Abstract
Large, cooperative assemblies of proteins that wrap and/or loop genomic DNA may “epigenetically” shift configurational equilibria that determine developmental pathways. Such is the case of the λ bacteriophage which may exhibit virulent (lytic) or quiescent (lysogenic) growth. The lysogenic state of λ prophages is maintained by the λ repressor (CI), which binds to tripartite operator sites in each of the OL and OR control regions located about 2.3 kbp apart on the phage genome and represses lytic promoters. Dodd and collaborators have suggested that an initial loop formed by interaction between CI bound at OR and OL provides the proper scaffold for additional CI binding to attenuate the PRM promoter and avoid over production of CI. Recently, the looping equilibrium as a function of CI concentration was measured using tethered particle motion analysis, but the oligomerization of CI in looped states could not be determined. Scanning force microscopy has now been used to probe these details directly. An equilibrium distribution of looped and unlooped molecules confined to a plane was found to be commensurate to that for tethered molecules in solution, and the occupancies of specific operator sites for several looped and unlooped conformations were determined. Some loops appeared to be sealed by oligomers of 6–8, most by oligomers of 10–12, and a few by oligomers of 14–16.
Keywords: Lambda repressor, DNA looping, atomic force microscopy
Introduction
From viruses to humans, transcription is regulated by proteins that bind the DNA. It is becoming increasingly clear that, in most cases, genes are controlled by large, cooperative assemblages of proteins that wrap and loop the DNA. These protein-induced configurational changes often represent real “epigenetic switches” in which shifting the equilibrium towards one configuration versus the other commits the system to one developmental pathway instead of another. Such is the case of the λ bacteriophage and, it is suspected, of most temperate bacteriophages which may adopt a quiescent lifestyle (the lysogenic growth) or a virulent lifestyle (the lytic growth). After infection, repressor protein often binds to multipartite operators and mediate cooperative, long-range interactions which repress the lytic genes maintaining a stable lysogenic state, until adverse environmental conditions (DNA damage, poisoning, starvation, etc) induce a cascade of events that leads to repressor dissociation from the double helix and efficient switch to lysis. The λ epigenetic switch is not only a paradigm of transcriptional regulation, but is also at the basis of our understanding of phage lysogeny [1].
The lysogenic state of λ prophages is maintained by the λ repressor, or CI protein [2]. During lysogeny, dimers of CI bind to the OL and OR control regions, located about 2.3 kbp apart on the phage genome and repress the PL and PR, promoters for the lytic genes. Each control region contains three binding sites for CI, OL1, OL2, OL3 and OR1, OR2, OR3 [3–5]. CI binds to these operators with an intrinsic affinity OL1 > oR1> OL3> OL2 > OR2 > OR3 [6, 7]. By studying the oL and oR regions separately and in isolation from the rest of the λ chromosome [8], it was found that pairs of dimers interact when bound to adjacent or nearby operators, forming tetramers. These cooperative interactions improve the specificity and strength of CI binding to OR1 and OR2, and OL1 and OL2, respectively, so that the binding affinity ranking becomes OR1~OL1~OR2~OL2 > OL3 > OR3. Biochemical and genetic studies have identified the contacts between amino acids in the C-terminal domain that mediate these interactions. These contacts have been confirmed from the crystal structure of the isolated CTD tetramer [9] and are thought to contribute significantly to the stability of lysogeny. Occupancy of OR2 by CI also activates transcription of the CI gene from the PRM promoter, constituting a positive autoregulatory mechanism [10–12] to generate the amount of CI required for repression of the lytic genes, as described above. At very high concentrations, CI was observed to also bind to OR3 and repress its own transcription from PRM [13]. This negative autoregulation has been suggested to be important to prevent excessive accumulation of repressor to facilitate efficient switching to lysis when necessary. However, such negative regulation did not seem possible at physiological concentrations and the role of both OL3 and OR3 remained controversial. In fact, conventional wisdom held that OL3 was a “left over” of evolution.
Recently, it was suggested that CI molecules bind cooperatively not only to adjacent sites, but also to sites separated by over 2000 bp in the λ genome, inducing a regulatory loop in the phage DNA. This led to the hypothesis that the loop is first formed by interaction between two tetramers bound at OR1–OR2 and OL1–OL2, respectively. This octamer-mediated loop brings OL3 and OR3 into juxtaposition and favors their occupancy by CI dimers which can interact “head-to-head” in a long-range cooperative fashion, and lead to a CI octamer+tetramer-mediated loop. According to this hypothesis, the loop provides the right scaffold for CI binding to weak OR3 at lysogenic concentrations [3, 14, 15] and effective repression of PRM.
To evaluate this hypothesis, the stoichiometry of CI securing the regulatory loop in wild-type lambda DNA is a fundamental piece of information that has not been previously reported. We used atomic force microscopy, AFM, to image the CI-mediated loop and characterize the looping probability and the stoichiometry of the protein closure. Solutions of DNA and repressor were deposited on a flat, positively charged surface, rinsed and dried, and imaged using scanning force microscopy. In the resulting topographs, discrete bumps corresponding almost exclusively to specifically bound protein were found on both looped and unlooped DNA molecules. A looping equilibrium commensurate with that measured using tethered particle motion was measured as well as the volumes of individual protein particles securing loops between OL and OR. Virtually no CI tetramers were found to secure DNA loops. Instead higher order oligomers of 6–8 and especially 10–12 accounted for the majority of the CI particles associated with DNA loops. The data are consistent with the model in which multipartite operators collect CI dimers that multimerize to stabilize loops formed through random encounters between OL and OR.
