An active transposable element is originally targeted for DNA methylation and epigenetic silencing.
Abstract
Transposable elements (TEs) are mobile fragments of DNA that are repressed in both plant and animal genomes through the epigenetic inheritance of repressed chromatin and expression states. The epigenetic silencing of TEs in plants is mediated by a process of RNA-directed DNA methylation (RdDM). Two pathways of RdDM have been identified: RNA Polymerase IV (Pol IV)-RdDM, which has been shown to be responsible for the de novo initiation, corrective reestablishment, and epigenetic maintenance of TE and/or transgene silencing; and RNA-dependent RNA Polymerase6 (RDR6)-RdDM, which was recently identified as necessary for maintaining repression for a few TEs. We have further characterized RDR6-RdDM using a genome-wide search to identify TEs that generate RDR6-dependent small interfering RNAs. We have determined that TEs only produce RDR6-dependent small interfering RNAs when transcriptionally active, and we have experimentally identified two TE subfamilies as direct targets of RDR6-RdDM. We used these TEs to test the function of RDR6-RdDM in assays for the de novo initiation, corrective reestablishment, and maintenance of TE silencing. We found that RDR6-RdDM plays no role in maintaining TE silencing. Rather, we found that RDR6 and Pol IV are two independent entry points into RdDM and epigenetic silencing that perform distinct functions in the silencing of TEs: Pol IV-RdDM functions to maintain TE silencing and to initiate silencing in an RNA Polymerase II expression-independent manner, while RDR6-RdDM functions to recognize active Polymerase II-derived TE mRNA transcripts to both trigger and correctively reestablish TE methylation and epigenetic silencing.
Transposable elements (TEs) constitute large percentages of both animal and plant genomes. TEs are major targets of multiple endogenous gene-silencing pathways that act to limit their expression and ability to generate new insertions and mutations (for review, see Girard and Hannon, 2008). To study TE silencing, the process has been divided into three distinct mechanisms: the de novo initiation/triggering of silencing, the corrective reestablishment of silencing of TEs that were recently transcriptionally reactivated, and the epigenetic maintenance of TE silencing.
In the Arabidopsis (Arabidopsis thaliana) genome, nearly all TEs are found in a transcriptionally silenced state (Lippman et al., 2004). This transcriptional gene silencing is maintained by symmetrical DNA methylation, which is propagated through mitotic cell divisions (for review, see Law and Jacobsen, 2010). In contrast to animals, plants do not erase the DNA methylation patterns of their gametes; therefore, CG and CHG (where H = A, T, or C) symmetrical DNA methylation patterns established in one generation are inherited and act to maintain TE silencing in the next generation through a process termed transgenerational epigenetic inheritance (Mathieu et al., 2007; Becker et al., 2011). In addition to the maintenance of symmetrical methylation, methylation of TEs is continually reinforced through a process of RNA-directed DNA methylation (RdDM; for review, see Law and Jacobsen, 2010; Haag and Pikaard, 2011). This pathway involves RNA Polymerase IV (Pol IV), a plant-specific DNA-dependent RNA polymerase that transcribes heterochromatic regions such as TEs into non-protein-coding transcripts. These transcripts are converted into double-stranded RNA by the RNA-dependent RNA Polymerase RDR2 and cleaved into 24 nucleotide (nt) small interfering RNAs (siRNAs) by Dicer-like3 (DCL3). The resulting 24 nt siRNAs associate with Argonaute4 (AGO4) and AGO6 to target nascent TE transcripts in the nucleus produced by another plant-specific DNA-dependent RNA polymerase, RNA Polymerase V (Pol V). Targeting of Pol V transcripts by AGO protein complexes mediated through siRNA complementarity results in the recruitment of the DNA methyltransferase DRM2 to methylate cytosines at the Pol V-transcribed locus (for review, see Law and Jacobsen, 2010). DNA targets of RdDM can be identified through CHH context DNA methylation, as this asymmetrical methylation pattern is not replicated. The Pol IV-RDR2-DCL3-AGO4/6-Pol V-DRM2 pathway (herein referred to as the Pol IV-RdDM pathway) acts as a loop reinforcing methylation states at regions of heterochromatin and silenced TEs.
Pol IV is thought to transcribe regions of the genome that are already DNA methylated (Zheng et al., 2009; Wierzbicki et al., 2012). Therefore, the initiation of TE silencing solely by the Pol IV-RdDM pathway presents a chicken-and-egg dilemma: if Pol IV transcription is guided to previously heterochromatic and methylated TEs, how can TE silencing be initiated by Pol IV transcription? A possible resolution to this conundrum has been potentially illuminated by several recent studies that have begun to uncover factors involved in a distinct small RNA-directed DNA methylation pathway that acts independently of the Pol IV-RdDM machinery (Eamens et al., 2008; Garcia et al., 2012; Pontier et al., 2012; Stroud et al., 2013). This pathway utilizes 21-22 nt siRNAs dependent on the RNA-dependent RNA Polymerase RDR6 and DCL2 to target methylation and maintain the silencing of a single AtCopia18A long terminal repeat (LTR) retrotransposon fragment on chromosome 5, and an AtREP1 Helitron family TE on chromosome 1, as well as several intergenic regions (Pontier et al., 2012). Importantly, this RDR6-dependent DNA methylation pathway (herein referred to as the RDR6-RdDM pathway) was able to function in the absence of Pol IV, suggesting that it operates on siRNAs derived from RNA Polymerase II (Pol II) transcripts. When transcriptionally active, Pol II-derived TE mRNAs can be posttranscriptionally degraded into siRNAs that retarget complementary transcripts for further degradation in the cyclic RNA interference (RNAi) pathway (Sijen and Plasterk, 2003; Chung et al., 2008). In Arabidopsis, RDR6, DCL2, DCL4, and AGO1 degrade some TE mRNAs to produce siRNAs of 21 to 22 nts (McCue et al., 2012). Therefore, the RDR6-RdDM pathway could provide a link between posttranscriptional gene silencing/RNAi mediated by RDR6-dependent 21-22 nt siRNAs and the DNA methylation responsible for the initiation of Pol IV-RdDM and TE transgenerational silencing.
The Pol IV-RdDM pathway has been previously shown to be necessary for the initiation of transgene silencing (Aufsatz et al., 2002; Chan et al., 2004; Greenberg et al., 2011), the corrective reestablishment of TE silencing (Teixeira et al., 2009; Ito et al., 2011), and the maintenance of some TE silencing (Herr et al., 2005; Huettel et al., 2006). In this report, we did not focus on deciphering the precise RDR6-RdDM molecular mechanism; rather, we aimed to identify the major genome-wide targets of RDR6-RdDM while also determining if RDR6-RdDM is involved in the initiation, corrective reestablishment, and/or maintenance of TE silencing. If involved in these processes, we aimed to determine the relative contributions of the RDR6-RdDM and Pol IV-RdDM pathways. We particularly focused on RDR6, a protein identified as a necessary component of the Pol IV-RdDM-independent DNA methylation of transacting siRNA (tasiRNA)-generating loci as well as the single Copia and Helitron elements (Pontier et al., 2012; Wu et al., 2012). We have discovered that Pol IV-RdDM and RDR6-RdDM function differently, with RDR6-RdDM functioning only in the Pol II expression-dependent initiation and corrective reestablishment of TE silencing.
RESULTS
Genome-Wide Identification of RDR6-Dependent TE siRNAs
A recent report suggested that 21 nt siRNAs and RDR6-RdDM function to maintain the transcriptional silencing of two individual TEs (Pontier et al., 2012). To identify which TEs are targets of RDR6-RdDM on a genome-wide scale, we performed deep sequencing of small RNAs from both wild-type Columbia (Col) and rdr6 mutant inflorescences. We obtained 3,129,843 genome-matched 18 to 28 nt reads from wild-type Col and 1,479,287 from rdr6. In addition, we sequenced small RNAs from genomes with active TEs. Mutations in the swi/snf family chromatin-remodeling protein DDM1 result in global loss of heterochromatin and genome-wide transcriptional reactivation of TEs (Gendrel et al., 2002; Lippman et al., 2004). We sequenced small RNAs from a homozygous ddm1 single mutant (3,528,426 reads) and a ddm1 rdr6 double mutant (2,677,800). Therefore, we could determine which TEs have RDR6-dependent siRNAs in both TE-silenced (wild-type Col) and TE-activated (ddm1) backgrounds.
