A transposon tagging system was used to develop 509 activation-tagged lines through micropropagation of derived transgenic launchpads.
Abstract
Tomato (Solanum lycopersicum) is a model organism for Solanaceae in both molecular and agronomic research. This project utilized Agrobacterium tumefaciens transformation and the transposon-tagging construct Activator (Ac)/Dissociator (Ds)-ATag-Bar_gosGFP to produce activation-tagged and knockout mutants in the processing tomato cultivar M82. The construct carried hygromycin resistance (hyg), green fluorescent protein (GFP), and the transposase (TPase) of maize (Zea mays) Activator major transcript X054214.1 on the stable Ac element, along with a 35S enhancer tetramer and glufosinate herbicide resistance (BAR) on the mobile Ds-ATag element. An in vitro propagation strategy was used to produce a population of 25 T0 plants from a single transformed plant regenerated in tissue culture. A T1 population of 11,000 selfed and cv M82 backcrossed progeny was produced from the functional T0 line. This population was screened using glufosinate herbicide, hygromycin leaf painting, and multiplex polymerase chain reaction (PCR). Insertion sites of transposed Ds-ATag elements were identified through thermal asymmetric interlaced PCR, and resulting product sequences were aligned to the recently published tomato genome. A population of 509 independent, Ds-only transposant lines spanning all 12 tomato chromosomes has been developed. Insertion site analysis demonstrated that more than 80% of these lines harbored Ds insertions conducive to activation tagging. The capacity of the Ds-ATag element to alter transcription was verified by quantitative real-time reverse transcription-PCR in two mutant lines. The transposon-tagged lines have been immortalized in seed stocks and can be accessed through an online database, providing a unique resource for tomato breeding and analysis of gene function in the background of a commercial tomato cultivar.
Activation-tagged mutant populations have proven to be a useful resource in functional genomics, with topics of research including nutrient accumulation (Lee et al., 2009, 2011, 2012), fruit development (Sapir et al., 2008; Giménez et al., 2010; Gomez et al., 2011), stress resistance (Aboul-Soud et al., 2009; Wang et al., 2009; Kang et al., 2011), and morphology (van der Graaff et al., 2000; Busov et al., 2003; Huang et al., 2010). The development of many populations has involved a transfer DNA (T-DNA) tag containing four tandem copies of the enhancer region taken from the P35S promoter within Cauliflower mosaic virus (Walden et al., 1994). Insertion of this element into gene-flanking regions offers both the possibility of constitutive increases in transcription and amplification of expression profiles dictated by native promoters (Weigel et al., 2000). Although mutants can be generated through plant transformation, the use of the maize (Zea mays) transposon-tagging system has been more efficient for mutant population construction (Kondou et al., 2010). Mutant populations have been produced using the maize Activator (Ac)/Dissociator (Ds) system in Arabidopsis (Arabidopsis thaliana; Kuromori et al., 2004), rice (Oryza sativa; Qu et al., 2008), and poplar (Populus spp.; Fladung and Polak, 2012) and the Enhancer-Inhibitor/Supressor-mutator system for forward and reverse genetics in Arabidopsis (Speulman et al., 1999, 2000) and rice (Kumar et al., 2005; Krishnan et al., 2009). Use of the Ac/Ds system enables the recovery of many independent mutants from a single transformed line and facilitates the rapid identification of putatively mutagenized sequences through the isolation of Ds insertion flanking regions (Kuromori et al., 2004).
In addition to its importance as a food crop, tomato (Solanum lycopersicum) serves as a functional model organism for other crops in Solanaceae and plant molecular biology. The recent publication of the first whole-genome sequence for tomato (Sato et al., 2012) ensures that its role as a research organism will continue to grow. A history of research detailing independently developed activation-tagged mutant lines for various applications in tomato has already been published (Jones et al., 1994; Meissner et al., 2000; Mathews et al., 2003; Giménez et al., 2010). However, tomato still lacks the large, readily available populations of T-DNA-tagged mutants that have accelerated research in other model crops. Many previous efforts to generate mutant populations have focused on the popular research cultivar Micro-Tom (Meissner et al., 1997; Saito et al., 2011). While economical to maintain, the diminutive size and yield of cv Micro-Tom make it less useful in studying traits that directly relate to agronomic performance under field conditions. A proven alternative to cv Micro-Tom is the determinate processing cultivar M82, a selection of UC-82 (Stevens et al., 1982). The advantages of using this cultivar include prolific seed production, a mature plant size conducive to greenhouse cultivation, and the ability to correlate research findings with mechanized tomato production in commercial settings. The cultivar has also seen wide use in research, including collections of ethyl methanesulfonate mutants (Menda et al., 2004) and introgression lines (Eshed and Zamir, 1995).
In an effort to expand mutant resources available to researchers and increase the versatility of tomato as a model crop, we report the development of a highly functional mutant population harboring the Ac/Ds transposon-based activation-tagging system in tomato ‘M82’. This population has already produced some novel mutant lines and could potentially be propagated into a resource capable of saturating the tomato genome with activation-inducing Ds elements. Toward this goal, we report here the analysis of the transposon system within this population in addition to the T-DNA insertion sites for the available mutant lines.
RESULTS
T0 Line Generation and Analysis
A laborious transformation protocol involving hundreds of primary leaf explants regenerated three tomato ‘M82’ lines harboring the Ac/Ds-ATag-Bar_gosGFP construct (Trijatmiko, 2005; Fig. 1). Of these, one line did not set fruit after transfer to the greenhouse, and another harbored only the Ac portion of the construct. The remaining line, T0 No. 7, harbored all the elements of the T-DNA construct. Thermal asymmetric interlaced (TAIL)-PCR was performed on this line with intact T-DNA using primers situated at the right border of the Ds-ATag element, and the resulting product was sequenced. BLAST searching this sequence against the tomato genome (November 2011) indicated that the T-DNA had inserted into the single intron of 60S ribosomal protein L29 (Solyc05g053440.2.1) on tomato pseudochromosome 5 (Bombarely et al., 2011). The presence of the T-DNA insertion in this gene had no obvious effect on the overall phenotype compared with wild-type cv M82 plants. Subsequent analysis demonstrated that T1 progeny homozygous for the insertion did not have a lethal or deleterious phenotype. Further investigation was made into the expression of 60S ribosomal protein L29 and the nearest genes on either side, DNA-directed RNA polymerase subunit (Solyc05g053430.2.1) and late embryogenesis abundant protein1 (Solyc05g053450.2.1), respectively. The expression of 60S ribosomal protein L29 in tomato was predicted by comparison with orthologs in potato (Solanum tuberosum) group Phureja (PGSC0003DMG400027161) and Arabidopsis (At3g06700). RNA-seq data from Solanum spp. indicated widespread transcription of the native ortholog in every tissue type and treatment (Massa et al., 2011). Graphic expression of microarray data for At3g06700 revealed greater expression in shoot apex tissue in comparison with other tissue types (Winter et al., 2007). The cis-regulatory elements corresponding to pollen-specific expression were identified (Higo et al., 1999) within the promoter regions of both neighboring genes, each of which closely flanks Solyc05g053440.2.1. In addition, nonquantitative reverse transcription (RT)-PCR demonstrated readily detectable expression of the transposase gene in whole inflorescence tissue.
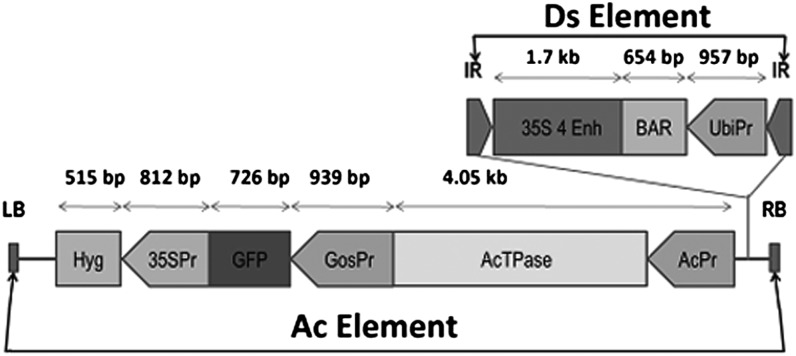
Figure 1.
The Ac/Ds-ATag-Bar_gosGFP construct. Elements are as follows: LB, left border; Hyg, hygromycin resistance; 35SPr, cauliflower mosaic virus 35S promoter; GosPr, maize Gos2 promoter; AcTPase, maize transposase derived from GenBank accession X05424.1; AcPr, maize transposase native promoter; IR, inverted repeat; 35S 4 Enh, four tandem copies of cauliflower mosaic virus 35S enhancer; BAR, glufosinate resistance; UbiPr, ubiquitin promoter; RB, right border.