Materials and Methods
1555 bp DNA fragments were produced by PCR amplification of segments of plasmids pDL944 and pDL965 using 5'-CGCAATTAATGTGAGTTAGCTCACTCATTAGGCACCCCAGGC-3' and 5'-GCATTGCTTATCAATTTGTTGCAACGAACAGGTCACTATCAGTC-3' as forward and reverse primers. These fragments contained respectively wild-type or mutant lambda operator regions (OL and OR) and including the associated promoters PL, PRM and PR. The distance between the midpoints of operator sites OL3 and OR3 was 393 bp. pDL965 contains CC to AT mutations in OL3 and OR3, which abrogate CI binding (Lewis et al. manuscript in preparation and [16]). PCR using the same plasmid templates was also used to generate 505 or 392 bp DNA fragments that contained only one group of binding sites (OR or OL).
Another 732 bp DNA fragment containing two high affinity lac operators Oid (5’-TGTGAGCGCTCACA-3’) and O1 (5’-AATTGTGAGCGGATAACAATT-3’) [17, 18] separated by 70 bp was provided by Opher Gileadi (Quantomix Ltd, Rehovot, Israel). It was produced by PCR using the plasmid pOid-O1 from the Müller-Hill laboratory as a template and 5’-GCCACCTCTGACTTAAGCGTCG-3’ and 5’-TTGAGGGGACGTCGACAGTATC-3’ as forward and reverse primers.
The wild-type CI protein (7.25 ug/ul) was purified from pEA305 in the laboratory of Sankar Adhya. 20 nM CI and 2 to 4 nM DNA were gently mixed in a buffer containing 50mM HEPES, 150mM NaCl and 0.1 mM EDTA (pH 7.0) and incubated at RT for 10 min. Shortly before deposition, a 10 µl drop of 0.1 µg/ml poly-L-ornithine (1 kDa MW, product #P5666, Sigma-Aldrich, St. Louis, MO) was incubated on freshly cleaved mica for one minute at RT. The poly-L-ornithine-coated mica was then washed with 0.4 ml HPLC water and dried with compressed air. Then 5 µl of the solution containing DNA and protein was quickly diluted with 40 µl of buffer, and a 10 µl droplet of this solution was deposited on the poly-L-ornithine-coated mica and incubated for one minute at RT. The droplet was rinsed away with 0.4 ml HPLC water and dried gently with compressed air. The sample was left overnight in a dessicator at RT before imaging.
Images were acquired with a NanoScope MultiMode AFM microscope (Digital Instrument, Santa Barbara, CA) operated in tapping mode using a 50–60 mV oscillation amplitude of uncoated, etched silicon tips with a resonance frequency of 75 kHz (NSC18, MirkoMasch, San Jose, CA). Areas of 1×1 µm2 were scanned at a rate of 1.2 Hz and a resolution of 512×512 pixels.
After filtering images to remove scan line offsets and bowing, DNA molecules were interactively traced with NeuronJ [19], a plug-in function for ImageJ [20]. To measure the volume of protein particles, a basal threshold was established above (typically 0.08 nm) the background. The mean value of all pixels below this threshold was calculated and used as the base for the measurement. The volumes of isolated protein particles were determined as the sum of the pixel heights above the base within the area of the particle protruding above the basal threshold. For DNA-bound protein particles, a second “DNA” threshold was chosen just above the DNA. The volume of protein particles was determined as the sum of the pixel heights above the base within the area of the particle protruding above the “DNA” threshold.
Results and Discussion
Dried, 1555 bp-long DNA molecules (Fig. (1), upper and middle rows) containing both OL and OR averaged 510 nm in length in scanning force micrographs. The measured pitch of the DNA on poly-L-ornithine was therefore 0.327 nm/bp which is quite close to that of the B-form structure [21].
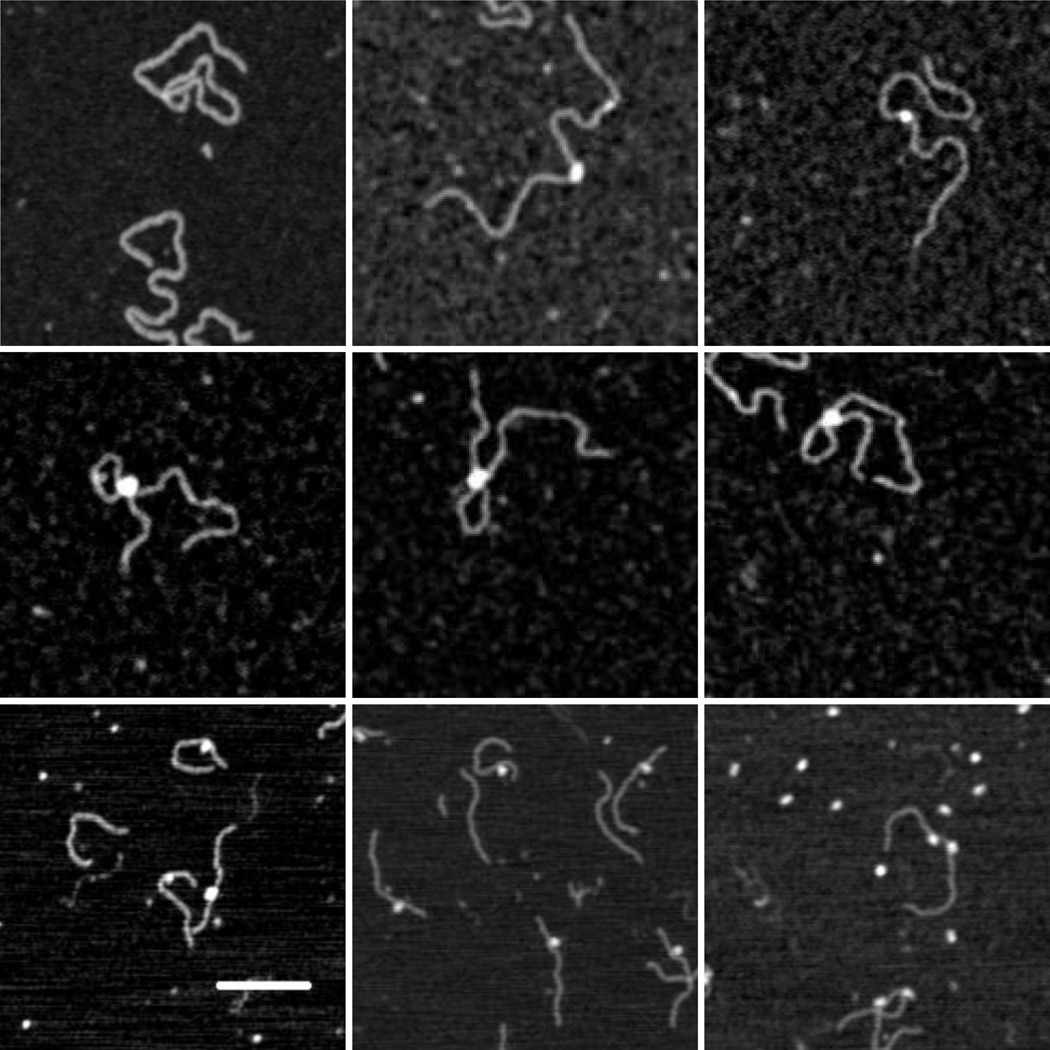
Figure 1.
