WRKY transcription factors modulate the expression of nuclear genes encoding mitochondrial and chloroplast proteins via direct promoter binding and coordinate common stress responses.
Abstract
The expression of a variety of nuclear genes encoding mitochondrial proteins is known to adapt to changes in environmental conditions and retrograde signaling. The presence of putative WRKY transcription factor binding sites (W-boxes) in the promoters of many of these genes prompted a screen of 72 annotated WRKY factors in the Arabidopsis (Arabidopsis thaliana) genome for regulators of transcripts encoding mitochondrial proteins. A large-scale yeast one-hybrid screen was used to identify WRKY factors that bind the promoters of marker genes (Alternative oxidase1a, NADH dehydrogenaseB2, and the AAA ATPase Ubiquinol-cytochrome c reductase synthesis1), and interactions were confirmed using electromobility shift assays. Transgenic overexpression and knockout lines for 12 binding WRKY factors were generated and tested for altered expression of the marker genes during normal and stress conditions. AtWRKY40 was found to be a repressor of antimycin A-induced mitochondrial retrograde expression and high-light-induced signaling, while AtWRKY63 was identified as an activator. Genome-wide expression analysis following high-light stress in transgenic lines with perturbed AtWRKY40 and AtWRKY63 function revealed that these factors are involved in regulating stress-responsive genes encoding mitochondrial and chloroplast proteins but have little effect on more constitutively expressed genes encoding organellar proteins. Furthermore, it appears that AtWRKY40 and AtWRKY63 are particularly involved in regulating the expression of genes responding commonly to both mitochondrial and chloroplast dysfunction but not of genes responding to either mitochondrial or chloroplast perturbation. In conclusion, this study establishes the role of WRKY transcription factors in the coordination of stress-responsive genes encoding mitochondrial and chloroplast proteins.
The expression of many nuclear genes encoding mitochondrial and chloroplast proteins is highly adaptive to changes in the environment. Furthermore, it is suspected that interplay exists between the regulatory mechanisms of both organelles. Nevertheless, very few transcription factors have been isolated that regulate transcripts of nuclear genes encoding organellar proteins (herein, we refer to genes for organellar proteins that are encoded in the nucleus, unless otherwise stated). Examples of transcription factors that regulate genes encoding mitochondrial proteins are the B3 domain transcription factors Abscisic acid insensitive3 (ABI3), Fusca3, basic leucine zipper53 (bZIP53), which are important for regulating the expression of iron-sulfur proteins of complex II (Roschzttardtz et al., 2009). Also, a class of TCP transcription factors were found to regulate the expression of genes encoding mitochondrial proteins during the diurnal cycle (Giraud et al., 2010), while the Circadian Clock Associated1 transcription factor was found to regulate the diurnal control of chloroplastic genes such as thioredoxins (Barajas-López et al., 2011). Also for chloroplasts, the GATA-type transcription factors GNC and CGA1 were shown to modulate the expression of genes encoding chloroplast proteins such as Genomes Uncoupled4 (GUN4) and HEMA1 (a glutamyl-tRNA reductase involved in chlorophyll synthesis; Hudson et al., 2011). One example of a transcription factor that is known to regulate genes encoding both mitochondrial and chloroplast proteins is Abscisic Acid Insensitive4 (ABI4; Koussevitzky et al., 2007; Giraud et al., 2009). ABI4 appears to be on the interface of external signals (such as abscisic acid [ABA] and sugars) and retrograde signals coming from chloroplasts and mitochondria.
A previous study identified a set of nuclear genes encoding mitochondrial proteins that are highly responsive to a wide range of stresses (Van Aken et al., 2009). These genes included alternative respiration pathway components such as Alternative oxidase1a (AOX1a) and NADH dehydrogenaseB2 (NDB2), AAA ATPases (Ubiquinol-cytochrome c reductase synthesis1 [BCS1]), mitochondrial substrate carriers, heat shock proteins, and a variety of other functions. Past studies showed that a variety of signals can lead to the activation of these genes (Ho et al., 2008; Giraud et al., 2012). This includes broad-spectrum redox triggers such as hydrogen peroxide (H2O2) but also more specific signals inhibiting mitochondrial function such as antimycin A (AA) and rotenone (Dojcinovic et al., 2005; Giraud et al., 2009). Also, pathogen signals including hormones such as salicylic acid and abiotic stress signals such as ABA have been found to regulate genes encoding mitochondrial proteins (Van Aken et al., 2007; Ho et al., 2008). In silico analysis of the promoter regions of stress-responsive genes encoding mitochondrial proteins revealed that most contain a TTGAC motif, the core binding site for WRKY-type transcription factors (W-box; Rushton et al., 1995; Van Aken et al., 2009). This observation suggests that WRKY transcription factors play a key role in the expression of genes encoding mitochondrial proteins.
WRKY transcription factors are a mostly plant-specific class of proteins that all contain at least one highly conserved Trp-Arg-Lys-Thr (WRKY) or related amino acid sequence and an additional zinc finger motif. Many WRKY factors have been reported to be involved in regulating stress responses (Eulgem and Somssich, 2007; Rushton et al., 2010) and in Arabidopsis (Arabidopsis thaliana), 72 WRKY proteins have been annotated, varying in length from 109 (AtWRKY43) to potentially 1,895 (AtWRKY19) amino acids. This diversity suggests a wide variety of upstream signaling factors and downstream target genes in plants, indicating a highly complex regulatory network with likely redundancies that controls gene expression.
WRKY transcription factors were found to be tightly correlated with biotic stresses (Rushton et al., 1996) and have been shown to be important regulators of microbe- or pathogen-associated molecular pattern-triggered immunity and effector-triggered immunity (Eulgem and Somssich, 2007; Rushton et al., 2010). In barley (Hordeum vulgare), HvWRKY1 and HvWRKY2 are involved in powdery mildew resistance, whereas NaWRKY3 and NaWRKY6 coordinate the response to herbivory in the native tobacco species Nicotiana attenuata (Shen et al., 2007; Skibbe et al., 2008). In Arabidopsis, AtWRKY23 is involved in nematode infection (Grunewald et al., 2008), and a complex interaction between AtWRKY18, AtWRKY40, and AtWRKY60 in resistance to a variety of pathogens such as Botrytis cinerea (Xu et al., 2006) and Golovinomyces orontii (Shen et al., 2007) has been described extensively. Therefore, it is believed that WRKY transcription factors are closely involved in the signaling networks regulated by the phytohormones salicylic acid and jasmonic acid, often in opposite directions (Xu et al., 2006). Several reports have appeared linking WRKY transcription factor function to abiotic stresses. For example, overexpression of OsWRKY11 led to enhanced heat and drought tolerance, while in Arabidopsis, AtWRKY25, AtWRKY26, and AtWRKY33 interact functionally in the regulation of salt and heat stress tolerance (Jiang and Deyholos, 2009; Li et al., 2011).
Recently, the involvement of WRKY factors in response to ABA has become apparent. A complex interaction of AtWRKY40, AtWRKY18, and AtWRKY60 with the C-terminal domain of the chloroplastic ABA receptor (GUN5) was reported (Shang et al., 2010). In this model, AtWRKY40 is bound as a repressor to the promoters of ABA signaling factors, and in the presence of ABA, it relocalizes to the cytosolic tail of the ABA receptor, relieving the repression of ABA signaling. This places AtWRKY40 upstream of several ABA signaling transcription factors such as ABI4, ABI5, DREB1/2A, and MYB2. Another study identified AtWRKY63 in a screen for ABA hypersensitivity (Ren et al., 2010). AtWRKY63 would operate downstream of AtWRKY40 and ABI5 and stimulate the expression of Abscisic acid response-element binding factor2 (ABF2) and RD29A. Therefore, it is clear that WRKY transcription factors play an important role in hormone-mediated responses triggered by environmental changes.
As putative WRKY binding sites are overrepresented in the promoters of genes encoding stress-responsive mitochondrial proteins and WRKY transcription factors are known to contribute to responses that trigger the expression of these genes, WRKY factors are plausible candidates to regulate the expression of genes encoding mitochondrial proteins. Therefore, three widely stress-responsive genes were selected as a starting point for identifying which WRKY transcription factors are specifically involved in regulating mitochondrial stress responses: AOX1a, NDB2, and BCS1 (Van Aken et al., 2009). Our results indicate that W-boxes play a clear role in the expression of these genes; furthermore, WRKY transcription factors were identified that directly bind their promoters. Transgenic approaches show that AtWRKY40, AtWRKY57, and AtWRKY63 are regulators of stress responses of mitochondrially targeted proteins.
RESULTS
W-Boxes in the Promoters of AOX1a, NDB2, and BCS1 Are Required for Normal Promoter Activity
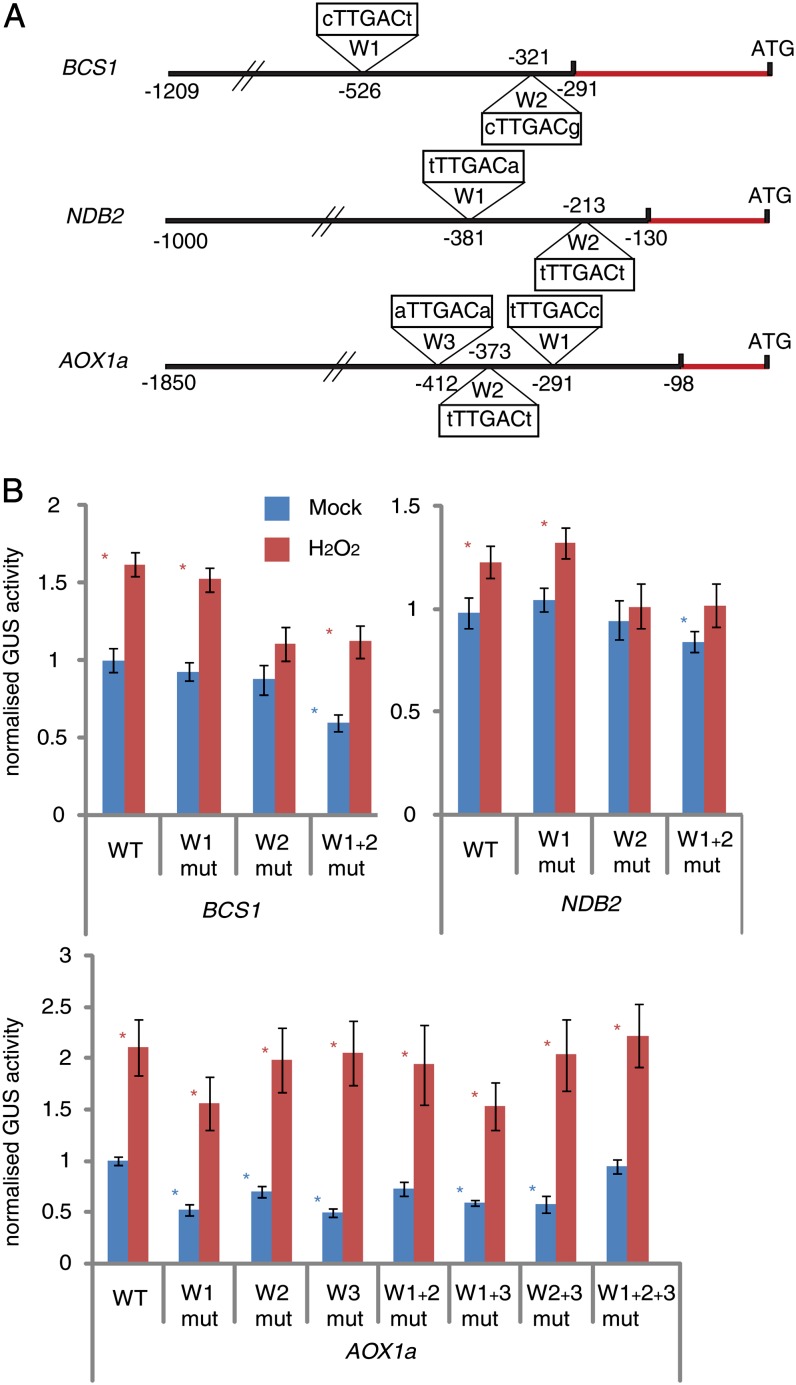
The function of the putative TTGAC W-boxes in the promoters of BCS1, NDB2, and AOX1a was tested to establish if the putative W-boxes play a role in regulating transcriptional activity. The 2-kb promoter of AOX1a contains three TTGAC sequences, and the 1.5-kb promoters of NDB2 and BCS1 promoters contain two TTGAC motifs (Fig. 1A). An additional putative W-box was present 3.3 kb upstream of the BCS1 start codon, but given the long distance, this was not further analyzed experimentally. To assess promoter activity, the 1.5- to 2-kb promoter sequences were cloned in front of a GUS reporter gene using the pLUS reporter vector (Ho et al., 2008). The putative W-boxes were mutated using site-directed mutagenesis to repeats of A (Supplemental Table S2) either singly or in different combinations. Cell cultures were mock treated to assess basal promoter activity and treated with H2O2 to assess the general stress responsiveness of the promoters. GUS activity in mock- and H2O2-treated cells was measured and normalized for transformation efficiency using a luciferase reporter gene (Fig. 1B). The levels of promoter activity induction by H2O2 were comparable to previous reports using this system (Ho et al., 2008). The observation that increases in promoter activity after H2O2 treatment are lower than the maximal fold change induction of the respective transcript levels (Ho et al., 2008) may be due to the fact that the transient transformation is in itself a stress, and thus even “mock” treatments will have an elevated promoter activity. The results show that in the AOX1a promoter, mutation of each W-box significantly reduced promoter activity under mock conditions by up to 50%, and mutation of W-box W1 also reduced inducibility by H2O2. This is in agreement with a previous study showing that deletion of an 18-bp region overlapping with AOX1a W1 resulted in strongly reduced promoter induction by AA and monofluoroacetate (Dojcinovic et al., 2005). In this study, specific mutation of the AOX1a W1 TTGAC sequence also resulted in a more than 2-fold decrease in promoter induction by monofluoroacetate and a mild decrease in AA induction (Dojcinovic et al., 2005). AOX1a W1 was also identified as a putative regulatory element in another study but was not functionally characterized (Ho et al., 2008). In the BCS1 promoter, mutation of W2 abolished induction by H2O2, and mutation of both W1 and W2 almost halved normal promoter activity compared with the wild-type promoter. H2O2 induction could be observed after mutation of both W1 and W2, but the resulting induced promoter activity was significantly lower than in the wild-type promoter and similar to that after deletion of BCS1 W2. Similarly, in the NDB2 promoter, mutation of W1 boxes resulted in no observable changes and mutation of W2 abolished induction by H2O2. Simultaneous deletion of W1 and W2 resulted in a significant reduction of basal promoter activity and no induction by H2O2. Mutations in multiple W-boxes resulted in reduced or unchanged promoter activity compared with the wild type, indicating a complex interplay of these elements. In conclusion, these experiments indicate a complex interplay between the putative W-boxes in the promoters of stress-responsive mitochondrial genes, pointing at both positive and negative roles in regulating promoter activity.