Analysis of T1 Families
Using axillary shoots as a means of propagation, we micropropagated the original, single transformant harboring a complete launchpad to generate a population of 25 clonal T0 plants. Progeny of each of these plants were identified by the assigned identity of each clone (a–z) of the T0 parent. Analysis of these T0 clones and their progeny revealed four distinct groups of plants based on the frequency of transposition. The first group (7d, 7e, 7h, 7k, 7n, 7o, 7q, 7v, 7w) demonstrated no incidence of Ds transposition from the original insertion site on pseudochromosome 5. This was confirmed by both the lack of segregation between the Ac and Ds elements in 1,363 T1 progeny analyzed and sequencing of the nontransposed Ds element through TAIL-PCR performed on selections of 12 progeny from each family. No instance of the Ds element remaining beside Ac in the original T-DNA insertion site was identified among the progeny of T0 families belonging to the other three groups. The second group (7a, 7b, 7f, 7i, 7p, 7r, 7s, 7t, 7u, 7z) was characterized by the recovery of many independent transpositions of Ds among the T1 progeny, with some common insertions, presumed to derive from somatic transposition events prior to meiosis, unique among the progeny of single T0 families within the group. Several phenotypic mutants were identified among the T1 progeny of this group. The third group (7c, 7g, 7m, 7x, 7y) was defined by the presence of a single common insertion on pseudochromosome 8 that was prevalent in the progeny of all the families within the group. Additional somatic and many germinal transposition events were also identified in the progeny of this group, included several phenotypic mutants. In addition, a fourth trend was observed in a single line (7l), where the Ds element, transposed or not, could not be found in any progeny. This apparent loss of the Ds element during excision was unique to 7l of the micropropagated derivatives of transgenic line 7 but had been observed in one of two additional independently transformed T0 lines generated over the course of the project. RT-PCR analysis demonstrated seeming equivalent transposase expression within T0 lines from all four groups.
T1 Progeny Screening
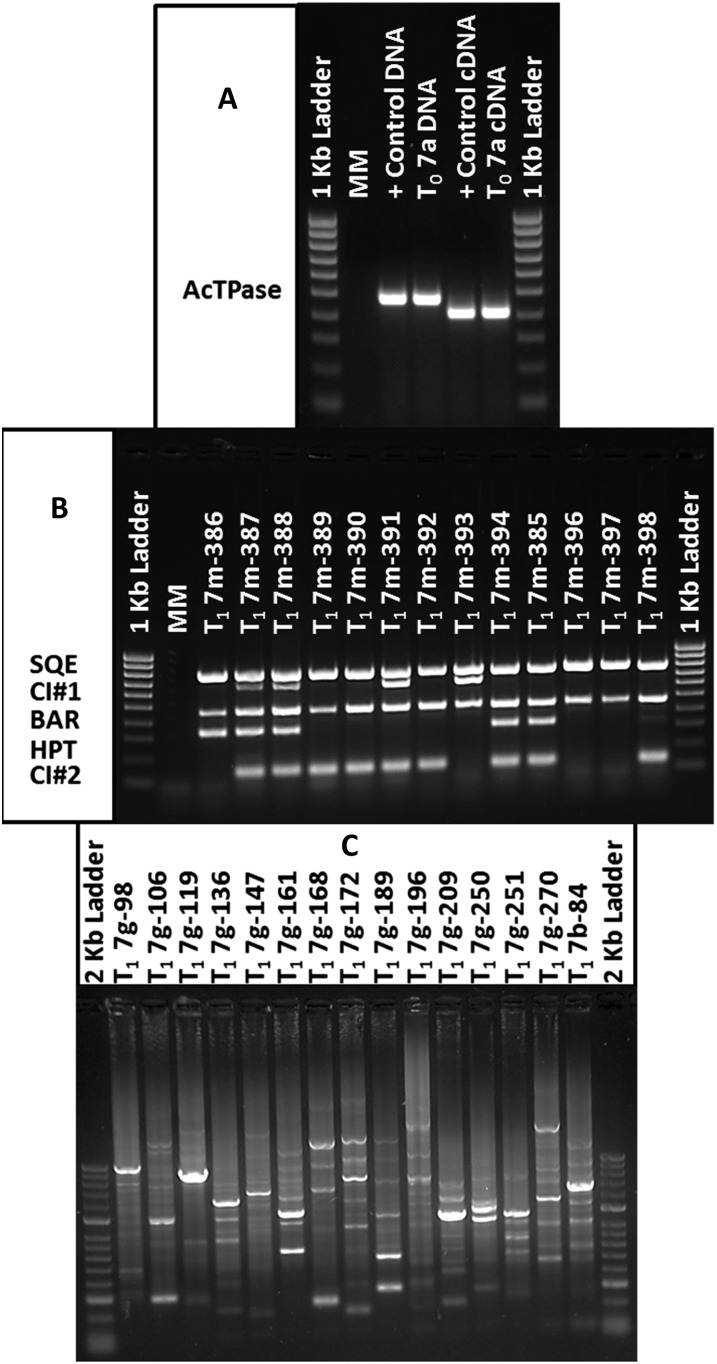
A functional progeny-screening protocol was developed based on selectable marker genes within the T-DNA construct. GFP expression was insufficient for selection at any stage of development during tissue culture and T1 progeny analysis. Exploiting herbicide resistance conveyed by the BAR gene within the Ds element, spray applications of Liberty herbicide were highly effective in selection against wild-type and Ac-only seedlings (Fig. 2, A and B). Due to the ineffectiveness of GFP as a functional marker in the tomato population, a hygromycin leaf-painting procedure was developed and refined to distinguish “Ds-only” individuals from Ac/Ds and Ac + Ds seedlings. Necrotic spots visible on both the upper and lower leaf surfaces 5 to 7 d after painting revealed the absence of the Ac element and its hygromycin resistance gene (Fig. 2, C and D). Leaf damage was strictly limited to areas painted with hygromycin solution, with no damage appearing on other parts of tested seedlings. Necrotic regions did not change size after screening and remained readily identifiable until natural leaf senescence. The putative Ds-only transposants (herbicide resistant, hygromycin sensitive) were verified by multiplex PCR (Fig. 3B). A standard PCR multiplex utilizing three primer pairs was developed to identify T-DNA sequences conferring glufosinate resistance (BAR), hygromycin resistance (Hyg), and the native tomato gene squalene epoxidase (SQE). This multiplex combination was adjusted to include zygosity primers and the T-DNA primer, Ds5′-3, in order to identify previously sequenced common insertions of the Ds-ATag element. Samples amplifying for zygosity products during multiplex testing were excluded from further evaluation. The percentage of Ds-only T1 progeny recovered among the total progeny of each family varied from 5% to 25% (Table I). After common insertions were excluded, 9% to 64% of these Ds-only progeny were found to harbor independent insertions of Ds (Table I). The screening protocol was found to be 74% effective in identifying Ds-only individuals using the combination of herbicide spraying and hygromycin leaf painting. The protocol of combining zygosity primers with the PCR multiplex to identify common insertions prior to performing more expensive TAIL-PCR varied in efficiency by family and the reliability of the zygosity primer designed for the common insertion site.
Figure 2.
Phenotypic screening of T1 progeny. A, Herbicide-resistant 6-week-old tomato seedling (left) next to a sensitive seedling (right) 7 d after spraying with 0.05% (v/v) Liberty (glufosinate) solution. B, Closeup of an herbicide-resistant leaf (left) next to a sensitive leaf (right) removed from the seedlings in A. C, Adaxial side of four leaves detached from different seedlings sensitive to hygromycin 6 d after painting with 100 mg L−1 hygromycin + two drops of polysorbate 20. Reaction to hygromycin was localized to the region of a leaf that had been painted. D, Abaxial side of a leaf sensitive to hygromycin displaying the diagnostic necrotic spots that distinguished hygromycin sensitivity from common leaf damage due to pathogens or mechanical injury.
Figure 3.
