Altering the expression of a rice transcription factor regulates chloroplast development and modifies plant size, tiller number, and grain production.
Abstract
Chloroplast biogenesis has been well documented in higher plants, yet the complex methods used to regulate chloroplast activity under fluctuating environmental conditions are not well understood. In rice (Oryza sativa), the CYTOKININ-RESPONSIVE GATA TRANSCRIPTION FACTOR1 (Cga1) shows increased expression following light, nitrogen, and cytokinin treatments, while darkness and gibberellin reduce expression. Strong overexpression of Cga1 produces dark green, semidwarf plants with reduced tillering, whereas RNA interference knockdown results in reduced chlorophyll and increased tillering. Coexpression, microarray, and real-time expression analyses demonstrate a correlation between Cga1 expression and the expression of important nucleus-encoded, chloroplast-localized genes. Constitutive Cga1 overexpression increases both chloroplast biogenesis and starch production but also results in delayed senescence and reduced grain filling. Growing the transgenic lines under different nitrogen regimes indicates potential agricultural applications for Cga1, including manipulation of biomass, chlorophyll/chloroplast content, and harvest index. These results indicate a conserved mechanism by which Cga1 regulates chloroplast development in higher plants.
Chloroplast evolution transformed the earth by increasing atmospheric oxygen and providing a vital energy source to support higher life (Gould et al., 2008). The endosymbiotic incorporation of the chloroplast is perhaps one of the most significant evolutionary events in history. Genome sequences of cyanobacteria and Arabidopsis (Arabidopsis thaliana) leave little doubt that plant chloroplasts originated from a cyanobacterium (Raven and Allen, 2003). Since that time, there has been an intricate exchange of DNA between the chloroplast and plant nucleus, resulting in a highly complex system regulating chloroplast development and activity (Raven and Allen, 2003; Gould et al., 2008). The molecular aspects involved in chloroplast biogenesis (Kessler and Schnell, 2009; Okazaki et al., 2010; Pogson and Albrecht, 2011), chlorophyll biosynthesis (Eckhardt et al., 2004; Tanaka and Tanaka, 2007; Reinbothe et al., 2010), photosynthesis (Laisk et al., 2009), and carbon metabolism (Stitt et al., 2010) have been quite well documented in recent decades. However, plants rely on their immediate environment for the acquisition of resources (e.g. light, water, nutrients) and must be able to adjust chloroplast activity according to their availability. Due to this complexity, our understanding of how plants regulate chloroplast development with fluctuating environmental conditions is lacking.
While increases in food production have permitted global population expansion over the past century, further increases are necessary in order to sustain projected future growth (Tilman et al., 2002). Escalating food consumption rates combined with environmental issues are placing pressure on both farmers and scientists to increase yields in a sustainable fashion. Understanding how chloroplast activity relates to environmental conditions is crucial for agricultural production. In plants, carbon capture is largely limited by nitrogen (N) availability (Lawlor, 2002). N fertilizer is the most essential element determining crop production, directly influencing chlorophyll content, biomass, and yield (Lawlor, 2002; Tilman et al., 2002). However, the fertilization process comes with significant economic and environmental costs (Tilman et al., 2002). In C3 plants, the majority of assimilated N is found in the chloroplast, where it is invested in the photosynthetic machinery and, in particular, the carbon incorporation enzyme Rubisco (Nunes-Nesi et al., 2010). The inefficiency of Rubisco in accepting either carbon dioxide or oxygen as a substrate provides an opportunity to increase C3 photosynthesis and yield (Evans and von Caemmerer, 2011; Raines, 2011). Increasing CO2 fixation in crops will require detailed understanding of plant carbon-N balance and must also involve improvements in both N use efficiency and water use efficiency (Lawlor, 2002; Nunes-Nesi et al., 2010; Raines, 2011).
Rice (Oryza sativa) is a staple cereal crop for more than one-half of the world’s population. The short stature and sturdy stalks of the semidwarf “Green Revolution” varieties of rice provided resistance to flattening by wind or rain (Peng et al., 1999; Evenson and Gollin, 2003). Furthermore, these cultivars were resistant to lodging and able to utilize greater amounts of N fertilizer and increase yields (Peng et al., 1999; Evenson and Gollin, 2003). The mutations leading to these phenotypes in rice were found to be in genes responsible for the biosynthesis of active GAs, an important class of phytohormone (Peng et al., 1999; Sasaki et al., 2002). GA controls many important aspects of development, including seed germination, stem elongation, and flowering (Greenboim-Wainberg et al., 2005). Evidence indicates that there is significant cross talk between GA signaling and another class of phytohormone, cytokinins (Gan et al., 2007; Weiss and Ori, 2007; Steiner et al., 2012). Cytokinins are key regulators of plant growth and development, including cell division, chloroplast biogenesis, differentiation, stress tolerance, and organ senescence (Argueso et al., 2010). Nitrate signaling is intrinsically linked to cytokinin, influencing both cytokinin synthesis and transport (Sakakibara et al., 2006; Hirose et al., 2008). Furthermore, cytokinin signaling mediates many of the effects of applied N and can mimic these responses in the absence of N (Sakakibara, 2003; Sakakibara et al., 2006). Both cytokinin application and the modification of genes involved in cytokinin signaling influence chloroplast development, rice plant architecture, and yield (Ashikari et al., 2005; Hirose et al., 2007). As such, understanding the links between N, cytokinin, and GA signaling is also important for making improvements in agricultural crops.
The CYTOKININ-RESPONSIVE GATA TRANSCRIPTION FACTOR1/GNC-like (CGA1/GNL) transcription factor was originally identified in Arabidopsis (At4g26150) due to rapidly increased expression following cytokinin application and similarity to a paralogous gene produced through genome duplication (Reyes et al., 2004; Kiba et al., 2005; Naito et al., 2007; Mara and Irish, 2008). The GATA family of transcription factors exhibits a significant degree of conservation between the dicot model organism Arabidopsis and rice, a monocot cereal with many member-retaining paralogs following genome duplication events (Reyes et al., 2004). In Arabidopsis, the expression patterns of Arabidopsis CGA1 as well as the paralogous transcription factor GATA, NITRATE-INDUCIBLE, CARBON METABOLISM-INVOVLED (GNC) have been well documented in recent years (Bi et al., 2005; Manfield et al., 2007; Naito et al., 2007; Mara and Irish, 2008; Richter et al., 2010; Hudson et al., 2011). Both are primarily expressed in green tissues and follow circadian-regulated expression patterns (Manfield et al., 2007; Naito et al., 2007; Mara and Irish, 2008). Light, N, and cytokinin all act to increase the expression CGA1, while both darkness and GA have also been shown to significantly reduce expression (Naito et al., 2007; Richter et al., 2010). Transgenic Arabidopsis plants with altered expression of CGA1 have been shown to exhibit differences in germination, chlorophyll content, chloroplast number, leaf size, flowering time, and senescence (Mara and Irish, 2008; Richter et al., 2010; Hudson et al., 2011). This includes recent reports showing that ectopic overexpression promotes chloroplast biogenesis in cells where they are not typically found (Köllmer et al., 2011; Chiang et al., 2012). These data indicate that GNC and CGA1 function as key transcriptional regulators of chloroplast biogenesis in Arabidopsis.
In this work, we demonstrate that the conserved GATA transcription factor Cga1 (Os02g12790) regulates chloroplast development and plant architecture in rice (Oryza sativa). Rice Cga1 expression shows a similar expression pattern to the Arabidopsis ortholog. Transgenic rice with altered expression of Cga1 exhibits differences in chlorophyll, chloroplast number, and starch content, which has also been reported in Arabidopsis. However, we also observed a dosage-dependent influence on phenotype, with strong overexpression causing a semidwarf phenotype, similar to the GA mutant Green Revolution varieties. We present novel evidence that altering Cga1 expression in rice significantly influences tillering, biomass, and yield. We demonstrate changes in the expression of important nucleus-encoded, chloroplast-localized genes involved in chlorophyll binding, photosynthesis, and amino acid and starch biosynthesis in the Cga1 transgenics. Altering expression of the rice homolog to the FILAMENTOUS TEMPERATURE SENSITIVE-Z (FtsZ) gene involved in chloroplast division provides a potential mechanism for controlling chloroplast number. Growing the transgenic lines under different N conditions indicates that Cga1 is able to maintain chloroplast development under reduced N conditions, leading to an increased harvest index despite reduced plant size.
