Cadmium-induced oxidative stress leads to nuclear relocation of glyceraldehyde-3-phosphate dehydrogenase.
Abstract
NAD-dependent glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is a ubiquitous enzyme involved in the glycolytic pathway. It has been widely demonstrated that mammalian GAPDH, in addition to its role in glycolysis, fulfills alternative functions mainly linked to its susceptibility to oxidative posttranslational modifications. Here, we investigated the responses of Arabidopsis (Arabidopsis thaliana) cytosolic GAPDH isoenzymes GAPC1 and GAPC2 to cadmium-induced stress in seedlings roots. GAPC1 was more responsive to cadmium than GAPC2 at the transcriptional level. In vivo, cadmium treatments induced different concomitant effects, including (1) nitric oxide accumulation, (2) cytosolic oxidation (e.g. oxidation of the redox-sensitive Green fluorescent protein2 probe), (3) activation of the GAPC1 promoter, (4) GAPC1 protein accumulation in enzymatically inactive form, and (5) strong relocalization of GAPC1 to the nucleus. All these effects were detected in the same zone of the root tip. In vitro, GAPC1 was inactivated by either nitric oxide donors or hydrogen peroxide, but no inhibition was directly provided by cadmium. Interestingly, nuclear relocalization of GAPC1 under cadmium-induced oxidative stress was stimulated, rather than inhibited, by mutating into serine the catalytic cysteine of GAPC1 (C155S), excluding an essential role of GAPC1 nitrosylation in the mechanism of nuclear relocalization, as found in mammalian cells. Although the function of GAPC1 in the nucleus is unknown, our results suggest that glycolytic GAPC1, through its high sensitivity to the cellular redox state, may play a role in oxidative stress signaling or protection in plants.
Plants are exposed to several biotic and abiotic stresses. Among the latter, drought, salinity, extreme temperatures, anoxia, excess light, xenobiotics, and heavy metals dramatically affect plant growth and development as well as crop yield (Jaspers and Kangasjärvi, 2010; Urano et al., 2010; Kosová et al., 2011). Plants have developed sophisticated signaling mechanisms that link the perception of the stress signals to specific responses, such as up- or down-regulation of stress-responsive genes (Yamaguchi-Shinozaki and Shinozaki, 2006). Reactive oxygen and nitrogen species (ROS and RNS, respectively) and phosphorylation cascades of mitogen-activated protein kinases (MAPKs) play a prominent role in these processes (Rodriguez et al., 2010).
ROS and RNS are also produced under physiological conditions and their levels held in check by low-Mr reductants (e.g. glutathione and ascorbate; Foyer and Noctor, 2011) and enzymatic antioxidant systems (e.g. catalases, peroxidases, thioredoxins, and glutaredoxins; Meyer et al., 1999; Apel and Hirt, 2004; Foyer and Noctor, 2005). Various physiological processes are regulated by ROS and RNS, particularly in roots (e.g. transition from cell proliferation to cell differentiation [Tsukagoshi et al., 2010], apical growth of root hairs [Foreman et al., 2003], and lateral root formation [Correa-Aragunde et al., 2004]), but also in leaves (e.g. stomata movements [Kwak et al., 2003]).
However, when plants are exposed to different types of stress, the production of ROS and RNS can overwhelm the scavenging capacity of the cell, thereby leading to accumulation of these reactive molecules with the establishment of oxidative stress conditions (Apel and Hirt, 2004; Møller et al., 2007; Valderrama et al., 2007; Corpas et al., 2008). Interestingly, multiple MAPK modules that mediate oxidative stress responses are not only induced by ROS, but can also regulate ROS levels by stimulating catalase activities (Rodriguez et al., 2010). Moreover, ROS and RNS do interact in vivo. For instance, programmed cell death was induced by a combination of hydrogen peroxide (H2O2) and nitric oxide (NO; Delledonne et al., 2001), and MAPKs were shown to regulate NO levels upon H2O2 induction (Wang et al., 2010).