Analysis of transgenic and transposon-tagged lines by RT-PCR, multiplex PCR, and TAIL-PCR. A, Expression of transposase in whole inflorescence RNA. The PCR product spans transposase intron 2 (DNA product = 476 bp, cDNA product = 405 bp). Positive control DNA and cDNA extracted from transgenic potato inflorescence are shown. B, Screening of 13 T1 progeny (lanes 3–14) by multiplex PCR and zygosity testing. SQE, Squalene epoxidase (distinguishes the wild types from failed reactions); CI#1, common insertion 1 from a somatic transposition event early in development (defines T0 group 2); BAR, glufosinate herbicide resistance gene (Ds element); HPT, hygromycin resistance gene (Ac element); CI#2, common insertion 2 from somatic transposition event unique to T0 7m. Plants in lanes 13 and 14 represent potentially unique transposants destined for TAIL-PCR. Lanes 1 and 16 are the 1-kb ladder; lane 2 is master mix without DNA (negative control). C, Visualization of primary TAIL-PCR products. Lanes 2 to 16 (T1 7g-98–T1 7b-84) are amplification products obtained from 16 independently identified Ds-only, BAR-positive, hyg-sensitive T1 plants; lanes 1 and 17 are the 2-kb ladder.
Table I. T1 progeny screening applied to 15 T0 families derived by self-pollination and two cross pollinations of wild-type cv M82 with T0 lines 7i and 7o.
All T0 plants harbored a nonmobile Ac element and a mobile Ds element. The progeny were screened for resistance to Liberty herbicide (BAR+) to identify those with the Ds element, followed by sensitivity to hygromycin leaf painting (hyg−) of herbicide-resistant seedlings to identify those with the Ac element. Screening efficiency measured the percentage of seedlings identified as putative transposants (herbicide resistant, hygromycin sensitive) that were verified as Ds only by multiplex PCR. Totals represent only a subset of T1 progeny screened during the project. For χ2, expected was 3:1 BAR+/BAR− for T0 selfs, 1:1 for wild type × T0 crosses. **P < 0.01; ns, not significant, P > 0.05. Ds only (%) = (Ds only/total) × 100. Screening efficiency = (Ds only/hyg−) × 100. NA, Not applicable.
| T0 Family | BAR+ | BAR− | Total | BAR+:1 BAR− | χ2 | hyg− | Ds Only | Ds Only | Independent Ds Only | Independent Ds Only | Screening Efficiency |
|---|---|---|---|---|---|---|---|---|---|---|---|
| % | % | ||||||||||
| 7a | 498 | 474 | 972 | 1.1 | 292** | 49 | 46 | 5 | 28 | 61 | 94 |
| 7b | 609 | 187 | 796 | 3.3 | 0.9ns | 144 | 68 | 9 | 42 | 62 | 47 |
| 7c | 578 | 58 | 636 | 10.0 | 85** | 141 | 102 | 16 | 34 | 33 | 72 |
| 7g | 438 | 22 | 460 | 19.9 | 99** | 91 | 81 | 18 | 39 | 48 | 88 |
| 7h | 172 | 128 | 300 | 1.3 | 49** | 0 | 0 | NA | 0 | NA | NA |
| 7i | 804 | 443 | 1,247 | 1.8 | 73** | 185 | 138 | 11 | 46 | 33 | 75 |
| Wild-type × 7i | 240 | 400 | 640 | 0.6 | 40** | 52 | 42 | 7 | 25 | 60 | 81 |
| 7k | 253 | 65 | 318 | 3.9 | 3.3ns | 0 | 0 | NA | 0 | NA | NA |
| 7m | 641 | 25 | 666 | 25.6 | 159** | 134 | 90 | 14 | 22 | 24 | 67 |
| 7o | 81 | 34 | 115 | 2.4 | 1ns | 0 | 0 | NA | 0 | NA | NA |
| Wild-type × 7o | 267 | 293 | 560 | 0.9 | 1.1ns | 0 | 0 | NA | 0 | NA | NA |
| 7p | 310 | 21 | 331 | 14.8 | 60** | 93 | 83 | 25 | 39 | 47 | 89 |
| 7r | 264 | 105 | 369 | 2.5 | 2.2ns | 59 | 40 | 11 | 18 | 45 | 68 |
| 7s | 62 | 42 | 104 | 1.5 | 13** | 26 | 18 | 17 | 6 | 33 | 69 |
| 7t | 45 | 3 | 48 | 15.0 | 8** | 12 | 11 | 23 | 7 | 64 | 92 |
| 7u | 107 | 2 | 109 | 53.5 | 30** | 30 | 27 | 25 | 12 | 44 | 90 |
| 7z | 202 | 23 | 225 | 8.8 | 26** | 38 | 33 | 15 | 3 | 9 | 87 |
| Total | 5,571 | 2,325 | 7,896 | 1,054 | 779 | 321 | 74 | ||||
The segregation ratio of resistant-to-sensitive genotypes varied by T0 family and ranged from 1.1:1 to 54:1 for selfed progeny of primary transgenic lines and from 0.6:1 to 0.9:1 for T1 progeny of wild type × T0 crosses (Table I). χ2 tests for the expected 3:1 segregation in T0 selfs, or 1:1 in crosses to the wild type, revealed that the expected segregation ratio only occurred in four (7b, 7k, 7o, and 7r) of 15 selfed families and in one (7o) of two crosses to the wild type. Families 7c, 7g, 7m, 7p, 7t, 7u, and 7z exhibited an excess of resistant progeny, whereas families 7a, 7h, 7i, and 7s, and wild type × 7i crosses demonstrated an excess of sensitive progeny. These trends do not coincide with the previously established groups of families based on transposition of Ds. This inconsistency was expected, due to the ability of Ds to transpose to new locations independently in each T0 plant, leading to an unpredictable segregation of the Ds element among T1 progeny.
Characteristics of Transposition
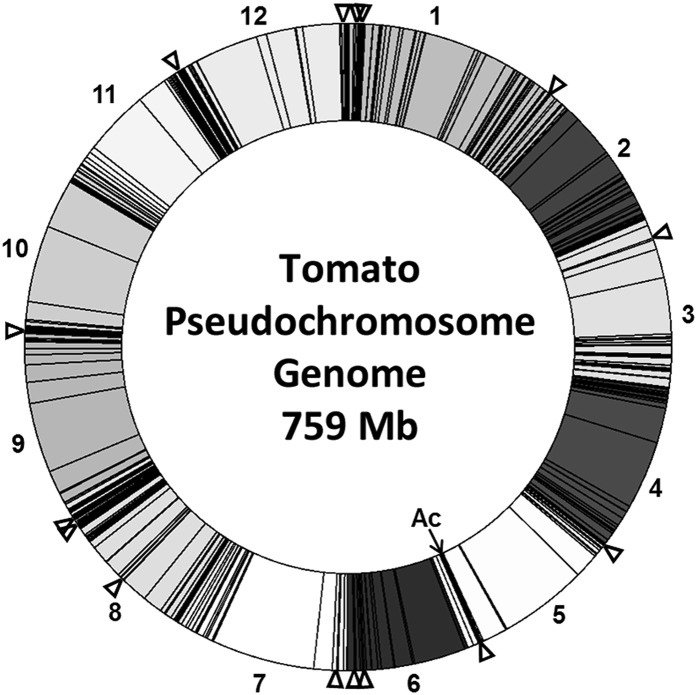
Amplification of genomic regions flanking transposed Ds insertions revealed incidences of insertion onto every pseudochromosome in the tomato genome (Fig. 4; Bombarely et al., 2011). As a whole, the construct engendered global transposition by distributing Ds elements throughout the genome. When the distribution of insertions was broken down by T0 family, the presence of a common insertion of Ds by somatic transposition on a particular chromosome occurred in conjunction with a greater incidence of independent insertions on that same chromosome (Table II). On average, 29% of independent Ds insertions in the T1 progeny of a given family were found on a pseudochromosome harboring a known common insertion. Analysis of genomic regions flanking these insertion sites revealed a greater incidence (64%) of insertion into genes and their associated regulatory regions rather than into intergenic sequences (36%; Table III). Insertions into intergenic regions were divided into those occurring greater than or less than 5 kb from the nearest described gene or putative gene model. Insertions greater than 5 kb from the nearest gene have been considered less likely to enhance transcription based on primary analysis of T2 lines.
Figure 4.
Graphic distribution of 509 transposed Ds elements to tomato pseudochromosomes (Tomato WGS Chromosomes SL2.40). Self and wild type × T0 progeny from every T0 family are included. Both common and unique insertions are represented by a single band. The original T-DNA insertion site during transformation (Ac element) is denoted by ←Ac on chromosome 5. Locations of common somatic insertions are denoted by triangles.
Table II. Distribution of transposed Ds elements to tomato pseudochromosomes (Tomato WGS Chromosomes SL2.40), sorted by T0 family, including T0 self and wild type × T0 progeny.