To characterize the TE content of our small RNA libraries, we began by separating the small RNA libraries into sizes of 21, 22, and 24 nts, because these are the sizes with known biogenesis and function (for review, see Chen, 2010). The overall distribution of perfectly genome-matching small RNAs of these small RNA size classes is similar in wild-type Col and rdr6 (Fig. 1A). As a control, we quantified the number and size of tasiRNAs in each library (Fig. 1B), and as predicted, we found that libraries without a functional RDR6 protein do not produce tasiRNAs (Allen et al., 2005), validating the library quality. We next classified the TE-derived siRNAs and again found very similar size distributions between wild-type Col and rdr6 (Fig. 1C), in agreement with previous studies that found that RDR6 is generally not involved in TE siRNA production in a TE-silenced background (Kasschau et al., 2007). Due to the short length of TE siRNAs and their repetitive nature, assigning the single TE locus that produced each TE siRNA is not possible. Instead, we categorized the TE class, family, and subfamily from which each TE siRNA was derived (for details, see “Materials and Methods”). We compared wild-type Col and rdr6 on the TE subfamily level to identify particular TE subfamilies that have RDR6-dependent 21 or 22 nt siRNAs. Of the 318 TE subfamilies analyzed, we detected only two (0.63%) that display a 3-fold or greater difference between wild-type Col and rdr6 (Fig. 1D, red dots). These two TE subfamilies are RomaniAT5, an LTR retrotransposon, and AtRep10C, a short nonautonomous derivative of the Helitron1 family. We analyzed the siRNA size distribution of the RomaniAT5 and AtRep10C subfamilies and found that RomaniAT5 has RDR6-dependent 22 nt siRNAs, while AtRep10C has RDR6-dependent 21 nt siRNAs (Fig. 1E). Using genomic polymorphisms, we determined that one individual TE copy (At5TE55255) of the 123 copies of the AtRep10C subfamily generates 95% of all AtRep10C subfamily RDR6-dependent 21 nt siRNAs. In our comparison between wild-type Col and rdr6 TE siRNAs, we did not detect a change in siRNA levels of the TEs AtRep1 and AtCopia18A (Fig. 1D, green dots), which represent the subfamilies of the two individual elements identified by Pontier et al. (2012) as regulated by RDR6-RdDM. AtRep10C is a nonautonomous member of the larger autonomous AtRep1 element family, and together, these data demonstrate that RDR6-RdDM regulates only a few of the individual elements of this larger Helitron family, perhaps due to the position in the genome rather than to TE identity. We conclude that, in a TE-silenced epigenome such as wild-type Col, very few TEs produce RDR6-dependent siRNAs.
Figure 1.
Identification of TEs that produce RDR6-dependent siRNAs. A, Accumulation of 21, 22, and 24 nt small RNAs in the four genotypes sequenced. Library size is normalized by calculating reads per million of 18 to 28 nt genome-matched small RNAs. An increase in ddm1 21 nt siRNAs is apparent in the total small RNA pool. B, TasiRNAs were investigated as a library control. TasiRNAs of specifically the 21 nt size accumulate only in genotypes with a functional RDR6 protein. C, Accumulation of TE-derived siRNAs. Few 21 or 22 nt siRNAs accumulate from TEs in any genotype except for ddm1. D, Comparison of 21 and 22 nt TE siRNA accumulation between wild-type Col and rdr6 single mutants. Each point represents a TE subfamily. Only two TE subfamilies (AtRep10C and RomaniAT5) have a 3-fold or more difference between wild-type Col and rdr6 (red dots). The AtRep1 and AtCopia18A subfamilies (green dots) show no dependence on RDR6 for the accumulation of their siRNAs. E, Small RNA accumulation of the AtRep10C and RomaniAT5 TE subfamilies. F, Fifteen TE subfamilies have 3-fold or higher levels of 21 or 22 nt siRNAs in ddm1 compared with wild-type Col. These siRNAs are dependent on RDR6, as they are lost in ddm1 rdr6 double mutants. G, TE siRNA distribution when the 15 subfamilies identified in F are removed from the analysis. All of the 318 TE subfamilies (with the exception of the 15 subtracted) display low levels of 21 and 22 nt siRNAs and high accumulation of 24 nt siRNAs in all four genotypes. H, The combined 21 and 22 nt siRNA accumulation of the identified 15 subfamilies with increased RDR6-dependent siRNA production in ddm1. The relative contribution of each subfamily is shown.
Although RDR6-dependent TE siRNA production is very low when TEs are epigenetically silenced, we find that RDR6 plays a much larger role in TE siRNA production when TEs are transcriptionally active in the ddm1 mutant background. In the total small RNA pool, ddm1 mutants display increased amounts of 21 nt small RNAs compared with wild-type Col (Fig. 1A). These 21 nt small RNAs are RDR6 dependent, as their levels revert back to wild-type Col levels in ddm1 rdr6 double mutants (Fig. 1A). The increase in ddm1 21 nt small RNAs detected in the total small RNA sample is produced from TE siRNAs (Fig. 1C). We found that in ddm1 mutants, 90% of the abundant TE 21 and 22 nt siRNAs are dependent on RDR6, while in wild-type Col, only 3% of the low-level TE 21 and 22 nt siRNAs are RDR6 dependent (Fig. 1C). The dependence of these TE 21 and 22 nt siRNAs on an RNA-dependent RNA polymerase defines them as secondary siRNAs amplified from primary TE transcripts. Upon characterization of the TE families and subfamilies, we identified 15 of 318 TE subfamilies (4.7%) that are responsible for all of the RDR6-dependent TE siRNAs when TEs are transcriptionally active in the ddm1 mutant epigenome. If only these 15 subfamilies are analyzed, a 77-fold increase in 21 nt siRNAs and a 10-fold increase in 22 nt siRNAs compared with wild-type Col are observed, both of which are RDR6 dependent (Fig. 1F). When these 15 subfamilies are subtracted from the analysis, no increase in 21 and/or 22 nt siRNAs in ddm1 mutants is observed (Fig. 1G). These 15 TE subfamilies are listed in Figure 1H with their relative contributions to the pool of 21 and 22 nt siRNAs in ddm1. The majority (96%) of RDR6-dependent TE siRNAs are from Athila family retrotransposons (Athila0, Athila1, Athila2, Athila3, Athila4, Athila5, Athila6A, and Athila6B; Fig. 1H). Several AtENSPM family CACTA-like DNA transposons (AtENSPM2, AtENSPM5, and AtENSPM6), two AtCOPIA LTR retrotransposon subfamilies (AtCOPIA93 and AtENSPM52), and the individual subfamilies of the AtGP1 LTR retrotransposon and Vandal3 Mutator superfamily DNA transposons also produce RDR6-dependent siRNAs in ddm1 mutants (Fig. 1H). The AtCOPIA93 subfamily is also called Évadé and has been shown to produce 21 and 22 nt siRNAs only when transcriptionally active (Mirouze et al., 2009). These identified TE subfamilies represent high-copy (Athila2 has 413 copies/fragments present in the reference Arabidopsis genome), medium-copy (AtENSPM2 has 114 copies/fragments), and low-copy (AtCopia52 has seven copies/fragments) TE subfamilies. Although only 15 subfamily elements display RDR6-dependent siRNA accumulation, these 15 TE subfamilies occupy 24.8% of the total TE space and 4.85% of the entire Arabidopsis genome. Therefore, we conclude that RDR6 plays a larger role in the production of TE siRNAs in ddm1 mutants, presumably when the TEs are transcriptionally active, and we have used this genome-wide small RNA sequencing data to generate candidate TEs regulated by RDR6-RdDM activity.
RDR6-RdDM and Pol IV-RdDM Can Target the Same Transcriptionally Active TE Region for CHH Hypermethylation
We next aimed to determine if TE transcriptional activation leads to expression-dependent DNA methylation specifically through the production of 21 and 22 nt siRNAs and RDR6-RdDM. We concentrated on whether TEs with RDR6-dependent siRNAs are targets of RDR6-RdDM by comparing TE methylation in ddm1 single mutants (with 21 and 22 nt siRNAs) with ddm1 rdr6 double mutants, which lose 21 and 22 nt siRNAs (Fig. 1F). A problem that potentially complicates our analysis stems from the observation that some 24 nt siRNAs from the Pol IV-RdDM pathway also decrease in ddm1 rdr6 double mutants (Fig. 1F). Therefore, we searched our small RNA sequencing data sets for a TE subfamily that has RDR6-dependent 21 and 22 nt siRNAs in ddm1 in single mutants and 24 nt siRNAs that are not RDR6 dependent. By focusing on this TE subfamily, we were able to separate methylation induced by 21 and 22 nt siRNAs and RDR6-RdDM from methylation induced by 24 nt siRNAs and Pol IV-RdDM. We identified the TE subfamily AtENSPM6, which has increased RDR6-dependent 21 and 22 nt siRNAs in ddm1 compared with wild-type Col (Fig. 2A) and is one of the most minor contributors to the production of RDR6-dependent siRNAs in ddm1, accounting for only 0.19% of the total increase (Fig. 1H). We further analyzed AtENSPM6 to identify a specific region of the element that displays increased levels of RDR6-dependent 21 and 22 nt siRNAs in ddm1 compared with wild-type Col and consistent levels of 24 nt siRNAs in ddm1 and ddm1 rdr6. We identified the first and second exons of AtENSPM6 for further analysis (Fig. 2B, red box). Examination of this region shows equal amounts of 24 nt siRNAs in ddm1 and ddm1 rdr6 (Fig. 2C). Bisulfite sequencing of DNA methylation patterns for this region identified strong CHH hypermethylation in ddm1 single mutants: from 9% CHH methylation in wild-type Col to 63% in ddm1 (Fig. 2D). CHH hypermethylation of TEs is known to occur when TEs are transcriptionally reactivated, particularly at Gypsy family LTR retrotransposons such as Athila (Stroud et al., 2013), either in TE-reactivated mutants (Saze and Kakutani, 2007) or when they are transcriptionally activated in the pollen vegetative nucleus of wild-type plants (Schoft et al., 2009; Slotkin et al., 2009; Calarco et al., 2012; Ibarra et al., 2012). In ddm1 rdr6 double mutants, CHH hypermethylation of AtENSPM6 is significantly reduced to 32% (P < 0.01; Fig. 2D), demonstrating that the specific loss of the 21 and/or 22 nt siRNAs is responsible for a portion of the CHH hypermethylation. This analysis of AtENSPM6 demonstrates that activation of TEs can lead to their methylation through the RDR6-RdDM pathway and that locating sites of RDR6-dependent 21 and 22 nt siRNA production can be used to successfully identify TE or other candidate genomic locations of RDR6-RdDM activity. In addition to the change in CHH context methylation levels, the CG and CHG methylation levels of AtENSPM6 in ddm1 single mutants (79% CG and 73% CHG) also decrease in ddm1 rdr6 (52.5% CG and 47.4% CHG; Fig. 2D). CG and CHG methylation are likely higher than CHH methylation in each genotype tested due to siRNA-directed targeting of new methylation to cytosines in any sequence context, followed by S-phase replication of DNA methylation patterns only in the symmetrical CG and CHG contexts.
