Abstract
The ability to efficiently isolate undifferentiated human induced pluripotent stem cells (UD-hiPSCs) as colonies from contaminating non-pluripotent cells is a crucial step in the stem cell field to maintain hiPSC survival, purity, and karyotype stability. Here we demonstrate significant differences in ‘adhesive signature’ among UD-hiPSCs, parental cells, partially reprogrammed cells, and differentiated progeny. The distinct adhesive signature of hiPSCs was exploited to rapidly (~10 min) and efficiently isolate fully reprogrammed bona fide hiPSCs as intact colonies from heterogeneous reprogramming cultures and differentiated progeny using microfluidics. hiPSCs were isolated in a label-free fashion and enriched to > 95–99% purity and survival without adversely affecting the transcriptional profile, differentiation potential or karyotype of the pluripotent cells. This rapid and label-free strategy is applicable to isolate UD-hPSCs (hiPSCs, hESCs) from heterogeneous cultures during reprogramming and routine cultures and can be expanded to purify stem cells of specific lineages, such as neurons and cardiomyocytes.
Keywords: reprogramming, cell adhesion, fibronectin, integrin, focal adhesion, microfluidics
Generation of human induced pluripotent stem cells (hiPSCs) from somatic cells using reprogramming factors represents a promising strategy to produce autologous cells for regenerative therapies and novel models of human development and disease1–3. The reprogramming process is inefficient (0.001–2%)4 and hiPSC cultures are often heterogeneous because of the presence of undifferentiated stem cells, parental and partially reprogrammed cells, and differentiated derivatives5. Reprogrammed hiPSC colonies are typically hand-picked and residual parental cells and partially reprogrammed cells introduce variability, pathogenic contamination, and potential immunogenicity6. Non-hiPSC contamination is also problematic in directed differentiation protocols to generate specific lineages7–11.
hiPSC survival and stemness require compact colonies with E-cadherin-mediated cell-cell adhesion12. Current methods for maintenance of hiPSC cultures rely on manual isolation alone or in combination with enzymatic dissociation13–17. Such methods are tedious, time-intensive, require skilled labor, and are heavily dependent on morphologic recognition of undifferentiated cells. Whereas many reagents have been developed for enzymatic passaging, such methods are not selective for hPSCs and therefore unwanted cells are often transferred during passaging14,18. Furthermore, enzymatic methods cause karyotypic abnormalities compared to manual passaging13–15,19 and require re-aggregation of dissociated hPSCs for improved survival20. Although FACS sorting21 can enrich the purity of undifferentiated populations, this method requires single cell dissociation of hPSCs, which induces cell death12, and re-plated cells fail to form compact colonies (Supplementary Fig. 1a).
Current hiPSC purification methods remain a bottleneck in the stem cell field, thus there is a great need to develop unbiased, high-throughput technologies that can efficiently separate colonies of UD-hiPSCs from contaminating non-reprogrammed and partially reprogrammed cells, feeder cells, or differentiated cells while avoiding tedious manual isolation, enzymatic dissociation into single cells, and labeling with antibodies. In this study, we demonstrate a unique “adhesive signature” for hPSCs dictated by their phenotypic state as a multifactorial function of ECM-bound integrins, assembly of focal adhesions, and resulting cell-ECM adhesion strength. We present a platform technology that exploits differences in the adhesion strength among non-reprogrammed and partially reprogrammed cells, UD-hiPSCs, and differentiated cells to selectively isolate UD-hiPSCs using microfluidics and applicable to differentiated progeny.
RESULTS
Changes in ‘adhesive signature’ with induced reprogramming
During reprogramming, hiPSCs undergo significant changes in morphology resulting in an epithelial phenotype indicative of the pluripotent state (Fig. 1a). IMR90 and dermal fibroblasts, common somatic cell sources1,2 for reprogramming, exhibited an elongated morphology without direct cell-cell adhesions (Fig. 1b) and lack of pluripotent markers (Fig. 1c) compared to reprogrammed UD-hiPSCs which existed as tightly packed colonies (Fig. 1d,e and Supplementary Fig. 1b). Because of the low reprogramming efficiency that remains a critical limitation in hiPSC generation4, < 1% fibroblasts were converted to fully reprogrammed hiPSCs22 positive for OCT4, SSEA4, TRA-1-60, NANOG, and TRA-1-81 (Fig. 1e and Supplementary Fig. 2a,b). Residual non-reprogrammed and partially reprogrammed cells were less spread and expressed some, but not all, pluripotency markers (Fig. 1e).
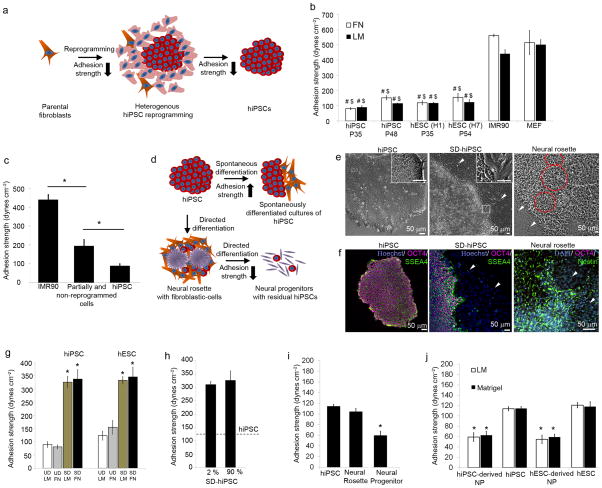
Figure 1. Adhesive signature of hiPSCs undergoing reprogramming and differentiation.
(a) Schematic representing changes in adhesion of hiPSCs with reprogramming. Orange: Parental fibroblast; pink: partially reprogrammed cells; red: UD-hiPSCs. (b–e) Morphological changes and expression of pluripotency markers SSEA4 and OCT4 in parental fibroblasts, reprogrammed cultures with UD-hiPSCs (white arrowheads, white box) and non-reprogrammed/partially reprogrammed cells (red arrowheads, red box). (f) Immunostaining showing vinculin localization to focal adhesions and actin in IMR90 cells, partially reprogrammed culture with residual parental and transduced cells, and UD-hiPSCs on laminin. (g) Adhesion strength for UD-hiPSCs, hESCs, IMR90, and MEF on fibronectin and laminin. (h) Adhesion strength for non-reprogrammed and partially reprogrammed cells compared to parental cells and UD-hiPSCs. (i) Schematic representing changes in adhesive signature of hiPSCs with differentiation. Orange: SD-hiPSCs; purple: neural cells. (j–k) Morphological differences and expression of SSEA4 and OCT4 in UD-hiPSCs, SD-hiPSCs (white arrowheads), and Nestin expressed by neural rosettes (red circles). (l) Adhesion strength for SD-hiPSCs and hESCs compared to undifferentiated cells cultured on fibronectin and laminin. (m) Recruitment of vinculin to focal adhesions in SD-hiPSCs (magenta arrowhead) distinct from UD-hiPSCs (white arrowhead). Adhesion strength for (n) SD-hiPSCs with 2% and 90% differentiation, cultured on laminin, (o) UD-hiPSCs, hiPSC-derived neural rosettes and neural progenitors (NP) on laminin, and (p) NP derived from hiPSCs and hESCs on laminin and Matrigel. All bar graphs represent average ± s.d. (*P < 0.05, #P < 0.05 stem cells vs. IMR90, $P < 0.05 stem cells vs. MEFs, n = 3).