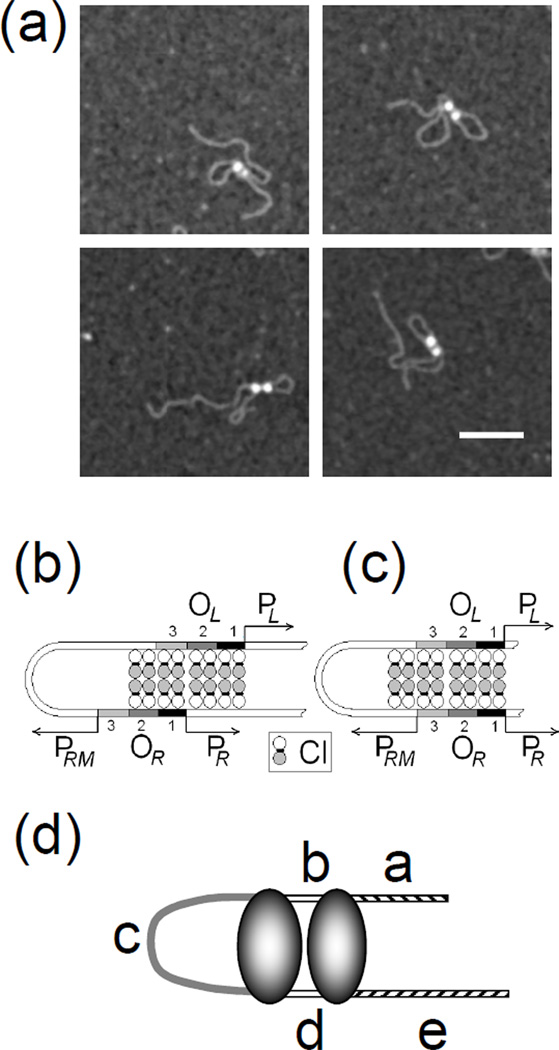
AFM images of CI and DNA: (upper left) 1555 bp DNA containing OL and OR, (upper middle and right) CI protein bound to 1555 bp DNA, (middle row) CI- mediated loops in 1555 bp DNA, (bottom left) CI bound to DNA containing OL (wild-type), (bottom center) CI protein bound to DNA containing OL (O3-), (bottom right) lac repressor bound to Oid and O1 containing DNA. The white bar represents 100 nm.
Specific binding to operator sites
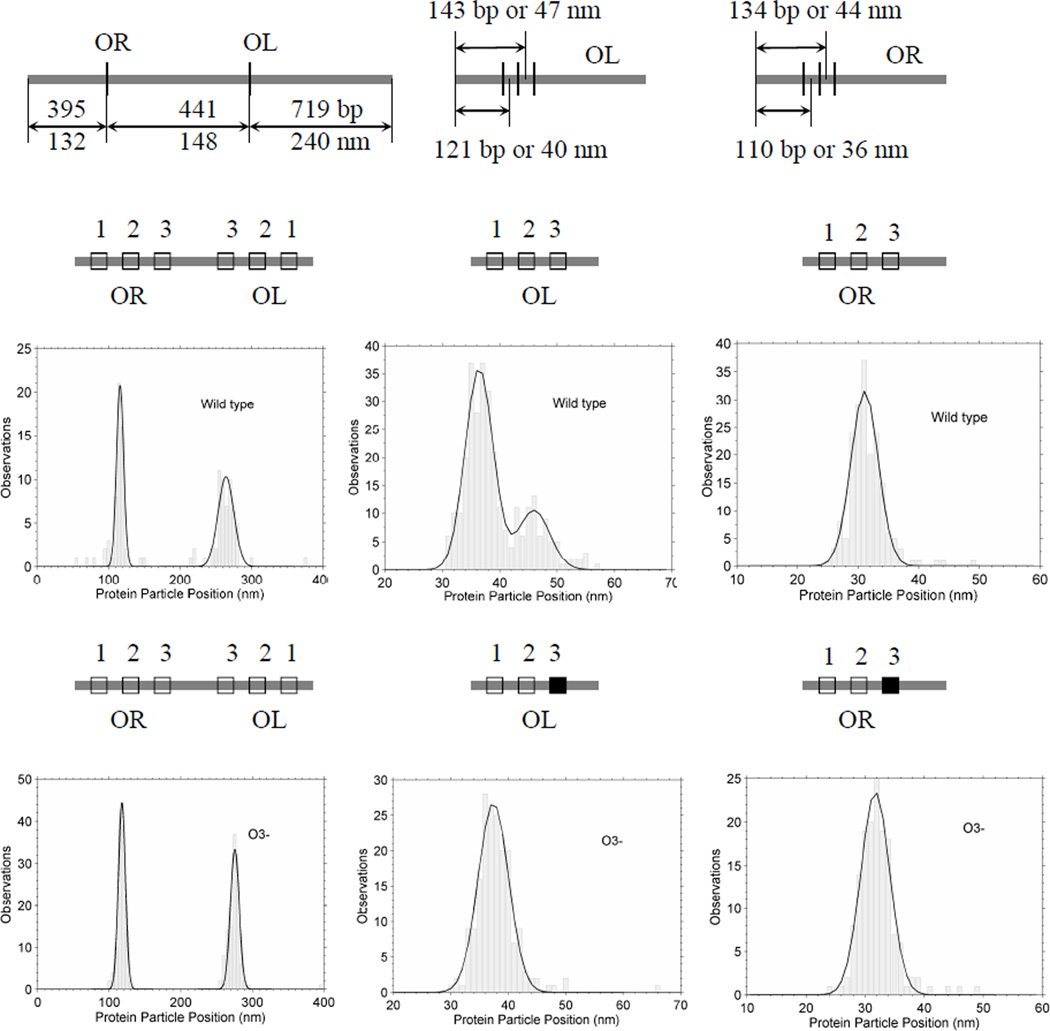
To assay the specificity of CI binding, the positions of CI particles were measured along unlooped DNA. Figure 2 shows schematic diagrams of the molecules used, along with the positions of the right and left operator regions. The positions of the center of bound CI particles were measured and frequency distributions are shown for DNA containing both wild-type operator regions (Fig. (1), upper center and left; Fig. (2), middle-left). There was almost no non-specific binding with the vast majority of particles located near the OL and OR regions118 and 265 nm from one end of the molecules.
Figure 2.
AFM measurements of the positions of CI particles bound to DNA. Schematic diagrams of the DNA constructs with wild-type or O3- operators in the OL and OR regions appear just above histograms of the AFM-determined positions of CI particles bound to the indicated DNA fragments.
Weak affinity for the oR3 operator site
The peak at OL was noticeably broader than that corresponding to OR. It is well accepted that CI dimers on adjacent operator sites may bind cooperatively; two dimers could occupy either O1 and O2 or O2 and O3. Given the experimentally determined affinities of the operator sites [22], this is likely to have occurred at OL but not OR, because the affinity of CI dimers for OL3 is greater than that for OR3. This interpretation was supported by experiments using DNA with mutations in the third binding sites (O3-) that abrogated the binding of CI dimers to OL3 and OR3. As in the case of the wild-type DNA, CI binding to the OR region of O3- DNA produced a narrow peak at 119 nm (Fig. 2, bottom-left). However, with respect to this peak, CI binding to the OL region of O3- DNA shifted to give a narrow peak at 275 nm, in which the cooperative binding of CI to OL2 and OL3 seemed to have disappeared.
To further demonstrate the weak affinity for the OR3 site, experiments were done with short fragments containing either OR or OL (Fig. (1), bottom left and center). In histograms of particle locations on the wild-type OL containing fragment, there are two peaks separated by 9.5 nm (Fig. (2), middle-center). This distance is slightly larger than the value expected for cooperatively bound dimers bridging either sites OL1 and OL2 or OL2 and OL3 (20 bp or 6.7 nm). However, the peak located at 47 nm, which corresponds to the OL3 site, disappeared for DNA with the OL3- mutation (Fig. (3), bottom-center) while the peak at OR (32 nm) remained unchanged (Fig. (2), compare middle-right and bottom-right). The simplest interpretation is that no significant binding to OR3 occurred with or without mutation while OL3 binding was observed only for the wild-type operator.
