Chlamydomonas reinhardtii has two-tiered gene regulation under sulfur deficiency, and ARS73a is involved in the regulation of second-tier genes.
Abstract
During sulfur (S) deprivation, the unicellular alga Chlamydomonas reinhardtii exhibits increased expression of numerous genes. These genes encode proteins associated with sulfate (SO42−) acquisition and assimilation, alterations in cellular metabolism, and internal S recycling. Administration of the cytoplasmic translational inhibitor cycloheximide prevents S deprivation-triggered accumulation of transcripts encoding arylsulfatases (ARS), an extracellular polypeptide that may be important for cell wall biosynthesis (ECP76), a light-harvesting protein (LHCBM9), the selenium-binding protein, and the haloperoxidase (HAP2). In contrast, the rapid accumulation of transcripts encoding high-affinity SO42− transporters is not affected. These results suggest that there are two tiers of transcriptional regulation associated with S deprivation responses: the first is protein synthesis independent, while the second requires de novo protein synthesis. A mutant designated ars73a exhibited low ARS activity and failed to show increases in ECP76, LHCBM9, and HAP2 transcripts (among others) in response to S deprivation; increases in transcripts encoding the SO42− transporters were not affected. These results suggest that the ARS73a protein, which has no known activity but might be a transcriptional regulator, is required for the expression of genes associated with the second tier of transcriptional regulation. Analysis of the ars73a strain has helped us generate a model that incorporates a number of complexities associated with S deprivation responses in C. reinhardtii.
Sulfur (S) is an essential macronutrient that is integral to proteins, lipids, electron carriers, and redox controllers (Leustek et al., 2000). Most organisms, including Chlamydomonas reinhardtii, have a limited capacity to store S and thus depend on a continual supply of an external source of this nutrient. The majority of S that is taken up from the soil solution by microbes and plants is in the form of the sulfate anion (SO42−). Assimilation of SO42− involves reduction to sulfide and subsequent incorporation into the amino acids Cys and Met (Grossman and Takahashi, 2001).
The ability of microorganisms to acclimate to limited amounts of nutrients, including S, is critical to their survival in the natural environment. C. reinhardtii exhibits both general and specific responses when experiencing S deprivation. The general responses are common to a number of stress conditions and include the cessation of cell division, the accumulation of storage starch, and a decrease in metabolic processes including photosynthesis. In contrast, the specific responses are those associated with the deprivation of a single nutrient and include an elevated rate of SO42− uptake, the synthesis of extracellular arylsulfatases (ARS), and an increased capacity to assimilate SO42− by increasing the levels of enzymes needed for Cys biosynthesis (de Hostos et al., 1988; Yildiz et al., 1994; Ravina et al., 2002). Changes in genome-wide transcript accumulation as C. reinhardtii experiences S deprivation were recently reported (González-Ballester et al., 2010). The results of that study suggest that there are marked alterations in the activities of pathways associated with the biosynthesis of S compounds and that specific mechanisms have evolved to limit the synthesis of proteins with high-S amino acid content; this process has been termed S sparing (Fauchon et al., 2002; González-Ballester et al., 2010). Changes in the levels of a number of specific proteins encoded by S-responsive transcripts have also been observed (Takahashi et al., 2001; Pootakham et al., 2010).
ARS, an activity first detected approximately 3 h after the transfer of cells to medium lacking S (de Hostos et al., 1988), is secreted into the periplasmic space of C. reinhardtii cells, where it hydrolyzes soluble SO42− esters in the medium, releasing free SO42− for uptake and assimilation. The identification and characterization of ARS polypeptides led to the cloning of two ARS-encoding genes, ARS1 and ARS2. Transcripts from both of these genes accumulate in response to S deprivation (de Hostos et al., 1989; Ravina et al., 2002; Zhang et al., 2004). S deprivation of C. reinhardtii also elicits an increase in SO42− uptake, which is a consequence of the de novo synthesis of specific SO42− transport systems (Yildiz et al., 1994). The SO42− transporters encoded by SULTR2 (for Sulfate Transporter2), SLT1 (for SAC1-like transporter1), and SLT2 are strongly up-regulated at the transcript and protein levels almost immediately following the imposition of S deprivation (Pootakham et al., 2010). The initial rate of SO42− uptake increases as early as 1 h following the removal of S from the medium and becomes maximal after approximately 6 h. An increase in the affinity of the transport system for SO42− could also be detected within 1 h of S deprivation (Yildiz et al., 1994). Interestingly, S-starved cells show increased SO42− uptake prior to the detection of ARS activity, suggesting that the control of these two processes is differentially sensitive to the level of S in the environment.
C. reinhardtii also has mechanisms to conserve and recycle intracellular S during S-limiting conditions. The degradation of proteins and lipids that are not essential under S-deficient conditions can supply cells with a limited amount of S (Ferreira and Teixeira, 1992). S-starved C. reinhardtii cells degrade most of the chloroplast sulfolipid to redistribute S for protein synthesis and other processes (Sugimoto et al., 2007). Four prominent extracellular polypeptides, ECP56, ECP61, ECP76, and ECP88, are synthesized in response to S deprivation (Takahashi et al., 2001; González-Ballester et al., 2010). While the functions of these polypeptides have not been established, they contain almost no S-containing amino acids and exhibit features similar to those of cell wall, Hyp-rich glycoproteins. These findings suggest that the amino acids of S-rich cell wall proteins present during S-replete growth can be replaced by the ECPs; the S-containing amino acids of the S-rich cell wall proteins would become available for recycling (Takahashi et al., 2001). S deprivation also triggers a potential change in the subunit composition of light-harvesting complexes, favoring the synthesis of complexes containing polypeptides with few S amino acids (Nguyen et al., 2008; González-Ballester et al., 2010).
A number of S starvation-elicited responses appear to be controlled at the level of transcript abundance and gene activity. Transcripts encoding SO42− transporters, ARS, ECPs, LHCBM9 (for light-harvesting protein), and enzymes involved in SO42− assimilation [e.g. ATP sulfurylase, Ser O-acetyl transferase, O-acetyl Ser (thiol)lyase] are markedly elevated in cells deprived of S (de Hostos et al., 1989; Yildiz et al., 1996; Takahashi et al., 2001; Ravina et al., 2002; Zhang et al., 2004). Phenotypic screening for mutants unable to properly acclimate to S limitation led to the identification of three key regulators of S deprivation responses, SAC1 (for sulfur acclimation), SNRK2.1 (for Snf1-related protein kinase), and SNRK2.2 (Davies et al., 1994; Pollock et al., 2005; González-Ballester et al., 2008). SAC1 encodes an integral membrane protein similar to an Na+/SO42− cotransporter. However, SAC1 is a positive regulator critical for the activation of many genes involved in scavenging and assimilating S from the environment, including those encoding ARS, ECPs, a number of proteins important for SO42− assimilation (Davies et al., 1996; Ravina et al., 2002; Zhang et al., 2004), and proteins associated with the potential restructuring of the photosynthetic apparatus (Davies et al., 1994; Wykoff et al., 1998). The SNRK2.2 gene, previously designated SAC3 (Davies et al., 1999), encodes a Ser/Thr kinase that acts as a negative regulator of S deprivation-responsive genes. A snrk2.2 mutant exhibits low constitutive ARS activity and expresses elevated basal levels of S deprivation-responsive genes during S-replete growth (Davies et al., 1994, 1999; Ravina et al., 2002; González-Ballester et al., 2008). SNRK2.2 is also required for the proper down-regulation of chloroplast transcriptional activity in S-deprived cells (Irihimovitch and Stern, 2006). Finally, another Ser/Thr kinase, SNRK2.1, has recently been shown to play a central role in controlling S deprivation responses. The snrk2.1 mutant fails to activate the expression of most S deprivation-responsive genes and cannot maintain normal basal levels of expression of some genes under nutrient-replete conditions (González-Ballester et al., 2008, 2010).
In this study, we discovered that cycloheximide, an inhibitor of translation on 80S cytosolic ribosomes, did not block the induction of SO42− transporter genes (accumulation of the mRNA was not affected) but did block the induction of a subset of S deprivation-responsive genes, most of which are associated with S scavenging and recycling. This finding suggests the sequential, temporal regulation of subsets of S deprivation-responsive genes and that this tiered regulatory response may be dependent upon the accumulation of a specific protein(s) synthesized during the early stages of S deprivation. In a previous screen for mutants defective in the synthesis of active ARS (Pollock et al., 2005), a mutant designated ars73a was identified. The phenotype of this mutant is congruent with its involvement in the second tier responses of C. reinhardtii to S deprivation.
RESULTS
Identification of the ars73a Mutant
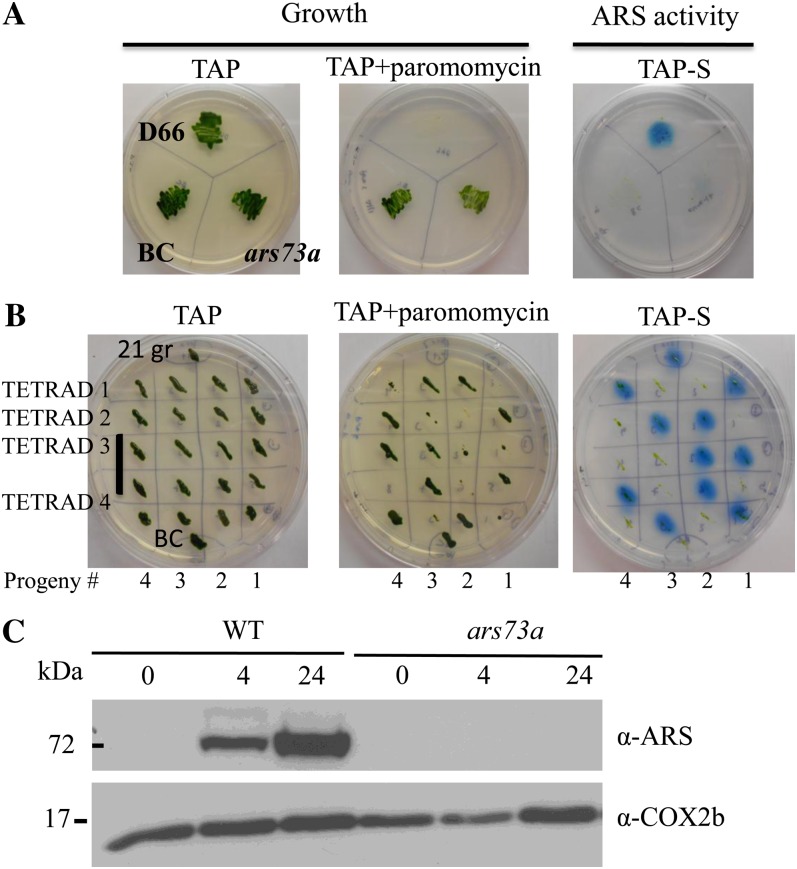
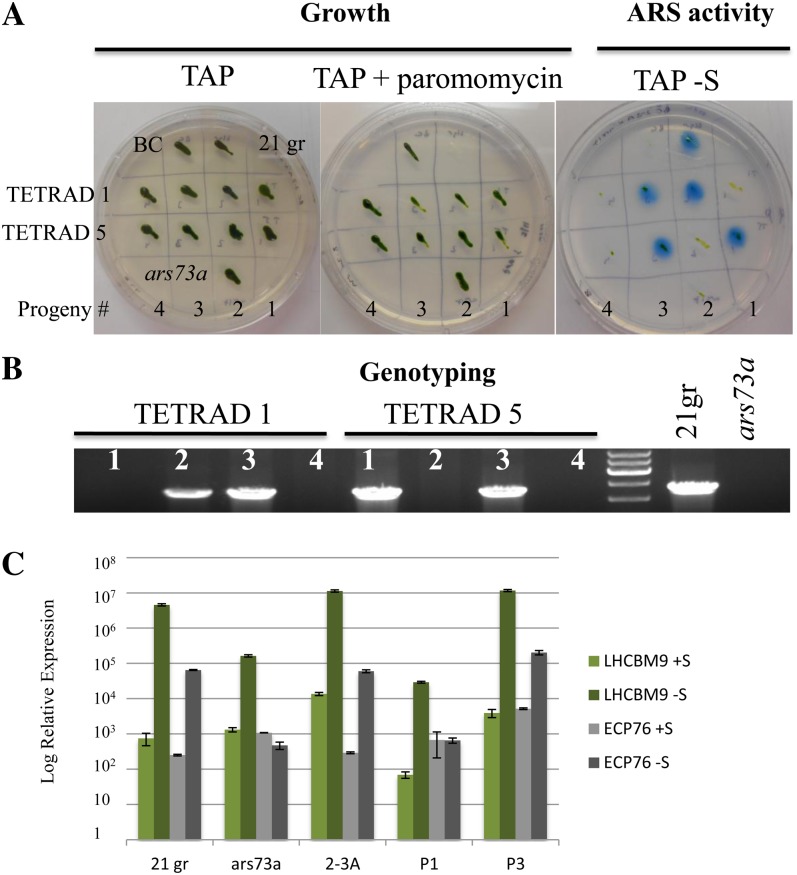
The acclimation of C. reinhardtii cells to S deprivation can be monitored by assaying the cells for the production of extracellular ARS activity. Wild-type C. reinhardtii cells grown on solid medium containing a limited amount of SO42− export the ARS enzyme into the periplasmic space or into the agar medium (in strains lacking a cell wall). The activity of this enzyme can be detected using the chromogenic substrate 5-bromo-4-chloro-3-indolyl sulfate; hydrolysis of this substrate by ARS results in the formation of a blue precipitate. This assay was employed in a screen to identify insertional mutant strains (using the AphVIII gene as an insertion marker that confers transformants with paromomycin resistance) that cannot synthesize ARS following S deprivation (Pollock et al., 2005). One of the mutants identified in this screen, designated ars73a, exhibited almost no ARS activity following growth on Tris-acetate-phosphate medium without sulfur (TAP-S; Fig. 1A, both ars73a and BC; the latter is the backcrossed ars73a strain), while high levels of activity are detected in the parental strain. The mutant displayed no obvious phenotype when grown on S-replete medium (e.g. cell growth was similar to that of the parental strain) except for a loss of motility, which was unlinked to the insertion of the AphVIII gene (see below).
