Abstract
We develop here a novel approach to barcode large numbers of cells through cell-surface expression of programmable zinc-finger DNA-binding domains (sZFs). We show sZFs enable double-stranded DNA to sequence-specifically label living cells, and also develop a sequential tagging approach to in situ image >3 cell types using just 3 fluorophores. Finally we demonstrate their broad versatility through ability to serve as surrogate reporters and facilitate selective cell capture and targeting.
The ability to construct and interrogate complex tissues and cellular libraries at single cell resolution requires methods that enable highly multiplexed in situ probing of living cells. Here while the use of fluorescent proteins has revolutionized probing of biological phenomena, their multiplexed use is limited to combinations that can be spectrally resolved. To expand the repertoire of probing tools, we explored the possibility of using DNA binding domains such as zinc finger proteins (ZFs) and transcription activator-like effectors (TALEs). Our motivation stemmed from the observation that as a receptor-ligand pair the ZF-DNA or TALE-DNA interaction is very unique in that both the receptor (ZF or TALE protein) and the ligand (DNA) are highly programmable, and hence the space of engineerable orthogonal interactions is huge. Consequently they can be leveraged for engineering macromolecular interactions beyond genome targeting1–4. Specifically, here we exploit the programmability of this interaction to devise a scheme to barcode and image large numbers of cell types by anchoring zinc finger proteins to the outside of the cell membrane and thus making them accessible to DNA based probes provided in the extra-cellular medium.
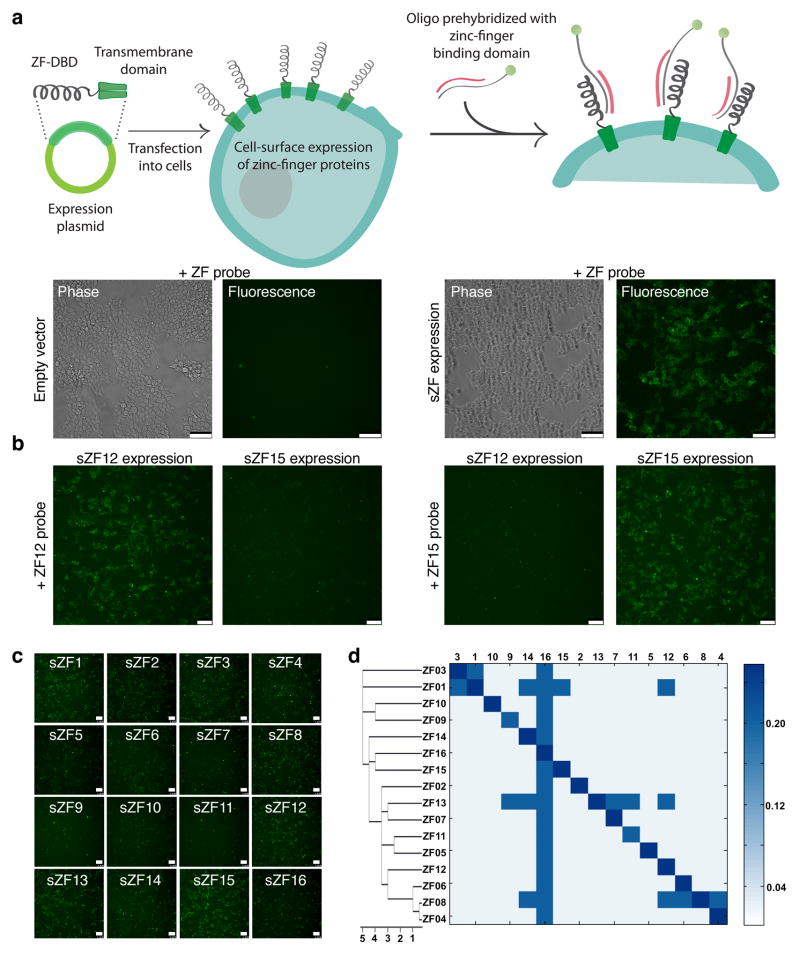
To express zinc-finger DNA binding domains on the cell surface, we fused at their N-terminus an Ig κ-chain leader sequence and at the C-terminus a platelet derived growth factor (PDGF) transmembrane domain (refer Methods)5. To test the ability of surface zinc finger (sZF) expressing cells to bind DNA we exposed them to fluorophore tagged DNA molecules. sZF expressing cells strongly bound the DNA while control cells exhibited very low binding signals, implying functional zinc-finger proteins were successfully expressed on the cell surface (Fig. 1a). Two aspects of this sZF-DNA interaction were of note: First, sZFs were observed to bind to both single6 and double stranded DNA molecules (Supplementary Fig. 1a), however the former interaction was abrogated in the presence of competitor dsDNA (here Salmon Sperm DNA). Second, sZFs also non-specifically bound to dsDNA, but again in the presence of competitor dsDNA binding to only their cognate target dsDNA was retained (Supplementary Fig. 1b). Similar results were obtained using FACS based assays too (Supplementary Fig. 2). Thus in the presence of competitor dsDNA, sZF expressing cells specifically bind their target dsDNA probe and hence each zinc-finger protein uniquely barcodes the cell type expressing them (Fig. 1b).
