A subunit of the oligosaccharyltransferase complex is important for efficient glycosylation of specific glycoproteins, including the receptor kinase EFR involved in innate immunity and the endo-β-1,4-glucanase KORRIGAN1 required for cellulose biosynthesis.
Abstract
Asparagine-linked glycosylation of proteins is an essential cotranslational and posttranslational protein modification in plants. The central step in this process is the transfer of a preassembled oligosaccharide to nascent proteins in the endoplasmic reticulum by the oligosaccharyltransferase (OST) complex. Despite the importance of the catalyzed reaction, the composition and the function of individual OST subunits are still ill defined in plants. Here, we report the function of the highly conserved OST subunit OST3/6. We have identified a mutant in the OST3/6 gene that causes overall underglycosylation of proteins and affects the biogenesis of the receptor kinase EF-TU RECEPTOR involved in innate immunity and the endo-β-1,4-glucanase KORRIGAN1 required for cellulose biosynthesis. Notably, the ost3/6 mutation does not affect mutant variants of the receptor kinase BRASSINOSTEROID-INSENSITIVE1. OST3/6 deficiency results in activation of the unfolded protein response and causes hypersensitivity to salt/osmotic stress and to the glycosylation inhibitor tunicamycin. Consistent with its role in protein glycosylation, OST3/6 resides in the endoplasmic reticulum and interacts with other subunits of the OST complex. Together, our findings reveal the importance of Arabidopsis (Arabidopsis thaliana) OST3/6 for the efficient glycosylation of specific glycoproteins involved in different physiological processes and shed light on the composition and function of the plant OST complex.
Asn-linked glycosylation of proteins is a universal cotranslational and posttranslational modification of proteins entering the secretory pathway. In all eukaryotes, a hallmark of N-glycosylation is the en bloc transfer of a common preassembled oligosaccharide (Glc3Man9GlcNAc2) from the lipid carrier dolichol pyrophosphate to selected Asn residues in the sequence Asn-X-Ser/Thr (where X ≠ P) within nascent polypeptides. This transfer takes place in the lumen of the endoplasmic reticulum (ER) and is catalyzed by the oligosaccharyltransferase (OST), a heteromeric membrane protein complex (Kelleher and Gilmore, 2006). In yeast (Saccharomyces cerevisiae) and mammals, OST consists of a catalytically active subunit (STAUROSPORIN AND TEMPERATURE SENSITIVE3 [STT3]) and several different noncatalytic subunits that contribute to N-glycosylation by regulation of the substrate specificity, stability, or assembly of the complex (Yan and Lennarz, 2002; Mohorko et al., 2011). In yeast, the OST complex is composed of a single STT3 protein and eight additional subunits (Knauer and Lehle, 1999a). Five of them (Stt3p, Ost1p, Ost2p, Wbp1p, and Swp1p) are essential for the viability of yeast cells. In mammals, the overall composition and organization of the OST complex is more complex, and different subunit compositions have been described (Shibatani et al., 2005; Mohorko et al., 2011; Roboti and High, 2012). Apart from the presence of several noncatalytic subunits required for maximal enzyme activity, mammals can utilize two distinct STT3 isoforms (STT3A and STT3B) that differ in their catalytic activity and acceptor substrate selectivity and perform cotranslational as well as posttranslational glycosylation of proteins (Ruiz-Canada et al., 2009).
Yeast Ost3p and Ost6p are two nonessential OST subunits, which share 21% sequence identity and a similar membrane topology with an N-terminal lumenal domain and a transmembrane domain composed of four helices (Kelleher and Gilmore, 2006). The two isoforms are present in distinct OST complexes and consequently define two different OST subcomplexes with site-specific glycosylation efficiency (Knauer and Lehle, 1999b; Schwarz et al., 2005; Yan and Lennarz, 2005b; Schulz et al., 2009). Disruption of both subunits results in a severe underglycosylation defect, while the deficiency of either Ost3p or Ost6p leads only to a modest reduction of the in vivo glycosylation efficiency on a distinct set of glycoproteins (Karaoglu et al., 1995; Knauer and Lehle, 1999b; Schulz and Aebi, 2009). A recent study demonstrated that Ost3p and Ost6p have oxidoreductase activity mediated by a thioredoxin-like fold including a CxxC active-site motif (Schulz et al., 2009), which indicates that Ost3p and Ost6p might prevent fast protein folding to achieve optimal glycosylation efficiency on a subset of proteins in yeast.
The transfer of the preassembled oligosaccharide in plants involves a similar OST multisubunit complex, which is still poorly described. In Arabidopsis (Arabidopsis thaliana), two proteins, termed STT3A and STT3B, with homology to the yeast and mammalian catalytic subunits have been identified (Koiwa et al., 2003). STT3A-deficient plants are viable but display a protein underglycosylation defect that affects the biogenesis of heavily glycosylated proteins, such as the pattern recognition receptor EF-TU RECEPTOR (EFR; Nekrasov et al., 2009; Saijo et al., 2009; Häweker et al., 2010). In contrast, STT3B deficiency does not lead to an obvious underglycosylation defect, and EFR function is not compromised (Koiwa et al., 2003; Nekrasov et al., 2009; Häweker et al., 2010). However, Arabidopsis stt3a stt3b double knockout plants are gametophytic lethal (Koiwa et al., 2003). These findings highlight the importance of the catalytic OST subunit for protein glycosylation in plants and show that the two putative catalytic subunits have partially overlapping as well as substrate-specific functions. In another study, it was found that depletion of Arabidopsis Defective Glycosylation1 (DGL1), which is a homolog of the essential yeast subunit Wbp1p, is embryo lethal (Lerouxel et al., 2005). A mutant line with a weak dgl1 allele displayed reduced protein N-glycosylation efficiency consistent with a crucial role of DGL1 in plant OST function. In addition, Arabidopsis proteins have been identified that display significant homology to yeast Swp1p/mammalian ribophorin II (HAPLESS6 [HAP6]) and OST2/defender-against apoptotic death (DAD1 and DAD2; Gallois et al., 1997; Danon et al., 2004; Johnson et al., 2004). Arabidopsis DAD1 could rescue the apoptotic phenotype of a mutant hamster cell line, indicating a conserved function between animals and plants (Gallois et al., 1997). On the other hand, neither for Arabidopsis DAD1/DAD2 nor for HAP6 has a specific role in protein glycosylation been described so far, and their incorporation into the OST complex remains to be shown. Other homologs of mammalian or yeast OST subunits are present in plants (Supplemental Table S1) but have not been functionally characterized yet, and the overall composition of the Arabidopsis OST complex as well as the distinct role of its subunits are unknown (Pattison and Amtmann, 2009).
In this study, we identified in a forward genetic screen (Nekrasov et al., 2009) an Arabidopsis mutant impaired in the response to the bacterial elongation factor Tu (EF-Tu), which is recognized by the pattern recognition receptor EFR (Zipfel et al., 2006). The mutant line harbors a defect in a gene encoding a protein homologous to yeast OST3/6 proteins. OST3/6 deficiency causes reduced N-glycosylation site occupancy on distinct proteins and affects the biogenesis of EFR, resulting in the impairment of innate immunity. Moreover, we provide evidence that OST3/6 plays an important role for abiotic stress tolerance and for the functionality of the endoglucanase KORRIGAN1 (KOR1) essential for cellulose biosynthesis. However, OST3/6 does not appear to be required for the ER quality control of the mutant variants of the brassinosteroid receptor BRASSINOSTEROID-INSENSITIVE1-5 (BRI1-5) and BRI1-9.
RESULTS
The Arabidopsis elf18-Insensitive Mutant elfin23-7 Carries a Mutation in OST3/6
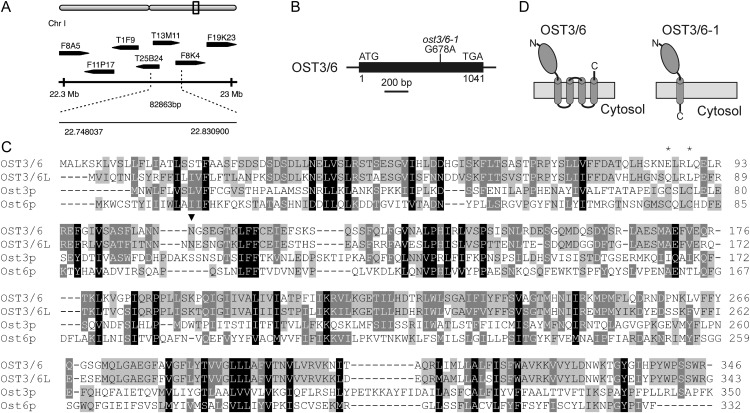
The pattern recognition receptor EFR senses bacteria through recognition of the pathogen-associated molecular pattern (PAMP) EF-Tu, resulting in PAMP-triggered immunity (Zipfel et al., 2006). A forward genetic screen aimed at the discovery of proteins involved in the biogenesis and function of EFR resulted in the identification of elf18-insensitive (elfin) mutants (Li et al., 2009; Nekrasov et al., 2009). One mutant, elfin23-7, was mapped to an approximately 83-kb region on the bottom of chromosome I (Fig. 1A). Sequencing of candidate genes in this region identified a single base pair change (G678A) in the intronless open reading frame of At1g61790, introducing a premature stop codon (Fig. 1B).
Figure 1.
ost3/6-1 carries a mutation in the OST3/6 gene. A, Schematic representation of the approximately 83-kb interval on the bottom of chromosome I where the elfin23-7 mutation was mapped. The dotted lines indicate the positions of the flanking markers F8K4-2 and AC005850-7476. B, Schematic presentation of the OST3/6 gene structure. The open reading frame, which is present on a single exon, is depicted as a black box. The single point mutation in the ost3/6-1 mutant is indicated. C, Sequence alignment of OST3/6 (At1g61790) and OST3/6-LIKE1 (OST3/6L; At1g11560) with Saccharomyces cerevisiae Ost3p (systematic name, YOR085W) and Ost6p (YML019W). The alignment was performed with ClustalW (http://www.ch.embnet.org/software/ClustalW.html) using default settings. Similar amino acid residues present in all four sequences are shaded in black, similar residues present in three sequences are shaded in dark gray, and similar residues present in two sequences are shaded in light gray. The absence of the CxxC active-site motif in the Arabidopsis sequences is indicated by asterisks. The single conserved N-glycosylation site in the putative lumenal domain is marked by an arrowhead. D, Schematic presentation of the predicted protein topology for OST3/6 and for the truncated OST3/6-1 form, which is produced in ost3/6-1. The model is based on the prediction of signal peptides using the SignalP 4.0 server (http://www.cbs.dtu.dk/services/SignalP/) and transmembrane helices using the TMHMM server version 2.0 (http://www.cbs.dtu.dk/services/TMHMM/).
At1g61790 encodes a 346-amino acid protein with a predicted N-terminal signal peptide, a lumenal domain, and four C-terminal transmembrane helices (Fig. 1D). An amino acid sequence alignment (Fig. 1C) and in silico prediction of a conserved membrane protein topology (Fig. 1D; Supplemental Table S2) suggest that At1g61790 encodes a plant homolog of Ost3p/Ost6p, which are known subunits of the yeast OST complex. Consequently, we renamed elfin23-7 as ost3/6-1 and refer to the gene product of At1g61790 as OST3/6.
Arabidopsis Contains Two OST3/6 Homologs
Apart from OST3/6, the Arabidopsis genome contains another intronless gene (At1g11560) coding for a putative ortholog of yeast Ost3p/Ost6p that is here referred to as OST3/6-LIKE1. The amino acid sequence of OST3/6 displays 65% identity (80% similarity) to OST3/6-LIKE1, and the overall protein topology is conserved between OST3/6 and OST3/6-LIKE1. Extraction of data from microarray experiments (Obayashi et al., 2007) reveals that OST3/6 is widely expressed in Arabidopsis, and OST3/6 transcript expression correlates well with the expression of other genes coding for OST subunits like STT3A and DAD1 (Supplemental Fig. S1; Supplemental Table S3). In contrast, OST3/6-LIKE1 displays organ-specific expression with low levels in whole seedlings, higher expression levels in stems and stamens, and no clear coexpression with genes for other OST subunits (Supplemental Fig. S2).