Implication of ROS and RNS (chiefly H2O2 and NO) in redox signaling probably stems from their capability to induce posttranslational modifications of target proteins (Foyer and Noctor, 2005; Foyer and Noctor, 2009; Yu et al., 2012). Cysteines that are made reactive by their particular protein environment can be primarily oxidized to sulfenic acids (-SOH) by H2O2 and further oxidized to sulfinic or sulfonic forms (-SO2H and -SO3H, respectively) over prolonged oxidative stress (Poole et al., 2004). Alternatively, sulfenic cysteines can form disulfide bonds with other protein cysteines (-SS-) or with glutathione (-SSG, glutathionylation; Zaffagnini et al., 2012a, 2012b). NO can also modify reactive cysteines, giving rise to nitrosothiol groups (-SNO; Besson-Bard et al., 2008; Yu et al., 2012). While sulfinic and sulfonic modifications are irreversible, all other types of redox modifications (-SOH, -SSG, -SS-, and -SNO) can be recovered by physiological redox active systems involving glutaredoxins, thioredoxins, and low-Mr reductants such as reduced glutathione (GSH) and NADPH. In this manner, glutathione, the major thiol-based redox buffer of plant cells (Noctor et al., 2011), may regulate redox signaling pathways (Zaffagnini et al., 2012b). For example, protein Tyr phosphatase, known to regulate the intensity and duration of signals involving MAPK modules (Gupta and Luan, 2003; Rodriguez et al., 2010), is itself regulated by redox modification of active site cysteines (Dixon et al., 2005).
Not surprisingly, several proteomic studies have recently reported that H2O2, NO-donors, and biotic and abiotic stresses applied to plants may affect the redox state of reactive protein cysteines, in addition to affecting their transcriptomic and proteomic profiles (Chen et al., 2003; Polverari et al., 2003; Parani et al., 2004; Lindermayr et al., 2005; Herbette et al., 2006; Roth et al., 2006; Sarry et al., 2006; Besson-Bard et al., 2008; Romero-Puertas et al., 2008; Astier et al., 2011). Because of the high reactivity of its catalytic Cys, cytosolic glyceraldehyde 3-P dehydrogenase (GAPDH) has been identified as a target of different types of redox modifications (Hancock et al., 2005, 2006; Lindermayr et al., 2005; Holtgrefe et al., 2008; Bedhomme et al., 2012).
Plant GAPDHs are abundant and ubiquitous enzymes playing essential metabolic roles in glycolysis and photosynthetic carbon assimilation. However, a pea (Pisum sativum) chloroplast GAPDH isoform has been shown to have uracil DNA glycosylase activity (Wang et al., 1999), and in mammals, GAPDH was shown to fulfill additional nonmetabolic functions, including posttranscriptional regulation, maintenance of DNA integrity, and apoptosis (Sirover, 2011; Tristan et al., 2011). In rat cells, stress conditions may cause nitrosylation of GAPDH catalytic Cys and interaction with Seven in absentia homolog1 (Siah1; an E3 ubiquitin ligase), whose nuclear localization signal drives the GAPDH-Siah1 complex into the nucleus, where the ubiquitinating activity of Siah1 promotes apoptosis (Hara et al., 2005). On the other hand, nitrosylated GAPDH was found to mediate transnitrosylation of nuclear proteins, an activity that could further propagate the redox signal (Kornberg et al., 2010). However, alternative mechanisms have been proposed in which GAPDH translocation to the nucleus is independent on both nitrosylation and Siah1 (Sirover, 2011).
Here, we studied the behavior of the two Arabidopsis (Arabidopsis thaliana) cytosolic GAPDH isoforms GAPC1 (At3g04120) and GAPC2 (At1g13440) under oxidative stress conditions imposed by the common soil pollutant cadmium (Sanità di Toppi and Gabbrielli, 1999). The expression of both genes was stimulated by cadmium, but GAPC1 was the more responsive one. Through the generation of transgenic plants expressing either a transcriptional or a translational GAPC1 reporter, we demonstrated that the oxidative stress induced by cadmium treatment stimulated a transient increase of GAPC1 promoter activity and a steady accumulation of the GAPC1 protein in an inactive state. In vitro studies underlined the sensitivity of Arabidopsis GAPC1 activity to treatment with H2O2 and NO donors, whereas no inhibitory effect was observed in the presence of cadmium under conditions tested. Interestingly, in Arabidopsis roots, the cadmium-dependent oxidative stress induced a massive accumulation of GAPC1 in the nucleus. This process could be mimicked and amplified by mutation of GAPC1 catalytic Cys, thus demonstrating that nitrosylation of GAPC1 is not a requisite for nuclear relocalization, as previously observed in mammalian cells (Hara et al., 2005). To our best knowledge, this is the first demonstration that GAPDH can be translocated to the nucleus of plant cells under oxidative stress conditions.