Figure 1.
ARS activity and genetic linkage analysis. A, Phenotypic analysis of the ars73a mutant determined by assaying strains for ARS activity on solid medium. The parental, wild-type strain D66, the ars73a mutant, and the backcrossed mutant (BC; backcrossed to 21gr) were spread on TAP-S solid medium, and after the cells grew for 7 d, they were sprayed with the chromogenic substrate 5-bromo-4-chloro-3-indolyl sulfate. Note that the ars73a mutant has almost no ARS activity, while essentially no activity is detected in the BC strain. Only the original mutant and the mutant derived from the backcross grew on paromomycin, as expected. B, Linkage analysis of the BC ars73a strain. The BC mutant was crossed to 21gr mt+, and 15 full tetrads were analyzed for both paromomycin resistance and ARS activity. All of the paromomycin-resistant progeny exhibited an ars− phenotype. Results from four of the tetrads are shown. Tetrad 3 has eight progeny, while all of the others have four progeny. C, Western-blot analysis of the ARS polypeptide in the wild type (WT; 21gr) and the ars73a mutant. C. reinhardtii cells were grown in TAP medium and then transferred to TAP-S medium. Samples were collected just before and 4 and 24 h (indicated above the blot) after the removal of S from the medium. The COX2b subunit of cytochrome c oxidase served as a loading control. Molecular masses (in kD) are shown on the left.
To determine whether the ars− mutant phenotype (dramatic reduction in ARS activity) cosegregated with the introduced AphVIII gene, the original ars73a strain was crossed to a wild-type C. reinhardtii strain. Approximately 500 random progeny were scored for growth on paromomycin-containing medium and ARS activity on medium devoid of S. The loss of ARS activity always cosegregated with the paromomycin resistance phenotype, demonstrating a strong linkage between the AphVIII insertion and the loss of ARS activity. Linkage was also analyzed using the backcrossed ars73a mutant. The original ars73a mt+ was backcrossed three times to 21gr (another wild-type strain), the ars73a mt− progeny from the third backcross were crossed to 21gr mt+, and 15 full tetrads were analyzed to determine if the paromomycin resistance phenotype was linked to the ars− phenotype. All 15 tetrads showed a 1:1 segregation of the paromomycin-resistant to paromomycin-sensitive phenotype, indicating a single insertion of the AphVIII gene. Furthermore, the resistant phenotype always cosegregated with the loss of ARS activity. The phenotype of the progeny of four of the analyzed tetrads is shown in Figure 1B. These results demonstrate linkage between the ars− phenotype and insertion of the marker gene. Furthermore, the paralyzed flagella phenotype, which was observed in the original mutant strain, segregated from the paromomycin-resistant and ars− phenotypes, demonstrating that a lesion causing flagella dysfunction in the original mutant background was not linked to the AphVIII insertion. Finally, western-blot analysis demonstrated that ars73a did not synthesize any ARS polypeptide following S deprivation of the cells (Fig. 1C).
Mapping Insertion Sites in the ARS73a Gene
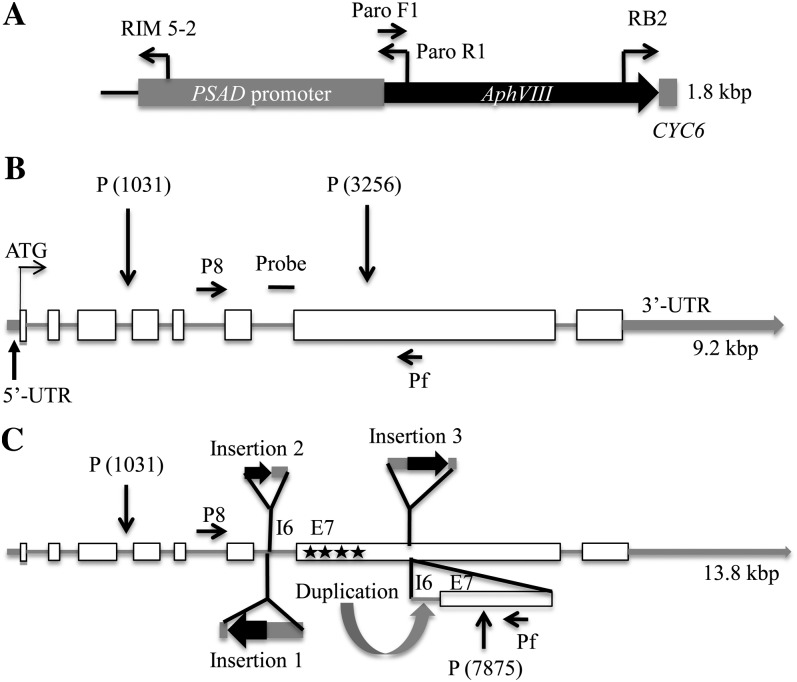
Initially, adaptor ligation-mediated PCR and inverse PCR methods (Pollock et al., 2003) were used to locate the AphVIII marker gene (Fig. 2A) in the genome of the ars73a mutant. The sequence of the PCR product (Pollock et al., 2005) indicated that the marker gene insertion was in the sixth intron of gene model C_523864 (Chlre v4; http://genome.jgi-psf.org/chlamy/chlamy.info.html). To further characterize the insertion site and to determine whether the mutant harbors a single copy of the AphVIII gene (e.g. a single restriction fragment with an inserted paromomycin cassette), we performed DNA-blot hybridizations to genomic DNA digested with PstI (this enzyme does not cut in the AphVIII gene) using the entire AphVIII cassette as a hybridization probe as well as a probe that hybridizes to most of intron 6 and 283 nucleotides of exon 7 of ARS73a (designated “probe” in Fig. 2B). The AphVIII cassette (Fig. 2A) also contains a 57-bp intron sequence of the CYC6 gene (at the 3′ end of AphVIII) and the PSAD gene promoter. DNA-blot hybridization with these probes revealed a unique hybridizing band of approximately 6.8 kb that is only present in the mutant (hybridization results using the AphVIII cassette probe are shown in Supplemental Fig. S1), suggesting that the mutant has a single AphVIII insertion; the hybridization signal at approximately 5 kb, which is observed in both the wild-type and mutant strains, is a genomic fragment containing the endogenous PSAD gene. The paromomycin cassette is approximately 1.8 kb, and the sequence has no PstI restriction site; therefore, a single insertion into the 2.2-kb PstI fragment of wild-type cells [predicted size of the fragment based on the genomic sequence and represented in Fig. 2B between P(1031) and P(3256)] would yield a hybridizing band of approximately 4 kb. The larger molecular mass of the hybridizing fragment (approximately 6.8 kb) suggested that the fragment contained more than one cassette and/or had experienced a rearrangement near the insertion site.
Figure 2.
Maps showing the insertion cassette and the ARS73a genomic regions in wild-type cells and the ars73a mutant. A, Schematic representation of the AphVIII cassette with the positions of the primers used for genome walking, the location of the PSAD promoter at the 5′ end, and the location of the CYC6 sequence at the 3′ end. B, Schematic representation of the ARS73a gene model in wild-type cells. Exons are represented with white boxes, introns with gray lines, and UTRs with thick gray lines. The position of the probe used for DNA-blot hybridizations is shown as a horizontal line. P8 and Pf are the primers used for genotyping. C, Schematic representation of insertions in the genomic DNA of the ars73a mutant. Genome walking was performed using PCR from both sides of AphVIII (present in insertions 1 and 3) and into the ARS73a sequence (see “Materials and Methods”); the resulting PCR products were sequenced and aligned to the sequence on the genome browser. Alignment of the sequenced reads suggested the presence of three insertions in the gene. Insertion 1 (in intron 6) was a complete cassette inserted in the opposite orientation relative to ARS73a. Insertions 2 (also in intron 6) and 3 (in exon 7) were in the same orientation as the ARS73a gene, but they represented partial cassette sequences. Duplication of part of intron 6 (I6) and exon 7 (E7) is shown with a curved arrow. PstI sites that are not susceptible to digestion are marked with black stars. The 6.8-kb PstI (P) fragment from P(1031) to P(7875) is the hybridizing band shown in the DNA-blot hybridization in Supplemental Figure S1. Drawings are approximately to scale. For the precise positions of the insertions, see text and Supplemental Figure S3.
Extensive PCR and full sequence data (of the 6.8-kb fragment from the mutant) demonstrated the presence of multiple insertions as well as a duplication of part of the genomic DNA. An insertion of the full-length cassette (in the opposite orientation relative to ARS73a) in intron 6 of ARS73a (insertion 1 in Fig. 2C) was immediately adjacent to a small insertion (insertion 2 in Fig. 2C) derived from the 3′ end of the cassette, followed by a third insertion (insertion 3 in Fig. 2C) of a 5′ truncated cassette in exon 7. This last insertion contains the complete AphVIII coding sequence but is missing part of the PSAD promoter (0.7 kb at the 5′ region of the cassette). The region of the ARS73a gene that was duplicated (indicated with a curved arrow labeled “Duplication”) was from nucleotides 2,564 to 4,013 of ARS73a; the full ARS73a gene sequence is given in Supplemental Figure S2. Finally, the four PstI sites present within the 6.8-kb PstI fragment from mutant genomic DNA, all of which are in a 600-bp region of exon 7 (Fig. 2C, E7 and four adjacent stars), do not appear to be susceptible to endonuclease digestion. This loss of susceptibility could be a consequence of RNA interference-mediated methylation around the site of insertion. Multiple AphVIII insertions may elicit the synthesis of double-stranded RNA or RNA hairpins that could promote methylation (Huettel et al., 2007). An unusual CAG methylation found in plants can prevent PstI digestion (Gruenbaum et al., 1981), although this has not been experimentally examined for C. reinhardtii. A map that shows the complex structure of the 6.8-kb PstI fragment with the various insertions is presented in Figure 2C. The full nucleotide sequence of the 6.8-kb fragment (from the ars73a mutant) is given in Supplemental Figure S3. We cannot rule out the possibility that other fragments of the cassette integrated into other regions of the genome, although if this were the case the integrated fragments would likely be very small, since no other restriction fragments were observed to hybridize to the AphVIII cassette.
The results presented in this section clearly demonstrate a disruption of the ARS73a gene, with insertions in both intron 6 and exon 7. These insertions in the mutant strain are likely to completely eliminate the synthesis of a functional ARS73a protein. Furthermore, these results emphasize that some lesions generated during the insertional inactivation of genes could result in complex changes in the genomic DNA in the neighborhood of the insertion.
Cycloheximide Impacts the Expression of a Subset of S Deprivation-Responsive Genes
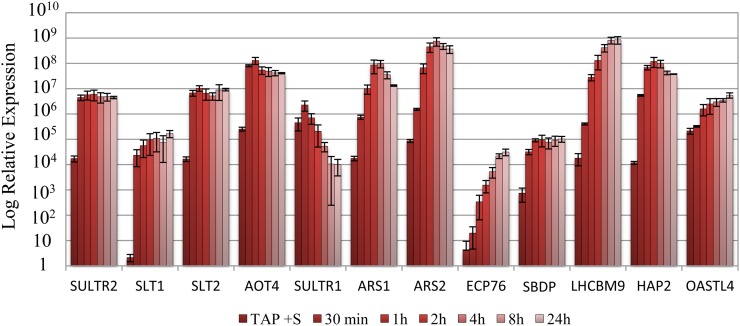
When C. reinhardtii is deprived of S, several transcripts, including those encoding the high-affinity SO42− transporters, ARS, and enzymes integral to SO42− assimilation, increase in abundance. We explored the induction kinetics of subsets of S deprivation-responsive genes in both wild-type cells and the ars73a mutant. Transcripts encoding the SO42− transporters (SULTR2, SLT1, and SLT2), a putative amino acid transporter (AOT4), and the selenobinding protein (SBDP) increased and reached peak levels very quickly (within approximately 30 min) following the imposition of S deprivation; the levels of these transcripts remained high over the 24-h deprivation period imposed on the cells (Fig. 3). In contrast, transcripts encoding arylsulfatases (ARS1 and ARS2), O-acetyl-Ser (thiol)lyase (OASTL4), an extracellular polypeptide (ECP76), a light-harvesting polypeptide (LHCBM9), and a putative haloperoxidase (HAP2) gradually increased during the first 4 h of S deprivation, while the mRNA encoding SULTR1 gradually declined (Fig. 3).
Figure 3.
Changes in transcript abundances for various S deprivation-responsive genes following the removal of S from the medium. Levels of transcripts encoding SULTR2, SLT1, SLT2, AOT4, SULTR1, ARS1, ARS2, ECP76, SBDP, LHCBM9, HAP2, and OASTL4 were measured by RT-qPCR. RNA samples were isolated from wild-type cells (21gr) grown in S-replete medium and after being transferred to S-depleted medium for 30 min and 1, 2, 4, 8, and 24 h. [See online article for color version of this figure.]