Figure 1.
Locations and activities of W-boxes in the promoters of stress-responsive genes encoding mitochondrial protein. A, Locations of W-boxes with the TTGAC core sequence in the promoters of three widely stress-responsive genes encoding mitochondrial proteins. The red line indicates the 5′ UTR. Numbers indicate base positions upstream of the start codon. B, Relative activities of the promoter sequences fused to the GUS reporter gene compared with the promoter sequence with single or multiple W-boxes mutated. The mock-treated wild-type (WT) promoter activity was normalized to 1. Blue asterisks indicate significant differences (P < 0.05) compared with the wild-type promoter for the mock treatments, and red asterisks indicate significant differences (P < 0.05; n = 9) of the H2O2 treatments compared with the mock treatment of the same construct. Error bars indicate se.
WRKY Transcription Factors Bind the Promoters of Mitochondrial Stress-Responsive Genes
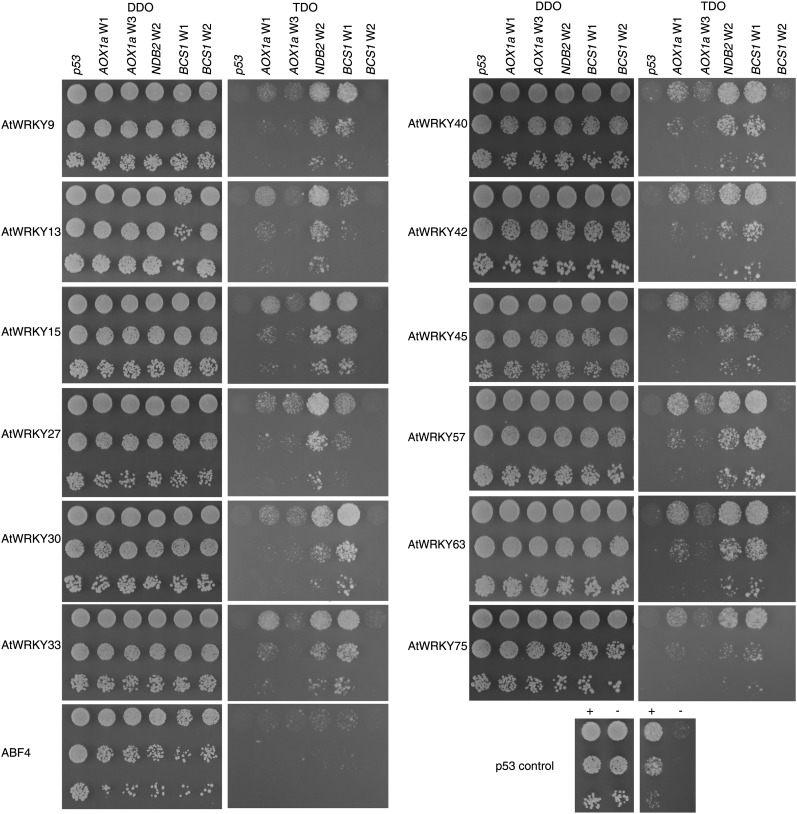
To identify which of the 72 AtWRKY transcription factors present in Arabidopsis are responsible for binding and potentially regulating the promoters of mitochondrial stress-responsive genes, the WRKY transcription factors were cloned into the pGADT7-rec2 yeast one-hybrid prey vector. AtWRKY16, AtWRKY19, AtWRKY24, AtWRKY52, and AtWRKY64 were not cloned despite several attempts. The genes for AtWRKY16, AtWRKY19, and AtWRKY52 are exceptionally long (more than 3.8 kb), which may explain the difficulty in amplification. The 50-bp promoter regions surrounding selected W-boxes of AOX1a, NDB2, and BCS1 were cloned into the pHIS2 yeast one-hybrid bait vector (Supplemental Table S1). For AOX1a, W1 and W3 were selected, for NDB2, W2 was selected, and for BCS1, W1 and W2 were selected, as their mutation had the strongest effects on promoter activity (Fig. 1).
Given the large number of combinations between the 67 transcription factors and selected bait promoter sequences, a 96-well plate assay was optimized and adapted to the yeast one-hybrid transformation and screening procedure (Supplemental Fig. S1). As self-activation and false-positive interactions form a potential difficulty in yeast one-hybrid interaction screens, background binding of all cloned WRKY factors against an active promoter sequence that does not contain the TTGAC W-box sequence (pHIS2-p53) was tested. No significant binding of any of the WRKY factors could be observed with the pHIS2-p53 negative control, while the p53 protein showed a strong positive interaction, allowing the screen against the W-boxes to proceed (Supplemental Fig. S1). Additionally, some WRKY transcription factors were recalcitrant toward transformation, likely because their expression causes lethal defects within the yeast host cells (Supplemental Fig. S1). Interaction assays were performed using selected W-boxes of AOX1a, NDB2, and BCS1 as bait (Supplemental Table S1). Several WRKY transcription factors displayed clear growth on the selective interaction medium as compared with the pHIS2-p53 negative control. Based on the yeast one-hybrid screen results, a selection of 12 WRKY transcription factors was made for further confirmation using spotting assays in a dilution series (Fig. 2). Positive interactions of specific WRKY factors were observed with AOX1a W1 and W3, NDB2 W2, and BCS1 W1, although none of the WRKY factors showed a positive interaction with the BCS1 W2 W-box (Fig. 2; Supplemental Table S1). As an additional negative control, the transcription factor ABF4 that is known to bind the AGCTC motif (Choi et al., 2000) was included to demonstrate that binding of WRKY transcription factors to the TTGAC motif is specific. These experiments revealed that, overall, AOX1a W1, NDB2 W2, and BCS1 W1 are bound most strongly. This may be explained by the observation in previous studies that WRKY transcription factors appear to bind most strongly to TTGAC(C/T) motifs. The strongest binding sites based on our yeast one-hybrid screen, AOX1a W1, NDB2 W2, and BCS1 W1, are effectively followed by a C or T, whereas the weakest binding sites, AOX1a W3 and BCS1 W2, are followed by an A or G. The BCS1 W2 shows similarity to the AS-1 element found in the cauliflower mosaic virus 35S promoter and may potentially bind bZIP transcription factors (Hara et al., 2000). Most of the 12 selected WRKY factors show binding to these three elements, but the relative strength of the interactions varies for each specific protein.
Figure 2.
Yeast one-hybrid analysis of selected WRKY transcription factors against W-boxes of AOX1a, NDB2, and BCS1 promoters and a negative control (promoter region without the W-box). Non-WRKY transcription factor ABF4 was also included as a negative control for bait self-activation. Three 10-fold dilutions were spotted vertically for each assay on DDO (transformation control) and TDO (interaction) media, including p53 positive and negative controls.
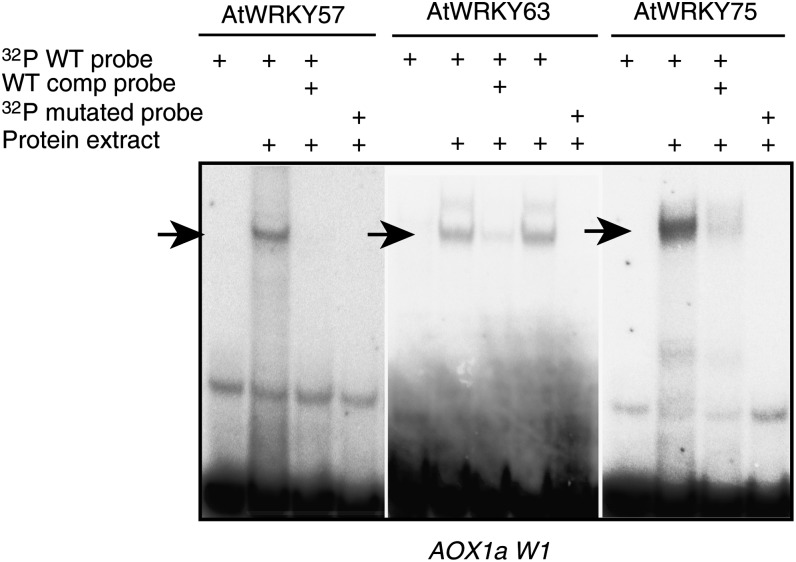
To further confirm the binding of WRKY factors, electromobility shift assays were performed (Fig. 3). AtWRKY57, AtWRKY63, and AtWRKY75 were found to express most efficiently in Escherichia coli BL12 DE3 Rosetta 2 cells, whereas the other WRKY proteins showed little or no expression of soluble protein. Purified proteins were incubated with radiolabeled double-stranded DNA probes containing wild-type or mutated W-boxes. In an additional reaction, a surplus of unlabeled wild-type competitor probe was added to verify the specificity of binding. The W1 W-box of the AOX1a promoter was used as a representative for binding to confirm the yeast one-hybrid results. Figure 3 demonstrates that AtWRKY57, AtWRKY63, and AtWRKY75 are able to bind the AOX1a promoter and that their binding is abolished when the W-box is mutated, indicating that the binding is specific for the W-box. Binding was also abolished when an excess of unlabeled competitor probe was added. In conclusion, our results show that the promoters of mitochondrial stress response genes can effectively be bound by multiple WRKY transcription factors, indicating a complex regulatory interplay between different transcription factors.
Figure 3.
Electromobility shift assays of purified WRKY proteins against radiolabeled probes with wild-type and mutated W1 W-box for the AOX1a promoter sequence. Radiolabeled 30-bp probes were incubated with purified WRKY proteins in the presence or absence of unlabeled promoter probe. As a negative control, the promoter probe with mutated W-box was included. comp, Competitor; WT, wild type. Arrows indicate specific shifts that are abolished by adding competitor or using mutated probes.