The ost3/6-1 Mutant Affects EFR Biogenesis
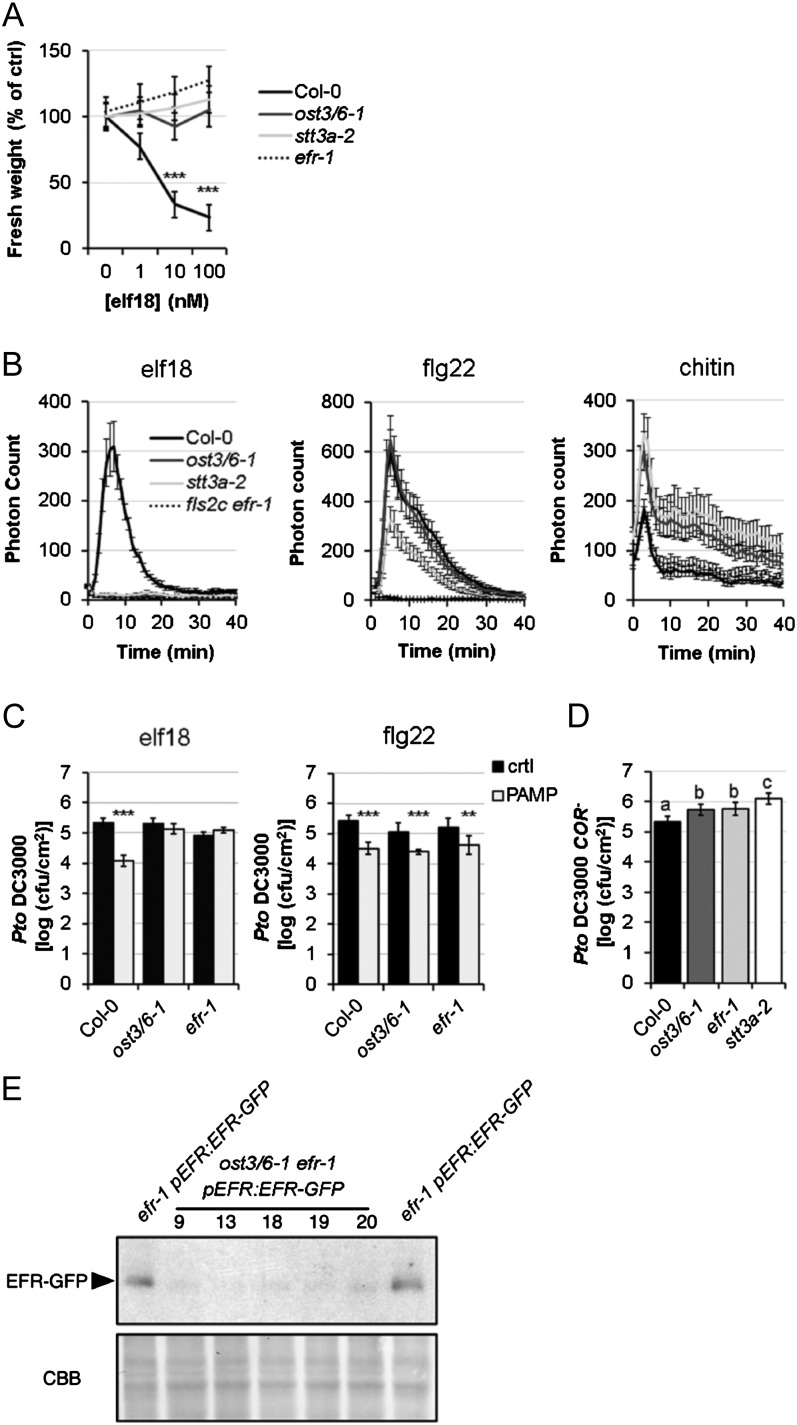
Measurement of seedling growth inhibition in response to elf18 treatment suggested that the biogenesis and/or the function of the Leu-rich repeat receptor kinase (LRR-RK) EFR are compromised in the ost3/6-1 mutant (Fig. 2A). In contrast, seedling growth inhibition mediated by flg22 that is recognized by the related LRR-RK FLAGELLIN SENSING2 (FLS2; Gómez-Gómez and Boller, 2000) was similar to wild-type plants (data not shown), showing that ost3/6-1 is sensitive to flg22 treatment similar to efr-1 and the previously characterized stt3a-2 mutant (Nekrasov et al., 2009; Häweker et al., 2010). Consistent with this findings, the oxidative burst triggered by elf18 was almost abolished (Fig. 2B), while oxidative bursts triggered by two other PAMPs, flg22 and chitin (recognized by the LysM-RK CHITIN ELICITOR RECEPTOR KINASE1 [CERK1]; Miya et al., 2007; Wan et al., 2008), were not drastically altered (Fig. 2B). To test whether elf18-induced resistance to the bacterial pathogen Pseudomonas syringae pv tomato (Pto) DC3000 is also affected in ost3/6-1, we pretreated plants with elf18 and investigated the inhibition of pathogen growth. Notably, elf18-induced bacterial resistance was severely impaired in the mutant (Fig. 2C), while pretreatment with flg22 did not alter the growth of Pto DC3000 in ost3/6-1 (Fig. 2C). Interestingly, the ost3/6-1 mutation led to an increase in susceptibility toward the hypovirulent strain Pto COR− upon spray infection comparable to that caused by loss of EFR (Fig. 2D; Nekrasov et al., 2009).
Figure 2.
Seedling growth inhibition triggered by elf18 on 2-week-old seedlings of the Col-0 wild type, ost3/6-1, stt3a-2, and fls2c efr-1. A, Fresh weights are represented relative to the untreated control. Values are averages from four independent biological repeats ± 2× se (n = 24–36, average of 34). Means and differences were calculated by two-way ANOVA (Holm-Sidak pairwise multiple comparison; significant differences are indicated with asterisks [***P < 0.001]). B, Oxidative burst in leaf discs from 4-week-old Col-0 wild-type, ost3/6-1, stt3a-2, and fls2c efr-1 plants in response to 100 nm elf18, 100 nm flg22, or 1 mg mL−1 chitin. Results are averages ± se (n = 8). Similar results were observed in at least three independent experiments. C, PAMP-induced resistance measured on 4-week-old Col-0 wild-type, ost3/6-1, and efr-1 plants preinfiltrated with water, 1 μm elf18 (left graph), or 1 μm flg22 (right graph), and 24 h later syringe infiltrated with Pto DC3000 (105 colony-forming units [cfu] mL−1). Bacteria were quantified at 2 d post inoculation as log(cfu cm−2). Results are averages ± 2× se (n = 12). ANOVA was performed using Holm-Sidak one-way ANOVA, and significant differences are indicated with asterisks (**P < 0.01, ***P < 0.001). D, Spray infection of 4-week-old Col-0 wild-type, ost3/6-1, efr-1, and stt3a-2 plants with Pto DC3000 COR− (107 cfu mL−1). Bacteria were quantified at 3 d post inoculation as log(cfu cm−2). Results are averages from five independent biological repeats ± 2× se. Means and differences were calculated by two-way ANOVA (Holm-Sidak pairwise multiple comparison; a, b, and c indicate significant differences). E, EFR-GFP levels in efr-1/pEFR:EFR-GFP and different ost3/6-1 efr-1/pEFR:EFR-GFP lines determined by immunoblot analysis with anti-GFP-HRP. Similar results were observed in two independent experiments. CBB, Coomassie Brilliant Blue.
Since EFR is heavily glycosylated (Nekrasov et al., 2009; Saijo et al., 2009; Häweker et al., 2010), we hypothesized that N-glycosylation of EFR is altered in the ost3/6-1 mutant, potentially leading to altered accumulation and/or function of EFR. We generated ost3/6-1 efr-1 double mutants expressing GFP-tagged EFR under the control of the native promoter. Immunoblot analysis monitoring the EFR-GFP levels revealed that EFR-GFP accumulation is reduced in independent homozygous ost3/6-1 efr-1/pEFR:EFR-GFP lines when compared with efr-1/pEFR:EFR-GFP (Fig. 2E). These observations show that OST3/6 function is required for EFR biogenesis similar to previously described mutants with defects in glycosylation or ER quality control (Li et al., 2009; Lu et al., 2009; Nekrasov et al., 2009; Saijo et al., 2009; von Numers et al., 2010).
ost3/6-1 Displays a Protein Underglycosylation Defect
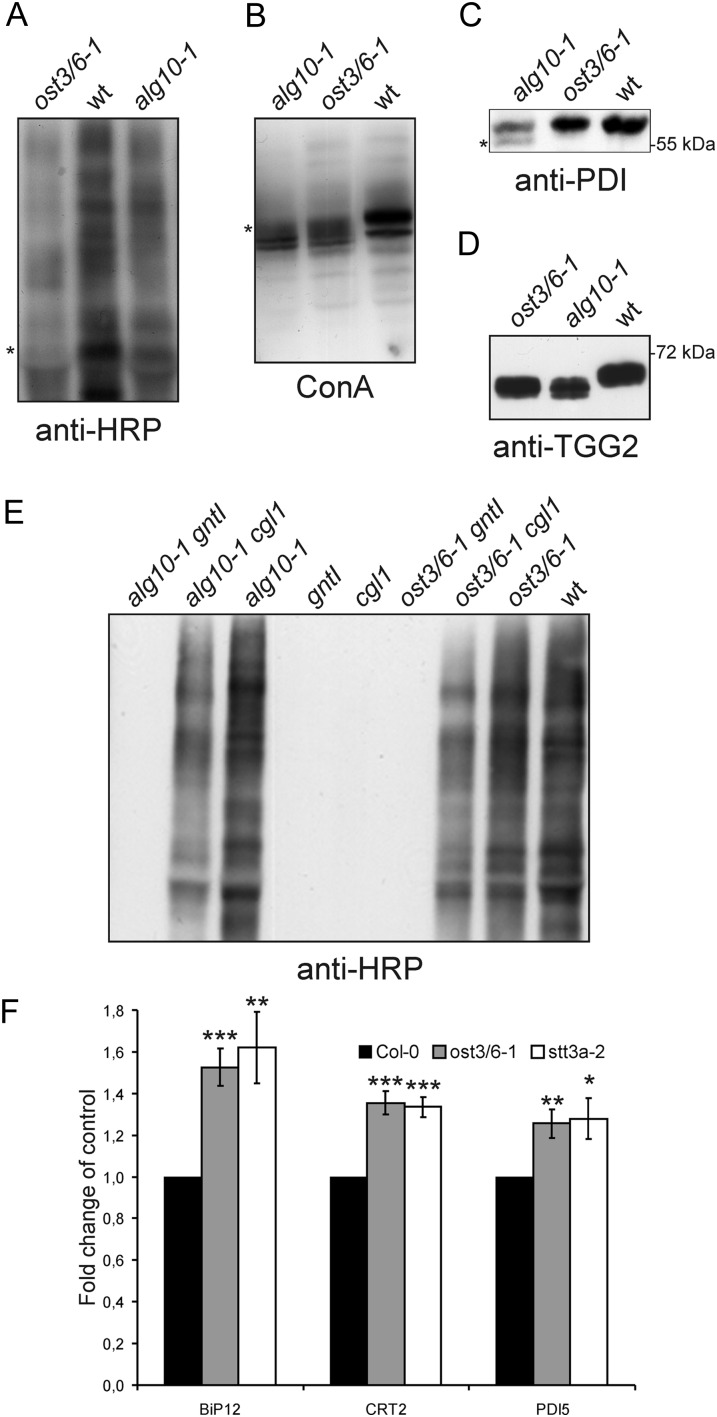
The introduction of a premature stop codon by the ost3/6-1 mutation suggests that a truncated 225-amino acid protein is generated that is not fully functional. Consequently, we hypothesized that a deficiency in OST3/6 causes alterations in protein N-glycosylation leading to protein underglycosylation, a defect that has already been described previously for other Arabidopsis OST subunit mutants (Koiwa et al., 2003; Lerouxel et al., 2005). First, we analyzed the overall N-glycosylation profile of glycoproteins extracted from rosette leaves. The total N-glycan pool was unchanged (Supplemental Fig. S3), showing that the mutant does not affect the biosynthesis or processing of N-glycans. However, SDS-PAGE separation and subsequent immunoblot analysis with antibodies against complex N-glycans revealed an overall reduced signal intensity, indicating the presence of lower amounts of complex N-glycans (Fig. 3A). A lectin blot with concanavalin A, which binds mainly to terminal mannosyl and glucosyl residues of glycoproteins, showed that the mobility of individual bands is altered (Fig. 3B). Immunoblotting with protein-specific antibodies demonstrated that the ER-resident protein disulfide isomerase (PDI), which harbors two N-glycosylation sites and is commonly used as a reporter to monitor underglycosylation, is not affected in ost3/6-1 (Fig. 3C). By contrast, in the alg10-1 mutant, which is deficient in the last biosynthetic step of the lipid-linked oligosaccharide assembly and consequently displays reduced glycosylation efficiency (Farid et al., 2011), faster migrating PDI forms were detectable upon SDS-PAGE separation. Another glycoprotein, β-thioglucoside glucohydrolase2 (TGG2), containing four Asn-linked glycans (Liebminger et al., 2012), displayed higher mobility in ost3/6-1 compared with the wild type, indicating that TGG2 is hypoglycosylated in ost3/6-1 (Fig. 3D). The restoration of complex N-glycan formation due to reduced or completely impaired glycosylation of a misfolded N-acetylglucosaminyltransferase I (CGL1-GnTI) protein is another sensitive tool to monitor alterations in glycosylation efficiency in Arabidopsis mutants (Frank et al., 2008; Farid et al., 2011). We crossed ost3/6-1 to cgl1 plants and analyzed the generation of complex N-glycans in the ost3/6-1 cgl1 double mutant. Consistent with a role of OST3/6 in the glycosylation of proteins, we detected the restoration of complex N-glycosylation in ost3/6-1 cgl1, which was not observed in ost3/6-1 gntI control plants that contain a gntI null allele instead of the leaky cgl1 allele (Fig. 3E). This finding provides evidence that N-glycosylation site occupancy of CGL1-GnTI is also reduced in the ost3/6-1 genetic background.