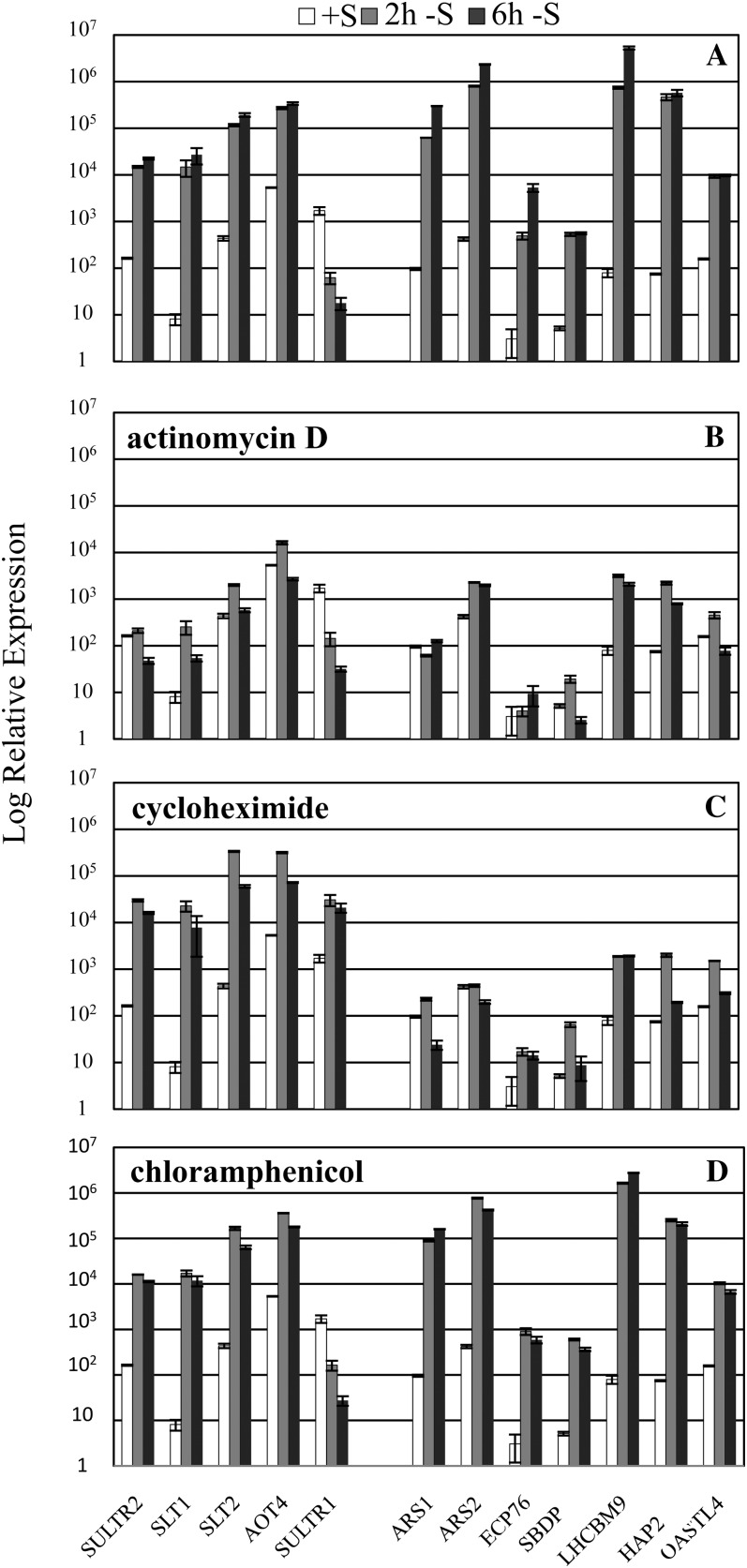
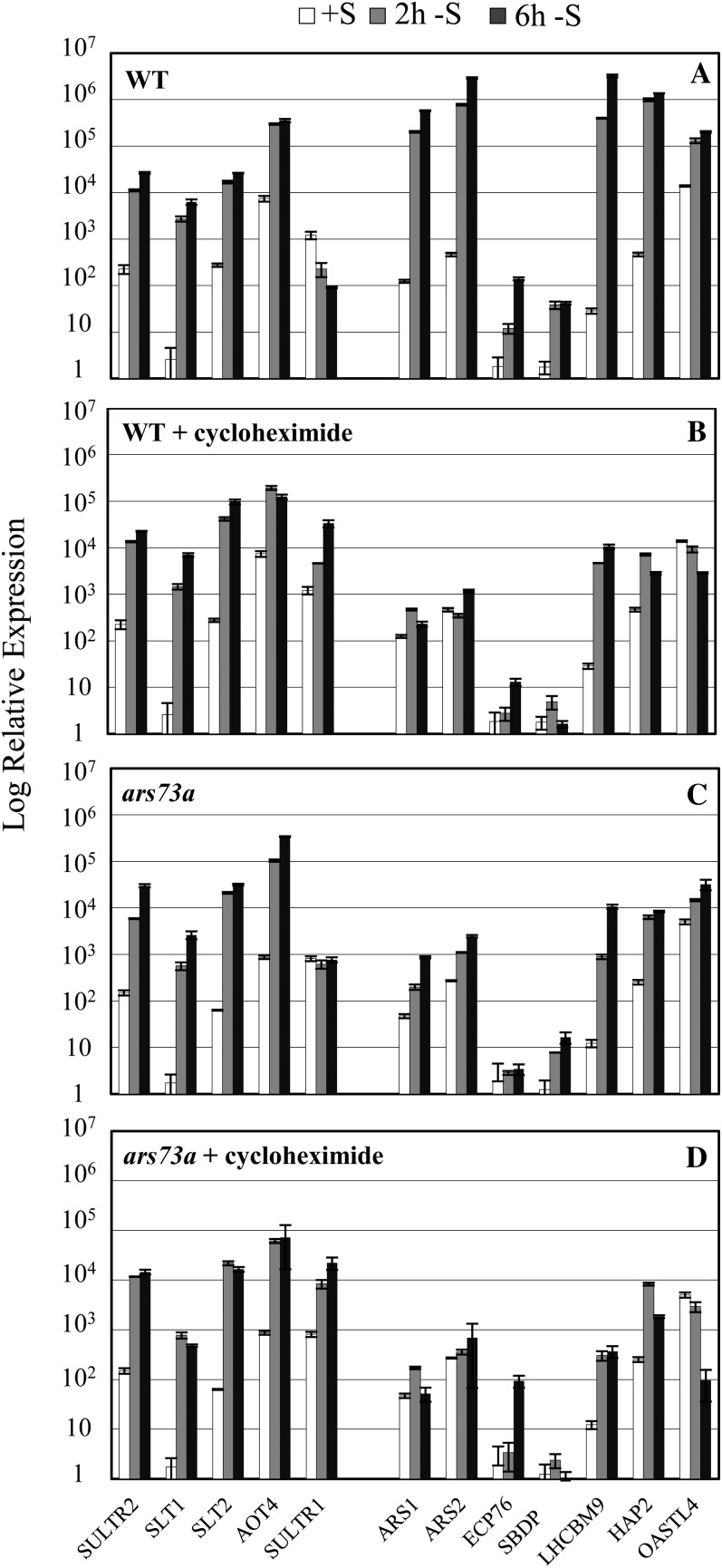
As expected, the accumulation of all transcripts was markedly inhibited by administration of the transcriptional inhibitor actinomycin D to the culture at the time of S removal (compare Fig. 4, A and B; note the log scale on the y axis). Incomplete inhibition of transcript accumulation may be a consequence of the completion of transcription that had already been initiated and/or a decrease in the degradation rate in the presence of the antibiotic. The results are strikingly different when cycloheximide, which inhibits translation on 80S cytoplasmic ribosomes, was added to the culture medium as the cells were depleted of S. The inhibition of cytosolic protein synthesis by cycloheximide strongly inhibited the S deprivation-elicited increase in the levels of transcripts encoding ARS1, ARS2, ECP76, LHCBM9, SBDP, and HAP2 (compare Fig. 4, A and C); the levels of these transcripts were similar in actinomycin D- and cycloheximide-treated cells. Indeed, the cells were behaving as if the S deprivation-dependent increase in mRNAs normally observed for this set of transcripts was blocked. In contrast, increases in the level of transcripts encoding the high-affinity SO42− transporters (SULTR2, SLT1, and SLT2) and a putative amino acid transporter (AOT4) were not altered in the presence of cycloheximide (but were strongly altered when actinomycin D was added to the medium).
Figure 4.
Effects of transcriptional and translational inhibitors on the accumulation of S deprivation-responsive transcripts. Levels of transcripts encoding SULTR2, SLT1, SLT2, AOT4, SULTR1, ARS1, ARS2, ECP76, SBDP, LHCBM9, HAP2, and OASTL4 were measured by RT-qPCR. Wild-type cells (21gr) were grown in TAP medium and then transferred to TAP-S medium either in the absence of an inhibitor (A) or in the presence of the transcriptional inhibitor actinomycin D (B), the cytosolic translational inhibitor cycloheximide (C), or the organellar translational inhibitor chloramphenicol (D). Inhibitors were added to the cultures at the same time as S was removed. Samples were taken prior to starvation and at 2 and 6 h after removal of S from the medium, which is shown with white, gray, and black bars, respectively.
Although cycloheximide did not significantly impact the expression of SULTR2, SLT1, and SLT2, it did cause an elevated accumulation of the SULTR1 transcript (Fig. 4C). This result may reflect the translational inhibitor-mediated stabilization of the transcripts, possibly by direct inhibition of turnover or by protection of the transcripts in ribosomal complexes (Lopez et al., 1998). The mRNA level of the housekeeping gene CBLP was shown to remain constant during S deprivation (Chang et al., 2005); however, an increase in the CBLP transcript level was also observed when cycloheximide was administered to cells following the removal of S from the medium (data not shown), supporting the argument that cycloheximide can generally impact mRNA stability. Finally, chloramphenicol, a translational inhibitor of 70S ribosome (inhibits translation in mitochondria and plastids), had essentially no impact on the expression of S deprivation-responsive genes (Fig. 4D).
The results concerning the impact of inhibitors of translation on transcript abundance suggest that there are at least two tiers of transcriptional regulation in response to S deprivation. The first tier of regulation controls the transcription of the SO42− transporters and does not require protein synthesis following the imposition of deprivation conditions. In contrast, a second, protein synthesis-dependent tier of regulation controls many genes encoding enzymes involved in SO42− assimilation and restructuring of the cell during S deprivation.
Expression of S Deprivation-Responsive Genes in ars73a
To study the responses of ars73a to S deprivation, transcript abundances from several genes previously shown to be controlled in wild-type cells by the S status of the medium (SULTR1, SULTR2, SLT1, SLT2, AOT4, ARS1, ARS2 for ARS, ECP76, SBDP, LHCBM9, HAP2, and OASTL4), as presented in Figure 3, were analyzed in the ars73a mutant (Fig. 5). As expected from measurements of ARS activity in the ars73a mutant, ARS1 and ARS2 transcripts did not markedly increase (2–3 orders of magnitude less than in wild-type cells) after transferring the mutant to medium devoid of S. The levels of the ECP76, LHCBM9, HAP2, and SBDP transcripts were also markedly diminished in the mutant relative to the wild-type strain (compare Fig. 5, A and C). In contrast, the abundance of transcripts for the three inducible SO42− transporters (SULTR2, SLT1, and SLT2) and the putative amino acid transporter (AOT4) were not significantly affected in the mutant (compare Fig. 5, A and C). Even more remarkable, the pattern of gene expression observed in the mutant was very similar to that of wild-type cells treated with cycloheximide (compare Fig. 5, B and C). For some S deficiency-induced genes (e.g. ARS1, ARS2, SBDP, and LHCBM9), a further reduction (although generally small) in transcript levels occurred following the addition of cycloheximide to ars73a cells deprived of S (Fig. 5D), suggesting the possible participation of another regulatory factor(s) in controlling the activity of these genes.
Figure 5.
Transcript abundance analyzed by RT-qPCR in wild-type cells (WT; 21gr) and the ars73a mutant. Accumulation of SULTR2, SLT1, SLT2, AOT4, SULTR1, ARS1, ARS2, ECP76, SBDP, LHCBM9, HAP2, and OASTL4 transcripts in wild-type cells (A and B) and the ars73a mutant (C and D) was monitored. RNA samples were collected prior to and 2 and 6 h after the cells were transferred from TAP to TAP-S medium in the presence (B and D) or absence (A and C) of the cytosolic translational inhibitor cycloheximide.
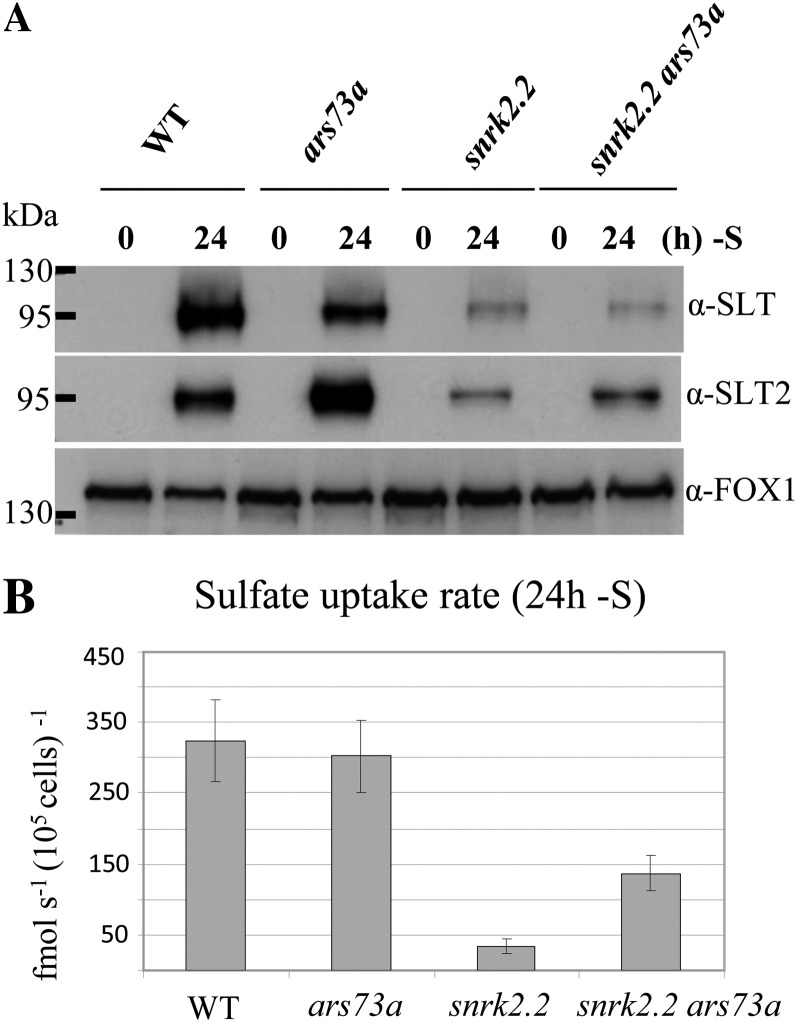
In accord with the transcript abundance measurements, SULTR2, SLT1, and SLT2 polypeptides accumulated to approximately the same extent in wild-type cells and the ars73a mutant during S deprivation (for epistasis analysis, see below). Furthermore, there was essentially no accumulation of ARS proteins in ars73a upon S deprivation (Fig. 1C), as expected from measurements of both ARS activity and transcript levels.