Figure 3.
Pairs of CI particles bound to adjacent O1 and O3 sites were observed in AFM images (upper). The white bar represents 100 nm. (lower) The mean volume of these particle was 174 ± 70 nm3.
Multiple operators may recruit dimers
Among the hundreds of molecules in the recorded topographs, there were a few DNA molecules with small protein particles bound in adjacent positions that were commensurate with the distance between the O1 and O3 operator sites (Fig. (3)). Based on the calibration that was performed and is described below, these particles with a mean volume of 174 nm3 were identified as CI oligomers of 2–4 monomers. According to the DNA construct, the center-to-center distance from OL1 to OL3 is 44 bp which corresponds to 14.7 nm and 47 bp (15.7 nm) for OR1 to OR3. Since the distance between pairs of adjacent particles found in the OR or OL region was 15.4 and 14.0 nm respectively, the experiment indicated non-cooperative binding to the O1 and O3 binding sites. These observations suggested that perhaps the presence of three operator sites in each region enhances the probability of capturing CI dimers such that a sufficient number of proteins accumulate and stands ready to secure a loop when a random collision between OR and OL occurs. However, one cannot exclude that these species might have been looped molecules that did not survive deposition and washing during sample preparation.
Looping equilibrium
Indeed, the deposition process was reported to affect the measured equilibrium for protein-DNA complexes with 3D topology that distorts upon binding to the surface [23]. Although the operator sites to which CI binds to secure the DNA loop lie closely spaced, there is a slight helical shift between the O2 and O3 operator sites. This might add some three-dimensionality to a looped structure. However scoring 884 or 354 molecules with specifically bound CI particles as either “looped” or “unlooped” for wildtype or O3- DNA at a 20 nM concentration of CI led to 43.9 and 17.8% estimated looping probabilities respectively (Table 1). These are fairly close to the 40 and 10% probabilities measured using tethered particle motion for wild-type DNA segments in the presence of the same concentration of CI [16]. Successful measurement of the looping equilibrium suggested that the molecular species in the AFM images were relevant to CI-mediated looping and should be characterized further.
Table 1.