Underlined numbers indicate that a common insertion (somatic transposition) occurred on the respective pseudochromosome in a given family; common insertions were counted as only a single independent transposition (e.g., for family 7a, there were 10 independent insertions on pseudochromosome 1, including one that was found in numerous seedlings).
| T0 Family | Pseudochromosome |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | Total | |
| 7a | 10 | 1 | 9 | 4 | 1 | 1 | 4 | 0 | 5 | 2 | 0 | 4 | 41 |
| 7b | 26 | 3 | 1 | 7 | 2 | 4 | 2 | 10 | 7 | 4 | 3 | 3 | 72 |
| 7c | 5 | 2 | 2 | 0 | 1 | 3 | 0 | 19 | 1 | 1 | 16 | 3 | 53 |
| 7f | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 4 | 0 | 0 | 4 | 0 | 12 |
| 7g | 9 | 4 | 7 | 8 | 5 | 4 | 5 | 30 | 2 | 5 | 19 | 5 | 103 |
| 7i | 58 | 9 | 5 | 6 | 5 | 5 | 5 | 4 | 4 | 3 | 2 | 5 | 111 |
| 7m | 12 | 2 | 1 | 5 | 1 | 0 | 0 | 8 | 2 | 1 | 2 | 2 | 36 |
| 7p | 5 | 1 | 2 | 2 | 1 | 10 | 3 | 6 | 0 | 2 | 5 | 6 | 43 |
| 7r | 1 | 1 | 1 | 0 | 8 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 14 |
| 7s | 0 | 0 | 2 | 0 | 3 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 6 |
| 7t | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 1 | 1 | 6 |
| 7u | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 1 | 0 | 0 | 4 | 1 | 9 |
| 7z | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 3 |
| Total | 129 | 23 | 31 | 33 | 28 | 29 | 22 | 86 | 21 | 20 | 57 | 30 | 509 |
Table III. Insertion site classification of 509 identified Ds transposition events based on Tomato WGS Chromosomes SL2.40.
Insertion site preference is displayed as a percentage of the 509 total insertions. Coding = exons, unigenes, cDNA clones, and unknown protein models; promoter = up to 1 kb upstream from the nearest gene model; 3′ UTR (untranslated region) = up to 500 bp downstream from the nearest gene model; intergenic = distance to the nearest gene model, upstream or downstream.
| Parameter | Promoter | Coding | Intron | 3′ UTR | Intergenic |
|
|---|---|---|---|---|---|---|
| <5 kb | >5 kb | |||||
| Ds insertions | 75 | 139 | 80 | 32 | 95 | 88 |
| Percentage | 15 | 27 | 16 | 6 | 19 | 17 |
T2 Analysis
In order to observe the effect of the activation tag on gene expression, we selected two T1 lines for T2 analysis using zygosity primers and quantitative real-time RT-PCR. T1 line Wt7i-15 harbored a single intergenic insertion of Ds adjacent to the gene, Solyc12g010240.2.1, annotated as similar to CRABS CLAW (Fragment), and Solyc01g010250.2.1, annotated as 6-phosphogluconolactonase (Tables IV and V; Fig. 5) and expressed a dominant mutant phenotype (Fig. 6B). T1 line 7i-203 had no obvious mutant phenotype and harbored a Ds insertion in the single intron of Solyc12g006160.1.1, a gene annotated as homologous to Downward leaf-curling protein, placing the element 7.3 kb downstream of Solyc12g006170.1.1, annotated as WRKY transcription factor2 (Tables IV and V). Over 20 T2 progeny for each line were observed and screened for the segregation of Ds, with the dominant mutant phenotype in line Wt7i-15 with the Ds element. For hemizygous or homozygous Ds T2 plants of line Wt7i-15, RT-PCR revealed a 981- to 1,137-fold increase in Solyc12g010240.2.1 gene expression as well as a 5- to 7-fold increase in transcripts of Solyc01g010250.2.1. For line 7i-203, a 2- to 6-fold increase was observed in Solyc12g006160.1.1, with a 0- to 2-fold increase for Solyc12g006170.1.1. No significant changes in transcription were observed when comparing progeny homozygous for the Ds element with their hemizygous siblings. The dominant mutant phenotype of line Wt7i-15 was visually identical when comparing homozygous and hemizygous siblings, while wild-type plants demonstrated restoration of the cv M82 phenotype.
Table IV. Quantitative real-time RT-PCR analysis of T2 progeny for two T1 mutant lines harboring a single insertion of the Ds element.
Segregation of the Ds element among T2 progeny of both lines was determined by zygosity primer screening. For χ2, expected was 1:2:1 Ds for T1 selfs. ns, Not significant.
| T1 Line | Homozygous | Hemizygous | Wild Type | Transgenic:1 Wild Type | χ2 |
|---|---|---|---|---|---|
| Wt7i-15 | 6 | 13 | 4 | 4.75 | 0.74ns |
| 7i-203 | 3 | 17 | 5 | 4.0 | 3.56ns |
Table V. Quantitative real-time RT-PCR analysis of T2 progeny for two T1 mutant lines harboring a single insertion of the Ds element.
Fold change in transcription is shown for genes of interest bordering Ds insertions. All fold changes compare a Ds element zygosity genotype with sibling wild-type genotypes using the comparative cycle threshold method. Significant differences were calculated using Tukey’s honestly significant difference test. Mean Ct values within columns followed by the same letter are not significantly different (P < 0.05). Ct, Cycle threshold.
| Zygosity | T1 Line |
|||||||
|---|---|---|---|---|---|---|---|---|
| Wt7i-15 |
7i-203 |
|||||||
| CRABS CLAW (Fragment) |
6-Phosphogluconoactonase |
Downward Leaf-Curling Protein |
WRKY Transcription Factor2 |
|||||
| Mean Ct | Fold Change | Mean Ct | Fold Change | Mean Ct | Fold Change | Mean Ct | Fold Change | |
| Homozygous | 16.32A | 1,137 | 20.08A | 7 | 21.21A | 6 | 22.75A | 2 |
| Hemizygous | 16.38A | 981 | 20.25A | 5 | 22.23B | 2 | 22.98AB | 1.5 |
| Wild type | 26.53B | 1 | 22.91B | 1 | 23.60C | 1 | 23.43B | 1 |
Figure 5.
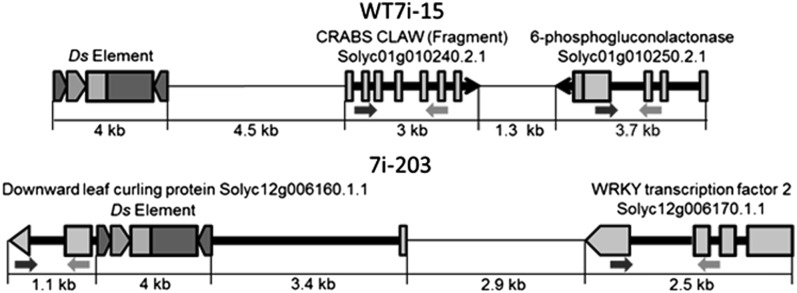
Insertion sites of the Ds element for mutant lines Wt7i-15 and 7i-203 (Tomato WGS Chromosomes SL2.40). −, Both Ds element inserted. Primer sites for quantitative real-time RT-PCR are denoted by arrows below the exons.
Figure 6.
Phenotypes for seven T1 activation-tagged mutants. A, 7c-79: rolled leaves, no lateral branching, weak stem. B, Wt7i-15: twisted, soft leaves. C, 7g-300: crumpled leaves. D, 7i-271: twisted leaves, extreme dwarfism. E, 7g-25: tightly rolled, leathery leaves, dwarf, aborted flowers. F, Wt7i-48: extensive yellow variegation, aborted flowers. G, 7i-413: leaves, internodes, inflorescences, fruit, and seeds proportionally reduced in size.
Phenotypic Mutants
Fifteen T1 progeny from several actively transposing families were visually identified as having unique mutant phenotypes during screening. Most of these phenotypes involved leaf structural changes, yellow variegation, and changes in plant size. Loss of fertility due to flower abortion was also observed in several mutant lines. Among the most interesting of these lines was 7c-69 (Fig. 6A), exhibiting elongated internodes, no lateral branching, crumpled leaves, and a weak stem. An equally complex phenotype was identified in 7i-271 (Fig. 6D), which demonstrated short internodes, reduced leaves, and slow growth. A third phenotype cataloged was that of 7g-25 (Fig. 6E), which was observed to have tightly rolled, pale leaves with a leathery texture, a shortened stature, and aborted flowers. Further analysis of these lines would be required to verify if activation tags on transposed Ds elements interact with neighboring genes. Efforts to preserve sterile phenotypic mutants through in vitro propagation and cryogenic tissue stocks have been successful in most cases. In addition to these mutants, the T1 population exhibited many potential metabolic or more subtle mutant phenotypes that may be selected for detailed analysis based on insertion site. The population also contained knockout mutants that may exhibit altered phenotypes once Ds insertions can be evaluated in homozygous genotypes in the T2 generation. Somaclonal variation cannot be ruled out as the cause of some of these striking phenotypes until phenotypic segregation with the activation tag has been demonstrated.
