A directed genetic screen produced auxin receptors with increased activity both in vitro and in the plant.
Abstract
The phytohormone auxin regulates virtually every aspect of plant development. The hormone directly mediates the interaction between the two members of the auxin coreceptor complex, a TRANSPORT INHIBITOR RESPONSE (TIR1)/AUXIN SIGNALING F-BOX protein and an AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) transcriptional repressor. To learn more about the interaction between these proteins, a mutant screen was performed using the yeast (Saccharomyces cerevisiae) two-hybrid system in Arabidopsis (Arabidopsis thaliana). Two tir1 mutations were identified that increased interaction with Aux/IAAs. The D170E and M473L mutations increase affinity between TIR1 and the degron motif of Aux/IAAs and enhance the activity of the SCFTIR1 complex. This resulted in faster degradation of Aux/IAAs and increased transcription of auxin-responsive genes in the plant. Plants carrying the pTIR1:tir1 D170E/M473L-Myc transgene exhibit diverse developmental defects during plant growth and display an auxin-hypersensitive phenotype. This work demonstrates that changes in the leucine-rich repeat domain of the TIR1 auxin coreceptor can alter the properties of SCFTIR1.
The plant hormone indole-3-acetic acid (IAA) is the most important natural auxin, with the ability to regulate virtually every aspect of plant development (Woodward and Bartel, 2005; Möller and Weijers, 2009; Sundberg and Østergaard, 2009; Takahashi et al., 2009; Overvoorde et al., 2010; Vernoux et al., 2010). Auxin signaling is mediated by at least three protein families: the TRANSPORT INHIBITOR RESPONSE/AUXIN SIGNALING F-BOX (TIR1/AFB) proteins, the AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) transcriptional repressors, and the AUXIN RESPONSE FACTOR (ARF) transcription factors. At low auxin levels, the transcriptional activity of the ARF proteins is inhibited through an interaction with an Aux/IAA protein and the corepressor TOPLESS (Reed, 2001; Tiwari et al., 2001; Weijers et al., 2005; Guilfoyle and Hagen, 2007; Szemenyei et al., 2008). Auxin promotes the recruitment of the Aux/IAA protein to the SCFTIR1/AFB E3 ubiquitin ligase, leading to degradation of the Aux/IAA proteins by the ubiquitin-proteasome pathway, thus allowing ARF-dependent transcription (Ruegger et al., 1998; Gray et al., 2001; Mockaitis and Estelle, 2008).
In Arabidopsis (Arabidopsis thaliana), 29 Aux/IAAs have been identified, most of which share a similar protein structure (Reed, 2001). Domain I of the Aux/IAAs binds TOPLESS and is required for transcriptional repression (Tiwari et al., 2004; Szemenyei et al., 2008). The core region of domain II is a six-amino acid sequence (VGWPPV/I) called the degron, which is required for proteolytic degradation of the Aux/IAA proteins (Worley et al., 2000; Ramos et al., 2001; Dreher et al., 2006). Several dominant or semidominant gain-of-function mutants, mutated within the conserved degron sequence, have been isolated (Reed, 2001). Later studies showed that these mutations stabilize the affected Aux/IAAs, leading to auxin resistance (Worley et al., 2000; Ramos et al., 2001). Domains III and IV mediate homodimerization and heterodimerization, including with the ARF proteins (Mockaitis and Estelle, 2008).
TIR1 is a member of a small family of F-box proteins that contains five additional AFB proteins, AFB1 to AFB5 (Dharmasiri et al., 2005b). All six members function as auxin receptors (Dharmasiri et al., 2005a, 2005b; Greenham et al., 2011). The TIR1 structure in complex with auxin, ASK1 (for Arabidopsis SKP1-LIKE1), and the degron motif from IAA7 revealed that the 18 Leu-rich repeat (LRR) domain forms an auxin-binding pocket and binding surface for the Aux/IAA protein (Tan et al., 2007). Auxin acts as a “molecular glue” to direct the interaction between TIR1 and the degron motif of Aux/IAAs (Tan et al., 2007). Recent studies have shown that different combinations of TIR1/AFB and the Aux/IAA proteins form coreceptor complexes with a wide range of auxin-binding affinities, which may function in different auxin-regulated processes (Navarro et al., 2006; Parry et al., 2009; Vidal et al., 2010; Calderón Villalobos et al., 2012).
To learn more about the interactions between auxin, TIR1/AFBs, and Aux/IAAs, we performed a mutant screen based on the yeast (Saccharomyces cerevisiae) two-hybrid system (Prigge et al., 2010; Calderón Villalobos et al., 2012). Two TIR1 mutations, D170E and M473L, were identified that increase the interaction between TIR1 and Aux/IAAs. We present data showing that these two mutations increase the affinity between TIR1 and the degron motif of Aux/IAAs and further enhance the degradation of Aux/IAAs in the plant. Plants expressing the mutant genes exhibit an auxin-hypersensitive phenotype.
RESULTS
Identification of TIR1 Mutations with Increased Affinity for Aux/IAAs
Previous studies showed that the LexA yeast two-hybrid system can be used to study the interaction between auxin receptors TIR1/AFB and their substrates the Aux/IAAs (Prigge et al., 2010; Calderón Villalobos et al., 2012). The TIR1/AFB proteins interact with the Aux/IAAs in an auxin concentration-dependent manner. However, the interaction does not occur between the TIR1/AFB proteins and degron-mutated Aux/IAAs even in the presence of auxin (Prigge et al., 2010; Calderón Villalobos et al., 2012).
Using error-prone PCR, random mutations were introduced into full-length TIR1 to create an extensive library of TIR1 mutants. The TIR1 mutant library was then fused to the LexA DNA-binding domain (DBD) and introduced into a strain expressing the Aux/IAA-12 (IAA12) protein fused to the B42 activation domain (AD). IAA12 was chosen because it has a relatively low affinity for TIR1, making it easer to identify mutations that increase the interation. By screening the mutant library, we searched for TIR1 mutations that increased the interaction with AD-IAA12 upon auxin treatment. Twelve yeast colonies were recovered in the screen, and the tir1 plasmids were sequenced.