Figure 1.
Simplex labeling of cell-surface zinc finger expressing cells. (a) Schematic of approach to express zinc fingers on the cell surface and their labeling with dsDNA probes is depicted in the top panel. The lower panels demonstrates that live sZF expressing cells strongly bind DNA while control cells exhibit very low binding signals implying functional zinc-finger proteins were successfully expressed on the cell surface. (b) sZF expressing cells also specifically bind their target dsDNA probe, here evidenced by the ability of sZF12 expressing cells to only bind the ZF12 probe and likewise for sZF15 cells too. Thus each zinc-finger protein uniquely barcodes the cell type expressing them (c) A total of 16 zinc finger proteins were tested using this approach. Assaying intensity of the fluorescence signal from the bound dsDNA probe, we conclude that different ZFs exhibit different binding affinities for their target dsDNA. (d) Next we tested each sZF for its ability to bind its own target dsDNA sequence and also target dsDNA sequences corresponding to the other zinc finger proteins, i.e., a total of 16×16 interactions were probed to generate a cross reactivity profile. It is evident from the heat plot that most zinc fingers bind their target dsDNA specifically, however some strong binders (refer (c) above) show a degree of cross-reactivity (ZFs 1, 8, 13). Interestingly, all the zinc fingers were also observed to bind the ZF16 target dsDNA, likely in part to the relative poly-G rich content of this sequence. The ZFs in this plot were clustered based on their target sequence similarity (refer Supplementary Table 1). Overall, sZFs enable sequence-specific labeling of cells by dsDNA molecules. The scale bar is 100microns.
A total of 16 zinc finger proteins7 were tested using this approach (protein sequences and target dsDNA sequences are provided in Supplementary Table 1). Several aspects of sZF-dsDNA interactions emerged from this analysis. First, different sZFs have different binding affinities for their target dsDNA (Fig. 1c). Specifically, while some bound, as assayed by both fluorescence intensity and duration of binding, their targets strongly (ZFs 1, 3, 8, 12, 13, 15, 16), some were moderately strong binders (ZFs 2, 4, 5, 6, 7, 10, 14), while others were only weak binders (ZFs 9, 11). Next we evaluated the sZF cross reactivity profile for these 16 ZFs (Fig. 1d). We found that while most zinc fingers bound their target dsDNA specifically, some showed a significant degree of cross-reactivity (ZFs 1, 8, 13). The strong ZF binders were particularly susceptible to this phenomenon8. Interestingly, almost all the zinc fingers were observed to bind the ZF16 target dsDNA, likely in part to the high poly-G rich content of this sequence. Based on the above ZFs 2, 3, 4, 5, 6, 7, 10, 12, 14, 15 were found to be orthogonal to each other and were moderate to strong binders and thus good candidates for barcoding cells.