Database
Information on the transposants is presented in an online database at http://hortmutants.vbi.vt.edu. Transposants (n = 505) are grouped by their specific launchpad (a, b, c, f, g, i, m, p, r, s, t, u, and z). For each line, the TAIL-PCR sequence is presented along with the best BLAST hit to the tomato genome assembly on the Sol Genomics Network to create a temporary feature to visualize the location of insertion (yellow shading indicates the alignment of the entire TAIL-PCR sequence, with a blue bar indicating the insertion site).
DISCUSSION
Although the Agrobacterium tumefaciens-mediated transformation efficiency was only a fraction of 1%, it was possible to exploit the transposition frequency of a single T0 line to initiate the development of a functional resource for activation tagging in tomato. The practice of using micropropagation to produce many clonal plants from a single tissue culture regenerant proved valuable, as it multiplied T1 seed production by up to 25 times. This strategy also capitalized on the behavior of transposase in Ac/Ds-ATag-Bar_gosGFP by isolating chimeric tissue from the original transformant into separate plantlets, allowing germinal transposition from multiple sites of Ds integration. The selection of a self-fertile, true breeding tomato cultivar allowed crossing to nontransgenic cv M82, thus maximizing T1 seed production. Pollen could be collected from transgenic flowers and distributed to multiple nontransgenic plants, all while still obtaining transgenic self-progeny.
Modifications made to the transformation protocol yielded several transgenic lines after established procedures failed to produce results when applied to cv M82. Even with modification, the protocol resulted in a callus and shoot regeneration phase lasting upward of a full year. The long callus phase subsequent to transformation likely resulted in undesirable somaclonal variation, as evidenced by two out of three regenerated T0 lines being useless due to sterility and loss of the transposable element. More work is needed to develop transformation protocols specific to cv M82 for it to become a competitive model organism. This barrier against routine transformation makes ongoing populations as reported in this paper of even greater importance in expanding current mutant resources in tomato.
The lack of observable GFP expression proved to be a shortcoming of the Ac/Ds-ATag-Bar_gosGFP construct in our cv M82 transgenic tomato. This was unexpected, as Quadrana et al. (2011) demonstrated clear GFP expression in tomato ‘Moneymaker’ leaves and fruit after transformation with pBIN-35S-gpf; however, they selected two high-expression lines among 25 primary transgenic plants. This deficiency of the Ac/Ds-ATag-Bar_gosGFP construct may be due to issues with the compatibility of monocot Gos promoter in a dicot system, since the construct was designed to work in rice, where more than 5,000 stable insertions have been generated using the markers (E. Guiderdoni, personal communication). The loss of GFP was compensated by the development of an in planta hygromycin leaf-painting procedure, for which a direct precedence in the literature could not be found for tomato. A similar procedure was described in Arabidopsis to screen for transposase-containing lines using a hygromycin painting assay (Speulman et al., 1999). With refinement, this procedure provided effective screening when used in conjunction with well-established glufosinate herbicide-screening protocols. Even though the effectiveness of hygromycin leaf painting has not been evaluated in other crops, the unique and easily identifiable sensitivity reaction demonstrated in tomato give it the potential to be a widely applicable screening procedure. While the described herbicide and hygromycin screening protocol proved effective, both were sensitive to improper application. Herbicide sprays were easily rendered ineffective if solutions were not sufficiently absorbed by leaves prior to regular watering. More importantly, application of herbicide to older seedlings (more than 6 weeks; Fig. 2) often resulted in less obvious sensitive phenotypes, from which plants could recover to the point of being indistinguishable from resistant seedlings if phenotypic data were not collected within 7 d following treatment. Application of herbicide to 3- to 5-week-old seedlings consistently resulted in complete necrosis of sensitive individuals, which rendered screening easier and more efficient. While hygromycin leaf painting proved less influenced by watering and evaluation time, sensitive phenotypes could be easily confused with other forms of leaf damage that may occur in a greenhouse setting. Screening results from T1 progeny in which complications arose that confounded effective data collection due to the aforementioned factors were excluded from efficiency calculations.
The germinal transposition of the Ds element in the T1 population produced during this project is encouraging for implementation of a high-throughput system to produce large populations of activation-tagged mutants. Variation for germinal transposition among the different clonally propagated copies of our single Ac/Ds transgenic plant was extensive, as reported for Ds excision among F1 plants from Ds-only to Ac-only tomato after backcrossing the F1 to wild-type tomato (Carroll et al., 1995; Břza et al., 2000). Most importantly, the Ds element globally transposed to all 12 tomato chromosomes from its initial launchpad site on pseudochromosome 5 (Fig. 4). Such widespread distribution has been observed before in tomato, with some indication of a preference of transposition to sites on the same chromosome as the launchpad (Healy et al., 1993; Thomas et al., 1994; Bříza et al., 1995, 2002; Carroll et al., 1995). When the distribution of these transposed insertions was taken into consideration with other data from T1 progeny screening, further insights into the nature of transposition in these lines were revealed. Given that no incidences of the Ds element remaining in the original T-DNA insertion site were detected within thousands of progeny from actively transposing families, it can be deduced that unique transposition events were the result of multiple movements of the Ds element during the lifespan of the T0 plant. This scenario is further supported by the family-specific detection of disproportionately more unique insertions onto chromosomes also harboring a common insertion (Table II). The implications of these data are that a given Ds element insertion site could be effectively utilized as a new launchpad for future insertions, simply by reintroducing the transposase gene through cross pollination of Ds-only plants with Ac-only plants; the transactivation system described by Schmitz and Theres (1994) effectively demonstrates this model. Transposition of cis-acting elements to the entire genome has been demonstrated in rice (Kumar et al., 2005; Qu et al., 2008). Although the first transposition of the Ds-ATag in our original transgenic plant would have occurred via cis-activation, once the Ds-ATag had moved to another chromosome, the action of the Ac-TPase to relaunch the Ds-ATag would have been trans in most cases, as Ac-TPase would have remained stationary on pseudochromosome 5. Under either strategy, the result is genomic saturation, whether by cis-activation of many transgenic plants with insertion sites on different chromosomes or by trans-activation of Ds-ATag elements occurring on every chromosome. With careful selection, lines containing a Ds element on a target chromosome or euchromatic region can be utilized to produce more unique insertions and mutants. More conveniently, Ds elements conferring a screenable phenotype due to an adjacent tagged gene can be used as excision markers to select for new insertions positively with glufosinate selection for inserted Ds. These mutants would contain Ds activation tags preferentially distributed to the targeted genomic region neighboring the originally selected Ds insertion. The viability of this strategy was partially confirmed, as a population of T2 plants derived from a single Ac/Ds T1 individual was found to exhibit novel Ds insertions sites that, to our knowledge, had not been identified previously in the parent plant.
The reliable generation of unique transposition events in this mutant population may in part be a function of a fortuitous insertion of the original T-DNA vector harboring the conjoined Ac and Ds elements. In theory, transposase expression should be consistent with the Ac promoter driving the transcription of its coding sequence. In the case of the Ac/Ds-ATag-Bar_gosGFP construct, the promoter for transposase was the native maize Ac promoter, which has proven conducive to use in transposon-tagging applications (Jones et al., 1994). It is also known that regions flanking a T-DNA insertion site may influence transgene expression, as observed with promoter- and enhancer-trapping schemes (Topping et al., 1994; Wu et al., 2003). Expression profiling data for the Arabidopsis ortholog of the 60S ribosomal protein L29 into which the T-DNA inserted dictated widespread transcription of the gene, with the greatest expression occurring in shoot tips. The influence of a promoter region driving the somatic expression of transposase is consistent with the frequent somatic transpositions of Ds detected in each T0 family. Furthermore, the frequency of germinal transposition may have been enhanced by pollen-specific cis-regulatory elements on adjacent genes. The potential influence of these elements is consistent with the clear tendency for frequent germinal transposition of Ds to occur in T0 families where an initial transposition event had already taken place. These putative interactions between genomic elements and transposase expression are supported by the current body of evidence concerning the influence of genomic elements on gene expression within T-DNA insertions (Mirza, 2005; Filipecki and Malepszy, 2006).