Genes Encoding Mitochondrial Proteins Respond Differentially in Transgenic Lines with Altered WRKY Expression
In the previous sections, we demonstrated that the promoters of stress-responsive genes encoding mitochondrial proteins contain active W-boxes and are bound by WRKY transcription factors. Therefore, it is plausible that WRKY transcription factors play a part in controlling the expression of these genes. To study the effects of WRKY transcription factors, we generated transgenic Arabidopsis lines with reduced and increased expression for the selection of 12 WRKY genes (Fig. 2; Table I; Supplemental Fig. S2). For all 12 genes, overexpression was confirmed by quantitative reverse transcription (QRT)-PCR, and a clear reduction in expression was confirmed in the insertion or RNA interference mutant lines, with the exception of AtWRKY30, for which no transfer DNA (T-DNA) mutant lines are available, and AtWRKY15, for which the T-DNA is present in the 5′ untranslated region (UTR).
Table I. WRKY transgenic lines.
Overview of transgenic overexpression and loss-of-function lines for selected WRKY genes. For loss-of-function lines, the line names and the position of each insertion are indicated. n/a, Not available.
| Gene | Overexpression Line | T-DNA Line 1 | Insertion Site | T-DNA Line 2 | Insertion Site |
|---|---|---|---|---|---|
| AtWRKY9 | Yes | SALK_067122 | Intron 4 of 4 | ||
| AtWRKY13 | Yes | SALK_064346 | Exon 2 of 3 | SALK_032911 | Exon 1 of 3 |
| AtWRKY15 | Yes | GABI_097A12 | 5′ UTR | ||
| AtWRKY27 | Yes | SALK_048952 | Exon 2 of 3 | ||
| AtWRKY33 | Yes | SALK_006602 | Exon 3 of 5 | ||
| AtWRKY40 | Yes | CSHL_ET5883 | Exon 2 of 4 | ||
| AtWRKY42 | Yes | SALK_121674 | Exon 3 of 6 | ||
| AtWRKY45 | Yes | RATM11-0634-1_H | Intron 1 of 1 | ||
| AtWRKY57 | Yes | SALK_076716 | Intron 1 of 2 | GABI_078H12 | Intron 1 of 2 |
| AtWRKY63 | Yes | SALK_007496 | Exon 3 of 3 | n/a | |
| AtWRKY75 | Yes | amiRNA line | |||
| AtWRKY30 | Yes | n/a |
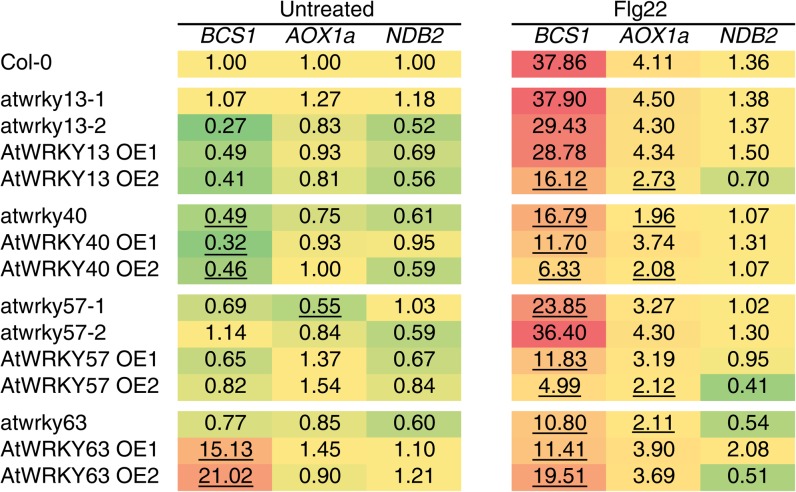
Expression profiles for the genes encoding mitochondrial proteins (AOX1a, NDB2, and BCS1) were investigated in the mutant lines for the 12 selected WRKY transcription factors under both normal and stress conditions. To cover the wide spectrum of signaling pathways that affect the expression of stress-responsive mitochondrial genes, we selected AA for mitochondrial retrograde pathways (Dojcinovic et al., 2005), Flg22 for biotic signaling (Ho et al., 2008), and ABA for abiotic stress signaling (Giraud et al., 2009). Untreated plants and randomized plants treated with AA, Flg22, or ABA were collected in biological duplicates for all 24 transgenic lines, including ecotype Columbia (Col-0). The expression of AOX1a, NDB2, and BCS1 was analyzed by QRT-PCR. Furthermore, the ABA response marker RD29A and the universal reactive oxygen species marker gene UPOX, which encodes a mitochondrial protein of unknown function, were also included (Gadjev et al., 2006). The results of this large-scale expression analysis are shown in Supplemental Figure S3. AOX1a and NDB2 expression was strongly induced by AA and responded more moderately to Flg22 and ABA. BCS1 responded strongly to Flg22 and AA but not ABA, whereas UPOX was highly induced by AA and slightly by ABA. RD29A expression only responded to ABA treatment. Detailed transcript analysis revealed that the expression of the marker genes was different in several WRKY transgenic lines compared with wild-type plants (Supplemental Fig. S3). In brief, AtWRKY13 and AtWRKY40 appeared to affect the response to AA, AtWRKY63 appeared to affect the basal expression of BCS1, while AtWRKY40, AtWRKY57, and AtWRKY63 affected the response to Flg22 (Supplemental Fig. S3).
Based on the initial screening assay for altered responses to stress treatments (Supplemental Fig. S3), AtWRKY13, AtWRKY40, AtWRKY57, and AtWRKY63 were retained for confirmation in a repeat experiment under selected conditions using two independent overexpression lines and, where available, two independent knockout alleles (Figs. 4–6). Strikingly, under untreated conditions, BCS1 transcript levels were significantly higher in both AtWRKY63 overexpression lines, whereas AOX1a and NDB2 levels were unchanged. This confirms that BCS1 induction can be uncoupled from AOX1a induction and can respond to different signals at least under these conditions (Ho et al., 2008). Upon Flg22 treatment, the induction of BCS1 and AOX1a expression was approximately 4- and 2-fold repressed in atwrky63 knockout plants (Fig. 4). Expression levels of BCS1 after Flg22 treatment remained similar to the already elevated levels in untreated conditions, while no change in the induction of AOX1a was observed in AtWRKY63 overexpression lines. These observations imply that AtWRKY63 is an activator of biotic responses and BCS1 responds specifically to these signals. For AtWRKY57, up to 7.5-fold reduction (P < 0.05) of BCS1 expression upon Flg22 stimulation was observed in the overexpression lines. atwrky57 knockout lines showed no or little change in induction, indicating that AtWRKY57 can act as a repressor of biotic responses. In atwrky40 knockout plants, a more than 2-fold reduction in the Flg22 response of BCS1 and AOX1a was observed. Interestingly, there was also a strongly reduced induction of BCS1 in AtWRKY40 overexpression lines, suggesting that reprogramming has resulted in an upset balance of repression and activation in these mutants.
Figure 4.
QRT-PCR analysis of transcripts encoding mitochondrial proteins in transgenic WRKY overexpression or knockout/knockdown Arabidopsis lines. Wild-type and mutant plants with increased or reduced expression of WRKY transcription factors were grown in soil for 3 weeks and then treated with Flg22 for 45 min in biological duplicate. Samples were harvested, and mRNA levels were measured using QRT-PCR. Expression values are shown and color coded for visual representation: yellow, unchanged; green, reduced; red, induced. Underlined values indicate statistically significant differences (Student’s t test, P < 0.05) compared with the expression in Col-0 following the same treatment.
Figure 6.
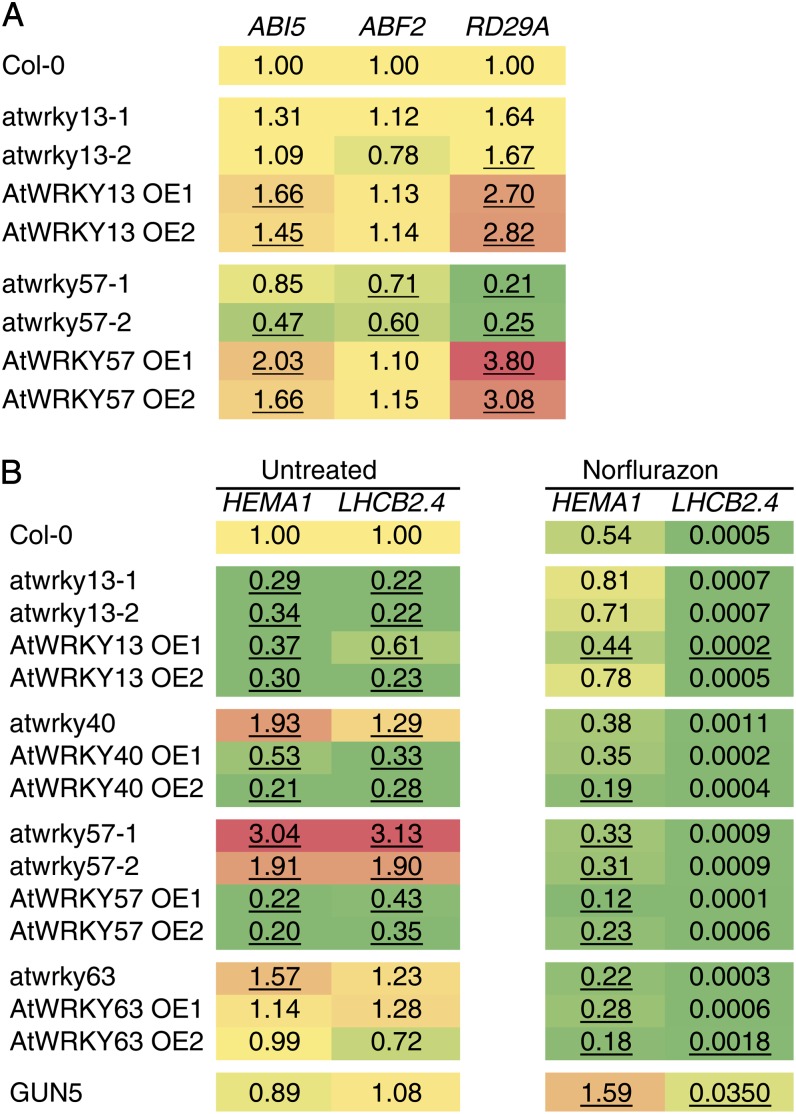
QRT-PCR analysis of transcripts encoding chloroplast proteins in transgenic WRKY overexpression or knockout/knockdown lines. A, Expression of ABA-related genes in 3-week-old AtWRKY13 and AtWRKY57 mutant lines. B, Wild-type and mutant plants were grown on MS plates with or without norflurazon for 5 d. Samples were harvested and analyzed using QRT-PCR in duplicate. Expression values are shown and color coded for visual representation: yellow, unchanged; green, reduced; red, induced. Underlined values indicate statistically significant differences (P < 0.05) compared with the expression in Col-0 within the same treatment.