Figure 2.
CHH hypermethylation of the transcriptionally active AtENSPM6 TE is dependent on both the RDR6-RdDM and Pol IV-RdDM pathways. A, Profile of siRNAs derived from AtENSPM6 subfamily TEs. B, Small RNA profile of the first 5,000 bp of the 8,825-bp AtENSPM6 consensus element. The number of reads per million of 21, 22, and 24 nt siRNAs are mapped for wild-type Col, ddm1, and ddm1 rdr6. The scale of each track is 1 to 100 reads per million. The red box represents the region further interrogated in C to E. TIR, Terminal inverted repeat. C, Profile of siRNAs derived from the red-boxed region of AtENSPM6 in B. D, Bisulfite sequencing of DNA methylation levels of the AtENSPM6 region from the red box in B. Methylation in the CG (red), CHG (blue), and CHH (green) sequence contexts are shown (where H = C, T, or A). The number of clones sequenced and the C→T conversion efficiency for each bisulfite reaction (as judged by sequencing of a nonmethylated genic exon) are shown for each sample. This region of AtENSPM6 undergoes hypermethylation in ddm1 mutants. This hypermethylation is reduced in ddm1 double mutants with the RDR6-RdDM pathway (ddm1 rdr6), in the pol IV mutant of the Pol IV-RdDM pathway (ddm1 nrpD1), and in the shared component Pol V (ddm1 nrpE1). E, qRT-PCR analysis of the steady-state polyadenylated transcript accumulation of AtENSPM6. ns, Not a statistically significant difference.
In addition to being targeted for RDR6-RdDM methylation, this same region of AtENSPM6 is also targeted by 24 nt siRNAs and CHH methylation via Pol IV-RdDM. Mutations in the largest subunit of RNA Pol IV (nrpD1) in double mutant combination with ddm1 also show reduced methylation compared with ddm1 single mutants (Fig. 2D). Mutations in either the Pol IV-RdDM or RDR6-RdDM pathway each produce roughly one-half of the level of AtENSPM6 CHH methylation (both 32%) compared with when the TE is active in ddm1 mutant plants (63%; Fig. 2D). This demonstrates that RDR6-RdDM and Pol IV-RdDM function on the same TE to establish CHH hypermethylation upon TE transcriptional activation. Mutations in the largest subunit of RNA Pol V (nrpE1) in combination with ddm1 show a more severe reduction compared with ddm1 rdr6 and ddm1 nrpD1 (Fig. 2D), corroborating previous findings that suggest that Pol V is a shared component of both the RDR6-RdDM and Pol IV-RdDM pathways (Wu et al., 2012).
Examination of the AtENSPM6 steady-state polyadenylated expression levels demonstrates that the methylation of this coding region of the AtENSPM6 element examined in Figure 2, B to D, does not correlate with or control the element’s transcriptional activity. AtENSPM6 activation in a ddm1 single mutant displays the same expression level as ddm1 nrpD1 and ddm1 nrpE1 double mutants (Fig. 2E), which have significantly less AtENSPM6 methylation (Fig. 2D). ddm1 rdr6 double mutants have increased expression levels compared with the ddm1 single or other ddm1 double mutants (Fig. 2E); however, this change is not statistically significant. Rather than acting through methylation and on the transcriptional level, in this case, RDR6 may be acting posttranscriptionally to degrade the activated AtENSPM6 mRNA into 21 and 22 nt siRNAs. Therefore, although targeted for methylation by both Pol IV-RdDM and RDR6-RdDM, once activated in ddm1, RDR6-RdDM and Pol IV-RdDM play no role in the repression of AtENSPM6 expression. This is similar to the tasiRNA-generating loci, where methylation induced by RDR6-RdDM plays no role in altering gene expression levels (Wu et al., 2012), as this methylation is likely too distant from the promoter of the tasiRNA-generating loci or AtENSPM6 to exert an influence on transcriptional rates.
Methylation Induced by RDR6-RdDM and Pol IV-RdDM Controls the Expression of the Athila6 TE
To determine if RDR6-RdDM has any role in regulating the expression of the TEs that produce RDR6-dependent 21 and 22 nt siRNAs, we next searched to identify a TE subfamily with RDR6-dependent 21 and 22 nt siRNAs near its TE 5′ terminus, the site of TE promoter elements and transcript initiation. Steady-state expression levels of AtENSPM6 are not influenced by the methylation status of its coding region (Fig. 2, D and E), and the 5′ 500 bp that includes the 5′ terminal inverted repeat and transcriptional regulatory elements (Banks et al., 1988) produce very few siRNAs in wild-type Col and none in ddm1 mutants (Fig. 2B). Therefore, we focused on the TE family that is the major producer of RDR6-dependent siRNAs in ddm1 mutants: the Athila LTR retrotransposon family. We focused on the Athila6 subfamily, as it is the top contributor to the 21 and 22 nt siRNA pool in ddm1 mutants (Fig. 1H). When transcriptionally activated, Athila6 21 and 22 nt siRNAs increase 72.5-fold compared with wild-type Col and constitute 15% of the total small RNA library in ddm1 mutants (Fig. 3A).
Figure 3.
The expression-dependent CHH hypermethylation of the Athila6 TSS is dependent on both RDR6-RdDM and Pol IV-RdDM. A, Accumulation of Athila6 subfamily-derived siRNAs. B, Accumulation of siRNAs from the Athila6 LTR TSS region assayed for DNA methylation by bisulfite sequencing in C. C, Bisulfite sequencing of the Athila6 LTR gag/pol TSS demonstrates that components of both the RDR6-RdDM and Pol IV-RdDM pathways are required to attain the full CHH hypermethylation present in ddm1 single mutants. Two biological replicates of ddm1 rdr6 were performed. D, Small RNA northern blot of the Athila6 LTR TSS region. All 24 nt siRNA production is dependent on RDR2 and Pol IV/NRPD1. All 21 and 22 nt siRNA production is dependent on RDR6. miR161 is used as a loading control. E, qRT-PCR analysis of the steady-state polyadenylated gag/pol transcript levels of Athila6 demonstrate that the expression level increases in ddm1 double mutants that lose hypermethylation compared with ddm1 single mutant plants.
To characterize the methylation status of the Athila6 transcriptional regulatory region, we first mapped the transcriptional start site (TSS) of the gag/pol protein-coding transcript of the Athila6 subfamily (Supplemental Fig. S1) and subsequently used the surrounding 350 bp of this 5′ LTR region for analysis of DNA methylation. When transcriptionally active in ddm1 mutants, this TSS region undergoes a 24-fold increase in 21 nt siRNAs, a 4.9-fold increase in 22 nt siRNAs, and a 6.2-fold increase in 24 nt siRNAs, all of which are dependent on RDR6 (Fig. 3B). We performed bisulfite sequencing of the Athila6 LTR TSS and found in wild-type Col high levels of CG methylation (93.9%), intermediate levels of CHG methylation (54.7%), and low levels of CHH methylation (15.9%; Fig. 3C). When transcriptionally activated in ddm1 mutants, this methylation pattern is drastically altered: the CG methylation is reduced, and the CHH methylation increases 4.3-fold compared with wild-type Col. Like AtENSPM6, we found that this CHH hypermethylation is dependent on both the RDR6-RdDM pathway and the Pol IV-RdDM pathway (Fig. 3C). In ddm1 rdr6 double mutants, the CHH methylation is significantly reduced to wild-type Col levels. As a control, we assayed sibling plants from a segregating family, both of which were ddm1 homozygous mutants but differed in the presence or absence of a functional RDR6 gene, and found a statistically significant (P < 0.001) dependence on RDR6 for establishing CHH hypermethylation (Fig. 3C). ddm1 double mutants with Pol IV-RdDM components, such as rdr2 and pol IV/nrpD1, also displayed significant reduction in CHH methylation (13.8% and 21.0%, respectively). Importantly, ddm1 nrpD1 double mutants lose all Athila6 TSS 24 nt siRNAs (Fig. 3D), confirming that there is an alternative pathway besides the production of 24 nt siRNAs for the establishment of the 21% CHH methylation in this double mutant. The least amount of methylation in any sequence context was detected in the ddm1 ago6 (6.2%) and ddm1 nrpE1 (2.7%) double mutants (Fig. 3C). The CHH methylation detected in the ddm1 nrpE1 double mutant is the minimum baseline that can be detected in these samples, as this is very close to our experimentally determined conversion efficiency of 98.3% (Fig. 3C). In addition to Pol V, AGO6 has been identified as a downstream and shared component of both the RDR6-RdDM and Pol IV-RdDM pathways (Wu et al., 2012). CHH hypermethylation of the Athila LTR also occurs when Athila is transcriptionally activated in the wild-type pollen vegetative nucleus, and in these experiments, CHH methylation was dependent only on Pol V and not Pol IV (Schoft et al., 2009). Due to the high levels of Athila 21 and 22 nt siRNAs detected in pollen (Slotkin et al., 2009), we suggest that Pol IV-RdDM plays no role in methylating Athila and possibly other TEs in the pollen grain; rather, these TEs are methylated by RDR6-RdDM.
