Beneficial soil-borne Pseudomonas rhizobacteria influence root development and promote growth, independent of their ability to stimulate systemic immunity.
Abstract
Plant roots are colonized by an immense number of microbes, referred to as the root microbiome. Selected strains of beneficial soil-borne bacteria can protect against abiotic stress and prime the plant immune system against a broad range of pathogens. Pseudomonas spp. rhizobacteria represent one of the most abundant genera of the root microbiome. Here, by employing a germ-free experimental system, we demonstrate the ability of selected Pseudomonas spp. strains to promote plant growth and drive developmental plasticity in the roots of Arabidopsis (Arabidopsis thaliana) by inhibiting primary root elongation and promoting lateral root and root hair formation. By studying cell type-specific developmental markers and employing genetic and pharmacological approaches, we demonstrate the crucial role of auxin signaling and transport in rhizobacteria-stimulated changes in the root system architecture of Arabidopsis. We further show that Pseudomonas spp.-elicited alterations in root morphology and rhizobacteria-mediated systemic immunity are mediated by distinct signaling pathways. This study sheds new light on the ability of soil-borne beneficial bacteria to interfere with postembryonic root developmental programs.
Eukaryotic organisms have evolved in the context of complex communities with commensal microbes. Like animals and humans, plants have their own microbiome that safeguards optimal plant functioning and fitness under diverse environmental conditions (Berendsen et al., 2012). The plant microbiome is predominantly hosted by the roots, which secretes up to 20% of the plant’s photosynthetically fixed carbon into the rhizosphere, rendering this small zone around the root system one of the most energy-rich habitats on Earth (Bisseling et al., 2009). One gram of bulk soil may contain up to one billion microbes. However, epiphytic and endophytic microbial communities are highly distinct from microbial populations in the bulk soil, suggesting that plants have evolved to recruit specific microbes from their environment (Bais et al., 2004). Exciting new discoveries combining metagenomics, PhyloChip analysis, and quantitative plant genetics have revealed a core microbiome within the roots and rhizosphere of plants (Mendes et al., 2011; Brown et al., 2012; Bulgarelli et al., 2012; Lundberg et al., 2012; Sessitsch et al., 2012).
Selected strains of soil-borne beneficial microbes, collectively referred to as plant growth-promoting bacteria (PGPR) and plant growth-promoting fungi (PGPF), have long been demonstrated to promote plant growth, improve host nutrition, and protect plants from various forms of abiotic stress and soil-borne diseases (Ryu et al., 2003; Lugtenberg and Kamilova, 2009; Yang et al., 2009; Blom et al., 2011; Schwachtje et al., 2011). Similar to the immunostimulatory properties of human probiotics, root colonization by selected PGPR and PGPF strains primes the whole-plant body to efficiently defend itself against a broad range of pathogens and even insects. This form of systemic resistance is called induced systemic resistance (ISR) and widely occurs in monocotyledonous and dicotyledonous plant species (Van Wees et al., 2008; De Vleesschauwer and Höfte, 2009; Zamioudis and Pieterse, 2012). Colonization of the roots by ISR-inducing rhizobacteria and fungi does not directly activate the plant immune system but primes the aboveground plant parts for an accelerated defense response upon pathogen or insect attack, thus providing a cost-effective protection against plant diseases (Conrath et al., 2006).
Many PGPR and PGPF strains have long been known to cause alterations in the root system architecture of host plants by promoting the formation of secondary roots and thus improving the root’s exploratory capacity. However, only recently have the molecular mechanisms underpinning those phenomena started to be dissected at the genetic and molecular levels (López-Bucio et al., 2007; Contreras-Cornejo et al., 2009; Felten et al., 2009). The root system architecture is defined by three main processes: (1) indeterminate growth of the main root, a process orchestrated by the root meristem; (2) lateral root (LR) formation; and (3) root hair (RH) formation. Postembryonic root development is controlled by cell divisions in the meristematic zone, cell expansion in the elongation zone, and functional differentiation into specialized cell types in the differentiation zone of the root. The population of mitotic cells in the root meristem originates from stem cells whose identity is controlled by an organizing center called the quiescent center (QC; Bennett and Scheres, 2010). Auxin gradients such as those established by the PIN-FORMED (PIN) auxin efflux facilitator network and a genetic program regulated by WUSCHEL-RELATED HOMEOBOX5 (WOX5), SCARECROW (SCR), SHORT-ROOT, and PLETHORA transcription factor proteins are crucial for stem cell maintenance and function (Di Laurenzio et al., 1996; Helariutta et al., 2000; Aida et al., 2004; Blilou et al., 2005; Sarkar et al., 2007).
LR and RH constitute important traits of the root architecture that facilitate plant anchorage and increase the root’s exploratory capacity for water and minerals. LR originate from xylem pole pericycle cells that are primed in the basal meristem to become LR founder cells. LR initiation occurs in more distal parts of the root, where an initial anticlinal and asymmetrical division of a pair of adjacent founder cells is followed by a series of cell divisions to form higher order lateral root primordia (LRP). At later stages, the LR emerges from the parental root by concurrent expansions of cells within the LRP and cell wall modifications in the surrounding tissues (Casimiro et al., 2003; Benková and Bielach, 2010). Local auxin accumulation and signaling has a critical role during LR formation by regulating developmental processes from founder cell specification to LR emergence (Dubrovsky et al., 2008). RH originate from a subset of epidermal cells, referred to as hair (H) cells, positioned in the cleft between two cortical cells. Epidermal cells that are located over a single cortical cell do not develop into RH and are generally referred to as nonhair (N) cells (Dolan et al., 1994; Galway et al., 1994). Several signaling components that determine the H and N fate specification in the root epidermis have been identified. N fate is controlled by a transcriptional complex formed by WEREWOLF (WER)-GLABRA3 (GL3), ENHANCER OF GLABRA3 (EGL3), and TRANSPARENT TESTA GLABRA (TTG1), which promote the expression of GL2 in N cells to specify hairless cell differentiation. The MYB-like transcription factor proteins CAPRICE (CPC), TRIPTYCHON, and ENHANCER OF CAPRICE, on the other hand, act redundantly in H cells to promote the hair fate. In H cells, CPC competes with WER for binding to the GL3-EGL3-TTG1 complex. Consequently, the expression of GL2 is reduced and the H fate is adopted by those cells (Schiefelbein, 2003; Ueda et al., 2005; Ishida et al., 2008; Grebe, 2012).
Soil-borne Pseudomonas spp. represent one of the most abundant genera of the root microbiome (Mendes et al., 2011; Brown et al., 2012; Bulgarelli et al., 2012; Lundberg et al., 2012; Sessitsch et al., 2012). In recent years, significant progress has been made in our understanding of the molecular mechanisms underpinning rhizobacteria-mediated systemic resistance. It is now well established that the ISR signaling pathway triggered by Pseudomonas spp. rhizobacteria is controlled by the plant hormones jasmonic acid (JA) and ethylene (ET) and depends on the transcriptional (co)activators NONEXPRESSOR OF PR GENES1 (NPR1), MYB72, and MYC2 (Pieterse et al., 1998; Pozo et al., 2008; Van der Ent et al., 2008). However, little is known about the plant growth-promoting activities of those strains and the molecular mechanisms underpinning morphological alteration of the root system architecture to rhizobacteria-derived signals. Recent data that report on hormonal cross talk between auxin and JA/ET in primary root (Ortega-Martínez et al., 2007; Chen et al., 2011) and LR development (Ivanchenko et al., 2008; Sun et al., 2009) further raise questions about a plausible interrelationship between JA/ET-dependent rhizobacteria-mediated ISR and rhizobacteria-induced alterations in root morphology. In this study, by using a germ-free system, we dissected the root developmental program triggered by selected soil-borne Pseudomonas spp. bacteria. By studying cell type-specific responses and employing genetic and pharmacological approaches, our study reveals the essential role of auxin transport and signaling in Pseudomonas spp.-stimulated root phenotypic plasticity. We further provide evidence that rhizobacteria-induced alterations in the root system architecture and ISR are governed by distinct signaling pathways.
RESULTS
ISR-Inducing Rhizobacteria Promote Plant Growth and Affect the Root System Architecture
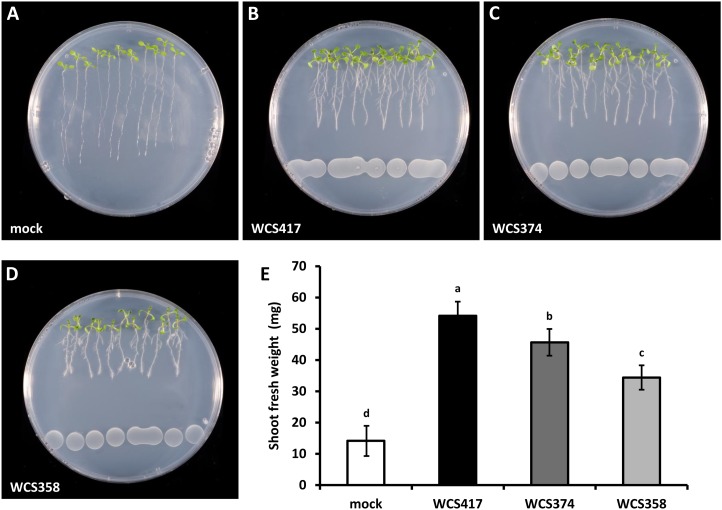
Three well-characterized PGPR, Pseudomonas fluorescens WCS417, P. fluorescens WCS374, and Pseudomonas putida WCS358 (hereafter called WCS417, WCS374, and WCS358, respectively), which have been previously shown to differentially trigger ISR, were tested for their ability to promote plant growth. The WCS417 strain was originally isolated from the wheat (Triticum aestivum) rhizosphere in a field soil suppressive to take-all disease (Lamers et al., 1988), whereas the WCS374 and WCS358 strains were originally isolated from the potato (Solanum tuberosum) rhizosphere (Geels and Schippers, 1983). Both WCS417 and WCS358 are capable of triggering ISR in Arabidopsis (Arabidopsis thaliana; Pieterse et al., 1996; Van Wees et al., 1997). WCS374 does not stimulate systemic immunity in Arabidopsis, but it does trigger ISR in radish (Raphanus sativus; Leeman et al., 1995; Van Wees et al., 1997). The plant growth-promoting effects of each strain were investigated on Arabidopsis ecotype Columbia (Col-0) seedlings growing vertically on agar-solidified medium. To test whether diffusible and/or volatile compounds affect plant growth, bacterial suspensions of each strain were applied at a 5-cm distance from the root tip of 4-d-old seedlings. All bacterial strains tested were capable of stimulating plant biomass production (Fig. 1, A–D). After 8 d of cocultivation, we measured increases of 3.9- and 3.2-fold in the shoot fresh weight of seedlings growing in the presence of WCS417 and WCS374, respectively, whereas WCS358 stimulated shoot biomass by 2.4-fold (Fig. 1E). Thus, selected strains of rhizobacteria have the potential to promote growth in Arabidopsis independent of their ability to trigger ISR in this plant. Besides stimulating shoot fresh weight, the aforementioned rhizobacteria triggered a number of developmental alterations, as evidenced by the reduction of primary root length and the increased number of LR in seedlings exposed to rhizobacteria-derived compounds (Fig. 1; Supplemental Fig. S1).