RESULTS
Expression of Cga1 in Rice
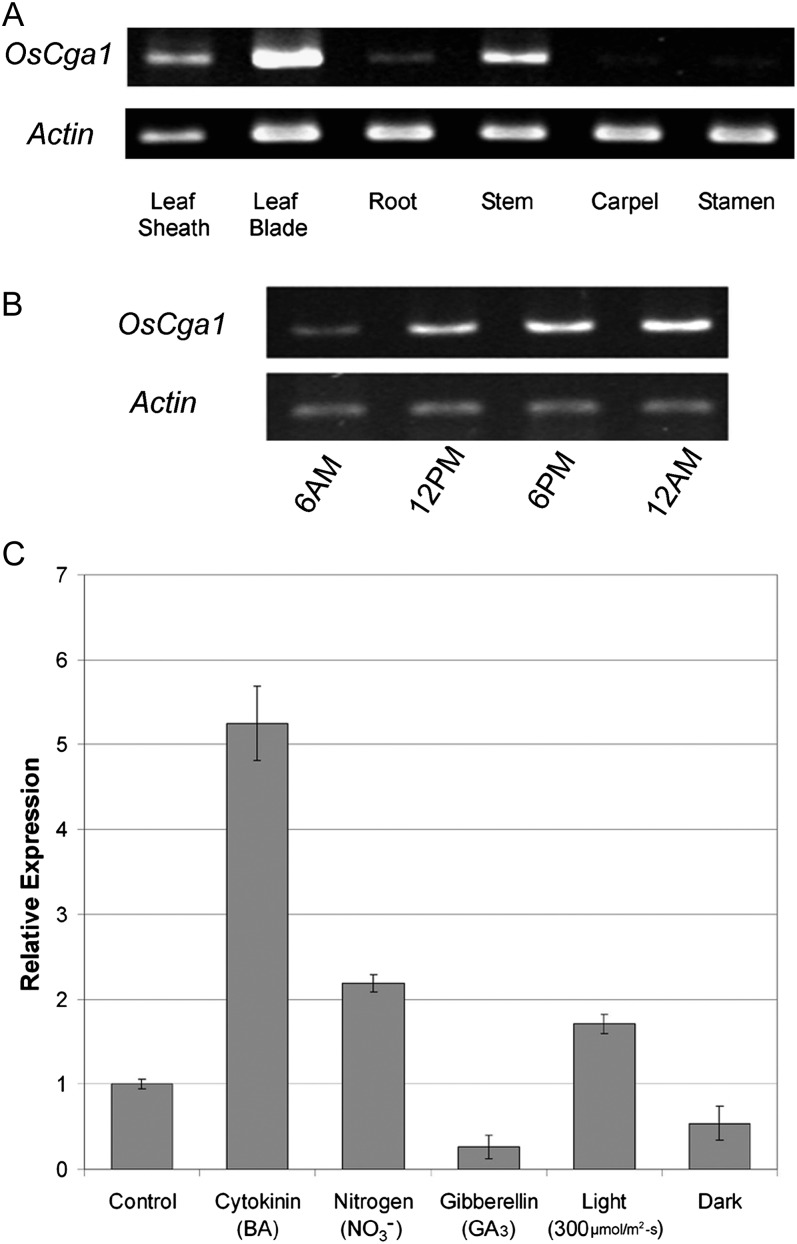
We analyzed rice tissues and established patterns of expression for Cga1 in wild-type Kaybonnet rice. Cga1 exhibited the strongest expression in green leaf tissue, with little and no expression in roots and floral organs, respectively (Fig. 1A). In Arabidopsis, it was reported that the floral homeotic transcription factors APETALA3 and PISTILLATA directly inhibit AtCGA1 transcription in floral organs (Mara and Irish, 2008). Furthermore, it was theorized that inhibition of AtCGA1 expression may be required in order to prevent chlorophyll biosynthesis in nongreen tissues (Mara and Irish, 2008; Hudson et al., 2011). Significant expression of rice Cga1 in chloroplast-containing tissues potentially indicates a conserved role in regulating chloroplast development.
Figure 1.
Expression of rice Cga1 (Os02g12790). A, Expression of Cga1 in selected rice tissues. B, Expression of Cga1 at 6-h intervals throughout the course of the day (long day; 16 h of light). C, Real-time PCR of OsCga1 expression levels following periods of darkness or light and following treatment with 10 mm nitrate, 10 µmol of BA cytokinin, or 10 µmol of GA3.
We also analyzed the expression of Cga1 following a number of treatments. Previous work indicated that rice Cga1 expression follows a circadian oscillation pattern in rice (Filichkin et al., 2011). We also observed differences in Cga1 expression throughout the course of the day (Fig. 1B). Light was found to significantly increase Cga1 expression, whereas periods of darkness reduced expression (Fig. 1, B and C). Cga1 was highly up-regulated (approximately 5-fold) by the synthetic cytokinin benzyladenine (BA; Fig. 1C). The gene was originally named based on its rapidly induced expression following cytokinin application in Arabidopsis (Kiba et al., 2005; Naito et al., 2007), and this also seems to apply in rice. Nitrate (NO3−) also significantly increased Cga1 expression, although to a lesser extent than BA (Fig. 1C). GA application was found to reduce Cga1 expression, indicating a role in regulating the cross talk between cytokinin and GA. Control over the expression of rice Cga1 is highly similar to results obtained in the Arabidopsis ortholog, which indicates conservation between the dicot model and a moncot crop species.
Transgenic Modification of Cga1 Expression Alters Plant Architecture and Chloroplast Development
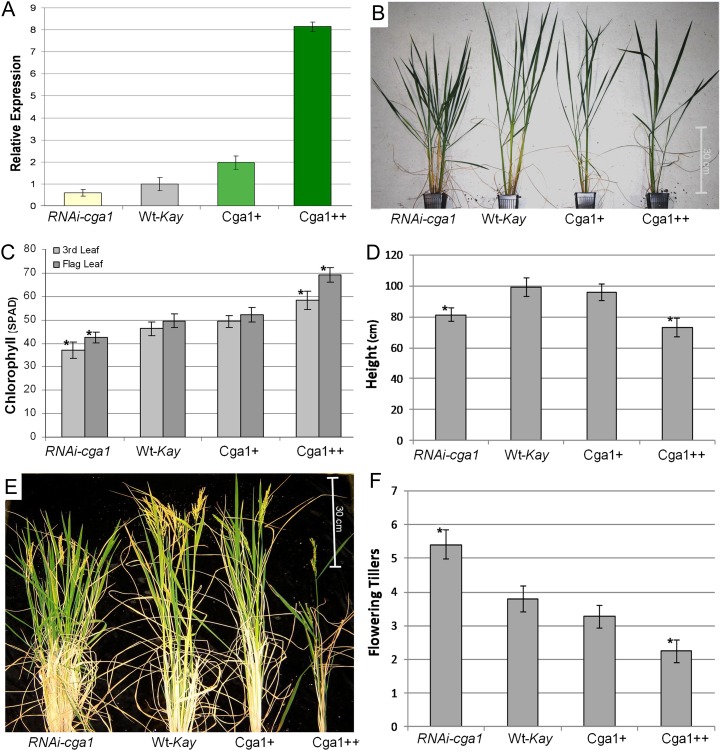
We created transgenic rice Kaybonnet lines with modified Cga1 expression. Multiple transgenic RNA interference (RNAi)-cga1 lines were created showing reduced expression. All RNAi lines showed similar phenotypes, including reduced chlorophyll, reduced height, and increased tillering (Supplemental Fig. S1). The RNAi lines selected for comparative analysis had around 70% expression of Cga1 compared with the wild type (Fig. 2A). We also analyzed two unique overexpression lines with different amounts of Cga1 expression. One of these lines exhibited a moderate increase in expression (Cga1+; approximately 2-fold), while the other line (Cga1++) showed very strong overexpression, with around 8-fold more transcript than wild-type controls (Fig. 2A). The moderate overexpression line showed only a slight reduction in size and appeared quite similar to wild-type controls (Fig. 2B). As seen in Arabidopsis, altered expression of rice Cga1 resulted in significant differences in chlorophyll content (Fig. 2C). We found this to occur in a dose-dependent fashion, with strong overexpression drastically increasing chlorophyll throughout development, whereas moderate overexpression results in only a marginal increase and is not significantly different from wild-type controls (Fig. 2C). Differences in plant architecture were also evident in the transgenic lines (Fig. 2D). At maturity, all the transgenic lines show reductions in overall plant height compared with wild-type plants (Fig. 2, D and E). Reduced Cga1 expression significantly increased the production of flowering tillers, mild overexpression plants largely resembled wild-type controls, while strong overexpression resulted in a semidwarf phenotype with reduced tiller production (Fig. 2, D and F).
Figure 2.
Transgenic modification to Cga1 expression alters chlorophyll content and plant architecture. A, Expression of Cga1 in the selected transgenic lines compared with wild-type Kaybonnet controls (Wt-Kay) measured using qRT-PCR. B, Preflowering transgenic OsCga1 lines at 60 d after germination exhibit differences in plant architecture. C, Chlorophyll at young (third leaf) and late (flag leaf) stages of development (n = 36+; *P < 0.05). D, Height of mature transgenic plants (n = 40; *P > 0.05). E, Mature transgenic plants (150 d after germination) compared with the wild type. F, Number of flowering tillers (n = 80; *P < 0.05). All data are means ± sd.