Our results from both Athila6 and AtENSPM6 suggested that RDR6-RdDM and Pol IV-RdDM function independently to establish CHH hypermethylation on transcriptionally active TEs. To determine if these are the only two pathways responsible for establishing TE methylation, we constructed a ddm1 nrpD1 rdr6 triple mutant, inactivating critical components of both the RDR6-RdDM and Pol IV-RdDM pathways at the same time. The ddm1 nrpD1 rdr6 triple mutant has very low levels of Athila6 LTR TSS methylation in all sequence contexts (10.6% CG, 2.2% CHG, and 1.6% CHH; Fig. 3C), lower than both the ddm1 rdr6 and ddm1 nrpD1 double mutants that inactivate RDR6-RdDM and Pol IV-RdDM individually. Therefore, we conclude that RDR6 and Pol IV are the only two entry points into RdDM responsible for the expression-dependent CHH hypermethylation of Athila6, and these pathways work independently and additively on their target(s) to establish TE methylation levels.
In contrast to AtENSPM6, the methylation status of the Athila6 TSS has a direct effect on the steady-state transcript accumulation of the gag/pol protein-coding transcript. Activation of expression is detected in ddm1 single mutants and increases (P < 0.001) when hypermethylation is reduced in ddm1 rdr2, ddm1 nrpD1, and ddm1 nrpE1 double mutants (Fig. 3E). This suggests that the transcript increase (P < 0.001) in ddm1 rdr6 compared with ddm1 single mutants (Fig. 3, C and D) is due to the loss of hypermethylation in the double mutant rather than or in addition to RDR6 acting posttranscriptionally. Athila6 expression was particularly high in ddm1 nrpE1 double mutants, 10-fold higher compared with ddm1 single mutants. This expression level likely represents close to the maximum transcript accumulation potential of Athila6 when virtually uninhibited by repressive DNA methylation.
RDR6-RdDM Functions to Initiate Expression-Dependent TE Methylation
The process of TE silencing has been divided into three distinct mechanisms: the de novo initiation of silencing, the corrective reestablishment of silencing of recently reactivated TEs, and the epigenetic maintenance TE of silencing. After identifying RDR6-RdDM target TEs, we set out to determine if the RDR6-RdDM pathway functions to initiate, reestablish, or maintain TE silencing. If it does play a role in these separate processes, we aimed to identify the relative contributions of Pol IV-RdDM and RDR6-RdDM to these functions. We have focused on the Athila6 retrotransposon, as hypermethylation of the LTR TSS functions to regulate TE expression (Fig. 3, C–E). Even though other studies have suggested that additional 21 and 22 nt siRNA-generating proteins, such as RDR1, play a role in DNA methylation (Pontier et al., 2012; Stroud et al., 2013), we find no alteration of Athila6 siRNA levels between ddm1 single mutants and ddm1 rdr1 double mutants (Fig. 3D). Therefore, we have focused our studies on RDR6, which is responsible for the production of all Athila6 LTR TSS 21 and 22 nt siRNAs, as well as Pol IV, which is responsible for the production of 24 nt siRNAs (Fig. 3D).
We wanted to assess if RDR6-RdDM plays a role in the initiation of TE silencing, and if so, its relative contribution compared with Pol IV-RdDM. To test the initiation of TE silencing, we transformed plants with a transgene containing a 946 bp fragment of Athila6. We placed the Athila6 fragment under the control of an estradiol-inducible promoter to test if Pol II expression is necessary for the function of either RdDM pathway. We transformed wild-type Col as well as rdr6 and pol IV/nrpD1 mutants, which do not have detectable levels of Athila6 expression (Fig. 4A), with our inducible Athila6 (ind-Athila) transgene. After a mock induction of T1 generation plants, very low levels of ind-Athila expression were detected, which were just above the limit of detection (Fig. 4A). With estradiol induction of expression, wild-type Col plants with ind-Athila undergo a 13.7-fold increase in expression, rdr6 undergoes an 18.8-fold increase, and pol IV/nrpD1 undergoes a 129-fold increase compared with the mock-induced plants (Fig. 4A). We compared the expression of the induced genotypes and found that wild-type Col has the lowest expression level, rdr6 is intermediate (although not statistically significantly higher than wild-type Col), and pol IV/nrpD1 has the highest expression of the ind-Athila transgene (P < 0.05; Fig. 4A). As a control, we estradiol treated nontransgenic wild-type Col plants and determined that estradiol treatment does not activate the expression of endogenous Athila6 elements (Fig. 4A).
Figure 4.
RDR6-RdDM-dependent initiation of TE silencing is dependent on expression. A, qRT-PCR analysis of the expression of Athila6 in plants without the ind-Athila transgene, with the mock-induced transgene, and with estradiol induction of transgene expression. wt, Wild type. B, Small RNA northern blot of Athila6 siRNAs. The production of 21 and 22 nt siRNAs from the estradiol-induced ind-Athila transgene is dependent on RDR6. miR161 is shown as a loading control. C, Bisulfite sequencing of the Athila6 region of the ind-Athila transgene in mock- and estradiol-induced conditions. The non-TE-containing inducible RAN1 (ind-RAN1) transgene is used as a control to distinguish background transgene silencing from TE silencing.
We next determined that, similar to Pol II-derived reactivated endogenous Athila6 transcripts (e.g. in ddm1 mutants), estradiol-induced transcripts of ind-Athila6 are processed into 21 and 22 nt siRNAs (Fig. 4B). This processing is dependent on RDR6, as rdr6 mutant plants with estradiol-induced Athila6 transgene expression fail to produce 21 and 22 nt siRNAs (Fig. 4B). We were unable to determine if our ind-Athila transgene also produces Athila6 24 nt siRNAs, as the endogenous 24 nt siRNAs present in wild-type Col plants mask this production (Figs. 3, A and D, and 4B; McCue et al., 2012). There are nine endogenous Athila6 TEs that match the ind-Athila transgene with 95% or greater nucleotide identity over the length of the TE portion of the transgene. The 24 nt siRNAs these endogenous Athila6 elements produce are dependent on Pol IV and RDR2, as pol IV/nrpD1 and rdr2 mutants fail to accumulate Athila6 24 nt siRNAs (McCue et al., 2012). Using our ind-Athila transgene system, we were able to control TE expression and 21 and 22 nt siRNA production to determine the role of RDR6-RdDM on the initiation of TE methylation.
To determine if RDR6-RdDM plays a role in the initiation of TE methylation, we performed bisulfite sequencing of the promoter and Athila6 portion of the ind-Athila transgene in T1 generation plants using estradiol-induced inflorescence tissue as well as mock-induced sibling plants. To differentiate between TE and transgene silencing, we bisulfite sequenced a wild-type Col line with the same transgene backbone expressing the RAN1 gene (At5g20010) and no repetitive DNA. We found that even without the induction of expression, this transgene does accumulate some methylation (7.5% CG, 13.0% CHG, and 6.3% CHH). However, the symmetrical CG and CHG methylation level was one-third or less of the methylation of mock-induced ind-Athila in wild-type Col, demonstrating that ind-Athila is specifically recognized as a repetitive TE fragment and methylated beyond the levels of a standard genic transgene even without high levels of expression (Fig. 4C). Without induction of expression, ind-Athila methylation levels in wild-type Col are higher in the symmetrical CG and CHG contexts (22.3% CG and 33.8% CHG) compared with the asymmetrical CHH context (5.2% CHH), suggesting that methylation was initiated on the transgene earlier in development compared with the floral bud stage we assayed, and we are detecting only the replicated symmetrical methylation from this previous event. In rdr6 mutants, the methylation level of mock-induced ind-Athila (31.2% CG, 34.9% CHG, and 7.7% CHH) was similar to wild-type Col, demonstrating that without TE expression, RDR6 plays no role in the initiation of silencing and de novo methylation (Fig. 4C). Alternatively, pol IV/nrpD1 mutants display less CG and CHG methylation even without the induction of expression (9.4% CG and 13.2% CHG), demonstrating that Pol IV-RdDM functions in a Pol II expression-independent manner (or at least with very little expression) to initiate TE methylation.