Figure 1.
Effects of molecules released by selected strains of ISR-inducing Pseudomonas spp. bacteria on plant growth and root system architecture of Arabidopsis Col-0 seedlings. A to D, Representative images of seedlings growing on control plates and plates containing WCS417, WCS374, or WCS358 bacteria. Surface-sterilized seeds were sown on 1× Murashige and Skoog agar-solidified medium supplemented with 0.5% Suc. At 4 d post germination, 240 μL of bacterial suspension was spotted on the opposite side of the plate at a 5-cm distance from the root tip. Photographs were taken after 8 d of cocultivation. E, Shoot biomass production measured after 8 d of cocultivation with the indicated bacterial strains. Data represent mean fresh weights ± sd of three groups of seedlings each consisting of 10 excised shoots. Different letters indicate statistically significant differences (Tukey’s HSD test; P < 0.05). The experiment was repeated twice with similar results.
WCS417 Inhibits Primary Root Elongation
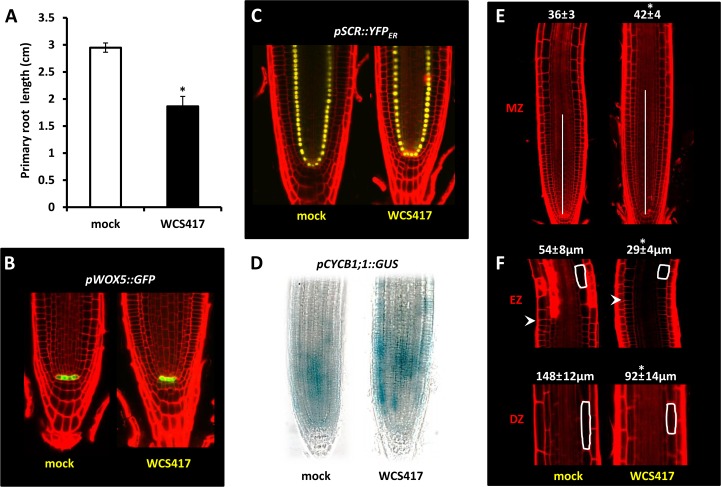
To analyze in detail the molecular mechanisms underpinning Pseudomonas spp.-stimulated effects on root development, we focused our analysis on developmental responses to the model strain WCS417. After 8 d of cocultivation, the primary root length of seedlings exposed to WCS417 bacteria was reduced by approximately 40% compared with mock-treated roots, suggesting a suppressive effect of WCS417 on primary root elongation (Fig. 2A). We reasoned that the short-root phenotype of WCS417-treated roots may be derived from effects on the organization of the root meristem. To test this, we used the QC-localized marker pWOX5::GFP (Sarkar et al., 2007) and the endodermis/QC-localized marker pSCR::YFPER (Sabatini et al., 1999) in order to detect any apparent differences in the meristem identity of WCS417-treated roots. Expression patterns of these marker genes were similar to those of mock-treated seedlings, suggesting that WCS417 does not have profound impact on the overall organization of the root meristem (Fig. 2, B and C). Likewise, confocal microscopy of the QC-localized markers pQC46::YFPER (Sabatini et al., 1999) and pQC25::CFPER (Sabatini et al., 1999), and the distal stem cell and columella differentiation markers Q1630 (Sabatini et al., 2003) and J2341 (Sabatini et al., 1999), respectively, further confirmed the neutral effects of WCS417 on QC and stem cell identity (Supplemental Fig. S2).
Figure 2.
Effects of WCS417 on primary root development of Arabidopsis. A, Primary root length of Col-0 seedlings cocultivated or not (mock) with WCS417 bacteria (n = 20). B and C, Representative confocal images showing the expression patterns of the QC-localized marker pWOX5::GFP and the endodermis/QC-localized marker pSCR::YFPER under control and WCS417-induced conditions. D, pCYCB1;1::GUS expression in roots under control (mock) and WCS417-induced conditions. E, Meristem size of mock- and WCS417-treated roots. Values represent average numbers ± sd of cortical cells in the meristem zone (MZ; n = 15). F, Cortical cell elongation in the elongation zone (EZ) and differentiation zone (DZ) of roots growing in the presence or absence of WCS417. Values represent average lengths ± sd of 45 cells in each developmental zone (n = 15). Asterisks indicate statistically significant differences compared with mock-treated roots (Student’s t test; P < 0.05). The experiment was repeated twice with similar results.
Next, we tested whether reduced meristem size could account for the observed WCS417-mediated reduction in primary root growth. To this end, we used the cell cycle marker pCYCB1;1::GUS (Colón-Carmona et al., 1999), which allows the visualization of cells at the G2/M phase of the cell and reports for cell division rates in the root meristem. We observed enhanced expression of the reporter in the meristematic zone of WCS417-treated roots, indicating that WCS417 promotes rather than inhibits cell division (Fig. 2D). Accordingly, the number of meristematic cells (as defined by the number of isodiametric epidermal cells in the meristematic zone) was found to be significantly higher in WCS417-compared with mock-treated roots (Fig. 2E). To test the possibility that the reduction of root length in WCS417-treated roots is due to effects on cell elongation, the length of root epidermal cells in the elongation and differentiation zones was assessed after 8 d of cocultivation with WCS417. This number was found to be reduced by approximately 40% in WCS417-treated roots compared with mock-treated roots (Fig. 2F). Collectively, our data indicate that reduced primary root elongation in response to WCS417 is due to inhibitory effects on cell expansion rather than on the organization and function of the root meristem.
WCS417 Promotes LR Formation
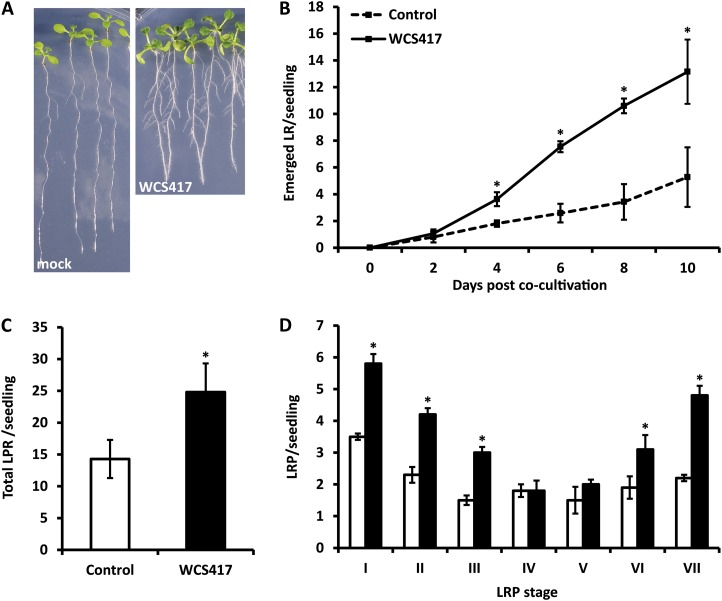
One of the most prominent WCS417-mediated morphological alterations in the root system architecture is the stimulation of LR formation (Fig. 3A). In order to follow the kinetics of LR production in response to WCS417, LR formation was assessed over a 10-d period. We measured a 2-fold increase in the number of emerged LR on WCS417-treated roots after 4 d of cocultivation, whereas WCS417-treated roots formed approximately 3-fold more LR compared with mock-treated roots after 10 d (Fig. 3B). To further investigate the stage of LRP that is affected by WCS417, the developmental stage of each LRP on control and WCS417-treated roots was classified according to Malamy and Benfey (1997). We used GUS expression patterns of the pCYCB1;1::GUS reporter in order to precisely localize LRP across the primary root and accurately define each LRP stage. After 8 d of cocultivation, the average number of total LRP in roots exposed to WCS417 was 1.5-fold higher than in control roots (Fig. 3C). Interestingly, in clear contrast to mock-treated roots, in which the developmental stages of LRP were almost uniformly distributed over the seven classes, in WCS417-treated roots, a clear overrepresentation of early (I–III) and late (VI–VII) LRP stages could be observed (Fig. 3D). Collectively, these data indicate that WCS417 promotes LR formation by stimulating both LR initiation and LR outgrowth.
Figure 3.