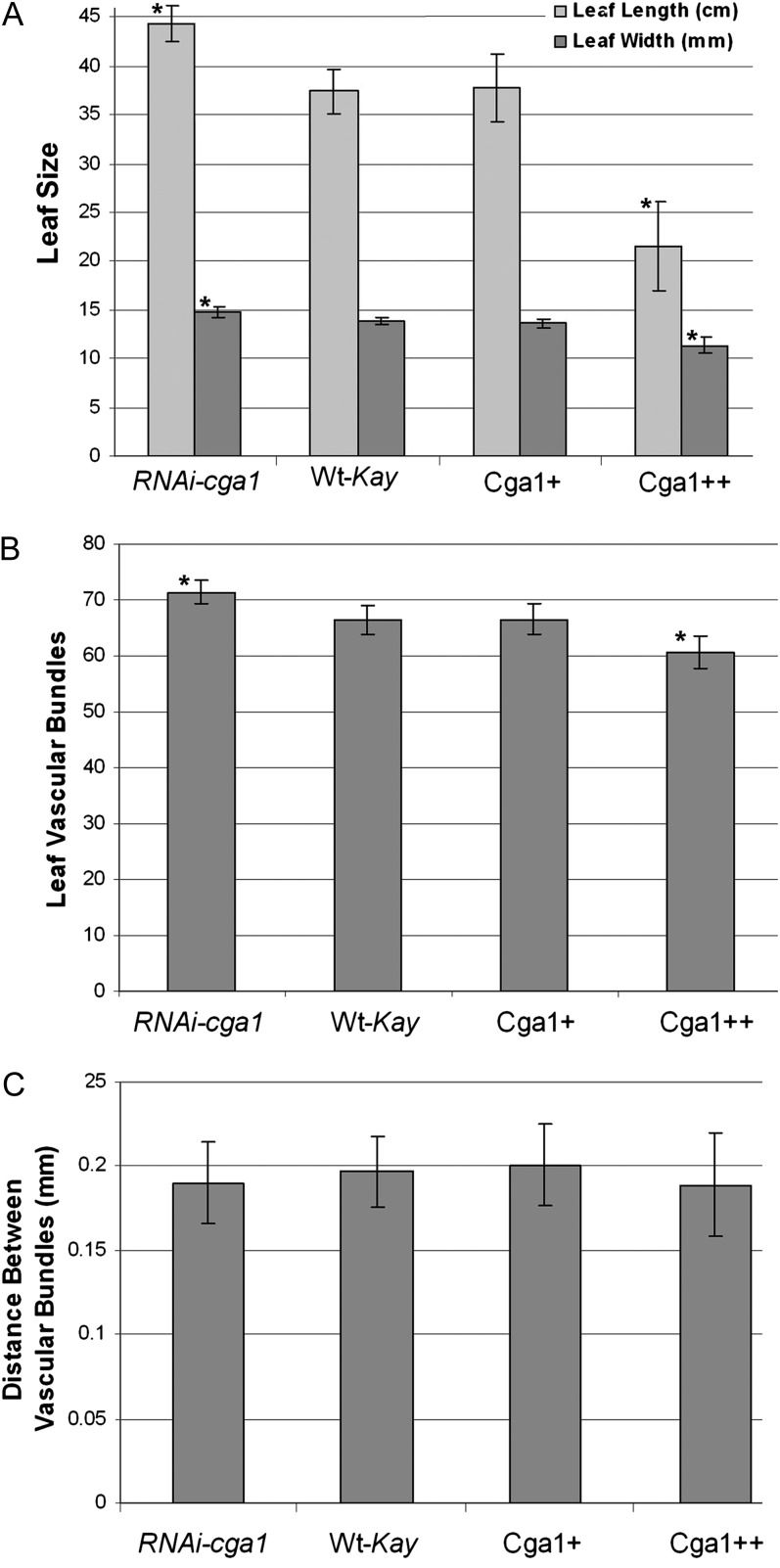
Despite overall reductions in plant height, the length and width of leaf blades were found to be influenced by Cga1 expression (Fig. 3A). Leaf size was also reported to be different in Arabidopsis plants with altered CGA1 expression (Hudson et al., 2011). When light is limiting, plants naturally expand their leaves, whereas leaves with high photosynthetic activity are typically smaller and show decreased cell expansion (Rahim and Fordham, 1991). Differences in leaf size can be indicative of altered cell expansion, yet microscopic examination of the leaves of Cga1 transgenics did not indicate this to be the case. When considering the entire leaf, RNAi-cga1 resulted in a greater number of vascular bundles spanning the width of the leaf compared with wild-type controls, while strong overexpression caused a significant reduction (Fig. 3B). Despite this, the distance between the vascular bundles was not found to be significantly different in the transgenic lines (Fig. 3C). This demonstrates increased leaf development and indicates altered production of the total number of cells within each leaf rather than differences in leaf cell expansion.
Figure 3.
Cga1 expression alters leaf development. A, Length and width of leaves from Cga1 transgenic lines compared with wild-type Kaybonnet controls (Wt-Kay; n = 40; *P < 0.05). B, Number of vascular bundles spanning the width of the leaf blade (n = 12; *P < 0.05). C, Distance between vascular bundles (n = 12; *P > 0.05).
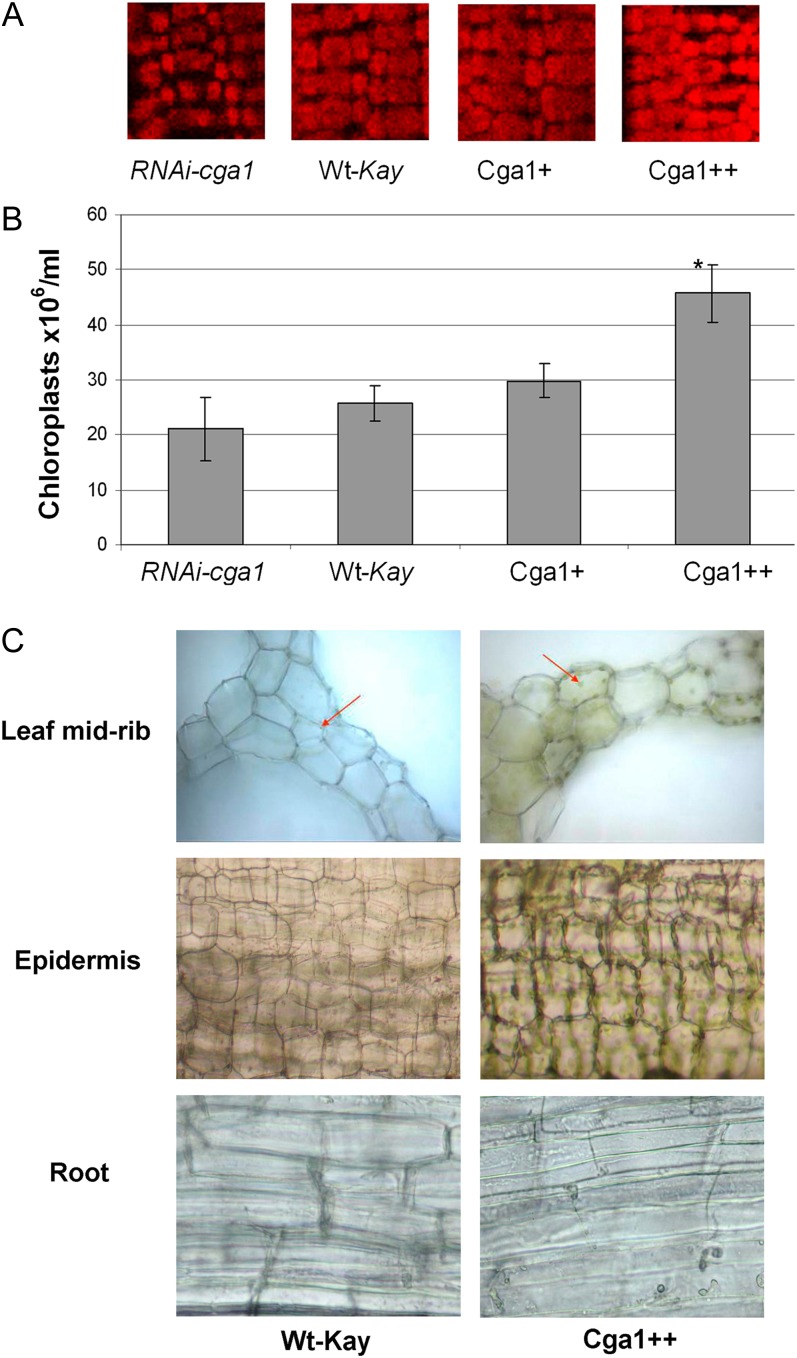
Confocal images of the leaf blade measuring chlorophyll autofluorescence of mesophyll cells demonstrate obvious differences in chloroplast development between the RNAi-cga1 and Cga1++ transgenic lines compared with the wild type (Fig. 4A). The density of chloroplasts in these regions makes it difficult to obtain quantifiable numbers of mesophyll cell chloroplasts using microscopy. In Arabidopsis, we found that the increased chlorophyll in the AtCGA1 transgenics was largely the result of an overall difference in chloroplast number (Hudson et al., 2011). This was also the case for the rice transgenics. Leaf extractions confirmed differences in the overall number of chloroplasts (Fig. 4B). Most notably, the strong Cga1++ overexpression line showed nearly a 2-fold increase in leaf chloroplasts. Analysis of regions lacking chloroplasts in wild-type plants provides further evidence for the regulation of chloroplast development (Fig. 4C). Internal cells in the midvein of the leaf from Cga1++ plants contain significant chloroplasts in comparison with the wild type (Fig. 4C). Epidermal tissues of the leaf sheath taken near the base of the plant also indicate increased chloroplast development with Cga1 overexpression (Fig. 4C). Furthermore, aberrant chloroplast development in root tissue of the Cga1++ plants indicates that high levels of Cga1 are sufficient to induce chloroplast production in cells where they are not normally found (Fig. 4C). These results imply that Cga1 functions to regulate chloroplast development and to alter aspects of plant architecture in rice.
Figure 4.
Cga1 increases chloroplast development, A, Confocal microscopy of leaf blade from the Cga1 transgenics compared with wild-type Kaybonnet controls (Wt-Kay). B, Chloroplast number counted using a hemocytometer (n = 40+; *P < 0.05). C, Light microscopy showing increased chloroplast development in regions where they are not prevalent in wild-type plants. In the structural cells of the leaf midrib (top), the red arrows point to an immature etioplast in the wild type and a fully developed chloroplast in Cga1++. Epidermal cells of the leaf sheath (middle) sampled 1 cm from the base of the plant demonstrate increased chloroplast development in Cga1++. Root cells (bottom) also display aberrant chloroplast development in Cga1++ plants.