The adhesive strength of a cell to its ECM is dependent on ECM-ligated integrins and their association to cytoskeletal elements23. Using flow cytometry (Supplementary Fig. 3a) and adhesion inhibition studies (Supplementary Fig. 3b,c), we determined that parental fibroblasts expressed predominantly α5β1 integrin whereas UD-hiPSCs expressed high levels of α6β1 integrin, regardless of whether the cells were cultured on fibronectin, laminin or Matrigel, consistent with hESC studies24. Fibroblasts possessed actin stress fibers and vinculin and talin were enriched at focal adhesions (Fig. 1f and Supplementary Fig. 4a). In contrast, hiPSCs exhibited significantly fewer actin fibers with diffused vinculin and talin throughout the cytoplasm or localized to cell-cell junctions (Fig. 1f and Supplementary Fig. 4b). Non-pluripotent cells in reprogramming cultures exhibited mixed regions of well-defined focal adhesions in spread cells and round cells without distinct focal adhesions (Fig. 1f). Based on these differences in adhesive structures, we hypothesized that alterations in the “adhesive signature” of cells related to integrin binding and cytoskeletal components accompany induced pluripotency and differentiation of hiPSCs.
The steady-state cell-ECM adhesion strength for hiPSCs and IMR90 cells was evaluated using a spinning disk device23 (Supplementary Fig. 5a). Adhesion strength analysis revealed seven-fold lower adhesion strength to fibronectin for hiPSCs compared to parental fibroblasts (Fig. 1g). Analyses among fibroblastic parental and feeder cells, hESCs, and hiPSCs revealed significantly lower adhesion strength to fibronectin, laminin, and Matrigel for hPSCs compared to fibroblasts (P < 0.02, Fig. 1g and Supplementary Fig. 5b), indicating the shift in adhesive properties between pre- and post-reprogramming for hiPSCs, equivalent to those observed with hESCs. These results were independent of passage number, underlying matrix, and parental fibroblast source (Supplementary Fig. 5c). Using micropatterned hiPSC colonies, we found that adhesion strength of hiPSCs was independent of colony size (Supplementary Fig. 6). We next examined the adhesion strength of non-reprogrammed/partially reprogrammed cells that expressed some but not all pluripotency markers (e.g., OCT4+, SSEA4−). These cells exhibited higher adhesion strength compared to UD-hiPSCs but lower than parental cells (Fig. 1h). The differences in adhesive force correlate to increased focal adhesion assembly in parental cells compared to hiPSCs. Collectively, these results indicate striking differences in the adhesive signatures of hiPSCs and hESCs compared to parental and non-reprogrammed/partially reprogrammed cells that can be exploited to identify fully reprogrammed hiPSCs from partially or non-reprogrammed cells.
Distinct adhesive properties of differentiated hiPSCs
We next determined the adhesive signature of hiPSCs undergoing spontaneous or directed differentiation (Fig. 1i). Unlike UD-hiPSCs (Fig. 1j,k), colonies with spontaneous differentiation exhibited mixed regions of mesenchymal–epithelial morphologies and fibroblastic cells lost pluripotency markers (Fig. 1j,k). We performed adhesion strength analyses on SD-hiPSCs (~10% TRA-1-60+) and detected significant increases in the adhesion strength to ECM of SD-hiPSCs compared to UD-hiPSC (Fig. 1l, P < 0.006). Similar differences in adhesion strength were observed for SD-hESCs compared to UD-hESCs. SD-hiPSCs displayed actin stress fibers and localized vinculin and talin to focal adhesions (Fig. 1m and Supplementary Fig. 4c) compared to undifferentiated colonies. Differences in adhesion strength between undifferentiated and differentiated cells were independent of the levels of spontaneous differentiation (Fig. 1n).
We also examined the adhesive signature of directed differentiated progeny. Early-stage multi-potent neural stem cells (neural rosettes10) exhibited a radial pattern of epithelial morphology (Fig. 1j), and staining for Nestin (Fig. 1k) and Musashi (Supplementary Fig. 7a) was distinct from UD-hiPSCs although adhesion strength values were comparable (Fig. 1o). Rosettes, however, exhibited significantly lower adhesion strength compared to contaminating fibroblast-like cells (P < 0.05). Rosettes were manually isolated and differentiated to neural progenitors (NPs) and neurons (Supplementary Fig. 7b). NPs exhibited adhesion strength comparable to neurons but 50% lower relative to UD-hiPSCs (Fig. 1o,p) and ~6-fold lower than spontaneously differentiated fibroblastic cells (Fig. 1j), independent of hPSC type and matrix (Fig. 1p). These analyses demonstrate that hPSCs, progenitors, and terminally differentiated cells exhibit distinct adhesive signatures.
Hydrodynamic isolation of fully reprogrammed hiPSCs
We exploited the unique adhesive signatures between pre- and post-reprogrammed states of hiPSCs to develop a novel strategy to isolate undifferentiated hPSCs from a heterogeneous cell population. Adhesive force-based separation of multiple distinct cell populations via a simple microfluidic system represents a promising, label-free separation technique that requires minimal cell processing and can detach cells in their native cell-cell microenvironment. We termed this technology μSHEAR (micro Stem cell High-Efficiency Adhesion-based Recovery). High-throughput microfluidic devices are being adapted in routine cell culture25–27 and offer advantages over conventional hydrodynamic sorting, including laminar flow with a million-fold less buffer volume and recovery of detached cells28.
μSHEAR devices were fabricated for a range of culture surface areas (Fig. 2a and Supplementary Fig. 8a). Within the microfluidic device, cells remained viable, retained their distinct morphologies, and hiPSCs remained undifferentiated (Supplementary Fig. 8b,c). Application of laminar flow in the microfluidic device generated fluid shear stresses on adherent cells. hiPSC colonies detached at a shear stress of 85–125 dynes cm−2 within 4 min of applying fluid flow and were completely detached in 10–14 min (Fig. 2b,c) irrespective of underlying ECM (Supplementary Fig. 8d,e), whereas fibroblasts with stronger adhesive properties remained attached. To quantify the efficiency of UD-hiPSC purification, recovered cells were incubated with StainAlive-DyLight488-conjugated TRA-1-60 antibody to stain for live UD-hiPSCs and Cell Tracker Red dye (CMPTX). Flow cytometry analysis of the recovered cells revealed significant enrichment of hiPSCs when detached at 85–125 dynes cm−2 with up to 99% purity (Fig. 2d) compared to the initial baseline purity of 39% UD-hiPSCs (all cells trypsinized). Exposure to higher fluid forces (250–350 dynes cm−2) resulted in contamination with 18% of IMR90 cells compared to cultures exposed to 85–125 dynes cm−2 with less than 1% fibroblast contamination. We observed high proportions of IMR90 cells in detached populations exposed to 750–850 dynes cm−2, similar to trypsinized samples under no flow conditions (Fig. 2d). The μSHEAR isolation efficiency was independent of hiPSC purity in the initial coculture (Fig. 2e,f and Supplementary Fig. 9). Fewer than 3% of residual cells in devices post-fluid detachment were hiPSCs, indicating high recovery yield of hiPSCs by μSHEAR (Fig. 2e,f). Similar results were observed with hESCs and feeder-layer MEFs cocultures (Fig. 2e,f and Supplementary Fig. 8f). These results demonstrate that adhesion strength differences can be exploited to selectively purify hPSCs as intact colonies from a mixed fibroblast-hPSC culture. Additionally, we selectively enriched hiPSCs from cocultures where hiPSCs were allowed to proliferate with parental cells for 5–7 days within microfluidic devices (Fig. 2g,h and Supplementary Movie 1). We also tested the high-throughput potential of μSHEAR across 0.5–9 cm2 culture areas and found 95–99% enrichment efficiency, demonstrating the scalable potential of μSHEAR to routine culture platforms. Loading and recovery in μSHEAR is facile because cells are loaded in < 30 s and no time is spent pre-cleaning the cultures. Self-contained disposable microfluidic devices ensure sterility and cell recovery takes ~5 min, similar to a routine centrifugation steps following enzymatic dissociation.