Percentages of CI-mediated loops in wild-type and o3- DNA molecules visualized using AFM.
| Wild-type 10 min incubation |
o3- 10 min incubation |
|
|---|---|---|
| Number of molecules | 884 | 354 |
| % Looped | 43.9% | 17.6% |
Volume calibration
Given the possibility for oligomerization of CI, the number of CI dimers securing a DNA loop may play an important role in the dynamics of loop formation. However, there are few experimental methods apart from direct visualization with which to determine this oligomerization on looped molecules. AFM is well suited for this type of analysis, since the volume of the particle at the closure of a DNA loop can be measured directly in the topographs. However a calibration to relate the measured volume to the molecular weight, and hence the oligomerization of the protein, is essential.
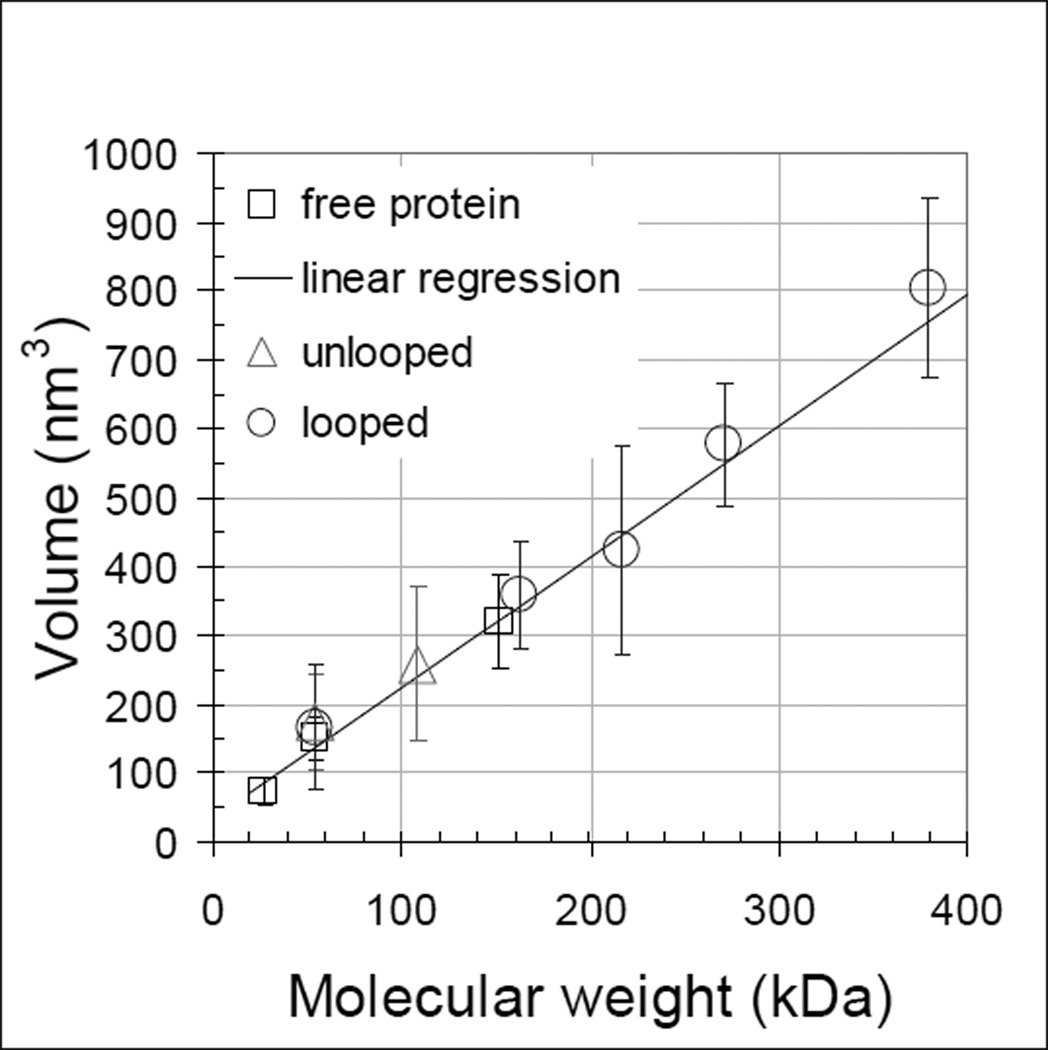
Several calibration curves have been produced previously for tapping mode images of proteins with both silicon nitride [24] and etched silicon probes [25, 26]. Both the convolution of the probe shape and the compression that results from the tapping force affect the relationship, and linear fits to volume vs. molecular weight calibrations have slopes ranging from 1.2 to 1.75 for probes with spring constants near 40 N/m and area thresholds set low or at half-height. For the experiments reported here, lac repressor (lacI) was a convenient reference which maintains a tetrameric state both free and bound to the DNA [27] while free CI was expected to partition into a 7:1 ratio of monomeric and dimeric forms at a concentration of 20 nM. The distributions of protein particles measured for CI and lacI without DNA exhibited peaks at 75, 150 and 320 nm3 (supplementary figures S1 and S2). For the etched silicon probes with a 3.5 N/m spring constant that were used in these experiments, a calibration considering monomeric and dimeric CI and tetrameric lacI proteins deposited on poly-L-ornithine-coated-mica gave a slope of 1.9 (Fig. (4)). This higher value most likely reflects both the softer cantilever which reduces compression and the low threshold used to delimit the area of individual proteins.
Figure 4.
AFM measurements of the volume of protein particles both free and bound to DNA. Standard deviations are indicated for all points. Linear regression of volume measurements of unbound λ and lac repressor proteins (dark squares) gave the calibration line (dark). The volumes of CI protein particles were measured on unlooped (grey triangles) and looped (grey circles) DNA and CI oligomerization values were assigned to the nearest dimer multiple using the calibration line. Numerical values for the plot are given in supplementary table S3.