RESULTS
Cadmium Induces Accumulation of NO and Cytosolic Oxidation in Arabidopsis Root Tips
Cadmium, a common soil pollutant (Sanità di Toppi and Gabbrielli, 1999), has been reported to induce accumulation of NO and ROS in plant cells (Cho and Seo, 2005; Besson-Bard et al., 2009; De Michele et al., 2009; Cuypers et al., 2011). In our hands, 1-week-old Arabidopsis seedlings exposed to different concentrations of cadmium (0.1–0.4 mm) for 24 h displayed a browning region in the root tip, particularly apparent in differentiation and transition zones (Supplemental Fig. S1). However, treatment with 0.1 mm cadmium could be prolonged for up to 72 h without affecting cell viability, although root tip morphology was clearly altered and root hair formation was induced (Supplemental Fig. S1). Treatments with 0.1 mm cadmium for up to 72 h were therefore adopted in the following experiments aimed at testing the capability of cadmium to induce oxidative stress conditions in Arabidopsis seedlings.
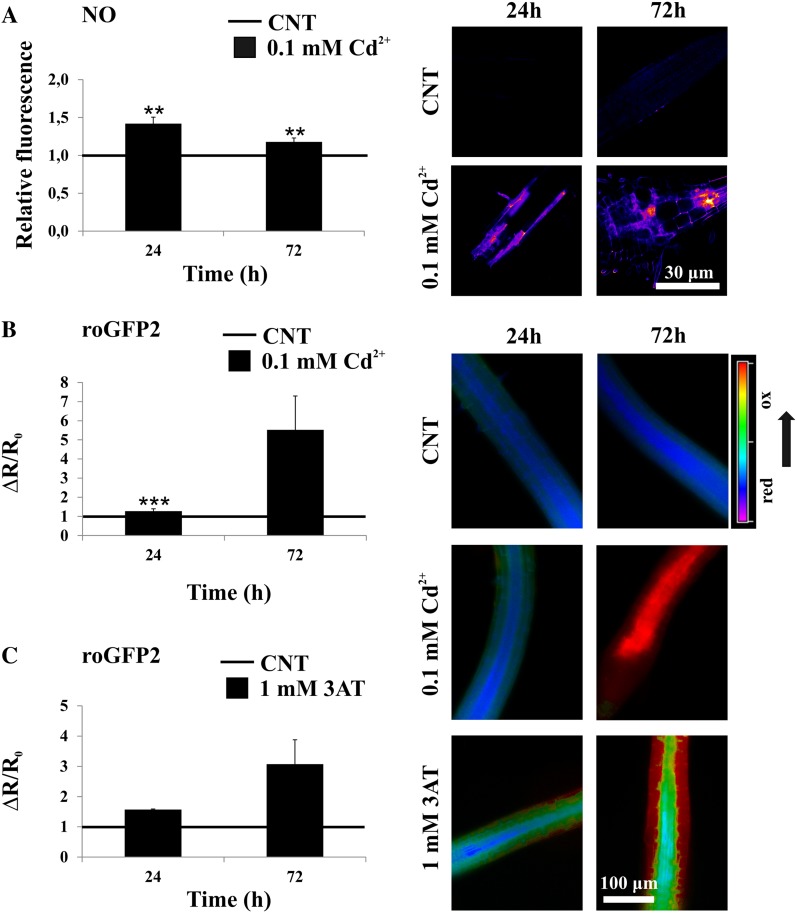
In vivo analysis of NO levels was performed with 4-amino-5-methylamino-2′,7′-difluorofluorescein (DAF-FM) diacetate as a membrane-permeable NO-sensitive indicator (Kojima et al., 1998; Zottini et al., 2007). Cadmium treatment induced NO accumulation at 24 h and was still detected at 72 h (Fig. 1A). The effect of cadmium on the cellular redox state was assessed by using Arabidopsis transgenic seedlings expressing free redox-sensitive (ro)GFP2 (localized in the cytosol and nucleus [Meyer et al., 2007]). The roGFP2 is a redox-sensitive fluorescent probe that can exist in either reduced (dithiol, -SH HS-) or oxidized (disulfide, -SS-) forms, each with a different excitation spectrum. In vivo, the redox state of roGFP2 depends on the redox potential of glutathione (itself a function of the [GSH]2/[GSSG] ratio; Meyer et al., 2007), but possibly also by other molecules (e.g. ROS and RNS) that might react with roGFP2 redox-sensitive cysteines. After 24 h of cadmium treatment, a slight increase in the 405/488 nm probe ratio reflected an increased oxidation of the roGFP2 probe. After 72-h cadmium treatment, this increase was dramatic (Fig. 1B). A similar effect was observed by treating the roGFP2-seedlings with the catalase inhibitor 3-amino-1,2,4-triazole (3-AT; Fig. 1C; Gechev et al., 2005) or with exogenous H2O2 (data not shown). In spite of the clear effect, it is difficult to assess whether the oxidation of the roGFP2 probe upon treatments with cadmium, 3-AT, or H2O2 might be mediated by a partial oxidation of the glutathione pool or alternatively achieved by other means (e.g. direct oxidation by H2O2). In any case, the results of Figure 1 clearly showed that cadmium treatments poise the cellular redox state to more oxidizing conditions. In addition to causing the oxidation of the redox probe roGFP2, these conditions are likely to cause oxidation of several reactive cysteines present in the cell.