Figure 3.
The ost3/6-1 mutant displays a protein underglycosylation defect. A, Immunoblot analysis of total proteins extracted from ost3/6-1, wild-type Col-0 (wt), and alg10-1 leaves. Protein extracts were analyzed using anti-HRP antibody, which recognizes complex N-glycans with β-1,2-Xyl and core α-1,3-Fuc residues. B, Protein extracts from seedlings were analyzed using the lectin concanavalin A (ConA). C, Protein extracts from the indicated seedlings were subjected to SDS-PAGE, and blots were analyzed with anti-PDI antibody. The asterisk indicate the presence of faster migrating underglycosylated protein variants. D, Protein extracts from young leaves were analyzed by immunoblotting with anti-TGG2 antibody. The occurrence of double bands indicates the presence of different underglycosylated TGG2 variants. E, Protein extracts from seedlings of wild-type Col-0 (wt) and from the indicated single (alg10-1, gntI, cgl1, ost3/6-1) and double (alg10-1 gntI, alg10-1 cgl1, ost3/6-1 gntI, ost3/6-1 cgl1) mutants were analyzed with anti-HRP antibody. F, OST3/6 deficiency leads to a significant up-regulation of some ER stress genes. qRT-PCR analysis of BiP12, CRT2, and PDI5 expression is shown for wild-type Col-0, ost3/6-1, and stt3a-2 seedlings. Values are averages ± se from at least three independent biological samples. All PCRs were performed at least twice with similar results. Asterisks denote significant differences compared with the expression in Col-0 (*P < 0.05, **P < 0.01, and ***P < 0.005).
Underglycosylation caused by genetic lesions or chemical treatment with tunicamycin triggers the unfolded protein response (UPR; Koiwa et al., 2003; Martínez and Chrispeels, 2003). To monitor induction of the UPR in ost3/6-1, we performed quantitative reverse transcription (qRT)-PCR and monitored the expression of genes coding for ER-resident proteins that are typically activated during ER stress (Martínez and Chrispeels, 2003). As for stt3a-2, the expression of genes for binding proteins (At5g28540 [BiP1] and At5g42020 [BiP2]), a calreticulin (At1g09210 [CRT2]), and a PDI (At1g21750 [PDI5]) was significantly increased in ost3/6-1, suggesting that the defect in ost3/6-1 triggers the activation of the UPR (Fig. 3F), similar to STT3A-deficient plants.
OST3/6 Deficiency Affects the Biogenesis of KOR1
The deficiency of OST3/6 in ost3/6-1 affects the biogenesis and function of EFR but seems to have little effect on the response mediated by the related heavily glycosylated LRR-RK FLS2. To investigate whether proper OST3/6 function is required for the biogenesis or functionality of mutant variants of the LRR-RK BRI1 (Li and Chory, 1997), we crossed ost3/6-1 to the weak bri1 alleles bri1-9 and bri1-5 and analyzed the suppression or enhancement of the bri1-9 or bri1-5 dwarf phenotype (Noguchi et al., 1999). The bri1-9 and bri1-5 plants contain mutant alleles of the BRI1 gene and express structurally altered but still functional BRI1 proteins that are retained in the ER by different quality control processes, including glycan-dependent mechanisms (Jin et al., 2007; Hong et al., 2008, 2012). The ost3/6-1 bri1-9 and ost3/6-1 bri1-5 double mutants were indistinguishable from bri1-9 and bri1-5, revealing that underglycosylation in ost3/6-1 neither rescues the bri1-9/bri1-5 phenotypes nor enhances their growth phenotypes (Fig. 4A). This finding suggests that the mutated BRI1 variants are not substrates of OST3/6.
Figure 4.
OST3/6 is required for KOR1 accumulation. A, ost3/6-1 does not suppress the dwarf phenotype of bri1-9 and bri1-5 mutants. Representative soil-grown single and double mutant plants are shown. B, Proteins were extracted from leaves and subjected to SDS-PAGE and immunoblotting using anti-KOR1 antibody. The presence of the KOR1-specific band is marked by the asterisk. C, ost3/6-1 enhances the growth phenotype of rsw2-1. Plants were grown on MS medium supplemented with 1.5% (w/v) Suc and grown for 14 d.
Another candidate protein that is sensitive to alterations in glycosylation is the endo-β-1,4-glucanase KOR1 (Nicol et al., 1998), which plays a yet unclear role during cell wall formation and cellulose synthesis. KOR1 has eight potential glycosylation sites in its extracellular domain, and genetic interactions of the weak KOR1 allele rsw2-1 (Lane et al., 2001) with different N-glycan-processing mutants indicate that N-glycans play a crucial role for KOR1 (Kang et al., 2008; Liebminger et al., 2009). We analyzed KOR1 expression by immunoblotting in ost3/6-1 and found that KOR1 accumulation is considerably reduced in the mutant (Fig. 4B). Next, we tested if OST3/6 deficiency enhances the phenotype of rsw2-1. In comparison with rsw2-1, the ost3/6-1 rsw2-1 double mutant displayed a severe growth phenotype with very short roots similar to stt3a rsw2-1 (Kang et al., 2008; Fig. 4C). In summary, our data show that OST3/6 function is required for KOR1 biogenesis, presumably by assisting in the transfer of oligosaccharides to selected KOR1 glycosylation sites.
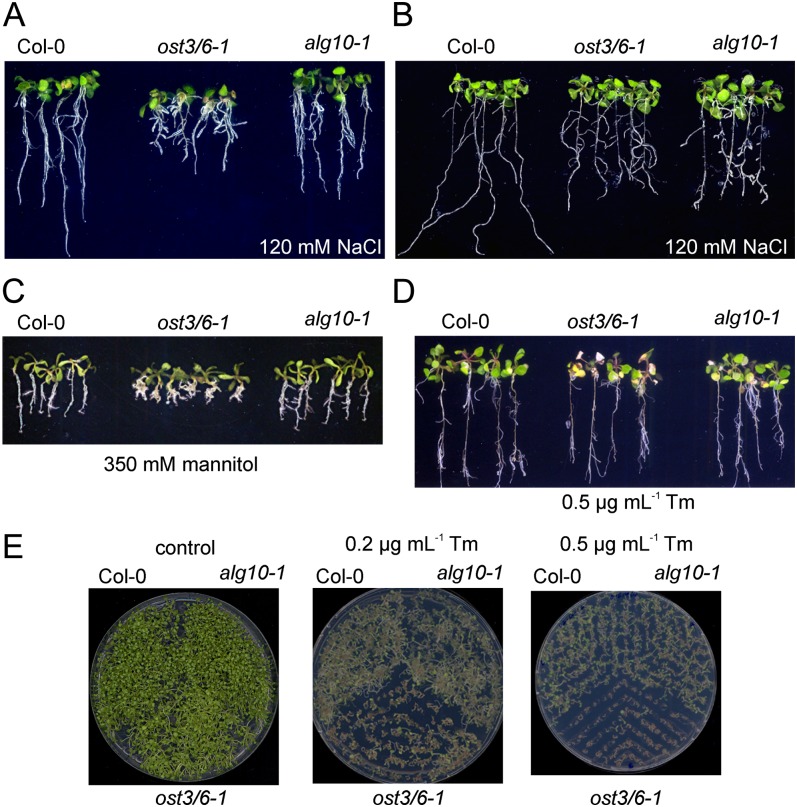
ost3/6-1 Is Hypersensitive to NaCl, Mannitol, and Tunicamycin
Although the ost3/6-1 plants display a severe underglycosylation defect, these plants do not show any developmental or morphological phenotype under normal growth conditions (Fig. 4A). Underglycosylation mutants are frequently more sensitive to salt stress and tunicamycin, an inhibitor of protein glycosylation (Koiwa et al., 2003; Zhang et al., 2009; Farid et al., 2011). Germination of ost3/6-1 on medium containing 120 mm NaCl (Fig. 5A) resulted in decreased root growth. Increased sensitivity to NaCl was also observed when 6-d-old seedlings were transferred to Murashige and Skoog (MS) medium supplemented with 120 mm NaCl and grown for an additional 14 d (Fig. 5B). These effects were even more pronounced under higher salt concentrations (Supplemental Fig. S4). When ost3/6-1 was grown for 2 weeks on MS medium supplemented with 350 mm mannitol, swelling of roots and enhanced lateral root formation were observed (Fig. 5C). Transfer of 6-d-old ost3/6-1 seedlings to MS plates containing 0.5 µg mL−1 tunicamycin resulted in the formation of an increased number of pale seedlings (Fig. 5D). A germination assay confirmed that ost3/6-1 is much more sensitive to tunicamycin than wild-type and alg10-1 plants (Fig. 5E).
Figure 5.
ost3/6-1 is more sensitive to abiotic stresses. A, Seeds of the Col-0 wild-type, ost3/6-1, and alg10-1 were germinated on MS medium supplemented with 120 mm NaCl and grown for 14 d. B, Seedlings were grown on MS medium for 6 d, transferred to MS medium supplemented with 120 mm NaCl, and grown for an additional 14 d. C, Seeds were spread on MS medium supplemented with 350 mm mannitol and grown for 14 d. D, Seven-day-old seedlings were transferred to MS plates supplemented with 0.5 µg mL−1 tunicamycin (Tm) and grown for 14 d. E, Seeds were directly germinated on MS plates supplemented with the indicated concentrations of tunicamycin or on a control plate without any tunicamycin and grown for 14 d.