The volumes of lacI and CI oligomers bound to DNA were also measured. The lacI DNA contains two lac repressor binding sites, Oid and O1. The specificity of particle binding was verified by tracing DNA segments as described for the CI data shown in Figure 2. The average volume of particles binding on linear DNA was 355 ± 73 nm3. Since lac repressor was expected to remain tetrameric in the conditions of the experiment (5 nM) [27], this volume was associated with an oligomer weighing 155 kDa. The difference between the measured volumes for protein free and bound to the DNA was about 30 nm which corresponds well to the volume of a segment of DNA the length of the lacI binding site, 21 bp.
The average volume of CI particles on unlooped DNA measured 259 nm3. Employing the calibration curve and considering that the molecular weight of CI monomer is 26–28 kDa [28, 29] indicated that the average particles in the experiment could have corresponded to CI tetramers (240 nm3 from the calibration curve). Of course the standard deviation of these measurements was larger than those of lac repressor, because the λ operator regions contain three adjacent binding sites, so that several stoichiometries of CI binding were possible. In fact some higher molecular weight particles were observed that are difficult to reconcile with the idea that a looped DNA scaffold is required to promote “head-to-head” binding between CI tetramers to give octamers [14, 30]. One interpretation is that specific binding nucleated adjacent non-specific binding.
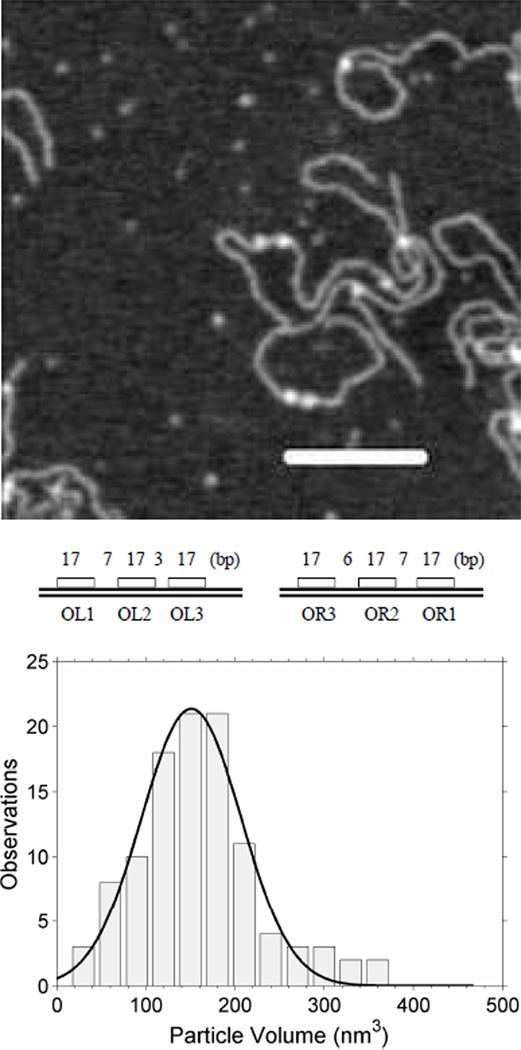
Loop closures are prevalently dodecamers
Similarly large volume, high molecular weight CI particles were commonly found securing looped DNA molecules. In Fig. (5), the lower panel shows measurements of DNA segments corresponding to the length: from one end to the OR site, of the loop, and from OL to the other end of the DNA. The narrowly distributed measurements and the good correspondence with the expected values based on the DNA construct indicated loops secured by specifically bound CI. The volumes of these CI particles were distributed as shown in the upper panel of Fig. (5). The curve exhibits three central peaks in the distribution that roughly correspond to oligomers of (from right to left): 6–8, 10–12, and 14–16. This interpretation was developed using the calibration shown in Figure 4 and assigning molecular weights to the nearest multiple of a dimer, since CI binds DNA as a dimer. The rightmost and leftmost peaks were negligibly small and were not considered further.
Figure 5.
Oligomerization of CI securing DNA loops. (upper) AFM measurements of the volumes of single CI particles securing DNA loops. (lower) The lengths of segments in the looped DNA correspond well with those expected from the design of the construct.
Oligomers of 10–12 monomers were observed most frequently securing loop closures. Such oligomers would nearly or fully saturate the operator sites in the juxtaposed OL and OR regions and are consistent with the loop stabilization conferred by “ocamer+tetramer” protein binding that was also found using modeling of tethered particle motion data [16]. A significant number of oligomers of 6–8 monomers were also observed at loop closures, but very little tetrameric CI, which corresponds well with the weaker loop stabilization afforded by oligomers lacking contacts between O3 regions [16]. Oligomers of more than 12 monomers constituted a minor fraction which suggested that CI specifically bound to operators in one region might nucleate adjacent binding of non-specifically bound CI. These additional CI dimers might further stabilize the closure through interaction with corresponding dimers from the opposite region.
Alternative loop closures
A small number of DNA loops (3.2%) contained two adjacent CI particles (Fig. (6)). The average volume of these particles was 425 nm3 which identified them as CI octamers. By tracing the DNA in a subset of particularly distinct two-particle-loops, two types of conformer were established. One type was modeled with directly juxtaposed operators in which one octamer apparently included four specifically bound dimers at O1 and O2 (or O2 and O3), and another consisting of two specifically bound CI dimers at O3 (or O1) flanked by two non-specifically bound dimers to form a second octamer (Fig. (6c)). Whether non-specifically bound dimers preferentially flanked O1 or O3 could not be determined. The other type of conformer was modeled with staggered OR and OL regions leaving OR3 unoccupied (Fig. (6b)) and CI oligomers bridging non-specific sites adjacent to OR1. Table 2 shows the results of measuring segments in these looped molecules as schematically shown in Fig. (6d). For such a small number of cases, statistically significant differences could not be established, but, as expected from the schematic diagrams, segments a and e were longer in the directly juxtaposed conformation while c was longer in the staggered conformation. These few conformers might represent early intermediates in the looping process that result from collisions between OL and OR regions that are nearly saturated with CI dimers. Such intermediates may include CI tetramers that bind ”semi-specifically” between OL1 and a non-specific site adjacent to OL1. Subsequent shifting to create complete juxtaposition of all of the specific operators would be expected to increase the stability of the loop and sterically repress the CI promoter, PRM, near OR3.