Effects of WCS417 on LR formation in Arabidopsis Col-0 seedlings. A, Representative images of seedlings after 8 d of growth on control or WCS417-containing plates. B, Time course of LR formation in response to WCS417. At time point 0, WCS417 bacteria were spotted on the plates. C and D, LRP density (C) and distribution of LRP (D) in seven developmental classes (Malamy and Benfey, 1997) as defined by the pCYCB1;1::GUS activity after 8 d of cocultivation. White bars represent the mock treatment, and black bars represent the WCS417 treatment. For each experiment, values represent means ± sd of at least 20 seedlings. Asterisks indicate statistically significant differences compared with mock-treated roots (Student’s t test; P < 0.05). The experiment was repeated twice with similar results. [See online article for color version of this figure.]
WCS417 Promotes RH Development
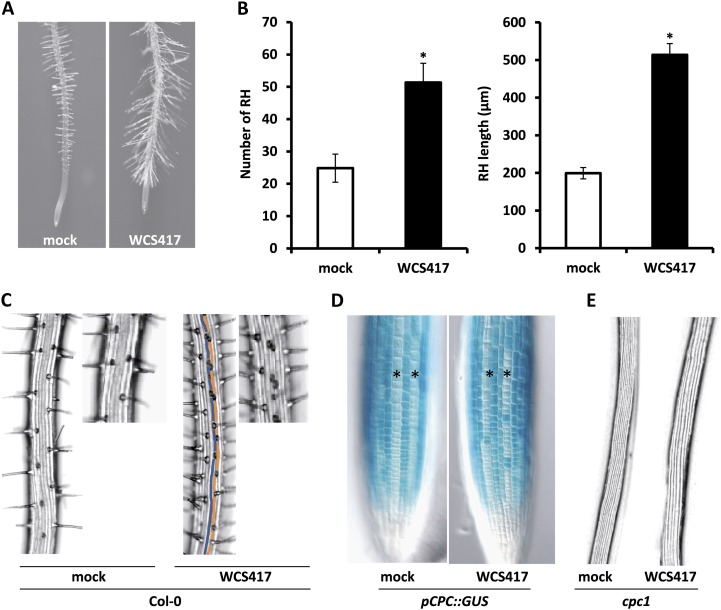
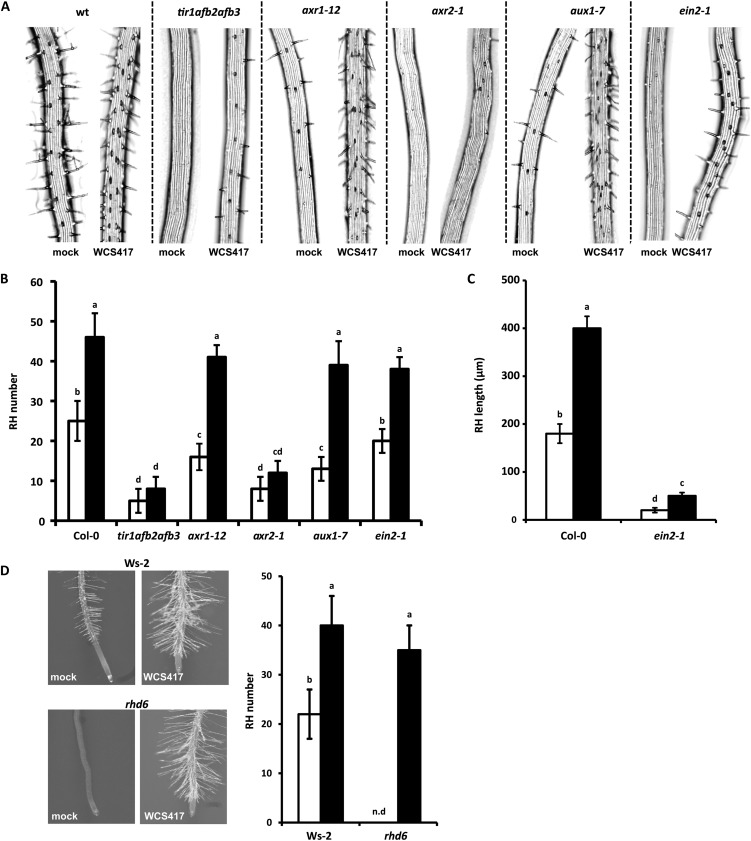
In addition to positive effects on LR formation, WCS417 has a strong impact on RH development, as evidenced by the increased RH density and length in WCS417-treated roots (Fig. 4A). In particular, at day 8 of cocultivation, we measured a 2-fold increase in the RH number and a 2.5-fold increase in the average RH length of WCS417-exposed roots (Fig. 4B). A close inspection of RH topology in WCS417-treated roots revealed accelerated RH formation in cells located in H positions but also RH formation in adjacent epidermal cell files (Fig. 4C). This latter finding correlates with the expression of the RH-specific marker CPC, as revealed by the expression pattern of the corresponding pCPC::GUS reporter line (Wada et al., 1997; Fig. 4D). We then tested whether WCS417 is able to rescue the RH-defective phenotype of the cpc1 mutant lacking the central transcription factor that promotes differentiation in hair-forming cells. In this mutant background, WCS417 was unable to promote RH formation in H or in adjacent positions, indicating that WCS417-induced RH formation depends on a canonical RH fate specification pattern (Fig. 4E).
Figure 4.
Effects of WCS417 on Arabidopsis RH formation. A, Representative images of Col-0 root tips showing RH formation after 8 d of growth on control (mock) or WCS417-containing plates. B, RH density expressed as the average RH number ± sd in the root segment located 1.0 cm above the root tip (n = 20) and average RH length ± sd (80 RH; n = 20 roots) of Col-0 seedlings growing on control or WCS417-containing plates. Asterisks indicate statistically significant differences compared with mock-treated roots (Student’s t test; P < 0.05). The experiment was repeated twice with similar results. C, Binocular views of RH distribution on mock- and WCS417-treated Col-0 roots. Note that in contrast to mock-treated roots, formation occurs in adjacent epidermal cell files (false-colored blue and orange lines) in WCS417-treated roots. D, pCPC::GUS expression patterns in the root tips of mock- and WCS417-treated seedlings. Asterisks indicate N epidermal files. E, Functional CPC1 is required for WCS417-induced RH formation. Binocular views are shown for segments located 1.0 cm above the root tip of the cpc1 mutant under control and WCS417-induced conditions.
To further test whether the ectopic formation of RH in WCS417 treatment is due to cell fate respecification from the N to the H fate, or is due to an increased number of cells allocated in the H position, cross sections of mock- and WCS417-treated roots were examined. WCS417 increased the number of cortical cells and, accordingly, the number of epidermal cells located in H positions (Supplemental Table S1), suggesting that the induction of RH is conditionally mediated rather than via a cell-autonomous pathway. Collectively, these data indicate that WCS417 promotes RH initiation in cells located in H files and further increases the number of RH via the allocation of more epidermal cells in H positions.
WCS417 Enhances Auxin-Regulated Gene Expression
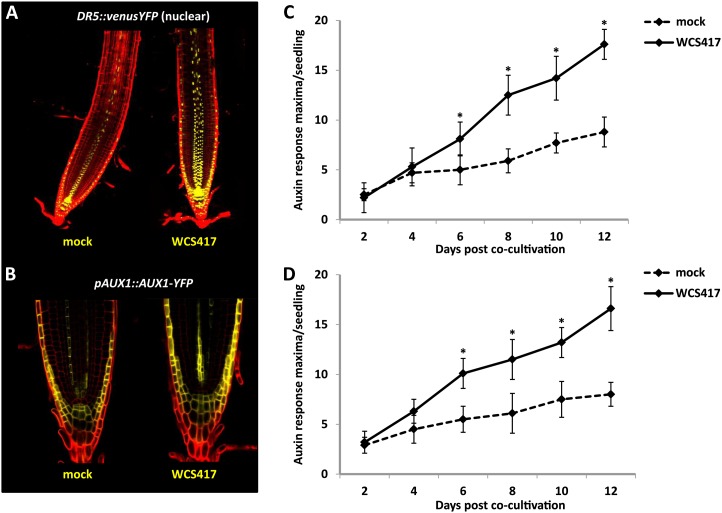
The effects of WCS417 on the primary root length and the abundance of LR and RH resemble those described for exogenous auxin-induced developmental alterations. In order to investigate whether WCS417-treated roots exhibit an enhanced auxin response, transgenic plants expressing the synthetic auxin reporter DR5::vYFPNLS (Laskowski et al., 2008) and the auxin-regulated pAUX1::AUX1-YFP reporter (Swarup et al., 2001) were subjected to confocal imaging. In mock-treated DR5::vYFPNLS-expressing seedlings, a strong auxin response was detected toward the root tip in the root meristem of primary root tips and at the distal end of the vasculature. Similar spatial expression patterns were also observed in the roots of seedlings growing in the presence of WCS417. However, the yellow fluorescent protein fluorescence toward and upward of the root meristem was remarkably enhanced (Fig. 5A). Likewise, AUX1-YFP more strongly accumulated at the plasma membrane of meristematic and LR cap cells of WCS417-treated roots, pointing to stimulatory effects of WCS417 on auxin-regulated gene expression (Fig. 5B). Imaging of both reporter lines over time with a binocular fluorescence microscope further revealed an increased density of auxin-response maxima along the primary root of WCS417-treated seedlings (Fig. 5, C and D).
Figure 5.
Effects of WCS417 on auxin distribution in the Arabidopsis root. A and B, Representative confocal images of DR5::vYFPNLS and pAUX1::AUX1-YFP expression in the Arabidopsis root at 8 d of cocultivation with WCS417. C and D, Time-course analysis of the density of auxin-response maxima across the primary root, marked by the DR5::vYFPNLS and pAUX1::AUX1-YFP activity. Data represent means ± sd of fluorescent spots in the primary root of at least 20 seedlings. Asterisks indicate statistically significant differences compared with mock-treated roots (Student’s t test; P < 0.05). The experiment was repeated twice with similar results.