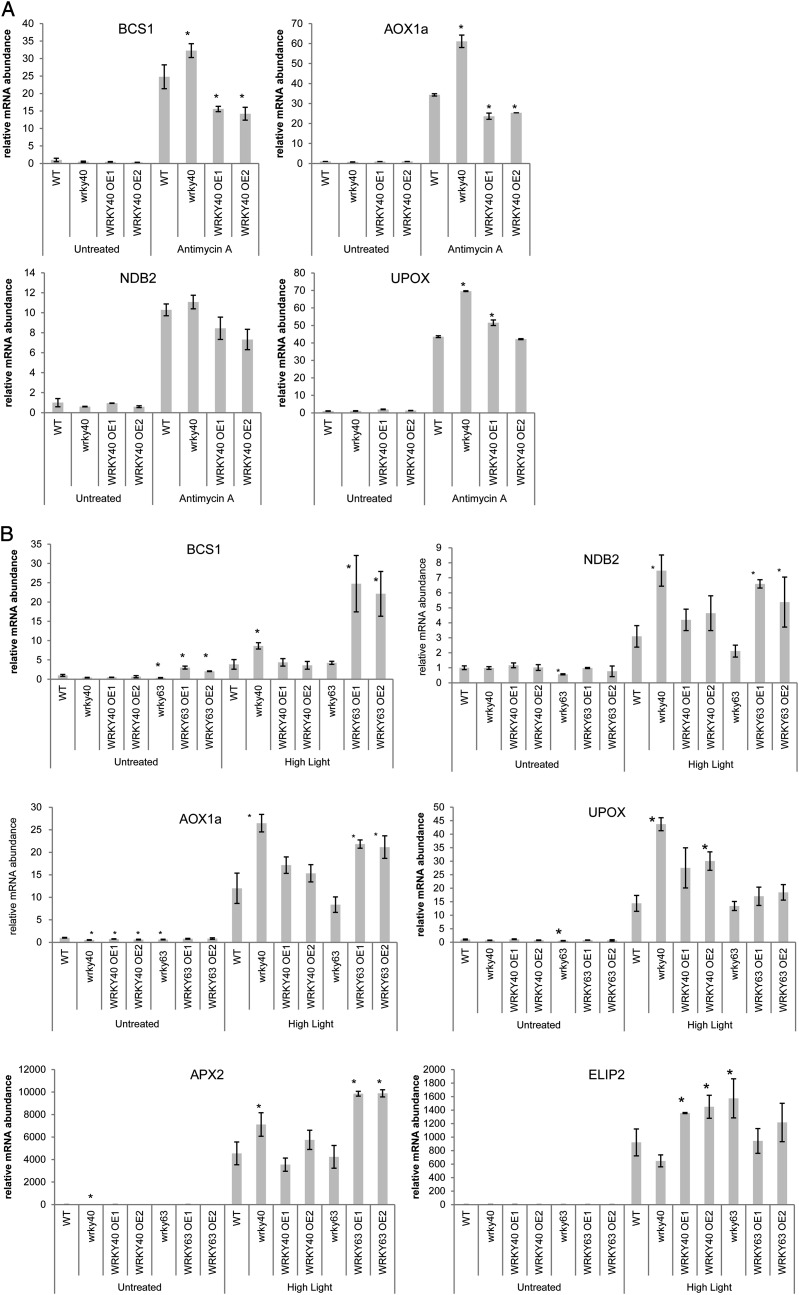
Based on the initial screening (Supplemental Fig. S3), transgenic lines AtWRKY40 and AtWRKY13 were treated with AA. As expected, AA increased marker gene expression in the wild type, but strikingly, a significantly higher induction was observed in atwrky40 plants for BCS1, AOX1a, and UPOX. Conversely, less induction of BCS1 and AOX1a was observed in both AtWRKY40 overexpression lines (Fig. 5A). These experiments indicate that AtWRKY40 can act as a repressor during stress responses to AA. The effect of AtWRKY13 on AA responses did not appear to be consistent in the repeat experiments (Supplemental Table S4).
Figure 5.
QRT-PCR analysis of transcripts encoding mitochondrial proteins in transgenic WRKY overexpression or knockout/knockdown lines. Wild-type (WT) and mutant plants with increased or reduced expression of WRKY transcription factors were grown in soil for 3 weeks and then treated with AA for 4 h in biological duplicate or with HL for 1 h in biological triplicate. Samples were harvested, and mRNA levels were measured using QRT-PCR. Asterisks indicate statistically significant differences (P < 0.05) compared with the expression in Col-0 within the same treatment.
Whereas AA was used for inducing mitochondrial retrograde signaling, the transgenic lines for AtWRKY13, AtWRKY40, AtWRKY57, and AtWRKY63 were also treated with high light (HL; 1 h at 1,000 µE m–2 s–1) to induce chloroplast retrograde signaling (Estavillo et al., 2011). Single replicates were initially tested for changes in marker gene expression (Supplemental Table S3), and based on these results, transgenic lines for AtWRKY40 and AtWRKY63 were selected for further analysis in biological triplicate (Fig. 5B). Significant changes in the expression of AOX1a, BCS1, NDB2, and UPOX were observed in the transgenic lines for AtWRKY40 and AtWRKY63. As control marker genes for the HL treatment, Ascorbate peroxidase2 (APX2) and Early light-inducible protein2 (ELIP2) were used (Estavillo et al., 2011). As observed for the AA treatment, atwrky40 plants showed a significantly stronger induction (more than 2-fold) of the mitochondrial stress marker genes by HL. This further supports the role of AtWRKY40 as a repressor of mitochondrial stress-responsive genes. Overexpression of AtWRKY40 did not consistently change marker gene expression under the conditions used for the mitochondrial marker genes, suggesting that AtWRKY40 activity is limited by other factors and may be part of a multiprotein complex, as described previously (Xu et al., 2006). Interestingly, the two marker genes for HL were also significantly affected by perturbation of AtWRKY40 levels: APX2 behaved similarly to the mitochondrial stress response marker genes (i.e. more strongly induced in atwrky40 plants), while ELIP2, which encodes a chloroplast protein, behaved in the opposite fashion (i.e. more strongly induced in the AtWRKY40 overexpression lines).
For AtWRKY63, both overexpression lines again showed a significant induction of BCS1 expression in untreated conditions, whereas the basal expression of BCS1 was down-regulated in atwrky63 lines (Fig. 5B). Moreover, after the HL treatment, a significantly higher induction of AOX1a, BCS1, NDB2, UPOX, and also APX2 were observed in the AtWRKY63 overexpression lines. These expression patterns are consistent with a role of AtWRKY63 as a positive regulator of mitochondrial stress responses. In conclusion, the identified WRKY transcription factors are capable of modulating gene expression patterns of stress-responsive genes encoding mitochondrial proteins, with AtWRKY40 acting as a repressor and AtWRKY63 as an activator.
AtWRKY13 and AtWRKY57 Affect Basal Expression Levels of ABA-Related Genes
Previous reports have shown the involvement of several WRKY transcription factors in the regulation of ABA responses (Ren et al., 2010; Shang et al., 2010; Rushton et al., 2012). Therefore, the expression of ABA signaling-related genes was analyzed in transgenic lines for selected WRKY transcription factors (Fig. 6A; Supplemental Fig. S3). In the large-scale transcript analysis, AtWRKY13 and AtWRKY57 overexpression lines showed increased basal expression of the ABA response marker gene RD29A (Supplemental Fig. S3), while AtWRKY40 and AtWRKY63 showed no significant effects (Supplemental Fig. S3; Supplemental Table S4). In repeat experiments with independent transgenic lines, AtWRKY13 overexpression lines consistently showed nearly 3-fold higher expression levels of RD29A and a minor induction of ABI5 (a bZIP transcription factor; Finkelstein and Lynch, 2000). ABF2 was unaffected. AtWRKY57 overexpression lines showed a similar 3- to 4-fold induction of RD29A and about a 2-fold induction of ABI5 (Fig. 6). In agreement with a role of AtWRK57 as an activator of ABA-related gene expression, RD29A was 4- to 5-fold down-regulated in both atwrky57 mutant lines.
AtWRKY13, AtWRKY40, and AtWRKY57 Affect the Early Expression of Genes Encoding Chloroplast Proteins
The four WRKY transcription factors identified here as affecting transcripts encoding mitochondrial proteins were investigated for a role in the expression of genes encoding chloroplast proteins. Wild-type, AtWRKY13, AtWRKY40, AtWRKY57, and AtWRKY63 overexpression, and knockout seedlings were grown under continuous-light conditions for 5 d on Murashige and Skoog (MS) and MS + 5 μm norflurazon (a chlorophyll synthesis inhibitor) medium in an assay identical to the GUN plastid-to-nucleus signaling screen (Koussevitzky et al., 2007). Light-harvesting chlorophyll B-binding protein2.4 (LHCB2.4; At3g27690) and HEMA1 were selected as marker genes for chloroplast function and stress responses (McCormac and Terry, 2004; Koussevitzky et al., 2007; Fig. 6). For AtWRKY63 transgenic lines, no consistent changes were observed under untreated conditions; however, both AtWRKY40 and AtWRKY57 were found to be basal repressors of the chloroplast function reporter genes, with knockout lines showing induced expression and overexpression lines showing strong repression. Conversely, atwrky13 knockout lines showed strong repression of the chloroplast reporter genes, indicating that AtWRKY13 is an activator of chloroplast reporter genes. Interestingly, AtWRKY13 overexpression lines also showed reduced expression of LHCB2.4 and HEMA1, suggesting a perturbed steady state of expression in these mutants. Treatment with norflurazon triggered the previously reported reduction in the expression of LHCB2.4 and HEMA1 in wild-type plants, which was alleviated in the GUN5 mutant that was included as a control (Koussevitzky et al., 2007). In this study, none of the WRKY transgenic lines showed a classical GUN phenotype (Fig. 6).
WRKY Transcription Factors Directly Bind the Promoters of Genes Encoding Chloroplast Proteins
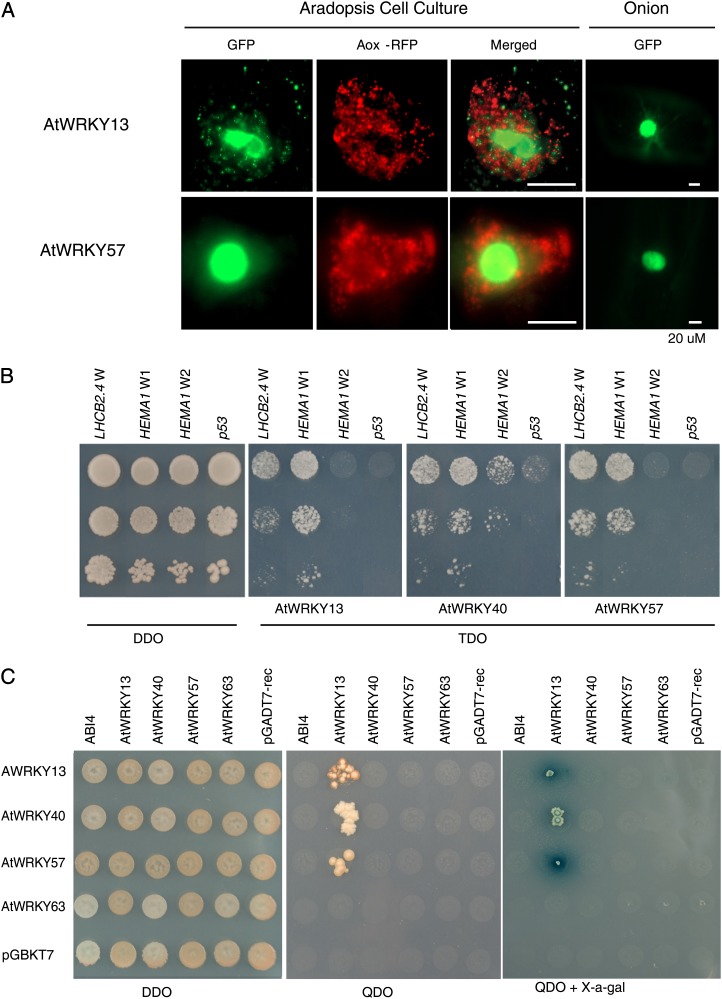
Being transcription factors, the subcellular localization of AtWRKY factors is expected to be nuclear. Previous reports indicated the presence of AtWRKY40 in the nucleus as well as associated with chloroplast membranes, whereas AtWRKY63 was found to be exclusively nuclear (Ren et al., 2010; Shang et al., 2010). As the subcellular localization of AtWRKY13 and AtWRKY57 was not determined previously, GFP fusion protein assays confirmed their nuclear localization (Fig. 7A). We showed that a range of AtWRKY transcription factors regulate the expression of genes encoding chloroplast proteins (Fig. 6). Therefore, the occurrence of putative W-boxes was investigated in the promoters of chloroplast protein marker genes LHCB2.4 and HEMA1. The promoter of LHCB2.4 contained one W-box (1,465 bp upstream of ATG) and the promoter of HEMA1 contained two putative W-boxes (774 [W-box 2] and 164 [W-box 1] bp upstream of ATG) in the 1.5-kb promoter regions. To confirm if the regulation of genes encoding chloroplast proteins by AtWRKY13, AtWRKY40, and AtWRKY57 is through direct promoter interactions, the LHCB2.4 and HEMA1 W-box promoter regions were cloned into the yeast one-hybrid reporter system and interactions with AtWRKY13, AtWRKY40, and AtWRKY57 were assessed. Figure 7B shows that AtWRKY13, AtWRKY40, and AtWRKY57 clearly bind the LHCB2.4 W-box and HEMA1 W-box 1. Only AtWRKY40 showed a weak interaction with HEMA1 W-box 2. Given the previous reports of the involvement of ABI4 in the regulation of genes both encoding mitochondrial and chloroplast proteins (Koussevitzky et al., 2007; Giraud et al., 2009), we assessed whether direct interactions of WRKY transcription factors occurred with ABI4 using the yeast two-hybrid protein-protein interaction method. ABI4, AtWRKY13, AtWRKY40, AtWRKY57, and AtWRKY63 were cloned into the yeast two-hybrid reporter vectors and screened for binding (Fig. 7C). No binding of WRKY proteins with ABI4 was observed; however, direct binding of AtWRKY13 with itself, AtWRKY40, and AtWRKY57 was demonstrated.