In a previous study, it was found that the lew3 mutant, which displays underglycosylation of proteins due to a defect in the biosynthesis of the oligosaccharide precursor, is more sensitive to NaCl, tunicamycin, and abscisic acid (ABA; Zhang et al., 2009). To investigate differences in ABA sensitivity, we performed a germination assay in the presence of two different ABA concentrations. In contrast to lew3, ost3/6-1 germination was not impaired by ABA (Supplemental Fig. S4).
Complementation of ost3/6-1 Restores Glycosylation, Salt Tolerance, and elf18 Sensitivity
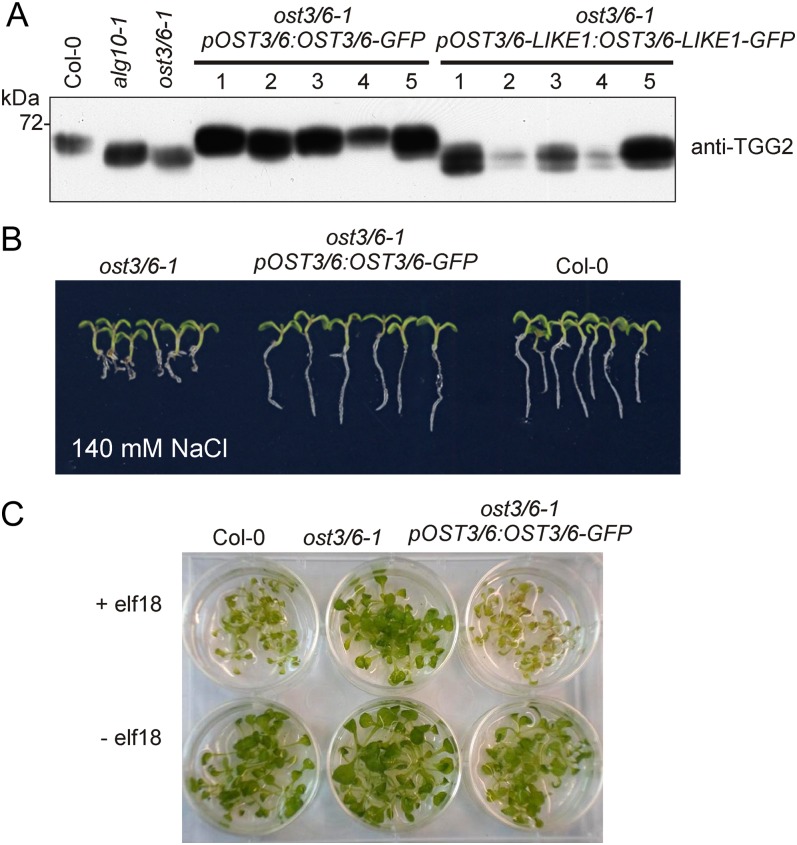
We could not identify additional ost3/6 alleles in our forward genetic screen, and there are no OST3/6 transfer DNA knockout mutants available to corroborate that the detected phenotypes are caused by OST3/6 deficiency. To confirm that the observed phenotypes are indeed caused by the molecular lesion in ost3/6-1, we expressed OST3/6-GFP under the control of its native promoter in ost3/6-1. Immunoblot analysis using anti-TGG2 antibody revealed that the underglycosylation defect was restored in the ost3/6-1/pOST3/6:OST3/6-GFP transgenic plants (Fig. 6A). Consistent with this finding, the transgenic seedlings were as tolerant to increased salt concentrations as wild-type plants (Fig. 6B), and elf18-induced seedling growth inhibition was triggered in ost3/6-1 expressing pOST3/6:OST3/6-GFP (Fig. 6C). Restoration of TGG2 glycosylation was also observed when OST3/6-GFP was expressed under the control of the ubiquitin10 gene promoter in ost3/6-1 (Supplemental Fig. S5). By contrast, expression of neither pOST3/6-LIKE1:OST3/6-LIKE1-GFP (Fig. 6A) nor pUBQ10:OST3/6-LIKE1-GFP (Supplemental Fig. S5) in ost3/6-1 resulted in the compensation of the TGG2 glycosylation defect. It is interesting that no OST3/6-LIKE1-GFP could be detected in these transgenic lines by immunoblotting, suggesting that OST3/6-LIKE1-GFP is not properly expressed at all or is highly unstable compared with OST3/6.
Figure 6.
Complementation of the ost3/6-1 mutant restores TGG2 underglycosylation, the salt stress phenotype, and elf18 sensitivity. A, Protein extracts from independent stably transformed Arabidopsis ost3/6-1 lines and the indicated control plants were subjected to SDS-PAGE and immunoblotting with anti-TGG2 antibody. TGG2 from ost3/6-1 plants expressing pOST3/6:OST3/6-GFP shows a migration position similar to TGG2 from wild-type plants. The occurrence of double bands indicates the presence of partially glycosylated TGG2 variants. B, Six-day-old seedlings were transferred to MS plates supplemented with 140 mm NaCl and grown for 14 d. The root growth phenotype of ost3/6-1 is restored in the presence of pOST3/6:OST3/6-GFP. C, Seedling growth inhibition was triggered by treatment with elf18. Five-day-old Arabidopsis seedlings were transferred to liquid 1× MS supplemented with 1.5% (w/v) Suc and incubated with 50 nm elf18 or without elf18 for 5 d.
OST3/6-1 Is Located in the ER and Interacts with OST4B and STT3A
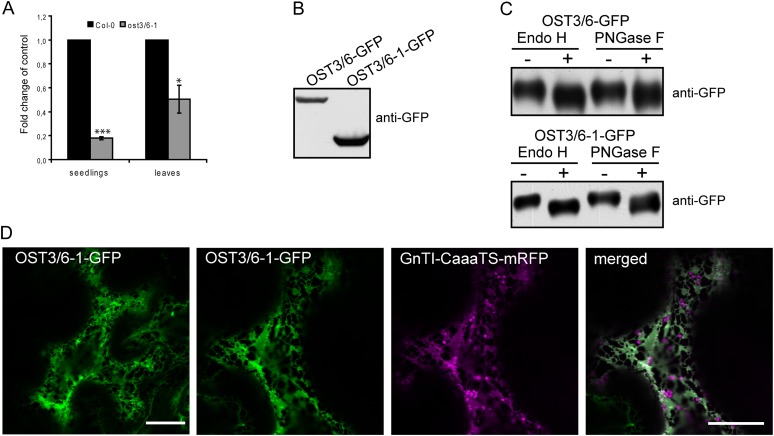
To analyze the molecular defect caused by the ost3/6-1 mutation in more detail, we performed qRT-PCR analysis. OST3/6 transcript levels were clearly reduced in seedlings and leaves (Fig. 7A), suggesting that the nonsense mutation in ost3/6-1 leads to OST3/6 mRNA decay. To assess whether the truncated protein can be produced in plants, we generated a construct where the 225-amino acid OST3/6-1 protein is fused to GFP and transiently expressed the OST3/6-1-GFP fusion protein in Nicotiana benthamiana. Immunoblot analysis with antibodies against GFP confirmed the stable production of the truncated variant when expressed under the control of heterologous regulatory sequences (Fig. 7B). Next, we tested whether the topology of OST3/6-1 is altered due to the absence of the C-terminal domain (Fig. 1C). To this end, we analyzed the glycosylation status of OST3/6 and OST3/6-1. OST3/6 contains a single N-glycosylation site at position Asn-108 in the large lumenal domain that is also present in OST3/6-1. Digestion with endoglycosidase H (Endo H) or peptide:N-glycosidase F (PNGase F) caused shifts in mobility upon SDS-PAGE separation, indicative of OST3/6 glycosylation with an oligomannosidic N-glycan (Fig. 7C). The presence of Endo H-sensitive glycoforms revealed that the large N-terminal domain of OST3/6-1 is also facing the ER lumen, demonstrating that the protein topology is not altered due to the protein truncation (Fig. 7C). Moreover, the presence of an oligomannosidic N-glycan further indicates that the truncated variant is like OST3/6 retained in the ER and not processed by Golgi-located glycosylation enzymes. Confocal microscopy analysis confirmed that OST3/6-1-GFP is predominantly located in the ER when transiently expressed in N. benthamiana (Fig. 7D). Together, these findings suggests that the reduced mRNA levels in ost3/6-1 contribute significantly to the observed OST3/6 deficiency, while the distinct role of the C-terminal domain with three additional transmembrane helices for OST3/6 function remains elusive.
Figure 7.
OST3/6 transcript levels are reduced in ost3/6-1, but the truncated OST3/6-1 can be stably expressed in plants. A, qRT-PCR analysis of OST3/6 expression in seedlings and leaves from 4-week-old soil-grown Col-0 wild-type and ost3/6-1 plants. Values are averages ± se from at least three independent biological samples. All PCRs were performed at least twice with similar results. Asterisks denote significant differences compared with the expression in the wild type (*P < 0.05, ***P < 0.005). B, OST3/6-GFP and OST3/6-1-GFP were transiently expressed in N. benthamiana leaves, and proteins were extracted 24 h post infiltration and subjected to immunoblotting using anti-GFP antibody. C, Extracted proteins were digested with Endo H and PNGase F and analyzed by SDS-PAGE and immunoblotting with anti-GFP antibody. D, OST3/6-1-GFP was expressed either alone (left panel) or in combination with the ER-retained marker GnTI-CaaaTS-mRFP in N. benthamiana leaf epidermal cells. Analysis of fluorescent proteins was done by confocal laser scanning microscopy. Bars = 20 µm.
OST3/6 Localizes to the ER
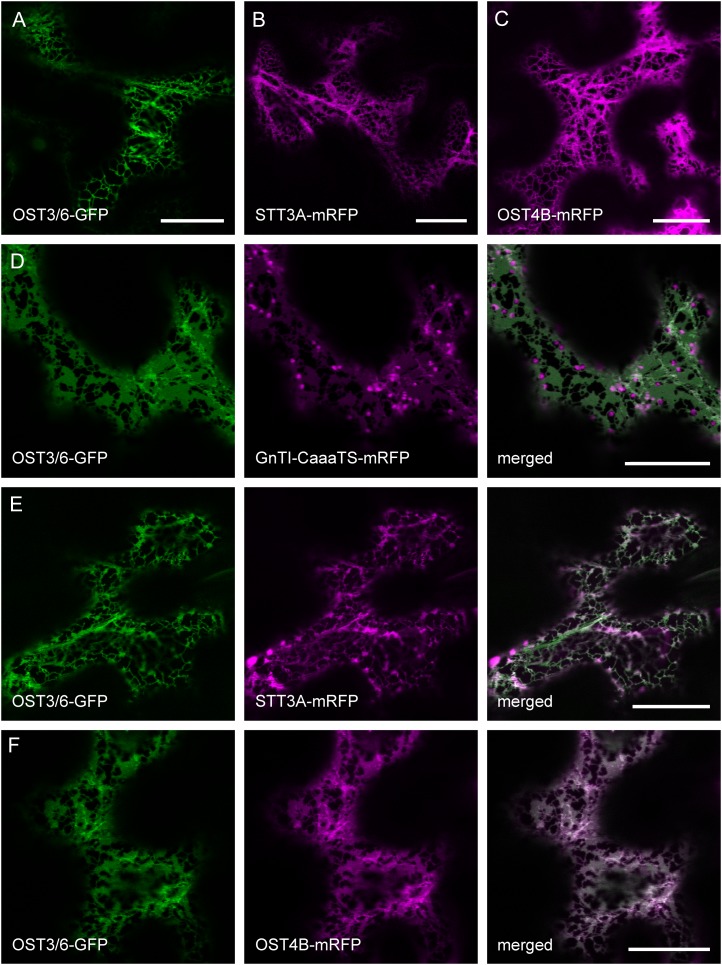
A role of OST3/6 in protein glycosylation suggests that it localizes to the ER. To test this hypothesis, we analyzed transgenic ost3/6-1/pOST3/6:OST3/6-GFP plants by confocal laser scanning microscopy. However, the fluorescence intensity was quite low, and no distinct subcellular location could be seen. Consequently, we expressed transiently the GFP-tagged OST3/6 protein under the control of the UBQ10 promoter in N. benthamiana leaves. Analysis of the OST3/6-GFP fusion protein by confocal microscopy of epidermal cells revealed a reticular distribution pattern resembling ER localization (Fig. 8A). To confirm this finding, we coexpressed OST3/6-GFP with GnTI-CaaaTS-monomeric red fluorescent protein (mRFP), which carries a mutated form of the Golgi-marker GnTI and localizes mainly to the ER (Schoberer et al., 2009). In addition, we performed colocalization experiments with other tagged OST subunits like STT3A-mRFP and Arabidopsis OST4B-mRFP, a putative ortholog of yeast Ost4p (Fig. 8, B and C). All of the expressed proteins displayed ER-like labeling and colocalization with OST3/6-GFP, consistent with a possible function of OST3/6 as an OST subunit in the ER (Fig. 8, D–F).