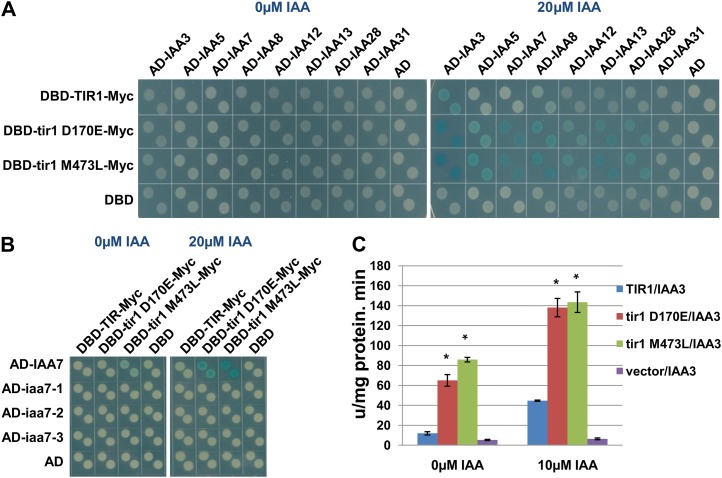
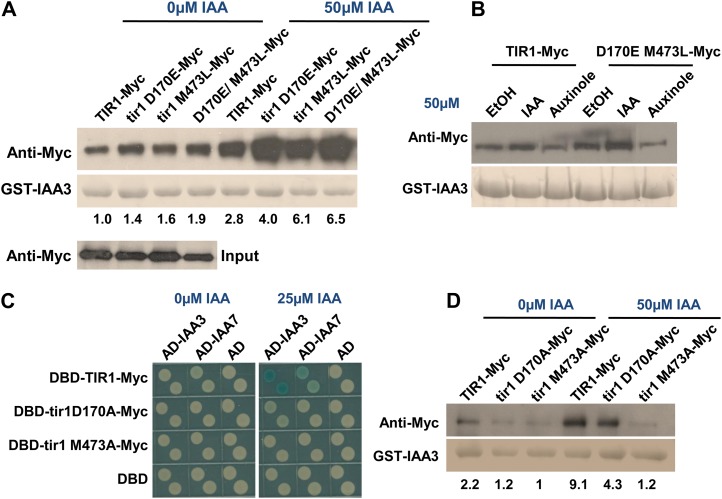
Two TIR1 mutations, D170E and M473L, were identified that increase the strength of the interaction between TIR1 and IAA12 compared with control yeast colonies as measured by β-galactosidase activity (Fig. 1A). To determine if this behavior was specific for IAA12, we tested the mutant TIR1 proteins against a number of additional Aux/IAA proteins. In every case, the mutations increased the level of β-galactosidase activity (Fig. 1A). Furthermore, the TIR1 mutant proteins did not interact with degron-mutated IAA7 mutants in the presence or absence of auxin (Fig. 1B). The results of protein blots show that the TIR1 mutants are expressed at a similar level to the wild type, suggesting that the enhanced interaction in yeast may be caused by changes in affinity between TIR1 and Aux/IAAs (Supplemental Fig. S1A). To address this possibility, glutathione S-transferase (GST)-tagged IAA3 was purified from Escherichia coli and an in vitro pull-down assay was performed with mutant and wild-type TIR1-Myc synthesized by Transcription/Translation (TnT)-coupled wheat germ extract. For this experiment, we also generated a double mutant protein carrying both the D170E and M473L substitutions. The results show that GST-IAA3 pulls down more mutant protein than the wild-type TIR1 both in the absence and presence of auxin (Fig. 2A). In addition, the effects of each mutation were additive, such that the double mutant protein had even higher affinity for IAA3. To further verify this result, the tir1 D170E/M473L-Myc construct regulated by the native TIR1 promoter was generated and introduced into a pHS:AXR3NT-GUS transgenic line. This line can express domains I and II of AXR3/IAA17 (AXR3NT) upon heat shock and is used as a reporter of auxin-dependent degradation of Aux/IAAs (Gray et al., 2001). After heat shock and treatment with MG132, an inhibitor of the 26S proteasome, total protein extract was prepared and incubated with and without auxin. TIR1-Myc was pulled down by anti-c-Myc beads, and the amount of AXR3NT-GUS in the complex was determined. The results show that tir1 D170E/M473L-Myc pulls down much more AXR3NT-GUS than the control (Supplemental Fig. S1B). These experiments indicate that the mutations enhance the affinity between TIR1 and Aux/IAA proteins. However, the effect of the mutations can be abolished with an antiauxin compound, auxinole (Hayashi et al., 2012). Auxinole binds to TIR1 and blocks the formation of the TIR1-IAA-Aux/IAA complex. This result shows that GST-IAA3 cannot pull down additional tir1 D170E/M473L-Myc in the presence of auxinole, similar to the TIR1 control (Fig. 2B).
Figure 1.
Identification of TIR1 mutants with altered binding to the Aux/IAA proteins. A, The TIR1 mutations D170E and M473L increase the interaction between TIR1 and different Aux/IAAs upon auxin treatment. B, The mutant proteins do not interact with degron-mutated IAA7 proteins. The degron motif of IAA7 is VGWPPV, while it is mutated to VGWSPV in iaa7-1, to VGWPSV in iaa7-2, and to VGWSSV in iaa7-3. C, Measurement of β-galactosidase activity in yeast strains with and without auxin treatment. Asterisks indicate statistically significant differences between the mutants and the wild-type control (Student’s t test, P < 0.01). Error bars are se.
Figure 2.
Functions of TIR1 Asp-170 and Met-473 in Aux/IAA binding. A, In vitro pull-down assay shows that GST-IAA3 pulls down more D170E and M473L protein than wild-type TIR1 protein. B, The antiauxin compound auxinole abolishes the interaction between tir1 D170E/M473L-Myc and GST-IAA3. EtOH, Ethanol. C, Interaction between tir1D170A and tir1M473A with Aux/IAAs assessed by yeast two-hybrid assay. The D170A mutation reduces the interaction with IAA3 and IAA7, while tir1 M473A does not interact even after auxin treatment. D, Interaction between TIR1 mutants and GST-IAA3 assessed by pull-down assay. Numbers indicate the fold change relative to the control sample.
Asp-170 and Met-473 Are Required for the Function of TIR1
The structure of TIR1 shows that Asp-170 and Met-473 are outside of the auxin-binding pocket. Asp-170 is located on the top of the LRR domain, while Met-473 is in a helix region within the hydrophobic core of this domain (Supplemental Fig. S2A). Interestingly, Asp-170 is conserved among the six TIR1/AFB proteins. In addition, the residue corresponding to TIR1 Asp-170 in AFB4 (Asp-250) is mutated to Asn in the afb4-2 mutant (Greenham et al., 2011). To determine the effects of the Asp-to-Asn substitution on TIR1, we made this mutant protein and tested it in the pull-down assay. The tir1 D170N protein has reduced interaction with GST-IAA3 (Supplemental Fig. S2B).