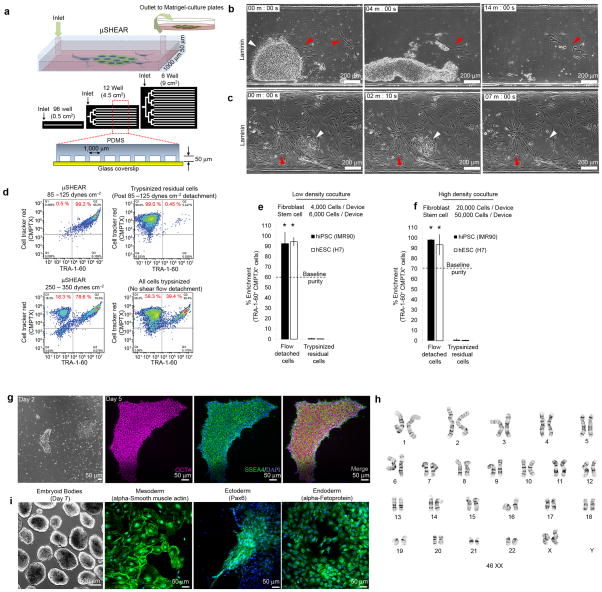
Figure 2. Adhesion strength-based isolation of undifferentiated pluripotent stem cells in microfluidic devices.
(a) Schematic of μSHEAR device cross-section and scale-up. Selective isolation of UD-hiPSCs at 85–125 dynes cm−2 shear stress when cocultured with (b) low and (c) high density IMR90 cells on laminin. (d) Flow cytometry plots showing detached hiPSCs (TRA-1-60+CMPTX+) and IMR90 cells (TRA-1-60−CMPTX+) at 85–125 and 250–350 dynes cm−2. Post-μSHEAR, residual cells in devices were trypsinized and analyzed. (e–f) Flow cytometry-based measurements of enrichment of hiPSCs and hESCs isolated at 85–125 dynes cm−2 from a coculture of IMR90 and MEF cells, respectively. Table presents enrichment with varying levels of hiPSCs in a fibroblast-hiPSC coculture. (g) Selective isolation of UD-hiPSC colonies after 5 days of coculture with IMR90 fibroblasts on laminin and (h) enrichment of hiPSCs detached at 85–125 dynes cm−2 from 5 day cultures. (i) Isolation of PBMC-derived hiPSCs cocultured with PBMCs. Loosely adherent PBMCs were removed at 10 dynes cm−2 and hiPSCs were harvested by increasing the flow to 100 dynes cm−2. (j) Flow cytometry measurements of enrichment of blood cell-derived hiPSCs detached at 100 dynes cm−2. (k) Detached UD-hiPSC colonies adhered to Matrigel and retained expression of SSEA4 and OCT4. (l) Karyotype analysis of hiPSCs exposed to two isolations 7–8 days apart. (m) μSHEAR-isolated hiPSCs generated embryoid bodies (day 14) that differentiated into all three germ layers (day 21). Graph shows average ± s.d. (*P < 0.05, n = 3).
We also demonstrated efficient separation of hiPSCs from other parental sources, such as blood29. Because peripheral blood mononuclear cell (PBMCs) remain loosely adherent and become adhesive post-reprogramming to hiPSCs (Supplementary Fig. 10), we performed negative selection of hiPSCs by first exposing the culture to a shear stress of 10 dynes cm−2 to remove PBMCs. All PMBCs were removed from the devices within 1 min of exposure to flow (Fig 2i), and thereafter hiPSC were collected as colonies with ~99% purity by increasing the flow (Fig. 2j and Supplementary Movie 2).
A critical consideration for μSHEAR-based hiPSC isolation is whether recovered colonies can be efficiently re-cultured, retain their pluripotency, and maintain a stable karyotype. Colonies recovered by μSHEAR initially adhered as small colonies (Fig. 2k) with the ability to self-renew without any signs of differentiation (Supplementary Fig. 11). The isolated colonies retained their pluripotent phenotype (Fig. 2k and Supplementary Fig. 11) and exhibited no chromosomal abnormalities (Fig. 2l). μSHEAR-isolated hiPSC colonies readily generated embryoid bodies and differentiated into mesoderm, ectoderm, and endoderm derivatives (Fig. 2m).
Isolation of hiPSCs from heterogeneous reprogramming cultures
Typical reprogramming cultures are comprised of highly heterogeneous populations of non-reprogrammed and imperfectly/partially reprogrammed cells22, and there is no label-free, high-throughput method to identify nascent hiPSCs. We anticipated that adhesive signature differences could be exploited to selectively isolate hiPSCs at early stages of reprogramming. Using μSHEAR, hiPSC colonies were effectively isolated (94 ± 3.6% purity) at 100 dynes cm−2 prior to detachment of residual non-reprogrammed and partially reprogrammed cells (Fig 3a,b and Supplementary Fig. 12a,b). We observed only 0.05% residual hiPSCs while non-hiPSCs constituted 99.9% of the culture in μSHEAR device (Fig 3b and Supplementary Fig. 12c). Isolated hiPSCs and residual cells were stained for pluripotency markers to establish that the μSHEAR-isolated cells were indeed bona fide hiPSCs. Coincident expression of TRA-1-60, TRA-1-81, DNMT3B, REX1, OCT4, SSEA4, GDF3, hTERT, and NANOG denote the fully reprogrammed state of hiPSCs22. Although partially reprogrammed cells express some pluripotency markers, only fully reprogrammed hiPSCs express TRA-1-60, DNMT3B and REX122. μSHEAR-isolated hiPSCs expressed all markers characteristic of bona fide hiPSCs, whereas residual cells expressed OCT4, hTERT, and GDF3 but not TRA-1-60, TRA-1-81, DNMT3B, REX1, SSEA4, and NANOG (Fig 3c–h). We also analyzed epigenetic changes in methylation patterns30,31 of endogenous OCT4, NANOG, and SOX2 genes. μSHEAR-isolated hiPSCs displayed unmethylated OCT4, SOX2 and NANOG, similar to patterns in hand-picked hiPSCs and unmethylated genomic DNA controls (Supplementary Fig. 12d). These results demonstrate equivalent methylation patterns for OCT4, SOX2, and NANOG between bona fide manually passaged and μSHEAR-isolated hiPSCs. Finally, μSHEAR-isolated hiPSCs formed teratomas when implanted into immunodeficient mice (Fig. 3i and Supplementary Fig. 13). These studies demonstrate that fully reprogrammed, bona fide hiPSCs can be selectively isolated from residual parental fibroblasts and partially reprogrammed cells using μSHEAR.