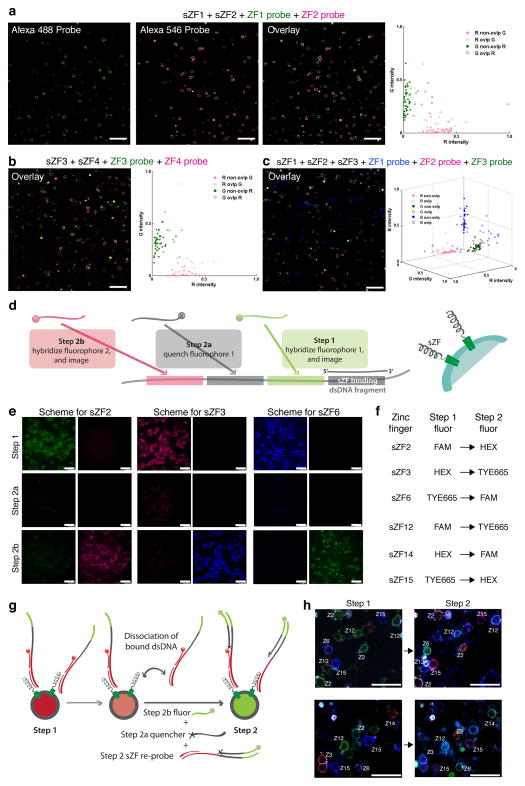
If sZFs are to serve as efficacious barcodes compatible with analysis of structured tissues, they must enable differential labeling of cells in complex mixtures that is detectable in microscopic images. To investigate this we designed experiments to image and analyze mixtures of sZF expressing cell populations. Specifically, cells expressing either sZF1, sZF2, sZF3 or sZF4 were mixed in pairs (sZF1+sZF2; and sZF3+sZF4) or in a pool of three (sZF1+sZF2+sZF3), and were probed using appropriate combinations of fluorophore labeled target dsDNA molecules. We then developed a suite of MatLab GUI applications to analyze the resulting images and compute quantitative measures of the specificity of binding of sZFs to their corresponding oligos at both the whole cell and single pixel level (processing flow for images is depicted in Supplementary Fig. 3). Qualitative inspection and quantitative analysis of the images confirms that the sZF-dsDNA interactions are sequence specific (Figs. 2a, 2b, & 2c, Supplementary Table 2, and Supplementary Figs. 3–9).
Figure 2.
Multiplex labeling of cell-surface zinc finger expressing cells. Cells expressing either of sZF1, sZF2, sZF3 or sZF4 were mixed in pairs (sZF1+sZF2 (a); and sZF3+sZF4 (b)) or in a pool of three (sZF1+sZF2+sZF3 (c)), and were probed using appropriate combinations of Alexa488, Alexa546 and Alexa647 labeled target dsDNA molecules. Qualitative and quantitative inspection of the images shows that these cells bind one labeled oligonucleotide probe and not the others confirming the sZF-dsDNA interactions are sequence specific in a multiplex setting too. (d) Schematic approach of a sequential tagging technique for imaging of >3 barcoded cell types using 3 resolvable fluorophores is depicted. Each sZF has a corresponding probe comprising two parts: a dsDNA portion that specifically binds the zinc finger protein, and a single-stranded portion that is designed to include several hybridization sites. These hybridization sites provide a unique sequence code for the sZF, which is decoded by probing the sites sequentially as follows: in step 1, a fluorophore tagged complementary oligonucleotide is hybridized to its target site enabling a first fluorescence readout; in step 2, two adjacent complementary oligonucleotides are annealed, the first bearing a quencher that suppresses the step 1 fluorescence signal, and the second bearing another fluorophore that enables a second fluorescent readout and so on. (e) A basic demonstration of the scheme in a simplex setting is provided where sZF expressing cells are sequentially probed - each sZF identity here is encoded by two colors, specifically sZF2 by green in step 1 and red in step 2, sZF3 by red in step 1 and blue in step 2 and similarly for sZFs 6, 12, 14 and 15 (f). (g) Schematic of protocol for sequential imaging in a multiplex setting is depicted. Of note, in addition to the step specific quencher and fluorescent oligonucleotides that hybridize to the single-stranded portions of the bound sZF probes, we also freshly re-probed the sZFs at each step to compensate for loss of fluorescence signal due to dissociation of dsDNA probes from the zinc fingers in the time interval between imaging steps. (h) Demonstration of this labeling approach in a multiplex setting is provided. Here 6 sZFs are individually expressed in cells that are subsequently mixed and then sequentially imaged. Two example sets imaged using 3 fluorophores is shown. These results confirm that a sequential tagging scheme can successfully identify the various constituent sZF barcoded cells in complex mixtures. The scale bar is 100microns.