Cga1 Modifies the Expression of Important Chloroplast-Localized Genes
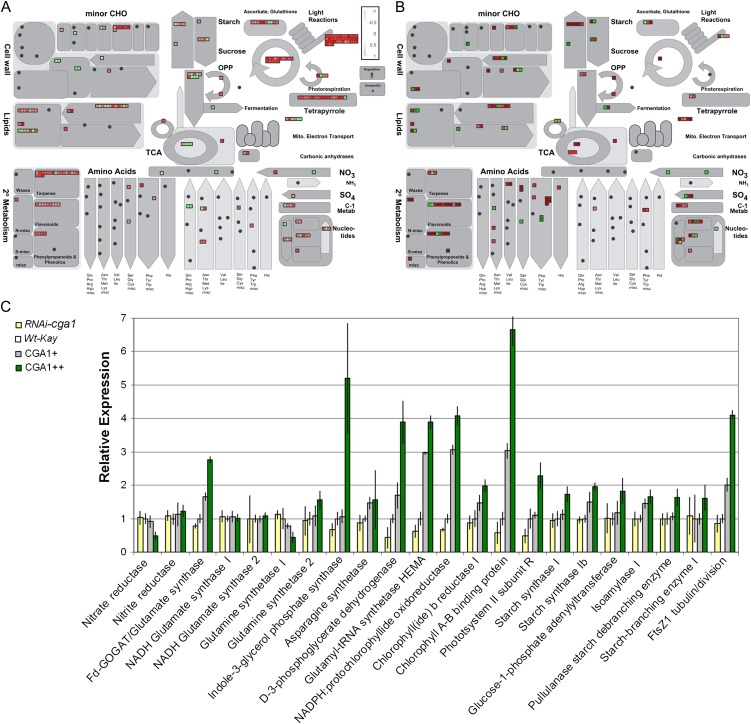
Coexpression analysis of Cga1 was performed using the RiceArray coexpression platform (http://www.ricearray.org/coexpression/coexpression.shtml). We plotted the output (Supplemental Data S1) using MapMan software (mapman.gabipd.org), which demonstrates that Cga1 displays similar patterns of expression as important chloroplast, photosynthesis, and carbon metabolism genes (Fig. 5A). Genes involved in amino acid synthesis and secondary metabolism also showed significant coexpression with Cga1 (Fig. 5A). We performed microarray analysis comparing the transcript profiles of the strong Cga1++ overexpression line and wild-type controls (Supplemental Data S2). Several genes up-regulated in the microarray shared a high level of coexpression with Cga1 (Fig. 5B). The expression of many interesting candidate genes in the Cga1 transgenic lines was analyzed using quantitative real-time PCR (Fig. 5C). Ferredoxin-dependent (Fd) GOGAT controls the initial stage of chloroplast N assimilation and, in Arabidopsis, was found to be modulated by Cga1 expression (Hudson et al., 2011). Corroborating this, rice Fd-GOGAT is reduced in the RNAi-cga1 plants and increased in the Cga1 overexpression lines (Fig. 5C). Mutation of rice Fd-GOGAT has been shown to cause a pale green phenotype similar to the RNAi-cga1 plants (Jung et al., 2008b). The chloroplast-localized Gln synthetase (GS) was also found to be increased in the strong Cga1++ overexpression line. In contrast, most nonchloroplast genes upstream in the N assimilation process were found to be expressed at similar levels to controls (Fig. 5C). Nitrate reductase and cytosolic GS actually showed a slight inverse relationship to Cga1 expression (Fig. 5C). In Arabidopsis, similar regulation over N and chlorophyll-related genes was demonstrated. Regulation of GS/Fd-GOGAT without altering N assimilation upstream of the chloroplast indicates specific control over chloroplast processes. Asn synthetase, indole-3-glycerol phosphate synthase, and D3 phosphoglycerate dehydrogenase, involved in the synthesis of N-containing amino acids (Asn, Tyr, and Ser, respectively), were also found to be increased with Cga1 expression (Fig. 5C). Again, similar to results obtained in Arabidopsis (Richter et al., 2010; Hudson et al., 2011), the expression of key chlorophyll biosynthesis genes (Glu-transfer RNA reductase, Protochlorophyllide oxidoreductase, Chlorophyllide b reductase) was altered in the rice Cga1 transgenics (Fig. 5C), which correlates to phenotypic analysis (Fig. 2). Conserved regulation of these genes indicates a role for CGA1 in directing assimilated N toward chlorophyll biosynthesis.
Figure 5.
Potential targets of Cga1 revealed by coexpression, microarray, and qRT-PCR of transgenic lines. A, Coexpression analysis of Cga1 (Os02g12790) presented using MapMan. B, Microarray analysis of Cga1++ versus wild-type Kaybonnet controls presented using MapMan. C, Real-time quantitative PCR confirms the altered expression of genes involved in chloroplast N assimilation, N amino acid synthesis, chlorophyll biosynthesis, chlorophyll binding, photosynthesis, starch biosynthesis, and chloroplast division (mean ± sd). Wt-Kay, Wild-type Kaybonnet controls.
While differences in chlorophyll and starch content were reported in Arabidopsis, we did not previously analyze chloroplast genes beyond N assimilation and chlorophyll biosynthesis. Microarray analysis in rice indicated a much broader regulation over chloroplast-localized processes than previously thought. Free chlorophyll is known to photooxidatively damage cells (Kusaba et al., 2007), yet we found a chlorophyll-binding gene to be enhanced with Cga1 overexpression (Fig. 5, B and C). We also found at least one photosynthesis machinery gene (PSII subunit R) to also be significantly altered in the Cga1 transgenics (Fig. 5, B and C). Furthermore, a number of starch biosynthesis genes (starch synthases, Glc-1-P adenyltransferase, isoamylase, starch-branching enzymes) were found to be increased with Cga1 overexpression, although most did not show any reduction in the RNAi-cga1 line (Fig. 5C). Still, regulation of these genes indicates that chlorophyll is being incorporated into a functional photosynthetic apparatus in the chloroplast in the Cga1 overexpression line. These results confirm a broader regulation of chloroplast-localized gene expression by Cga1, including genes involved in carbon metabolism.
While these genes might be expected to be increased in plants that contain more chloroplasts, they do not account for the increased chloroplast phenotype. In Arabidopsis, we analyzed the expression of chloroplast division factors known to alter chloroplast number, including plastid division and accumulation and replication of cholorplast genes (Miyagishima et al., 2006; Glynn et al., 2008; Okazaki et al., 2009). However, these genes are not expressed in a similar fashion (Jen et al., 2006) and were not found to be significantly altered in Arabidopsis CGA1 transgenic lines (Hudson et al., 2011). Instead, both Arabidopsis and rice show significant coexpression with tubulin-like Fts proteins found in the chloroplast thylakoid membrane that show homology to bacterial cell division genes (Jen et al., 2006; Jung et al., 2008a; Karamoko et al., 2011). Chloroplast division in plant cells requires the coordinated action of the tubulin-like FtsZ ring inside the chloroplast and the dynamin-like ring outside (Osteryoung et al., 1998; Osteryoung and McAndrew, 2001; Liu et al., 2010b; Pogson and Albrecht, 2011). The rice FtsZ gene (Os04g56970) is predicted to be involved in chloroplast division and was found to be significantly regulated by Cga1 expression in both microarray and quantitative reverse transcription (qRT)-PCR (Fig. 5C). Cga1-regulated modification of FtsZ provides a potential mechanism for increasing chloroplast number in response to the amounts of light and N perceived by the plant. While genetic manipulations of both ring systems have been shown to alter chloroplast number, the ability of Cga1 to influence chloroplast biogenesis without significantly inhibiting chloroplast function could have significant implications with respect to carbon fixation.
Cga1 Expression Influences Starch Production, Panicle Development, and Grain Filling
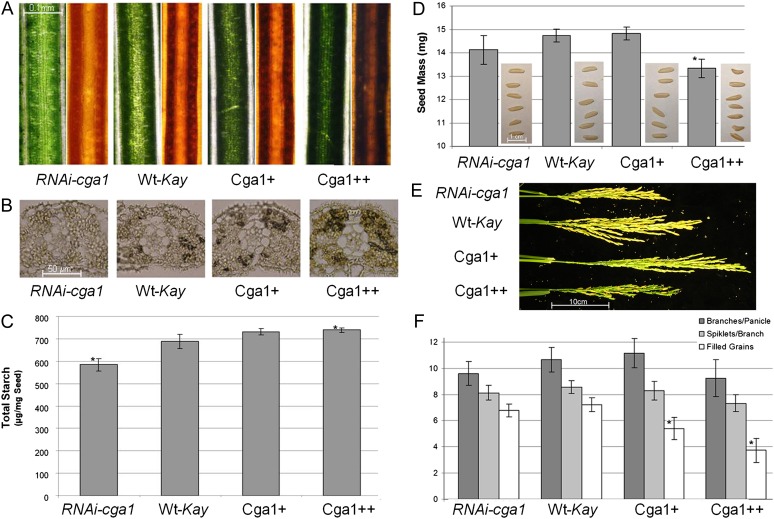
Differences in the expression of genes involved in photosynthesis and starch production indicate potential differences in carbon assimilation with altered Cga1 expression. Starch is the major carbohydrate reservoir in plants and is found in granular form within chloroplasts. Along with increased starch gene expression in the Cga1 overexpression lines, we found starch production to be significantly different in the Cga1 transgenic lines (Fig. 6). Visible differences in chlorophyll observed in fresh leaf blade tissue corresponded to differences in leaf starch following staining with Lugol’s iodine (IKI; Fig. 5A). Microtome sections of wax-embedded leaf tissue also demonstrate differences in the production of starch granules in the transgenic lines (Fig. 6B). While these sections again indicate similar cellular size and development, the increase in chloroplast number was notable in the strong Cga1 overexpression line following wax embedding and sectioning (Fig. 6B). Ethanol dehydration/rehydration series typically render tissues relatively clear of pigments, yet sections from the Cga1++ line retain a slightly green color, and mesophyll cells are filled with chloroplasts (Fig. 6B).
Figure 6.
Cga1 expression influences starch content and grain production. A, Light microscopy of leaf blades showing fresh samples (right) and following IKI staining (left) for starch. B, Wax-embedded leaf tissue sections (12 µm) stained with IKI. C, Seed starch measured using the Megazyme Total Starch kit (Wilcoxon; n = 5–6; *P < 0.05). D, Seed mass (n = 100; *P > 0.05) and images of seeds produced from transgenic lines. E, Panicles from the primary tiller showing differences in architecture as well as delayed senescence of Cga1 overexpression lines compared with the wild-type Kaybonnet control (Wt-Kay). F, Panicle architecture and grain filling in the Cga1 transgenics compared with the wild type (n = 40+; *P < 0.05). All data are means ± sd.
