Abstract
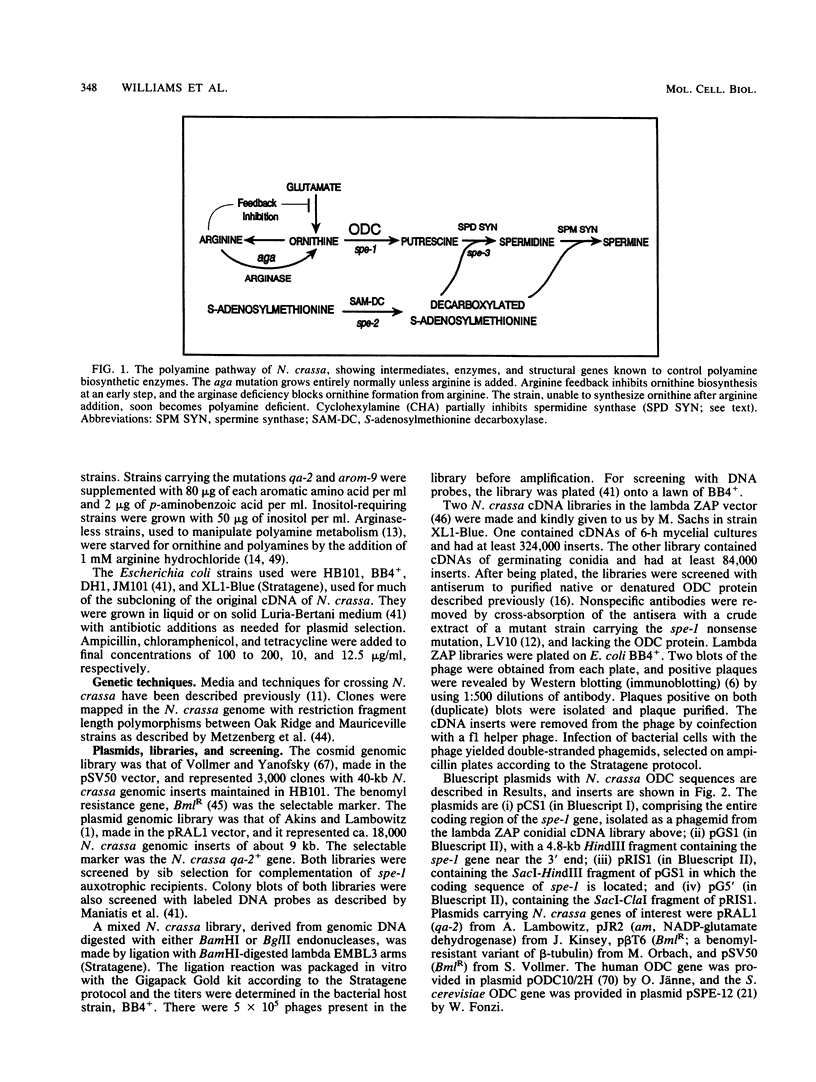
Ornithine decarboxylase (ODC), which initiates the biosynthesis of the polyamines putrescine, spermidine, and spermine, is encoded by the spe-1 gene of the fungus Neurospora crassa. This gene and its cDNA have been cloned and sequenced. The gene has a single 70-nucleotide intron in the coding sequence. The cDNA, comprising the entire coding region, recognizes a single 2.4-kb mRNA in Northern (RNA) blots. The mRNA transcript, defined by S1 mapping, has an extremely long, 535-base leader without strong secondary-structure features or an upstream reading frame. The translational start of the protein is ambiguous: a Met-Val-Met sequence precedes the Pro known to be the N terminus of the ODC polypeptide. The polypeptide encoded by the N. crassa spe-1 gene (484 amino acids) has 46% amino acid identity with that of Saccharomyces cerevisiae (466 amino acids) and 42% with that of mouse (461 amino acids). Alignment of the longer N. crassa sequence with S. cerevisiae and mouse sequences creates gaps in different sites in the S. cerevisiae and mouse sequences, suggesting that N. crassa ODC is closer to an ancestral form of the enzyme than that of either yeast or mouse ODC. N. crassa ODC, which turns over rapidly in vivo in the presence of polyamines, has two PEST sequences, found in most ODCs and other proteins with rapid turnover. In striking contrast to other eucaryotic organisms, the variation in the rate of ODC synthesis in response to polyamines in N. crassa is largely correlated with proportional changes in the abundance of ODC mRNA. Spermidine is the main effector of repression, while putrescine has a weaker effect. However, putrescine accumulation appears to increase the amount of active ODC that is made from a given amount of ODC mRNA, possibly by improving its translatability. Conversely, prolonged starvation for both putrescine and spermidine leads to the differentially impaired translation of ODC mRNA.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akins R. A., Lambowitz A. M. General method for cloning Neurospora crassa nuclear genes by complementation of mutants. Mol Cell Biol. 1985 Sep;5(9):2272–2278. doi: 10.1128/mcb.5.9.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arfin S. M., Bradshaw R. A. Cotranslational processing and protein turnover in eukaryotic cells. Biochemistry. 1988 Oct 18;27(21):7979–7984. doi: 10.1021/bi00421a001. [DOI] [PubMed] [Google Scholar]
- Barnett G. R., Seyfzadeh M., Davis R. H. Putrescine and spermidine control degradation and synthesis of ornithine decarboxylase in Neurospora crassa. J Biol Chem. 1988 Jul 15;263(20):10005–10008. [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabant M., McConlogue L., van Daalen Wetters T., Coffino P. Mouse ornithine decarboxylase gene: cloning, structure, and expression. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2200–2204. doi: 10.1073/pnas.85.7.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette W. N. "Western blotting": electrophoretic transfer of proteins from sodium dodecyl sulfate--polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981 Apr;112(2):195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- Cavener D. R., Ray S. C. Eukaryotic start and stop translation sites. Nucleic Acids Res. 1991 Jun 25;19(12):3185–3192. doi: 10.1093/nar/19.12.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm D. A convenient moderate-scale procedure for obtaining DNA from bacteriophage lambda. Biotechniques. 1989 Jan;7(1):21–23. [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Cobianchi F., Wilson S. H. Enzymes for modifying and labeling DNA and RNA. Methods Enzymol. 1987;152:94–110. doi: 10.1016/0076-6879(87)52013-4. [DOI] [PubMed] [Google Scholar]