Exploring additional ZFs, or extending this approach to TALEs9, 10 will further expand and refine the list of orthogonal interaction pairs that can be exploited for cellular barcoding. Regardless, one is still limited by the small number of spectrally distinct fluorophores available for simultaneous cell imaging. To address this problem we next devised a sequential live-cell hybridization and imaging approach (suitable for adherent cells). It uses a modified two-part DNA probe that presents a double-stranded portion that binds the sZF and a single-stranded portion containing barcode sequences that can be read-out by serial hybridizations (approach in Fig. 2d): this approach is fast and does not use enzymes or chemical reactions and is thus compatible with use on live cells. Extending this scheme to n steps enables barcoding of 3n cell types using just 3 fluorophores. A basic demonstration of the scheme in a simplex setting is provided in Fig. 2e where sZF expressing cells are sequentially probed - each sZF identity here is encoded by two colors, for instance sZF2 by green in step 1 and red in step 2, sZF3 by red in step 1 and blue in step 2 and similarly for sZFs 6, 12, 14 and 15 (Fig. 2f). We were also able to mix up to six individually labeled cells and identify their barcode in situ using two hybridization cycles (Figs. 2g, 2h). In these experiments, the zinc finger-binding probes were also re-supplied for each round of sequencing by hybridization. This re-probing compensated for the loss of fluorescence signal due to the dissociation of dsDNA probes from the sZFs in the interval between imaging steps, and also aided in active displacement of the existing probes, thus mitigating effects of any incomplete quenching in the previous step (refer also Supplementary Figs. 10, 11 respectively for the dissociation kinetics of dsDNA probes and confirmation of the genotype-to-labeling association in these experiments). Use of toe-hold mediated strand exchange11 to displace bound DNA probes can be exploited to further refine this technique. Overall, our results in Fig. 2e and Fig. 2h above suggest that such a sequential tagging scheme can successfully identify the various constituent cells in complex mixtures of barcoded cell types using just 3 spectrally distinct fluorophores.
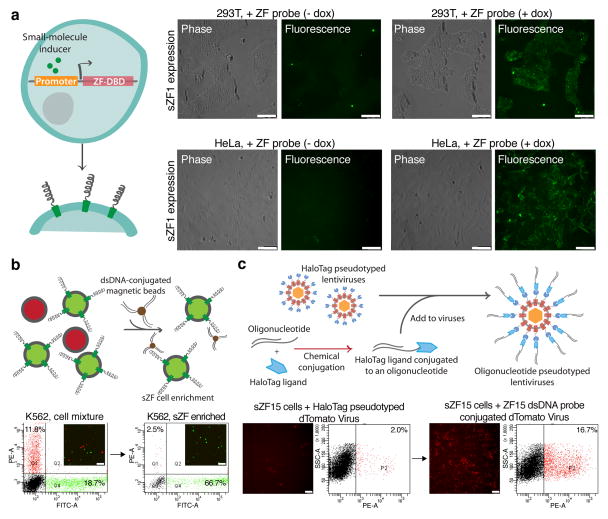
Finally we explored the versatility of sZFs through three applications. First we used sZFs as surrogate reporters of endogenous cellular activity. For this, lentiviral vectors with small molecule (tetracycline and cumate) inducible promoters for driving sZF expression were constructed. Stable transductions of 293T and HeLa cells were performed, and upon small molecule induction sZF expression could indeed be readily detected by the ability of the cells to bind dsDNA molecules (Fig. 3a, Supplementary Fig. 12). This temporally inducible expression of barcodes can also be exploited to minimize effects of sZF expression on cell physiology and toxicity (Supplementary Fig. 13). Second, we exploited the fact that sZFs are expressed on cell surfaces where they are physically accessible and can thus provide convenient handles for DNA mediated cell capture. Specifically, sZF expressing cells were successfully enriched from a mixed population of K562 cells by performing a pull-down using either dsDNA probe-conjugated magnetic beads (Fig. 3b), or on dsDNA arrays (Supplementary Fig. 14). Third, we demonstrated sZF mediated selective gene delivery by pseudotyping12 lentiviruses with dsDNA probes (Supplementary Fig. 15). Specifically, these modified lentiviruses successfully delivered genes to sZF barcoded cells, but in the absence of the DNA pseudotyping, lentiviral delivery efficiency was significantly diminished (Fig. 3c). Taken together, these three applications demonstrate that sZFs have uses beyond direct labeling that include state-probing, capture, and targeting of cells.