Figure 8.
OST3/6-GFP displays a reticulate fluorescence pattern indicating ER localization. A, N. benthamiana leaf epidermal cells expressing OST3/6-GFP alone. B, N. benthamiana leaf epidermal cells expressing STT3A-mRFP alone. C, N. benthamiana leaf epidermal cells expressing OST4B-mRFP alone. D, Expression of OST3/6-GFP in combination with the ER-retained marker protein GnTI-CaaaTS-mRFP. E and F, Expression of OST3/6-GFP in combination with the OST subunit STT3A-mRFP (E) and coexpression of OST3/6-GFP with the OST subunit OST4B-mRFP (F). Analysis of fluorescent proteins was done by confocal laser scanning microscopy. Bars = 20 µm.
OST3/6 Interacts with Other OST Subunits
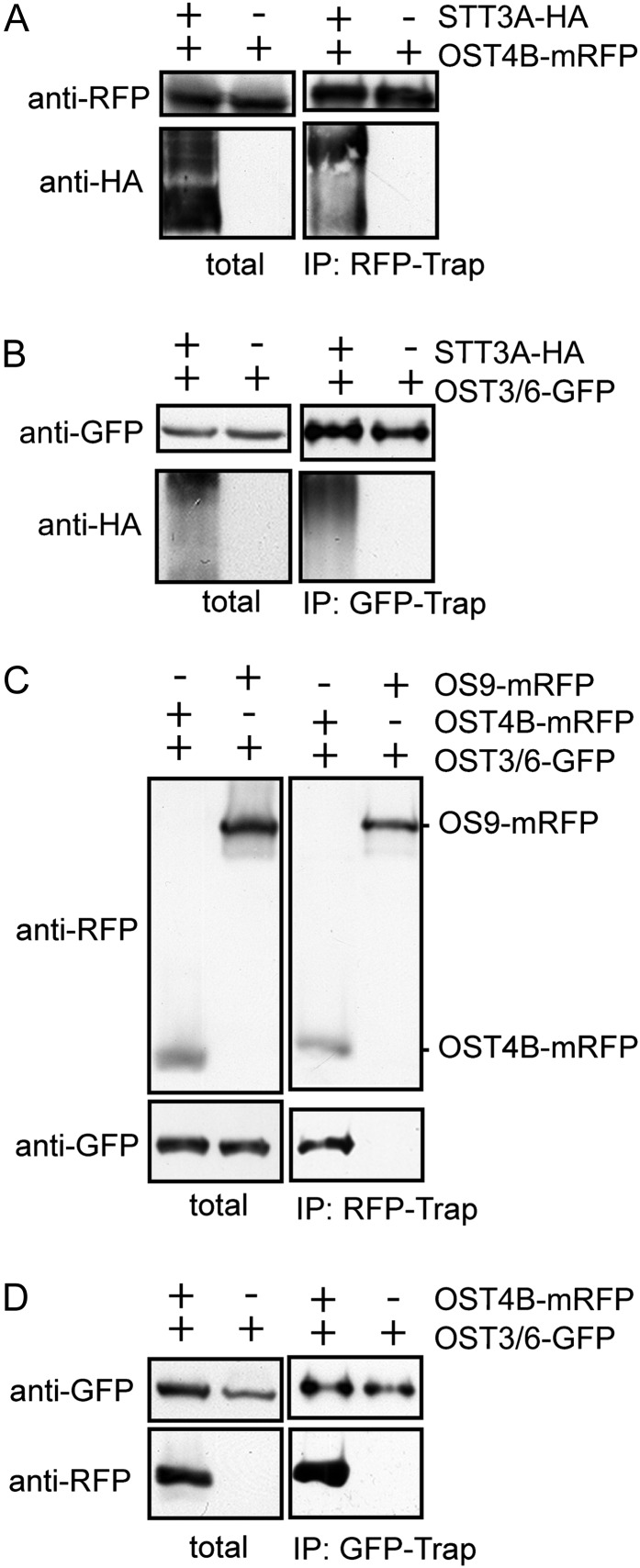
In yeast, it has been proposed that Ost4p acts as a bridge and mediates the interaction of Ost3p with the catalytic subunit Stt3p (Kim et al., 2003). In addition, Ost4p was found to regulate the recruitment of either Ost3p or Ost6p into two distinct yeast OST subcomplexes (Spirig et al., 2005). To further investigate whether OST3/6 is part of such a complex and interacts with other OST subunits, we transiently coexpressed OST3/6-GFP with tagged forms of STT3A (STT3A-HA) and OST4B (OST4B-mRFP) in N. benthamiana leaves and performed coimmunoprecipitation (Co-IP) experiments. In our first Co-IP experiment, we coexpressed STT3A-HA with OST4B-mRFP. Immunoblotting revealed the copurification of STT3A-HA (Fig. 9A). In subsequent experiments, we observed the binding of OST3/6-GFP to STT3A-HA (Fig. 9B) and the interaction of OST3/6-GFP with OST4B-mRFP (Fig. 9, C and D). Notably, the interaction is not compromised when the truncated OST3/6-1-GFP form is coexpressed instead of OST3/6 (Supplemental Fig. S6). In summary, our data suggest that OST3/6 is part of an OST3/6-OST4B-STT3A subcomplex that has been described previously in yeast (Karaoglu et al., 1997; Kim et al., 2003).
Figure 9.
OST3/6 interacts with STT3A and OST4B. All fusion proteins were transiently expressed in N. benthamiana leaves, and purification was done 24 h post infiltration. A, Co-IP of OST4B-mRFP and STT3A-HA or OST4B-mRFP alone. Tagged proteins were purified by RFP-Trap. The presence of OST4B-mRFP in the extract (total) and eluate (IP:RFP-Trap) was detected by anti-RFP antibody; the presence of copurified STT3-HA was monitored using anti-HA antibody. B, Co-IP of OST3/6-GFP with STT3-HA. OST3/6-GFP alone was used as a control. Purification was done with GFP-Trap, and copurified proteins were monitored using anti-HA antibody. C, Co-IP of OST4B-mRFP with OST3/6-GFP. As a control, the ER-resident OS9-mRFP was coexpressed with OST3/6-GFP. mRFP-tagged proteins were purified with RFP-Trap, and input as well as copurified proteins were monitored with anti-RFP and anti-GFP antibodies. D, Co-IP of OST3/6-GFP and OST4B-mRFP. OST3/6-GFP purification served as a control. OST3/6-GFP was purified with GFP-Trap, and copurification of OST4B-mRFP was analyzed by immunoblotting with an anti-RFP antibody.
DISCUSSION
Despite some recent progress in characterization of N-glycan biosynthesis and N-glycan processing, our understanding of the key glycosylation reaction mediated by the OST complex is quite limited in plants. Here, we provide several lines of evidence that OST3/6 is part of the Arabidopsis OST complex and has a supportive role in the glycosylation of a distinct set of glycoproteins, including the pattern recognition receptor EFR and the β-1,4-glucanase KOR1: (1) OST3/6 localizes to the ER; (2) OST3/6 interacts with the OST subunits STT3A and OST4B; (3) OST3/6 mRNA expression correlates well with the expression of STT3A and other putative OST subunits like DAD1, DAD2, ribophorin I, and HAP6 (note that OST4A and OST4B are not present on the Affymetrix chip; therefore, their microarray data are not available for comparison); (4) OST3/6 deficiency results in a severe underglycosylation defect that has also been observed for other mutants with defects in OST subunits (Koiwa et al., 2003; Lerouxel et al., 2005); (5) OST3/6 deficiency impairs EFR biogenesis and function, resulting in insensitivity toward the bacterial PAMP elf18 comparable to STT3A-deficient plants (Nekrasov et al., 2009; Saijo et al., 2009); (6) KOR1 function is affected in ost3/6-1; and (7) ost3/6-1 plants are hypersensitive to tunicamycin and salt/osmotic stress and display an activation of ER stress genes similar to stt3a plants (Koiwa et al., 2003; Kang et al., 2008).
Arabidopsis OST3/6 Is a Putative Ortholog of Yeast Ost3p and Ost6p
Protein glycosylation catalyzed by OST is an essential cotranslational and posttranslational protein modification in all eukaryotes (Yan and Lennarz, 2005a; Kelleher and Gilmore, 2006; Mohorko et al., 2011). In comparison with yeast and mammals, the composition of the plant OST complex and the function of individual subunits in the heteromeric protein complex are not well known. A recent N-glycoproteome comparison between different model organisms including Arabidopsis revealed common characteristics of glycosylation site selection (Zielinska et al., 2012), which highlights the conserved nature of this essential protein modification across species. In plants, a distinct role in the glycosylation of proteins has been shown for STT3A and DGL1 (Koiwa et al., 2003; Lerouxel et al., 2005). Other proteins with homology to mammalian/yeast OST subunits have been identified, but their involvement in the transfer of oligosaccharides to proteins has not been reported (Gallois et al., 1997; Danon et al., 2004; Johnson et al., 2004). The importance of this protein complex for plants is also underlined by the fact that the dgl1 null alleles as well as plants completely deficient in STT3 (stt3a stt3b double knockouts) are not viable.
Besides the strikingly conserved membrane topology, Arabidopsis OST3/6 contains a predicted thioredoxin-like fold domain (IPR012336; amino acids 46–182) that is characteristic for yeast and mammalian OST3/OST6 proteins (Fetrow et al., 2001). However, OST3/6 lacks the CxxC active-site motif in the lumenal domain, which is required for oxidoreductase activity and is assumed to play a role in oxidative folding of glycoproteins (Schulz et al., 2009). Mutation of the CxxC motif in Ost3p and Ost6p reduced the glycosylation efficiency on distinct glycosylation sites of yeast glycoproteins. Notably, the mutated SxxS yeast variants were partially functional, demonstrating that the CxxC active-site motif is only essential for a subset of glycosylation sites. In plants, the CxxC-dependent oxidoreductase function might not be required at all or adjacent residues and sequence motifs might perform that task. Alternatively, additional proteins like specific folding catalysts (e.g. PDIs) could work in close association with the plant OST complex and link oxidative protein folding to substrate-specific glycosylation. The role of the thioredoxin-like fold and the CxxC motif for mammalian members of the OST3/6 family is also widely unknown, and the biological function of the mammalian orthologs is somewhat controversial. Human MagT1/IAP and N33/TUSC3, which display sequence identity to the well-characterized yeast Ost3p/Ost6p proteins and to Arabidopsis OST3/6 are associated with the OST complex (Kelleher et al., 2003) but might also have additional functions, like the transport of magnesium, that seem unrelated to protein glycosylation (Zhou and Clapham, 2009; Mohorko et al., 2011). Interestingly, homozygous mutants in the human gene N33/TUSC3 coding for an OST3/6 family protein caused a defect specifically in neuronal cells leading to mental retardation (Molinari et al., 2008). However, the molecular targets and the involvement of glycosylation have not been elucidated.