- Davis R. H., Hynes L. V., Eversole-Cire P. Nonsense mutations of the ornithine decarboxylase structural gene of Neurospora crassa. Mol Cell Biol. 1987 Mar;7(3):1122–1128. doi: 10.1128/mcb.7.3.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. H., Krasner G. N., DiGangi J. J., Ristow J. L. Distinct roles of putrescine and spermidine in the regulation of ornithine decarboxylase in Neurospora crassa. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4105–4109. doi: 10.1073/pnas.82.12.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. H., Lawless M. B., Port L. A. Arginaseless Neurospora: genetics, physiology, and polyamine synthesis. J Bacteriol. 1970 May;102(2):299–305. doi: 10.1128/jb.102.2.299-305.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGangi J. J., Seyfzadeh M., Davis R. H. Ornithine decarboxylase from Neurospora crassa. Purification, characterization, and regulation by inactivation. J Biol Chem. 1987 Jun 5;262(16):7889–7893. [PubMed] [Google Scholar]
- Eversole P., DiGangi J. J., Menees T., Davis R. H. Structural gene for ornithine decarboxylase in Neurospora crassa. Mol Cell Biol. 1985 Jun;5(6):1301–1306. doi: 10.1128/mcb.5.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fonzi W. A. Regulation of Saccharomyces cerevisiae ornithine decarboxylase expression in response to polyamine. J Biol Chem. 1989 Oct 25;264(30):18110–18118. [PubMed] [Google Scholar]
- Fonzi W. A., Sypherd P. S. The gene and the primary structure of ornithine decarboxylase from Saccharomyces cerevisiae. J Biol Chem. 1987 Jul 25;262(21):10127–10133. [PubMed] [Google Scholar]
- Fu Y. H., Marzluf G. A. nit-2, the major nitrogen regulatory gene of Neurospora crassa, encodes a protein with a putative zinc finger DNA-binding domain. Mol Cell Biol. 1990 Mar;10(3):1056–1065. doi: 10.1128/mcb.10.3.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. H., Paietta J. V., Mannix D. G., Marzluf G. A. cys-3, the positive-acting sulfur regulatory gene of Neurospora crassa, encodes a protein with a putative leucine zipper DNA-binding element. Mol Cell Biol. 1989 Mar;9(3):1120–1127. doi: 10.1128/mcb.9.3.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geever R. F., Huiet L., Baum J. A., Tyler B. M., Patel V. B., Rutledge B. J., Case M. E., Giles N. H. DNA sequence, organization and regulation of the qa gene cluster of Neurospora crassa. J Mol Biol. 1989 May 5;207(1):15–34. doi: 10.1016/0022-2836(89)90438-5. [DOI] [PubMed] [Google Scholar]
- Ghoda L., Phillips M. A., Bass K. E., Wang C. C., Coffino P. Trypanosome ornithine decarboxylase is stable because it lacks sequences found in the carboxyl terminus of the mouse enzyme which target the latter for intracellular degradation. J Biol Chem. 1990 Jul 15;265(20):11823–11826. [PubMed] [Google Scholar]
- Ghoda L., van Daalen Wetters T., Macrae M., Ascherman D., Coffino P. Prevention of rapid intracellular degradation of ODC by a carboxyl-terminal truncation. Science. 1989 Mar 17;243(4897):1493–1495. doi: 10.1126/science.2928784. [DOI] [PubMed] [Google Scholar]
- Grens A., Scheffler I. E. The 5'- and 3'-untranslated regions of ornithine decarboxylase mRNA affect the translational efficiency. J Biol Chem. 1990 Jul 15;265(20):11810–11816. [PubMed] [Google Scholar]
- Gupta M., Coffino P. Mouse ornithine decarboxylase. Complete amino acid sequence deduced from cDNA. J Biol Chem. 1985 Mar 10;260(5):2941–2944. [PubMed] [Google Scholar]
- Heby O., Persson L. Molecular genetics of polyamine synthesis in eukaryotic cells. Trends Biochem Sci. 1990 Apr;15(4):153–158. doi: 10.1016/0968-0004(90)90216-x. [DOI] [PubMed] [Google Scholar]