Figure 3.
sZFs enable state-probing, capture and targeting of cells. (a) State-probing: sZFs were investigated for their ability to also serve as surrogate reporters of endogenous cellular activity. Stable transduction of 293T and HeLa cells by a lentiviral vector with a tetracycline inducible promoter to drive sZF expression was performed. Upon small molecule induction sZF expression could indeed be readily detected by the ability of the cells to bind dsDNA molecules. (b) Capture: Here a mixed population of K562 cells comprising sZF expressing (labeled green) and non-sZF expressing (labeled red) cell types was selectively enriched for the sZF expressing sub-population using a dsDNA conjugated magnetic bead based pull down. (c) Targeting: The schematic depicts the approach to create oligonucleotide conjugated lentiviruses by tethering dsDNA probes to HaloTag protein pseudotyped lentiviruses through the HaloTag-protein/HaloTag-ligand interaction. These lentiviruses pseudotyped with sZF specific dsDNA probes are successfully delivered to sZF barcoded cells, but in the absence of the DNA pseudotyping lentiviral delivery efficiency is significantly diminished, thus demonstrating selective targeting of only the appropriately pseudotyped gene delivery vehicle. The scale bar is 100microns.
In summary, sZF barcoding enables specific and quantifiable cellular labeling (Supplementary Figs. 4–8 and 11), and is suited for applications requiring tracking of heterogeneous mixtures of cells. It can also be synergized with existing methods for multiplexed cell probing such as elemental isotope labeled antibody based mass cytometry13 and combinatorial fluorescent protein expression14. Since a threshold amount of sZF protein expression is needed to discernibly label a cell, for certain applications such as sZFs as surrogate reporters (Fig. 3a), they will only quantitate presence of endogenous activity that is above a certain level. Here brighter probes such as quantum dots could be used to amplify the signal. Overall, the versatility of sZFs makes them a powerful tool enabling a gamut of synthetic biology applications: ranging from the possibility of doing highly multiplexed tracking of endogenous gene activity for studying complex pathways and interacting gene networks15; to tissue engineering through control of physical cell arrangement and cell-cell associations using DNA mediated interactions16, 17, and engineering of designer macromolecular associations18–20.
Online Methods
Plasmid construction
The zinc finger DNA binding domains were synthesized as gBlocks from IDT. To express ZFs on the cell surface, we fused at their N-terminus an Ig κ-chain leader sequence and at the C-terminus the platelet derived growth factor (PDGF) transmembrane domain (pDisplay system from Invitrogen). Additional endoplasmic reticulum import sequences based on the serotonin receptor 5HT3A, and transmembrane domains from Neurokinin-1 receptor (NK1R) or beta-2 adrenergic receptor were also tried with similar success (relevant DNA fragments were cloned from NEB plasmids N9184S and N9216S). The lentiviral plasmids for inducible tetracycline expression and cumate expression were obtained respectively from Addgene (plasmids 20321, 20342) and System Biosciences (QM800A-1), and the sZF fusion constructs were directly cloned into these. Small molecule inducers doxycycline and cumate were used at 1μg/ml and 30μg/ml concentrations respectively. All reagents developed in this study are available via Addgene (http://www.addgene.org/browse/pi/765/articles/).