In sum, our results suggest that (1) there are two tiers of S deprivation responses, one independent of (first tier) and the other requiring (second tier) protein synthesis, and (2) the ARS73a protein is required for manifestation of the majority of second tier regulation.
Rescue of the Mutant Phenotype
Over a period of several months, we noted that the ars73a mutant exhibited some suppression of the ars− phenotype: low levels of ARS activity were observed in colonies of S-deprived mutant cells. Therefore, we backcrossed the mutant with 21gr to remove putative suppressor mutations, regenerating progeny with essentially no ARS activity. We then cotransformed the backcrossed strain with BAC39M22 and the plasmid pSP124S; the latter contains the bacterial ble gene, which provides a selection (resistance to zeocin) for potential cotransformants. BAC39M22 is small (approximately 42 kb of genomic DNA from chromosome 6 plus the approximately 12-kb pBACmn vector) and contains the ARS73a genomic locus as well as part of an upstream gene (model C_523863, encoding a putative amino acid transporter) and a downstream gene (model C_523865, encoding a protein with significant homology to ARS73a; an alignment of the two sequences is presented in Supplemental Fig. S4). To specifically test if disruption of the ARS73a gene was responsible for the mutant phenotype, BAC39M22 DNA was digested with SmaI, which does not cut within ARS73a but does cleave the downstream and upstream genes, and the digested DNA plus pSP124S were introduced into ars73a by electroporation; transformants were selected on Tris-acetate-phosphate (TAP) medium containing 5 to 10 μg mL−1 zeocin. Zeocin-resistant colonies were picked, transferred onto solid TAP medium, and allowed to grow for approximately 1 week. Cells from each colony were then transferred onto solid TAP-S medium, allowed to grow for 7 d, and then assayed for ARS activity. Eleven strains out of approximately 500 zeocin-resistant transformants tested exhibited ARS activity. These strains were genotyped by PCR using primers between exon 4 and exon 7 of ARS73a (the primer pairs flank all the insertions) to identify strains with wild-type and mutant copies of ARS73a. Based on this analysis, of the 11 ARS+ strains 10 contained a copy of the wild-type ARS73a gene. The transformant that did not have a wild-type copy of the gene exhibited much lower ARS activity relative to the 10 other ARS+ transformants, suggesting the occurrence of low-frequency second-site suppression that shows partial restoration of ARS activity, which is consistent with what we observed for the original ars73a mutant population. We also transformed the mutant with the pSP124S vector alone and observed some transformants (less than 1%) with very low-level ARS activity (none of the suppressor strains ever had high ARS activity). The ARS activity in these strains was always less than 10% of that observed in wild-type cells or in ars73a mutant cells harboring a wild-type copy of ARS73a. These findings provide further evidence for suppression of the ars− phenotype at a significant frequency. Future analyses of these suppressor strains might help elucidate processes that contribute to the expression of S-responsive genes.
To further confirm that the ARS73a gene was responsible for rescue of the mutant phenotype, we crossed one of the rescued ars73a mutants to the original ars73a mutant and analyzed 17 zygotes for segregation of paromomycin resistance, ARS activity, and the presence of the introduced copy of ARS73a (the wild-type copy of the gene introduced into the rescued mutant). One-half of the progeny showed high levels of ARS activity, and the other one-half had no ARS activity. Furthermore, based on PCR genotyping (generation of a 2,250-bp product using primers that amplify the gene sequence from exon 4 to exon 7, containing the entire region with the insertions), only those progeny that exhibited ARS activity retained the wild-type copy of ARS73a; progeny devoid of ARS activity had lost the wild-type copy of the gene, which was manifest as a lack of a PCR product (the region between the primers in the mutant strain is too long to be amplified with the extension time used; they would generate a 6.9-kb product). An analysis of two tetrads of the cross described above is shown in Figure 6. All of the progeny of the tetrads were paromomycin resistant, with only two of the four expressing ARS activity (Fig. 6A). The same colonies that exhibited ARS activity also harbored the introduced, wild-type copy of the ARS73a gene (Fig. 6B). The rescued strain (for ARS activity), 2-3A, and its rescued progeny, P3, also showed normal expression of LHCBM9 and ECP76 (Fig. 6C), while the original ars73a mutant and P1, a mutant progeny, had lower transcript levels. Increased mRNA levels following S deprivation were observed for all rescued strains; an increase in ECP76 transcript levels was especially notable. These results demonstrate that the ARS73a gene product is a major element that controls second tier regulation of the S deprivation responses in C. reinhardtii.
Figure 6.
Analysis of progeny from the complemented strain (2-3A). A, A rescued mt− strain was crossed to ars73a mt+, and the progeny were analyzed for both growth on paromomycin and ARS activity, which is shown for two tetrads (TETRAD 1 and TETRAD 5). B, All of the progeny that displayed ARS activity harbored a wild-type copy of ARS73a, as indicated by the presence of a 2,250-bp PCR product. Progeny devoid of ARS activity did not have a wild-type copy of ARS73a (no PCR product detected; see text). PCR results from genomic DNA isolated from 21gr (wild-type cells) and the ars73a mutant are shown as controls. C, Analysis of the rescued strain (2-3A) and its rescued progeny (P3 of TETRAD 1) for levels of LHCBM9 and ECP76 transcripts after 6 h of S deprivation. Similar transcript analyses for progeny that are not rescued (P1 of TETRAD 1), for 21gr, and for ars73a are also shown.
ARS73a Transcripts
We performed 5′ and 3′ RACE to identify the ends of the ARS73a mRNA. Based on these results, the 3′ untranslated region (UTR) is 24 nucleotides shorter than what has been reported in the C. reinhardtii database (http://genome.jgi-psf.org/chlamy/chlamy.info.html), while the 5′ RACE product terminated 299 bp upstream of the predicted translation start site.
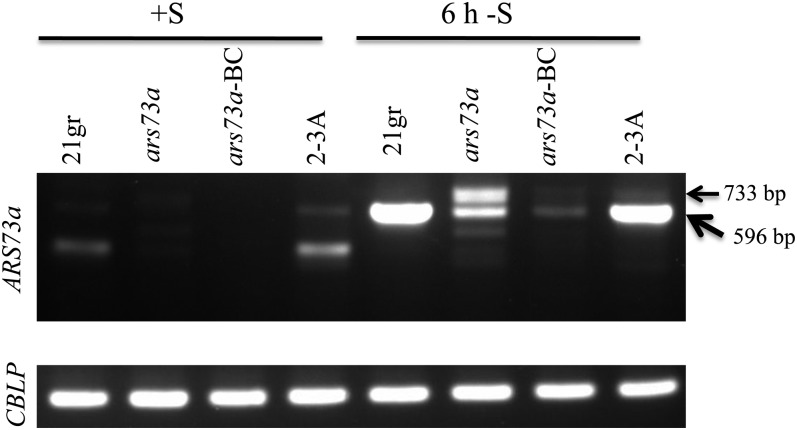
To assay for the presence of ARS73a mRNA in ars73a and the 21gr wild-type strain, reverse transcription (RT)-PCR was performed with a pair of primers, one of which anneals to exon 4 and the other to exon 7; the expected size of the wild-type PCR product is 596 bp, while the amplified product in the ars73a mutant would potentially include part of insertion 1 (which is inserted into intron 6).
Under nutrient-replete conditions, 21gr and the rescued strain (2-3A) had low but detectable levels of ARS73a mRNA, represented by a faint band at 596 bp. The level of this band increased dramatically when the cells were shifted to S deprivation conditions (Fig. 7). The identity of the 596-bp band amplified from RNA extracted from cells grown under nutrient-replete and S deprivation conditions was confirmed by sequencing the isolated band. A lower molecular mass RNA observed in nutrient-replete 21gr and the 2-3A rescued strain, also present in low abundance, was not sequenced; the identity of this product is not known. In contrast, little mRNA was detected in either the original or backcrossed ars73a mutant when the cells were grown under nutrient-replete conditions (Fig. 7). However, when the mutant cells were shifted to S deprivation conditions, they accumulated low levels of two transcripts: one transcript is approximately 596 bp (the expected size) and the other is 733 bp (Fig. 7). The two different transcripts in the mutant cells were sequenced. The 596-bp product had the expected sequence. The higher molecular mass product contained a partial AphVIII sequence in the opposite orientation relative to ARS73a (in accord with our finding that the insertion in intron 6 is in the opposite orientation relative to ARS73a; Fig. 2C); this sequence was located between exons 6 and 7. We also observed a partial deletion of exon 6 in the aberrant 733-bp mRNA (data not shown). The sequences from the amplified RNA segments identified in mutant cells are presented in Supplemental Figure S5.
Figure 7.
ARS73a mRNA in 21gr (wild type), ars73a, ars73a-BC (backcrossed ars73a), and the rescued strain (2-3A). The cells were grown under S-replete conditions or exposed to 6 h of S-depleted conditions prior to RNA isolation. First-strand cDNA was amplified with primers flanking the insertion in intron 6 (amplifying exons 4–7). The expected product derived from the wild-type transcript is 596 bp. A longer 733-bp product is observed in the ars73a mutant and the backcrossed mutant under S deprivation conditions. The CBLP transcript was used as a loading control. The sequences of these two mRNAs are given in Supplemental Figure S5.
It is interesting that wild-type cells and the rescued ars73a mutant accumulated low levels of normal ARS73a mRNA under S-replete conditions, which increased dramatically when the cells were deprived of S. Such results suggest that the cells have a basal level of ARS73a mRNA (and maybe protein) at the onset of the acclimation process and that this low-level expression may be required for acclimation to occur. Although the mutant accumulated some mRNA under S-depleted conditions (with the backcrossed strain accumulating less than the original mutant), since these mRNAs from the mutant would be aberrant (given the three insertions in ARS73a in mutant cells), they might be rapidly degraded and are not likely to encode a functional protein.
We also examined the accumulation of transcripts from the gene (C_523865) downstream of ARS73a and found that it was the same in D66 and the ars73a mutant under both S-replete and S-depleted conditions, with no aberrant mRNAs detected based on a number of different primer pairs used for RT-PCR analyses (data not shown). These results suggest that the insertions in ars73a did not impact expression of the downstream gene. The control for all of these experiments was RT-PCR of the transcript encoding CBLP, a C. reinhardtii G-protein β-subunit-like protein. There was no difference in the level of this transcript under nutrient-replete and S deprivation conditions or between the mutant and the D66 parental strain.
Regulation of ARS73a Expression
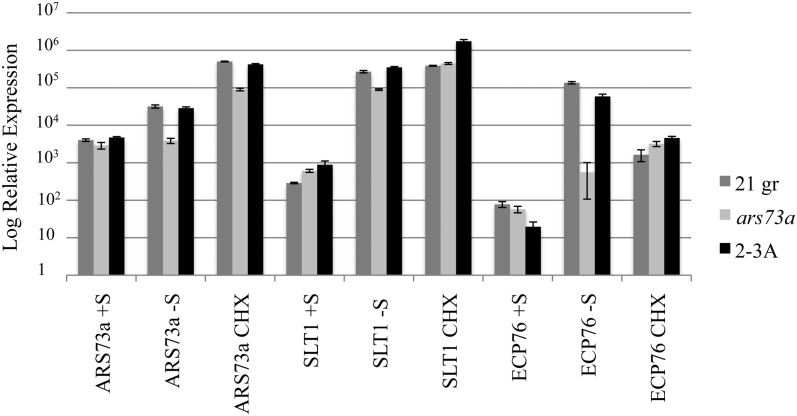
We also examined whether the ARS73a gene exhibited a first or a second tier response to S deprivation. RT-quantitative PCR (qPCR) was performed to determine ARS73a transcript abundances in S-replete, S-depleted for 6 h, or S-depleted and cycloheximide-treated cultures (Fig. 8). The ARS73a transcript level was elevated in S-depleted conditions in 21gr and the complemented strain (2-3A) but not in ars73a. While this experiment showed an increase in the transcript level after 6 h of S deprivation, in other experiments we observed increased transcript accumulation after 30 min of deprivation (data not shown). Cycloheximide treatment did not block this increased transcript accumulation, suggesting that up-regulation of ARS73a does not require de novo protein synthesis. Therefore, we concluded that the ARS73a gene itself is up-regulated as a first tier gene during S deprivation conditions.
Figure 8.
ARS73a transcript regulation. RNA was isolated from cultures of 21gr (wild type), ars73a, and 2-3A (complemented strain) grown in S-replete medium (+S) or 6 h after they were transferred to S-depleted medium without cycloheximide (−S) or with cycloheximide (CHX). Levels of transcripts for ARS73a, SLT1 (as a control for first tier genes), and LHCBM9 (as a control for second tier genes) were measured by RT-qPCR.