Quantification of starch from rice grains also indicated similar differences in starch content in the Cga1 transgenics (Fig. 6C). In addition to differences in seed composition, we also observed differences in grain size. Grains from the Cga1++ overexpression line were not only smaller, but many also showed seed shape deformities (Fig. 6D). Overexpression of the Cga1 ortholog in Arabidopsis caused similar defects and resulted in reduced germination (Richter et al., 2010; Hudson et al., 2011). In contrast, RNAi-cga1 rice produced seeds that appeared slightly narrower than wild-type seed, although they did not show a significant decrease in weight (Fig. 6D). While moderate overexpression of Cga1 did not influence seed size or weight, delayed senescence was evident in the panicles of both the moderate Cga1+ line and the strong Cga1++ overexpression line, although significantly more so in the latter (Fig. 6E). Altered senescence was also observed in Arabidopsis CGA1 transgenic lines (Hudson et al., 2011). Transgenic rice lines showed some evidence of altered panicle architecture, although there was a large variation between panicles on each plant and these differences were not found to be significant (Fig. 6D). Still, moderate overexpression resulted in panicle architecture similar to wild-type controls, while both the strong Cga1++ overexpression line and the RNAi-cga1 plants showed reduced branch and spikelet production per panicle (Fig. 6, E and D). Although differences in panicle architecture were not found to be significant, Cga1 overexpression did cause a significant reduction in grain filling (Fig. 6F). While a relatively small percentage of spikelets from the wild-type and RNAi-cga1 lines did not develop filled grains, overexpression impaired grain filling most notably in the lower panicles, which developed later. The majority of N reassimilated to the seed comes from the breakdown of chloroplasts (Masclaux-Daubresse et al., 2008, 2010). As such, we speculate that increased chloroplast activity, as made evident by delayed senescence, inhibits nutrient remobilization to the seed and prevents proper grain filling, especially in the lower panicles.
Response of Cga1 Transgenics to Reduced N
N directly influences the chlorophyll content, tillering, and biomass of most agricultural crops. Because rice is an important agricultural crop, we assessed the performance of Cga1 transgenics under three different N regimes. The full-nitrogen (FN) condition consisted of 1 g of slow-release fertilizer per plant along with four monthly applications of a nutrient solution containing N at 200 µL L−1 (NH4NO3). We determined a sufficient-nitrogen (SN) condition to be where there was a significant reduction (approximately 50%) in wild-type biomass, but no visible signs of N stress could be observed. Two liters of modified Hoagland solution containing 10 mm NO3− was supplied weekly for SN, while the limiting-nitrogen (LN) condition used was 3 mm NO3−. Reduction in the amount of N supplied led to significantly lower chlorophyll content in all lines except the strong Cga1++ overexpression line (Table I). Despite a slight reduction in chlorophyll, the overexpression lines maintain increased chlorophyll under reduced N conditions compared with wild-type controls. The strong Cga1++ overexpression line maintains very high chlorophyll even under LN. Like most characteristics, the moderate overexpression line is not significantly different from the wild type under FN, yet it did show significantly more chlorophyll in SN and LN conditions (Table I). In contrast, RNAi-cga1 produced less chlorophyll under all conditions and showed a more drastic decrease with reduced N. These results again confirm those reported for Arabidopsis transgenics grown under different N conditions.
Table I. Influence of N on Cga1 transgenics.
Chlorophyll, tillering, biomass, seed yield, and harvest index (seed mass/total biomass) of the Cga1 transgenics are shown compared with the wild-type Kaybonnet control (Wt-Kay) under three different N regimes: FN, SN, and LN (n ≤ 40). Asterisks indicate significant differences (P < 0.0) from the wild type under the same N condition.
| Parameter |
RNAi-cga1 |
Wt-Kay |
Cga1+ |
Cga1++ |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FN | SN | LN | FN | SN | LN | FN | SN | LN | FN | SN | LN | |
| Chlorophyll (SPAD) | 42.4* | 37.4* | 30.1* | 49.2 | 42.1 | 38.1 | 51.9 | 46.8* | 43.3* | 60.2* | 58.8* | 56.2* |
| Flowering tillers | 5.41* | 4.06* | 3.16* | 3.79 | 2.86 | 2.34 | 3.27* | 2.54 | 2.12 | 2.24* | 1.76* | 1.32* |
| Dry biomass (g) | 22.65 | 11.74 | 3.89* | 21.99 | 11.15 | 5.32 | 21.43 | 9.70 | 5.61 | 10.6* | 4.22* | 1.54* |
| Seed mass (g) | 7.64* | 4.23 | 1.07* | 9.26 | 4.51 | 1.89 | 9.09 | 4.12 | 2.06 | 3.07* | 1.87* | 0.66* |
| Harvest index | 0.34* | 0.36* | 0.28* | 0.42 | 0.40 | 0.36 | 0.43 | 0.44 | 0.37 | 0.29* | 0.44 | 0.43* |
As expected, biomass and growth characteristics demonstrate decreases under reduced N conditions (Table I). However, the transgenic lines respond differently to reductions in N. RNAi-cga1 plants produce more flowering tillers under all N conditions, while strong Cga1++ overexpression produces less (Table I). Although RNAi-cga1 plants produce slightly more overall biomass in FN and SN conditions, LN resulted in decreased biomass compared with wild-type controls (Table I). Moderate overexpression of Cga1 did not cause significant differences compared with wild-type controls, although there was a slight reduction in tillering biomass and seed production under FN and SN conditions (Table I). In contrast, strong overexpression of Cga1 resulted in less than 50% of the overall dry biomass under all N conditions. The RNAi-cga1 lines also showed both reduced yields and harvest index (seed mass/total biomass) under all N conditions. However, the strong Cga1++ line actually shows the highest harvest index with reduced N. The reduced biomass, tillering, and grain-filling problems lead to a significantly reduced yield for Cga1++ plants in FN. However, they retain increased harvest index with reductions in N. With reduced N, the number of secondary tillers producing flowers was decreased. Since grain-filling issues were primarily observed in these lower panicles, this reduced the number of grains not being filled effectively, increasing the harvest index. This indicates that plants overexpressing Cga1 may be able to maintain chloroplast development and harvest index when N conditions are limiting.
DISCUSSION
Cga1 Regulates Chloroplast Development and Activity
In this work, we demonstrate that rice Cga1 expression is up-regulated by light, N, and cytokinin, leading to increased chloroplast development. These results confirm those obtained for the orthologous gene in Arabidopsis, indicating some degree of evolutionary conservation for Cga1 in higher plants. In addition to the previously reported phenotypes in Arabidopsis, we demonstrate broad regulation over chloroplast development and provide evidence for modifications to rice architecture. Furthermore, regulation of the rice FtsZ chloroplast division gene by Cga1 provides a potential mechanism for regulating chloroplast biogenesis. We had previously hypothesized a similar influence in Arabidopsis (Hudson et al., 2011) based on coexpression with chloroplast FtsH proteases that have been shown to cause variegation or albinism when mutated (Adam et al., 2006). However, these genes are found in both green and nongreen chloroplasts, and rather than altering chloroplast number, they appear to arrest chloroplast development, generating white plastids in developmental sectors (Adam et al., 2006; Liu et al., 2010a). There are multiple FtsH genes that have been shown to demonstrate functional redundancy (Zaltsman et al., 2005). While Arabidopsis contains at least two FtsZ homologs (AtFtsZ1-1 and AtFtsZ2-1; Stokes et al., 2000), rice appears to contain only a single FtsZ copy of this ancestral bacterial division gene. Increased production of FtsZ genes results in a significantly reduced number of enlarged chloroplasts, indicating a severe inhibition of chloroplast division (Stokes et al., 2000; Yoder et al., 2007; Schmitz et al., 2009). Transgenic Arabidopsis plants overexpressing FtsZ1 showed increased chloroplasts in mesophyll cells, but these plants were chlorotic and died as seedlings (Stokes and Osteryoung, 2003). Still, it has been suggested that under specific circumstances, elevated AtFtsZ1-1 levels could increase the frequency of chloroplast division (Stokes and Osteryoung, 2003). It is likely that increased chloroplast division along with broad up-regulation of important nucleus-encoded chloroplast genes are required to produce functional chloroplasts without causing significant developmental aberrations, as seen in FtsZ transgenics. The presence of chloroplasts in cells where they are not normally found confirms that Cga1 expression is sufficient to increase chloroplast number in both Arabidopsis and rice.
Starch constitutes the majority (90%) of milled rice seed (Bao et al., 2008). As such, understanding the process involved in regulating starch production is important for making improvements in this crop. The GS/GOGAT cycle controls N assimilation in the chloroplast, and from this point all other N-containing biological molecules are produced (Coschigano et al., 1998; Tabuchi et al., 2007). Modifying the GS/GOGAT cycle genes has been shown to lead to changes in chloroplast activity but does not appear to influence primary N assimilation (Coschigano et al., 1998; Kissen et al., 2010). N is not only a key determinant for carbon fixation and organic acid production but also influences sugar and starch levels in plants (Coruzzi and Bush, 2001; Coruzzi and Zhou, 2001). This highlights the intricate carbon-N balance and how it must be adjusted according to environmental conditions. With sufficient N and chloroplast activity, excess sugars produced during photosynthesis will be stored in the chloroplast as starch. Starch primarily serves as a transient sink to accommodate excess photosynthate that cannot be converted to Suc and exported (Paul and Pellny, 2003). Increased chloroplast number and starch production resulting from increased expression of Cga1 indicates enhanced carbon acquisition, even under reduced N conditions.
Conserved Regulation of Cga1
With the genomes of many plant species now sequenced, making interspecies gene comparisons increases our ability to understand evolutionary processes in plants. Although Arabidopsis has been a useful model for studying plant genetics, demonstrating the functionality of orthologous genes in relevant crop species is a major challenge facing plant biologists. Some genes may be unique to an individual family or species, while others could be conserved throughout higher plants. Like many important housekeeping genes, sequence analysis of the GATA family indicates significant conservation in higher plants (Reyes et al., 2004). Sequence similarity alone does not necessarily imply redundancy; however, in addition to sequence similarity within the coding region, the rice Cga1 promoter region also contains conserved regions containing similar circadian, light, and hormone signaling motifs to those found in Arabidopsis (www.dna.affrc.go.jp/PLACE/). Expression results (Fig. 1) also imply that regulation of Cga1 expression occurs through a similar conserved regulatory system involving input from N, cytokinin, GA, and light signaling pathways.