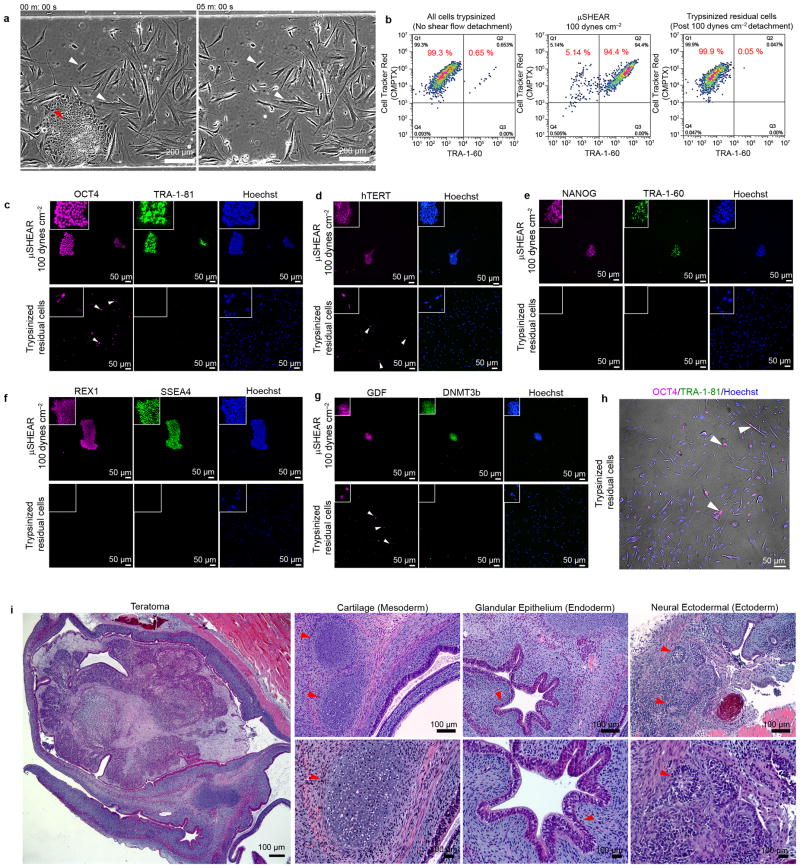
Figure 3. μSHEAR-based isolation of bona fide hiPSCs from a heterogeneous reprogramming culture.
(a) A heterogeneous reprogramming culture seeded into μSHEAR device. Colonies of hiPSCs were selectively detached within 5 min of flow at 100 dynes cm−2 shear stress. (b) Flow cytometry plots showing detached hiPSCs (TRA-1-60+CMPTX+) and non-reprogrammed/partially reprogrammed cells (TRA-1-60−CMPTX+). At 100 dynes cm−2 shear stress, hiPSCs were isolated with 95% purity from a heterogeneous reprogramming culture with initial 0.65% hiPSC purity. Following μSHEAR isolation, residual cells in the devices were trypsinized and the residual cells consisted of 99.9% non-hiPSCs. μSHEAR-isolated hiPSCs and residual culture from the devices were re-plated on Matrigel and stained for bona fide hiPSC markers for fully reprogrammed cells: (c) OCT4 and TRA-1-81. Isolated hiPSCs expressed both markers whereas residual cells contained several cells expressing OCT4 but negative for TRA-1-81. (d) Merged phase contrast and OCT4, TRA-1-81 staining for the residual cells indicate presence of OCT4+TRA-1-80− cells with elongated and spread morphology, distinct from hiPSCs. (e–h) NANOG, TRA-1-60, REX1, SSEA4, hTERT, GDF, and DNM3Tb. Isolated hiPSCs expressed all bona fide markers. Residual cells were negative for NANOG, TRA-1-81, REX1, SSEA4, and DNMT3b but expressed hTERT and GDF. (i) Representative hematoxylin-eosin (H/E) stained sections from a formalin-fixed teratoma produced from μSHEAR-isolated hiPSCs. μSHEAR-isolated formed differentiated tissues representing all three embryonic germ layers including: cartilage (mesoderm), glandular epithelium (endoderm), and neural tissues (ectoderm).
Selective isolation of UD-hiPSCs from differentiated cells
Spontaneous differentiation of hPSCs is a significant common problem that requires frequent (i.e. daily) intensive manual maintenance of cultures to remove differentiated cells and preserve the undifferentiated phenotype of pluripotent cells16,17,21,32,13. Similarly, current methods to terminally differentiate cells into mature neurons9,10 and cardiomyocytes are highly variable and often lead to heterogeneous populations including residual hiPSCs7,8. We exploited the unique adhesive signature of undifferentiated hiPSCs to effectively separate undifferentiated PSCs from differentiated progeny (Fig. 1l). SD-hiPSC cultures with varying levels of differentiation were dissociated and cultured overnight in μSHEAR devices with hiPSCs (Supplementary Fig. 14a). UD-hiPSCs were selectively isolated as intact colonies before detaching SD-hiPSCs with > 97% purity and yield irrespective of the levels of SD-hiPSCs (Fig. 4a, Supplementary Fig. 14b), demonstrating the ability of μSHEAR to function as a simple, robust purification method. Selective purification was not observed with commonly used enzymatic agents (Fig. 4a). Enrichment efficiencies for hESCs were comparable to hiPSCs (Supplementary Fig. 14c). A major advantage of μSHEAR is the ability to isolate hiPSC as colonies and label-free, whereas existing TRA-1-60 antibody-based purification (e.g., EasySep and FACS) requires dissociation of colonies into single cells and labeling with molecular probes. Although EasySep and μSHEAR demonstrated 97% purification efficiency, μSHEAR achieved improved purification with less variability in TRA-1-60 expression compared to EasySep (Fig. 4b). When plated, EasySep or FACS-purified cells did not form compact epithelial colonies (Supplementary Fig. 1a) and exhibited poor survival (< 40%, Fig. 4b) compared to μSHEAR (> 80% survival).
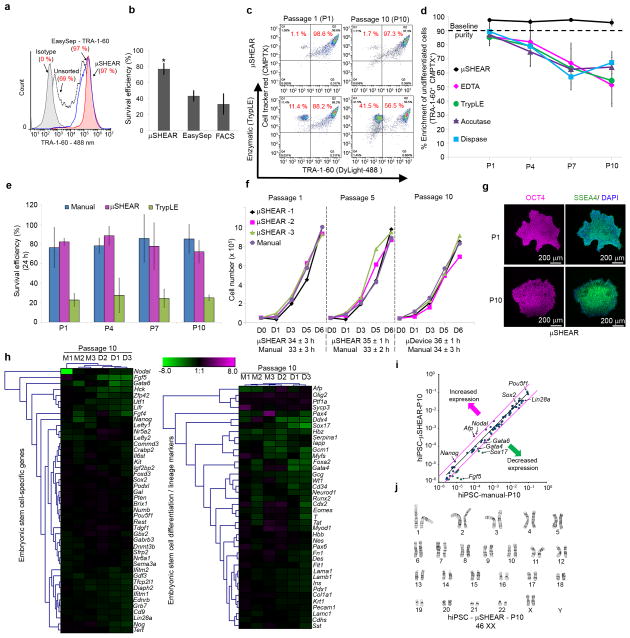
Figure 4. Adhesive differences in spontaneously differentiated pluripotent stem cells enable adhesive force-based enrichment of undifferentiated cells.