Classification of Ds insertions by their flanking regions demonstrated insertion site preferences reasonably consistent with results from screens of over 10,000 tagged lines in Arabidopsis, favoring insertion into genes and euchromatin (Kuromori et al., 2004). Of the 509 identified Ds insertions, 40% were in regions suitable for functional activation tagging (promoter, 3′ untranslated region, less than 5 kb intergenic; Table III). An additional 27% of the Ds insertions were within annotated or putative coding regions. While such locations may be less accommodating to activation tagging, these lines provide a potential source of knockout mutants for further study. The 16% of Ds insertions into introns are of unknown value, as such insertions may result in reduced function or activation-tagged phenotypes. Analysis of progeny derived from T1 mutant line 7i-203 by quantitative real-time RT-PCR revealed a 2- to 6-fold increase in amplicons for a gene harboring an insertion of Ds into an intron, indicating that at least some knockout insertions may result in activation tag phenotypes. The remaining 17% of transposed Ds elements were inserted into intergenic regions that were greater than 5 kb from the nearest described gene model, rendering these lines of unknown value. T2 analysis of mutant line Wt7i-15 demonstrated an over 1,000-fold increase in the transcription of a gene 4.3 kb upstream, indicating that these lines may still have mutant phenotypes. However, less than 10-fold change in expression was quantified for a gene 8.8 kb upstream. For the progeny of T1 mutant line 7i-203, little change in transcription was observed for a gene 6.3 kb upstream. Other insertions in this category are embedded into repetitive regions, limiting the opportunity for interaction with native genes. The implication of these insertion site preferences is that up to 83% of unique, activation-tagged progeny are potential phenotypic mutants.
The tendency for Ds to transpose to new locations near a previous insertion site on a given chromosome holds promise for the development of new ATag lines. Under this strategy, a group of single-insertion, Ds-only lines would be selected as potential launchpads based on the genomic locations of the Ds insertions (e.g. in close proximity to genes of interest, or possibly on a sparsely saturated chromosome). These Ds-only plants would then be crossed to sibling Ac-only plants, generating Ac + Ds plants in the subsequent generation. These new Ac + Ds lines could be self-pollinated, with the expectation of transposition of Ds to preferentially saturate the region of the genome surrounding the parental Ds location. Continued global transposition of Ds would yield novel insertions into nontarget regions. Further research is required to evaluate the feasibility and limitations of this strategy. Finally, the development of an effective progeny-screening protocol makes this population and its derivatives capable of functioning in a high-throughput system. These factors make the transgenic population developed during the course of this project a unique, new resource for functional genomics of tomato with a new array of phenotypic mutants available to augment those that have already been characterized from earlier transposon-tagging schemes (Bishop et al., 1996; van der Biezen et al., 1996; Keddie et al., 1998).
MATERIALS AND METHODS
Plant Transformation and Selection
The activation-tagging construct Ac/Ds-ATag-Bar_gosGFP (Fig. 1) in the vector plasmid PMOG22 was selected for use in tomato (Solanum lycopersicum) transformation based on its active transposition in rice (Oryza sativa; Trijatmiko, 2005). Primary leaf explants taken from cv M82 seedlings grown for 21 d in vitro in Magenta boxes (Magenta Plastics; four to five seedlings per box with 40 mL of Murashige and Skoog basal medium [Murashige and Skoog, 1962]) in a growth chamber were selected and transformed according to protocols published by Khoudi et al. (2009), with several modifications made during transformation and regeneration. Modified media formulations are listed in Supplemental Table S1. Selection on hygromycin was not initiated for the first 3 weeks following transformation. The concentration of zeatin in shoot regeneration and shoot elongation media was doubled in all cases. The concentration of cefotaxime in regeneration and elongation media was increased to 500 mg L−1, while carbenicillin was excluded. As a further modification, regenerating shoot masses were kept on shoot regeneration medium until a readily identifiable apical meristem was formed. Only masses with identifiable meristems were transferred to elongation medium. Once a shoot had regenerated from a putatively transformed callus and elongated to have multiple nodes, the apical portion was removed to rooting medium and the base with a single node was retained until an axillary branch had grown sufficiently to repeat the process. By this method of vegetative propagation in tissue culture, each transgenic plant was represented by a population of consecutively lettered shoots. Plantlets were transferred to rooting medium (Murashige and Skoog basal medium + vitamins, 4.43 g L−1; Suc, 30 g L−1; indole-3-acetic acid, 2 mg L−1; cefotaxime, 250 mg L−1; Phytagel, 2 g L−1) for 7 d or until roots emerged. Rooted cuttings were transferred to hydrated autoclaved peat plugs (Jiffy Products of America) in Magenta boxes for 7 d to promote acclimation to potting medium before transfer to a greenhouse facility. All tissue cultures were incubated in a growth chamber under 16-h days at 25°C under 110 μE m−2 s−1 fluorescent light and 8-h nights at 20°C.
T1 Population Generation
T0 plants were grown to maturity in soilless mix (equal volumes of peat moss, coarse vermiculite, and perlite + 30 kg m−3 gypsum) in 2.5-L pots and allowed to self-pollinate with the assistance of daily flower vibration by a pollinator wand. Crosses were made between nontransformed cv M82 plants and transformed T0 lines, with the transgenic plant acting as the pollinator in all cases. Seeds and their accompanying ovary flesh were cut from whole ripened fruit and further cleaned by soaking in approximately 100 mL of 1 m HCl in a 125-mL flask under 20 min of constant blending by a stir bar. Seeds were rinsed with tap water, then dried for 24 h at 37°C prior to storing in 5-mL capsules with silica bead desiccant.
Selection of Transposants
All seedlings were grown in greenhouses at 25°C under a 16-h photoperiod using natural and fluorescent light. T1 seeds obtained from T0 selfs or wild type × T0 crosses were soaked in tap water for 12 h and sown in soilless mix into flats of 48- or 96-cell packs. Emerging seedlings were screened with glufosinate herbicide (Bayer CropScience) after 3 to 4 weeks by spraying with an aqueous 0.05% (v/v) Liberty solution. Reactions to the herbicide were evaluated after 5 to 7 d, and sensitive seedlings were discarded. Glufosinate-resistant seedlings were screened for hygromycin resistance by painting a primary or secondary leaf on each plant with a solution of 100 mg L−1 hygromycin + two drops of polysorbate 20. Reactions to the antibiotic were evaluated after 5 to 7 d based on the presence of characteristic necrotic regions indicative of sensitivity to hygromycin. Hygromycin-sensitive seedlings were identified as putative transposants and transplanted in 4-L nursery pots for flowering and fruit collection.
DNA Extraction and PCR Screening
A single expanded leaf was collected from each glufosinate-resistant, hygromycin-sensitive seedling and frozen with liquid nitrogen in a 2-mL microcentrifuge tube containing a single No. 2 steel shot pellet (Ballistic Products). The tissue was ground using a Geno/Grinder 2000 (BT&C) at cryogenic temperature. The DNA was isolated from ground tissue using a modified cetyl-trimethyl-ammonium bromide protocol (Doyle and Doyle, 1987; Supplemental Materials and Methods S1). Isolated DNA was diluted 10-fold for PCR analysis. The genotype of putative transposants was confirmed in a multiplex PCR (Fig. 3), which amplified the hygromycin resistance gene from the Ac element (HPT5F and HPT5R primers; Supplemental Table S2), the glufosinate resistance gene from the Ds element (G38BARF and G38BARR primers; Supplemental Table S2), and a sequence from squalene epoxidase within the tomato genome (Sl_SQEF and Sl_SQER primers; Supplemental Table S2). For families containing Ds insertions common to many T1 seedlings, a single zygosity primer and the sequencing primer Ds5′-3 (Supplemental Table S2) from within the Ds element were included to identify transposants harboring the common insertion (Fig. 3B). Seedlings for which only the BAR gene and the squalene epoxidase gene were amplified were identified as putative unique transposants.