Epistasis Analysis of the Double Mutants sac1 ars73a, snrk2.1 ars73a, and snrk2.2 ars73a
To place the ARS73a protein into the context of the S deprivation network of regulators that have already been examined, we analyzed the phenotype of ars73a in the sac1, snrk2.1, and snrk2.2 genetic backgrounds. SAC1 encodes a putative transmembrane SO42− sensor (Davies et al., 1996). The SNRK2.1 and SNRK2.2 proteins both encode protein kinases, and the phenotypes of snrk2.1 and snrk2.2 mutants were described previously (Davies et al., 1999; González-Ballester et al., 2008).
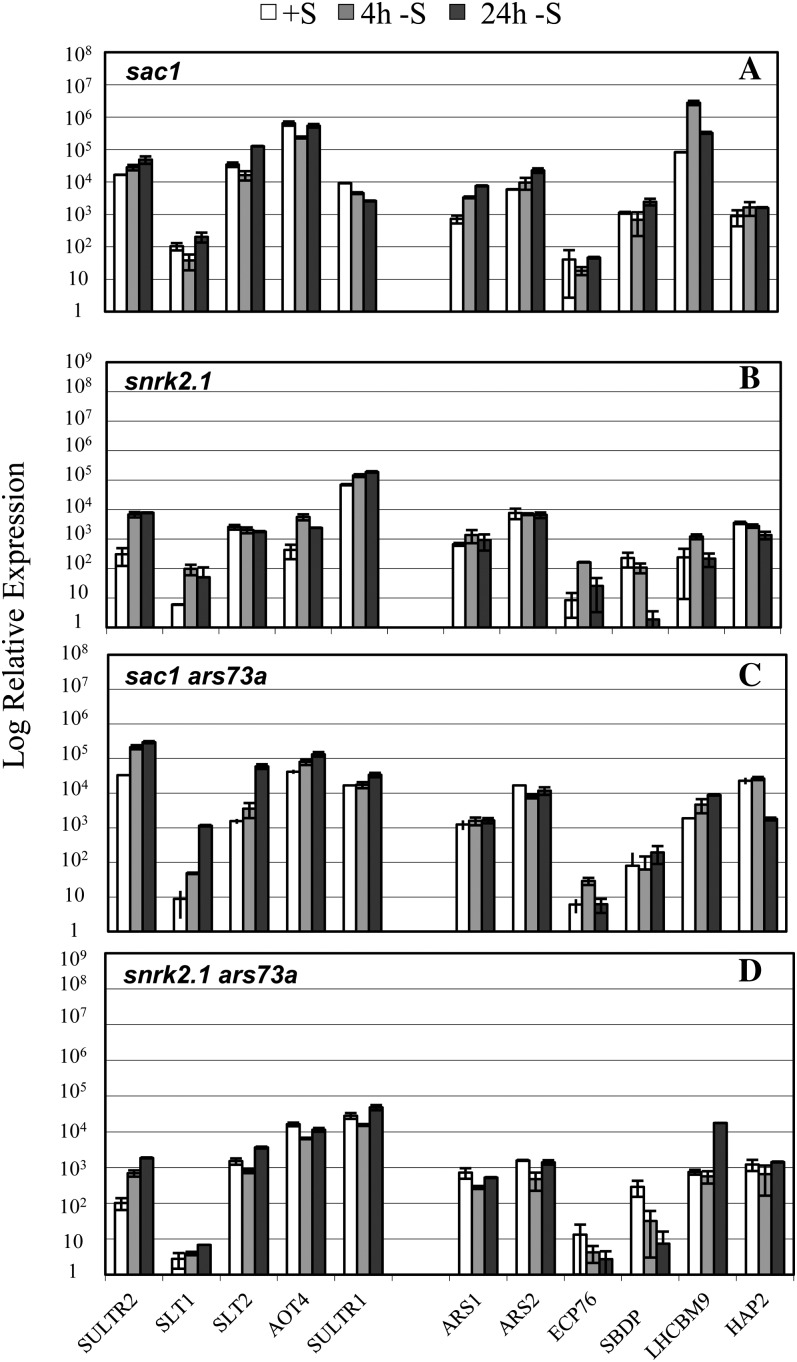
The sac1 mutant exhibits little increase in transcript levels from any of the S-responsive genes upon S deprivation (Fig. 9A; Zhang et al., 2004). Since the ars73a mutant also shows a strong depression of the level of second tier transcripts under S deprivation conditions (Fig. 5C), it is not possible to determine an epistasis relationship between SAC1 and ARS73a based on the levels of second tier transcripts. However, up-regulation of first tier genes is generally low in the sac1 ars73a double mutant (Fig. 9C) relative to ars73a (Fig. 5C); the double mutant does not express first tier genes to the same extent as either ars73a or wild-type cells upon S deprivation. These results suggest that SAC1 is epistatic to ARS73a with respect to the expression of first tier genes. Furthermore, the snrk2.1 phenotype is clearly epistatic to ars73a; the double mutant exhibits little expression of any of the S-responsive transcripts that were analyzed (Fig. 9D), which is similar to what was observed for the snrk2.1 single mutant (Fig. 9B). Interestingly, a whole-transcriptome analysis performed using RNA-seq showed that snrk2.1 had almost no ARS73a transcript under S-replete conditions and had truncations of the ARS73a transcript in the 5′ and 3′ UTRs under S-depleted conditions (see RNA-seq data on the University of California, Santa Cruz genome browser at http://genomes.mcdb.ucla.edu/; Supplemental Fig. S6). These results suggest that ARS73a expression may be controlled by the SNRK2.1 kinase.
Figure 9.
RT-qPCR analysis of transcript accumulation in sac1, snrk2.1, and the double mutants sac1 ars73a and snrk2.1 ars73a. Transcripts encoding SULTR2, SLT1, SLT2, AOT4, SULTR1, ARS1, ARS2, ECP76, SBDP, LHCBM9, and HAP2 were quantified by RT-qPCR. RNA samples were isolated from cells grown in S-replete medium (white bars) and at 4 h (gray bars) and 24 h (black bars) following the transfer of cells to medium devoid of S.
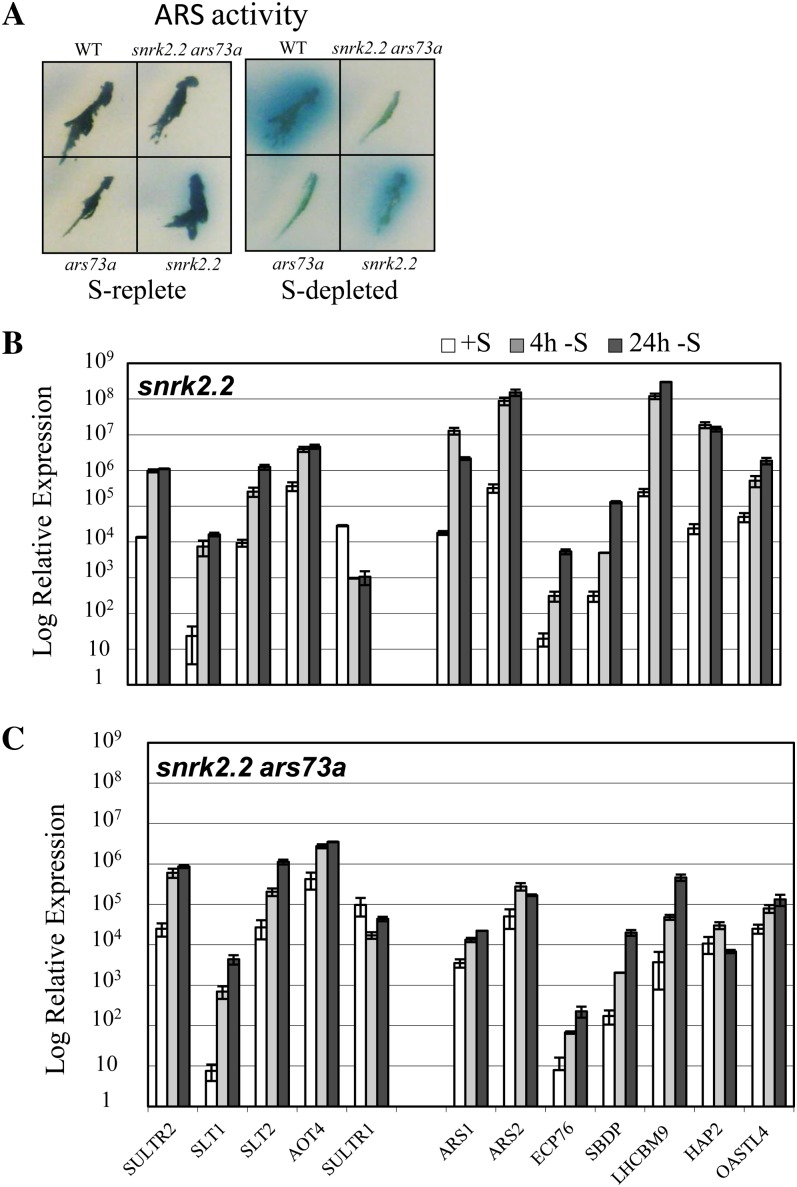
Finally, we generated a snrk2.2 ars73a double mutant. SNRK2.2 encodes a Ser/Thr kinase that acts as a negative regulator of S deprivation-responsive genes under nutrient-replete conditions (Davies et al., 1999). The snrk2.2 mutant has low constitutive ARS activity under nutrient-replete conditions. The double mutant has an ars− phenotype similar to ars73a; there is little constitutive accumulation of ARS activity under S-replete conditions, and ARS activity does not markedly increase under S-depleted conditions (Fig. 10A). Furthermore, transcripts from genes associated with the second tier of regulation do not accumulate to high levels when the double mutant is deprived of S (compare Fig. 10, B and C). These results suggest that ARS73a is epistatic to SNRK2.2 with respect to the regulation of second tier genes. The increases in SULTR2, SLT1, and SLT2 transcripts were similar in the snrk2.2 single mutant and the snrk2.2 ars73a double mutant (compare Fig. 10, B and C). This is not surprising, since ARS73a does not appear to control the SLT or SULTR genes (Figs. 5, A and C, and 6) while SNRK2.2 does (González-Ballester et al., 2008). It is interesting that while the snrk2.2 mutant shows low-level constitutive expression of the transporter mRNA in S-replete medium and normal induction during S deprivation (González-Ballester et al., 2008), only low levels of the transporter proteins accumulate in the membranes in both the snrk2.2 single mutant and the snrk2.2 ars73a double mutant (Fig. 11A), and both strains have low rates of SO42− uptake (Fig. 11B) under S-replete and S-depleted conditions. These results suggest a further role (in addition to the suppression of transporter and ARS genes) of the SNRK2.2 kinase in the biogenesis of the transporters (for model, see Fig. 12).
Figure 10.
Epistasis analysis of ARS73a and SNRK2.2. A, ARS activity in the wild type (WT; 21gr), the individual mutants snrk2.2 and ars73a, and the snrk2.2 ars73a double mutant on S-replete and S-depleted medium. B and C, RT-qPCR showing the levels of SULTR2, SLT1, SLT2, AOT4, SULTR1, ARS1, ARS2, ECP76, SBDP, LHCBM9, HAP2, and OASTL transcripts in the snrk2.2 single mutant (B) and snrk2.2 ars73a double mutant (C).
Figure 11.
Epistasis analysis of ARS73a and SNRK2.2. A, Accumulation of proteins encoding the high-affinity SLT SO42− transporters in the wild type (WT; 21gr), ars73a, snrk2.2, and snrk2.2 ars73a after 24 h of S starvation. A ferroxidase, FOX1, served as a loading control. B, SO42− uptake in wild-type C. reinhardtii cells (21gr) and the ars73a, snrk2.2, and snrk2.2 ars73a mutant strains. Uptake rates were determined at 250 μm SO42− (specific activity of 330 Ci mol−1). The values are averages of a single experiment with three technical replicates, although similar results were obtained in independent experiments, and the error bars represent 1 sd.
Figure 12.
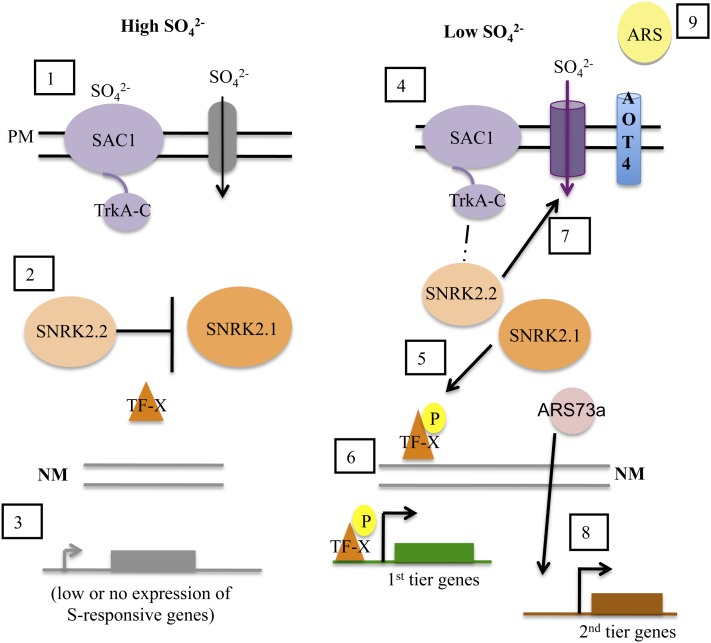
Model depicting regulatory events associated with the acclimation of C. reinhardtii cells to S deprivation. (1) Under S-replete conditions (high SO42−), the plasma membrane (PM)-located SAC1 senses SO42− levels in the environment by binding to SO42−. This disengages SAC1 from downstream regulatory events. (2) Cytosolic SNRK2.2 represses the transcription of the S-responsive genes, possibly by inhibiting the activity of the positive regulatory factor SNRK2.1 (e.g. by phosphorylation of SNRK2.1). This interaction ensures that (3) there is low or no expression from S-responsive genes. (4) Under S deprivation conditions (low SO42−), SAC1 is free of bound SO42−, which allows it to interact with cytosolic SNRK2.2 (hypothesized interaction). This interaction prevents the inhibition of SNRK2.1, which can then (5) become active under S deprivation conditions and phosphorylate a downstream transcription factor (denoted TF-X; it has not been identified). This phosphorylation leads to the activation of first tier genes (6). (7) The SAC1-associated SNRK2.2 functions to help route the SO42− transporters to the plasma membrane, integrate them into the membrane, or stabilize them once they are in the membrane. (8) The ARS73a transcript and protein are synthesized at high levels, and the protein (which may or may not be modified) directly (by involvement in transcription) or indirectly (by facilitating the translation of a transcription factor) is involved in the transcription of second tier genes. (9) One product of second tier gene activation is ARS, which is secreted to the extracellular space.