(a) μSHEAR-based isolation of UD-hiPSCs (white arrowhead) from SD-hiPSCs (red arrowhead) at 100 dynes cm−2 with high enrichment efficiency irrespective of SD-hiPSC contamination (table). Bottom panel shows non-selective detachment of cells using a trypsin-like enzyme (TrypLE). (b) Flow cytometry histograms indicating high purification and survival efficiency of hiPSCs compared to EasySep. (c) Flow cytometry scatter plots showing detached UD-hiPSCs (TRA-1-60+CMPTX+) and SD-hiPSCs (TRA-1-60−CMPTX+) across 10 passages using μSHEAR and TrypLE. (d) Enrichment efficiency of hiPSCs when repeatedly passaged by μSHEAR, EDTA, TrypLE, Dispase, or Accutase. hiPSCs from same batch (P0, 90% TRA-1-60+) were used and the recovered culture was propagated for 5–6 days. (e) Cell survival on Matrigel after passaging with μSHEAR, manual hand-picking, or TrypLE. (f) Growth curves for cells on Matrigel using μSHEAR or hand-picking and starting with an equivalent number of cells at day 0 for each passage (5×104 cells). (g) Immunostaining for SSEA4 and OCT4 showing μSHEAR-isolated UD-hiPSC cultured on Matrigel retained undifferentiated characteristics across 10 passages. (h) Heat-map of expression of stem cell-related and differentiation genes in hiPSCs at P10 showing no differences in expression between μSHEAR and manual hand-picking, compared to the starting P0 population. (i) Relative expression comparison for stem cell-related genes in isolated hiPSCs at P10. Magenta lines indicate two-fold change in gene expression threshold. (j) Karyotype analysis of hiPSCs at P10. Data report average ± s.d. (*P < 0.05, n = 3).
We next assessed the efficacy of μSHEAR for culture maintenance over the course of 10 passages (5–7 days apart) starting with ~10% spontaneously differentiated population that was not cleaned prior to separation. μSHEAR-based isolation resulted in repeated high purity (> 97%) whereas routinely used solutions and enzymatic methods failed to selectively enrich undifferentiated cells and levels of spontaneous differentiation continuously increased with passaging (Fig. 4c–d). The use of defined culture substrates, such as E-cadherin (e.g., StemAdhere), also used for hiPSC passaging33, were not selective for UD-hiPSCs as SD-hiPSCs still remained attached (Supplementary Fig. 15). These results demonstrate the high selectivity of μSHEAR over existing hPSC passaging methods. Because μSHEAR isolates cells as colonies, survival efficiency was significantly higher than TrypLE-based passaging (< 30%) and comparable to manual passaging (Fig. 4e). The doubling time of μSHEAR-purified hiPSCs (~34 h) over 10 passages was equivalent to the doubling time of hiPSCs passaged manually (Fig. 4f).
When cultured on Matrigel, μSHEAR-purified hiPSCs detached at 85–125 dynes cm−2 appeared as undifferentiated colonies (Supplementary Fig. 16a,b). In contrast, application of a high shear stress (750 dynes cm−2) resulted in complete detachment of all cells and recovered colonies contained differentiated cells (Supplementary Fig. 16a), thus confirming that selective detachment of UD-hiPSCs occurs only at a shear stress range where differences in adhesive strength between phenotypic states can be exploited. The recovered undifferentiated colonies retained pluripotency over 10 passages (Fig. 4g and Supplementary Fig. 16c) and expression profiles of stem-cell related genes were similar at P10 to P0 for both μSHEAR and manual hand-picked hiPSCs (Fig. 4h). The overall expression profiles for differentiation and lineage specific genes were equivalent or down-regulated for both μSHEAR and manual passaged hiPSCs compared to starting P0 cells (Fig. 4h). Hierarchical clustering analysis indicated that μSHEAR-passaged hiPSCs clustered closely with manually passaged cells. Scatter plot analysis of gene expression at P10 revealed a high degree of similarity between μSHEAR and manual passaging (Fig. 4i). Karyotype analysis at P10 demonstrated that μSHEAR-passaged hiPSCs exhibited no chromosomal abnormalities (Fig. 4j). Epigenetic analysis revealed no changes in the methylation status of OCT4, SOX2 and NANOG of μSHEAR-passaged hiPSCs compared to hand-picked hiPSCs (Supplementary Fig. 16d).
We also applied μSHEAR to terminally differentiated cells such as neurons. Because of their lower adhesion strength compared to hiPSCs, neurons were detached at 60 dynes cm−2 whereas hiPSCs remained adherent to the substrate. The isolated neurons exhibited excellent viability, neurite growth (Supplementary Fig. 17), and expression of MAP2 and β-III tubulin. Similarly, we successfully isolated hiPSC-derived cardiomyocytes (Supplementary Movie 3) from hiPSCs with > 95% purity (Supplementary Fig. 18a,b). Recovered hiPSC adhered as colonies whereas the residual cardiomyocytes expressed α-smooth muscle actin and exhibited spontaneous contractile activity (Supplementary Fig. 18c,d and Supplementary Movie 4).
DISCUSSION
We demonstrate that unique differences in adhesion strength can be exploited to purify undifferentiated hPSCs from contaminating cell types in a rapid, facile, efficient and label-free manner using the μSHEAR technology. μSHEAR purifies hPSCs in a more rapid and efficient manner with enhanced hiPSC survival than conventional methods that are either non-selective or result in dissociated single cells with reduced survival efficiency. μSHEAR allows the application of a wide range of shear forces with small working volumes and precise magnitudes of shear force. In addition, the microfluidic strategy provides for direct visualization of the detachment process of stem cell colonies which allows for the application of a gradient of shear forces and could be exploited to serially isolate individual colonies from the same chamber and sorted downstream individually; this latter point cannot be achieved with any bulk passaging methods, regardless of the specific reagents or purities of hPSC cultures. Because of the high efficiency, flexibility, and high-throughput nature of μSHEAR, this technology will facilitate more integrated and complex cell isolation procedures, such as separating completely reprogrammed hiPSCs from partially reprogrammed cells and in-line biochemical, genomic, proteomic and metabolomic analyses.
ONLINE METHODS
hiPSC Reprogramming and Culture
hiPSCs (IMR90) were derived and validated by ArunA Biomedical using the viPS Vector Kit (Thermo Scientific Open Biosystems) consisting of 6 lentiviral vectors encoding OCT4, NANOG, SOX2, LIN28, KLF4 and c-MYC driven by the EF1alpha promoter. IMR90 human fetal lung fibroblasts (female, ATCC) were transduced (MOI 10 each vector) for reprogramming per manufacturer’s protocol. Transduced fibroblasts were seeded onto inactivated MEFs or Matrigel to form colonies and emerging hiPSC colonies were manually passaged by mechanical dissociation on day 30. hiPSCs demonstrated well defined borders, high nuclear to cytoplasmic ratio, prominent nucleoli, alkaline phosphatase activity, positive expression of cell surface marker SSEA4, embryoid body formation and teratoma formation. To transition hiPSCs to a feeder-free culture system, colonies were manually passaged by mechanical dissociation onto Matrigel (1:100 dilution; BD Biosciences) in mTeSR1 medium (STEMCELL Technologies). hiPSCs used in the study were between passage 26–48, routinely cultured as feeder-free undifferentiated colonies (UD-hiPSC) in mTeSR1 medium on Matrigel, enzymatically passaged with dispase (1 mg mL−1), followed by scraping. For μSHEAR experiments, reprogramming was performed on Matrigel and the entire culture was introduced into the devices after 27–30 days. Human dermal fibroblast-derived hiPSCs (11b, healthy male donor) were obtained from Harvard Stem Cell Institute and cultured as above. Human peripheral blood mononuclear cell-derived hiPSCs were a gift from Dr. Joseph Wu (Stanford University, USA) and were derived using Sendai virus. hESCs used in the study were at passage 35 (H1, Wicell), passage 54 (H7, Wicell) and cultured in mTeSR1 medium on Matrigel. IMR90 human fetal lung fibroblasts (passage 15–20), human dermal fibroblasts (primary adult skin, Cell Applications) and MEFs (primary isolated, passage 2) were cultured in DMEM with 1% L-glutamine, 1% non-essential amino acids, 10% FBS, and 1% penicillin/streptomycin. Immunostaining and flow cytometer measurements were performed using antibodies listed in the Supplementary Table 1.