Upon estradiol induction of expression, RDR6-RdDM functions to establish CHH methylation. When ind-Athila is estradiol treated in wild-type Col plants, the CHH methylation triples from 5.2% in mock-induced wild-type Col to 15.5% in induced wild-type Col (Fig. 4C). This demonstrates that externally activating TE expression and increasing transcription from the locus do not directly remove DNA methylation. The expression-dependent doubling of CHH methylation levels found in wild-type Col is not detected in estradiol-induced rdr6 mutants (3.7% CHH), demonstrating that RDR6-RdDM is required for Pol II expression-dependent CHH methylation (Fig. 4C). In addition, there is a reduction in the total methylation levels (CG and CHG contexts) in estradiol-induced rdr6 mutants compared with estradiol-induced wild-type Col plants (Fig. 4C). Methylation in all sequence contexts is lowest in pol IV/nrpD1 estradiol-induced plants (5.7% CG, 9.9% CHG, and 2.8% CHH), demonstrating that Pol IV-RdDM plays a larger cumulative role in the de novo initiation of TE methylation compared with RDR6-RdDM.
The RDR6-RdDM Pathway Functions to Correctively Reestablish Methylation and Silencing
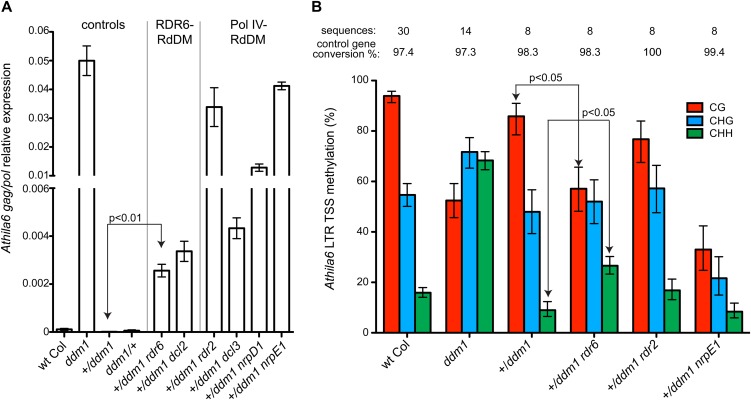
We next aimed to determine if the RDR6-RdDM pathway functions to correctively reestablish the silencing of recently reactivated TEs. The Pol IV-RdDM pathway has been previously identified as necessary to correctively resilence TEs that were transcriptionally active in a ddm1 mutant (Teixeira et al., 2009). To test the corrective reestablishment of silencing of recently reactivated TEs, we conducted a series of genetic crosses between mutant ddm1 and wild-type DDM1 plants (Table I). For the majority of TEs, the transcriptionally reactivated state in ddm1 mutants is inherited, and TEs retain transcriptional activity during the subsequent formation of ddm1/+ heterozygous progeny (Kakutani et al., 1999). In contrast, we find that Athila6 resilencing of gag/pol expression occurs very efficiently and quickly in an F1 ddm1/+ heterozygote (Fig. 5A), which is likely due to the abundant siRNAs these TEs produce when active (Fig. 3A). This resilencing occurs as early as in the +/ddm1 F1 seedling (Supplemental Fig. S2). We used ddm1 double mutants to perform a TE resilencing assay with parents that were both homozygous for a mutation in a second gene that produces a protein involved in either the RDR6-RdDM or Pol IV-RdDM pathway (Table I). We found that Athila6 expression is not fully resilenced in mutant backgrounds of either the Pol IV-RdDM pathway or the RDR6-RdDM pathway (Fig. 5A). The Pol IV-RdDM pathway mutants (rdr2, dcl3, and pol IV/nrpD1) play a larger role in this resilencing, as their Athila6 expression levels are on average higher than the RDR6-RdDM pathway mutants. However, the RDR6-RdDM pathway mutants (rdr6 and dcl2) do show statistically significant (P < 0.01) expression that is greater than the control cross of wild-type Col × ddm1, demonstrating that both the Pol IV-RdDM and RDR6-RdDM pathways play roles in the resilencing of Athila6 expression.
Table I. Crosses performed to test the role of RDR6-RdDM and Pol IV-RdDM components in TE resilencing.
Figure 5.
RDR6-RdDM functions in the corrective reestablishment of TE silencing. A, qRT-PCR analysis of Athila6 gag/pol transcript accumulation in the progeny of the crosses detailed in Table I. Active Athila6 TEs in a ddm1 background are efficiently resilenced upon crossing to wild-type (wt) plants. Mutations in the Pol IV-RdDM and RDR6-RdDM pathways result in only partial resilencing of the Athila6 gag/pol transcript. B, Bisulfite sequencing of the Athila6 LTR around the gag/pol TSS demonstrates that RDR6 and Pol V are required for the efficient return of methylation levels to their wild-type states of high CG and low CHH. Plants examined are the progeny of the crosses detailed in Table I.
We next determined if the lack of full resilencing in an rdr6 mutant background is due to incomplete reestablishment of DNA methylation. We performed bisulfite DNA sequencing on the progeny of the crosses in Table I and found that the expression levels in Figure 5A inversely correlate with the methylation levels of the Athila6 LTR TSS. The high level of CG and low level of CHH methylation found in wild-type Col are restored in the progeny of wild-type Col × ddm1 (86% CG and 9% CHH; Fig. 5B). However, when this cross is performed in an rdr6 background, CG and CHH methylation levels are only partially restored (57% CG and 27% CHH; Fig. 5B). These data demonstrate that RDR6 is necessary for the complete remethylation and resilencing of Athila6 TEs. In the pol V/nrpE1 mutant background, which encodes what is likely a shared component of the RDR6-RdDM and Pol IV-RdDM pathways, ddm1 heterozygous progeny from Table I display very low CG and CHH methylation levels (33% CG and 8% CHH; Fig. 5B), resulting in the highest levels of Athila6 expression (Fig. 5A). The combined data from our crosses performed in Table I demonstrate that at least for Athila6, RDR6-RdDM and Pol IV-RdDM both function to correctively reestablish TE DNA methylation. However, RDR6-RdDM plays a minor role compared with Pol IV-RdDM in the reestablishment of TE transcriptional silencing.
RDR6-RdDM Does Not Globally Act to Maintain TE Silencing
We next aimed to determine if RDR6 and RDR6-RdDM play roles in maintaining the repression of TE expression in a wild-type epigenome with transcriptionally silenced TEs. There are few RDR6-dependent TE siRNAs when TEs are transcriptionally silenced (Fig. 1D; Kasschau et al., 2007), and only a few individual TE copies have been reported to transcriptionally activate in rdr6 single mutants (Pontier et al., 2012). In addition, we found that in a wild-type Col epigenome restrictive to TE activity, RDR6 plays no role in the maintenance of TE methylation or the expression of either AtENSPM6 (Fig. 2, D and E) or Athila6 (Fig. 3, C–E). Recent analysis of the global methylation change in rdr6 seedlings has shown that genes, and not TEs, are the targets of RDR6-RdDM in a wild-type Col epigenome (Stroud et al., 2013). We used publicly available gene expression microarray data to assay global TE expression in three biological replicates of wild-type Col and rdr6 single mutant plants (Allen et al., 2005). We characterized the microarray expression values of 1,155 TEs (see “Materials and Methods”) and found that 95% of TEs examined have no or less than 2-fold expression difference in rdr6 compared with wild-type Col, and no TE showed a greater than 2.5-fold increase in expression in rdr6 mutants (Fig. 6A), suggesting that RDR6-RdDM plays no global role in the maintenance of transcriptionally silencing TE expression. Pontier et al. (2012) identified two individual TEs (AtRep1 and AtCopia18A) for which RDR6-RdDM functions to maintain cytosine methylation patterns and silenced expression. We did not detect any RDR6-dependent siRNAs for these TE subfamilies (Fig. 1D), and these TEs are not present on the ATH1 microarray. However, we did identify two TE subfamilies (AtRep10C and RomaniAT5) that have RDR6-dependent 21 and 22 nt siRNAs in a wild-type Col epigenome (Fig. 1, D and E). We focused on the individual AtRep10C element At5TE55255 and the RomaniAT5 subfamily to attempt to detect any role of RDR6-RdDM in the maintenance of TE silencing. We found that both the AtRep10C element At5TE55255 and the RomaniAT5 subfamily display no loss of DNA methylation or statistically significant increase in steady-state expression levels in rdr6 mutants (Fig. 6, B–E). In contrast to symmetrical DNA methylation and Pol IV-RdDM, our data demonstrate that RDR6-RdDM has no or very little function in maintaining TE silencing.
Figure 6.
RDR6-RdDM does not function to maintain TE silencing. A, Comparison of TE expression levels between wild-type (wt) Col and rdr6 mutant inflorescences. Microarray gene expression values for 1,155 individual TEs are shown (see “Materials and Methods”). In this box plot, the center line represents the median fold change; the top and bottom box borders represent the 75% and 25% percentile fold change, while the whiskers represent the 90% and 10% percentiles. Individual outlier TEs are shown as points; however, no single TE has a greater than 2.5-fold increase (red line) in expression in rdr6 single mutants compared with wild-type Col. The gene ARF4 (At5g60450) is shown as a positive control for a gene with increased expression in rdr6 mutants, as this gene is targeted by RDR6-dependent tasiRNAs (Allen et al., 2005). B, Bisulfite sequencing of the DNA methylation levels of the AtRep10C TE At5TE55255. No reduction of methylation is detected in rdr6 mutants. C, qRT-PCR analysis of the steady-state polyadenylated expression from the AtRep10C TE At5TE55255. No increase in expression is detected in rdr6 mutants. D, Bisulfite sequencing of the DNA methylation levels of the RomaniAT5 TE subfamily. E, qRT-PCR analysis of the steady-state polyadenylated expression from the RomaniAT5 TE subfamily. Expression reactivated in a ddm1 mutant is shown for comparison. The expression difference between wild-type Col and rdr6 is not statistically significant (ns; P > 0.05).