To further explore the function of residues Asp-170 and Met-473, the tir1 mutations D170A and M473A were generated and tested in the two-hybrid system. The results show that DBD-tir1 D170A-Myc displays a reduced interaction with AD-Aux/IAAs in the presence of auxin, while DBD-tir1 M473A-Myc does not interact with AD-Aux/IAAs compared with the control (Fig. 2C). Similar results were obtained by in vitro pull-down assay. The D170A substitution reduced the recovery of TIR1, while the M473A-Myc protein did not respond to auxin (Fig. 2D). These results indicate that Asp-170 and Met-473 are required for TIR1 function.
The TIR1 Mutants Enhance the Interaction with the Degron Motif of Aux/IAAs
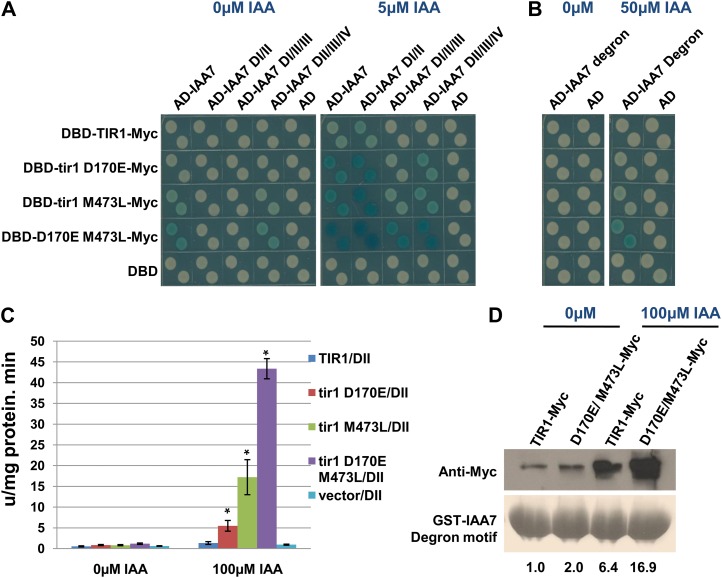
Structural studies have established the spatial relationship between TIR1 and the degron motif of IAA7. However, the structures of other domains of IAA7 and their potential interactions with TIR1 are not known. To further study the effect of D170M and M473L, the interaction between the TIR1 mutants and different fragments of IAA7 was determined by yeast two-hybrid test. The results show that the mutations increase the interaction between TIR1 and different IAA7 fragments, including DI/II, DI/II/III, and DII/III/IV, suggesting that the mutations may enhance the interaction with DII of IAA7 (Fig. 3A). To address this possibility, a yeast two-hybrid experiment was performed to test the interaction between the degron motif from IAA7 and the tir1 D170E M473L. The results show that the mutant protein interacts better with domain II of IAA7 upon auxin treatment than does the control (Fig. 3, B and C). Similar results were observed in pull-down assays. The tir1 D170E/M473L protein does not interact with GST-DI or GST-DIII/IV of IAA7 (Supplemental Fig. S1C). However, the GST-degron motif pulls down much more tir1 D170E/M473L-Myc than the TIR1-Myc control in the absence and presence of auxin (Fig. 3D). This indicates that the TIR1 mutations enhance the interaction with the degron motif of Aux/IAAs and that this effect is at least partially independent of auxin.
Figure 3.
The tir1 mutations increase binding to the IAA7 degron motif. A, Yeast two-hybrid interaction between TIR1 single and double mutant proteins and IAA7 fragments. B, Interaction between tir1 D170E/M473L and the IAA7 degron. C, β-Galactosidase activity in yeast cells expressing the mutants and the IAA7 degron. Error bars are se. Asterisks denote statistically significant differences between the mutants and the wild-type control (Student’s t test, P < 0.01). D, In vitro pull-down assay shows that the GST-degron motif pulls down more tir1 D170E/M473L-Myc protein than wild-type TIR1-Myc. The degron motif is from positions 83 to 92 of IAA7. Numbers indicate the fold change relative to the control sample.
To determine whether the effect of the mutations is specific to IAA, different auxins were tested, including 4-Cl-IAA, naphthylacetic acid, 2,4-dichlorophenoxyacetic acid (2,4-D), and picloram. The results show that each of these auxins except picloram dramatically increased the interaction. The behavior of picloram is consistent with earlier studies showing that TIR1 does not recognize picloram (Calderón Villalobos et al., 2012; Supplemental Fig. S1D).
The TIR1 Mutations D170E and M473L Enhance the Degradation of Aux/IAAs
Although neither Asp-170 nor Met-473 is in the F-box domain, it is possible that the mutations affect the interaction with the ASK1 adaptor protein. We tested this possibility in yeast. The results show that the TIR1 mutants exhibited a similar interaction with ASK1 as the control, suggesting that the mutations do not affect the formation of the TIR1-ASK complex (Supplemental Fig. S3).
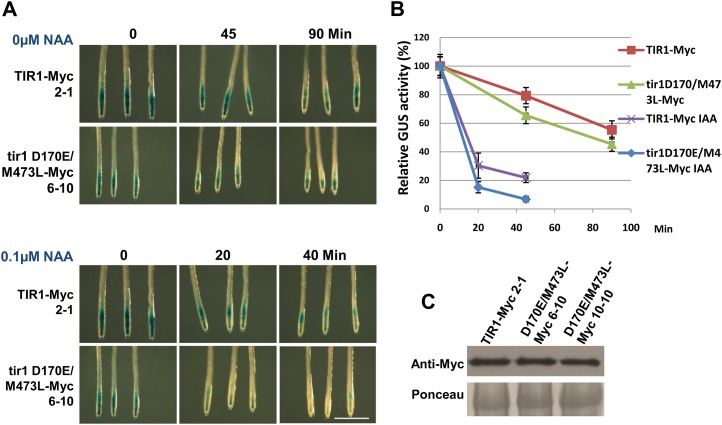
To explore the effects of the mutations on the degradation of Aux/IAAs, we used the pTIR1:tir1 D170E/M473L-Myc pHS:AXR3NT-GUS transgenic line. GUS activity was measured after heat-shock treatment. Compared with the control, the tir1 D170E/M473L-Myc transgenic seedlings display lower GUS activities after heat-shock treatment in the absence or presence of auxin, suggesting that the tir1 D170E/M473L-Myc transgenic plants degrade AXR3NT faster than the control (Fig. 4, A and B). A protein blot showed that the transgenic lines expressed TIR1 at a similar level (Fig. 4C).