Neural Cell and Cardiomyocyte Derivation
Neural rosettes and subsequent neural progenitor cells and differentiated neuronal cells were derived from feeder-free, pluripotent colonies of hiPSC (passage 40 or 52), based on methods previously described using hESC34. Briefly, hiPSC were enzymatically passaged 1:2 with dispase (1 mg mL−1) followed by cell scraping onto BD Matrigel (diluted 1:100) in mTeSR1 Medium (STEMCELL Technologies). Medium was changed every other day for 4 days. On day 5, medium was switched to neural derivation medium (DMEM/F-12 supplemented with N-2), 4 ng mL−1 FGF2, 2 mM l-glutamine and 50 U mL−1:50μg mL−1 penicillin-streptomycin) and changed every other day. After 1 week in neural derivation medium, neural rosettes were manually isolated and then propagated on Matrigel (1:200) in Neurobasal medium (Life Technologies) supplemented with B-27, 20 ng mL−1 FGF2, 10 ng mL−1 LIF, 2 mM L-glutamine and 50 U mL−1:50 μg mL−1 penicillin-streptomycin), with medium changed every other day. After 4 weeks of differentiation, neural progenitor cells were manually isolated from neural rosette cultures and propagated as an adherent monolayer on Matrigel in neural proliferation medium. After several manual passages with a cell scraper, confluent cultures of hiPSC-derived neural progenitor cells on Matrigel (1:200) were differentiated for 2 weeks to mature, β-III tubulin (TUJ1)/MAP2-positive neuronal cells by removing FGF2 from the neural proliferation medium and changing medium every 2–3 days. hiPSC (IMR90)-derived cardiomyocytes were a kind gift from Dr. Chunhui Xu (Emory University) and were derived as previously reported35. Cells were cultured in RPMI/B27 media and exhibited spontaneously contractile activity (Supplementary Movie 3).
Design and Fabrication of PDMS Micropatterned Arrays
PDMS micropattern arrays with 10, 20 μm, 56 μm, and 170 μm were fabricated from silicon array masters23,36. Microcontact printing on glass coverslips coated with Ti (100 Å) followed by Au (100 Å) was achieved using hexadecanethiol/(HO(CH2CH2O)3-(CH2)11SH) chemistry23. Coverslips were incubated with ECM proteins (fibronectin or laminin, 50 μg mL−1 in PBS)23. After blocking with 1% heat denatured bovine serum albumin (Sigma) for 30 min and eluting proteins for 2 h in PBS, single cell suspensions of IMR90 cells or hiPSCs were seeded in mTeSR1 medium with ROCK-inhibitors Y27362 (10 μM, Calbiochem) or Thiazovivin (2 μM, Stemgent). Briefly, hiPSCs were treated with 0.05% trypsin for 1 min and scraped as intact colonies. Cells were then prepared as single cells in mTeSR1 with Y27362-ROCK inhibitor and seeded as 100,000 cells mL−1 on the micropatterned islands overnight. For unpatterned surfaces, glass coverslips were incubated with ECM proteins as above and single cell suspensions were seeded as 60,000 cells mL−1 and cultured overnight.
Cell Adhesion Strength Measurements
Cell adhesion strength was measured using a spinning disk system23. Coverslips with adherent cells cultured overnight were spun in PBS with 2 mM dextrose for 5 min at a constant speed in a custom-built device in compliance with American Society for Testing and Materials (ASTM F2664-11). The applied shear stress (τ) is given by the formula τ = 0.8r(ρμω3)1/2, where r is the radial position, ρ and μ are the fluid density and viscosity, respectively, and ω is the spinning speed. After spinning, cells were fixed in 3.7% formaldehyde, permeabilized in 0.1% Triton X-100, stained with DAPI (Life Technologies), and counted at specific radial positions using a 10X objective lens in a Nikon TE300 microscope equipped with a Ludl motorized stage, Spot-RT camera, and Image Pro analysis system. Sixty-one fields were analyzed and cell cluster counts were normalized to the number of cell cluster counts at the center of the disk, where the applied force is zero. The fraction of adherent cell cluster (f) was then fit to a sigmoid curve f = 1/(1+exp[b(τ-τ50)]), where τ50 is the shear stress for 50% detachment and b is the inflection slope. τ50 represent the mean adhesion strength for a population of cells. The adhesion strength response was analyzed on micropatterned islands coated with fibronectin or laminin (50 μg mL−1) or Matrigel (1:80).
Focal Adhesion Assembly
Immunofluorescence staining of focal adhesion proteins was performed as previously described23. Briefly, cells were prewashed with ice-cold PBS with calcium and magnesium, incubated in ice-cold cytoskeleton stabilization buffer (50 mM NaCl, 150 mM sucrose, 3 mM MgCl2, 1 μg mL−1 aprotinin, 1 μg mL−1 leupeptin, 1 μg mL−1 pepstatin and 1 mM phenylmethylsulfonyl fluoride) for 1 min, followed by two time incubation (1 min each) in cytoskeleton buffer supplemented with 0.5% Triton X-100. Detergent-extracted cells were fixed in 4% paraformaldehyde in PBS, washed with PBS, incubated with a primary antibody against vinculin (Upstate) or talin (Sigma) and detected with Alexa Fluor 488-conjugated antibodies (Life Technologies).
Fabrication of Microfluidic Devices
Poly(dimethylsiloxane) (PDMS, Sylgard 184, Dow Corning, MI) microfluidic devices were fabricated as reported earlier using a negative photoresist (SU-8 2050, 50 μm thickness, MicroChem) and UV photolithography37. Patterned negative molds were then exposed to vapor-phase tridecafluoro-1,1,2,2-tetrahydrooctyl-1-trichlorosilane (United Chemical Technologies) in a vacuum desiccator to prevent adhesion of PDMS. A 5 mm thick layer of degassed PDMS mixture (10:1) was cast onto the mold and cured at 70 °C for 2 h. Cast PDMS devices were peeled-off and then punctured for inlet-outlet holes and bonded to glass coverslips by exposure to oxygen plasma for 20 s.