GATA factors have long been implicated in controlling light- and N-related gene expression (Kudla et al., 1990; Reyes et al., 2004; Richter et al., 2010). Cga1 expression appears to be controlled directly by the circadian clock in both Arabidopsis and rice (Manfield et al., 2007; Filichkin et al., 2011). Input from light, N, and cytokinin all act to significantly increase Cga1 expression beyond levels achieved through circadian regulation alone. The perception of light by phytochromes involves signal transduction through transcription factors known as protein interaction factors (PIFs). In Arabidopsis, Cga1 transcription is induced by light in a phytochrome-dependent fashion (Monte et al., 2004; Naito et al., 2007). GA treatment reduces CGA1 expression by repressing the activity of DELLA proteins, thus releasing PIFs to bind sites in the CGA1 promoter and repress transcription (Richter et al., 2010). Like the GATA family, PHYTOCHROME/PIF genes and their functions are also conserved between monocots and dicots (Leivar and Quail, 2011). Rice PhyA has been show to retain functionality in Arabidopsis (Kneissl et al., 2008). Furthermore, N assimilation and cytokinin-related genes are also quite conserved in higher plants. This includes the His kinase cytokinin receptors as well as downstream phosphotransfer proteins, type A and type B ARRs involved in cytokinin signal transduction (Ito and Kurata, 2006). It is unclear whether Cga1 expression increases as a direct result of N application or indirectly increased through N-induced cytokinin production. However, results obtained in Arabidopsis indicate a requirement for functional cytokinin receptors and downstream ARRs for increased expression to occur (Naito et al., 2007; Hudson et al., 2011; Chiang et al., 2012). The rapid increase in Cga1 expression following cytokinin application indicates a higher degree of response to cytokinin than to N or light (Fig. 1). Regardless, regulation of Cga1 by these signaling pathways provides a mechanism for plants to control chloroplast development based on environmental conditions. When light and N are prevalent, cytokinin will also be elevated, and subsequently, the expression of Cga1 will increase. Under low-light or low-N conditions, photosynthetic activity will be limited; therefore, chloroplast activity and development must also be entrained. Periods of darkness and/or low N have been shown to enhance GA activity, which will subsequently repress Cga1 expression by increasing PIF activity. Regulating the expression of Cga1 through inputs from light, N, cytokinin, and GA allows plants to modulate chloroplast development.
Cga1 Shows Potential for Utilization in Crop Improvement
Crop plant architecture determines planting density in the field and directly influences light harvest, disease resistance, and nutrient acquisition (Guo et al., 2011). Rice plant architecture is one of the most important factors influencing rice yield (Reinhardt and Kuhlemeier, 2002). Elite varieties of rice can produce higher grain yields largely due to alterations in plant architecture (Yuan, 1977; Khush, 2001). Differences in rice tillering, biomass production, and yield of the Cga1 transgenics could have significant agricultural implications. Phenotypes resulting from altered Cga1 expression are similar to elite-yielding varieties of rice that have been achieved through genetic modifications to cytokinin or GA signaling genes. The introduction of the Green Revolution semidwarf varieties contributed substantially to increased rice yields (Peng et al., 1999; Khush, 2003). Despite the fact that these semidwarf varieties exhibit reduced tillering, these lines showed increased harvest index and produced more grains per unit area (Peng et al., 1999; Sasaki et al., 2002). The most recent high-yielding rice varieties of “super” hybrid rice have also focused on reduced tillering capacity and improved lodging resistance (Peng et al., 2008). Specific regulation of Cga1 expression could be used to modify rice tillering and permit altered planting densities in the field.
Panicle architecture is also regulated at numerous levels, including genetic factors and hormone signaling, depending on environmental factors (McSteen, 2009). While N remobilization is vitally important for grain filling, cytokinin signaling has been shown to influence rice stem length and spikelet branching and, thus, at least in part controlling yield (Ashikari et al., 2005; Hirose et al., 2007; Tabuchi et al., 2007; Zhang et al., 2010). Overexpression of Cga1 shows similar potential for increasing yield via altering panicle architecture; however, constitutive overexpression resulted in detrimental effects with respect to grain filling. Grain-filling problems have also been reported in modern super rice varieties, which have numerous spikelets on a panicle, but they frequently fail to exhibit their high-yield potential due to the poor grain filling of secondary panicles (Yang and Zhang, 2010; Fu et al., 2011). This was also the case for Cga1 overexpression lines. Increased harvest index was only achieved at reduced N conditions, which would not be ideal for agriculture, since the overall yield is less than with high N. Cytokinins influence not only chloroplast development but also chloroplast degradation during the senescence process (Argueso et al., 2010). Because the panicles of the Cga1++ line remain dark green (Fig. 5F), we hypothesize that N is not being properly remobilized to the seed. Creating transgenics for commercial purposes would require the use of more specialized, tissue-specific promoters in order to prevent or remove the detrimental problems associated with senescence and grain filling.
Conservation of Cga1 between Arabidopsis and rice indicates that changes in its expression could be used to adjust chloroplast development, starch production, and overall biomass in many crops. It is not surprising that the rice Cga1 coding region shows a higher percentage of sequence similarity to many important agricultural species, including maize (Zea mays), wheat (Triticum aestivum), barley (Hordeum vulgare), sorghum (Sorghum bicolor), and grape (Vitis vinifera; blast.ncbi.nlm.nih.gov/), than to Arabidopsis. Limiting Cga1 expression might be used to increase tillering and biomass in species where this is desirable and starch production is of little importance. In contrast, specifically regulated overexpression may be useful for reducing planting densities and increasing chloroplast activity, especially under low N. As conditions in the field are often less than optimal and fertilizer applications are often uneven, assessing the yield potential of Cga1 transgenics in the field will be the goal of future work. Cultivation of rice typically involves copious amounts of both N and water. Utilization of cultivars able to perform better or maintain harvest index under reduced amounts of N and/or water is a major goal of rice research. Even a moderate enhancement of planting density or the ability to withstand low N could have a significant impact on agricultural production.
MATERIALS AND METHODS
Growth Conditions
Rice (Oryza sativa) was grown in growth chambers using standard long-day conditions with 16 h of white light (500 µmol m−2 s−1) in a 4:1:1 vermiculite:peat moss:LA4 Sunshine Soil mixture (SunGro Horticulture) containing 1 g of 13-13-13 slow-release fertilizer (NutriCote) with micronutrients and watered four times with an 18-9-18 solution (Plant Products; msds.plantprod.com/document/11072/en/label) at 200 µL L−1 N (NH4NO3). For N depletion conditions, we used a modified Hoagland solution as described previously (Bi et al., 2009), applying 2 L weekly with 10 mm NO3 as SN, whereas 3 mm NO3 was used as LN. Short-day treatment (10 h of light) was used to induce flowering during the 6th week.
In order to determine factors influencing Cga1 expression, wild-type plants were grown hydroponically for 2 weeks in 1× Murashige and Skoog plant salts (MP Biomedicals), removed from nutrients to deionized water in darkness for 24 h, and then resupplemented with N (10 mm NO3−) or 10 µmol of BA (Fluka), while GA3 (Sigma) was applied as a 10-µmol foliar spray. Samples were flash frozen after 2 h of treatment for RNA extraction.
Creation of Transgenic Rice Lines
The constructs used for overexpression and RNAi of Cga1 (Os02g12790) were made by Syngenta using a UBIQUITIN promoter in front of the endogenous rice complementary DNA (cDNA) sequence. Agrobacterium tumefaciens-mediated transformation was performed according to standard protocols, and the T1 transgenic seeds were harvested. Phosphomannose isomerase was used for genotyping the selectable phospho-Man isomerase marker, and expression levels were confirmed with qRT-PCR (Negrotto et al., 2000).
Chlorophyll, Chloroplast, and Starch Measurements
Chlorophyll levels were measured using the SPAD 502DL chlorophyll meter (Minolta). Chloroplasts were extracted using a Percoll gradient as described previously (Hudson et al., 2011) from 1 g of leaf tissue and counted using a standard 0.1-mm hemocytometer. Starch was stained using IKI in both fresh leaf samples and following standard wax embedding/sectioning. The starch content of seeds was determined using the Megazyme Total Starch Assay kit according to the manufacturer’s instructions (Megazyme International) with 100 mg of liquid N ground seed. Confocal microscopy was performed on fresh mounted tissue using a Leica CM-1000 microscope with LCS Lite software (Leica Microsystems).
Statistical Analysis
All statistics were performed using JMP Statistical Discovery Software 9.0 (www.jmp.com).
Microarray Hybridization and Analysis
Five micrograms of total RNA from each sample was used to synthesize double-stranded cDNAs from wild-type Kaybonnet rice and the Cga1++ strong overexpression line. Labeled copy RNA, synthesized from the cDNA, was hybridized to the Affymetrix rice whole-genome array. Data analysis was conducted using GeneSpring software (Agilent). The data were normalized with a default setting of the program. Differentially expressed genes in the transgenic line were identified with at least 1.5-fold change first, and then ANOVA was used to identify significance (Welch’s t test; P value cutoff at 0.05). Data have been submitted to the Gene Expression Omnibus repository (GSE35630) and can be found in Supplemental Data S2 along with the results of coexpression analysis.
Semiquantitative Reverse Transcription-PCR and Quantitative Real-Time PCR
RNA was extracted from 100 mg of leaf tissue using Trizol (Invitrogen), treated with DNase (Promega), and purified using the RNeasy Mini kit (Qiagen). Extracts were quantified using the Nanodrop ND-1000, and first-strand synthesis of cDNA was performed using qScript cDNA SuperMix (Quanta Biosciences) from 1 µg of total RNA. For semiquantitative reverse transcription-PCR, reactions were performed using GoTaq Flexi (Promega), and the expression of transgenic lines was quantified using ImageJ software. Rice ACTIN5 was used as an endogenous control. Quantitative real-time expression was performed using PerfeCTa SYBR Green SuperMix ROX (Quanta Biosciences) on the ABI7300 (Applied Biosystems) with ACTIN5 used as an endogenous control. Primers were selected from previous publications or designed using the Applied Biosystems software Primer Express 2.0.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Selection of RNAi-cga1 transgenic lines.