Figure 4.
The ProTIR1:tir1D170E/M473L-Myc line exhibits increased degradation of AXR3NT. A and B, GUS activity of the indicated lines after heat-shock treatment for 2 h. GUS staining and measurement of GUS activity were performed in 7-d-old seedlings. NAA, 1-Naphthaleneacetic acid. Bar = 1 mm. C, Plants carrying the ProTIR1:tir1 D170E/M473L-Myc transgene express similar TIR1 protein levels to the control.
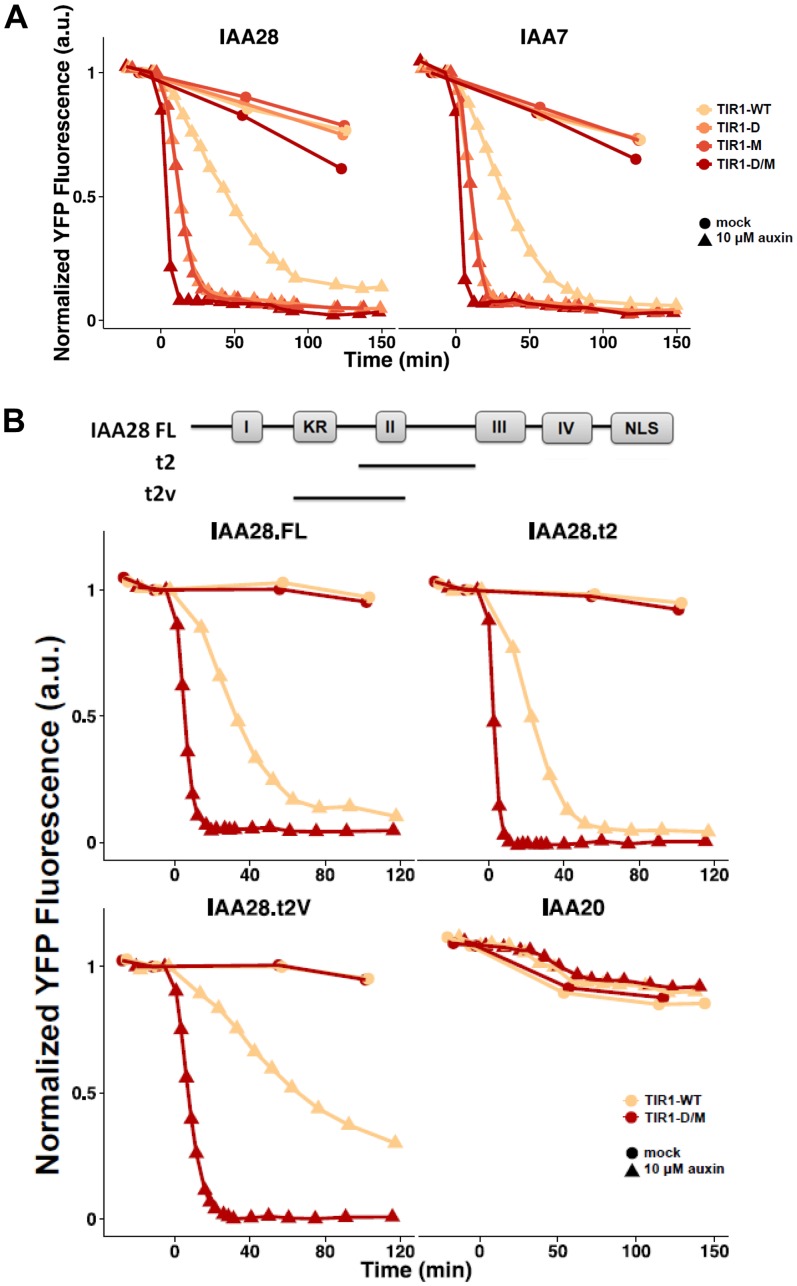
As a complement to this experiment, we also assessed the effects of the mutation on Aux/IAA degradation in yeast (Havens et al., 2012). Since eukaryotes share conserved components in the SCF-dependent degradation pathway, yellow fluorescent protein (YFP)-Aux/IAA fusion proteins are rapidly degraded by the SCFTIR1 complex upon auxin treatment in yeast (Nishimura et al., 2009; Havens et al., 2012). The amount of YFP-Aux/IAA fusion protein was determined by flow cytometry and used to calculate the Aux/IAA degradation rate (k5, according to the nomenclature of Havens et al. [2012]). Faster degrading Aux/IAAs have larger k5 values, while more stable Aux/IAAs have lower k5 values. Initially, we tested the degradation of IAA7 and IAA28. Our results show that both D170E and M473L dramatically enhance the degradation of both Aux/IAA proteins upon auxin treatment compared with the control (Fig. 5A; Table I). The mutations appear to act independently on degradation, since the effects are additive or synergistic in the double mutant. The D170E M473L double mutant also increased the rate of degradation of IAA1, IAA6, and IAA17, indicating that the mutations do not act selectively with respect to Aux/IAA protein (Supplemental Fig. S4).
Figure 5.
Degradation of YFP-IAA fusion proteins in yeast in the presence of wild-type and mutant TIR1 proteins after the addition of auxin. Yeast cells were imaged using time-lapse flow cytometry. Degradation curves were normalized to the starting fluorescence. a.u., Absorbance units; D, Asp; D/M, Asp/Met; M, Met; WT, wild type.
Table I. Degradation rates (k5 value) for various IAAs in the presence of wild-type (WT) and mutant TIR1 proteins.
k5 values were calculated as described in “Materials and Methods.”
| k5 | IAA7 | IAA28 | IAA28.FL | IAA28.t2 | IAA28.t2V | IAA20 |
|---|---|---|---|---|---|---|
| TIR1-WT | 0.023 | 0.016 | 0.024 | 0.043 | 0.009 | 0.001 |
| tir1 D170E | 0.100 | 0.081 | ||||
| tir1 M473L | 0.150 | 0.105 | ||||
| tir1 D170E/M473L | 0.418 | 0.232 | 0.290 | 0.730 | 0.210 | 0.001 |
We also examined the degradation of two truncated IAA28 proteins containing DII: t2, centered on the DII domain; and t2V, shifted N terminally to include the upstream KR sequence (Fig. 5B; Table I). Both proteins were rapidly degraded by D170E M473L, consistent with our two-hybrid and pull-down assays, indicating that the mutations affect the interaction with the DII domain. Finally, we examined the degradation of IAA20, which lacks the DII domain. This protein is stable, confirming that the DII region is required for degradation by both wild-type and mutant proteins (Fig. 5B).