μSHEAR-based Isolation
Prior to coating with ECM proteins, the microfluidic channels and tubes were cleaned with 70% ethanol and rinsed thoroughly with PBS. ECM proteins at 50 μg mL−1 (fibronectin or laminin) or 1:80 Matrigel were flowed through sterile devices and incubated for 1 h at room temperature. Small colonies of pluripotent stem cells and single cell suspensions of fibroblasts were premixed and pipetted into the inlet reservoir using a 200 μL pipette tip and were cultured in the device for 24 h at 37°C and 5% CO2 before detachment experiments. The device inlet was connected to a syringe pump using polyethylene tubing (BB31695-PE/4, Scientific Commodities) and outlet tubes emptied into collecting tubes. PBS was flowed at pre-determined flow rates through the device to match up the desirable fluid shear stress and cell detachment was monitored though a Nikon TE microscope. For this microfluidic flow configuration, the applied wall shear stress (τ) is defined by the formula τ = 12(μQ/wh2), where w and h are the width and height of the channel, respectively, μ is the fluid viscosity, and Q is the fluid flow rate28. Cell/colonies plated on Matrigel coated tissue culture plates in 10 μM ROCK inhibitor Y27362 (or 2 μM Thiazovivin) containing mTeSR1 media. For flow cytometry studies to determine purification efficiency, collected colonies or cells were immediately resuspended in a suspension of StainAlive-DyLight-488 mouse anti-human TRA-1-60 antibody (Stemgent), and CMPTX-Cell Tracker Red dye, stained for 45 min, washed and analyzed using Accuri flow cytometer (BD Biosciences).
Pluripotent Stem Cell Characterization
Karyotype analysis was performed on 20 metaphase spreads for each sample by CellLine Genetics (Madison, WI). To determine population doubling time and survival, detached colonies from μSHEAR were dissociated into single cells and plated in Matrigel-coated 12-well plates. At predetermined times, wells were washed and cells were counted. Embryoid bodies from detached and expanded hiPSCs were formed using ultra-high throughput forced aggregation method38 and after 24 h, cell aggregates were transferred to a suspension culture on a rotary orbital shaker (65 RPM). Differentiation was followed by plating EBs in cell chambers (BD Falcon) and after 21 days differentiated cells were fixed with 4% paraformaldehyde, permeabilized with 0.05% Triton-X, and stained with antibodies against α-fetoprotein, α-smooth muscle actin, and PAX6.
Gene Expression Analysis
RNA was isolated from iPSCs using QIAshredder and RNeasy Mini kits (Qiagen) according to manufacturer’s protocols. First strand cDNA synthesis was performed using the RT2 First Strand Kit (SABiosciences) followed by real-time PCR using the Human Embryonic Stem Cells PCR array (SABiosciences) according to manufacturer’s recommended protocols and using a BioRad MyCycler and BioRad MyIQ real time thermal cycler, respectively. Individual Ct values were first internally normalized to GAPDH and subsequently analyzed with Genesis software (Graz University of Technology) including log2 transformation and hierarchical clustering. Heat-map were generated for the expression of 84 embryonic stem cell-related genes for transcription factors, pluripotency and self-renewal, cytokines and growth factors, and embryonic stem cell differentiation/lineage marker genes extracted from gene expression microarrays. A log plot of the relative expression level of each gene (2−ΔCt) between manual (x-axis) and μSHEAR (y-axis) was generated for analysis.
Bisulfite Genomic Sequencing
Bisulfite treatment of gDNA was carried out using a Cells-to-CpG Bisulfite Conversion kit (Life Technologies) according to the manufacturer’s protocol and plotted as melting curves representing methylation status30,31. Converted gDNA was amplified by PCR using primers within the OCT4, SOX2, and NANOG promoters.
OCT4 forward: CCCTCCTCTAAAAAAC
OCT4 reverse: GGGTTGTAGTTGTGTTTATT
NANOG forward: AATTACAAAAATAAACCACC
NANOG reverse: TAGGTGGGGAATTAGAAAAT
Sox2forward: CATATACAACATAATAAAAA
Sox2 reverse: GTTTTTTTGGGTTATTTTG.
Teratoma Formation
The μSHEAR-isolated cells were expanded on Matrigel and then collected as pellets resuspended in DMEM-F12 at 7 million cells per 50 μL. Cells were injected intramuscularly in the hind limb of SCID mice (Harlan). Seven weeks after injection, tumors were dissected, weighted, and fixed with formalin. Paraffin-embedded tissue was sectioned and stained with hematoxylin and eosin (H/E) and imaged using a Nikon 80i microscope.
Statistics
Paired, two-tailed, Student’s t-test was performed to determine the significance of differences between 2 groups in adhesion blocking, adhesion strength, and μSHEAR assays. For integrin profiling one-way analysis of variance (ANOVA) was performed followed by Bonferroni correction using OriginPro 8.5.1. In all tests, P < 0.05 was regarded as statistically significant. All experiments were repeated in triplicates unless otherwise stated and bar graph data represents average ± s.d.
Supplementary Material
Acknowledgments
This work was supported by an ARRA supplement to the National Institutes of Health (NIH) grant R01 GM065918 (A.J.G.), NIH R43 NS080407 (J.M.C. and A.J.G.), the Stem Cell Engineering Center at Georgia Institute of Technology (T.C.M.), a Sloan Foundation Fellowship (H.L.), the National Science Foundation (NSF) DBI-0649833 (H.L.) and an ARRA sub-award under RC1CA144825 (H.L.), NSF CMMI-1129611 (J.F.), the Georgia Tech Emory Center for (GTEC) for Regenerative Medicine and Parker H. Petit IBB at Georgia Institute of Technology. We thank J. Wu (Stanford University) for providing blood cell-derived hiPSCs and C. Xu (Emory University) for providing hiPSC-derived cardiomyocytes. We thank T. Hookway for cardiomyocyte culture, A. Ortiz for teratoma studies, and A. Cheng for her help with focal adhesion assays. We thank J. Phillips, D. Dumbauld, Y. Wang, and A. Bratt-Leal for insightful discussions and suggestions.
Footnotes
AUTHOR CONTRIBUTION
A.S. and S.S. conducted all adhesion and microfluidic studies, collected data and performed data analysis. S.S., A.S., and H.L. developed microfluidic methods. J.M.C. and S.L.S. established and provided the IMR90-derived hiPSC cells and neural stem cells. T.L., W.C., and J.F. developed and provided micropatterns. M.T.C. and A.S. conducted microarray and epigenetic analysis. A.J.G. and T.C.M developed the concept, and together with A.S. contributed to the planning and design of the project. A.S., S.S., T.C.M, and A.J.G. wrote the manuscript and all authors discussed the results and commented on the manuscript.