Supplemental Data S1. Coexpression analysis of rice Cga1.
Supplemental Data S2. Microarray analysis of Cga1 overexpression compared with wild-type control.
Acknowledgments
We thank Caroline Hoy, Chris Lam, Greg Yuristy, and Kosala Ranathunge for help with plant care and collection.
Glossary
- N
nitrogen
- RNAi
RNA interference
- Fd
ferredoxin-dependent
- GS
Gln synthetase
- qRT
quantitative reverse transcription
- IKI
Lugol’s iodine
- FN
full-nitrogen
- SN
sufficient-nitrogen
- LN
limiting-nitrogen
- PIF
protein interaction factor
- cDNA
complementary DNA
- BA
benzyladenine
References
- Adam Z, Rudella A, van Wijk KJ. (2006) Recent advances in the study of Clp, FtsH and other proteases located in chloroplasts. Curr Opin Plant Biol 9: 234–240 [DOI] [PubMed] [Google Scholar]
- Argueso CT, Raines T, Kieber JJ. (2010) Cytokinin signaling and transcriptional networks. Curr Opin Plant Biol 13: 533–539 [DOI] [PubMed] [Google Scholar]
- Ashikari M, Sakakibara H, Lin S, Yamamoto T, Takashi T, Nishimura A, Angeles ER, Qian Q, Kitano H, Matsuoka M. (2005) Cytokinin oxidase regulates rice grain production. Science 309: 741–745 [DOI] [PubMed] [Google Scholar]
- Bao J, Jin L, Xiao P, Shen S, Sun M, Corke H. (2008) Starch physicochemical properties and their associations with microsatellite alleles of starch-synthesizing genes in a rice RIL population. J Agric Food Chem 56: 1589–1594 [DOI] [PubMed] [Google Scholar]
- Bi YM, Kant S, Clarke J, Gidda S, Ming F, Xu J, Rochon A, Shelp BJ, Hao L, Zhao R, et al. (2009) Increased nitrogen-use efficiency in transgenic rice plants over-expressing a nitrogen-responsive early nodulin gene identified from rice expression profiling. Plant Cell Environ 32: 1749–1760 [DOI] [PubMed] [Google Scholar]
- Bi YM, Zhang Y, Signorelli T, Zhao R, Zhu T, Rothstein S. (2005) Genetic analysis of Arabidopsis GATA transcription factor gene family reveals a nitrate-inducible member important for chlorophyll synthesis and glucose sensitivity. Plant J 44: 680–692 [DOI] [PubMed] [Google Scholar]
- Chiang YH, Zubo YO, Tapken W, Kim HJ, Lavanway AM, Howard L, Pilon M, Kieber JJ, Schaller GE. (2012) Functional characterization of the GATA transcription factors GNC and CGA1 reveals their key role in chloroplast development, growth, and division in Arabidopsis. Plant Physiol 160: 332–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coruzzi G, Bush DR. (2001) Nitrogen and carbon nutrient and metabolite signaling in plants. Plant Physiol 125: 61–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coruzzi GM, Zhou L. (2001) Carbon and nitrogen sensing and signaling in plants: emerging ‘matrix effects.’ Curr Opin Plant Biol 4: 247–253 [DOI] [PubMed] [Google Scholar]
- Coschigano KT, Melo-Oliveira R, Lim J, Coruzzi GM. (1998) Arabidopsis gls mutants and distinct Fd-GOGAT genes: implications for photorespiration and primary nitrogen assimilation. Plant Cell 10: 741–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt U, Grimm B, Hörtensteiner S. (2004) Recent advances in chlorophyll biosynthesis and breakdown in higher plants. Plant Mol Biol 56: 1–14 [DOI] [PubMed] [Google Scholar]
- Evans JR, von Caemmerer S. (2011) Enhancing photosynthesis. Plant Physiol 155: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evenson RE, Gollin D. (2003) Assessing the impact of the Green Revolution, 1960 to 2000. Science 300: 758–762 [DOI] [PubMed] [Google Scholar]
- Filichkin SA, Breton G, Priest HD, Dharmawardhana P, Jaiswal P, Fox SE, Michael TP, Chory J, Kay SA, Mockler TC. (2011) Global profiling of rice and poplar transcriptomes highlights key conserved circadian-controlled pathways and cis-regulatory modules. PLoS ONE 6: e16907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Huanga Z, Wang Z, Yanga J, Zhang J. (2011). Pre-anthesis non-structural carbohydrate reserve in the stem enhances the sink strength of inferior spikelets during grain filling of rice. Field Crops Res 123: 170–182 [Google Scholar]
- Gan Y, Liu C, Yu H, Broun P. (2007) Integration of cytokinin and gibberellin signalling by Arabidopsis transcription factors GIS, ZFP8 and GIS2 in the regulation of epidermal cell fate. Development 134: 2073–2081 [DOI] [PubMed] [Google Scholar]
- Glynn JM, Froehlich JE, Osteryoung KW. (2008) Arabidopsis ARC6 coordinates the division machineries of the inner and outer chloroplast membranes through interaction with PDV2 in the intermembrane space. Plant Cell 20: 2460–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SB, Waller RF, McFadden GI. (2008) Plastid evolution. Annu Rev Plant Biol 59: 491–517 [DOI] [PubMed] [Google Scholar]
- Greenboim-Wainberg Y, Maymon I, Borochov R, Alvarez J, Olszewski N, Ori N, Eshed Y, Weiss D. (2005) Cross talk between gibberellin and cytokinin: the Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell 17: 92–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Fourcaud T, Jaeger M, Zhang X, Li B. (2011) Plant growth and architectural modelling and its applications: preface. Ann Bot (Lond) 107: 723–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose N, Makita N, Kojima M, Kamada-Nobusada T, Sakakibara H. (2007) Overexpression of a type-A response regulator alters rice morphology and cytokinin metabolism. Plant Cell Physiol 48: 523–539 [DOI] [PubMed] [Google Scholar]
- Hirose N, Takei K, Kuroha T, Kamada-Nobusada T, Hayashi H, Sakakibara H. (2008) Regulation of cytokinin biosynthesis, compartmentalization and translocation. J Exp Bot 59: 75–83 [DOI] [PubMed] [Google Scholar]
- Hudson D, Guevara D, Yaish MW, Hannam C, Long N, Clarke JD, Bi YM, Rothstein SJ. (2011) GNC and CGA1 modulate chlorophyll biosynthesis and glutamate synthase (GLU1/Fd-GOGAT) expression in Arabidopsis. PLoS ONE 6: e26765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Kurata N. (2006) Identification and characterization of cytokinin-signalling gene families in rice. Gene 382: 57–65 [DOI] [PubMed] [Google Scholar]
- Jen CH, Manfield IW, Michalopoulos I, Pinney JW, Willats WG, Gilmartin PM, Westhead DR. (2006) The Arabidopsis co-expression tool (ACT): a WWW-based tool and database for microarray-based gene expression analysis. Plant J 46: 336–348 [DOI] [PubMed] [Google Scholar]
- Jung KH, Dardick C, Bartley LE, Cao P, Phetsom J, Canlas P, Seo YS, Shultz M, Ouyang S, Yuan Q, et al. (2008a) Refinement of light-responsive transcript lists using rice oligonucleotide arrays: evaluation of gene-redundancy. PLoS ONE 3: e3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung KH, Lee J, Dardick C, Seo YS, Cao P, Canlas P, Phetsom J, Xu X, Ouyang S, An K, et al. (2008b) Identification and functional analysis of light-responsive unique genes and gene family members in rice. PLoS Genet 4: e1000164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamoko M, El-Kafafi S, Mandaron P, Lerbs-Mache S, Falconet D. (2011) Multiple FtsZ2 isoforms involved in chloroplast division and biogenesis are developmentally associated with thylakoid membranes in Arabidopsis. FEBS Lett 585: 1203–1208 [DOI] [PubMed] [Google Scholar]
- Kessler F, Schnell D. (2009) Chloroplast biogenesis: diversity and regulation of the protein import apparatus. Curr Opin Cell Biol 21: 494–500 [DOI] [PubMed] [Google Scholar]
- Khush G. (2003) Productivity improvements in rice. Nutr Rev 61: S114–S116 [DOI] [PubMed] [Google Scholar]
- Khush GS. (2001) Green Revolution: the way forward. Nat Rev Genet 2: 815–822 [DOI] [PubMed] [Google Scholar]
- Kiba T, Naitou T, Koizumi N, Yamashino T, Sakakibara H, Mizuno T. (2005) Combinatorial microarray analysis revealing Arabidopsis genes implicated in cytokinin responses through the His→Asp phosphorelay circuitry. Plant Cell Physiol 46: 339–355 [DOI] [PubMed] [Google Scholar]
- Kissen R, Winge P, Tran DH, Jørstad TS, Størseth TR, Christensen T, Bones AM. (2010) Transcriptional profiling of an Fd-GOGAT1/GLU1 mutant in Arabidopsis thaliana reveals a multiple stress response and extensive reprogramming of the transcriptome. BMC Genomics 11: 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneissl J, Shinomura T, Furuya M, Bolle C. (2008) A rice phytochrome A in Arabidopsis: the role of the N-terminus under red and far-red light. Mol Plant 1: 84–102 [DOI] [PubMed] [Google Scholar]
- Köllmer I, Werner T, Schmülling T. (2011) Ectopic expression of different cytokinin-regulated transcription factor genes of Arabidopsis thaliana alters plant growth and development. J Plant Physiol 168: 1320–1327 [DOI] [PubMed] [Google Scholar]
- Kudla B, Caddick MX, Langdon T, Martinez-Rossi NM, Bennett CF, Sibley S, Davies RW, Arst HN., Jr (1990) The regulatory gene areA mediating nitrogen metabolite repression in Aspergillus nidulans: mutations affecting specificity of gene activation alter a loop residue of a putative zinc finger. EMBO J 9: 1355–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba M, Ito H, Morita R, Iida S, Sato Y, Fujimoto M, Kawasaki S, Tanaka R, Hirochika H, Nishimura M, et al. (2007) Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell 19: 1362–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laisk A, Nedbal L, Govindjee(2009) Photosynthesis In Silico: Understanding Complexity from Molecules to Ecosystems. Springer, Dordrecht, The Netherlands [Google Scholar]