tir1 Mutant Transgenic Lines Display Diverse Developmental Defects and Exhibit an Auxin-Hypersensitive Phonotype
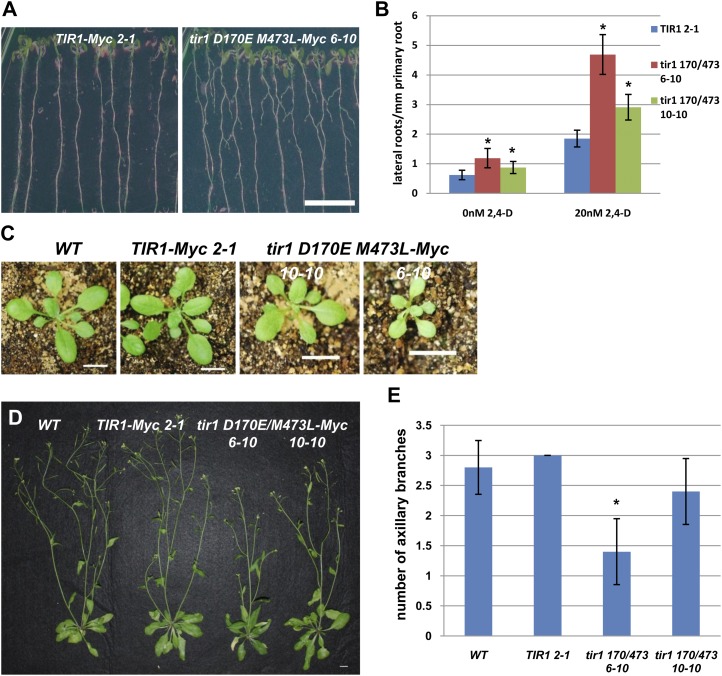
To explore the effect of the tir1 mutations on plant development, the phenotypes of two pTIR1::tir1 D170E/M473L-Myc transgenic lines, 6-10 and 10-10, were characterized. Levels of tir1 D170E/M473L-Myc are similar to the control (Fig. 4C). Compared with the control, the tir1 D170E/M473L-Myc plants initiate many more lateral roots and have smaller rosette leaves (Fig. 6, A–C). In addition, the 6-10 plants developed fewer axillary branches after bolting compared with the control (Fig. 6, D and E).
Figure 6.
ProTIR1:tir1 D170E/M473L plants exhibit growth defects. A, After 10 d of growth, ProTIR1:tir1 D170E/M473L-Myc 6-10 seedlings have more lateral roots than the wild type. B, Four-day-old seedlings were transferred onto medium with and without 2,4-D for an additional 6 d. The number of emerged lateral roots per mm of primary root length was determined. C, ProTIR1:tir1 D170E/M473L-Myc plants have smaller rosettes compared with the control line. Plants are 3 weeks old. D and E, ProTIR1:tir1 D170E/M473L 6-10 plants have fewer axillary branches compared with the control. WT, Wild type. Bar = 1 cm. Plants are 7 weeks old. For both B and E, error bars are se. Asterisks denote statistically significant differences between the tir1 D170E/M473L-Myc and TIR1-Myc control plants (Student’s t test, P < 0.05).
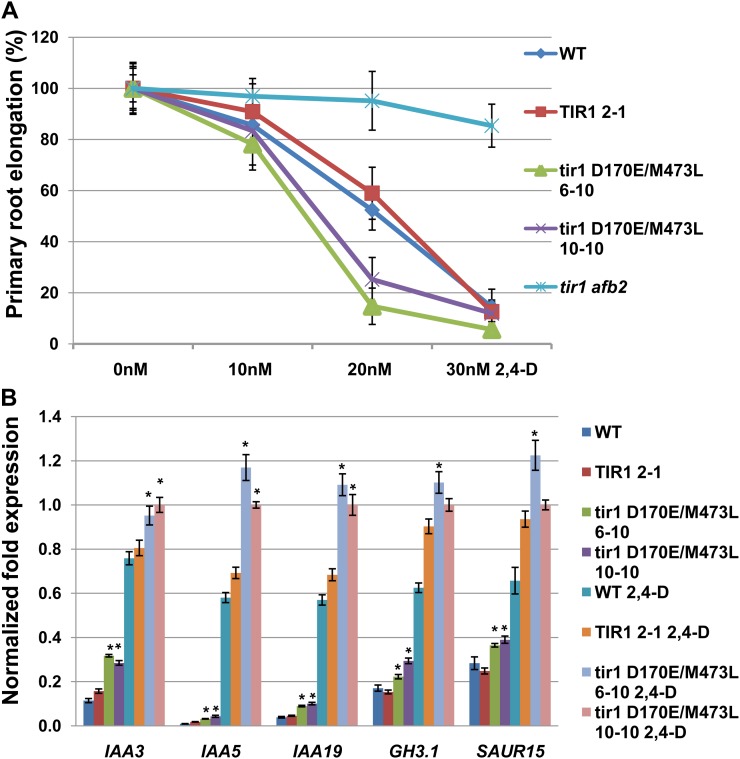
To characterize the auxin response in these lines, the effect of auxin on root growth was determined. Six-day-old seedlings were transferred onto fresh ATS (for Arabidopsis solution) medium with different concentrations of the synthetic auxin 2,4-D. After another 2 d of growth, the length of newly grown primary root was measured and expressed relative to growth on control plates. The result shows that primary root elongation is strongly inhibited in pTIR1:tir1 D170E/M473L-Myc transgenic seedlings at low concentrations of 2,4-D compared with the control (Fig. 7A). This suggests that tir1 D170E/M473L causes an auxin-hypersensitive phenotype in the plant.
Figure 7.
The ProTIR:tir1D170E/M473L lines are auxin hypersensitive. A, Root growth on increasing concentrations of 2,4-D relative to untreated controls. B, Expression of auxin-responsive genes in ProTIR1:tir1D170E/M473L-Myc plants. Seven-day-old seedlings were analyzed by quantitative PCR. Data shown are from three biological replicates. Error bars are se. Asterisks denote statistically significant differences between the tir1 D170E/M473L-Myc and TIR1-Myc control plants (Student’s t test, P < 0.05).