DISCUSSION
In this report, we used the AtENSPM6 TE to demonstrate that methylation can be established by RDR6-dependent 21 and 22 nt siRNAs through a separate pathway from Pol IV-RdDM. We found that RDR6 and Pol IV represent distinct entry points for differently sized siRNAs into an RdDM mechanism that shares the components AGO6 and Pol V, and these two entry points work independently and additively on TE targets to establish TE methylation. We used small RNA deep sequencing to determine that TEs only produce RDR6-dependent 21 and 22 nt siRNAs when they are transcriptionally active, and we have identified 15 TE subfamilies that constitute this pool of siRNAs and therefore are potential targets of RDR6-RdDM. The TE copy number does not correlate with siRNA production, and hence copy number does not account for the ability of a TE to produce RDR6-dependent siRNAs when transcriptionally active. Therefore, why only these 15 TE subfamilies generate RDR6-dependent siRNAs is currently unknown. The Athila superfamily of retrotransposons contributes the vast majority of RDR6-dependent 21 and 22 nt siRNAs, and is the major target of RDR6-RdDM when transcriptionally active. Athila may generate large quantities of these siRNAs due to the tight distribution of Athila to the centromere core, their high copy number, their highly nested configuration, and/or their ancient integration into the plant genome, each of which differs from the average Arabidopsis TE family including other LTR retrotransposons (for review, see Slotkin, 2010).
We demonstrated that both RDR6-RdDM and Pol IV-RdDM regulate Athila6 expression levels. However, RDR6-RdDM may not regulate the expression of all 15 potential RDR6-RdDM target TE subfamilies, as some methylation does not result in a change in expression, similar to the methylation established at tasiRNA-generating loci (Wu et al., 2010). For example, the AtENSPM6 TE is a direct target of RDR6-RdDM, but this methylation does not affect the TE transcript levels. These differences may be due to the region of each TE that is targeted by RdDM. Methylation of promoters and regulatory regions by RdDM will likely affect the TE expression state, whereas the methylation of Pol II-transcribed protein-coding regions may not. For example, the Athila6 promoter/TSS is targeted by RdDM, resulting in the regulation of expression, while methylation of the AtENSPM6 protein-coding region does not result in the regulation of expression. Retrotransposons may be more efficient targets for transcriptional repression by RDR6-RdDM compared with DNA transposons or genes, because their promoters and regulatory information must be converted into RNA for element duplication. This may account for why LTR retrotransposons, such as Athila, are targets of RDR6-RdDM expression regulation, because they contain a downstream transcribed LTR that is processed into 21 and 22 nt siRNAs that match the 5′ LTR containing the promoter and TSS.
In contrast to previous findings, we find that RDR6 and RDR6-RdDM play no role (or a very minor role) in the maintenance of TE silencing. Transcriptionally silenced TEs in a wild-type Col epigenome produce very few RDR6-dependent siRNAs, while even the two TE subfamilies we identified with RDR6-dependent siRNAs (AtREP10C and RomaniAT5; Fig. 1) do not show increased expression in rdr6 mutants. Furthermore, both AtENSPM6 and Athila6 methylation and expression are unaltered in rdr6 single mutants compared with wild-type Col. The individual TEs identified by Pontier et al. (2012) that are maintained in a silenced state by RDR6-RdDM may be slightly Pol II transcriptionally active but quickly turned over into siRNAs. We theorize that these and/or other individual TEs in the Arabidopsis genome may retain their activity in the repressive wild-type Col epigenome due to the influence of their local euchromatic context and adjacent genes. However, the vast majority of Arabidopsis TEs are Pol II transcriptionally inactive. In fact, both the AtENSPM6 and Athila6 TEs are so efficiently and deeply silenced in wild-type Col that their transcriptional silencing is not dependent on the Pol IV-RdDM pathway to continuously retarget methylation and silencing. Although these two TEs continue to be retargeted by the Pol IV-RdDM pathway to produce 24 nt siRNAs, their expression is only dependent on the maintenance of symmetrical DNA methylation (Figs. 2E and 3E; Kato et al., 2003; Lister et al., 2008), while other TEs are not as deeply silenced and therefore are still dependent on constant retargeting of DNA methylation by Pol IV-RdDM (Herr et al., 2005).
In contrast to the maintenance of TE silencing, RDR6-RdDM plays a significant role in the corrective reestablishment of TE silencing. When Athila6 is transcriptionally active, RDR6-RdDM functions to establish high levels of CHH methylation at the LTR TSS, apparently in an attempt to resilence this TE. However, in the ddm1 mutant background, this resilencing of expression cannot be established (Figs. 2 and 3). When a ddm1 mutant individual with transcriptionally active TEs is outcrossed, the resulting F1 hybrid now has a functional DDM1 protein, and methylation induced by both Pol IV-RdDM and RDR6-RdDM establishes resilencing of the TE (Fig. 5). Compared with RDR6-RdDM, Pol IV-RdDM plays a larger role in correctively reestablishing the transcriptional silencing of Athila6 as well as in the initiation of silencing of the ind-Athila6 transgene (see below). This larger dependence on Pol IV-RdDM may be due to the previous finding that in pol IV and rdr2 mutants, the AGO6 protein does not accumulate to its normal wild-type Col levels (Havecker et al., 2010). AGO6 is a shared component of the RDR6-RdDM and Pol IV-RdDM pathways (Wu et al., 2012), and when limited in pol IV and rdr2 mutants, an indirect dampening effect of RDR6-RdDM is expected. Therefore, the reduction of TE methylation and increase in TE expression in the Pol IV-RdDM pathway mutants nrpD1 and rdr2 are overestimations of the impact of the Pol IV-RdDM pathway, accounting for the observed larger contribution of Pol IV-RdDM compared with RDR6-RdDM in the initiation and corrective reestablishment of TE silencing. However, why similar levels of partial methylation in +/ddm1 rdr2 compared with +/ddm1 rdr6 progeny from Figure 5 result in different expression levels is currently unknown. Another puzzling question remains: when transcriptionally activated in ddm1 mutants, why are Athila6 24 nt siRNAs dependent on RDR6 while all other TE 24-nucleotide siRNAs (including from AtENSPM6) are RDR6 independent? We hypothesize that Athila6 methylation induced by RDR6-RdDM indirectly helps establish the Pol IV-RdDM template. We speculate that without the initiation of CHH methylation via RDR6-RdDM at the Athila6 TSS, the low level of methylation in this region results in a loss of Pol IV targeting to this region. Therefore, without RDR6, the drop in methylation results in less Pol IV transcription and fewer 24 nt siRNAs. Conversely, increased methylation by RDR6-RdDM may generate more Pol IV transcription from these loci, creating more 24 nt siRNAs.
In addition to the corrective reestablishment of TE silencing, RDR6-RdDM functions in the de novo initiation of TE silencing. By analyzing the initiation of silencing of ind-Athila6 (Fig. 4), we can separate the distinct activities of Pol IV-RdDM from RDR6-RdDM. Pol IV-RdDM functions independently of ind-Athila6 expression, or at least with very low detectable expression levels. This silencing is likely due to the abundant 24 nt siRNAs that the endogenous Athila6 elements consistently produce, and silencing must be dependent on homology between the siRNAs and the target TE. In contrast, RDR6-RdDM functions only after the induction of expression from ind-Athila6. Similar to the corrective reestablishment of TE silencing, pol IV mutants have a more severe effect on the initiation of element silencing, but again, this could be due to the influence of Pol IV on AGO6 protein accumulation (see above). Whether RDR6-RdDM function is dependent on homology to the already present and silenced TEs in the genome is currently unknown. Alternatively, RDR6-RdDM may function in a homology-independent manner by somehow recognizing Pol II-derived TE transcripts and initiating silencing at these loci.
Two General Mechanisms to Initiate TE Silencing
The distinction in the epigenetic state of their targets (silenced TEs versus transcriptionally active TEs) allows us to propose a model of separate Pol IV-RdDM and RDR6-RdDM functions. The distinction is best illustrated by considering two examples of a transcriptionally active TE entering a genome. In the first instance, the active TE enters the genome through cross hybridization within a single species or from two closely related species. If these two species share a recent common ancestor (and, therefore, can mate), their TEs will share homology to each other. If one of the two parent species contains a TE copy that has been previously silenced and whose silencing is maintained by the Pol IV-RdDM pathway, the complementarity between the silenced TE Pol IV-dependent 24 nt siRNAs and the other parent’s transcriptionally active TEs will result in homology-dependent trans-silencing of the active TEs. Where and when in the F1 hybrid plant this silencing occurs remains an open question, but if we use the model of the corrective reestablishment experiment that we performed (with one parent having transcriptionally active TEs while the other has the same TEs in a silenced state; Fig. 5), the F1 silencing occurs very early during embryogenesis. In this way, the previously silenced TEs in a plant genome act as a battle history, cellular memory, and inoculum for any incoming homologous active TEs (Jensen et al., 1999). We theorize that Pol IV-RdDM plays a larger role in our corrective reestablishment and initiation of TE-silencing experiments due to at least one parent in each of these experiments having silenced Athila6 elements and producing Pol IV-dependent Athila6 24 nt siRNAs.