WCS417-Triggered LR Formation Depends on Auxin Signaling
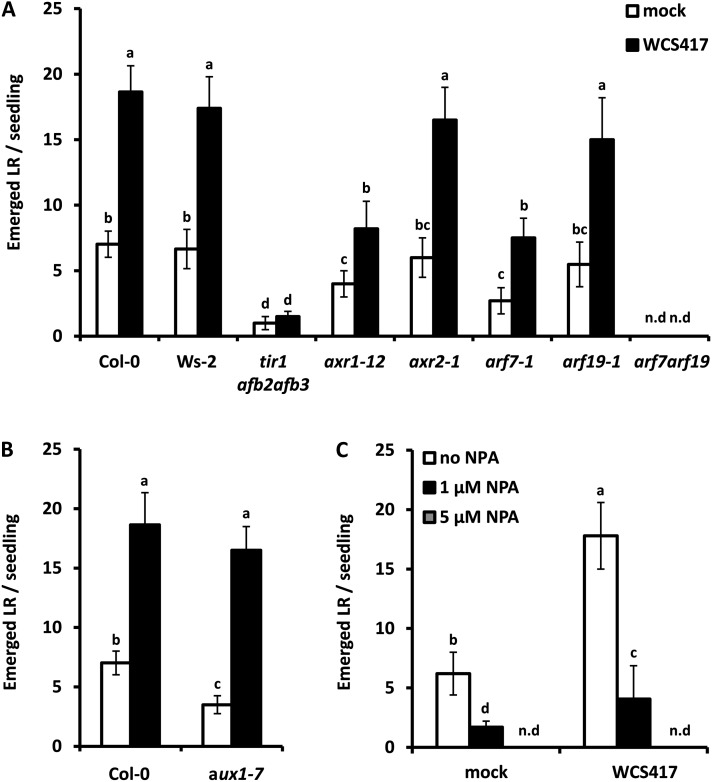
In order to address the role of auxin signaling in WCS417-induced LR formation, we assessed LR development in the auxin perception triple mutant tir1afb2afb3 and the auxin signaling mutants axr1-12 and axr2-1. The tir1afb2afb3 genotype carries mutant alleles of TIR1, AFB2, and AFB3 that encode the corresponding F-box auxin receptors (Dharmasiri et al., 2005); axr1-12 carries a mutant allele of AXR1 that encodes a subunit of the RUB1-activating enzyme that regulates the protein degradation activity of Skp1-Cullin-F-box complexes (Leyser et al., 1993); the auxin-resistant axr2-1 mutant carries a dominant gain-of-function mutation of IAA7 that encodes a member of the auxin/indole-3-acetic acid (IAA) repressors of auxin-inducible gene expression (Timpte et al., 1994). Stimulation of LR formation by WCS417 was also analyzed in the single arf7-1 and arf19-1 mutants and in the arf7arf19 double mutant, carrying mutant alleles of the auxin-response factors ARF7 and AFR19 essential for LR development (Okushima et al., 2007; Fig. 6A). The tir1afb2afb3 mutant, in which LR formation is severely compromised, was insensitive to WCS417-stimulated LR formation. The axr1-12 mutant, which formed a significantly reduced number of LR under noninduced conditions, responded to WCS417 by producing more LR, but the LR number reached only approximately 40% of that in WCS417-stimulated Col-0 roots. By contrast, no differences could be detected in LR formation in the axr2-1 mutant, both under basal and WCS417-induced conditions, when compared with the wild-type background. Regarding the role of ARF7 and ARF19 transcription factors, LR formation was compromised in the arf7-1 mutants and completely abolished in the arf7arf19 double mutant, suggesting that WCS417-triggered LR formation operates via a canonical auxin-response pathway (Fig. 6A).
Figure 6.
Influence of auxin signaling and transport on WCS417-mediated LR formation in the Arabidopsis root. A, LR formation under control and WCS417-induced conditions in wild-type roots (Col-0 and Wassilewskija-2 [Ws-2]) and roots of auxin perception (tir1afb2afb3; in the Col-0 and Ws-2 backgrounds) and signaling (axr1-12, axr2-1, arf7-1, arf19-1, and arf7arf19; in the Col-0 background) mutants after 8 d of cocultivation. B, Assessment of LR formation in the auxin influx mutant aux1-7 after 8 d of cocultivation. White bars represent the mock treatment, and black bars represent the WCS417 treatment. C, Effects of the polar auxin transport inhibitor NPA on WCS417-induced LR formation. Values represent means ± sd of at least 20 seedlings. Different letters indicate statistically significant differences (Tukey’s HSD test; P < 0.05). The experiment was repeated twice with similar results. n.d, Not detected.
In order to address the role of auxin transport in WCS417-induced LR formation, the LR phenotype of the auxin influx mutant aux1-7 was assessed (Fig. 6B). Although this mutant formed 50% less LR compared with Col-0 under control conditions, WCS417-treated roots developed a comparable number of LR to the wild type, reaching approximately 18 LR per seedling after 8 d of cocultivation. To further address the role of polar auxin transport in WCS417-mediated LR formation, LR formation was quantified in the presence of the polar auxin transport inhibitor 1-N-naphthylphthalamic acid (NPA). At 1 μm NPA, the ability of WCS417 to stimulate LR formation was severely affected, whereas in the presence of 5 μm NPA, LR formation in response to WCS417 was completely abolished (Fig. 6C). Thus, it can be concluded that a functional auxin efflux machinery is required for WCS417-induced LR formation. Previously, it was reported that the stimulation of LR in Arabidopsis by the ectomycorrhizal fungus Laccaria bicolor requires PIN2-mediated auxin transport (Felten et al., 2009). To investigate the role of PIN proteins, the LR phenotype of the triple pin2pin3pin7 mutant was analyzed in order to test whether basipetal auxin transport is involved in WCS417-mediated LR formation. No differences could be detected in LR production under basal and WCS417-induced conditions when compared with the wild type (Supplemental Fig. S3), suggesting that L. bicolor and WCS417 utilize different components of the efflux machinery to confer alterations in the root system architecture.
WCS417 Promotes RH Development via Auxin Signaling
Auxin and ET have been demonstrated to play key roles in regulating processes related to RH initiation and outgrowth (Rahman et al., 2002). We set out to investigate the role of auxin signaling in WCS417-mediated RH formation by evaluating the RH phenotypes of the auxin perception triple mutant tir1afb2afb3, the auxin signaling mutants axr1-12 and axr2-1, and the auxin influx mutant aux1-7. Under noninduced conditions, aux1-7 and axr1-12 produced significantly fewer RH, whereas RH formation was severely affected in axr2-1 and tir1afb2afb3 (Fig. 7, A and B). In response to WCS417, the absolute increase in the RH number of the axr1-12 and aux1-7 mutants did not differ significantly from that of Col-0, whereas both the tir1afb2afb3 and axr2-1 mutants failed to initiate new RH (Fig. 7, A and B), suggesting that WCS417-triggered RH initiation requires a functional auxin perception machinery and AXR2-mediated signaling.
Figure 7.
Influence of auxin and ET signaling on WCS417-induced RH formation in the Arabidopsis root. A, Representative binocular views of segments located 1.0 cm above the root tip of seedlings of the wild type (wt), various auxin mutants, and the ET-signaling mutant ein2-1 growing under control and WCS417-induced conditions. B, Quantification of the data presented in A. Values represent average RH numbers ± sd in the root segment located 1.0 cm above the root tip (n = 15). White bars represent the mock treatment, and black bars represent the WCS417 treatment. C, RH length in Col-0 and the ein2-1 mutant in the absence (white bars) or presence (black bars) of WCS417. Values represent average lengths ± sd of RH located in the 1.0-cm root segment above the root tip (80 RH; n = 20 roots). D, RH density in Wassilewskija-2 (Ws-2; the wild type) and the RH-defective mutant rhd6 in the absence (white bars) or presence (black bars) of WCS417. Values represent average RH numbers ± sd in the root segment located 1.0 cm above the root tip (n = 15). Data in all panels were obtained after 8 d of cocultivation. Different letters indicate statistically significant differences (Tukey’s HSD test; P < 0.05). The experiment was repeated twice with similar results. n.d, Not detected.
In order to address the role of ET signaling, we assessed RH development in the ET signaling mutant ein2-1. Although no clear difference could be detected in the RH number of ein2-1 when compared with wild-type roots under basal or induced conditions (Fig. 7, A and B), RH elongation was significantly compromised in both conditions (Fig. 7C). RH formation in response to WCS417 was further assessed in the rhd6 mutant that is defective in RH initiation and is known to be rescued by exogenous application of synthetic auxins (Masucci and Schiefelbein, 1994). After 8 d of cocultivation with WCS417, the rhd6 mutant responded similarly to wild-type roots (Fig. 7D), indicating that WCS417 is capable of rescuing the RH phenotype stemming from this mutation.