Next, we measured the expression of auxin-responsive genes in the tir1 D170E/M473L-Myc transgenic lines using real-time quantitative PCR. Our results show that expression of the tir1 D170E/M473L proteins increases the transcription of auxin-responsive genes in the plant. For example, the tir1 D170E/M473L-Myc transgenic seedlings displayed a significantly increased IAA3 RNA level compared with the wild-type TIR1 control with or without 2,4-D treatment (Fig. 7B). Similar effects can be observed for other auxin-responsive genes as well (Fig. 7B).
The Functions of the Mutations May Be Unique for the TIR1 Protein
TIR1 belongs to a small family of F-box proteins that contains five additional AFB proteins, AFB1 to AFB5 (Dharmasiri et al., 2005b). As noted previously, Asp-170 is conserved in all six proteins, and the afb4-2 mutation affects this residue (Greenham et al., 2011). However, we find that the corresponding Asp-to-Glu substitution in AFB1 and AFB2 does not increase binding to the Aux/IAAs (Supplemental Fig. S5, B and C). In the case of AFB2, the mutation decreased the interaction. Either Met or Leu can be found in other family members at positions homologous to TIR1 Met-473. The M468L mutation in AFB2 does not affect the interaction with the Aux/IAAs. Thus, the effects of TIR1 D170E and M473L are dependent on the TIR1 context.
DISCUSSION
The ubiquitin-proteasome pathway is one of the most important proteolytic pathways in eukaryotes. In this pathway, the small protein ubiquitin is attached to protein substrates, and the ubiquitin-protein conjugates are recognized and degraded by the 26S proteasome (Fang and Weissman, 2004). The SCF complexes are a major class of ubiquitin ligase enzyme (Gagne et al., 2002; Petroski and Deshaies, 2005). In this complex, the F-box protein plays a critical role in the determination of substrate specificity (Petroski and Deshaies, 2005). The plant hormone auxin directly induces rapid degradation of the Aux/IAA family of transcriptional repressors by SCFTIR1/AFB E3 ubiquitin ligase (Gray et al., 2001; Dharmasiri et al., 2005a, 2005b; Kepinski and Leyser, 2005; Tan et al., 2007). In Arabidopsis, TIR1 is a member of a small group of F-box proteins that also includes AFB1 through AFB5 and the jasmonic acid receptor CORONATINE INSENSITIVE1 (COI1; Dharmasiri et al., 2005b). The TIR1 protein plays a critical role in regulating most aspects of the auxin response.
In this study, two TIR1 mutations, D170E and M473L, were isolated that increase the interaction between TIR1 and the Aux/IAA proteins (Fig. 1, A and C). Further studies show that the TIR1 mutations increase the affinity for the degron motif of the Aux/IAAs (Fig. 2A; Supplemental Fig. S1B). Both Asp-170 and Met-473 are located outside of the TIR1 auxin-binding pocket and do not directly contact auxin or the degron motif of Aux/IAAs (Tan et al., 2007). However, when they are mutated to Ala, they either reduce the interaction with IAA7 for D170A or abolish the interaction in the case of M473A, indicating that these two positions are essential for TIR1 function (Fig. 2, C and D).
At this point, it is not clear how the two mutations impact Aux/IAA binding. It is possible that they induce structural changes in the auxin-binding pocket that result in more efficient contact between TIR1 and the degron motif. For example, the two oxygens in the carboxyl group of Asp-170 form hydrogen bonds with two backbone amide groups (Ala-195 and Ser-196) in an adjacent loop. The D170E substitution may strengthen these interactions, while D170A would eliminate them. Because Met-473 is within the core of the LRR domain, mutation to Ala is likely to affect folding of the domain. Met-473 is buried within the LRR domain and, hence, cannot contact other interacting proteins. Substitution of Leu for Met may result in a subtle change in the LRR domain, somehow facilitating interaction with the degron.
Experiments with the pHS:AXR3NT-GUS transgenic line show that pTIR1:tir1 D170E/M473L-Myc plants degrade AXR3 faster than the wild type, consistent with the observation that the mutations increase the affinity with Aux/IAAs and further enhance the function of the SCFTIR1 complex in yeast (Figs. 4 and 5). The root elongation of the pTIR1:tir1 D170E/M473L-Myc line is more sensitive to low concentrations of auxin treatment; meanwhile, an increased transcription of auxin-responsive genes has been observed even without auxin treatment, indicating that tir1 D170E/M473L enhances auxin signal transduction in the plant (Fig. 7).
Previous studies demonstrated that the TIR1-IAA-degron module is a powerful tool to study protein function. The auxin-inducible degron system can be used to control the degradation of target proteins upon auxin treatment in a reversible manner in nonplant cells such as yeast, chicken, mouse, hamster, monkey, and human cells (Nishimura et al., 2009; Kanke et al., 2011; Holland et al., 2012). This system is particularly useful for investigating the functions of proteins in specific developmental processes (Holland et al., 2012). In our yeast analysis, the tir1 D170E/M473L protein increases the rate of degradation of YFP-Aux/IAA7/IAA28 approximately 15- to 25-fold compared with wild-type TIR1 (Fig. 6). Our results suggest that it may be possible to modify the TIR1/AFB proteins from Arabidopsis or other plant species to increase the flexibility of the auxin-inducible degron system.
MATERIALS AND METHODS
Generation of a Mutant Library and Mutant Screen
Full-length TIR1 complementary DNA (cDNA) was mutagenized by error-prone PCR with a mutation ratio of approximately two mutations per tir1 molecule. The PCR products were ligated into the pGILDA vector and transformed into Escherichia coli competent cells to generate a library of approximately 1.3 × 104 colonies. The library was transformed into the pB42AD:IAA12 yeast (Saccharomyces cerevisiae) strain (EGY48), and yeast colonies were screened on synthetic dropout medium lacking uracil, His, Trp, and Leu, supplemented with 10 μm IAA. The number of total yeast colonies screened was about 2.5 × 105. Yeast colonies that grew fastest were isolated and tested on 5-bromo-4-chloro-indolyl-β-d-galactopyranoside plates. The pGILDA:tir1-Myc plasmids from positive colonies were extracted and sequenced.
Plant Materials and Conditions
All Arabidopsis (Arabidopsis thaliana) mutants used in this study were generated in the Columbia-0 ecotype. Seeds were surface sterilized for 20 min in 30% commercial bleach, plated on ATS medium supplemented with 0.8% agar, and stratified for 2 to 4 d at 4°C. ATS medium consists of 1% Suc, 5 mm KNO3, 2.5 mm KPO4, 2 mm MgSO4, 2 mm Ca(NO3)2, 50 μm Fe-EDTA, and 1 mL L−1 micronutrients. All seedling experiments were performed under long-day conditions (16 h of light and 8 h of dark) in a growth chamber (80 μmol m−2 s−1, 22°C), unless otherwise stated. Plants in soil were grown in long-day conditions at 22°C.