Figure 7.
Subcellular localization and yeast one/two-hybrid assays for WRKY proteins. A, Full-length coding sequences were fused to a GFP coding sequence and transformed into Arabidopsis suspension cells and onion epidermal cells. AOX-RFP was included as a mitochondrial marker. Bars = 20 μm. B, Binding of WRKY transcription factors to W-boxes present in the promoters of genes encoding chloroplast proteins examined using yeast one-hybrid screening. A p53 promoter sequence not containing a W-box was included as a negative control. Transformation is indicated by growth on DDO medium, and interaction is indicated by growth on TDO medium. Cultures were diluted 10-fold in a vertical pattern. C, Screening of protein-protein interactions between WRKY transcription factors and ABI4 using the yeast two-hybrid system. pGADT7-rec and pGBKT7 are included as empty vector controls. Growth on DDO medium indicates the presence of bait and prey vectors, and interaction is indicated by growth on quadruple dropout medium (QDO) with and without 5-bromo-4-chloro-3-indolyl α-d-galactopyranoside (X-α-gal).
Genome-Wide Expression Analysis Reveals the Roles of AtWRKY40 and AtWRKY63 in Arabidopsis Stress Responses
The starting point of this study was to assess whether the putative W-boxes in the promoters of highly stress-responsive genes encoding mitochondrial proteins were functionally involved in the regulation of gene expression. In the previous sections, we identified AtWRKY40 and AtWRKY63 as regulators of marker genes such as AOX1a. To obtain a clear picture of which other genes and stress response pathways are modulated by the identified WRKY transcription factors, we performed a genome-wide microarray expression analysis. The transgenic lines atwrky40, AtWRKY40 OE1, atwrky63, and AtWRKY63 OE1 were compared with wild-type plants under untreated and HL conditions in biological triplicate using the Affymetrix ATH1 GeneChip platform. The HL treatment itself caused profound changes in gene expression in wild-type plants (Table II), and the changes in the selected marker genes (Fig. 5B) due to perturbed levels of AtWRKY40 or AtWRKY63 were confirmed by the microarray analysis (Supplemental Tables S4 and S5). Compared with wild-type plants, all four mutant lines displayed significantly altered expression profiles [PPDE(<P) > 0.95, where PPDE is posterior probability of differential expression] under both untreated and HL-treated conditions (Table II), demonstrating the role of AtWRKY40 and AtWRKY63 in modulating the expression of more than just the initially selected marker genes. Comparison of the expression profiles revealed a severalfold higher number of overlapping significantly changed probe sets than randomly expected, indicating a functional relatedness between AtWRKY40 and AtWRKY63 (Table III). Eighteen probe sets were more than 2-fold significantly changed in all four mutants compared with the wild type under untreated conditions, with the expected number of overlapping probe sets by random selection being close to zero (Table III).
Table II. Microarray analysis of WRKY mutants during HL stress.
The number of significantly differentially expressed probe sets is shown compared with wild-type plants [PPDE(<P) > 0.95] with a range of fold change cutoffs. Numbers in parentheses indicate up- and down-regulated genes compared with the wild type. UT, Untreated.
| Sample | PPDE(<P) > 0.95 | Fold Change > 1.5 | Fold Change > 2 |
|---|---|---|---|
| Compared with wild-type UT | |||
| wrky40 UT | 2,585 (1,446, 1,139) | 2,309 (1,286, 1,023) | 1,336 (723, 613) |
| WRKY40 OE1 UT | 953 (358, 595) | 897 (324, 573) | 583 (180, 403) |
| wrky63 UT | 512 (267, 245) | 490 (257, 233) | 315 (165, 150) |
| WRKY63 OE1 UT | 1,499 (975, 524) | 1,353 (914, 439) | 744 (580, 164) |
| Compared with wild-type HL | |||
| wrky40 HL | 4,239 (2,408, 1,831) | 2,690 (1,535, 1,155) | 1,271 (681, 590) |
| WRKY40 OE1 HL | 805 (391, 414) | 739 (366, 373) | 470 (250, 220) |
| wrky63 HL | 253 (124, 129) | 194 (92, 102) | 71 (34, 37) |
| WRKY63 OE1 HL | 1,698 (1,007, 691) | 1,544 (965, 579) | 960 (700, 260) |
| Col-0 HL versus Col-0 UT | 9,105 (3,919, 5,186) | 6,313 (2,662, 3,651) | 3,611 (1,687, 1,924) |
Table III. Comparison of overlap between WRKY mutant lines.
χ2 analysis is shown compared with the random distribution of overlapping 2-fold significantly changed genes and compared with the wild type in untreated and HL-treated conditions. UT, Untreated.
| Sample | Line 1 | Line 2 | Line 3 | Line 4 | Observed Overlap | Expected Overlap | P |
|---|---|---|---|---|---|---|---|
| Two-fold UT versus wild-type UT | |||||||
| wrky40 versus WRKY40 OE1 | 1,336 | 583 | 175 | 46 | <0.001 | ||
| wrky40 versus WRKY63 OE1 | 1,336 | 744 | 153 | 58 | <0.001 | ||
| WRKY40 OE1 versus wrky63 | 583 | 315 | 107 | 11 | <0.001 | ||
| wrky63 versus WRKY63 OE1 | 315 | 744 | 109 | 14 | <0.001 | ||
| All four lines | 1,336 | 583 | 315 | 744 | 18 | 0 | <0.001 |
| Two-fold HL versus wild-type HL | |||||||
| wrky40 versus WRKY40 OE1 | 1,271 | 470 | 164 | 35 | <0.001 | ||
| wrky40 versus WRKY63 OE1 | 1,271 | 960 | 327 | 72 | <0.001 | ||
| WRKY40 OE1 versus wrky63 | 470 | 71 | 21 | 2 | <0.001 | ||
| wrky63 versus WRKY63 OE1 | 71 | 960 | 29 | 4 | <0.001 | ||
| All four lines | 1,271 | 470 | 71 | 960 | 9 | 0 | <0.001 |
A survey of the direction of change (i.e. up- or down-regulation) in the four mutant lines demonstrated that significantly more genes than expected were up-regulated in atwrky40 plants and down-regulated in AtWRKY40 OE1 in untreated conditions (P < 0.001), consistent with the suggested role of AtWRKY40 being mainly a repressor (Supplemental Table S6). In AtWRKY63 OE1 plants, significantly more genes were up-regulated compared with the wild type, consistent with a role as activator. The same trends were observed in response to HL treatment for atwrky40 and AtWRKY63 overexpression (Supplemental Table S6). In general, the atwrky63 mutation caused the fewest changes compared with the wild type, and no bias could be detected toward up- or down-regulation, indicating that other redundant factors may partially complement the loss of AtWRKY63. Of the 18 probe sets that were commonly changed in the four mutant lines, 10 followed the expected pattern consistent with a role of AtWRKY63 as an activator (down-regulated in atwrky63 and up-regulated in AtWRKY63 OE1), and of these, seven followed the expected pattern of AtWRKY40 being a repressor (up-regulated in atwrky40 and down-regulated in AtWRKY40; Supplemental Fig. S4). AtWRKY40 and AtWRKY63 have previously been described to affect ABA signaling (Ren et al., 2010; Shang et al., 2010).To estimate the effect of perturbing AtWRKY40 and AtWRKY63 on ABA-responsive genes, the expression characteristics of the approximately 3,000 genes that are thought to be affected by ABA were analyzed in the microarray data set (Nemhauser et al., 2006). In agreement with previous findings, a significantly higher proportion of these genes were affected in the four analyzed lines compared with random expectation (Supplemental Table S6) under both untreated and HL-treated conditions. Again, the basal expression of ABI5 and RD29A was not different in AtWRKY40 and AtWRKY63 mutant lines compared with wild-type plants (Supplemental Table S3; Supplemental Fig. S3), suggesting that ABA responses are controlled by multiple pathways. Interestingly, the mild but significant induction of RD29A by HL treatment was completely repressed in atwrky40 plants (Supplemental Table S3).
AtWRKY40 and AtWRKY63 Modulate the Mitochondrial Stress Response
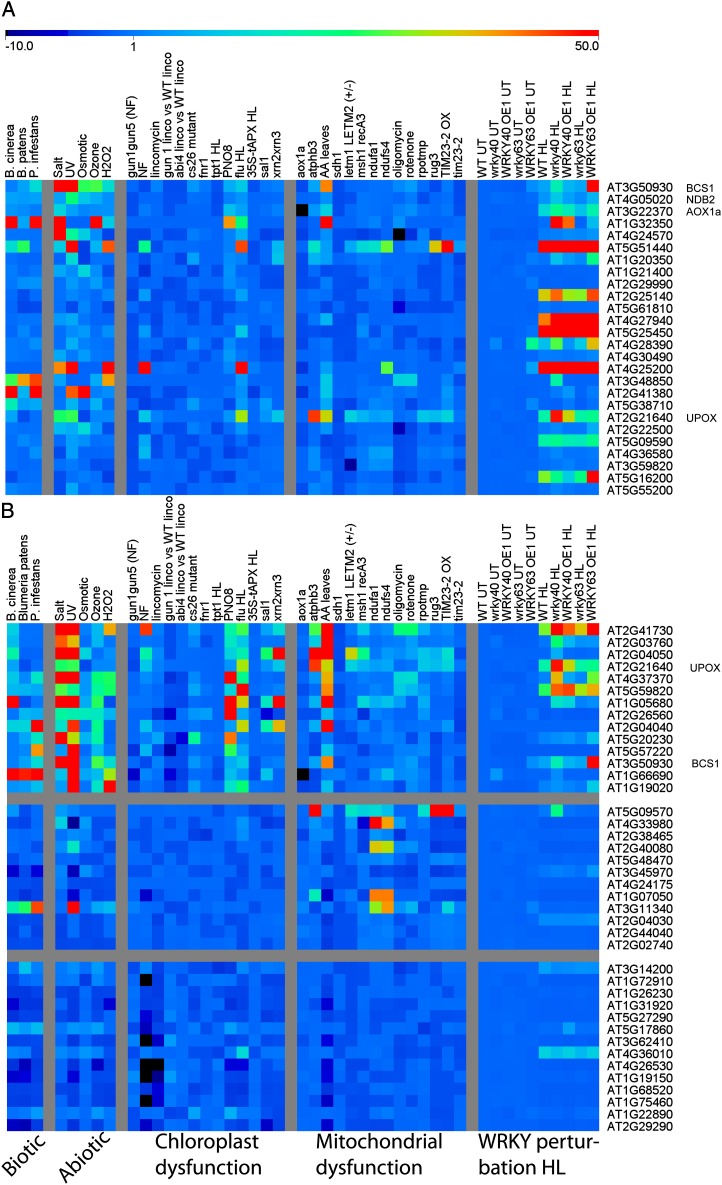
Subsequently, we analyzed the expression profiles of the 26 genes encoding mitochondrial proteins that were defined previously as the core mitochondrial stress response and that display an overrepresentation of W-boxes (Van Aken et al., 2009). Of these 26 genes, 23 responded significantly to the HL treatment in wild-type plants (Fig. 8A). Moreover, the responsiveness to HL of 20 of these 23 genes was significantly affected in the WRKY mutant lines: 16 genes responded differently from HL compared with the wild type in atwrky40, 12 in AtWRKY63 OE1, six in AtWRKY40 OE1, and one in atwrky63 plants (Fig. 8A; Table IV). Furthermore, 11 genes were commonly affected in atwrky40 and AtWRKY63 OE1. To rule out the possibility that these observations could be the result of random distribution, χ2 statistical tests indicated up to 5.7 times more changes (P < 0.01) as compared with random sampling for all lines (Table IV). Next, we assessed whether the changes caused by WRKY perturbations were biased toward genes encoding energy organelle proteins in general. Therefore, we assembled lists of all the genes that putatively encode energy organelle proteins and tested if there was a significant overrepresentation of these genes in the gene sets that were altered by AtWRKY40 or AtWRKY63 perturbation (Supplemental Table S6). Interestingly, this analysis revealed that 1.7 to 3.8 times fewer (P < 0.01) genes than expected encoding chloroplast or mitochondrial proteins were significantly changed in the WRKY mutant lines compared with wild-type plants. This was the case for all four mutant lines under untreated conditions and all except atwrky63 under HL conditions. Changes in the expression of genes encoding peroxisome proteins were also significantly underrepresented in the AtWRKY63 OE1 plants.
Figure 8.