Bacterial Determinants Involved in Rhizobacteria-Induced Root Developmental Programs
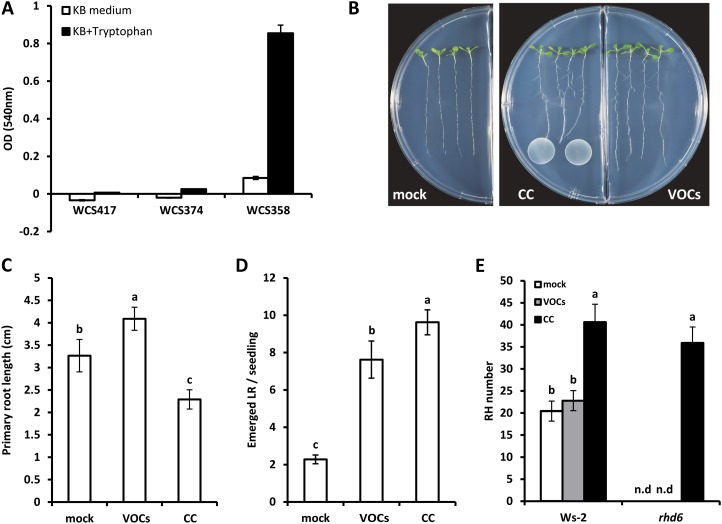
To investigate whether ISR-inducing rhizobacteria produce auxin, the total content of auxin was colorimetrically evaluated in the culture supernatant of WCS417, WCS347, and WCS358 bacteria growing in standard King’s medium B (KB) or KB supplemented with the auxin biosynthetic precursor Trp. As shown in Figure 8A, the WCS358 strain produces significant amounts of auxin, and this was enhanced by approximately 10-fold in Trp-enriched medium. However, we could not detect auxin in the culture supernatant of the WCS417 and WCS347 bacteria growing either in standard or Trp-supplemented medium (Fig. 8A). Consistent with these data, our preliminary analyses on the genomes of these bacterial strains suggest that the two major biosynthetic routes of bacterial IAA production, the indolepyruvic acid and the indole-3-acetamide pathways, are absent from the WCS417 and WCS347 rhizobacteria, whereas WCS358 likely synthesizes auxin via the indole-3-acetamide route (R.L. Berendsen, unpublished data). These results indicate that soil-borne beneficial bacteria may utilize other molecules than bacterial auxin to stimulate root developmental programs. Considering the well-documented role of rhizobacterial volatile organic compounds (VOCs) in promoting shoot growth (Ryu et al., 2003; Blom et al., 2011), we set out to investigate whether the volatile blend of WCS417 is capable of stimulating root developmental programs in the root of Arabidopsis. First, we confirmed previous findings regarding the ability of WCS417 VOCs to stimulate shoot growth (Blom et al., 2011; Supplemental Fig. S4). Subsequently, we assessed primary root elongation and LR formation under control and VOC-induced conditions by employing a split-plate assay in which bacteria and plants are separated into two parts by a septum that only allows bacterial VOCs to reach the seedlings (Fig. 8B). We found that the volatile blend of WCS417 moderately induced primary root elongation (Fig. 8C) and further stimulated LR formation to a comparable magnitude as WCS417 bacteria cocultivated to the same part of the plate with seedlings (Fig. 8D). Thus, VOCs are not involved in WCS417-induced inhibition of primary root length but are key determinants of WCS417-stimulated LR formation. We further found that WCS417 VOCs were not capable of rescuing the RH-defective phenotype of the rhd6 mutant (Fig. 8E), further supporting that multiple rhizobacterial determinants affect processes involved in Arabidopsis root development.
Figure 8.
A, Colorimetric assessment of auxin production in the culture supernatants of the WCS417, WCS374, and WCS358 strains in the presence or absence of Trp. OD, Optical density. B, Representative images of mock-treated seedlings and seedlings exposed to VOCs of WCS417 (right part of the plate) or cocultivated with WCS417 (CC; left part of the plate). C and D, Primary root length (C) and LR formation (D) of mock-treated seedlings and seedlings exposed to VOCs of WCS417 or cocultivated with WCS417 (CC). E, RH formation in the 1.0-cm segment located above the root tip of wild-type seedlings and seedlings of the rhd6 mutant. Phenotypic analysis in B to D was done after 6 d of exposure to VOCs or cocultivation with WCS417. Error bars indicate sd (n = 20). Different letters indicate statistically significant differences (Tukey’s HSD test; P < 0.05). The experiment was repeated twice with similar results. n.d, Not detected. [See online article for color version of this figure.]
ISR and WCS417-Mediated Root Developmental Plasticity Are Stimulated through Different Signaling Pathways
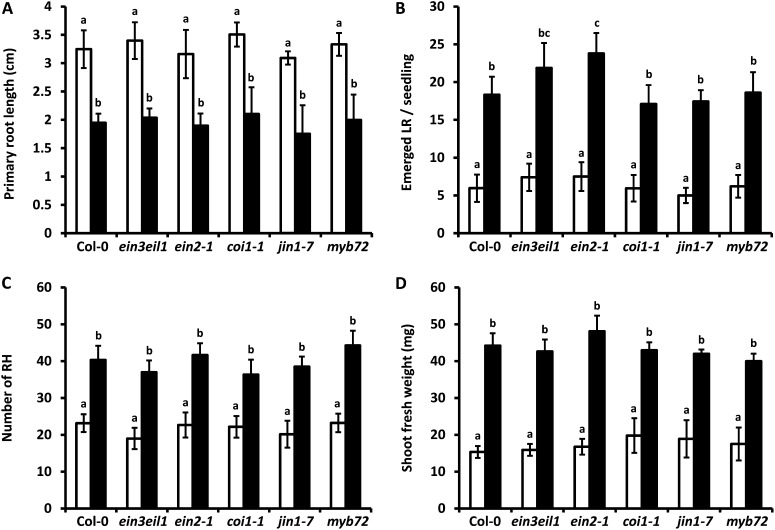
The crucial role of ET and JA signaling in systemic immune responses that are triggered by beneficial Pseudomonas spp. bacteria, including WCS417, is well documented (Van Wees et al., 2008; Van der Ent et al., 2009; Pieterse et al., 2012). In addition, both ET and JA have been shown to regulate aspects of root development, as both hormones play a role in the inhibition of primary root elongation and differentially influence LR formation through cross talk with auxin transport and signaling. In particular, ET, which has a negative impact on LR formation (Ivanchenko et al., 2008; Negi et al., 2008), was shown to inhibit primary root growth by triggering local auxin biosynthesis in the root tip via WEI2/ASA1 (Růzicka et al., 2007; Stepanova et al., 2007; Swarup et al., 2007), a rate-limiting enzyme in the biosynthetic pathway of IAA. On the other hand, jasmonates have been suggested to positively regulate LR formation through WEI2/ASA1 (Sun et al., 2009) and to inhibit primary root elongation in a MYC2-dependent manner (Chen et al., 2011). Considering the crucial role of the ET and JA signaling pathways in WCS417-mediated ISR, we were prompted to investigate whether WCS417 exerts its effects on auxin signaling in the roots by recruiting either of these hormone signaling pathways. To this end, we assessed primary root elongation and LR formation in the ET signaling mutants ein3eil1 and ein2-1, in the JA-response mutants coi1-1 and jin1-7 (myc2), and in the ISR signaling mutant myb72-1, which are all defective in their ability to mount ISR in response to WCS417 (Knoester et al., 1999; Pozo et al., 2008; Van der Ent et al., 2008). In all these mutants, we measured a significant reduction in the primary root length after 8 d of cocultivation that was comparable to that of Col-0 (Fig. 9A). Furthermore, all mutants responded to WCS417 similarly to the wild type by increasing the average LR number by 3-fold after 8 d of cocultivation (Fig. 9B). Likewise, neither WCS417-triggered RH formation nor WCS417-mediated shoot growth promotion was affected in the ISR mutants tested (Fig. 9, C and D). Therefore, it can be concluded that WCS417-stimulated developmental changes related to primary root elongation and LR and RH formation are mediated in an ET- and JA-independent manner and that WCS417 directly influences auxin signaling in the Arabidopsis root. These results also indicate that the induction of ISR and plant growth promotion are unlinked traits.
Figure 9.
WCS417-induced developmental plasticity and plant growth promotion in ISR-impaired Arabidopsis mutants after 8 d of cocultivation. Wild-type Col-0, the ET signaling mutants ein3eil1 and ein2-1, the JA-related mutants coi1-1 and jin1-7 (myc2), and the ISR signaling mutant myb72-1 were tested in the absence (white bars) and presence (black bars) of WCS417. A, Primary root length. B, LR density. C, RH density in the 1.0-cm segment located above the root tip. D, Shoot biomass production. Error bars indicate sd (n = 20). Different letters indicate statistically significant differences (Tukey’s HSD test; P < 0.05). The experiment was repeated twice with similar results.
DISCUSSION
In natural and agricultural ecosystems, the soil environment greatly influences plant health and productivity. In the soil, plant roots interact in a complex way with communities of beneficial microbes. Those microbes in the root-soil interface and within the root compartments structure a functional microbiome that provides important ecosystem services and promotes stress resistance against various forms of biotic and abiotic insults (Berendsen et al., 2012). In recent years, significant progress has been made in our understanding of how broad-spectrum immunity develops in response to root colonization by Pseudomonas spp. bacteria (Van Wees et al., 2008; Zamioudis and Pieterse, 2012). In this study, to our knowledge for the first time, we report on the ability of selected Pseudomonas spp. strains to interfere with postembryonic root developmental programs as well as the cellular and signaling responses that are triggered by rhizobacteria-derived semiochemicals. By employing a germ-free experimental system, we demonstrate that the model strain WCS417 enhances the auxin response in the root of Arabidopsis and stimulates the host’s endogenous programs related to primary root, LR, and RH development.
Pseudomonas spp. Rhizobacteria Target the Auxin Signaling Pathway
The essential role of the ET and JA signaling pathways in systemic immune responses activated upon root colonization by soil-borne beneficial microbes is well documented (Van Wees et al., 2008; Van der Ent et al., 2009; Pieterse et al., 2012). Both ET and JA may influence root development by interfering with processes related to auxin transport and signaling (Růzicka et al., 2007; Stepanova et al., 2007; Swarup et al., 2007; Chen et al., 2011). Here, we demonstrate that WCS417 does not recruit either of these pathways to induce developmental programs in the root of Arabidopsis, suggesting that WCS417-induced alterations in root morphology and WCS417-triggered systemic immunity are mediated through distinct signaling pathways. The fact that the RH initiation defect of the rhd6 mutant can be rescued upon cocultivation with WCS417 suggests that WCS417 directly interferes with the auxin signaling pathways by producing molecules with auxin activity. These determinants are expected to be diffusible compounds, because the volatile blend of WCS417 is unable to induce RH formation in the rhd6 mutant. Despite the fact that WCS417 does not produce auxin, it may produce other molecules with auxin activity, such as diketopiperazines, quorum-sensing bacterial molecules recently demonstrated to functionally mimic the binding of IAA to its receptor (Ortiz-Castro et al., 2011). In addition to secreted molecules, the volatile blend of WCS417 also appears to have a key role in promoting LR formation in Arabidopsis. This, in turn, suggests that rhizobacteria-mediated shoot growth promotion (Ryu et al., 2003; Blom et al., 2011) and morphological responses of roots are linked to common bacterial determinants. The mechanisms by which WCS417 VOCs promote LR development remain elusive. As mentioned above, although the volatile blend of WCS417 does not possess auxin activity, it may enhance the auxin response by stimulating auxin biosynthesis in local tissues. Alternatively, certain ingredients of the volatile mixture may be suitable substrates for the auxin biosynthetic pathway. Carbon dioxide has been demonstrated to stimulate root developmental programs, including primary root elongation and LR formation (Crookshanks et al., 1998); therefore, its role as a signaling molecule in the interaction between plants and beneficial soil-borne microbes should also be considered. Collectively, our study indicates that multiple bacterial molecules are involved in WCS417-mediated root developmental programs and further highlights the auxin signaling pathways as a node of convergence for different bacterial determinants.