Plasmid Constructs and Generation of Transgenic Lines
The pTIR1:TIR1/tir1-Myc constructs were made by introducing a 2-kb 5′ upstream region of the TIR1 gene adjacent to the TIR1/tir1 cDNA into pGWB16 vector. In all cases, plasmids were introduced into Columbia-0 plants as described (Clough and Bent, 1998).
RNA Extraction and Real-Time Reverse Transcription-PCR
Total RNA was extracted from 7-d-old seedlings by the RNeasy Plant Mini Kit (Qiagen), and RNA yield was quantified using a Thermo Scientific NanoDrop 2000. One microgram of RNA was used for cDNA synthesis using the SuperScript III First-Strand Synthesis Kit (Invitrogen). Quantitative reverse transcription-PCR was performed as described previously (Greenham et al., 2011). Data were normalized to the reference PP2AA3 according to the comparative threshold cycle method.
Protein Extraction and Pull-Down Assays
To perform pull-down assays from plant extracts, tissue was harvested, ground to a powder in liquid nitrogen, and vortexed vigorously in extraction buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 10% glycerol, 0.1% Nonidet P-40, complete protease inhibitor [Roche], and 20 μm MG132). Cellular debris was removed by centrifugation, and total protein concentration was determined using the Bradford assay. Total protein extract (1 mg) was incubated with or without 50 μm IAA at 4°C for 4 h. TIR1-Myc was recovered with 20 μL of anti-c-Myc agarose beads (Clontech) and washed by extraction buffer five times. The protein sample was eluted by SDS-PAGE sample buffer, heated for 8 min at 90°C, cooled on ice for 2 min, and fractionated by SDS-PAGE. The AXR3NT-GUS protein was detected by immunoblotting with anti-GUS (Invitrogen) and visualized using the ECL Plus Western Blotting Detection System (Amersham).
The in vitro pull-down assay from TnT-coupled wheat germ extracts was described previously (Parry et al., 2009). Briefly, GST-Aux/IAA was expressed in E. coli and purified with glutathione agarose beads (Sigma). TIR1/tir1-Myc proteins were synthesized in the TnT-coupled wheat germ extract system (Promega). Twenty microliters of extract was incubated with more than 10 μg of GST-Aux/IAA beads in 200 μL of lysis buffer (50 mm Tris, pH 8.0, 200 mm NaCl, 10% glycerol, 0.1% Tween 20, and protease inhibitors) in the presence or absence of IAA at 4°C for 1 h. The pull-down reaction was then transferred to a Micro Bio-Spin Chromatography Column (Bio-Rad). After washing with lysis buffer, samples were eluted using reduced glutathione (Sigma) and separated by SDS-PAGE. Proteins were detected by anti-c-Myc-peroxidase antibody (Roche) and visualized.
Measurement of GUS and β-Galactosidase Activity
GUS staining was performed on 7-d-old seedlings. The seedlings were collected in GUS staining solution [100 mm Na2PO4, pH 7.0, 10 mm EDTA, 0.1% Triton X-100, 1.0 mm K3Fe(CN)6, and 2 mm 5-bromo-4-chloro-3-indolyl-β-glucuronic acid], vacuum infiltrated for 20 min, and stained overnight at 37°C. The seedlings were cleared in 70% (v/v) ethanol and imaged with a Nikon SMZ1500 dissecting microscope. 4-Methylumbelliferyl-β-d-glucuronide assays were performed to quantify GUS activity (Ge et al., 2010). To measure β-galactosidase activity, proteins were extracted from yeast cells using Y-PER Reagent (Thermo), and the yield was determined by Bradford assay. The assay was performed using 100 μL of protein extract, 200 μL of 4 mg mL−1 o-nitrophenyl-β-d-galactoside with 700 μL of Z-buffer (16.1 g of Na2HPO4·7H2O, 5.5 g of NaH2PO4·H2O, 0.75 g of KCl, and 0.246 g of MgSO4·7H2O in 1 L of water, pH 7.0) at 37°C. The reaction was stopped by adding 400 μL of 1 m Na2CO3, and the optical density at 420 nm of the supernatant was measured.
Root Inhibition Assay
For root growth assays, 6-d-old seedlings were transferred onto fresh ATS medium with different concentrations of 2,4-D for 2 d, and the length of the new primary root was measured using ImageJ software.
Aux/IAA Degradation Assays in Yeast
Yeast degradation assays were carried out as described (Havens et al., 2012). Briefly, yeast strains coexpressing stably integrated TIR1 variants and YFP-tagged IAA proteins were prepared by transferring a freshly grown colony from yeast peptone dextrose plates into synthetic complete medium. Flow cytometry was used to estimate the cell density (in events μL−1) and dilute cells such that cultures were in log phase 16 h later and for the duration of the experiment. All cultures were grown at 30°C with shaking. Preauxin measurements were taken to ascertain baseline expression, followed by the addition of auxin (10 μm IAA) or mock treatment (95% [v/v] ethanol). Measurements were acquired over the course of 120 to 150 min following auxin treatment, with intervals ranging from 3 min early in the YFP-IAA degradation phase to 20 min later in the degradation phase. Controls were measured every 1 h for the duration of the experiment.
Quantitative Analysis of the IAA Degradation Rate
Quantitative analysis of IAA degradation profiles obtained in yeast was conducted as described (Havens et al., 2012). Briefly, the auxin-induced degradation dynamics of YFP-Aux/IAA fusion proteins was characterized by a second-order differential equation model:
YFP-IAA degradation time courses were grouped according to TIR1 protein identity (wild type, D170E, M473L, or D170E/M473L) to compose smaller subsets of data. Within each group, two parameters, k3 and k5, approximate to the expression and degradation rates of an Aux/IAA protein, respectively, were allowed to vary in the estimation process while holding the rest of the parameters constant. This global fitting approach ensured that the degradation variability among the unique TIR1 and YFP-Aux/IAA pairs is represented in the estimated k5 values. The model fit residual was computed using 2 − norm of the difference between the measured and model-predicted YFP intensity at measurement times from 0 to 150 min.