Identification of Ds Insertion-Flanking Regions
Sequences flanking Ds insertions in unique transposants were amplified using primers and thermocycler protocols according to Tsugeki et al. (1996). The protocol was modified to include primer design improvements according to Liu and Chen (2007), namely the use of long arbitrary degenerate primers and the addition of a 21-nucleotide sequence corresponding to the AC1 primer used in the primary and secondary TAIL-PCRs. Only the preamplification and primary TAIL-PCR were carried out. Primary products were run on a 1% agarose/1× Tris-acetate ethylenediaminetetraacetic acid gel with 0.1% ethidium bromide (Fig. 3B). The brightest band(s) in each lane was excised and isolated (Supplemental Materials and Methods S1). Extracted bands were sequenced using the respective TAIL primer originally intended for use in secondary TAIL-PCR (Ds5′-3 and Ds3′-3; Supplemental Table S2). Resulting sequences were trimmed of T-DNA and aligned to the prepublication tomato genome (Bombarely et al., 2011) using a BLAST engine. Insertion sites were assumed to correspond to single alignment hits with a score of less than e−30 or to the location with the lowest e-value in cases of multiple alignments. The identities and orientations of gene models up to 10 kb upstream and downstream from Ds insertion sites were recorded. Insertions for which sequencing produced multiple, nearly identical hits and those that did not align to the current genome (Tomato WGS Chromosomes SL2.40) were not counted as unique mutant lines during further analysis.
Quantitative Real-Time RT-PCR
T2 progeny from two selected Ds-only T1 lines were analyzed using zygosity primers designed from the genome regions flanking the T-DNA insertion. Nine sibling T2 plants were selected from each line, including three individuals each of homozygous and hemizygous T-DNA insertions and three wild-type individuals. T2 plants were transplanted into 7-L pots and grown for 8 weeks until flowering. For each individual T2 plant, three tissue samples were collected for RNA extraction, including juvenile leaf tissue, whole inflorescences with open flowers, and apical growing tips. All samples were collected and RNA was extracted according to protocol. The synthesis of complementary DNA (cDNA) and quantitative real-time RT-PCR were carried out using the DyNAmo SYBR Green 2-Step qRT-PCR Kit (Thermo Fisher Scientific) according to the provided protocols; 20-µL reactions loaded with 100 ng of cDNA were run on the 7300 Real Time PCR System (Applied Biosystems, Life Technologies). Primers for four putatively activation-tagged genes were designed to have a melting temperature of 60°C and a product ranging between 100 and 150 bp. Primers for the clathrin adaptor complex medium subunit (SGN-U314153) were run as a control for each sample in order to normalize the resulting data (Expósito-Rodríguez et al., 2008). A separate reaction was run for each tissue type from every T2 plant analyzed, yielding three replicates of each tissue type from each T-DNA zygosity genotype for a given insertion. In total, each of the two Ds insertions analyzed was represented by 27 independent reactions. Reaction products were visualized on agarose gels to confirm the presence of a single band of the expected size. No variation in expression levels according to tissue type was observed for the four genes tested. A cycle threshold value for each Ds insertion zygosity genotype was found by averaging all three tissue types tested in each of the three replicate plants from each genotype, yielding a total of nine replicates. Fold change was calculated using the comparative cycle threshold method, due to difficulties encountered while determining amplification efficiency for the control gene. For each fold change calculation, transcript quantity in transgenic lines either homozygous or hemizygous for a given Ds insertion was compared with those in wild-type sibling lines.
Database
Data were entered into a MySQL database and served dynamically by Java servlets running in an Apache Tomcat Web server.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Tissue culture media formulations.
Supplemental Table S2. Primer sequences.
Acknowledgments
We thank Dr. Rudi Trijatmiko for use of the activation tag construct, Kendall Upham for assistance in sample processing and seed collection, Jeff Burr for maintaining greenhouse facilities, Norma Manrique for assistance with transformation, Nan Lu and Tatiana Boluarte for efforts to sequence Ac/Ds-ATag-Bar_gosGFP, and Megan LeBlanc for assistance with quantitative PCR assays.
Glossary
- RT
reverse transcription
- cDNA
complementary DNA
- T-DNA
transfer DNA
- TAIL
thermal asymmetric interlaced
References
- Aboul-Soud MAM, Chen X, Kang JG, Yun BW, Raja MU, Malik SI, Loake GJ. (2009) Activation tagging of ADR2 conveys a spreading lesion phenotype and resistance to biotrophic pathogens. New Phytol 183: 1163–1175 [DOI] [PubMed] [Google Scholar]
- Bishop GJ, Harrison K, Jones JDG. (1996) The tomato Dwarf gene isolated by heterologous transposon tagging encodes the first member of a new cytochrome P450 family. Plant Cell 8: 959–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombarely A, Menda N, Tecle IY, Buels RM, Strickler S, Fischer-York T, Pujar A, Leto J, Gosselin J, Mueller LA. (2011) The Sol Genomics Network (solgenomics.net): growing tomatoes using Perl. Nucleic Acids Res 39: D1149–D1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bríza J, Carroll BJ, Klimyuk VI, Thomas CM, Jones DA, Jones JDG. (1995) Distribution of unlinked transpositions of a Ds element from a T-DNA locus on tomato chromosome 4. Genetics 141: 383–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bríza J, Niedermeierová H, Pavingerová D, Thomas CM, Klimyuk VI, Jones JDG. (2002) Transposition patterns of unlinked transposed Ds elements from two T-DNA loci on tomato chromosomes 7 and 8. Mol Genet Genomics 266: 882–890 [DOI] [PubMed] [Google Scholar]
- Břza J, Pavingerova D, Niedermeierová H, Rakouský S. (2000) Germinal excision and reinsertion frequencies of the mobile element Ds transposed from two unlinked T-DNA loci in tomato. Biol Plant 43: 185–192 [Google Scholar]
- Busov VB, Meilan R, Pearce DW, Ma C, Rood SB, Strauss SH. (2003) Activation tagging of a dominant gibberellin catabolism gene (GA 2-oxidase) from poplar that regulates tree stature. Plant Physiol 132: 1283–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll BJ, Klimyuk VI, Thomas CM, Bishop GJ, Harrison K, Scofield SR, Jones JDG. (1995) Germinal transpositions of the maize element Dissociation from T-DNA loci in tomato. Genetics 139: 407–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19: 11–15 [Google Scholar]
- Eshed Y, Zamir D. (1995) An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141: 1147–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Expósito-Rodríguez M, Borges AA, Borges-Pérez A, Pérez JA. (2008) Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol 8: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipecki M, Malepszy S. (2006) Unintended consequences of plant transformation: a molecular insight. J Appl Genet 47: 277–286 [DOI] [PubMed] [Google Scholar]
- Fladung M, Polak O. (2012) Ac/Ds-transposon activation tagging in poplar: a powerful tool for gene discovery. BMC Genomics 13: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giménez E, Pineda B, Capel J, Antón MT, Atarés A, Pérez-Martín F, García-Sogo B, Angosto T, Moreno V, Lozano R. (2010) Functional analysis of the Arlequin mutant corroborates the essential role of the Arlequin/TAGL1 gene during reproductive development of tomato. PLoS ONE 5: e14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez MD, Urbez C, Perez-Amador MA, Carbonell J. (2011) Characterization of constricted fruit (ctf) mutant uncovers a role for AtMYB117/LOF1 in ovule and fruit development in Arabidopsis thaliana. PLoS ONE 6: e18760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy J, Corr C, DeYoung J, Baker B. (1993) Linked and unlinked transposition of a genetically marked Dissociation element in transgenic tomato. Genetics 134: 571–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed]
- Huang J, Tang D, Shen Y, Qin B, Hong L, You A, Li M, Wang X, Yu H, Gu M, et al. (2010) Activation of gibberellin 2-oxidase 6 decreases active gibberellin levels and creates a dominant semi-dwarf phenotype in rice (Oryza sativa L.). J Genet Genomics 37: 23–36 [DOI] [PubMed] [Google Scholar]