Figure 6.
Specifically bound protein particles may nucleate adjacent semi-specific binding to secure DNA loops. (a) A small number of DNA loops were secured by two CI particles. Black scale bar represents 100 nm. Possible CI binding to (b) staggered or (c) directly juxtaposed OL and OR regions. (d) Labeled segments of looped DNA molecules secured by two CI particles.
Table 2.
Segment lengths (nm) for DNA loops secured by two protein particles (shown in Figure 6).
| Segment | a | b | c | d | e |
|---|---|---|---|---|---|
| DNA molecule | Directly juxtaposed operators | ||||
| expected | 129.0 | 14.0 | 125.7 | 13.0 | 237.5 |
| 1 | 126.8 | 20.2 | 113.7 | 19.6 | 233.4 |
| 2 | 127.1 | 14.1 | 123.2 | 16.5 | 238.4 |
| 3 | 125.2 | 11.7 | 113.0 | 13.6 | 221.8 |
| 4 | 125.3 | 20.0 | 117.9 | 17.7 | 222.6 |
| 5 | 124.0 | 19.5 | 104.3 | 20.4 | 231.2 |
| mean | 125.7 | 17.1 | 114.4 | 17.6 | 229.5 |
| Staggered operators | |||||
| expected | 116.8 | 14.7 | 142.2 | 14.7 | 230.0 |
| 6 | 116.1 | 18.5 | 123.7 | 15.9 | 223.1 |
| 7 | 119.3 | 16.5 | 130.9 | 18.0 | 231.1 |
Conclusions
The data in this report strongly support the idea that CI binding to O3 operators greatly stabilizes looping of λ DNA fragments. Overwhelmingly specific binding was exhibited by 20 nM CI protein to the λ operator sites. The intrinsic order of this binding, OL1 > OR1> OL3> OL2 > OR2 > OR3, [6, 7] changes to OR1~OL1~OR2~OL2 > OL3 > OR3 when cooperative interactions are considered, and this cooperative ranking was reflected in the slight shift of positions of CI particles on unlooped DNA upon mutation of the OL3 but not the OR3 operators. The strong affinity of the polyamine–coated mica for DNA preserved the looped-unlooped equilibrium of the DNA-protein complexes to permit relevant measurements of the protein oligomerization. The volumes of particles securing DNA loops corresponded most frequently to CI oligomers of 10–12, less often to oligomers of 6–8 and occasionally to oligomers of 14–16 that likely include non-specifically bound CI. This underscores the important role of the O3 binding sites in loop stabilization. Finally, rare observations of dimers bound to adjacent operators, and adjacent CI octamers securing specific loops suggest that the tripartite binding sites in the operator regions enhance the targeting of CI to promote efficient looping and transcriptional repression at low protein concentrations.
Supplementary Material
Acknowledgements
This work was supported by an Emory University graduate student scholarship (HW), Emory University (LF), and the laboratory of Sankar Adhya at the National Institutes of Health, National Cancer Institute and the Center for Cancer Research. We thank Chiara Zurla and Carlo Manzo for discussion and suggestions and William J. Dunn for assistance with image analysis.
References
- 1.Ptashne M. A Genetic Switch. Cambridge, MA: Cell Press; 1986. [Google Scholar]
- 2.Ptashne M, Gann A. Genes & Signals. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2002. [Google Scholar]
- 3.Dodd IB, Shearwin KE, Perkins AJ, Burr T, Hochschild A, Egan JB. Cooperativity in long-range gene regulation by the lambda CI repressor. Genes & Development. 2004;18(3):344–354. doi: 10.1101/gad.1167904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maniatis T, Ptashne M. Multiple Repressor Binding At Operators In Bacteriophage-Lambda - (Nuclease Protection Polynucleotide Sizing Pyrimidine Tracts Supercoils E-Coli) Proceedings Of The National Academy Of Sciences Of The United States Of America. 1973;70(5):1531–1535. doi: 10.1073/pnas.70.5.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oppenheim AB, Kobiler O, Stavans J, Court DL, Adhya S. Switches in bacteriophage lambda development. Annual Review Of Genetics. 2005;39:409–429. doi: 10.1146/annurev.genet.39.073003.113656. [DOI] [PubMed] [Google Scholar]
- 6.Koblan KS, Ackers GK. Site-Specific Enthalpic Regulation Of Dna-Transcription At Bacteriophage-Lambda Or. Biochemistry. 1992;31(1):57–65. doi: 10.1021/bi00116a010. [DOI] [PubMed] [Google Scholar]
- 7.Senear DF, Brenowitz M, Shea MA, Ackers GK. Energetics Of Cooperative Protein Dna Interactions - Comparison Between Quantitative Deoxyribonuclease Footprint Titration And Filter Binding. Biochemistry. 1986;25(23):7344–7354. doi: 10.1021/bi00371a016. [DOI] [PubMed] [Google Scholar]
- 8.Ptashne M. A genetic switch: Phage Lambda revisited. 3rd edn. Cambridge, MA: Cold Spring Harbor Laboratory Press; 2004. [Google Scholar]
- 9.Bell CE, Frescura P, Hochschild A, Lewis M. Crystal structure of the lambda repressor C-terminal domain provides a model for cooperative operator binding. Cell. 2000;101(7):801–811. doi: 10.1016/s0092-8674(00)80891-0. [DOI] [PubMed] [Google Scholar]
- 10.Jain D, Nickels BE, Sun L, Hochschild A, Darst SA. Structure of a ternary transcription activation complex. Molecular Cell. 2004;13(1):45–53. doi: 10.1016/s1097-2765(03)00483-0. [DOI] [PubMed] [Google Scholar]