Cell culture
HEK 293T cells and HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) high glucose supplemented with 10% fetal bovine serum (FBS), penicillin/streptomycin (pen/strep), and non-essential Amino acids (NEAA). K562 cells were cultured in RPMI-1640 medium supplemented with 10% FBS, pen/strep and NEAA. All cells were maintained at 37°C and 5% CO2 in a humidified incubator. Transfections of the sZF expressing plasmids were performed using Lipofectamine 2000 as per the manufacturer’s protocols. For K562s, cells were resuspended in SF reagent and nucleofected according to manufacturer’s instruction (Lonza). All reagents above were obtained from Gibco/Invitrogen.
Cell labeling and DNA probes
All cell labeling was performed in the following buffer: PBS (-CaCl2, -MgCl2) supplemented with 5% BSA (fraction V, fatty acid free), 20μM ZnCl2, 1mM MgCl2 and 100μg/ml Salmon Sperm DNA. DNA probes (synthesized by IDT) had 4 phosphorothioate bonds on both the 5′ and 3′ ends to enhance protection against nucleases prevalent in extra-cellular media. The fluorophores (conjugated to the probes) used for simplex and multiplex labeling experiments (refer Figs. 1, 2, 3, Supplementary Figs. 1, 9, 10, 12, and Supplementary Table 3) were: Alexa Fluor 488, Alexa Fluor 546, and Alexa Fluor 647. The quencher-dye pairs used for the sequential labeling experiments (refer Fig. 2, Supplementary Fig. 11 and Supplementary Table 4) were: Black Hole Quencher-1 with FAM, Iowa Black FQ with Hex, and Iowa Black RQ with TYE 665. All imaging was conducted using a Leica AM TIRF MC microscope. All dsDNA probes were used at a 100–200nM final concentration, and cells were labeled for 5–10 minutes with these, following which they were washed with buffer twice and subsequently imaged. Standard cell culture media (containing 10% FBS) can also be used as a buffer to re-suspend probes and successfully label cells, however the dsDNA probes are rapidly degraded in this media, and it is hence not ideal for long term imaging applications. Cell capture experiments were performed using streptavidin coated Miltenyi beads conjugated to biotinylated dsDNA probes. The DNA arrays used in the study were synthesized using Amine-conjugated oligonucleotides (synthesized by IDT) by spotting them onto epoxy coated slides using an Arrayit spotter. DNA arraying and slide passivation were performed as per the manufacturer’s instructions.
Oligonucleotide pseudotyped lentiviruses
First a cell-surface HaloTag (sHaloTag) expressing plasmid was constructed by fusing at its N-terminus an Ig κ-chain leader sequence and at the C-terminus a VSVG transmembrane domain. This plasmid was next used to produce the HaloTag protein pseudotyped lentiviruses in 293T cells with the lentiviral and packaging plasmids transfected in the following ratio (per 150mm cell culture dish): 15ug dTomato expressing lentivirus vector, 15ug gag/pol plasmid, 7.5ug SINmu plasmid (gift from Pin Wang, USC), and 7.5ug of the above sHaloTag plasmid. Next, the HaloTag ligand succinimidyl ester (O4) building block was conjugated to an amine group bearing DNA probe. Finally, the harvested lentiviruses were conjugated to the above sZF specific dsDNA probes through the HaloTag-protein/HaloTag-ligand interaction (Promega) yielding the desired oligonucleotide pseudotyped lentiviruses.