- Jones DA, Thomas CM, Hammond-Kosack KE, Balint-Kurti PJ, Jones JD. (1994) Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 266: 789–793 [DOI] [PubMed] [Google Scholar]
- Kang HG, Kim J, Kim B, Jeong H, Choi SH, Kim EK, Lee HY, Lim PO. (2011) Overexpression of FTL1/DDF1, an AP2 transcription factor, enhances tolerance to cold, drought, and heat stresses in Arabidopsis thaliana. Plant Sci 180: 634–641 [DOI] [PubMed] [Google Scholar]
- Keddie JS, Carroll BJ, Thomas CM, Reyes MEC, Klimyuk V, Holtan H, Gruissem W, Jones JDG. (1998) Transposon tagging of the Defective embryo and meristems gene of tomato. Plant Cell 10: 877–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoudi H, Nouri-Khemakhem A, Gouiaa S, Masmoudi K. (2009) Optimization of regeneration and transformation parameters in tomato and improvement of its salinity and drought tolerance. Afr J Biotechnol 8: 6068–6076 [Google Scholar]
- Kondou Y, Higuchi M, Matsui M. (2010) High-throughput characterization of plant gene functions by using gain-of-function technology. Annu Rev Plant Biol 61: 373–393 [DOI] [PubMed] [Google Scholar]
- Krishnan A, Guiderdoni E, An G, Hsing YI, Han CD, Lee MC, Yu SM, Upadhyaya N, Ramachandran S, Zhang Q, et al. (2009) Mutant resources in rice for functional genomics of the grasses. Plant Physiol 149: 165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar CS, Wing RA, Sundaresan V. (2005) Efficient insertional mutagenesis in rice using the maize En/Spm elements. Plant J 44: 879–892 [DOI] [PubMed] [Google Scholar]
- Kuromori T, Hirayama T, Kiyosue Y, Takabe H, Mizukado S, Sakurai T, Akiyama K, Kamiya A, Ito T, Shinozaki K. (2004) A collection of 11 800 single-copy Ds transposon insertion lines in Arabidopsis. Plant J 37: 897–905 [DOI] [PubMed] [Google Scholar]
- Lee S, Jeon US, Lee SJ, Kim YK, Persson DP, Husted S, Schjørring JK, Kakei Y, Masuda H, Nishizawa NK, et al. (2009) Iron fortification of rice seeds through activation of the nicotianamine synthase gene. Proc Natl Acad Sci USA 106: 22014–22019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Persson DP, Hansen TH, Husted S, Schjoerring JK, Kim YS, Jeon US, Kim YK, Kakei Y, Masuda H, et al. (2011) Bio-available zinc in rice seeds is increased by activation tagging of nicotianamine synthase. Plant Biotechnol J 9: 865–873 [DOI] [PubMed] [Google Scholar]
- Lee S, Ryoo N, Jeon JS, Guerinot ML, An G. (2012) Activation of rice Yellow Stripe1-Like 16 (OsYSL16) enhances iron efficiency. Mol Cells 33: 117–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YGC, Chen Y. (2007) High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques 43: 649–650, 652, 654 [DOI] [PubMed] [Google Scholar]
- Massa AN, Childs KL, Lin H, Bryan GJ, Giuliano G, Buell CR. (2011) The transcriptome of the reference potato genome Solanum tuberosum Group Phureja clone DM1-3 516R44. PLoS ONE 6: e26801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews H, Clendennen SK, Caldwell CG, Liu XL, Connors K, Matheis N, Schuster DK, Menasco DJ, Wagoner W, Lightner J, et al. (2003) Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell 15: 1689–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner R, Chague V, Zhu Q, Emmanuel E, Elkind Y, Levy AA. (2000) Technical advance: a high throughput system for transposon tagging and promoter trapping in tomato. Plant J 22: 265–274 [DOI] [PubMed] [Google Scholar]
- Meissner R, Jacobson Y, Melamed S, Levyatuv S, Shalev G, Ashri A, Elkind Y, Levy A. (1997) A new model system for tomato genetics. Plant J 12: 1465–1472 [Google Scholar]
- Menda N, Semel Y, Peled D, Eshed Y, Zamir D. (2004) In silico screening of a saturated mutation library of tomato. Plant J 38: 861–872 [DOI] [PubMed] [Google Scholar]
- Mirza B. (2005) Influence of the nature of the T-DNA insertion region on transgene expression in Arabidopsis thaliana. Russ J Genet 41: 1322–1328 [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Qu S, Desai A, Wing R, Sundaresan V. (2008) A versatile transposon-based activation tag vector system for functional genomics in cereals and other monocot plants. Plant Physiol 146: 189–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrana L, Rodriguez MC, López M, Bermúdez L, Nunes-Nesi A, Fernie AR, Descalzo A, Asis R, Rossi M, Asurmendi S, et al. (2011) Coupling virus-induced gene silencing to exogenous green fluorescence protein expression provides a highly efficient system for functional genomics in Arabidopsis and across all stages of tomato fruit development. Plant Physiol 156: 1278–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Ariizumi T, Okabe Y, Asamizu E, Hiwasa-Tanase K, Fukuda N, Mizoguchi T, Yamazaki Y, Aoki K, Ezura H. (2011) TOMATOMA: a novel tomato mutant database distributing Micro-Tom mutant collections. Plant Cell Physiol 52: 283–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapir M, Oren-Shamir M, Ovadia R, Reuveni M, Evenor D, Tadmor Y, Nahon S, Shlomo H, Chen L, Meir A, et al. (2008) Molecular aspects of Anthocyanin fruit tomato in relation to high pigment-1. J Hered 99: 292–303 [DOI] [PubMed] [Google Scholar]
- Sato S, Tabata S, Hirakawa H, Asamizu E, Shirasawa K, Isobe S, Kaneko T, Nakamura Y, Shibata D, Aoki K. (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz G, Theres K. (1994) A self-stabilizing Ac derivative and its potential for transposon tagging. Plant J 6: 781–786 [DOI] [PubMed] [Google Scholar]
- Speulman E, Metz PLJ, van Arkel G, te Lintel Hekkert B, Stiekema WJ, Pereira A. (1999) A two-component enhancer-inhibitor transposon mutagenesis system for functional analysis of the Arabidopsis genome. Plant Cell 11: 1853–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speulman E, van Asperen R, van der Laak J, Stiekema WJ, Pereira A. (2000) Target selected insertional mutagenesis on chromosome IV of Arabidopsis using the En-I transposon system. J Biotechnol 78: 301–312 [DOI] [PubMed] [Google Scholar]
- Stevens MA, Dickinson GL, Aguirre MS (1982) UC 82: A High Yielding Processing Tomato. University of California, Davis
- Thomas CM, Jones DA, English JJ, Carroll BJ, Bennetzen JL, Harrison K, Burbidge A, Bishop GJ, Jones JDG. (1994) Analysis of the chromosomal distribution of transposon-carrying T-DNAs in tomato using the inverse polymerase chain reaction. Mol Gen Genet 242: 573–585 [DOI] [PubMed] [Google Scholar]
- Topping JF, Agyeman F, Henricot B, Lindsey K. (1994) Identification of molecular markers of embryogenesis in Arabidopsis thaliana by promoter trapping. Plant J 5: 895–903 [DOI] [PubMed] [Google Scholar]
- Trijatmiko KR (2005) Comparative Analysis of Drought Resistance Genes in Arabidopsis and Rice. Wageningen University, Wageningen, The Netherlands
- Tsugeki R, Kochieva EZ, Fedoroff NV. (1996) A transposon insertion in the Arabidopsis SSR16 gene causes an embryo-defective lethal mutation. Plant J 10: 479–489 [DOI] [PubMed] [Google Scholar]
- van der Biezen EA, Brandwagt BF, van Leeuwen W, Nijkamp HJ, Hille J. (1996) Identification and isolation of the FEEBLY gene from tomato by transposon tagging. Mol Gen Genet 251: 267–280 [DOI] [PubMed] [Google Scholar]
- van der Graaff E, Dulk-Ras AD, Hooykaas PJ, Keller B. (2000) Activation tagging of the LEAFY PETIOLE gene affects leaf petiole development in Arabidopsis thaliana. Development 127: 4971–4980 [DOI] [PubMed] [Google Scholar]
- Walden R, Fritze K, Hayashi H, Miklashevichs E, Harling H, Schell J. (1994) Activation tagging: a means of isolating genes implicated as playing a role in plant growth and development. Plant Mol Biol 26: 1521–1528 [DOI] [PubMed] [Google Scholar]
- Wang AR, Zhang CH, Zhang LL, Lin WW, Lin DS, Lu GD, Zhou J, Wang ZH. (2009) Identification of Arabidopsis mutants with enhanced resistance to Sclerotinia stem rot disease from an activation-tagged library. J Phytopathol 157: 63–69 [Google Scholar]
- Weigel D, Ahn JH, Blázquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrándiz C, Kardailsky I, Malancharuvil EJ, Neff MM, et al. (2000) Activation tagging in Arabidopsis. Plant Physiol 122: 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718 [DOI] [PMC free article] [PubMed]
- Wu C, Li X, Yuan W, Chen G, Kilian A, Li J, Xu C, Li X, Zhou DX, Wang S, et al. (2003) Development of enhancer trap lines for functional analysis of the rice genome. Plant J 35: 418–427 [DOI] [PubMed] [Google Scholar]