DISCUSSION
Plants and algae have evolved a number of different acclimation strategies to accommodate changing levels of S in the environment. The utilization and assimilation of SO42− by plants and other photosynthetic organisms and alterations in cellular processes linked to reduced S availability in the environment have recently been reviewed (Takahashi et al., 2011). Responses of C. reinhardtii to S deprivation have been studied for many years (de Hostos et al., 1988, 1989; Davies et al., 1994, 1996, 1999; Yildiz et al., 1994, 1996; Takahashi et al., 2001; Zhang et al., 2004; Chang et al., 2005; González-Ballester et al., 2008, 2010; Moseley et al., 2009; Pootakham et al., 2010), and one of the first responses of this alga to S limitation was shown to be the activation of genes encoding high-affinity SO42− transporters (Yildiz et al., 1994; González-Ballester et al., 2010; Pootakham et al., 2010). Increased expression from high-affinity SO42− transporters has been noted to occur within 30 min of the removal of S from the culture medium (Fig. 3; Pootakham et al., 2010). Other S deprivation responses, including the synthesis and secretion of ARS, require longer periods of acclimation to be detected; the ARS transcript is detected 30 min following the imposition of S deprivation, but it continues to increase for an additional approximately 3 h (Fig. 3).
To better understand the temporal expression of S-responsive genes and their regulatory features, we measured the expression levels of various genes that have been associated with the S deprivation responses, with or without inhibitors of transcription and translation. Interestingly, cycloheximide treatment had little impact on the accumulation of transcripts encoding the previously identified, inducible SO42− transporters (SULTR2, SLT1, and SLT2) and AOT4, a putative amino acid transporter. However, administration of cycloheximide immediately prior to the imposition of S deprivation appeared to block the accumulation of transcripts encoding ARS, ECP76, SBDP, LHCBM9, HAP2, and OASTL4. These results suggest that (1) there are at least two tiers of regulation associated with S deprivation responses; (2) no protein synthesis is required for the first tier, which involves the regulation of genes encoding the high-affinity SO42− transporters; and (3) protein synthesis is required for the second tier, which involves regulation of the ARS genes as well as genes associated with SO42− assimilation and cellular reorganization. Our results demonstrate that the regulatory elements for first tier control are already present in cells under nutrient-replete conditions. First tier regulation appears to be dependent on a putative external sensor of SO42−, designated SAC1 (Davies et al., 1996; Zhang et al., 2004), as well as the potential internal regulatory element SNRK2.1 (González-Ballester et al., 2008, 2010). Soon after C. reinhardtii cells are transferred to S deprivation conditions, the regulatory elements (e.g. SAC1 and SNRK2.1) become active, potentially through some protein modification and/or changes in ligand binding, and first tier genes are activated. In contrast, de novo protein synthesis is required for the second tier of gene regulation, although we are only just beginning to identify the control elements involved in this process.
The advantage of maintaining a tiered acclimation response is that it would allow the cell to make efficient use of its energy budget by only mobilizing second tier control processes under persistent nutrient limitation conditions. The initial activation of genes encoding SO42− transporters would allow for the efficient utilization of even low levels of environmental SO42−. If the availability of SO42− increases during this initial phase of acclimation, the cells would not have to implement additional strategies associated with scavenging, conservation, and metabolic control. The energetically/metabolically costly synthesis of proteins involved in second tier synthesis may not be necessary in many instances (e.g. short-term fluctuation in S availability). However, if the cells experience long-term S deprivation, then additional modes of scavenging, such as accessing SO42− bound to organic molecules in the environment through the synthesis of ARS enzymes and restructuring cellular processes through the synthesis of LHCBM9 and the various ECPs, may help sustain cell viability.
In the course of examining C. reinhardtii mutants that were unable to acclimate to S deprivation, we noted that a strain designated ars73a exhibited no ARS activity when it was starved for S, but it appeared to express C. reinhardtii genes encoding the SO42− transporters (Fig. 5), which were previously shown to be required for the efficient uptake of SO42− under S deprivation conditions (Pootakham et al., 2010). Further characterization of the mutant strain indicated that while first tier genes were up-regulated to a similar extent as in wild-type cells, there was aberrant expression of second tier genes; the increase in expression during S deprivation was markedly reduced relative to wild-type cells (Fig. 5). These results raised the possibility that the ARS73a protein is synthesized along with the SO42− transporters during first tier gene activation, followed by its participation in second tier regulation. Therefore, we analyzed ARS73a transcript abundance by RT-PCR with a primer pair that amplifies a region of the transcript between exon 4 and exon 7. Wild-type C. reinhardtii cells make the ARS73a transcript under both S-replete and S-depleted conditions, although the level significantly increases as the cells become S deprived (Fig. 7). RT-qPCR performed with primers that amplify the exon 2-exon 3 junction also showed that ARS73a transcription increases under S-depleted conditions (Fig. 8). These results are in accord with results reported in a genome-wide transcriptome analysis of C. reinhardtii cells that were deprived of S; an approximately 3-fold increase in ARS73a mRNA was noted for S-deprived wild-type cells (González-Ballester et al., 2010; Supplemental Fig. S6). Cycloheximide treatment did not block the increase in ARS73a transcript accumulation, which suggests that ARS73a is a first tier responsive gene (Fig. 8). Hence, the ARS73a protein may already be present in cells grown under nutrient-replete conditions, although it is possible that the protein level is also posttranscriptionally modulated; immunodetection of the protein would enable us to quantify ARS73a protein levels under the different conditions. If the protein is present in nutrient-replete cells, then it may increase in abundance under S deprivation conditions to reach a threshold level that causes gene activation, becomes activated in a protein synthesis-dependent manner when the cells are deprived of S (e.g. a specific modifying protein may be synthesized during first tier protein synthesis that activates ARS73a), or interacts with a partner protein that is synthesized during first tier gene activation. Alternatively, the ARS73a transcript, which is present in the cell at low levels under nutrient-replete conditions, may not be translated. Activation of specific proteins during S-deprivation may allow ARS73a mRNA to be properly processed and translated. The existence of aberrant ARS73a mRNAs that are truncated at both 5′ and 3′ UTRs in the snrk2.1 mutant when the cells are deprived of S lends some support to this possibility; SNRK2.1 or a protein regulated by SNRK2.1 is needed to properly process/translate ARS73a mRNA.
With the same primers used for evaluating ARS73a transcript levels in wild-type cells, we could barely detect ARS73a mRNA in the ars73a mutant maintained in nutrient-replete medium (the product generated by these primers spans the first AphVIII insertion site in the mutant). However, when the cells were shifted to S-depleted conditions, we observed low levels of two transcripts (Fig. 7). One of these transcripts was identical to the 596-bp RT-PCR product in wild-type cells, and the other was shown to be a fusion between the ARS73a mRNA and the sequence of the cassette used for insertional mutagenesis. These results suggest that a transcript is still being synthesized from the mutated gene, that the cassette inserted into intron 6 can be excised from the RNA (although inefficiently), and that aberrant processing of the RNA also occurs, resulting in transcripts that are fused with sequences from the cassette. However, given that an additional insertion of a “partial” cassette has integrated into the ARS73a gene downstream of the segment of the transcript amplified by the RT-PCR used to evaluate ARS73a mRNA accumulation, it is likely that this strain does not make normal ARS73a protein.
Based on the predicted amino acid sequence, ARS73a has no nuclear localization domain and no known motifs that would allow it to bind DNA and function as a transcription factor. The closest homologs to ARS73a are two proteins encoded by the Volvox gene models Vocar20015096m.g and Vocar20015036m.g. These proteins have strong identity to ARS73a over the first 20% of the protein; the identity is small over the remaining 80% of the protein (data not shown). Both Volvox proteins have ankyrin repeats (not present in ARS73a) but no known physiological functions. Interestingly, ARS73a does have regions with poly-Gln tracks. Proteins with poly-Gln tracks have been shown to be involved in transcriptional regulation in a number of organisms (Ding et al., 2006; Kim et al., 2008; Landeras-Bueno et al., 2011; Atanesyan et al., 2012). Interestingly, another gene that is immediately downstream of ARS73a, gene model C_523865, encodes a protein with significant sequence similarity to ARS73a (Supplemental Fig. S4). It is also important to note that the predicted gene model upstream of ARS73a encodes a putative amino acid transporter (AOT4) that is likely involved in S acclimation, since the expression of this gene is severely depressed in the snrk2.1 mutant under both S-replete and S-depleted conditions (González-Ballester et al., 2008, 2010). A mechanistic understanding of the way in which ARS73a impacts second tier regulation, and the possible role of the protein encoded by the related, downstream gene in this regulation, will require the determination of subcellular locations of these proteins and characterizations of their interactions with other proteins in the cell (and potentially with each other).
We have identified a number of regulatory elements important for controlling the responses of C. reinhardtii to S deprivation. These proteins, which include SAC1, SNRK2.1, and SNRK2.2, appear to be involved in the first tier of gene regulation. SAC1 has sequence similarity to a Na+/SO42− cotransporter but functions as a sensor protein, while both SNRK2.1 and SNRK2.2 are Ser/Thr kinases. The sac1 mutant cannot elicit much increased expression of high-affinity transporters and has lower SO42− transport activity (Pootakham et al., 2010), but it is also unable to properly up-regulate second tier gene expression. Furthermore, the sac1 mutant is defective in acclimation to S deficiency and dies after 2 d in S-depleted medium (Davies et al., 1994). The sac1 ars73a double mutant has a similar phenotype to that of the sac1 single mutant; this is not surprising, since the phenotype with respect to the expression of ARS is the same for both strains, while SAC1 and not ARS73a is required for expression of the transporter genes.
SNRK2.1 is involved in the regulation of many S-responsive genes, and the snrk2.1 mutant, like sac1, dies within 2 d of S deprivation (González-Ballester et al., 2008, 2010). The levels of both SAC1 and SNRK2.2 transcripts are not impacted in the snrk2.1 mutant, suggesting that these genes are not regulated by SNRK2.1 (González-Ballester et al., 2010). Interestingly, the snrk2.1 mutant has almost no ARS73a transcript under nutrient-replete conditions, and the transcript present in S-depleted cells has abnormalities: it appears to be truncated at both the 5′ and 3′ UTRs (Supplemental Fig. S6; http://genomes.mcdb.ucla.edu/). These data suggest that SNRK2.1 has a role in regulating ARS73a transcription and/or the stability of the transcript. The snrk2.1 ars73a double mutant has a phenotype similar to that of the snrk2.1 single mutant (Fig. 9A), demonstrating that the snrk2.1 lesion is epistatic to ars73a; the accumulation of all transcripts, those associated with both first and second tier regulation, is blocked in the double mutant, and the levels of the transcripts from all first and second tier genes are extremely low. These results suggest that SNRK2.1 is involved in the activation of ARS73a when the cells are depleted of S.
The snrk2.2 ars73a double mutant exhibits an ars− phenotype identical to that of the ars73a single mutant (Fig. 9, B and C); the double mutant shows essentially no ARS activity, unlike the snrk2.2 single mutant, which has constitutive low-level activity (Davies et al., 1999). These findings suggest that ARS73a is epistatic to SNRK2.2 with respect to expression of the ARS genes. Interestingly, the snrk2.2 single mutant was shown to be unable to accumulate SO42− transporter proteins in the plasma membrane, and the snrk2.2 ars73a double mutant had a similar phenotype (Fig. 11A). This phenotype of the double mutant is in accord with the finding that ARS73a is not involved in controlling the SO42− transporter genes (Figs. 5, A and C, and 11). However, these findings provide interesting insights into the role of SNRK2.2 in the biogenesis of the C. reinhardtii SO42− transporters (see the model in Fig. 12 and discussion below). The SNRK2.2 kinase may have a dual role (or a single function that leads to two phenotypes in mutant strains) in the acclimation of the cells to S-deficient conditions; it is involved in the repression of S-responsive genes under nutrient-replete conditions (the snrk2.2 mutant has low constitutive ARS activity under nutrient-replete conditions) and is required for the trafficking to and/or integration/stability of high levels of transporter protein into the plasma membrane (Fig. 11A); the mutant exhibits little increase in SO42− uptake rates and low-level accumulation of the transporter proteins in the plasma membrane under S deprivation conditions (Fig. 11B; Davies et al., 1994; Pootakham et al., 2010). Recently, phosphorylation of the yeast (Hansenula polymorpha) nitrate transporter Ynt1 was shown to be essential for its delivery to the plasma membrane when the cells were deprived of nitrogen (Navarro et al., 2008). Similarly, phosphorylation of the yeast plasma membrane ATPase was shown to be critical for its proper maturation and routing to the cell surface (DeWitt et al., 1998). In addition, Phosphate Transporter Traffic Facilitator1, a plant-specific SEC12-related protein, has been demonstrated to play a critical role in allowing the exit of a high-affinity Arabidopsis (Arabidopsis thaliana) phosphate transporter from the endoplasmic reticulum (González et al., 2005). Therefore, it is possible that the snrk2.2 mutant is defective for the maturation and subsequent routing of SO42− transporters to the plasma membrane, either because the transporters themselves or a protein involved in transporter biogenesis must be phosphorylated (by SNRK2.2) to function properly.