All OST3/6 family proteins contain a conserved membrane topology with a predicted signal peptide that is cleaved off, a large lumenal domain, and four transmembrane helices. The function of these conserved protein domains has not been investigated in detail for yeast or mammalian proteins. Our data for the truncated OST3/6-1 variant lacking three of the four helices indicate that, in plants, this C-terminal region is neither involved in subcellular localization and ER retention nor does it influence the overall topology of OST3/6 and its interaction with the OST subunits OST4B and STT3A (Supplemental Fig. S6). However, we cannot rule out that the C-terminal domain including the three transmembrane helices might be required for the interaction of OST3/6 with other constituents of the OST complex, ER chaperones, or other ER-protein complexes, such as interactions with components of the translocon (Shibatani et al., 2005). Whether this is the case remains to be determined by additional studies.
OST Complex and EFR Function
Recently, the importance of the catalytic subunit STT3A for the biogenesis and function of EFR was established (Nekrasov et al., 2009; Saijo et al., 2009; Häweker et al., 2010). Here, we identified another OST subunit that contributes to the efficient N-glycosylation of EFR and its subsequent function in response to pathogens. Strikingly and in accordance with data for stt3a, full OST3/6 function is dispensable for FLS2, another heavily glycosylated LRR-RK with high structural similarity to EFR. Moreover, the wild-type response to chitin suggests that the glycosylated LysM-RK CERK1 is also functional in ost3/6-1. Together, our data underpin previous studies that have identified ER quality-control components specifically required for EFR function (Li et al., 2009; Lu et al., 2009; Nekrasov et al., 2009; Saijo et al., 2009). However, not all OST subunits seem to be involved in EFR biogenesis, as single mutants of DAD1 and DAD2 did not change EFR accumulation and elf18-mediated responses (Häweker et al., 2010). While the specific molecular functions of DAD1/DAD2 are currently unknown, it was reported that Arabidopsis dad1 plants displayed reduced secretion of pathogenesis-related proteins like PR1 and consequently exhibited increased susceptibility to pathogens (Wang et al., 2005). Even though the role of DAD1 in this process is not clear, these data indicate that substrate-specific glycosylation controlled by different subunits of the OST complex plays a crucial role for the pathogen response in plants.
Apart from EFR, we also found that other glycoproteins, like CGL1-GnTI, TGG2, and KOR1, are underglycosylated. While the role of glycosylation for TGG2 is unclear (Liebminger et al., 2012), it has been shown previously that N-glycosylation is required for KOR1 in vitro enzymatic activity (Mølhøj et al., 2001), and genetic evidence suggests that complex N-glycans are also vital for KOR1 in vivo function (Kang et al., 2008). In contrast, the unaltered phenotype of ost3/6-1 bri1-9 or ost3/6-1 bri1-5 mutants strongly suggests that the two mutated variants of the brassinosteroid receptor BRI1-5/BRI1-9, which are ER-retained versions of the LRR-RK BRI1 (Jin et al., 2007; Hong et al., 2008), are not severely underglycosylated in OST3/6-deficient plants. Notably, PDI, an ER-resident glycoprotein, was found underglycosylated in dgl1 and stt3a mutants (Lerouxel et al., 2005) but not in the ost3/6-1 mutant, indicating that the different OST subunits also have nonoverlapping functions. The protein intrinsic determinants of this substrate-specific glycosylation are currently unknown. Despite some recent progress (Häweker et al., 2010; Sun et al., 2012), further work on the glycosylation status, in particular on the occupancy of individual glycosylation sites on structurally related LRR-RKs, is required to understand the mechanisms of site-specific protein N-glycosylation.
What Is the Function of OST3/6-LIKE1?
In yeast, Ost3p and Ost6p mediate the substrate-specific glycosylation of proteins, and the two proteins are incorporated in distinct OST complexes (Knauer and Lehle, 1999b; Schwarz et al., 2005; Spirig et al., 2005; Schulz et al., 2009). In Arabidopsis, the specific role of the two OST3/6 family proteins, OST3/6 and OST3/6-LIKE1, is not entirely clear. Since we were unable to identify loss-of-function alleles for both genes, we cannot exclude a partially overlapping function in some cells/organs or under certain stress conditions. However, microarray data indicate that OST3/6 plays a predominant role in most plant tissues and developmental stages of the plant. Coexpression analysis displays almost no correlation between OST3/6 and OST3/6-LIKE1. In contrast, OST3/6 expression correlates well with that of other OST subunits and other ER-resident proteins. This coexpression is even more pronounced under biotic stress conditions, consistent with the finding that OST3/6 plays an important role in pathogen defense reactions. Interestingly, in genomes of monocots like rice (Oryza sativa), only one OST3/6 family gene can be detected. Stable expression of OST3/6-LIKE1 under the control of its own promoter or under the control of the UBQ10 promoter did not restore the ost3/6-1 phenotype, and the GFP-tagged protein was not detectable on immunoblots. Consequently, it might well be that OST3/6-LIKE1 is very tightly regulated or that the OST3/6-LIKE1 protein is unstable and subjected to rapid degradation. In conclusion, these data argue against a redundant function of the OST3/6-LIKE1 protein in the glycosylation of proteins.
In this study, we identified a mutant with a deficiency in a highly conserved subunit of the Arabidopsis OST complex. Characterization of the ost3/6-1 mutant revealed new insights into the function of the important OST complex that mediates protein glycosylation in all eukaryotes. We do not know the precise role of OST3/6 within the complex, but our data support the idea that OST3/6 assists STT3 in selecting individual glycosylation sites on specific substrate proteins. Further experiments are needed to determine unambiguously the role of OST3/6 in this process and to characterize the sequence-intrinsic properties that make certain glycoproteins like EFR or KOR1 substrates for a distinct OST complex involving STT3A and OST3/6 proteins.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) and Nicotiana benthamiana were grown under long-day conditions (16-h-light/8-h-dark photoperiod) at 22°C and 24°C, respectively, as described previously (Farid et al., 2011). The mutants alg10-1, alg10-1 gntI, alg10 cgl1 (Farid et al., 2011), gntI (Kang et al., 2008), stt3a-2 (Koiwa et al., 2003), cgl1 (von Schaewen et al., 1993), efr-1 (Zipfel et al., 2006), fls2c efr-1 (Nekrasov et al., 2009), rsw2-1 (Lane et al., 2001), bri1-9, and bri1-5 (Noguchi et al., 1999) are all available from previous studies (Zipfel et al., 2006; Liebminger et al., 2009; Farid et al., 2011; Hüttner et al., 2012). Tunicamycin, ABA, and other chemicals for seedling germination and growth assays were purchased from Sigma-Aldrich.
elfin Forward Genetic Screen and Identification of the ost3/6-1 (elfin23-7) Mutant
Screening of elfin mutant plants was performed as described in detail previously (Nekrasov et al., 2009). To identify the ost3/6-1 (elfin23-7) mutation, elfin23-7 plants in the ecotype Columbia-0 (Col-0) background were crossed to wild-type plants of the Landsberg erecta ecotype. Genomic DNA from elfin23-7 seedlings in the segregating F2 population was extracted and used for subsequent PCR analysis. The mutation was mapped to the bottom of chromosome I using a combination of simple sequence length polymorphism markers generated by comparing the Monsanto Arabidopsis Landsberg erecta Sequence Collection (http://www.arabidopsis.org/browse/Cereon/index.jsp) with the Col-0 sequence and markers from the Arabidopsis Mapping Platform (http://amp.genomics.org.cn/).
Seedling Growth Inhibition Assays, Chemical Treatments, and Infection Assays
The flg22 and elf18 peptides were purchased from Peptron, and chitin was purchased from Yaizu Suisankagaku Industry. Seedling growth inhibition, oxidative burst, and infection assays were performed as described previously (Zipfel et al., 2006; Nekrasov et al., 2009). In brief, for the bacterial infection assays, Pseudomonas syringae pv tomato strains were grown overnight on solid L medium, and bacteria were then scraped off and dissolved in 10 mm MgCl2. The suspensions were adjusted to optical density at 600 nm (OD600) = 0.2 and diluted to OD600 = 0.02 (for spray-infection assays) and OD600 = 0.0002 (for induced-resistance assays). For the spray-infection assays, 0.4 µL mL−1 Silwet L-77 was added, and the sprayed plants were kept under high humidity for 3 d. For the induced-resistance assays, bacteria were syringe infiltrated using a needleless syringe into leaves either preinfiltrated with water or 1 µm PAMP for 24 h. Four-week-old short-day-grown plants were used for both assays.
Protein Gel-Blot Analysis
Plant material was ground in liquid nitrogen using a mixer mill, resuspended in 10 µL of phosphate-buffered saline per mg of plant material, and centrifuged at 16,000g for 10 min. An aliquot of the supernatant was mixed with SDS-PAGE loading buffer, denatured at 95°C for 5 min, and subjected to SDS-PAGE under reducing conditions. Protein gel blots were blocked in phosphate-buffered saline containing 0.1% (v/v) Tween 20 and 3% (w/v) bovine serum albumin. The membranes were probed with anti-horseradish peroxidase (HRP; Strasser et al., 2004), anti-PDI (Farid et al., 2011), anti-TGG2 (Ueda et al., 2006; kindly provided by Ikuko Hara-Nishimura), and anti-KOR1 antibodies (generated by immunization of rabbits with a synthetic peptide CSGEEEATGKIDKNT; Genscript). Endo H (New England Biolabs) and PNGase F (New England Biolabs) digestions were done as described in detail recently (Liebminger et al., 2012). The ost3/6-1 efr-1/pEFR:EFR-eGFP-HA lines were generated by crossing ost3/6-1 with efr-1/pEFR:EFR-eGFP-HA (Nekrasov et al., 2009). Immunoblot detection of EFR-GFP was performed as described previously (Nekrasov et al., 2009) with HRP-conjugated anti-GFP antibody (Insight Biotechnology).
RNA Isolation and qRT-PCR
RNA was extracted from 35 mg of 7-d-old seedlings grown on 0.5× MS medium supplemented with 1.5% Suc or from 35 mg of leaves from soil-grown 4-week-old plants using the SV Total RNA Isolation Kit (Promega) according to the manufacturer’s protocol. First-strand complementary DNA (cDNA) was synthesized from 2 µg of extracted RNA in a total volume of 40 µL using the iScript cDNA synthesis kit (Bio-Rad). Primers for quantitative PCR are listed in Supplemental Table S4. cDNA was quantified using iQ SYBR Green supermix (Bio-Rad) with an iCycler (Bio-Rad). The cycle threshold values were calculated using CFX Manager 2.1 software (Bio-Rad), and the relative expression values were determined using UBQ5 gene expression as a reference and the comparative cycle threshold method (Schmittgen and Livak, 2008). PCR was done at least twice, and at least three independent biological experiments were performed. Significant differences between wild-type and mutant genotypes were tested using a two-tailed Student’s t test (Graphpad Software).