- Hickok N. J., Seppänen P. J., Kontula K. K., Jänne P. A., Bardin C. W., Jänne O. A. Two ornithine decarboxylase mRNA species in mouse kidney arise from size heterogeneity at their 3' termini. Proc Natl Acad Sci U S A. 1986 Feb;83(3):594–598. doi: 10.1073/pnas.83.3.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hölttä E., Hovi T. Polyamine depletion results in impairment of polyribosome formation and protein synthesis before onset of DNA synthesis in mitogen-activated human lymphocytes. Eur J Biochem. 1985 Oct 1;152(1):229–237. doi: 10.1111/j.1432-1033.1985.tb09188.x. [DOI] [PubMed] [Google Scholar]
- Ito K., Kashiwagi K., Watanabe S., Kameji T., Hayashi S., Igarashi K. Influence of the 5'-untranslated region of ornithine decarboxylase mRNA and spermidine on ornithine decarboxylase synthesis. J Biol Chem. 1990 Aug 5;265(22):13036–13041. [PubMed] [Google Scholar]
- Kameji T., Pegg A. E. Inhibition of translation of mRNAs for ornithine decarboxylase and S-adenosylmethionine decarboxylase by polyamines. J Biol Chem. 1987 Feb 25;262(6):2427–2430. [PubMed] [Google Scholar]
- Katz A., Kahana C. Isolation and characterization of the mouse ornithine decarboxylase gene. J Biol Chem. 1988 Jun 5;263(16):7604–7609. [PubMed] [Google Scholar]
- Kiebler M., Pfaller R., Söllner T., Griffiths G., Horstmann H., Pfanner N., Neupert W. Identification of a mitochondrial receptor complex required for recognition and membrane insertion of precursor proteins. Nature. 1990 Dec 13;348(6302):610–616. doi: 10.1038/348610a0. [DOI] [PubMed] [Google Scholar]
- Kozak M. The scanning model for translation: an update. J Cell Biol. 1989 Feb;108(2):229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Manzella J. M., Blackshear P. J. Regulation of rat ornithine decarboxylase mRNA translation by its 5'-untranslated region. J Biol Chem. 1990 Jul 15;265(20):11817–11822. [PubMed] [Google Scholar]
- Mehdi H., Ono E., Gupta K. C. Initiation of translation at CUG, GUG, and ACG codons in mammalian cells. Gene. 1990 Jul 16;91(2):173–178. doi: 10.1016/0378-1119(90)90085-6. [DOI] [PubMed] [Google Scholar]
- Orbach M. J., Porro E. B., Yanofsky C. Cloning and characterization of the gene for beta-tubulin from a benomyl-resistant mutant of Neurospora crassa and its use as a dominant selectable marker. Mol Cell Biol. 1986 Jul;6(7):2452–2461. doi: 10.1128/mcb.6.7.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orbach M. J., Sachs M. S., Yanofsky C. The Neurospora crassa arg-2 locus. Structure and expression of the gene encoding the small subunit of arginine-specific carbamoyl phosphate synthetase. J Biol Chem. 1990 Jul 5;265(19):10981–10987. [PubMed] [Google Scholar]
- Paluh J. L., Orbach M. J., Legerton T. L., Yanofsky C. The cross-pathway control gene of Neurospora crassa, cpc-1, encodes a protein similar to GCN4 of yeast and the DNA-binding domain of the oncogene v-jun-encoded protein. Proc Natl Acad Sci U S A. 1988 Jun;85(11):3728–3732. doi: 10.1073/pnas.85.11.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulus T. J., Cramer C. L., Davis R. H. Compartmentation of spermidine in Neurospora crassa. J Biol Chem. 1983 Jul 25;258(14):8608–8612. [PubMed] [Google Scholar]
- Paulus T. J., Davis R. H. Regulation of polyamine synthesis in relation to putrescine and spermidine pools in Neurospora crassa. J Bacteriol. 1981 Jan;145(1):14–20. doi: 10.1128/jb.145.1.14-20.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg A. E. Recent advances in the biochemistry of polyamines in eukaryotes. Biochem J. 1986 Mar 1;234(2):249–262. doi: 10.1042/bj2340249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson L., Holm I., Heby O. Regulation of ornithine decarboxylase mRNA translation by polyamines. Studies using a cell-free system and a cell line with an amplified ornithine decarboxylase gene. J Biol Chem. 1988 Mar 5;263(7):3528–3533. [PubMed] [Google Scholar]