With the new information generated by these studies of the ars73a mutant, we have been able to develop a more detailed model of the way in which S levels regulate acclimation responses in C. reinhardtii. This model, shown in Figure 12, integrates the tiered responses that were observed in these studies and the role of ARS73a in those responses. The genes regulated during the first tier of the control, which does not require protein synthesis, include those encoding the high-affinity SO42− transporters. The sensor protein SAC1 and the Ser/Thr kinase SNRK2.1 are absolutely necessary for full activation of first tier genes. Expression of the genes involved in SO42− scavenging (e.g. ARS) and restructuring of the cell (e.g. ECP76, LHCBM9) requires protein synthesis in a second tier regulatory response. ARS73a is critical for second tier gene expression. Furthermore, SNRK2.2 is required for both complete suppression of first tier gene expression during S-replete conditions and stable accumulation of the transporter proteins in the cytoplasmic membrane; these two phenotypic phenomena may or may not be independent. A detailed diagram of our regulatory model is shown in Figure 12, with a thorough description of the different regulatory events that occur under nutrient-replete and S deprivation conditions presented in the legend.
MATERIALS AND METHODS
Strains and Growth Conditions
The following Chlamydomonas reinhardtii strains were used in this work: the wild-type strains 21gr (CC-1690, available from the Chlamydomonas Center) and D66 (CC-4425, available from the Chlamydomonas Center; Pollock et al., 2005), snrk2.1 (González-Ballester et al., 2008), and sac1 and snrk2.2 (Davies et al., 1994, 1999). Cells were cultured in either S-replete or S-depleted TAP medium under continuous illumination (80 µmol photons m−2 s−1) on a rotating platform (200 rpm) at 25°C. TAP-S medium was prepared as described previously (Davies et al., 1994). For S starvation experiments, cells were grown to midlogarithmic phase (2–4 × 106 cells mL−1) in standard TAP medium, washed once with TAP-S medium (2,500g for 5 min), and resuspended in TAP-S to the original cell density.
Cell Treatment
Cycloheximide, which inhibits protein synthesis on cytoplasmic 80S ribosomes, was used at a final concentration of 10 µg mL−1, as described previously (Kawazoe et al., 2000). Chloramphenicol was used at a concentration of 300 µg mL−1 and actinomycin D at a concentration of 100 µg mL−1.
Assay for ARS Activity
The ARS activity of colonies growing on solid medium was assayed by spraying the colonies with 300 to 500 µL of 10 mm 5-bromo-4-chloro-3-indolyl sulfate (Sigma-Aldrich) in 0.1 m Tris-HCl, pH 7.5, as described by Davies et al. (1996). A blue precipitate formed around each colony producing active ARS protein within 1 to 2 h of exposure of the colonies to the chromogenic substrate.
SO42− Uptake Assay
SO42− transport assays were performed as described previously (Yildiz et al., 1994).
Southern-Blot Analyses
Genomic DNA was isolated from 100-mL liquid cultures of 21gr and the ars73a mutant using a standard phenol-chloroform extraction protocol (Sambrook et al., 1989). Approximately 10 µg of genomic DNA was digested overnight with 10 units of the restriction endonuclease PstI (New England Biolabs). The fragments resulting from the digestion were separated by agarose (0.8%–1%) gel electrophoresis, blotted overnight in 20× SSC onto nitrocellulose membranes (GeneScreen; DuPont-New England Nuclear), and the transferred DNA was cross-linked to the membrane by UV illumination. An alkaline phosphatase-labeled probe was synthesized by chemical cross linking of a thermostable alkaline phosphatase to the nucleic acid template. Probe synthesis and hybridization were performed using the Amersham AlkPhos Direct Labeling and Detection Systems following the manufacturer’s protocol (Amersham Biosciences).
RNA Isolation and Quantification
Total RNA was extracted from frozen cell pellets using the RNeasy Mini Kit (Qiagen) and treated with RNase-free DNase I (Qiagen) to remove residual genomic DNA. First-strand complementary DNA (cDNA) was synthesized from 2 μg of total RNA using oligo(dT)12-18 for priming the SuperScript III reverse transcriptase reaction, as described in the manual (Invitrogen). RT-qPCR was performed with a Roche LightCycler 480. PCR, in a final volume of 20 µL, used 10 µL of LightCycler 480 SYBR Green Master Mix (Roche) or SensiMix SYBR-NoROX (Bioline), 5 µL of a 1:50 cDNA dilution, 400 nm of each primer, and distilled water to make up the remainder of the 20-µL volume. The Roche mix conditions used for amplification in the thermocycler were as follows: preincubation at 95°C for 5 min, followed by 50 cycles of denaturation at 95°C for 10 s, annealing at 60°C for 20 s, elongation at 72°C for 20 s, and measurement of fluorescence after 80°C for 5 s (the last step was incorporated into the protocol to avoid background signals resulting from the formation of primer dimers). The Bioline mix preincubation time was increased to 10 min, and denaturation steps were for 15 s, as suggested by the manufacturer. Similar results were obtained with both mixes. A melt-curve analysis program (60°C–99°C, heating rate of 2.2°C s−1, and continuous fluorescence measurements) was used to evaluate the specificity of each amplification reaction. All reactions were performed in triplicate with at least two biological replicates. The CBLP gene was used as the control housekeeping gene (Chang et al., 2005). The primer pairs used for RT-qPCR were as follows: 5′-CTTCTCGCCCATGACCAC-3′ and 5′-CCCACCAGGTTGTTCTTCAG-3′ for CBLP; 5′-GCAATGTGGCAGGGGCATGGTT-3′ and 5′-GAGCACCGACCTGGATTACCAGCT-3′ for ARS1; 5′-CTTAATTGCATGCGCGCCGTCA-3′ and 5′-TCAGAACACCAACGCAAGTTTCCAG-3′ for ARS2; 5′-TGGCCATGCTTATCGTCATCTATG-3′ and 5′-TCGATGCGCATGACCAGGAT-3′ for SULTR1; 5′-ACGTGGCATGCAGCTCAT-3′ and 5′-CTTGCCACTTTGCCAGGT-3′ for SULTR2; 5′-ACGGGTTCTTCGAGCGAATTGC-3′ and 5′-CGACTGCTTACGCAACAATCTTGG-3′ for SLT1; 5′-GTACGGAGTTCCTTACGCGC-3′ and 5′-TTCTTCGCCACCGATGAGC-3′ for SLT2; 5′-CCCAGTCTTTTGGCGGCAAG-3′ and 5′-GGCCTACTCGCTACCGTACC-3′ for SLT3; 5′-CCTCGCTCTCCTCGCTGCTG-3′ and 5′-CGGCCGACTTGGGTAATTGC-3′ for ECP76; 5′-GGACGGCAGCATCATGGTGAGC-3′ and 5′-TCCACACGCCCTTGACCTTGAG-3′ for SBDP; 5′-CCGCCTGGCCATGTTCTCGTC-3′ and 5′-CCGTGGGCCCTATCCGTGGTA-3′ for LHCBM9; 5′-AAGGATTGTGTCAGGCTCGTCTCG-3′ and 5′-TTCCCACCGCAGCCTAACCACA-3′ for HAP2; 5′-GGCCCTCCTGGAAGCTGAATCA-3′ and 5′-CCCCACCCCACCCATTAGTAGTC-3′ for OASTL4; 5′-TTGGCGGTGAGGTAGGAACAGACG-3′ and 5′-TGCCGCCAGACAGGGAACCAC-3′ for AOT4; and 5′-GTTGGAGTTCCTGGACTGGTTTT-3′ and 5′-GATGGGCAGGCTAGGGAAAGT-3′ for ARS73a.
RT-PCR for ARS73a and Downstream Gene Model
The primer pair used for RT-PCR to amplify the region of ARS73a from exon 4 to exon 7 was R3 (5′-AGGAGTACGCGGTGCGCTGG-3′) and Pb (5′-CAGCCCACTGGACTGGTGGAGCAGCAT-3′). The primers used to detect transcripts for the gene model C_523865 were F1 (5′-TGCAGCCGGCCACCATGCACA-3′) and R1 (5′-CGGGCGCTGGCGCCAGCCCA-3′).
Backcrossing and Generation of the Double Mutants sac1 ars73a, snrk2.1 ars73a, and snrk2.2 ars73a
ars73a was backcrossed three times to the wild-type strain 21gr according to the protocol of Harris (1989) to remove most background mutations generated by the transformation procedure and to obtain a mutant strain in the 21gr genetic background. To generate the snrk2.2 ars73a double mutant, snrk2.2 mt+ (in the 21gr genetic background) was crossed to the third backcrossed progeny of ars73a mt−. The genotype of the progeny was determined by colony PCR using the following primers: 5′-CGTACAAGGCCCATGCGTGAGTC-3′ and 5′-TCGCCGAAAATGACCCAGAGC-3′ for identifying the snrk2.2 allele and 5′-TACCGGCTGTTGGACGAGTTCTTCTG-3′ (RB2) and 5′-CAGTTCGTGACATTCATTCTGACGG-3′ for identifying the ars73a allele. A similar method was used to generate the snrk2.1 ars73a and sac1 ars73a double mutants. The primers used to identify the snrk2.1 allele were 5′-CAGGATGCGGGACAGCAGGTC-3′ and RB2, while those used to identify the sac1 allele were 5′-GTCGTTGGGCATCTTCATCGC-3′ and 5′-GCGACTCGTTGAACTTCTCC-3′.
Generation of Antibody
Antibodies to ARS were prepared by Covance Research Products. Antibodies that recognize both ARS1 and ARS2 were generated in rabbits against a peptide region common to ARS1 and ARS2 (SDKPQNSKVGLHVD) but not well conserved in other ARS-like proteins. The peptide was conjugated to keyhole limpet hemocyanin via a terminal Cys and injected into rabbits. The preparation of antibodies against the SO42− transporters was described previously (Pootakham et al., 2010).
Protein Isolation, SDS-PAGE, and Immunoblot Analysis
C. reinhardtii cells (2–4 × 106 cells mL−1 in 100 mL) were collected by centrifugation (3,000g, 5 min) and resuspended in 0.1 m Na3PO4 buffer (pH 7.0). Chlorophyll was extracted from cells into 80% acetone-20% methanol, and its concentration was determined spectrophotometrically (Arnon, 1949) after removal of cell debris by centrifugation. Quantities of cells with equal chlorophyll content (200–300 µg) were pelleted, resuspended in an ice-cold homogenization buffer (0.25 m Suc, 0.1 m HEPES, pH 7.5, 15 mm EGTA, 5% glycerol, and 0.5% polyvinylpyrrolidone) containing a protease inhibitor cocktail (Sigma-Aldrich), and then disrupted by agitation with glass beads (425–600 µm). The lysate was centrifuged briefly (2,000g, 5 min) to remove cell debris. For total protein preparations (used for immunoblots), 1 volume of loading buffer (6.25 mm Tris-HCl, pH 6.8, 5% SDS, 6 m urea, 500 mm dithiothreitol, 10% glycerol, and 0.002% bromophenol blue) was added to samples prior to incubation at 100°C for 5 min. To prepare a microsomal fraction (used for immunoblots with anti-SLT and anti-SLT2 antibodies), the supernatant was centrifuged at 100,000g for 50 min. The microsomal pellet was resuspended in homogenization buffer containing 1% Triton X-100. An equal volume of loading buffer was added to samples prior to incubation at 42°C for 15 min. Solubilized polypeptides were resolved by SDS-PAGE on a 10% polyacrylamide gel and transferred to polyvinylidene difluoride membranes using a wet-transfer method. Blots were blocked in 5% milk in Tris-buffered saline solution with 0.1% Tween 20 prior to a 2-h incubation in the presence of primary antibodies. Dilutions of the primary antibodies used were as follows: 1:3,000 anti-SLT, 1:1,000 anti-SLT2, 1:1,000 anti-FOX1, and 1:2,000 anti-ARS. A 1:10,000 dilution of horseradish peroxidase-conjugated anti-rabbit IgG (Promega) was used as a secondary antibody, and the peroxidase activity was detected by an ECL assay (Amersham Biosciences).
Location of the Paromomycin Cassette in Genomic DNA of the ars73a Mutant
The AphVIII gene (Pollock et al., 2005) was transformed into C. reinhardtii cells by electroporation (Shimogawara et al., 1998), and three closely spaced insertions in ARS73a were identified (see below). The neighborhood of the insertion was thoroughly analyzed by PCR-based genome walking from the AphVIII gene insertions (see below) and sequencing of the genomic fragments generated during the analysis. We used genomic walking from the site of marker insertion 1 (in intron 6, but in the opposite orientation of ARS73a; Fig. 2) to the ARS73a 5′ UTR using the primers ParoF1 (5′-GTTGTTTGTCAAGGTGGCAGCT-3′; AphVIII coding sequence) and P5 (5′-AACGCTTTCCATCGCACGTGGAAGA-3′; 5′ UTR of ARS73a). A specific product was only observed when genomic DNA from the mutant was used as a template (Supplemental Fig. S7). This 3.3-kb amplification product was sequenced using the primers ParoF1 and RB2 (5′-TACCGGCTGTTGGACGAGTTCTTCTG-3′), which anneal to the AphVIII cassette, plus P5, R3 (5′-AGGAGTACGCGGTGCGCTGG-3′), F2 (5′-TATCTGCACGCACCCTGCCA-3′), F3 (5′-CCCCATTGCCTGTTCAAGCCGT-3′), and R1 (5′-CAGTTCGTGACATTCATTCTG-3′), which anneal to the 5′ region of ARS73a. Full sequence analyses of the region revealed another insertion of the AphVIII gene (designated insertion 3) downstream of the first insertion in exon 7 but in the same orientation as ARS73a (Fig. 2). We amplified a fragment extending from insertion 3 using ParoF1 and Pf (5′-GCATGCTAATCGTGTGCGCTG-3′). Again, a product (2.5 kb) was only obtained from mutant genomic DNA (Supplemental Fig. S7), and this product was sequenced using various primers including ParoF1 and RB2 in AphVIII and Pf, Pb (5′-CAGCCCACTGGACTGGTGGAGCAGCAT-3′), PD (5′-TGGTACTGCGTCAGCTCGTCCA-3′), and PE (5′-TACAGTAATTCACTGCAGATGGCCG-3) in ARS73a. We also performed PCR using RIM5-2 (primes in the 5′ region of insertion 1) and PF1 (5′-TAACTCATAATGACACAGCGC-3′) and PF4 (5′-GTGGACAGGTTCATCCAACATA-3′), both of which prime in intron 6 of ARS73a; the sequence revealed an additional insertion of a small sequence from the cassette that contains only 0.3 kb of the 3′ end of the AphVIII cassette (designated insertion 2). To amplify the region between the two AphVIII insertions, we performed PCR using the primers ParoR1 (5′-AGCTGCCACCTTGACAAACAAC-3′; reverse complement of ParoF1) and P3 (5′-ATAATGCAAGCGCTCTGCAACA-3′), which anneals to a sequence in intron 6 of ARS73a. We also amplified the region between insertions using RB2 (primes in the 3′ end of insertion 2) and ParoR1. A specific PCR product was generated using genomic DNA from the mutant as template, and the products were sequenced (data not shown). A map showing the entire region of the ARS73a gene in the mutant strain, with the three insertions and a partial ARS73a gene duplication, is presented in Figure 2, while the entire sequence of the region is given in Supplemental Figure S3.