References
- 1.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 2.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 3.Robinton DA, Daley GQ. The promise of induced pluripotent stem cells in research and therapy. Nature. 2012;481:295–305. doi: 10.1038/nature10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao Y, et al. Two supporting factors greatly improve the efficiency of human iPSC generation. Cell Stem Cell. 2008;3:475–479. doi: 10.1016/j.stem.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Enver T, et al. Cellular differentiation hierarchies in normal and culture-adapted human embryonic stem cells. Hum Mol Genet. 2005;14:3129–3140. doi: 10.1093/hmg/ddi345. [DOI] [PubMed] [Google Scholar]
- 6.Rodin S, et al. Long-term self-renewal of human pluripotent stem cells on human recombinant laminin-511. Nat Biotechnol. 2010;28:611–615. doi: 10.1038/nbt.1620. [DOI] [PubMed] [Google Scholar]
- 7.Boheler KR, et al. Differentiation of pluripotent embryonic stem cells into cardiomyocytes. Circ Res. 2002;91:189–201. doi: 10.1161/01.res.0000027865.61704.32. [DOI] [PubMed] [Google Scholar]
- 8.Burridge PW, et al. A universal system for highly efficient cardiac differentiation of human induced pluripotent stem cells that eliminates interline variability. PLoS One. 2011;6:e18293. doi: 10.1371/journal.pone.0018293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho MS, et al. Highly efficient and large-scale generation of functional dopamine neurons from human embryonic stem cells. Proc Natl Acad Sci U S A. 2008;105:3392–3397. doi: 10.1073/pnas.0712359105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee G, et al. Isolation and directed differentiation of neural crest stem cells derived from human embryonic stem cells. Nat Biotechnol. 2007;25:1468–1475. doi: 10.1038/nbt1365. [DOI] [PubMed] [Google Scholar]
- 11.Tang C, et al. An antibody against SSEA-5 glycan on human pluripotent stem cells enables removal of teratoma-forming cells. Nat Biotechnol. 2011;29:829–834. doi: 10.1038/nbt.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohgushi M, et al. Molecular pathway and cell state responsible for dissociation-induced apoptosis in human pluripotent stem cells. Cell Stem Cell. 2010;7:225–239. doi: 10.1016/j.stem.2010.06.018. [DOI] [PubMed] [Google Scholar]
- 13.Nandivada H, et al. Fabrication of synthetic polymer coatings and their use in feeder-free culture of human embryonic stem cells. Nat Protoc. 2011;6:1037–1043. doi: 10.1038/nprot.2011.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baker DE, et al. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat Biotechnol. 2007;25:207–215. doi: 10.1038/nbt1285. [DOI] [PubMed] [Google Scholar]
- 15.Mitalipova MM, et al. Preserving the genetic integrity of human embryonic stem cells. Nat Biotechnol. 2005;23:19–20. doi: 10.1038/nbt0105-19. [DOI] [PubMed] [Google Scholar]
- 16.Hartung O, Huo H, Daley GQ, Schlaeger TM. Clump passaging and expansion of human embryonic and induced pluripotent stem cells on mouse embryonic fibroblast feeder cells. Curr Protoc Stem Cell Biol. 2010;Chapter 1(Unit 1C):10. doi: 10.1002/9780470151808.sc01c10s14. [DOI] [PubMed] [Google Scholar]
- 17.McCracken KW, Howell JC, Wells JM, Spence JR. Generating human intestinal tissue from pluripotent stem cells in vitro. Nat Protoc. 2011;6:1920–1928. doi: 10.1038/nprot.2011.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bajpai R, Lesperance J, Kim M, Terskikh AV. Efficient propagation of single cells Accutase-dissociated human embryonic stem cells. Mol Reprod Dev. 2008;75:818–827. doi: 10.1002/mrd.20809. [DOI] [PubMed] [Google Scholar]
- 19.Maitra A, et al. Genomic alterations in cultured human embryonic stem cells. Nat Genet. 2005;37:1099–1103. doi: 10.1038/ng1631. [DOI] [PubMed] [Google Scholar]
- 20.Ellerstrom C, Strehl R, Noaksson K, Hyllner J, Semb H. Facilitated expansion of human embryonic stem cells by single-cell enzymatic dissociation. Stem Cells. 2007;25:1690–1696. doi: 10.1634/stemcells.2006-0607. [DOI] [PubMed] [Google Scholar]
- 21.Meng G, Liu S, Rancourt DE. Rapid isolation of undifferentiated human pluripotent stem cells from extremely differentiated colonies. Stem Cells Dev. 2011;20:583–591. doi: 10.1089/scd.2010.0400. [DOI] [PubMed] [Google Scholar]
- 22.Chan EM, et al. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat Biotechnol. 2009;27:1033–1037. doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- 23.Gallant ND, Michael KE, Garcia AJ. Cell adhesion strengthening: contributions of adhesive area, integrin binding, and focal adhesion assembly. Mol Biol Cell. 2005;16:4329–4340. doi: 10.1091/mbc.E05-02-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meng Y, et al. Characterization of integrin engagement during defined human embryonic stem cell culture. FASEB J. 2010;24:1056–1065. doi: 10.1096/fj.08-126821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blagovic K, Kim LY, Voldman J. Microfluidic perfusion for regulating diffusible signaling in stem cells. PLoS One. 2011;6:e22892. doi: 10.1371/journal.pone.0022892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gomez-Sjoberg R, Leyrat AA, Pirone DM, Chen CS, Quake SR. Versatile, fully automated, microfluidic cell culture system. Anal Chem. 2007;79:8557–8563. doi: 10.1021/ac071311w. [DOI] [PubMed] [Google Scholar]
- 27.Kim JY, et al. A cell culturing system that integrates the cell loading function on a single platform and evaluation of the pulsatile pumping effect on cells. Biomed Microdevices. 2008;10:11–20. doi: 10.1007/s10544-007-9105-z. [DOI] [PubMed] [Google Scholar]
- 28.Lu H, et al. Microfluidic shear devices for quantitative analysis of cell adhesion. Anal Chem. 2004;76:5257–5264. doi: 10.1021/ac049837t. [DOI] [PubMed] [Google Scholar]
- 29.Staerk J, et al. Reprogramming of human peripheral blood cells to induced pluripotent stem cells. Cell Stem Cell. 2010;7:20–24. doi: 10.1016/j.stem.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wojdacz TK, Dobrovic A, Algar EM. Rapid detection of methylation change at H19 in human imprinting disorders using methylation-sensitive high-resolution melting. Hum Mutat. 2008;29:1255–1260. doi: 10.1002/humu.20779. [DOI] [PubMed] [Google Scholar]
- 31.Wojdacz TK, Dobrovic A, Hansen LL. Methylation-sensitive high-resolution melting. Nat Protoc. 2008;3:1903–1908. doi: 10.1038/nprot.2008.191. [DOI] [PubMed] [Google Scholar]
- 32.Heng BC, Liu H, Cao T. Feeder cell density--a key parameter in human embryonic stem cell culture. In Vitro Cell Dev Biol Anim. 2004;40:255–257. doi: 10.1290/0407052.1. [DOI] [PubMed] [Google Scholar]
- 33.Nagaoka M, Si-Tayeb K, Akaike T, Duncan SA. Culture of human pluripotent stem cells using completely defined conditions on a recombinant E-cadherin substratum. BMC Dev Biol. 2010;10:60. doi: 10.1186/1471-213X-10-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dhara SK, et al. Human neural progenitor cells derived from embryonic stem cells in feeder-free cultures. Differentiation. 2008;76:454–464. doi: 10.1111/j.1432-0436.2007.00256.x. [DOI] [PubMed] [Google Scholar]
- 35.Laflamme MA, et al. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol. 2007;25:1015–1024. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- 36.Fu J, et al. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat Methods. 2010;7:733–736. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDonald JC, et al. Fabrication of microfluidic systems in poly(dimethylsiloxane) Electrophoresis. 2000;21:27–40. doi: 10.1002/(SICI)1522-2683(20000101)21:1<27::AID-ELPS27>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 38.Bratt-Leal AM, Carpenedo RL, Ungrin MD, Zandstra PW, McDevitt TC. Incorporation of biomaterials in multicellular aggregates modulates pluripotent stem cell differentiation. Biomaterials. 2011;32:48–56. doi: 10.1016/j.biomaterials.2010.08.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.