Heat maps representing the expression of marker genes of mitochondrial stress responses (A) and chloroplast or mitochondrial dysfunction (B). Relative expression is shown for a range of previously published microarray experiments involving abiotic stress, biotic stress, and chemical or genetic inhibition of chloroplast or mitochondrial functions and from this study for expression in AtWRKY40 and AtWRKY63 knockout/overexpression in untreated (UT) or HL conditions. WT, Wild type.
Table IV. Representation of marker genes for mitochondrial stress response and organellar dysfunction.
χ2 analysis is shown compared with a random distribution. exp, Expected; obs, observed.
| Sample | Changes versus Wild-Type HL | Present in Category | Observed Differential to Wild-Type HL | Expected Differential to Wild-Type HL | P | Ratio obs-exp |
|---|---|---|---|---|---|---|
| Mitochondrial stress response genes | ||||||
| wrky40 HL | 4,239 | 26 | 17 | 6.5 | <0.001 | 2.6 |
| WRKY40 OE1 HL | 805 | 26 | 7 | 1.2 | <0.001 | 5.7 |
| wrky63 HL | 253 | 26 | 2 | 0.4 | <0.01 | 5.2 |
| WRKY63 OE1 HL | 1,688 | 26 | 13 | 2.6 | <0.001 | 5.0 |
| Mitochondrial and chloroplast dysfunction | ||||||
| wrky40 HL | 4,239 | 13 | 10 | 3.2 | <0.001 | 3.1 |
| WRKY40 OE1 HL | 805 | 13 | 3 | 0.6 | <0.001 | 4.9 |
| wrky63 HL | 253 | 13 | 2 | 0.2 | <0.001 | 10.3 |
| WRKY63 OE1 HL | 1,688 | 13 | 7 | 1.3 | <0.001 | 5.4 |
| Mitochondrial dysfunction only | ||||||
| wrky40 HL | 4,239 | 12 | 3 | 3.0 | <0.05 | 1.0 |
| WRKY40 OE1 HL | 805 | 12 | 1 | 0.6 | <0.05 | 1.8 |
| wrky63 HL | 253 | 12 | 3 | 0.2 | <0.001 | 16.8 |
| WRKY63 OE1 HL | 1,688 | 12 | 1 | 1.2 | <0.05 | 0.8 |
| Chloroplast dysfunction only | ||||||
| wrky40 HL | 4,239 | 14 | 3 | 3.5 | <0.05 | 0.9 |
| WRKY40 OE1 HL | 805 | 14 | 2 | 0.7 | <0.05 | 3.0 |
| wrky63 HL | 253 | 14 | 0 | 0.2 | <0.05 | 0.0 |
| WRKY63 OE1 HL | 1,688 | 14 | 1 | 1.4 | <0.05 | 0.7 |
A previous study examined the gene expression patterns of genes encoding chloroplast and mitochondrial proteins in a set of nearly 1,300 ATH1 microarray chips, encompassing experiments related to light signaling, hormones, sugars, chloroplast function, reactive oxygen species, and general stresses (Leister et al., 2011). In this study, the genes encoding chloroplast and mitochondrial proteins were subdivided into functional subclasses. For chloroplast proteins, the subclasses photosynthesis, organellar gene expression, and tetrapyrrole metabolism were annotated; genes encoding mitochondrial proteins were subdivided into respiration and organellar gene expression. The AtWRKY40 and AtWRKY63 microarray data set after HL treatment was analyzed to determine if any of these categories were specifically affected by WRKY perturbations (Supplemental Table S6). For the purpose of this study, only nucleus-encoded genes were analyzed. Interestingly, the three chloroplast categories were represented either as randomly expected or even significantly underrepresented in the case of photosynthesis and tetrapyrrole metabolism in atwrky40 and AtWRKY63 OE1 plants (Supplemental Table S6). Similarly, the mitochondrial organellar gene expression category was represented either as randomly expected or significantly underrepresented in atwrky40 and AtWRKY63 OE1 plants. The mitochondrial respiration category was represented as randomly expected for all lines, except atwrky63, where an overrepresentation could be observed. This discrepancy could be caused by reduced statistical power due to the low number of statistically changed genes in atwrky63 genes in general (Table II).
The study by Leister and colleagues (2011) also ranked the genes encoding mitochondrial and chloroplast proteins by the number of treatments or mutations to which the genes were responsive. The categories “very highly responsive” (VHR; 127 genes) and “very weakly responsive” (VWR; 120 genes) were analyzed, and indeed, significantly more of the VHR genes were HL responsive than randomly expected, while significantly fewer of the VWR genes were HL responsive (P < 0.001; Supplemental Table S6). Furthermore, it was noted that a highly significant proportion of the VHR genes were altered in WRKY mutants compared with the wild type after HL treatment, whereas VWR genes were significantly less affected than expected by WRKY perturbations (except atwrky63). These analyses suggest that AtWRKY40 and AtWRKY63 are specifically involved in the regulation of the highly environment-responsive genes encoding mitochondrial and chloroplast proteins but are apparently less involved in more constitutively expressed genes encoding energy organellar proteins.
AtWRKY40 and AtWRKY63 Modulate Common Gene Expression Responses to Organellar Dysfunction
A recent study performed a meta-analysis on a wide range of microarray data sets that were specifically related to chemical or genetic inhibition of chloroplast or mitochondrial functions (Van Aken and Whelan, 2012). One of the main conclusions of this study was that perturbation of mitochondrial or chloroplast function triggered distinct transcriptional responses directed toward the impaired organelle. However, it was also found that a higher than expected overlap exists between the two organelle dysfunction responses, and most of the common genes are part of the general stress response. Three sets of marker genes were identified that respond specifically to either chloroplast or mitochondrial dysfunction or that respond to both organellar dysfunctions. Therefore, we analyzed the expression changes for these three marker gene sets in the AtWRKY40 and AtWRKY63 microarray data sets (Fig. 8B). For genes responding specifically to chloroplast dysfunction, no more significant changes could be observed in the WRKY mutant lines than what is randomly expected by HL treatment in wild-type plants. Likewise, no more significant changes could be observed for genes responding specifically to mitochondrial dysfunction in all WRKY mutant lines, except in atwrky63. However, three to 10 times more of the marker genes that respond to both chloroplast and mitochondrial perturbation than randomly expected were affected in all four WRKY mutant lines (P < 0.001; Table IV). The fact that WRKY factors specifically affect the genes responding in common to but not specifically to chloroplast or mitochondrial dysfunction is further supported by the observation that W-boxes are present in nearly all of the promoters of these genes (Van Aken and Whelan, 2012). In contrast, no overrepresentation of W-boxes is found in the promoters of the genes responding specifically to either chloroplast or mitochondrial dysfunction.
DISCUSSION
In this study, a series of WRKY transcription factors were identified that are involved in regulating the expression of stress-responsive genes encoding both mitochondrial and chloroplast proteins. The presence of over 70 different WRKY transcription factors in the Arabidopsis genome and the apparent lack of abnormal visual phenotypes in most WRKY knockout lines suggest a high level of redundancy between the different WRKY proteins. However, the clear changes in expression in single WRKY mutants indicate that individual proteins have discrete functions in the regulatory network (Figs. 4–6).
The initial goal of this study was to investigate if the overrepresentation of putative W-boxes in the promoters of the most strongly stress-responsive genes encoding mitochondrial proteins was functionally relevant to the expression patterns of these genes. Several lines of evidence now support this hypothesis. First, mutation of specific W-boxes significantly affects the promoter activity of the selected marker genes AOX1a, BCS1, and NDB2. Second, a number of WRKY factors are capable of directly binding to these promoter regions. And third, perturbing the activity of AtWRKY40 or AtWRKY63 significantly altered the expression pattern and stress responsiveness of a very large proportion of the 26 mitochondrial stress response marker genes after stress treatment (Van Aken et al., 2009). From the observed expression patterns, it is apparent that AtWRKY40 acts mainly as a repressor of these genes, whereas AtWRKY63 acts mostly as an activator. Interestingly, some genes respond in an opposite manner, such as ELIP2 encoding a chloroplast protein, indicating a complex network of direct and possibly indirect regulation. Using genome-wide expression analysis, we have further expanded the understanding of the role of these transcription factors. The results indicate that WRKY transcription factors are not significantly involved in the regulation of constitutive functions carried out by energy organelles such as photosynthesis, respiration, and organellar gene expression. In contrast, it appears that WRKY transcription factors are more involved in modulating the expression of genes that are specifically responsive when organellar function is inhibited either by a genetic defect or external factors. A proportion of the genes responsive to such cellular dysfunctions encode organellar proteins, as exemplified by AOX1a, BCS1, and other mitochondrial stress response genes, as well as the VHR gene set encoding chloroplast and mitochondrial proteins as identified by Leister et al. (2011). That study did not report an overrepresentation of W-boxes in the promoters of genes encoding organellar proteins. This may be explained by the fact that WRKY factors appear to regulate mainly stress-responsive genes encoding organellar proteins but not the large group of genes encoding organellar proteins that are less environmentally responsive. Furthermore, many stress-responsive genes not encoding energy organellar proteins (Fig. 8) appear to be modulated by WRKY transcription factors.
When specifically looking at how AtWRKY40 and AtWRKY63 regulate the expression of marker genes directly responsive to chloroplast or mitochondrial dysfunction (Van Aken and Whelan, 2012), a number of interesting patterns arose. The analysis showed that the WRKY factors are not significantly involved in the regulation of genes that specifically respond to either chloroplast or mitochondrial dysfunction. However, it was evident that the identified WRKY transcription factors are important regulators of the genes that are commonly affected by both chloroplast and mitochondrial perturbation. Moreover, these marker genes are widely affected during more general stresses (Fig. 8). The promoters of these genes (which include BCS1) almost all contain W-boxes, suggesting that the regulation is due to the direct binding of WRKY transcription factors.
Most of the identified WRKY factors seem to play multiple roles in seemingly disparate functions, ranging from response to ABA and biotic stress to the regulation of organellar stress-responsive proteins. One explanation may be the formation of multimeric protein complexes, where different combinations of interaction partners bind to different sets of promoters and have different effects. The formation of these multimeric WRKY protein complexes has been reported multiple times in the literature, with AtWRKY18, AtWRKY40, and AtWRKY60 being a well-studied example with roles in biotic stress and ABA signaling (Xu et al., 2006; Shang et al., 2010). Also, AtWRKY25 and AtWRKY33 are known to interact with roles in heat and osmotic stresses (Li et al., 2011). A genome-wide study using the yeast two-hybrid screening method identified additional homomultimeric interactions of AtWRKY18, AtWRKY36, and AtWRKY60 and heteromultimers AtWRKY17-18 and AtWRKY38-40 (Arabidopsis Interactome Mapping Consortium, 2011). Here, multimeric complexes were identified of AtWRKY13 with itself, AtWRKY40, and AtWRKY57 (Fig. 7). The precise makeup and stoichiometry of these complexes need further investigation. Given the complex interactions of repression and activation by different transcription factors, it is reasonable to speculate that they accomplish this in a coordinated or competitive manner. It is thus conceivable that AtWRKY40 and AtWRKY63 may achieve their opposing effects by sequestration or competition for binding sites.
Another explanation for the role of WRKY transcription factors in the regulation of divergent processes may be found in the upstream signaling events leading to their activation or inactivation. WRKY transcription factors have been found to be regulated by interactions with different types of proteins such as mitogen-activated protein kinases (Kim and Zhang, 2004; Mao et al., 2011) and calmodulin (Park et al., 2005; Arabidopsis Interactome Mapping Consortium, 2011). AtWRKY40 was shown to bind the cytosolic tail of the ABA receptor (GUN5) on the chloroplast surface upon ABA treatment (Shang et al., 2010). The differential binding with WRKY or non-WRKY interaction partners seems to be the most logical explanation for the observation that some WRKY factors are found to be both repressors and activators depending on the conditions and which downstream gene is being examined.