- Pitkin J., Davis R. H. The genetics of polyamine synthesis in Neurospora crassa. Arch Biochem Biophys. 1990 May 1;278(2):386–391. doi: 10.1016/0003-9861(90)90275-4. [DOI] [PubMed] [Google Scholar]
- Rogers S., Wells R., Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986 Oct 17;234(4774):364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- Rosenberg-Hasson Y., Bercovich Z., Ciechanover A., Kahana C. Degradation of ornithine decarboxylase in mammalian cells is ATP dependent but ubiquitin independent. Eur J Biochem. 1989 Nov 6;185(2):469–474. doi: 10.1111/j.1432-1033.1989.tb15138.x. [DOI] [PubMed] [Google Scholar]
- Rosenberg-Hasson Y., Bercovich Z., Kahana C. Characterization of sequences involved in mediating degradation of ornithine decarboxylase in cells and in reticulocyte lysate. Eur J Biochem. 1991 Mar 28;196(3):647–651. doi: 10.1111/j.1432-1033.1991.tb15861.x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T. L. Disparate evolution of yeasts and filamentous fungi indicated by phylogenetic analysis of glyceraldehyde-3-phosphate dehydrogenase genes. Proc Natl Acad Sci U S A. 1989 Sep;86(18):7063–7066. doi: 10.1073/pnas.86.18.7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder R. D., Edwards M. L. Effects of polyamine analogs on the extent and fidelity of in vitro polypeptide synthesis. Biochem Biophys Res Commun. 1991 May 15;176(3):1383–1392. doi: 10.1016/0006-291x(91)90440-i. [DOI] [PubMed] [Google Scholar]
- Tabor C. W., Tabor H. Polyamines in microorganisms. Microbiol Rev. 1985 Mar;49(1):81–99. doi: 10.1128/mr.49.1.81-99.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor C. W., Tabor H. Polyamines. Annu Rev Biochem. 1984;53:749–790. doi: 10.1146/annurev.bi.53.070184.003533. [DOI] [PubMed] [Google Scholar]
- Tsunasawa S., Stewart J. W., Sherman F. Amino-terminal processing of mutant forms of yeast iso-1-cytochrome c. The specificities of methionine aminopeptidase and acetyltransferase. J Biol Chem. 1985 May 10;260(9):5382–5391. [PubMed] [Google Scholar]
- Van Steeg H., Van Oostrom C. T., Hodemaekers H. M., Peters L., Thomas A. A. The translation in vitro of rat ornithine decarboxylase mRNA is blocked by its 5' untranslated region in a polyamine-independent way. Biochem J. 1991 Mar 1;274(Pt 2):521–526. doi: 10.1042/bj2740521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer S. J., Yanofsky C. Efficient cloning of genes of Neurospora crassa. Proc Natl Acad Sci U S A. 1986 Jul;83(13):4869–4873. doi: 10.1073/pnas.83.13.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L., Huang J. K., Blackshear P. J. Rat ornithine decarboxylase gene. Nucleotide sequence, potential regulatory elements, and comparison to the mouse gene. J Biol Chem. 1989 May 25;264(15):9016–9021. [PubMed] [Google Scholar]
- Winqvist R., Mäkelä T. P., Seppänen P., Jänne O. A., Alhonen-Hongisto L., Jänne J., Grzeschik K. H., Alitalo K. Human ornithine decarboxylase sequences map to chromosome regions 2pter----p23 and 7cen----qter but are not coamplified with the NMYC oncogene. Cytogenet Cell Genet. 1986;42(3):133–140. doi: 10.1159/000132266. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Daalen Wetters T., Macrae M., Brabant M., Sittler A., Coffino P. Polyamine-mediated regulation of mouse ornithine decarboxylase is posttranslational. Mol Cell Biol. 1989 Dec;9(12):5484–5490. doi: 10.1128/mcb.9.12.5484. [DOI] [PMC free article] [PubMed] [Google Scholar]