Auxin is an essential plant hormone in the maintenance of stem cell identity and function. Various experimental manipulations that alter the auxin gradient in the root tip of Arabidopsis, such as chemical inhibition of polar auxin transport and exogenous application of synthetic auxins, have profound effects on processes related to cell fate specification and tissue polarity (Sabatini et al., 1999). Our data on cell- and tissue-specific developmental markers demonstrate that WCS417 enhances the auxin response in the root tip of Arabidopsis, but this does not influence the pattern of auxin distribution and, accordingly, the overall organization of the root meristem. Differences in the transport mechanisms between different auxin analogs may explain these effects. For instance, in contrast to the synthetic auxin 2,4-dichlorophenoxyacetic acid, which is not redistributed by export carriers and predominantly induces DR5::GUS activity in the epidermal root layers (Sabatini et al., 1999), our data suggest that auxin-like molecules secreted by WCS417 are subjected to polar transport. However, consistent with the role of auxin in promoting cell division (Campanoni and Nick, 2005) and its inhibitory effects on cell elongation when in excess (Swarup et al., 2007), we observed enhanced expression of the pCYCB1;1::GUS reporter in the meristematic zone of WCS417-treated roots and reduced expansion of cells entering the elongation zone. Therefore, we conclude that auxin levels mounted in roots upon cocultivation with WCS417 are optimal to promote cell division yet inhibitory for cell elongation.
Auxin Transport and Signaling Are Essential for Pseudomonas spp.-Stimulated Root Developmental Programs
Our studies on WCS417-mediated LR formation provide important information on the mechanism by which candidate molecules with auxin activity are perceived and transported within the root. Treatment with the polar auxin transport inhibitor NPA compromised the ability of WCS417 to promote LR formation, suggesting that WCS417-produced molecules are subjected to polar transport. The PIN2, PIN3, and PIN7 efflux carriers that mediate basipetal auxin transport are not involved in this process. Hence, other members of the PIN or MULTIDRUG RESISTANCE/P-GLYCOPROTEIN family are likely to mediate efflux. Moreover, stimulation of LR formation by WCS417 does not require the influx carrier AUX1, a member of the amino acid permease family of proton-driven transporters that functions in the uptake of the auxin molecules IAA and 2,4-dichlorophenoxyacetic acid but not the lipophilic auxin 1-naphthaleneacetic acid (Marchant et al., 1999). Hence, it is tempting to speculate that auxin-like substances produced by WCS417 adopt a diffusion-based mode of entry into plant cells similar to 1-naphthaleneacetic acid and exported via the auxin efflux machinery.
In Arabidopsis, epidermal cell fate is determined by positional cues, whereas RH initiation and elongation are under the control of auxin and ET signaling (Schiefelbein, 2003). Here, we demonstrate that WCS417 increases RH density by affecting processes related both to cell fate specification and RH initiation. RH formation in adjacent epidermal files of WCS417-treated roots depends on CPC, the central transcription factor that promotes differentiation in hair-forming cells, but is not induced via a cell-autonomous pathway. Instead, it is based on an increase in the number of cortical cells that, in turn, generate additional epidermal files located in H positions. This effect is auxin dependent and correlates with the observation that WCS417 augments cell divisions in the root meristem. We further demonstrate that WCS417 bacteria utilize the intrinsic genetic program that involves AXR2-mediated signaling to promote RH initiation downstream of a preestablished H cell fate. Interestingly, RH elongation in WCS417-exposed roots was significantly compromised in the ET signaling mutant ein2, indicating that auxin acts upstream of ET during WCS417-induced RH outgrowth.
Biological Significance of Rhizobacteria-Mediated Root Developmental Plasticity
The composition of microbial communities in the rhizosphere significantly differs from that in the bulk soil and is highly dependent on low- and high-Mr compounds secreted from the roots, collectively referred to as root exudates (Bais et al., 2006). Root exudation represents a significant carbon cost for the plant. However, important biological processes mediated by root exudates, such as communication with symbiotic microorganisms (via secretion of semiochemicals) and alterations in the chemical and physical properties of the soil (via secretion of organic and inorganic substances) justify such an energy investment (Bais et al., 2004, 2006). Although the chemical composition of these compounds differs considerably among plant species, energy-rich molecules such as sugars and organic acids encompass a significant proportion. Importantly, most of the root exudation is considered to occur in the elongation zone of newly formed roots (Bais et al., 2006). Therefore, it is reasonable to speculate that enhanced LR formation in response to WCS417 and other beneficial microbes may be a conserved mechanism that soil microbes employ for their own benefit in order to enhance root exudation and thus increase the energy flow from the roots of host plants. On the other hand, considering that LR and RH formation are typical responses to nutrient-limited conditions (López-Bucio et al., 2003), it is expected that rhizobacteria-induced alterations in the root system architecture would greatly facilitate plant nutrition, thereby conferring significant ecological benefits to host plants.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) accessions Col-0 and Wassilewskija-2 were used as wild-type plant genotypes. The following mutants were used in this study: cpc-1 (Lee and Schiefelbein, 2002), tir1afb2afb3 (Dharmasiri et al., 2005), axr2-1 (Timpte et al., 1994), axr1-12 (Leyser et al., 1993), aux1-7 (Pickett et al., 1990), pin2pin3pin7 (Blilou et al., 2005), arf7-1, arf19-1, and arf7arf19 (Okushima et al., 2007), rhd6 (Masucci and Schiefelbein, 1994), ein3eil1 (Chao et al., 1997; Binder et al., 2007), ein2-1 (Guzmán and Ecker, 1990), jin1-7 (Lorenzo et al., 2004), coi1-1 (Feys et al., 1994), and myb72-1 (Van der Ent et al., 2008). Seeds were surface sterilized and sown on 1× Murashige and Skoog agar-solidified medium supplemented with 0.5% Suc at a density of eight to 10 per plate. After 2 d of stratification at 4°C, the petri dishes were transferred and positioned vertically in a growth chamber under a long-day photoperiod (16 h of light, with light intensity of 100 μmol m−2 s−1) at 22°C. For inhibition of polar auxin transport, NPA was added at a final concentration of 1 or 5 μm. For experiments involving bacterial VOCs, two-compartment circular plates with a center partition were used.
Cultivation of Rhizobacteria and Induction Treatments
Pseudomonas fluorescens WCS417, P. fluorescens WCS374, and Pseudomonas putida WCS358 were cultured on KB agar plates supplemented with 50 μg mL−1 rifampicin at 28°C. After 24 h of growth, cells were collected in 10 mm MgSO4, washed twice by centrifugation for 5 min at 5,000g, and finally resuspended in 10 mm MgSO4. The bacterial titer was adjusted to an optical density at 600 nm of 0.002 (106 colony-forming units mL−1), and 240 μL of bacterial suspension (or 10 mm MgSO4 as a control) was spotted at a 5-cm distance from the root tip of 4-d-old seedlings using a multichannel pipet. For experiments involving bacterial VOCs, 120 μL of bacterial suspension was applied in one part of the split plate.
GUS Histochemical Staining
Histochemical detection of GUS in the pCPC::GUS and pCYCB1;1::GUS reporter lines was performed in a GUS staining solution (50 mm sodium phosphate [pH 7], 10 mm EDTA, 0.5 mm K4[Fe(CN)6], 0.5 mm K3[Fe(CN)6], 0.5 mm 5-bromo-4-chloro-3-indolyl-β-glucuronic acid, and 0.01% Silwet L-77) at 37°C for defined periods. Stained roots were cleared in a mixture of chloral hydrate:glycerol:water (8:1:2) and observed with Nomarski optics.
Auxin Determination
Production of IAA was determined colorimetrically in the supernatants of WCS417, WCS374, and WCS358 cultures growing in standard KB or KB supplemented with Trp (0.5 g L−1), as described previously (Glickmann and Dessaux, 1995). Data were normalized to a bacterial optical density at 600 nm.
Fluorescence Microscopy
Local auxin-response maxima across the primary root of DR5::vYFPNLS and pAUX1::AUX1-YFP lines were observed with a Leica MZ16F fluorescence stereoscope at defined time points. Confocal laser-scanning microscopy in the DR5::vYFPNLS, pAUX1::AUX1-YFP, pWOX5::GFP, pSCR::YFPER, QC46::YFPER, QC25::CFPER, Q1630, and J2341 lines was performed using a Leica SP2 inverted microscope. As counterstain, roots were stained in 10 μg mL−1 propidium iodide solution for 2 min. Chromophores were excited using a 488-nm argon laser, and fluorescence was detected at 500 to 550 nm (GFP), 550 to 615 nm (yellow fluorescent protein), 465 to 500 nm (cyan fluorescent protein), and 570 to 620 nm (propidium iodide).