For AtWRKY40, a significant amount of functional information is available (Xu et al., 2006; Shen et al., 2007). Here, AtWRKY40 was found to be a novel repressor of retrograde expression in response to AA. AOX1a, BCS1, and UPOX expression in response to AA treatment was up to 80% more induced in atwrky40 plants than in wild-type plants. This is only the second report of transcription factors involved in mitochondrial retrograde signaling in plants, besides ABI4 (Giraud et al., 2009). ABI4 was found to keep the AOX1a promoter repressed under normal conditions, whereas AtWRKY40 appears to limit AOX1a expression in its induced state (Fig. 5). Also here, AtWRKY40 was found to be a regulator of responses to Flg22. atwrky40 knockout plants show a reduced induction by Flg22 of BCS1, and interestingly, both AtWRKY40 overexpression lines show an even stronger repression of BCS1. Looking at transcripts in response to HL, reduced expression of AtWRKY40 triggers an overall induction while AtWRKY40 overexpression results in an overall reduction (Fig. 5; Supplemental Table S6), although inverse correlations are also found, as exemplified by ELIP2 (Fig. 5). The fact that AtWRKY40 seems to have bidirectional roles in response to biotic stimuli has been observed earlier in that AtWRKY40 mutant plants have induced expression of PR1 but reduced expression of PDF1.2 (Pandey and Somssich, 2009). During early seedling establishment, AtWRKY40 was found to repress the expression of the GUN marker genes LHCB2.4 and HEMA1. Interestingly, once plants are more established (3 weeks old), AtWRKY40 appears to have no significant control over the basal expression of these genes (Supplemental Table S3), instead modulating how plants respond to stress stimuli.
An atwrky63 mutant was found in a screen for mutants in response to ABA (Ren et al., 2010), and in agreement with this report, a reduced induction of RD29A by ABA in atwrky63 plants was also seen (Supplemental Fig. S3). Interestingly, a role for AtWRKY63 as an activator in response to Flg22 treatment was also observed. atwrky63 mutant plants showed a reduced induction of BCS1 and AOX1a (Fig. 4), while AtWRKY63 overexpression plants showed severalfold induction of BCS1 already in untreated conditions. AtWRKY63 was also identified to play a significant role in the response to HL stress. AtWRKY63 appears to act as an activator for many of the stress-responsive genes encoding mitochondrial proteins, as evidenced by the often strong superinduction following HL stress. Some redundancy for this role as an activator for AtWRKY63 appears to be in place, as the atwrky63 knockout plants showed little change compared with wild-type plants. The identity of these redundant factors, whether belonging to the WRKY family or not, still needs to be determined in future work.
For AtWRKY57, a role as a potential repressor of Flg22 signaling could be observed, as the AtWRKY57 overexpression lines consistently showed a reduced induction of BCS1 and AOX1a upon stimulation. No consistent changes were observed in the atwrky57 knockout lines, pointing at potential functional redundancy with other proteins. AtWRKY57 also appears to stimulate the basal expression of the ABA marker genes RD29A and ABI5 and represses the expression of LHCB2.4 and HEMA1 during the first days of plant development.
The data presented in this study and the results shown by other groups expand the current network of cellular responses influenced by WRKY and other transcription factors. One of the findings that became apparent in this study is the coordination of transcripts responding to both chloroplast and mitochondrial dysfunction. Previously, ABI4 has been found to be a regulator of both organelles (Koussevitzky et al., 2007; Giraud et al., 2009). This study suggests that signaling events through ABI4 and WRKY transcription factors are not directly linked. First, no direct protein-protein interactions of ABI4 and WRKY transcription factors could be detected. Furthermore, abi4 mutant lines were reported to have a GUN phenotype, namely that the expression of chloroplast marker genes is not repressed as in wild-type plants by treatment with norflurazon (Koussevitzky et al., 2007). Such a GUN phenotype was not observed in wrky mutant lines; rather, an effect on the constitutive expression during seedling establishment was seen. Finally, ABI4 was found to be a repressor of AOX1a and needs to be relieved for AOX1a induction after the inhibition of mitochondrial function (Giraud et al., 2009), whereas AtWRKY40 keeps AOX1a induction levels in check once stress is occurring.
CONCLUSION
Although chloroplasts and mitochondria are physically separated from each other, their essential roles in energy metabolism necessitate a regulatory system that optimizes cross talk and interactions between the two organelles under both normal and stress conditions. Therefore, it makes sense that there are common transcription factors able to coordinate gene expression when these organelles are functionally compromised.
MATERIALS AND METHODS
Plant Growth Conditions
Arabidopsis (Arabidopsis thaliana) Col-0 seeds were sterilized using chlorine gas (100 mL of 12% NaOCl and 3 mL of 37% HCl). In vitro plants were grown on MS medium (4.3 g L−1 MS [Duchefa], 0.5 g L−1 MES, 20 g L−1 Suc, 8 g L−1 agar [LabM], and 1 mL L−1 Gamborg B5 vitamins [Duchefa]) at 22°C and 100 μE m–2 s–1 radiation in a 16-h-light/8-h-dark photoperiod. For stress treatments, plants were grown in soil for 3 weeks and sprayed with 50 μm AA (Sigma-Aldrich), 10 μm Flg22 (Biomatik), or 200 μm ABA (Sigma-Aldrich). Plants treated with AA were snap frozen after 4 h, those treated with Flg22 after 45 min, and those treated with ABA after 6 h. For the HL treatment, plants were exposed to 1,000 μE m–2 s–1 for 1 h and snap frozen. Arabidopsis Landsberg erecta cell cultures were grown under continuous-light conditions with shaking and treated with 100 mm H2O2.
T-DNA Insertion Lines and Overexpression Lines
T-DNA insertion lines as shown in Table I were obtained from the Arabidopsis Biological Resource Center. Plants were genotyped by PCR, and insertion positions were confirmed by sequencing. Primer sequences are listed in Supplemental Table S2. atwrky40 and atwrky18 atwrky40 atwrky60 mutant lines were kindly obtained from Prof. Da-Peng Zhang (Tsinghua University). AtWRKY75 RNA interference lines were kindly provided by Prof. Kashchandra Raghothama (Purdue University). WRKY overexpression lines were produced by cloning the coding sequences into cauliflower mosaic virus 35S expression vector pK7WG2 (Karimi et al., 2002) using the Gateway cloning system (Invitrogen) and dipped into Arabidopsis Col-0 plants according to Clough and Bent (1998). Homozygous plants with a single T-DNA locus were selected on MS medium containing 35 mg L−1 kanamycin. Overexpression was confirmed using QRT-PCR (for primer sequences, see Supplemental Table S2).
QRT-PCR
QRT-PCR was performed on snap-frozen Arabidopsis tissue. Total RNA isolation and complementary DNA (cDNA) synthesis were carried out as described previously (Lister et al., 2004). Transcript levels were assayed using the LightCycler 480 (Roche). From each cDNA preparation, transcripts were analyzed in duplicate and normalized to UBC as a housekeeping gene. QRT-PCR primers used for the genes AOX1a, BCS1, NDB2, UPOX, and UBC have been described previously (Clifton et al., 2005); additional primers are shown in Supplemental Table S2. Statistical analysis was performed using Student’s t test.
Cloning of Arabidopsis Promoter Regions
Promoter regions were cloned using standard protocols into pDRIVE (Qiagen) and subcloned into pLUS as described (Ho et al., 2008). The constructs were made as translational fusions with GUS, with the first ATG of the gene used as the start codon for GUS. The elements to be tested (Fig. 1) were mutated in the promoter via site-directed mutagenesis using the Stratagene Quikchange II site-directed mutagenesis kit.
Biolistic Transformation and Assays for Luc and GUS
Transformation was performed using the PDS-1000 system using the Hepta adaptor according to the manufacturer’s instructions (Bio-Rad), as described previously using Arabidopsis suspension cell cultures (Ho et al., 2008). se values are shown, and to determine statistical significance, Student’s t test was performed assuming unequal variances. For comparison of GUS activities of the motif deletions with that of the unmutated promoter, Student’s t test was also performed. Significance was defined as P ≤ 0.05. The following comparisons were carried out to determine the activity of each element as presented in Figure 1: (1) comparison of normalized GUS activity between wild-type promoter and mutated promoter (significance is indicated with blue asterisks); (2) comparison between mock-treated and stress-treated GUS values (significance is indicated with red asterisks).
Construction of Yeast One-Hybrid Vectors
Coding regions of WRKY transcription factors were cloned from Arabidopsis Col-0 cDNA into pDRIVE (Qiagen) or by recombination directly into pGADT7-rec2 (according to the Clontech handbook) with the Roche Expand High Fidelity PCR System (primers are shown in Supplemental Table S2). The PCR products in pDRIVE were then subcloned into the pGADT7-rec2 prey vector (Clontech). As a control, the binding capacity of the p53 transcription factor to a DNA sequence containing (p53+) or not containing (p53−) a p53-binding motif was used. For construction of the pHIS2 bait vectors, forward and reverse oligonucleotides (Supplemental Table S2) were annealed and subcloned into EcoRI-SacI-linearized pHIS2 vector. The 50-bp sequence surrounding the W-box elements was cloned into pHIS2 upstream of the HIS3 promoter region and the HIS3 reporter gene.
Yeast One-Hybrid Screen
Yeast one-hybrid transformation screens were performed in Saccharomyces cerevisiae strain Y187 using the Clontech Matchmaker One Hybrid kit (Clontech) and optimized for transformation on 96-well deep-well sterile plates (Axygen). For each yeast one-hybrid transformation, 50 μL of competent yeast cells was incubated with 100 ng of pHIS2 bait vector and 100 ng of pGADT7-Rec2 prey vector, 100 µg of Herring Testes Carrier DNA (Clontech), and 0.3 mL of polyethylene glycol/lithium acetate solution. Cells were transformed with 35 μL of dimethyl sulfoxide at 42°C for 15 min and then cooled on ice. Ten microliters of transformation was spotted onto synthetic dextrose (SD) medium –Leu –Trp (double dropout [DDO]) to select cotransformed cells and SD medium –His –Leu –Trp containing 150 mm 3-amino-1,2,4-triazole (triple dropout [TDO]; Sigma-Aldrich). Plates were incubated 4 d at 28°C. The pGADT7-rec2-p53 prey vector in combination with p53HIS2 was used as a positive control, and pGADT7-rec2-p53 with pHIS2 or pGADT7-rec2-ABF4 with p53pHIS2 were used as negative controls. For specific confirmations in Figure 3, transformed yeast cells were grown overnight in yeast peptone dextrose liquid medium to an optical density at 600 nm of 0.1 and diluted in a 10× dilution series. From each dilution, 10 µL was spotted on DDO and on TDO plates. Plates were incubated for 3 d at 28°C.
Electromobility Shift Assays
Oligonucleotide probes (30 bp; Supplemental Table S2) with W-box or mutated W-box were annealed by heating to 99°C and gradual cooling. Annealed probes were radiolabeled using [γ-32P]ATP (Perkin-Elmer) and polynucleotide kinase (Roche) and purified using Sephadex G-25 radiolabeled DNA Quick Spin columns (Roche). WRKY proteins were cloned into the glutathione S-transferase tag expression vector pDEST15 (Invitrogen) and transformed into Escherichia coli Rosetta 2 (DE3) pLysS competent expression cells. Proteins were expressed in 500 mL of culture for 4 h with 0.2 mm isopropyl-β-d-thiogalactoside at 18°C and 250 rpm. Cells were harvested, resuspended in extraction buffer (5× extraction buffer = 250 mm Tris-Cl, pH 8.5, 500 mm NaCl, 5 mm EDTA, and 1 mm dithiothreitol), and lysed by sonication. Lysate was centrifuged at 16,000g for 20 min to clarify. Filtered lysate was then incubated with glutathione-agarose beads (Scientifix) and washed with 10 volumes of extraction buffer containing Complete no-EDTA protease inhibitor cocktail (Roche). Purified proteins were eluted using extraction buffer containing 10 mm reduced glutathione. For gel-shift assays, 20-μL reactions were set up with 4 μL of 5× binding buffer (100 mm HEPES, pH 7.8, 0.5 m KCl, 5 mm MgCl2, 2.5 mm dithiothreitol, 5 mm EDTA, 0.25 mg mL−1 poly[dI-dC], and 50% glycerol), 1 fmol of radiolabeled probe (500 fmol of unlabeled probe for competitor reactions), and 1.5 μg of purified protein extract. Reactions were incubated for 20 min and separated on polyacrylamide gels (0.5× Tris-borate/EDTA, 2.5% glycerol, and 6% acrylamide) for 2 h at 200 V on a 16- × 20-cm2 Bio-Rad Protean II gel system. Gels were then dried on Whatman paper in a gel dryer, exposed overnight or longer, and visualized using phosphorimager detection plates.
