS, Ganguli AS, Yu S-H, Hangauer MJ, Hubbard SC, Kohler JJ, Bertozzi CR. Proceedings of the National Academy of Sciences. 2011;108:3141–3146. doi: 10.1073/pnas.1010045108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khidekel N, Ficarro SB, Clark PM, Bryan MC, Swaney DL, Rexach JE, Sun YE, Coon JJ, Peters EC, Hsieh-Wilson LC. Nat Chem Biol. 2007;3:339–348. doi: 10.1038/nchembio881. [DOI] [PubMed] [Google Scholar]
- 55.Wang Z, Park K, Comer F, Hsieh-Wilson LC, Saudek CD, Hart GW. Diabetes. 2009;58:309–317. doi: 10.2337/db08-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Z, Udeshi ND, O’Malley M, Shabanowitz J, Hunt DF, Hart GW. Molecular & Cellular Proteomics. 2010;9:153–160. doi: 10.1074/mcp.M900268-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sprung R, Nandi A, Chen Y, Kim SC, Barma D, Falck JR, Zhao Y. Journal of Proteome Research. 2005;4:950–957. doi: 10.1021/pr050033j. [DOI] [PubMed] [Google Scholar]
- 58.Nandi A, Sprung R, Barma DK, Zhao Y, Kim SC, Falck JR, Zhao Y. Analytical Chemistry. 2005;78:452–458. doi: 10.1021/ac051207j. [DOI] [PubMed] [Google Scholar]
- 59.Parker BL, Gupta P, Cordwell SJ, Larsen MR, Palmisano G. Journal of Proteome Research. 2010;10:1449–1458. doi: 10.1021/pr100565j. [DOI] [PubMed] [Google Scholar]
- 60.Klement E, Lipinszki Zn, Kupihár Zn, Udvardy A, Medzihradszky KF. Journal of Proteome Research. 2010;9:2200–2206. doi: 10.1021/pr900984h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang J, Campbell RE, Ting AY, Tsien RY. Nat Rev Mol Cell Biol. 2002;3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 62.Dias WB, Cheung WD, Wang Z, Hart GW. Journal of Biological Chemistry. 2009;284:21327–21337. doi: 10.1074/jbc.M109.007310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Klein AL, Berkaw MN, Buse MG, Ball LE. Molecular & Cellular Proteomics. 2009;8:2733–2745. doi: 10.1074/mcp.M900207-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tarrant MK, Rho H-S, Xie Z, Jiang YL, Gross C, Culhane JC, Yan G, Qian J, Ichikawa Y, Matsuoka T, Zachara N, Etzkorn FA, Hart GW, Jeong JS, Blackshaw S, Zhu H, Cole PA. Nat Chem Biol. 2012;8:262–269. doi: 10.1038/nchembio.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu SH, Boyce M, Wands AM, Bond MR, Bertozzi CR, Kohler JJ. Proceedings of the National Academy of Sciences. 2012;109:4834–4839. doi: 10.1073/pnas.1114356109. [DOI] [PMC free article] [PubMed] [Google Scholar]