Image processing and statistics
With the goal of quantitating sZF behavior and specificity, JPG images acquired as single z-slices from fluorescence confocal microscopy were processed using three in-house developed user-interactive MatLab (The Mathworks, Waltham MA) applications whose use is depicted in Supplementary Fig. 3. ImageNormalizer presents options for normalization of intensities and background subtraction in each channel to produce a standardized multi-channel TIF image used by other applications. ImageMasker enables users to identify image regions containing cell debris and dead cells that are to be excluded from subsequent analysis. SegmentOverlapAnalysis gives users interactive control over parameters used to identify regions of the images occupied by cells in each channel (i.e., image segmentation), and then allows the user to submit the segmented image to statistical analysis. Statistical analysis is performed on-line by a fourth MatLab function invoked by SegmentOverlapAnalysis but is not itself interactive. In brief, for the images analyzed here, normalization entailed intensity clipping at the 99.6th to the 99.8th percentile intensity in each channel with no subsequent background subtraction, an average of 37.0 ± 12.7 masks were created to mask out a total of 2.94% ± 1.43% of image area per image, and 47.1 ± 9.8 segments (Supplementary Table 2) were generated per channel (numbers are means ± standard deviations in all cases). Details can be found in Supplementary Information sections: Image analysis methods, statistics and results summary. To mitigate a potential bias in measuring sZF specificity by direct comparisons of intensity across channels caused by our use of intensity thresholds to segment images, we developed additional measures based on intensity correlations across channels, and overlaps between segments of different channels: In brief, if sZFs are specific, correlations should be negative and large numbers of segments should be seen in each channel that do not overlap segments from other channels. Data substantiating these and other quantitative observations relevant to sZF behavior and specificity are summarized in Supplemental Information (Supplementary Table 2 and Supplementary Figs. 4–8). All actual processing and statistical data files and figures generated by our image processing applications for the images, and the MatLab image processing applications themselves along with instructions on their use have been made freely available to the research community for non-commercial research on our web site: http://arep.med.harvard.edu/sZF_cell_barcode/.
Supplementary Material
Acknowledgments
This work was supported by NIH grant P50 HG005550.
Footnotes
Author Contributions
PM and GMC conceived the study and designed the experiments. PM performed experiments. JL, DL and LN developed reagents. JA developed the image analysis suite and performed associated analyses. PM, JA and GMC wrote the manuscript with support from all authors.
References
- 1.McMahon MA, Rahdar M, Porteus M. Nat Methods. 9:28–31. doi: 10.1038/nmeth.1811. [DOI] [PubMed] [Google Scholar]
- 2.Kim YG, Cha J, Chandrasegaran S. Proc Natl Acad Sci U S A. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rebar EJ, Pabo CO. Science. 1994;263:671–673. doi: 10.1126/science.8303274. [DOI] [PubMed] [Google Scholar]
- 4.Volna P, et al. Nucleic Acids Res. 2007;35:2748–2758. doi: 10.1093/nar/gkm182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chesnut JD, et al. J Immunol Methods. 1996;193:17–27. doi: 10.1016/0022-1759(96)00032-4. [DOI] [PubMed] [Google Scholar]
- 6.Brown RS. Curr Opin Struct Biol. 2005;15:94–98. doi: 10.1016/j.sbi.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 7.Maeder ML, et al. Mol Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pattanayak V, Ramirez CL, Joung JK, Liu DR. Nat Methods. 2011;8:765–770. doi: 10.1038/nmeth.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boch J, et al. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 10.Moscou MJ, Bogdanove AJ. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 11.Duose DY, et al. Nucleic Acids Res. 2012;40:3289–3298. doi: 10.1093/nar/gkr1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lei Y, Joo KI, Zarzar J, Wong C, Wang P. Virol J. 2010;7:35. doi: 10.1186/1743-422X-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bendall SC, et al. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livet J, et al. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- 15.Purnick PE, Weiss R. Nat Rev Mol Cell Biol. 2009;10:410–422. doi: 10.1038/nrm2698. [DOI] [PubMed] [Google Scholar]
- 16.Boyce M, Bertozzi CR. Nat Methods. 2011;8:638–642. doi: 10.1038/nmeth.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gartner ZJ, Bertozzi CR. Proc Natl Acad Sci U S A. 2009;106:4606–4610. doi: 10.1073/pnas.0900717106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barbas CF., III US 2007/0178499 A1. 2007
- 19.Delebecque CJ, Lindner AB, Silver PA, Aldaye FA. Science. 2011;333:470–474. doi: 10.1126/science.1206938. [DOI] [PubMed] [Google Scholar]
- 20.Dueber JE, et al. Nature biotechnology. 2009;27:753–759. doi: 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.