Generation of Binary Vectors for OST3/6-GFP, OST3/6-1-GFP, and OST3/6-LIKE1-GFP Expression
The full-length OST3/6 coding sequence including the promoter region was cloned in a two-step process into the binary expression vector p20F (Strasser et al., 2007). First, the OST3/6 promoter and part of the 5′ coding region were amplified using oligonucleotides At1g61790_5F and At1g61790_6R, and the PCR product was HindIII/BamHI digested and cloned into HindIII/BamHI-digested p20F. In the second cloning step, the OST3/6 coding region was amplified with At1g61790_1F and At1g61790_7R, SacI and BamHI digested, and cloned into the SacI/BamHI-digested p20F vector containing the OST3/6 fragment from the first cloning step. The resulting binary expression vector was designated pOST3/6:OST3/6-GFP. The corresponding pOST3/6-LIKE1:OST3/6-LIKE1-GFP construct was generated by PCR amplification of the OST3/6-LIKE1 promoter and coding sequence using At1g11560_3F and At1g11560_4R. The resulting PCR product was HindIII/BamHI digested and cloned into p20F.
For the pUBQ10:OST3/6-GFP construct, the full-length OST3/6 coding sequence was amplified by PCR using At1g61790_3F and At1g61790_7R, BamHI digested, and cloned into BamHI-digested vector p40. Binary expression vector p40 is derived from p20F by replacing the cauliflower mosaic virus 35S promoter with the Arabidopsis ubiquitin10 promoter (Grefen et al., 2010). The vector for expression of the truncated OST3/6-1 fused to GFP was generated by PCR amplification using the primers At1g61790_3F and At1g61790_10R. The PCR product was BamHI digested and cloned into BamHI-digested p20F. The pUBQ10:OST3/6-LIKE1-GFP construct was generated by PCR amplification of the full-length OST3/6-LIKE1 coding sequence using primers At1g11560_11F and At1g11560_4R. The PCR product was XbaI/BamHI digested and cloned into XbaI/BamHI-digested p40. All binary vectors were transformed into Agrobacterium tumefaciens strain UIA143 as described previously (Strasser et al., 2007).
Generation of STT3A and OST4B Constructs
The constructs for the expression of STT3A-mRFP and STT3A-HA were generated by PCR amplification of the STT3A coding region from Arabidopsis cDNA using primers STT3A_1F and STT3A_2R. The PCR product was subcloned, excised from the cloning vector by XbaI/BamHI digestion, and ligated into XbaI/BamHI-digested binary expression vector p31 (mRFP fusion) and vector p60 for the expression of a hemagglutinin (HA)-tagged protein (Hüttner et al., 2012). The binary expression vector for OST4B-mRFP was generated by PCR amplification of the OST4B coding region from Arabidopsis cDNA using primers At5g02502_3F and At5g02502_4R. The PCR product was XbaI/BamHI digested and cloned into p31.
Confocal Microscopy
Transient expression in N. benthamiana was done by infiltration of leaves as described previously (Schoberer et al., 2009). For coexpression experiments, resuspended agrobacteria were diluted to an OD600 of 0.05 for OST3/6-GFP, an OD600 of 0.05 to 0.10 for STT3A-mRFP, an OD600 of 0.05 for OST4B-mRFP, an OD600 of 0.05 for the ER marker protein GnTI-CaaaTS-mRFP, and an OD600 of 0.03 for OST3/6-1-GFP. Sampling and imaging of fluorescent proteins were performed 2 d after infiltration using a Leica TCS SP5 confocal microscope as described in detail recently (Schoberer et al., 2009). Postacquisition image processing was performed in Adobe Photoshop CS.
Protein Co-IP
Transient expression of fluorescent protein fusions was performed by infiltration of N. benthamiana leaves as described previously (Schoberer et al., 2009). For coexpression experiments, resuspended agrobacteria were diluted to an OD600 of 0.2 for OST3/6-1-GFP, OST4B-mRFP, and OS9-mRFP (Hüttner et al., 2012), OD600 of 0.3 for OST3/6-GFP, and OD600 of 0.4 for STT3A-HA. One gram (fresh weight) of infiltrated leaf tissue (for purification of STT3-HA, 2 g of infiltrated leaves was used due to lower expression levels of STT3-HA) was harvested, frozen in liquid nitrogen, ground to a fine powder using a mixer mill, and resuspended in 3 mL of radioimmunoprecipitation assay buffer (Sigma) supplemented with 1% (v/v) protease inhibitor cocktail (Sigma). The samples were placed on ice for 30 min with mixing every 10 min. After incubation, the samples were centrifuged at 3,500g for 7.5 min at 4°C, and the supernatant was centrifuged again for 7.5 min at 7,800g at 4°C and finally at 11,600g for 7.5 min at 4°C. The resulting pellet was discarded, and the cleared supernatant (referred to as “total”) was diluted with 2× dilution buffer containing 50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 0.5 mm EDTA, pH 8.0, and 1% (v/v) protease inhibitor cocktail. To equilibrate the GFP-Trap-A or RFP-Trap-A beads (Chromotek), 25 µL of slurry was resuspended in ice-cold radioimmunoprecipitation assay buffer and spun down at 600g for 1 min at 4°C. The supernatant was discarded, and the washing step was repeated twice. The beads were resuspended with 500 µL of ice-cold dilution buffer and transferred to the protein extracts followed by an incubation step with end-over-end mixing for 1 h at 4°C. The samples were centrifuged at 400g for 3 min at 4°C, and the beads were washed three times with dilution buffer. In a fourth wash step, the salt concentration in the dilution buffer was increased to 250 mm NaCl. Finally, the beads were resuspended in 90 µL of 2× SDS-PAGE loading buffer, loaded onto Micro Bio-Spin chromatography columns (Bio-Rad), and boiled for 5 min at 95°C. The eluate was again loaded onto Micro Bio-Spin chromatography columns and boiled for 5 min at 95°C. Eluted proteins (referred to as “IP:RFP-Trap” or “IP:GFP-Trap”) were subjected to SDS-PAGE and immunoblot analysis. For the detection of GFP fusion proteins anti-GFP (MACS Miltenyi Biotec), for mRFP-tagged fusion proteins anti-RFP (5F8; Chromotek), and for HA-tagged proteins anti-HA (Roche) antibodies were used.
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: At1g61790 (OST3/6), At1g11560 (OST3/6-LIKE1), At5g19690 (STT3A), At5g02502 (OST4B), At5g20480 (EFR), At5g49720 (KOR1), and At5g02410 (ALG10).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Network diagram showing the analysis of genes coexpressed with OST3/6 and OST3/6-LIKE1.
Supplemental Figure S2. Arabidopsis heat map (obtained from Genevestigator) showing the coexpression of OST3/6 with STT3A and DGL1.
Supplemental Figure S3. Matrix-assisted laser-desorption ionization time-of-flight mass spectrometry spectra of total N-glycans extracted from leaves of wild-type (Col-0) and ost3/6-1 plants.
Supplemental Figure S4. Phenotypic analysis of ost3/6-1 under salt stress and ABA treatment.
Supplemental Figure S5. ost3/6-1 underglycosylation of TGG2 is restored by the expression of pUBQ10:OST3/6-GFP but not by the expression of pUBQ10:OST3/6-LIKE1-GFP.
Supplemental Figure S6. The C-terminal region including three putative transmembrane helices from OST3/6 is dispensable for interaction with OST subunits STT3A and OST4B.
Supplemental Table S1. Overview of Arabidopsis, yeast (Saccharomyces cerevisiae), and human OST complex subunits.
Supplemental Table S2. Comparison of OST3/6 protein domain organization.
Supplemental Table S3. List of genes coexpressed with OST3/6.
Supplemental Table S4. List of all primer sequences used in this study.
Acknowledgments
We thank Ulrike Vavra for invaluable technical assistance, Karin Polacsek and Friedrich Altmann for N-glycan analysis, and Josef Glössl (all from the University of Natural Resources and Life Sciences, Vienna) for helpful discussions. Benjamin Schwessinger, Milena Roux, Martine Batoux, Jing Li, and Karen Morehouse (all from the Sainsbury Laboratory, Norwich) are acknowledged for their invaluable help in the different stages of the elfin genetic screen. We also thank Ikuko Hara-Nishimura (Department of Botany, Graduate School of Science, Kyoto University) for the kind gift of anti-TGG2 antibodies.
Glossary
- ER
endoplasmic reticulum
- OST
oligosaccharyltransferase
- PAMP
pathogen-associated molecular pattern
- LRR-RK
Leu-rich repeat receptor kinase
- Pto
Pseudomonas syringae pv tomato
- PDI
protein disulfide isomerase
- UPR
unfolded protein response
- qRT
quantitative reverse transcription
- MS
Murashige and Skoog
- ABA
abscisic acid
- Endo H
endoglycosidase H
- PNGase F
peptide:N-glycosidase F
- HA
hemagglutinin
- Co-IP
coimmunoprecipitation
- Col-0
ecotype Columbia-0
- OD600
optical density at 600 nm
- HRP
horseradish peroxidase
- cDNA
complementary DNA
- mRFP
monomeric red fluorescent protein
References
- Danon A, Rotari VI, Gordon A, Mailhac N, Gallois P. (2004) Ultraviolet-C overexposure induces programmed cell death in Arabidopsis, which is mediated by caspase-like activities and which can be suppressed by caspase inhibitors, p35 and Defender against Apoptotic Death. J Biol Chem 279: 779–787 [DOI] [PubMed] [Google Scholar]
- Farid A, Pabst M, Schoberer J, Altmann F, Glössl J, Strasser R. (2011) Arabidopsis thaliana alpha1,2-glucosyltransferase (ALG10) is required for efficient N-glycosylation and leaf growth. Plant J 68: 314–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetrow JS, Siew N, Di Gennaro JA, Martinez-Yamout M, Dyson HJ, Skolnick J. (2001) Genomic-scale comparison of sequence- and structure-based methods of function prediction: does structure provide additional insight? Protein Sci 10: 1005–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Kaulfürst-Soboll H, Rips S, Koiwa H, von Schaewen A. (2008) Comparative analyses of Arabidopsis complex glycan1 mutants and genetic interaction with staurosporin and temperature sensitive3a. Plant Physiol 148: 1354–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallois P, Makishima T, Hecht V, Despres B, Laudié M, Nishimoto T, Cooke R. (1997) An Arabidopsis thaliana cDNA complementing a hamster apoptosis suppressor mutant. Plant J 11: 1325–1331 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L, Boller T. (2000) FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Grefen C, Donald N, Hashimoto K, Kudla J, Schumacher K, Blatt MR. (2010) A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J 64: 355–365 [DOI] [PubMed] [Google Scholar]
- Häweker H, Rips S, Koiwa H, Salomon S, Saijo Y, Chinchilla D, Robatzek S, von Schaewen A. (2010) Pattern recognition receptors require N-glycosylation to mediate plant immunity. J Biol Chem 285: 4629–4636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Jin H, Tzfira T, Li J. (2008) Multiple mechanism-mediated retention of a defective brassinosteroid receptor in the endoplasmic reticulum of Arabidopsis. Plant Cell 20: 3418–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Kajiura H, Su W, Jin H, Kimura A, Fujiyama K, Li J. (2012) Evolutionarily conserved glycan signal to degrade aberrant brassinosteroid receptors in Arabidopsis. Proc Natl Acad Sci USA 109: 11437–11442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttner S, Veit C, Schoberer J, Grass J, Strasser R. (2012) Unraveling the function of Arabidopsis thaliana OS9 in the endoplasmic reticulum-associated degradation of glycoproteins. Plant Mol Biol 79: 21–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Yan Z, Nam KH, Li J. (2007) Allele-specific suppression of a defective brassinosteroid receptor reveals a physiological role of UGGT in ER quality control. Mol Cell 26: 821–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MA, von Besser K, Zhou Q, Smith E, Aux G, Patton D, Levin JZ, Preuss D. (2004) Arabidopsis hapless mutations define essential gametophytic functions. Genetics 168: 971–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JS, Frank J, Kang CH, Kajiura H, Vikram M, Ueda A, Kim S, Bahk JD, Triplett B, Fujiyama K, et al. (2008) Salt tolerance of Arabidopsis thaliana requires maturation of N-glycosylated proteins in the Golgi apparatus. Proc Natl Acad Sci USA 105: 5933–5938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaoglu D, Kelleher DJ, Gilmore R. (1995) Functional characterization of Ost3p: loss of the 34-kD subunit of the Saccharomyces cerevisiae oligosaccharyltransferase results in biased underglycosylation of acceptor substrates. J Cell Biol 130: 567–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaoglu D, Kelleher DJ, Gilmore R. (1997) The highly conserved Stt3 protein is a subunit of the yeast oligosaccharyltransferase and forms a subcomplex with Ost3p and Ost4p. J Biol Chem 272: 32513–32520 [DOI] [PubMed] [Google Scholar]