- Lawlor DW. (2002) Carbon and nitrogen assimilation in relation to yield: mechanisms are the key to understanding production systems. J Exp Bot 53: 773–787 [PubMed] [Google Scholar]
- Leivar P, Quail PH. (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yu F, Rodermel S. (2010a) An Arabidopsis pentatricopeptide repeat protein, SUPPRESSOR OF VARIEGATION7, is required for FtsH-mediated chloroplast biogenesis. Plant Physiol 154: 1588–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yu F, Rodermel S. (2010b) Arabidopsis chloroplast FtsH, var2 and suppressors of var2 leaf variegation: a review. J Integr Plant Biol 52: 750–761 [DOI] [PubMed] [Google Scholar]
- Manfield IW, Devlin PF, Jen CH, Westhead DR, Gilmartin PM. (2007) Conservation, convergence, and divergence of light-responsive, circadian-regulated, and tissue-specific expression patterns during evolution of the Arabidopsis GATA gene family. Plant Physiol 143: 941–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mara CD, Irish VF. (2008) Two GATA transcription factors are downstream effectors of floral homeotic gene action in Arabidopsis. Plant Physiol 147: 707–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux-Daubresse C, Daniel-Vedele F, Dechorgnat J, Chardon F, Gaufichon L, Suzuki A. (2010) Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann Bot (Lond) 105: 1141–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux-Daubresse C, Reisdorf-Cren M, Orsel M. (2008) Leaf nitrogen remobilisation for plant development and grain filling. Plant Biol (Stuttg) (Suppl 1) 10: 23–36 [DOI] [PubMed] [Google Scholar]
- McSteen P. (2009) Hormonal regulation of branching in grasses. Plant Physiol 149: 46–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima SY, Froehlich JE, Osteryoung KW. (2006) PDV1 and PDV2 mediate recruitment of the dynamin-related protein ARC5 to the plastid division site. Plant Cell 18: 2517–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte E, Tepperman JM, Al-Sady B, Kaczorowski KA, Alonso JM, Ecker JR, Li X, Zhang Y, Quail PH. (2004) The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc Natl Acad Sci USA 101: 16091–16098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito T, Kiba T, Koizumi N, Yamashino T, Mizuno T. (2007) Characterization of a unique GATA family gene that responds to both light and cytokinin in Arabidopsis thaliana. Biosci Biotechnol Biochem 71: 1557–1560 [DOI] [PubMed] [Google Scholar]
- Negrotto D, Jolley M, Beer S, Wenck AR, Hansen GH. (2000) The use of phosphomannose-isomerase as a selectable marker to recover transgenic maize plants (Zea mays L.) via Agrobacterium transformation. Plant Cell Rep 19: 798–803 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A, Fernie AR, Stitt M. (2010) Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Mol Plant 3: 973–996 [DOI] [PubMed] [Google Scholar]
- Okazaki K, Kabeya Y, Miyagishima SY. (2010) The evolution of the regulatory mechanism of chloroplast division. Plant Signal Behav 5: 164–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki K, Kabeya Y, Suzuki K, Mori T, Ichikawa T, Matsui M, Nakanishi H, Miyagishima SY. (2009) The PLASTID DIVISION1 and 2 components of the chloroplast division machinery determine the rate of chloroplast division in land plant cell differentiation. Plant Cell 21: 1769–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung KW, McAndrew RS. (2001) The plastid division machine. Annu Rev Plant Physiol Plant Mol Biol 52: 315–333 [DOI] [PubMed] [Google Scholar]
- Osteryoung KW, Stokes KD, Rutherford SM, Percival AL, Lee WY. (1998) Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial ftsZ. Plant Cell 10: 1991–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul MJ, Pellny TK. (2003) Carbon metabolite feedback regulation of leaf photosynthesis and development. J Exp Bot 54: 539–547 [DOI] [PubMed] [Google Scholar]
- Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, Beales J, Fish LJ, Worland AJ, Pelica F, et al. (1999) ‘Green Revolution’ genes encode mutant gibberellin response modulators. Nature 400: 256–261 [DOI] [PubMed] [Google Scholar]
- Peng S, Khush G, Virk P, Tang Q, Zou Y. (2008). Progress in ideotype breeding to increase rice yield potential. Field Crops Res 108: 32–38 [Google Scholar]
- Pogson BJ, Albrecht V. (2011) Genetic dissection of chloroplast biogenesis and development: an overview. Plant Physiol 155: 1545–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahim MA, Fordham R. (1991) Effect of shade on leaf and cell size and number of epidermal cells in garlic (Allium sativum). Ann Bot 67: 167–171 [Google Scholar]
- Raines CA. (2011) Increasing photosynthetic carbon assimilation in C3 plants to improve crop yield: current and future strategies. Plant Physiol 155: 36–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven JA, Allen JF. (2003) Genomics and chloroplast evolution: what did cyanobacteria do for plants? Genome Biol 4: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinbothe C, El Bakkouri M, Buhr F, Muraki N, Nomata J, Kurisu G, Fujita Y, Reinbothe S. (2010) Chlorophyll biosynthesis: spotlight on protochlorophyllide reduction. Trends Plant Sci 15: 614–624 [DOI] [PubMed] [Google Scholar]
- Reinhardt D, Kuhlemeier C. (2002) Plant architecture. EMBO Rep 3: 846–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes JC, Muro-Pastor MI, Florencio FJ. (2004) The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol 134: 1718–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter R, Behringer C, Müller IK, Schwechheimer C. (2010) The GATA-type transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from DELLA proteins and PHYTOCHROME-INTERACTING FACTORS. Genes Dev 24: 2093–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara H. (2003) Nitrate-specific and cytokinin-mediated nitrogen signaling pathways in plants. J Plant Res 116: 253–257 [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Takei K, Hirose N. (2006) Interactions between nitrogen and cytokinin in the regulation of metabolism and development. Trends Plant Sci 11: 440–448 [DOI] [PubMed] [Google Scholar]
- Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush GS, et al. (2002) Green Revolution: a mutant gibberellin-synthesis gene in rice. Nature 416: 701–702 [DOI] [PubMed] [Google Scholar]
- Schmitz AJ, Glynn JM, Olson BJ, Stokes KD, Osteryoung KW. (2009) Arabidopsis FtsZ2-1 and FtsZ2-2 are functionally redundant, but FtsZ-based plastid division is not essential for chloroplast partitioning or plant growth and development. Mol Plant 2: 1211–1222 [DOI] [PubMed] [Google Scholar]
- Steiner E, Efroni I, Gopalraj M, Saathoff K, Tseng TS, Kieffer M, Eshed Y, Olszewski N, Weiss D. (2012) The Arabidopsis O-linked N-acetylglucosamine transferase SPINDLY interacts with class I TCPs to facilitate cytokinin responses in leaves and flowers. Plant Cell 24: 96–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M, Lunn J, Usadel B. (2010) Arabidopsis and primary photosynthetic metabolism: more than the icing on the cake. Plant J 61: 1067–1091 [DOI] [PubMed] [Google Scholar]
- Stokes KD, McAndrew RS, Figueroa R, Vitha S, Osteryoung KW. (2000) Chloroplast division and morphology are differentially affected by overexpression of FtsZ1 and FtsZ2 genes in Arabidopsis. Plant Physiol 124: 1668–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes KD, Osteryoung KW. (2003) Early divergence of the FtsZ1 and FtsZ2 plastid division gene families in photosynthetic eukaryotes. Gene 320: 97–108 [DOI] [PubMed] [Google Scholar]
- Tabuchi M, Abiko T, Yamaya T. (2007) Assimilation of ammonium ions and reutilization of nitrogen in rice (Oryza sativa L.). J Exp Bot 58: 2319–2327 [DOI] [PubMed] [Google Scholar]
- Tanaka R, Tanaka A. (2007) Tetrapyrrole biosynthesis in higher plants. Annu Rev Plant Biol 58: 321–346 [DOI] [PubMed] [Google Scholar]
- Tilman D, Cassman KG, Matson PA, Naylor R, Polasky S. (2002) Agricultural sustainability and intensive production practices. Nature 418: 671–677 [DOI] [PubMed] [Google Scholar]
- Weiss D, Ori N. (2007) Mechanisms of cross talk between gibberellin and other hormones. Plant Physiol 144: 1240–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhang J. (2010) Grain-filling problem in ‘super’ rice. J Exp Bot 61: 1–5 [DOI] [PubMed] [Google Scholar]
- Yoder DW, Kadirjan-Kalbach D, Olson BJ, Miyagishima SY, Deblasio SL, Hangarter RP, Osteryoung KW. (2007) Effects of mutations in Arabidopsis FtsZ1 on plastid division, FtsZ ring formation and positioning, and FtsZ filament morphology in vivo. Plant Cell Physiol 48: 775–791 [DOI] [PubMed] [Google Scholar]
- Yuan LP. (1977) Hybrid rice breeding for super high yield. Hybrid Rice. 12: 1–6 [Google Scholar]
- Zaltsman A, Ori N, Adam Z. (2005) Two types of FtsH protease subunits are required for chloroplast biogenesis and photosystem II repair in Arabidopsis. Plant Cell 17: 2782–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Chen T, Wang Z, Yang J, Zhang J. (2010) Involvement of cytokinins in the grain filling of rice under alternate wetting and drying irrigation. J Exp Bot 61: 3719–3733 [DOI] [PubMed] [Google Scholar]