The second instance of TE silencing considers active TEs that are unique to the genome they enter. This may occur through horizontal transfer, which on an evolutionary time scale may be a somewhat common event for TEs in plant genomes (Diao et al., 2006; Roulin et al., 2008). Due to the unique TE sequence, the cell has no ability to silence the active TE based on homology. We speculate that RDR6-RdDM functions in the recognition of Pol II-derived transcripts from this new TE. How the RDR6-RdDM pathway recognizes TE transcripts is not understood, but evidence suggests that the mobilization of active TEs often produces spontaneous rearrangements or nested elements that drive the production of double-stranded RNA, triggering RNAi and siRNA production (Slotkin et al., 2005). Once produced, these 21 and 22 nt siRNAs can initiate TE DNA methylation through RDR6-RdDM. Since many organisms do not have Pol IV and 24 nt siRNAs, the Pol II-derived transcript identification mechanism may be the evolutionarily older mechanism responsible for surveillance of the transcriptome and targeting active TEs for the initiation of silencing, which in some cases in animals must be reset each generation (for review, see Smallwood and Kelsey, 2012). Once established by RDR6-RdDM, low levels of TE methylation in plants will be reinforced by the methylation-dependent Pol IV transcription of this TE locus and retargeting by Pol IV-RdDM. Over time, this TE will become deeply silenced and only dependent on symmetrical DNA methylation to propagate its transgenerational epigenetic silencing. It will produce the 24 nt siRNAs required for initiating the Pol IV-RdDM homology-dependent silencing of any incoming active TEs with sequence similarity.
MATERIALS AND METHODS
Plant Material
The mutant alleles of Arabidopsis (Arabidopsis thaliana) used in this study are described in Supplemental Table S1. Plants were grown under standard long-day conditions at 23°C. Inflorescence tissue was used in each experiment unless otherwise noted.
Expression Analysis by Quantitative Reverse Transcription-PCR
Three biological replicates were performed for each genotype. Each replicate consisted of a nonoverlapping pool of individuals. Quantitative reverse transcription (qRT)-PCR was performed and analyzed as described (McCue et al., 2012), with the exception that complementary DNA was generated using an oligo(dT) primer and Tetro Reverse Transcriptase (Bioline). Quantitative real-time PCR was performed using SensiMix (Bioline) on a Mastercycler ep realplex thermocycler (Eppendorf). The At1g08200 gene was used as a reference in all qRT-PCRs. qRT-PCR primers are listed in Supplemental Table S1.
Bisulfite Conversion and Sequencing
DNA was bisulfite converted using the EZ DNA Methylation-Gold kit (Zymo Research). Converted DNA was amplified using EpiTaq DNA polymerase (Takara) using the primers listed in Supplemental Table S1. PCR products were TOPO-TA cloned into pCR4 (Invitrogen) and sequenced. Analysis of individual DNA sequences was performed using the Kismeth analysis tool (Gruntman et al., 2008). For each bisulfite-converted DNA sample, an unmethylated exon of the gene At2g20610 was amplified, and at least three clones were sequenced to determine the C→T conversion efficiency, which is listed on each figure. Error bars represent Wilson score interval 95% confidence limits (Henderson et al., 2010). Differences between genotype methylation status were analyzed with a two-tailed Student’s t test. Each cloned sequence was given a methylated percentage based on the number of unconverted cytosines that were present out of total cytosines in the sequence. Each methylation context (CG, GHG, CHH, or total) was analyzed independently in this manner. Individual methylation percentages for each clone were grouped into populations by genotype, and these populations were compared with Student’s t test. This statistical approach is more stringent compared with the χ2 tests previously used for statistical comparisons of bisulfite data (Henderson et al., 2010), as it takes into account clone-to-clone methylation variation. All of our bisulfite methylation comparisons that were statistically significant (P < 0.05) using the Student’s t test were also statistically significant at P < 0.0001 using the χ2 approach.
Northern Blotting
Total RNA was isolated using Trizol reagent (Invitrogen), and small RNAs were concentrated with polyethylene glycol. Fourteen micrograms of small RNA-enriched RNA were loaded in each lane. Gel electrophoresis, blotting, and cross linking were performed as described (Pall et al., 2007). Athila probes were generated by randomly degrading a 32P-labeled in vitro RNA transcript. PCR primers used to generate the probes are listed in Supplemental Table S1. The miR161 control probe was generated by 5′ end labeling the DNA oligonucleotide shown in Supplemental Table S1.
Transgene Construction and Expression
The Athila6-inducible expression transgene (ind-Athila) was produced using the primers listed in Supplemental Table S1 from ddm1 oligo(dT)-primed complementary DNA. This fragment was cloned into vector pMDC7. Sequencing the transgene identified the Athila6A element At5g32197 as the specific element that was incorporated into this transgene. The inducible RAN1 control transgene was constructed using the same pMDC7 vector and expresses a hemagglutinin-tagged version of the RAN1 (At5g20010) gene. Induction was achieved using the 20 μm estradiol spray from Borghi (2010) or mock induction using the same spray without the inclusion of estradiol. T1 plants for the ind-Athila transgene were sprayed as they transitioned to flowering and then were sprayed every day for 5 d or more before inflorescence tissue collection. Biological replicates consisted of three nonoverlapping pools of T1 sibling plants. Each biological replicate consisted of at least three individual T1 plants. Primers used for qRT-PCR and bisulfite sequencing are listed in Supplemental Table S1. Bisulfite amplification utilized one transgene-anchored primer to ensure the amplification of ind-Athila only and not other endogenous Athila6 elements.
Small RNA Library Production, Sequencing, and Analysis
Inflorescence small RNA was isolated with Trizol reagent (Invitrogen) and concentrated using the mirVana microRNA Isolation Kit (Ambion). Libraries were produced using the TruSeq Small RNA Sample Preparation Kit (Illumina) as recommended by the manufacturer. Each library was barcoded and sequenced in the same lane of the Illumina Genome Analyzer IIx. The resulting sequences were demultiplexed, adapter trimmed, and filtered on length and quality, and transfer RNA/ribosomal RNA and low complexity reads were removed. Small RNAs were matched to the Arabidopsis genome, and sequences that did not perfectly align were discarded. Library size is normalized by calculating reads per million of 18 to 28 nt genome-matched small RNAs. Small RNAs were also matched to The Arabidopsis Information Resource TAIR10 (www.arabidopsis.org) and Repbase (www.girinst.org) annotations of the TE portion of the Arabidopsis genome using bowtie. To best handle multimapping sequences generated from repetitive regions of the genome, the bowtie modifiers “−−best–M1−−strata” were employed (Treangen and Salzberg, 2012). If more than one genome perfect match for a TE siRNA exists, only one random match is assigned per small RNA read. Counts of multimapping siRNAs were not amplified using our approach, and we have not overestimated read number for high-copy TEs. Small RNA tracks and display of the data in Figure 2B were performed using the Integrated Genome Browser (Nicol et al., 2009). The raw sequencing and genome-matched small RNAs analyzed are available from the National Center for Biotechnology Information Gene Expression Omnibus repository under number GSE41755.
Analysis of TE Expression from Microarrays
Microarray analysis of gene expression in wild-type Col and rdr6 single mutant inflorescences was performed by Allen et al. (2005; GSE2473). A total of 1,155 TE probes have previously been identified on the ATH1 gene expression microarray (Slotkin et al., 2009). Fold change in TE expression was calculated by the National Center for Biotechnology Information GEO2R.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Start site of the Athila6 gag/pol transcript.
Supplemental Figure S2. Athila6 reestablishment of silencing occurs early in plant development.
Supplemental Table S1. PCR primers, probes, and mutant alleles used in this study.
Acknowledgments
We thank Sarah H. Reeder, Xiao Zhou, Gregory Booton, and Yao Wan for their contributions, reagents, and statistical advice.