Rescue of the ars73a Mutant Phenotype
The ars73a mutant was backcrossed three times, and the mutant phenotype (ars−) of the backcrossed strain was rescued by the introduction of BAC39M22 (from the Clemson University Genomics Institute; http://www.genome.clemson.edu/cgi-bin/orders?page=productGroup&service=bacrc& productGroup=162), which has an approximately 42-kb insert containing the ARS73a gene; introduction of the bacterial artificial chromosome DNA was by cotransformation with pSP124 (ble gene encoding zeocin resistance; Lumbreras et al., 1998). Briefly, liquid cultures were chilled on ice for 5 min, and then 10% Tween 20 (1:2,000, v/v) was added to the cultures, which were then gently mixed. Cells were pelleted at 800g for 5 min at 4°C and resuspended to a final concentration of 2 × 108 cells mL−1 in TAP medium containing 40 mm Suc. Two hundred fifty microliters of the cell suspension was electroporated with 3 to 5 μg of SmaI-digested BAC39M22 (which cuts all of the genes on the plasmid except ARS73a) and 0.5 μg of BamHI-digested pSP124S. Electroporation was performed with the Bio-Rad GenePulser II at 25 μF and 0.9 kV. Following electroporation, the cells were transferred to 5 mL of TAP Suc medium and grown under low light (approximately 30 μmol photons m−2 s−1) overnight to allow for expression of the ble gene. Following this incubation, the cells were pelleted by centrifugation at 2,700g, resuspended in approximately 500 μL of TAP medium containing 20% starch, and equal volumes were spread onto two petri dishes containing solid TAP medium supplemented with 5 to 10 μg mL−1 zeocin. The cells were grown for 10 d prior to testing each zeocin-resistant colony for ARS activity.
Sequence data for ARS73a can be found in Phytozome v9.1 (www.phytozome.net) under accession number Cre06.g298800.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Insertion of the AphVIII cassette into the ARS73a locus.
Supplemental Figure S2. Complete nucleotide sequence of the ARS73a gene.
Supplemental Figure S3. Complete nucleotide sequence of the 6.8-kb PstI fragment containing the disrupted ARS73a gene in the ars73a mutant.
Supplemental Figure S4. Alignment of ARS73a (C_523864) with the protein encoded by the gene immediately downstream (C_523865).
Supplemental Figure S5. Partial mRNA sequence of ARS73a from exon 4 to exon 7 in D66 and ars73a.
Supplemental Figure S6. ARS73a transcript levels determined by RNA-seq analysis.
Supplemental Figure S7. DNA products generated by genome walking.
Acknowledgments
We thank the members of the Grossman and Bhaya laboratories for insightful discussions.
Glossary
- S
sulfur
- SO42−
sulfate
- ARS
arylsulfatases
- TAP-S
Tris-acetate-phosphate medium without sulfur
- TAP
Tris-acetate-phosphate
- UTR
untranslated region
- RT
reverse transcription
- qPCR
quantitative PCR
- cDNA
complementary DNA
References
- Arnon DI. (1949) Copper enzymes in isolated chloroplasts: polyphenoloxidase in Beta vulgaris. Plant Physiol 24: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanesyan L, Günther V, Dichtl B, Georgiev O, Schaffner W. (2012) Polyglutamine tracts as modulators of transcriptional activation from yeast to mammals. Biol Chem 393: 63–70 [DOI] [PubMed] [Google Scholar]
- Chang CW, Moseley JL, Wykoff D, Grossman AR. (2005) The LPB1 gene is important for acclimation of Chlamydomonas reinhardtii to phosphorus and sulfur deprivation. Plant Physiol 138: 319–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JP, Yildiz F, Grossman AR. (1994) Mutants of Chlamydomonas reinhardtii with aberrant responses to sulfur deprivation. Plant Cell 6: 53–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JP, Yildiz FH, Grossman AR. (1996) Sac1, a putative regulator that is critical for survival of Chlamydomonas reinhardtii during sulfur deprivation. EMBO J 15: 2150–2159 [PMC free article] [PubMed] [Google Scholar]
- Davies JP, Yildiz FH, Grossman AR. (1999) Sac3, an Snf1-like serine/threonine kinase that positively and negatively regulates the responses of Chlamydomonas to sulfur limitation. Plant Cell 11: 1179–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hostos EL, Schilling J, Grossman AR. (1989) Structure and expression of the gene encoding the periplasmic arylsulfatase of Chlamydomonas reinhardtii. Mol Gen Genet 218: 229–239 [DOI] [PubMed] [Google Scholar]
- de Hostos EL, Togasaki RK, Grossman A. (1988) Purification and biosynthesis of a derepressible periplasmic arylsulfatase from Chlamydomonas reinhardtii. J Cell Biol 106: 29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWitt ND, dos Santos CF, Allen KE, Slayman CW. (1998) Phosphorylation region of the yeast plasma-membrane H+-ATPase: role in protein folding and biogenesis. J Biol Chem 273: 21744–21751 [DOI] [PubMed] [Google Scholar]
- Ding YH, Liu NY, Tang ZS, Liu J, Yang WC. (2006) Arabidopsis GLUTAMINE-RICH PROTEIN23 is essential for early embryogenesis and encodes a novel nuclear PPR motif protein that interacts with RNA polymerase II subunit III. Plant Cell 18: 815–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauchon M, Lagniel G, Aude JC, Lombardia L, Soularue P, Petat C, Marguerie G, Sentenac A, Werner M, Labarre J. (2002) Sulfur sparing in the yeast proteome in response to sulfur demand. Mol Cell 9: 713–723 [DOI] [PubMed] [Google Scholar]
- Ferreira RMB, Teixeira ARN. (1992) Sulfur starvation in Lemna leads to degradation of ribulose-bisphosphate carboxylase without plant death. J Biol Chem 267: 7253–7257 [PubMed] [Google Scholar]
- González E, Solano R, Rubio V, Leyva A, Paz-Ares J. (2005) PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 is a plant-specific SEC12-related protein that enables the endoplasmic reticulum exit of a high-affinity phosphate transporter in Arabidopsis. Plant Cell 17: 3500–3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Ballester D, Casero D, Cokus S, Pellegrini M, Merchant SS, Grossman AR. (2010) RNA-seq analysis of sulfur-deprived Chlamydomonas cells reveals aspects of acclimation critical for cell survival. Plant Cell 22: 2058–2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Ballester D, Pollock SV, Pootakham W, Grossman AR. (2008) The central role of a SNRK2 kinase in sulfur deprivation responses. Plant Physiol 147: 216–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman A, Takahashi H. (2001) Macronutrient utilization by photosynthetic eukaryotes and the fabric of interactions. Annu Rev Plant Physiol Plant Mol Biol 52: 163–210 [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y, Naveh-Many T, Cedar H, Razin A. (1981) Sequence specificity of methylation in higher plant DNA. Nature 292: 860–862 [DOI] [PubMed] [Google Scholar]
- Harris EH (1989). The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. Academic Press, San Diego [DOI] [PubMed] [Google Scholar]
- Huettel B, Kanno T, Daxinger L, Bucher E, van der Winden J, Matzke AJ, Matzke M. (2007) RNA-directed DNA methylation mediated by DRD1 and Pol IVb: a versatile pathway for transcriptional gene silencing in plants. Biochim Biophys Acta 1769: 358–374 [DOI] [PubMed] [Google Scholar]
- Irihimovitch V, Stern DB. (2006) The sulfur acclimation SAC3 kinase is required for chloroplast transcriptional repression under sulfur limitation in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 103: 7911–7916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawazoe R, Hwang S, Herrin DL. (2000) Requirement for cytoplasmic protein synthesis during circadian peaks of transcription of chloroplast-encoded genes in Chlamydomonas. Plant Mol Biol 44: 699–709 [DOI] [PubMed] [Google Scholar]
- Kim DH, Kim GS, Yun CH, Lee YC. (2008) Functional conservation of the glutamine-rich domains of yeast Gal11 and human SRC-1 in the transactivation of glucocorticoid receptor Tau 1 in Saccharomyces cerevisiae. Mol Cell Biol 28: 913–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landeras-Bueno S, Jorba N, Pérez-Cidoncha M, Ortín J. (2011) The splicing factor proline-glutamine rich (SFPQ/PSF) is involved in influenza virus transcription. PLoS Pathog 7: e1002397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leustek T, Martin MN, Bick JA, Davies JP. (2000) Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Annu Rev Plant Physiol Plant Mol Biol 51: 141–165 [DOI] [PubMed] [Google Scholar]
- Lopez PJ, Marchand I, Yarchuk O, Dreyfus M. (1998) Translation inhibitors stabilize Escherichia coli mRNAs independently of ribosome protection. Proc Natl Acad Sci USA 95: 6067–6072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumbreras V, Stevens DR, Purton S. (1998) Efficient foreign gene expression in Chlamydomonas reinhardtii mediated by an endogenous intron. Plant J 14: 441–447 [Google Scholar]
- Moseley JL, González-Ballester D, Pootakham W, Bailey S, Grossman AR. (2009) Genetic interactions between regulators of Chlamydomonas phosphorus and sulfur deprivation responses. Genetics 181: 889–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro FJ, Martín Y, Siverio JM. (2008) Phosphorylation of the yeast nitrate transporter Ynt1 is essential for delivery to the plasma membrane during nitrogen limitation. J Biol Chem 283: 31208–31217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AV, Thomas-Hall SR, Malnoë A, Timmins M, Mussgnug JH, Rupprecht J, Kruse O, Hankamer B, Schenk PM. (2008) Transcriptome for photobiological hydrogen production induced by sulfur deprivation in the green alga Chlamydomonas reinhardtii. Eukaryot Cell 7: 1965–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock SV, Colombo SL, Prout DL, Jr, Godfrey AC, Moroney JV. (2003) Rubisco activase is required for optimal photosynthesis in the green alga Chlamydomonas reinhardtii in a low-CO2 atmosphere. Plant Physiol 133: 1854–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock SV, Pootakham W, Shibagaki N, Moseley JL, Grossman AR. (2005) Insights into the acclimation of Chlamydomonas reinhardtii to sulfur deprivation. Photosynth Res 86: 475–489 [DOI] [PubMed] [Google Scholar]
- Pootakham W, González-Ballester D, Grossman AR. (2010) Identification and regulation of plasma membrane sulfate transporters in Chlamydomonas. Plant Physiol 153: 1653–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravina CG, Chang C-I, Tsakraklides GP, McDermott JP, Vega JM, Leustek T, Gotor C, Davies J. (2002) The sac mutants of Chlamydomonas reinhardtii reveal transcriptional and posttranscriptional control of cysteine biosynthesis. Plant Physiol 130: 2076–2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Shimogawara K, Fujiwara S, Grossman AR, Usuda H. (1998) High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics 148: 1821–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K, Sato N, Tsuzuki M. (2007) Utilization of a chloroplast membrane sulfolipid as a major internal sulfur source for protein synthesis in the early phase of sulfur starvation in Chlamydomonas reinhardtii. FEBS Lett 581: 4519–4522 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Braby CE, Grossman AR. (2001) Sulfur economy and cell wall biosynthesis during sulfur limitation of Chlamydomonas reinhardtii. Plant Physiol 127: 665–673 [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Kopriva S, Giordano M, Saito K, Hell R. (2011) Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annu Rev Plant Biol 62: 157–184 [DOI] [PubMed] [Google Scholar]
- Wykoff DD, Davies JP, Melis A, Grossman AR. (1998) The regulation of photosynthetic electron transport during nutrient deprivation in Chlamydomonas reinhardtii. Plant Physiol 117: 129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz FH, Davies JP, Grossman AR. (1994) Characterization of sulfate transport in Chlamydomonas reinhardtii during sulfur-limited and sulfur-sufficient growth. Plant Physiol 104: 981–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz FH, Davies JP, Grossman AR. (1996) Sulfur availability and the SAC1 gene control adenosine triphosphate sulfurylase gene expression in Chlamydomonas reinhardtii. Plant Physiol 112: 669–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Shrager J, Jain M, Chang CW, Vallon O, Grossman AR. (2004) Insights into the survival of Chlamydomonas reinhardtii during sulfur starvation based on microarray analysis of gene expression. Eukaryot Cell 3: 1331–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]