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: TIR1 (At3G62980), AFB1 (At4G03190), AFB2 (At3G26810), IAA3 (At1G04240), IAA5 (At1G15580), IAA7 (At3G23050), IAA19 (At3G15540), IAA20 (At2G46990), IAA28 (At5G25890), GH3.1 (At2G14960), and GH3.3 (At2G23170).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Characterization of the TIR1 mutations D170E and M473L.
Supplemental Figure S2. The location of D170 and M473 in the TIR1 structure.
Supplemental Figure S3. The mutations do not affect the SCF complex assembly.
Supplemental Figure S4. The mutations enhance the function of the SCFtir1 complex in yeast.
Supplemental Figure S5. The effect of the mutations may be unique for the TIR1 protein.
Acknowledgments
We thank Prof. Ning Zheng and members of the Estelle and Nemhauser groups for helpful discussions.
Glossary
- IAA
indole-3-acetic acid
- Aux/IAA
auxin/indole-3-acetic acid
- LRR
Leu-rich repeat
- DBD
DNA-binding domain
- AD
activation domain
- GST
glutathione S-transferase
- 2,4-D
2,4-dichlorophenoxyacetic acid
- YFP
yellow fluorescent protein
- ATS
Arabidopsis solution
- cDNA
complementary DNA
References
- Calderón Villalobos LI, Lee S, De Oliveira C, Ivetac A, Brandt W, Armitage L, Sheard LB, Tan X, Parry G, Mao H, et al. (2012) A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol 8: 477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. (2005a) The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jürgens G, Estelle M. (2005b) Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 9: 109–119 [DOI] [PubMed] [Google Scholar]
- Dreher KA, Brown J, Saw RE, Callis J. (2006) The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18: 699–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S, Weissman AM. (2004) A field guide to ubiquitylation. Cell Mol Life Sci 61: 1546–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne JM, Downes BP, Shiu SH, Durski AM, Vierstra RD. (2002) The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci USA 99: 11519–11524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L, Peer W, Robert S, Swarup R, Ye S, Prigge M, Cohen JD, Friml J, Murphy A, Tang D, et al. (2010) Arabidopsis ROOT UVB SENSITIVE2/WEAK AUXIN RESPONSE1 is required for polar auxin transport. Plant Cell 22: 1749–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. (2001) Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414: 271–276 [DOI] [PubMed] [Google Scholar]
- Greenham K, Santner A, Castillejo C, Mooney S, Sairanen I, Ljung K, Estelle M. (2011) The AFB4 auxin receptor is a negative regulator of auxin signaling in seedlings. Curr Biol 21: 520–525 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Guilfoyle TJ, Hagen G. (2007) Auxin response factors. Curr Opin Plant Biol 10: 453–460 [DOI] [PubMed] [Google Scholar]
- Havens KA, Guseman JM, Jang SS, Pierre-Jerome E, Bolten N, Klavins E, Nemhauser JL. (2012) A synthetic approach reveals extensive tunability of auxin signaling. Plant Physiol 160: 135–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Neve J, Hirose M, Kuboki A, Shimada Y, Kepinski S, Nozaki H. (2012) Rational design of an auxin antagonist of the SCF(TIR1) auxin receptor complex. ACS Chem Biol 7: 590–598 [DOI] [PubMed] [Google Scholar]
- Holland AJ, Fachinetti D, Han JS, Cleveland DW. (2012) Inducible, reversible system for the rapid and complete degradation of proteins in mammalian cells. Proc Natl Acad Sci USA 109: E3350–E3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanke M, Nishimura K, Kanemaki M, Kakimoto T, Takahashi TS, Nakagawa T, Masukata H. (2011) Auxin-inducible protein depletion system in fission yeast. BMC Cell Biol 12: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O. (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451 [DOI] [PubMed] [Google Scholar]
- Mockaitis K, Estelle M. (2008) Auxin receptors and plant development: a new signaling paradigm. Annu Rev Cell Dev Biol 24: 55–80 [DOI] [PubMed] [Google Scholar]
- Möller B, Weijers D. (2009) Auxin control of embryo patterning. Cold Spring Harb Perspect Biol 1: a001545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JD. (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312: 436–439 [DOI] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. (2009) An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods 6: 917–922 [DOI] [PubMed] [Google Scholar]
- Overvoorde P, Fukaki H, Beeckman T. (2010) Auxin control of root development. Cold Spring Harb Perspect Biol 2: a001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry G, Calderon-Villalobos LI, Prigge M, Peret B, Dharmasiri S, Itoh H, Lechner E, Gray WM, Bennett M, Estelle M. (2009) Complex regulation of the TIR1/AFB family of auxin receptors. Proc Natl Acad Sci USA 106: 22540–22545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. (2005) Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 6: 9–20 [DOI] [PubMed] [Google Scholar]
- Prigge MJ, Lavy M, Ashton NW, Estelle M. (2010) Physcomitrella patens auxin-resistant mutants affect conserved elements of an auxin-signaling pathway. Curr Biol 20: 1907–1912 [DOI] [PubMed] [Google Scholar]
- Ramos JA, Zenser N, Leyser HM, Callis J. (2001) Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome-dependent. Plant Cell 13: 2349–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW. (2001) Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci 6: 420–425 [DOI] [PubMed] [Google Scholar]
- Ruegger M, Dewey E, Gray WM, Hobbie L, Turner J, Estelle M. (1998) The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev 12: 198–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg E, Østergaard L. (2009) Distinct and dynamic auxin activities during reproductive development. Cold Spring Harb Perspect Biol 1: a001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA. (2008) TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Miyazawa Y, Fujii N. (2009) Hormonal interactions during root tropic growth: hydrotropism versus gravitropism. Plant Mol Biol 69: 489–502 [DOI] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645 [DOI] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle TJ. (2004) Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ. (2001) AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13: 2809–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoux T, Besnard F, Traas J. (2010) Auxin at the shoot apical meristem. Cold Spring Harb Perspect Biol 2: a001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Araus V, Lu C, Parry G, Green PJ, Coruzzi GM, Gutiérrez RA. (2010) Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 4477–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Benkova E, Jäger KE, Schlereth A, Hamann T, Kientz M, Wilmoth JC, Reed JW, Jürgens G. (2005) Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J 24: 1874–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward AW, Bartel B. (2005) Auxin: regulation, action, and interaction. Ann Bot (Lond) 95: 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley CK, Zenser N, Ramos J, Rouse D, Leyser O, Theologis A, Callis J. (2000) Degradation of Aux/IAA proteins is essential for normal auxin signalling. Plant J 21: 553–562 [DOI] [PubMed] [Google Scholar]