Yeast Two-Hybrid Analysis
Full-length coding sequences were subcloned into the pGBKT7 and pGADT7-rec yeast two-hybrid vectors (Clontech). pGBKT7 vectors were transformed into Y187 yeast cells and pGADT7-rec vectors into AH109. Transformed cells were mated overnight in yeast peptone dextrose with gentle shaking at 28°C and spotted onto DDO and quadruple dropout (SD –Leu –Trp –His –adenine) media with or without 10 mm 5-bromo-4-chloro-3-indolyl α-d-galactopyranoside. Plates were incubated at 28°C for 1 week until interactions were clearly visible.
Subcellular Localization
Coding sequences were cloned into GFP fusion vectors as described (Carrie et al., 2008). AtWRKY13 was cloned into pDEST-NGFP and AtWRKY57 was cloned into pDEST-CGFP. As a mitochondrial marker, the AOX targeting signal of 42 amino acids fused to red fluorescent protein (RFP) was used (Carrie et al., 2008). Biolistic cotransformation of the GFP and RFP fusion vectors was performed on Arabidopsis cell culture and onion (Allium cepa) epidermal cells as reported previously (Carrie et al., 2008). GFP and RFP expression and targeting were visualized using a BX61 Olympus microscope using excitation wavelengths of 460/480 nm (GFP) and 535/555 nm (RFP) and emission wavelengths of 495 to 540 nm (GFP) and 570 to 625 nm (RFP). Images were captured using CellR imaging software.
Microarray Analysis
Analysis of the changes in transcript abundance between Col-0 and double mutants was performed using Affymetrix GeneChip Arabidopsis ATH1 Genome Arrays along with preliminary data quality assessment, as described previously (Zhang et al., 2012). In total, 16,993 probe sets were deemed present by the MAS5.0 algorithm. Once processed, Guanosine-Cytosine Robust Multiarray Average-normalized gene expression values were analyzed to identify differentially expressed genes by a regularized Student’s t test based on a Bayesian statistical framework using the software program Cyber-T (Kayala and Baldi, 2012). Changes were considered significant at a false discovery rate correction level of PPDE(<P) > 0.95. Representations of gene categories were calculated using χ2 statistical tests. Lists of energy organelle proteins and expression values of the selected marker genes in publicly available microarray experiments were collected as described by Van Aken and Whelan (2012).
The microarray data are publicly available from the Gene Expression Omnibus under accession number GSE46107.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Self-activation assay and example of a large-scale yeast one-hybrid screen of WRKY transcription factors.
Supplemental Figure S2. QRT-PCR expression analysis of AtWRKY transgenic lines.
Supplemental Figure S3. QRT-PCR expression analysis of AtWRKY transgenic lines.
Supplemental Figure S4. The expression of 18 probe sets commonly affected by AtWRKY40 and AtWRKY63 knockout/overexpression.
Supplemental Table S1. Yeast one-hybrid results.
Supplemental Table S2. Primers.
Supplemental Table S3. High-light QRT-PCR results.
Supplemental Table S4. Averaged Guanosine-Cytosine Robust Microarray Average-normalized microarray expression data.
Supplemental Table S5. Cyber-T microarray analysis.
Supplemental Table S6. Statistical analysis of gene class representations.
Acknowledgments
We thank the Arabidopsis Biological Resource Center for T-DNA insertional mutants, and we thank Ellen Paynter and Lin Xu for technical assistance.
Glossary
- ABA
abscisic acid
- H2O2
hydrogen peroxide
- QRT
quantitative reverse transcription
- UTR
untranslated region
- AA
antimycin A
- Col-0
ecotype Columbia
- HL
high light
- MS
Murashige and Skoog
- VHR
very highly responsive
- VWR
very weakly responsive
- SD
synthetic dextrose
- DDO
double dropout
- TDO
triple dropout
- RFP
red fluorescent protein
- cDNA
complementary DNA
- T-DNA
transfer DNA
- PPDE
posterior probability of differential expression
References
- Arabidopsis Interactome Mapping Consortium (2011) Evidence for network evolution in an Arabidopsis interactome map. Science 333: 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barajas-López JdD, Serrato AJ, Cazalis R, Meyer Y, Chueca A, Reichheld JP, Sahrawy M. (2011) Circadian regulation of chloroplastic f and m thioredoxins through control of the CCA1 transcription factor. J Exp Bot 62: 2039–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrie C, Murcha MW, Kuehn K, Duncan O, Barthet M, Smith PM, Eubel H, Meyer E, Day DA, Millar AH, et al (2008) Type II NAD(P)H dehydrogenases are targeted to mitochondria and chloroplasts or peroxisomes in Arabidopsis thaliana. FEBS Lett 582: 3073–3079 [DOI] [PubMed] [Google Scholar]
- Choi H, Hong J, Ha J, Kang J, Kim SY. (2000) ABFs, a family of ABA-responsive element binding factors. J Biol Chem 275: 1723–1730 [DOI] [PubMed] [Google Scholar]
- Clifton R, Lister R, Parker KL, Sappl PG, Elhafez D, Millar AH, Day DA, Whelan J. (2005) Stress-induced co-expression of alternative respiratory chain components in Arabidopsis thaliana. Plant Mol Biol 58: 193–212 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dojcinovic D, Krosting J, Harris AJ, Wagner DJ, Rhoads DM. (2005) Identification of a region of the Arabidopsis AtAOX1a promoter necessary for mitochondrial retrograde regulation of expression. Plant Mol Biol 58: 159–175 [DOI] [PubMed] [Google Scholar]
- Estavillo GM, Crisp PA, Pornsiriwong W, Wirtz M, Collinge D, Carrie C, Giraud E, Whelan J, David P, Javot H, et al. (2011) Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 23: 3992–4012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Somssich IE. (2007) Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol 10: 366–371 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ. (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadjev I, Vanderauwera S, Gechev TS, Laloi C, Minkov IN, Shulaev V, Apel K, Inzé D, Mittler R, Van Breusegem F. (2006) Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol 141: 436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E, Ng S, Carrie C, Duncan O, Low J, Lee CP, Van Aken O, Millar AH, Murcha M, Whelan J. (2010) TCP transcription factors link the regulation of genes encoding mitochondrial proteins with the circadian clock in Arabidopsis thaliana. Plant Cell 22: 3921–3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E, Van Aken O, Ho LH, Whelan J. (2009) The transcription factor ABI4 is a regulator of mitochondrial retrograde expression of ALTERNATIVE OXIDASE1a. Plant Physiol 150: 1286–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E, Van Aken O, Uggalla V, Whelan J. (2012) Redox regulation of mitochondrial function in plants. Plant Cell Environ 35: 271–280 [DOI] [PubMed] [Google Scholar]
- Grunewald W, Karimi M, Wieczorek K, Van de Cappelle E, Wischnitzki E, Grundler F, Inzé D, Beeckman T, Gheysen G. (2008) A role for AtWRKY23 in feeding site establishment of plant-parasitic nematodes. Plant Physiol 148: 358–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Yagi M, Kusano T, Sano H. (2000) Rapid systemic accumulation of transcripts encoding a tobacco WRKY transcription factor upon wounding. Mol Gen Genet 263: 30–37 [DOI] [PubMed] [Google Scholar]
- Ho LH, Giraud E, Uggalla V, Lister R, Clifton R, Glen A, Thirkettle-Watts D, Van Aken O, Whelan J. (2008) Identification of regulatory pathways controlling gene expression of stress-responsive mitochondrial proteins in Arabidopsis. Plant Physiol 147: 1858–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson D, Guevara D, Yaish MW, Hannam C, Long N, Clarke JD, Bi YM, Rothstein SJ. (2011) GNC and CGA1 modulate chlorophyll biosynthesis and glutamate synthase (GLU1/Fd-GOGAT) expression in Arabidopsis. PLoS ONE 6: e26765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Deyholos MK. (2009) Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol Biol 69: 91–105 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kayala MA, Baldi P. (2012) Cyber-T Web server: differential analysis of high-throughput data. Nucleic Acids Res 40: W553–W559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CY, Zhang S. (2004) Activation of a mitogen-activated protein kinase cascade induces WRKY family of transcription factors and defense genes in tobacco. Plant J 38: 142–151 [DOI] [PubMed] [Google Scholar]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J. (2007) Signals from chloroplasts converge to regulate nuclear gene expression. Science 316: 715–719 [PubMed] [Google Scholar]
- Leister D, Wang X, Haberer G, Mayer KF, Kleine T. (2011) Intracompartmental and intercompartmental transcriptional networks coordinate the expression of genes for organellar functions. Plant Physiol 157: 386–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Fu Q, Chen L, Huang W, Yu D. (2011) Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 233: 1237–1252 [DOI] [PubMed] [Google Scholar]
- Lister R, Chew O, Lee MN, Heazlewood JL, Clifton R, Parker KL, Millar AH, Whelan J. (2004) A transcriptomic and proteomic characterization of the Arabidopsis mitochondrial protein import apparatus and its response to mitochondrial dysfunction. Plant Physiol 134: 777–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao G, Meng X, Liu Y, Zheng Z, Chen Z, Zhang S. (2011) Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23: 1639–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormac AC, Terry MJ. (2004) The nuclear genes Lhcb and HEMA1 are differentially sensitive to plastid signals and suggest distinct roles for the GUN1 and GUN5 plastid-signalling pathways during de-etiolation. Plant J 40: 672–685 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J. (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126: 467–475 [DOI] [PubMed] [Google Scholar]
- Pandey SP, Somssich IE. (2009) The role of WRKY transcription factors in plant immunity. Plant Physiol 150: 1648–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CY, Lee JH, Yoo JH, Moon BC, Choi MS, Kang YH, Lee SM, Kim HS, Kang KY, Chung WS, et al. (2005) WRKY group IId transcription factors interact with calmodulin. FEBS Lett 579: 1545–1550 [DOI] [PubMed] [Google Scholar]
- Ren X, Chen Z, Liu Y, Zhang H, Zhang M, Liu Q, Hong X, Zhu JK, Gong Z. (2010) ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J 63: 417–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschzttardtz H, Fuentes I, Vásquez M, Corvalán C, León G, Gómez I, Araya A, Holuigue L, Vicente-Carbajosa J, Jordana X. (2009) A nuclear gene encoding the iron-sulfur subunit of mitochondrial complex II is regulated by B3 domain transcription factors during seed development in Arabidopsis. Plant Physiol 150: 84–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton DL, Tripathi P, Rabara RC, Lin J, Ringler P, Boken AK, Langum TJ, Smidt L, Boomsma DD, Emme NJ, et al. (2012) WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnol J 10: 2–11 [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Macdonald H, Huttly AK, Lazarus CM, Hooley R. (1995) Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of alpha-Amy2 genes. Plant Mol Biol 29: 691–702 [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ. (2010) WRKY transcription factors. Trends Plant Sci 15: 247–258 [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Torres JT, Parniske M, Wernert P, Hahlbrock K, Somssich IE. (1996) Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J 15: 5690–5700 [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Yan L, Liu ZQ, Cao Z, Mei C, Xin Q, Wu FQ, Wang XF, Du SY, Jiang T, et al. (2010) The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 22: 1909–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, Ulker B, Somssich IE, Schulze-Lefert P. (2007) Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315: 1098–1103 [DOI] [PubMed] [Google Scholar]
- Skibbe M, Qu N, Galis I, Baldwin IT. (2008) Induced plant defenses in the natural environment: Nicotiana attenuata WRKY3 and WRKY6 coordinate responses to herbivory. Plant Cell 20: 1984–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aken O, Pecenková T, van de Cotte B, De Rycke R, Eeckhout D, Fromm H, De Jaeger G, Witters E, Beemster GT, Inzé D, et al. (2007) Mitochondrial type-I prohibitins of Arabidopsis thaliana are required for supporting proficient meristem development. Plant J 52: 850–864 [DOI] [PubMed] [Google Scholar]
- Van Aken O, Whelan J. (2012) Comparison of transcriptional changes to chloroplast and mitochondrial perturbations reveals common and specific responses in Arabidopsis. Front Plant Sci 3: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aken O, Zhang B, Carrie C, Uggalla V, Paynter E, Giraud E, Whelan J. (2009) Defining the mitochondrial stress response in Arabidopsis thaliana. Mol Plant 2: 1310–1324 [DOI] [PubMed] [Google Scholar]
- Xu X, Chen C, Fan B, Chen Z. (2006) Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18: 1310–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Carrie C, Ivanova A, Narsai R, Murcha MW, Duncan O, Wang Y, Law SR, Albrecht V, Pogson B, et al. (2012) LETM proteins play a role in the accumulation of mitochondrially encoded proteins in Arabidopsis thaliana and AtLETM2 displays parent of origin effects. J Biol Chem 287: 41757–41773 [DOI] [PMC free article] [PubMed] [Google Scholar]