Phenotypic and Data Analysis
For shoot fresh weight measurements, seedlings were sectioned at the root-shoot junction, and the weight of three groups of 10 excised shoots was immediately measured on an analytical balance. For root length measurements, digital images of petri dishes of Arabidopsis seedlings were captured using a gel documentation system, and the primary root length of at least 20 seedlings was calculated with ImageJ software (http://rsb.info.nih.gov/nih-image/). The number of emerged LR (greater than 0.5 mm) of at least 20 seedlings was counted using a dissecting microscope. The LRP developmental stages were classified according to Malamy and Benfey (1997) as follows. Stage I: the LRI stage in which the first anticlinal divisions in the pericycle occur. In the longitudinal axis, approximately eight to 10 short pericycle cells are formed. Stage II: the formed LRP is divided into two layers (inner and outer layer) by a periclinal division. Stage III: the outer layer cells undergo periclinal divisions to create a three-layered LRP. Stage IV: the inner layer cells undergo periclinal divisions to form a LRP with four-cell layers. Stage V: the LRP is halfway through the parental cortex. Stage VI: the LRP has passed through the parent cortex layer and has penetrated the epidermis. Stage VII: the LRP appears to be just about to emerge from the primary root. For RH measurements, digital images were obtained from the primary root segment located 1.0 cm above the root tip with a microscope at a magnification of 40×. RH density and length were then quantified with ImageJ. For determination of RM size and cell length measurements, digital confocal images of propidium iodide-stained roots were analyzed with ImageJ software. RM size was assessed as the number of cortical cells between the QC and the first cell that was twice the length of the immediately preceding cell. Cell length measurements were performed on single cortical cells located in the elongation zone and differentiation zone. For measurements of cortical and H-positioned epidermal cells, plastic root sectioning was done as described previously (Hassan et al., 2010). Statistical analyses were done with ANOVA followed by Tukey’s honestly significant difference (HSD) test to allow for comparisons among all means or with Student's t test when two means were compared (SPSS version 14.0).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Primary root length and LR formation in Col-0 seedlings cocultivated for 8 d with WCS417, WCS374, and WCS358 bacteria (n = 20).
Supplemental Figure S2. Representative confocal images showing the expression pattern of the QC-localized markers pQC46::YFPER and pQC25::CFPER, and the distal stem cell and columella differentiation markers Q1630 and J2341, respectively, under mock- and WCS417-induced conditions after 8 d of cocultivation.
Supplemental Figure S3. LR formation under control and WCS417-induced conditions in wild-type roots and roots of the triple pin2pin3pin7 mutant after 8 d of cocultivation.
Supplemental Figure S4. Shoot biomass production measured after 6 and 10 d of exposure to the volatile blend of WCS417.
Supplemental Table S1. Numbers (average ± sd) of cortical and H-positioned epidermal cells per cross section of roots growing under standard (mock) and WCS417-induced conditions. Transverse root sections were performed after 8 d of cocultivation. At least 10 cross sections per condition were examined.
Acknowledgments
We thank Hans van Pelt for photography and excellent technical assistance.
Glossary
- PGPR
plant growth-promoting bacteria
- PGPF
plant growth-promoting fungi
- ISR
induced systemic resistance
- QC
quiescent center
- LR
lateral root
- RH
root hair
- LRP
lateral root primordia
- JA
jasmonic acid
- ET
ethylene
- Col-0
ecotype Columbia
- IAA
indole-3-acetic acid
- NPA
1-N-naphthylphthalamic acid
- KB
King’s medium B
- VOC
volatile organic compound
- HSD
honestly significant difference
- H
hair
- N
nonhair
References
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B. (2004) The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120 [DOI] [PubMed] [Google Scholar]
- Bais HP, Park SW, Weir TL, Callaway RM, Vivanco JM. (2004) How plants communicate using the underground information superhighway. Trends Plant Sci 9: 26–32 [DOI] [PubMed] [Google Scholar]
- Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM. (2006) The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol 57: 233–266 [DOI] [PubMed] [Google Scholar]
- Benková E, Bielach A. (2010) Lateral root organogenesis: from cell to organ. Curr Opin Plant Biol 13: 677–683 [DOI] [PubMed] [Google Scholar]
- Bennett T, Scheres B (2010) Root development: two meristems for the price of one? Curr Top Dev Biol 91: 67–102 [DOI] [PubMed] [Google Scholar]
- Berendsen RL, Pieterse CMJ, Bakker PAHM. (2012) The rhizosphere microbiome and plant health. Trends Plant Sci 17: 478–486 [DOI] [PubMed] [Google Scholar]
- Binder BM, Walker JM, Gagne JM, Emborg TJ, Hemmann G, Bleecker AB, Vierstra RD. (2007) The Arabidopsis EIN3 binding F-box proteins EBF1 and EBF2 have distinct but overlapping roles in ethylene signaling. Plant Cell 19: 509–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisseling T, Dangl JL, Schulze-Lefert P. (2009) Next-generation communication. Science 324: 691. [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Blom D, Fabbri C, Connor EC, Schiestl FP, Klauser DR, Boller T, Eberl L, Weisskopf L. (2011) Production of plant growth modulating volatiles is widespread among rhizosphere bacteria and strongly depends on culture conditions. Environ Microbiol 13: 3047–3058 [DOI] [PubMed] [Google Scholar]
- Brown SD, Utturkar SM, Klingeman DM, Johnson CM, Martin SL, Land ML, Lu T-YS, Schadt CW, Doktycz MJ, Pelletier DA. (2012) Twenty-one genome sequences from Pseudomonas species and 19 genome sequences from diverse bacteria isolated from the rhizosphere and endosphere of Populus deltoides. J Bacteriol 194: 5991–5993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgarelli D, Rott M, Schlaeppi K, Ver Loren van Themaat E, Ahmadinejad N, Assenza F, Rauf P, Huettel B, Reinhardt R, Schmelzer E, et al. (2012) Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488: 91–95 [DOI] [PubMed] [Google Scholar]
- Campanoni P, Nick P. (2005) Auxin-dependent cell division and cell elongation: 1-naphthaleneacetic acid and 2,4-dichlorophenoxyacetic acid activate different pathways. Plant Physiol 137: 939–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro I, Beeckman T, Graham N, Bhalerao R, Zhang HM, Casero P, Sandberg G, Bennett MJ. (2003) Dissecting Arabidopsis lateral root development. Trends Plant Sci 8: 165–171 [DOI] [PubMed] [Google Scholar]
- Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR. (1997) Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89: 1133–1144 [DOI] [PubMed] [Google Scholar]
- Chen Q, Sun J, Zhai Q, Zhou W, Qi L, Xu L, Wang B, Chen R, Jiang H, Qi J, et al. (2011) The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 23: 3335–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colón-Carmona A, You R, Haimovitch-Gal T, Doerner P. (1999) Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J 20: 503–508 [DOI] [PubMed] [Google Scholar]
- Conrath U, Beckers GJM, Flors V, García-Agustín P, Jakab G, Mauch F, Newman M-A, Pieterse CMJ, Poinssot B, Pozo MJ, et al. (2006) Priming: getting ready for battle. Mol Plant Microbe Interact 19: 1062–1071 [DOI] [PubMed] [Google Scholar]
- Contreras-Cornejo HA, Macías-Rodríguez L, Cortés-Penagos C, López-Bucio J. (2009) Trichoderma virens, a plant beneficial fungus, enhances biomass production and promotes lateral root growth through an auxin-dependent mechanism in Arabidopsis. Plant Physiol 149: 1579–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crookshanks M, Taylor G, Dolan L. (1998) A model system to study the effects of elevated CO2 on the developmental physiology of roots: the use of Arabidopsis thaliana. J Exp Bot 49: 593–597 [Google Scholar]
- De Vleesschauwer D, Höfte M (2009) Rhizobacteria-induced systemic resistance. In LC Van Loon, ed, Plant Innate Immunity, Vol 51. Academic Press/Elsevier Science, London, pp 223–281 [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. (2005) The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN. (1996) The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86: 423–433 [DOI] [PubMed] [Google Scholar]
- Dolan L, Duckett CM, Grierson C, Linstead P, Schneider K, Lawson E, Dean C, Poethig S, Roberts K. (1994) Clonal relationships and cell patterning in the root epidermis of Arabidopsis. Development 120: 2465–2474 [Google Scholar]
- Dubrovsky JG, Sauer M, Napsucialy-Mendivil S, Ivanchenko MG, Friml J, Shishkova S, Celenza J, Benková E. (2008) Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc Natl Acad Sci USA 105: 8790–8794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felten J, Kohler A, Morin E, Bhalerao RP, Palme K, Martin F, Ditengou FA, Legué V. (2009) The ectomycorrhizal fungus Laccaria bicolor stimulates lateral root formation in poplar and Arabidopsis through auxin transport and signaling. Plant Physiol 151: 1991–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys BJF, Benedetti CE, Penfold CN, Turner JG. (1994) Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6: 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galway ME, Masucci JD, Lloyd AM, Walbot V, Davis RW, Schiefelbein JW. (1994) The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev Biol 166: 740–754 [DOI] [PubMed] [Google Scholar]
- Geels FP, Schippers B. (1983) Selection of antagonistic fluorescent Pseudomonas spp. and their root colonization and persistence following treatment of seed potatoes. Phytopathol Z 108: 193–206 [Google Scholar]
- Glickmann E, Dessaux Y. (1995) A critical examination of the specificity of the Salkowski reagent for indolic compounds produced by phytopathogenic bacteria. Appl Environ Microbiol 61: 793–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebe M. (2012) The patterning of epidermal hairs in Arabidopsis: updated. Curr Opin Plant Biol 15: 31–37 [DOI] [PubMed] [Google Scholar]
- Guzmán P, Ecker JR. (1990) Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2: 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan H, Scheres B, Blilou I. (2010) JACKDAW controls epidermal patterning in the Arabidopsis root meristem through a non-cell-autonomous mechanism. Development 137: 1523–1529 [DOI] [PubMed] [Google Scholar]
- Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, Hauser MT, Benfey PN. (2000) The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101: 555–567 [DOI] [PubMed] [Google Scholar]
- Ishida T, Kurata T, Okada K, Wada T. (2008) A genetic regulatory network in the development of trichomes and root hairs. Annu Rev Plant Biol 59: 365–386 [DOI] [PubMed] [Google Scholar]
- Ivanchenko MG, Muday GK, Dubrovsky JG. (2008) Ethylene-auxin interactions regulate lateral root initiation and emergence in Arabidopsis thaliana. Plant J 55: 335–347 [DOI] [PubMed] [Google Scholar]
- Knoester M, Pieterse CMJ, Bol JF, Van Loon LC. (1999) Systemic resistance in Arabidopsis induced by rhizobacteria requires ethylene-dependent signaling at the site of application. Mol Plant Microbe Interact 12: 720–727 [DOI] [PubMed] [Google Scholar]
- Lamers JG, Schippers B, Geels FP (1988) Soil-borne diseases of wheat in the Netherlands and results of seed bacterization with pseudomonads against Gaeumannomyces graminis var. tritici, associated with disease resistance. In ML Jorna, LAJ Slootmaker, eds, Cereal Breeding Related to Integrated Cereal Production. Pudoc, Wageningen, The Netherlands, pp 134–139 [Google Scholar]
- Laskowski M, Grieneisen VA, Hofhuis H, Hove CA, Hogeweg P, Marée AFM, Scheres B. (2008) Root system architecture from coupling cell shape to auxin transport. PLoS Biol 6: e307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J. (2002) Cell pattern in the Arabidopsis root epidermis determined by lateral inhibition with feedback. Plant Cell 14: 611–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeman M, Van Pelt JA, Den Ouden FM, Heinsbroek M, Bakker PAHM, Schippers B. (1995) Induction of systemic resistance against fusarium wilt of radish by lipopolysaccharides of Pseudomonas fluorescens. Phytopathology 85: 1021–1027 [Google Scholar]
- Leyser HMO, Lincoln CA, Timpte C, Lammer D, Turner J, Estelle M. (1993) Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature 364: 161–164 [DOI] [PubMed] [Google Scholar]
- López-Bucio J, Campos-Cuevas JC, Hernández-Calderón E, Velásquez-Becerra C, Farías-Rodríguez R, Macías-Rodríguez LI, Valencia-Cantero E. (2007) Bacillus megaterium rhizobacteria promote growth and alter root-system architecture through an auxin- and ethylene-independent signaling mechanism in Arabidopsis thaliana. Mol Plant Microbe Interact 20: 207–217 [DOI] [PubMed] [Google Scholar]
- López-Bucio J, Cruz-Ramírez A, Herrera-Estrella L. (2003) The role of nutrient availability in regulating root architecture. Curr Opin Plant Biol 6: 280–287 [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R. (2004) JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugtenberg B, Kamilova F. (2009) Plant-growth-promoting rhizobacteria. Annu Rev Microbiol 63: 541–556 [DOI] [PubMed] [Google Scholar]
- Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, Tremblay J, Engelbrektson A, Kunin V, del Rio TG, et al. (2012) Defining the core Arabidopsis thaliana root microbiome. Nature 488: 86–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. (1997) Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44 [DOI] [PubMed] [Google Scholar]
- Marchant A, Kargul J, May ST, Muller P, Delbarre A, Perrot-Rechenmann C, Bennett MJ. (1999) AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J 18: 2066–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci JD, Schiefelbein JW. (1994) The rhd6 mutation of Arabidopsis thaliana alters root-hair initiation through an auxin-associated and ethylene-associated process. Plant Physiol 106: 1335–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, Schneider JHM, Piceno YM, DeSantis TZ, Andersen GL, Bakker PAHM, et al. (2011) Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332: 1097–1100 [DOI] [PubMed] [Google Scholar]
- Negi S, Ivanchenko MG, Muday GK. (2008) Ethylene regulates lateral root formation and auxin transport in Arabidopsis thaliana. Plant J 55: 175–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. (2007) ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19: 118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Martínez O, Pernas M, Carol RJ, Dolan L. (2007) Ethylene modulates stem cell division in the Arabidopsis thaliana root. Science 317: 507–510 [DOI] [PubMed] [Google Scholar]
- Ortiz-Castro R, Díaz-Pérez C, Martínez-Trujillo M, del Río RE, Campos-García J, López-Bucio J. (2011) Transkingdom signaling based on bacterial cyclodipeptides with auxin activity in plants. Proc Natl Acad Sci USA 108: 7253–7258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett FB, Wilson AK, Estelle M. (1990) The aux1 mutation of Arabidopsis confers both auxin and ethylene resistance. Plant Physiol 94: 1462–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CMJ, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SCM. (2012) Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol 28: 489–521 [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, van Wees SCM, Hoffland E, van Pelt JA, van Loon LC. (1996) Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis-related gene expression. Plant Cell 8: 1225–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CMJ, van Wees SCM, van Pelt JA, Knoester M, Laan R, Gerrits H, Weisbeek PJ, van Loon LC. (1998) A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10: 1571–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozo MJ, Van Der Ent S, Van Loon LC, Pieterse CMJ. (2008) Transcription factor MYC2 is involved in priming for enhanced defense during rhizobacteria-induced systemic resistance in Arabidopsis thaliana. New Phytol 180: 511–523 [DOI] [PubMed] [Google Scholar]
- Rahman A, Hosokawa S, Oono Y, Amakawa T, Goto N, Tsurumi S. (2002) Auxin and ethylene response interactions during Arabidopsis root hair development dissected by auxin influx modulators. Plant Physiol 130: 1908–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Růzicka K, Ljung K, Vanneste S, Podhorská R, Beeckman T, Friml J, Benková E. (2007) Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19: 2197–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu C-M, Farag MA, Hu CH, Reddy MS, Wei HX, Paré PW, Kloepper JW. (2003) Bacterial volatiles promote growth in Arabidopsis. Proc Natl Acad Sci USA 100: 4927–4932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P, et al. (1999) An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472 [DOI] [PubMed] [Google Scholar]
- Sabatini S, Heidstra R, Wildwater M, Scheres B. (2003) SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev 17: 354–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T. (2007) Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446: 811–814 [DOI] [PubMed] [Google Scholar]
- Schiefelbein J. (2003) Cell-fate specification in the epidermis: a common patterning mechanism in the root and shoot. Curr Opin Plant Biol 6: 74–78 [DOI] [PubMed] [Google Scholar]
- Schwachtje J, Karojet S, Thormählen I, Bernholz C, Kunz S, Brouwer S, Schwochow M, Köhl K, van Dongen JT. (2011) A naturally associated rhizobacterium of Arabidopsis thaliana induces a starvation-like transcriptional response while promoting growth. PLoS ONE 6: e29382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessitsch A, Hardoim P, Döring J, Weilharter A, Krause A, Woyke T, Mitter B, Hauberg-Lotte L, Friedrich F, Rahalkar M, et al. (2012) Functional characteristics of an endophyte community colonizing rice roots as revealed by metagenomic analysis. Mol Plant Microbe Interact 25: 28–36 [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Yun J, Likhacheva AV, Alonso JM. (2007) Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 19: 2169–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Xu Y, Ye S, Jiang H, Chen Q, Liu F, Zhou W, Chen R, Li X, Tietz O, et al. (2009) Arabidopsis ASA1 is important for jasmonate-mediated regulation of auxin biosynthesis and transport during lateral root formation. Plant Cell 21: 1495–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Friml J, Marchant A, Ljung K, Sandberg G, Palme K, Bennett M. (2001) Localization of the auxin permease AUX1 suggests two functionally distinct hormone transport pathways operate in the Arabidopsis root apex. Genes Dev 15: 2648–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Perry P, Hagenbeek D, Van Der Straeten D, Beemster GTS, Sandberg G, Bhalerao R, Ljung K, Bennett MJ. (2007) Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell 19: 2186–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpte C, Wilson AK, Estelle M. (1994) The axr2-1 mutation of Arabidopsis thaliana is a gain-of-function mutation that disrupts an early step in auxin response. Genetics 138: 1239–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda M, Koshino-Kimura Y, Okada K. (2005) Stepwise understanding of root development. Curr Opin Plant Biol 8: 71–76 [DOI] [PubMed] [Google Scholar]
- Van der Ent S, Van Wees SCM, Pieterse CMJ. (2009) Jasmonate signaling in plant interactions with resistance-inducing beneficial microbes. Phytochemistry 70: 1581–1588 [DOI] [PubMed] [Google Scholar]
- Van der Ent S, Verhagen BWM, Van Doorn R, Bakker D, Verlaan MG, Pel MJC, Joosten RG, Proveniers MCG, Van Loon LC, Ton J, et al. (2008) MYB72 is required in early signaling steps of rhizobacteria-induced systemic resistance in Arabidopsis. Plant Physiol 146: 1293–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wees SCM, Pieterse CMJ, Trijssenaar A, Van’t Westende YAM, Hartog F, Van Loon LC. (1997) Differential induction of systemic resistance in Arabidopsis by biocontrol bacteria. Mol Plant Microbe Interact 10: 716–724 [DOI] [PubMed] [Google Scholar]
- Van Wees SCM, Van der Ent S, Pieterse CMJ. (2008) Plant immune responses triggered by beneficial microbes. Curr Opin Plant Biol 11: 443–448 [DOI] [PubMed] [Google Scholar]
- Wada T, Tachibana T, Shimura Y, Okada K. (1997) Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277: 1113–1116 [DOI] [PubMed] [Google Scholar]
- Yang J, Kloepper JW, Ryu CM. (2009) Rhizosphere bacteria help plants tolerate abiotic stress. Trends Plant Sci 14: 1–4 [DOI] [PubMed] [Google Scholar]
- Zamioudis C, Pieterse CMJ. (2012) Modulation of host immunity by beneficial microbes. Mol Plant Microbe Interact 25: 139–150 [DOI] [PubMed] [Google Scholar]