- 11.Meyer BJ, Maurer R, Ptashne M. Gene-Regulation At The Right Operator (Or) Of Bacteriophage-Lambda .2. Or1, Or2, And Or3 - Their Roles In Mediating The Effects Of Repressor And Cro. Journal Of Molecular Biology. 1980;139(2):163–194. doi: 10.1016/0022-2836(80)90303-4. [DOI] [PubMed] [Google Scholar]
- 12.Nickels BE, Dove SL, Murakami KS, Darst SA, Hochschild A. Protein-protein and protein-DNA interactions of sigma(70) region 4 involved in transcription activation by lambda cl. Journal Of Molecular Biology. 2002;324(1):17–34. doi: 10.1016/s0022-2836(02)01043-4. [DOI] [PubMed] [Google Scholar]
- 13.Maurer R, Meyer BJ, Ptashne M. Gene-Regulation At The Right Operator (Or) Of Bacteriophage-Lambda .1. Or3 And Autogenous Negative Control By Repressor. Journal Of Molecular Biology. 1980;139(2):147–161. doi: 10.1016/0022-2836(80)90302-2. [DOI] [PubMed] [Google Scholar]
- 14.Dodd IB, Perkins AJ, Tsemitsidis D, Egan JB. Octamerization of lambda CI repressor is needed for effective repression of P-RM and efficient switching from lysogeny. Genes & Development. 2001;15(22):3013–3022. doi: 10.1101/gad.937301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dodd IB, Shearwin KE, Egan JB. Revisited gene regulation in bacteriophage lambda. Current Opinion In Genetics & Development. 2005;15(2):145–152. doi: 10.1016/j.gde.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 16.Zurla C, Manzo C, Dunlap D, Lewis DE, Adhya S, Finzi L. Direct demonstration and quantification of long-range DNA looping by the {lambda} bacteriophage repressor. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sadler JR, Sasmor H, Betz JL. A perfectly symmetric lac operator binds the lac repressor very tightly. Proc Natl Acad Sci U S A. 1983;80(22):6785–6789. doi: 10.1073/pnas.80.22.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simons A, Tils D, von Wilcken-Bergmann B, Muller-Hill B. Possible ideal lac operator: Escherichia coli lac operator-like sequences from eukaryotic genomes lack the central G X C pair. Proc Natl Acad Sci U S A. 1984;81(6):1624–1628. doi: 10.1073/pnas.81.6.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meijering E, Jacob M, Sarria JC, Steiner P, Hirling H, Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A. 2004;58(2):167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- 20.Abramoff MD, Magelhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11(7):36–42. [Google Scholar]
- 21.Claudio Rivetti SC. Accurate length determination of DNA molecules visualized by atomic force microscopy: evidence for a partial B- to A-form transition on mica. Ultramicroscopy. 2001;87(12) doi: 10.1016/s0304-3991(00)00064-4. [DOI] [PubMed] [Google Scholar]
- 22.Koblan KS, Ackers GK. Site-specific enthalpic regulation of DNA transcription at bacteriophage lambda OR. Biochemistry. 1992;31(1):57–65. doi: 10.1021/bi00116a010. [DOI] [PubMed] [Google Scholar]
- 23.Yang Y, Sass LE, Du C, Hsieh P, Erie DA. Determination of protein-DNA binding constants and specificities from statistical analyses of single molecules: MutS-DNA interactions. Nucleic Acids Res. 2005;33(13):4322–4334. doi: 10.1093/nar/gki708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneider SW, Larmer J, Henderson RM, Oberleithner H. Molecular weights of individual proteins correlate with molecular volumes measured by atomic force microscopy. Pflugers Arch. 1998;435(3):362–367. doi: 10.1007/s004240050524. [DOI] [PubMed] [Google Scholar]
- 25.Neaves KJ, Cooper LP, White JH, Carnally SM, Dryden DT, Edwardson JM, Henderson RM. Atomic force microscopy of the EcoKI Type I DNA restriction enzyme bound to DNA shows enzyme dimerization and DNA looping. Nucleic Acids Res. 2009;37(6):2053–2063. doi: 10.1093/nar/gkp042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ratcliff GC, Erie DA. A novel single-molecule study to determine protein--protein association constants. J Am Chem Soc. 2001;123(24):5632–5635. doi: 10.1021/ja005750n. [DOI] [PubMed] [Google Scholar]
- 27.Hsieh M, Brenowitz M. Comparison of the DNA association kinetics of the Lac repressor tetramer, its dimeric mutant LacIadi, and the native dimeric Gal repressor. J Biol Chem. 1997;272(35):22092–22096. doi: 10.1074/jbc.272.35.22092. [DOI] [PubMed] [Google Scholar]
- 28.Burz DS, Beckett D, Benson N, Ackers GK. Self-assembly of bacteriophage lambda cI repressor: effects of single-site mutations on the monomer-dimer equilibrium. Biochemistry. 1994;33(28):8399–8405. doi: 10.1021/bi00194a003. [DOI] [PubMed] [Google Scholar]
- 29.Maniatis T, Ptashne M. Multiple repressor binding at the operators in bacteriophage lambda. Proc Natl Acad Sci U S A. 1973;70(5):1531–1535. doi: 10.1073/pnas.70.5.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dodd IB, Shearwin KE, Perkins AJ, Burr T, Hochschild A, Egan JB. Cooperativity in long-range gene regulation by the lambda CI repressor. Genes Dev. 2004;18(3):344–354. doi: 10.1101/gad.1167904. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.