- Kelleher DJ, Gilmore R. (2006) An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology 16: 47R–62R [DOI] [PubMed] [Google Scholar]
- Kelleher DJ, Karaoglu D, Mandon EC, Gilmore R. (2003) Oligosaccharyltransferase isoforms that contain different catalytic STT3 subunits have distinct enzymatic properties. Mol Cell 12: 101–111 [DOI] [PubMed] [Google Scholar]
- Kim H, Yan Q, Von Heijne G, Caputo GA, Lennarz WJ. (2003) Determination of the membrane topology of Ost4p and its subunit interactions in the oligosaccharyltransferase complex in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 100: 7460–7464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauer R, Lehle L. (1999a) The oligosaccharyltransferase complex from yeast. Biochim Biophys Acta 1426: 259–273 [DOI] [PubMed] [Google Scholar]
- Knauer R, Lehle L. (1999b) The oligosaccharyltransferase complex from Saccharomyces cerevisiae: isolation of the OST6 gene, its synthetic interaction with OST3, and analysis of the native complex. J Biol Chem 274: 17249–17256 [DOI] [PubMed] [Google Scholar]
- Koiwa H, Li F, McCully MG, Mendoza I, Koizumi N, Manabe Y, Nakagawa Y, Zhu J, Rus A, Pardo JM, et al. (2003) The STT3a subunit isoform of the Arabidopsis oligosaccharyltransferase controls adaptive responses to salt/osmotic stress. Plant Cell 15: 2273–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DR, Wiedemeier A, Peng L, Höfte H, Vernhettes S, Desprez T, Hocart CH, Birch RJ, Baskin TI, Burn JE, et al. (2001) Temperature-sensitive alleles of RSW2 link the KORRIGAN endo-1,4-β-glucanase to cellulose synthesis and cytokinesis in Arabidopsis. Plant Physiol 126: 278–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerouxel O, Mouille G, Andème-Onzighi C, Bruyant MP, Séveno M, Loutelier-Bourhis C, Driouich A, Höfte H, Lerouge P. (2005) Mutants in DEFECTIVE GLYCOSYLATION, an Arabidopsis homolog of an oligosaccharyltransferase complex subunit, show protein underglycosylation and defects in cell differentiation and growth. Plant J 42: 455–468 [DOI] [PubMed] [Google Scholar]
- Li J, Chory J. (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938 [DOI] [PubMed] [Google Scholar]
- Li J, Zhao-Hui C, Batoux M, Nekrasov V, Roux M, Chinchilla D, Zipfel C, Jones JD. (2009) Specific ER quality control components required for biogenesis of the plant innate immune receptor EFR. Proc Natl Acad Sci USA 106: 15973–15978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebminger E, Grass J, Jez J, Neumann L, Altmann F, Strasser R. (2012) Myrosinases TGG1 and TGG2 from Arabidopsis thaliana contain exclusively oligomannosidic N-glycans. Phytochemistry 84: 24–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebminger E, Hüttner S, Vavra U, Fischl R, Schoberer J, Grass J, Blaukopf C, Seifert GJ, Altmann F, Mach L, et al. (2009) Class I α-mannosidases are required for N-glycan processing and root development in Arabidopsis thaliana. Plant Cell 21: 3850–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Tintor N, Mentzel T, Kombrink E, Boller T, Robatzek S, Schulze-Lefert P, Saijo Y. (2009) Uncoupling of sustained MAMP receptor signaling from early outputs in an Arabidopsis endoplasmic reticulum glucosidase II allele. Proc Natl Acad Sci USA 106: 22522–22527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez IM, Chrispeels MJ. (2003) Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell 15: 561–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, Narusaka Y, Kawakami N, Kaku H, Shibuya N. (2007) CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci USA 104: 19613–19618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohorko E, Glockshuber R, Aebi M. (2011) Oligosaccharyltransferase: the central enzyme of N-linked protein glycosylation. J Inherit Metab Dis 34: 869–878 [DOI] [PubMed] [Google Scholar]
- Mølhøj M, Ulvskov P, Dal Degan F. (2001) Characterization of a functional soluble form of a Brassica napus membrane-anchored endo-1,4-β-glucanase heterologously expressed in Pichia pastoris. Plant Physiol 127: 674–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari F, Foulquier F, Tarpey PS, Morelle W, Boissel S, Teague J, Edkins S, Futreal PA, Stratton MR, Turner G, et al. (2008) Oligosaccharyltransferase-subunit mutations in nonsyndromic mental retardation. Am J Hum Genet 82: 1150–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov V, Li J, Batoux M, Roux M, Chu ZH, Lacombe S, Rougon A, Bittel P, Kiss-Papp M, Chinchilla D, et al. (2009) Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. EMBO J 28: 3428–3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol F, His I, Jauneau A, Vernhettes S, Canut H, Höfte H. (1998) A plasma membrane-bound putative endo-1,4-beta-D-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J 17: 5563–5576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Yoshida S, Yuan H, Feldmann KA, Tax FE. (1999) Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol 121: 743–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T, Kinoshita K, Nakai K, Shibaoka M, Hayashi S, Saeki M, Shibata D, Saito K, Ohta H. (2007) ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res 35: D863–D869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattison RJ, Amtmann A. (2009) N-Glycan production in the endoplasmic reticulum of plants. Trends Plant Sci 14: 92–99 [DOI] [PubMed] [Google Scholar]
- Roboti P, High S. (2012) The oligosaccharyltransferase subunits OST48, DAD1 and KCP2 function as ubiquitous and selective modulators of mammalian N-glycosylation. J Cell Sci 125: 3474–3484 [DOI] [PubMed] [Google Scholar]
- Ruiz-Canada C, Kelleher DJ, Gilmore R. (2009) Cotranslational and posttranslational N-glycosylation of polypeptides by distinct mammalian OST isoforms. Cell 136: 272–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Tintor N, Lu X, Rauf P, Pajerowska-Mukhtar K, Häweker H, Dong X, Robatzek S, Schulze-Lefert P. (2009) Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J 28: 3439–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Schoberer J, Vavra U, Stadlmann J, Hawes C, Mach L, Steinkellner H, Strasser R. (2009) Arginine/lysine residues in the cytoplasmic tail promote ER export of plant glycosylation enzymes. Traffic 10: 101–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz BL, Aebi M. (2009) Analysis of glycosylation site occupancy reveals a role for Ost3p and Ost6p in site-specific N-glycosylation efficiency. Mol Cell Proteomics 8: 357–364 [DOI] [PubMed] [Google Scholar]
- Schulz BL, Stirnimann CU, Grimshaw JP, Brozzo MS, Fritsch F, Mohorko E, Capitani G, Glockshuber R, Grütter MG, Aebi M. (2009) Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency. Proc Natl Acad Sci USA 106: 11061–11066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz M, Knauer R, Lehle L. (2005) Yeast oligosaccharyltransferase consists of two functionally distinct sub-complexes, specified by either the Ost3p or Ost6p subunit. FEBS Lett 579: 6564–6568 [DOI] [PubMed] [Google Scholar]
- Shibatani T, David LL, McCormack AL, Frueh K, Skach WR. (2005) Proteomic analysis of mammalian oligosaccharyltransferase reveals multiple subcomplexes that contain Sec61, TRAP, and two potential new subunits. Biochemistry 44: 5982–5992 [DOI] [PubMed] [Google Scholar]
- Spirig U, Bodmer D, Wacker M, Burda P, Aebi M. (2005) The 3.4-kDa Ost4 protein is required for the assembly of two distinct oligosaccharyltransferase complexes in yeast. Glycobiology 15: 1396–1406 [DOI] [PubMed] [Google Scholar]
- Strasser R, Altmann F, Mach L, Glössl J, Steinkellner H. (2004) Generation of Arabidopsis thaliana plants with complex N-glycans lacking beta1,2-linked xylose and core alpha1,3-linked fucose. FEBS Lett 561: 132–136 [DOI] [PubMed] [Google Scholar]
- Strasser R, Bondili JS, Schoberer J, Svoboda B, Liebminger E, Glössl J, Altmann F, Steinkellner H, Mach L. (2007) Enzymatic properties and subcellular localization of Arabidopsis β-N-acetylhexosaminidases. Plant Physiol 145: 5–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Cao Y, Jansen Labby K, Bittel P, Boller T, Bent AF. (2012) Probing the Arabidopsis flagellin receptor: FLS2-FLS2 association and the contributions of specific domains to signaling function. Plant Cell 24: 1096–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H, Nishiyama C, Shimada T, Koumoto Y, Hayashi Y, Kondo M, Takahashi T, Ohtomo I, Nishimura M, Hara-Nishimura I. (2006) AtVAM3 is required for normal specification of idioblasts, myrosin cells. Plant Cell Physiol 47: 164–175 [DOI] [PubMed] [Google Scholar]
- von Numers N, Survila M, Aalto M, Batoux M, Heino P, Palva ET, Li J. (2010) Requirement of a homolog of glucosidase II beta-subunit for EFR-mediated defense signaling in Arabidopsis thaliana. Mol Plant 3: 740–750 [DOI] [PubMed] [Google Scholar]
- von Schaewen A, Sturm A, O’Neill J, Chrispeels MJ. (1993) Isolation of a mutant Arabidopsis plant that lacks N-acetyl glucosaminyl transferase I and is unable to synthesize Golgi-modified complex N-linked glycans. Plant Physiol 102: 1109–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Zhang XC, Neece D, Ramonell KM, Clough S, Kim SY, Stacey MG, Stacey G. (2008) A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20: 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Weaver ND, Kesarwani M, Dong X. (2005) Induction of protein secretory pathway is required for systemic acquired resistance. Science 308: 1036–1040 [DOI] [PubMed] [Google Scholar]
- Yan A, Lennarz WJ. (2005a) Unraveling the mechanism of protein N-glycosylation. J Biol Chem 280: 3121–3124 [DOI] [PubMed] [Google Scholar]
- Yan A, Lennarz WJ. (2005b) Two oligosaccharyl transferase complexes exist in yeast and associate with two different translocons. Glycobiology 15: 1407–1415 [DOI] [PubMed] [Google Scholar]
- Yan Q, Lennarz WJ. (2002) Studies on the function of oligosaccharyl transferase subunits: Stt3p is directly involved in the glycosylation process. J Biol Chem 277: 47692–47700 [DOI] [PubMed] [Google Scholar]
- Zhang M, Henquet M, Chen Z, Zhang H, Zhang Y, Ren X, van der Krol S, Gonneau M, Bosch D, Gong Z. (2009) LEW3, encoding a putative alpha-1,2-mannosyltransferase (ALG11) in N-linked glycoprotein, plays vital roles in cell-wall biosynthesis and the abiotic stress response in Arabidopsis thaliana. Plant J 60: 983–999 [DOI] [PubMed] [Google Scholar]
- Zhou H, Clapham DE. (2009) Mammalian MagT1 and TUSC3 are required for cellular magnesium uptake and vertebrate embryonic development. Proc Natl Acad Sci USA 106: 15750–15755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinska DF, Gnad F, Schropp K, Wiśniewski JR, Mann M. (2012) Mapping N-glycosylation sites across seven evolutionarily distant species reveals a divergent substrate proteome despite a common core machinery. Mol Cell 46: 542–548 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G. (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]