Glossary
- TE
transposable element
- RdDM
RNA-directed DNA methylation
- Pol IV
RNA polymerase IV
- siRNA
small interfering RNA
- Pol V
RNA polymerase V
- LTR
long terminal repeat
- Pol II
RNA polymerase II
- RNAi
RNA interference
- TAS
transacting small interfering RNA
- Col
Columbia
- TSS
transcriptional start site
- qRT
quantitative reverse transcription
- nt
nucleotide
- tasiRNA
transacting small interfering RNA
References
- Allen E, Xie Z, Gustafson AM, Carrington JC. (2005) MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 [DOI] [PubMed] [Google Scholar]
- Aufsatz W, Mette MF, van der Winden J, Matzke AJM, Matzke M. (2002) RNA-directed DNA methylation in Arabidopsis. Proc Natl Acad Sci USA (Suppl 4) 99: 16499–16506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks JA, Masson P, Fedoroff N. (1988) Molecular mechanisms in the developmental regulation of the maize Suppressor-mutator transposable element. Genes Dev 2: 1364–1380 [DOI] [PubMed] [Google Scholar]
- Becker C, Hagmann J, Müller J, Koenig D, Stegle O, Borgwardt K, Weigel D. (2011) Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature 480: 245–249 [DOI] [PubMed] [Google Scholar]
- Borghi L. (2010) Inducible gene expression systems for plants. Methods Mol Biol 655: 65–75 [DOI] [PubMed] [Google Scholar]
- Calarco JP, Borges F, Donoghue MTA, Van Ex F, Jullien PE, Lopes T, Gardner R, Berger F, Feijó JA, Becker JD, et al. (2012) Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell 151: 194–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SW-L, Zilberman D, Xie Z, Johansen LK, Carrington JC, Jacobsen SE. (2004) RNA silencing genes control de novo DNA methylation. Science 303: 1336. [DOI] [PubMed] [Google Scholar]
- Chen X. (2010) Small RNAs: secrets and surprises of the genome. Plant J 61: 941–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung W-J, Okamura K, Martin R, Lai EC. (2008) Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr Biol 18: 795–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao X, Freeling M, Lisch D. (2006) Horizontal transfer of a plant transposon. PLoS Biol 4: e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eamens A, Vaistij FE, Jones L. (2008) NRPD1a and NRPD1b are required to maintain post-transcriptional RNA silencing and RNA-directed DNA methylation in Arabidopsis. Plant J 55: 596–606 [DOI] [PubMed] [Google Scholar]
- Garcia D, Garcia S, Pontier D, Marchais A, Renou JP, Lagrange T, Voinnet O. (2012) Ago hook and RNA helicase motifs underpin dual roles for SDE3 in antiviral defense and silencing of nonconserved intergenic regions. Mol Cell 48: 109–120 [DOI] [PubMed] [Google Scholar]
- Gendrel A-V, Lippman Z, Yordan C, Colot V, Martienssen RA. (2002) Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297: 1871–1873 [DOI] [PubMed] [Google Scholar]
- Girard A, Hannon GJ. (2008) Conserved themes in small-RNA-mediated transposon control. Trends Cell Biol 18: 136–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg MVC, Ausin I, Chan SW-L, Cokus SJ, Cuperus JT, Feng S, Law JA, Chu C, Pellegrini M, Carrington JC, et al. (2011) Identification of genes required for de novo DNA methylation in Arabidopsis. Epigenetics 6: 344–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruntman E, Qi Y, Slotkin RK, Roeder T, Martienssen RA, Sachidanandam R. (2008) Kismeth: analyzer of plant methylation states through bisulfite sequencing. BMC Bioinformatics 9: 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag JR, Pikaard CS. (2011) Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat Rev Mol Cell Biol 12: 483–492 [DOI] [PubMed] [Google Scholar]
- Havecker ER, Wallbridge LM, Hardcastle TJ, Bush MS, Kelly KA, Dunn RM, Schwach F, Doonan JH, Baulcombe DC. (2010) The Arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell 22: 321–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Deleris A, Wong W, Zhong X, Chin HG, Horwitz GA, Kelly KA, Pradhan S, Jacobsen SE. (2010) The de novo cytosine methyltransferase DRM2 requires intact UBA domains and a catalytically mutated paralog DRM3 during RNA-directed DNA methylation in Arabidopsis thaliana. PLoS Genet 6: e1001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. (2005) RNA polymerase IV directs silencing of endogenous DNA. Science 308: 118–120 [DOI] [PubMed] [Google Scholar]
- Huettel B, Kanno T, Daxinger L, Aufsatz W, Matzke AJM, Matzke M. (2006) Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J 25: 2828–2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra CA, Feng X, Schoft VK, Hsieh TF, Uzawa R, Rodrigues JA, Zemach A, Chumak N, Machlicova A, Nishimura T, et al. (2012) Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science 337: 1360–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Gaubert H, Bucher E, Mirouze M, Vaillant I, Paszkowski J. (2011) An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472: 115–119 [DOI] [PubMed] [Google Scholar]
- Jensen S, Gassama MP, Heidmann T. (1999) Taming of transposable elements by homology-dependent gene silencing. Nat Genet 21: 209–212 [DOI] [PubMed] [Google Scholar]
- Kakutani T, Munakata K, Richards EJ, Hirochika H. (1999) Meiotically and mitotically stable inheritance of DNA hypomethylation induced by ddm1 mutation of Arabidopsis thaliana. Genetics 151: 831–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Carrington JC. (2007) Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol 5: e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Miura A, Bender J, Jacobsen SE, Kakutani T. (2003) Role of CG and non-CG methylation in immobilization of transposons in Arabidopsis. Curr Biol 13: 421–426 [DOI] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11: 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z, Gendrel A-V, Black M, Vaughn MW, Dedhia N, McCombie WR, Lavine K, Mittal V, May B, Kasschau KD, et al. (2004) Role of transposable elements in heterochromatin and epigenetic control. Nature 430: 471–476 [DOI] [PubMed] [Google Scholar]
- Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. (2008) Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu O, Reinders J, Caikovski M, Smathajitt C, Paszkowski J. (2007) Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell 130: 851–862 [DOI] [PubMed] [Google Scholar]
- McCue AD, Nuthikattu S, Reeder SH, Slotkin RK. (2012) Gene expression and stress response mediated by the epigenetic regulation of a transposable element small RNA. PLoS Genet 8: e1002474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirouze M, Reinders J, Bucher E, Nishimura T, Schneeberger K, Ossowski S, Cao J, Weigel D, Paszkowski J, Mathieu O. (2009) Selective epigenetic control of retrotransposition in Arabidopsis. Nature 461: 427–430 [DOI] [PubMed] [Google Scholar]
- Nicol JW, Helt GA, Blanchard SG, Jr, Raja A, Loraine AE. (2009) The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics 25: 2730–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pall GS, Codony-Servat C, Byrne J, Ritchie L, Hamilton A. (2007) Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Res 35: e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontier D, Picart C, Roudier F, Garcia D, Lahmy S, Azevedo J, Alart E, Laudié M, Karlowski WM, Cooke R, et al. (2012) NERD, a plant-specific GW protein, defines an additional RNAi-dependent chromatin-based pathway in Arabidopsis. Mol Cell 48: 121–132 [DOI] [PubMed] [Google Scholar]
- Roulin A, Piegu B, Wing RA, Panaud O. (2008) Evidence of multiple horizontal transfers of the long terminal repeat retrotransposon RIRE1 within the genus Oryza. Plant J 53: 950–959 [DOI] [PubMed] [Google Scholar]
- Saze H, Kakutani T. (2007) Heritable epigenetic mutation of a transposon-flanked Arabidopsis gene due to lack of the chromatin-remodeling factor DDM1. EMBO J 26: 3641–3652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoft VK, Chumak N, Mosiolek M, Slusarz L, Komnenovic V, Brownfield L, Twell D, Kakutani T, Tamaru H. (2009) Induction of RNA-directed DNA methylation upon decondensation of constitutive heterochromatin. EMBO Rep 10: 1015–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Plasterk RHA. (2003) Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature 426: 310–314 [DOI] [PubMed] [Google Scholar]
- Slotkin RK. (2010) The epigenetic control of the Athila family of retrotransposons in Arabidopsis. Epigenetics 5: 483–490 [DOI] [PubMed] [Google Scholar]
- Slotkin RK, Freeling M, Lisch D. (2005) Heritable transposon silencing initiated by a naturally occurring transposon inverted duplication. Nat Genet 37: 641–644 [DOI] [PubMed] [Google Scholar]
- Slotkin RK, Vaughn M, Borges F, Tanurdzić M, Becker JD, Feijó JA, Martienssen RA. (2009) Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136: 461–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood SA, Kelsey G. (2012) De novo DNA methylation: a germ cell perspective. Trends Genet 28: 33–42 [DOI] [PubMed] [Google Scholar]
- Stroud H, Greenberg MVC, Feng S, Bernatavichute YV, Jacobsen SE. (2013) Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152: 352–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira FK, Heredia F, Sarazin A, Roudier F, Boccara M, Ciaudo C, Cruaud C, Poulain J, Berdasco M, Fraga MF, et al. (2009) A role for RNAi in the selective correction of DNA methylation defects. Science 323: 1600–1604 [DOI] [PubMed] [Google Scholar]
- Treangen TJ, Salzberg SL. (2012) Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Rev Genet 13: 36–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT, Cocklin R, Mayampurath A, Lister R, Rowley MJ, Gregory BD, Ecker JR, Tang H, Pikaard CS. (2012) Spatial and functional relationships among Pol V-associated loci, Pol IV-dependent siRNAs, and cytosine methylation in the Arabidopsis epigenome. Genes Dev 26: 1825–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Mao L, Qi Y. (2012) Roles of dicer-like and argonaute proteins in TAS-derived small interfering RNA-triggered DNA methylation. Plant Physiol 160: 990–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Zhou H, Zhang Q, Zhang J, Ni F, Liu C, Qi Y. (2010) DNA methylation mediated by a microRNA pathway. Mol Cell 38: 465–475 [DOI] [PubMed] [Google Scholar]
- Zheng B, Wang Z, Li S, Yu B, Liu J-Y, Chen X. (2009) Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes Dev 23: 2850–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]