Figure 1.
NO production and redox status in Arabidopsis seedlings exposed to cadmium. A, Seven-day-old Arabidopsis wild-type seedlings treated with 0.1 mm cadmium for 24 and 72 h and stained with DAF-FM diacetate for NO quantification. The relative fluorescence values are related to the fluorescence level in control roots (set to 1). False-colored pictures are representative images of transition and elongation root tip zones considered for the data analyses reported in the graph of A. B, Normalized ratio values (405/488 nm) of cytosolic roGFP2 probe in 7-d-old Arabidopsis roGFP2 transgenic seedlings treated with 0.1 mm cadmium for 24 and 72 h. C, Normalized ratio values (405/488 nm) of cytosolic roGFP2 probe in 7-d-old Arabidopsis roGFP2 transgenic seedlings treated with 1 mm 3-AT for 24 and 72 h. False-colored pictures are representative ratiometric images (405/488-nm ratios) of transition and elongation root tip zones considered for the data analyses reported in the graphs of B and C. Asterisks indicate fluorescence levels or normalized ratios that are significantly different from those found in control roots as calculated by Student’s t test (***P < 0.005 and **P < 0.01).
In Root Tips, Cadmium Induced Transient Activation of GAPC1 Promoter and Steady Accumulation of GAPC1 Protein in Inactive State
Arabidopsis contains two genes coding for cytosolic GAPDHs, namely GAPC1 (At3g04120) and GAPC2 (At1g13440; http://www.arabidopsis.org). Our quantitative real-time reverse-transcription (RT)-PCR analysis of whole seedlings revealed that both genes were up-regulated after 24-h cadmium treatment, with GAPC1 being clearly more responsive to the treatment than GAPC2 (Fig. 2). Following experiments were thus focused on GAPC1.
Figure 2.
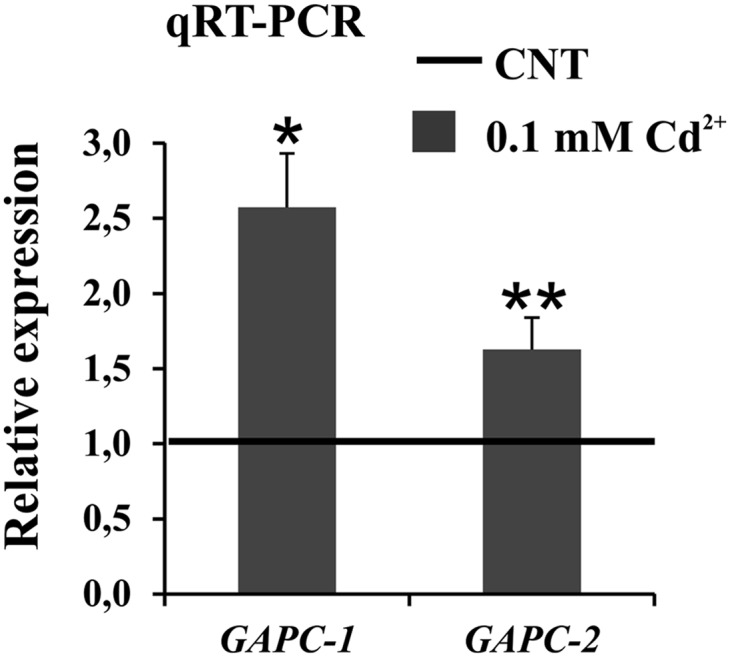
Quantitative real-time RT-PCR expression analysis of GAPC1 and GAPC2 genes in 7-d-old Arabidopsis wild-type seedlings treated with 0.1 mm cadmium for 24 h. The relative expression values of GAPC genes are related to the expression levels in control roots (set to 1). Values represent mean ± sd of relative quantification value of three experiments performed by using templates from three independent biological samples. Asterisks indicate expression levels that are significantly different from those found in control seedlings as calculated by Student’s t test (*P < 0.001 and **P < 0.01).
To investigate the effect of cadmium on GAPC1 expression at the cellular and subcellular level, two different Arabidopsis transgenic lines were generated: (1) a transcriptional reporter line harboring the GUS gene under the control of the GAPC1 promoter sequence (−633 to −1 from the ATG start codon; Supplemental Figs. S2A and S3) and (2) a translational reporter line harboring the chimeric GAPC1-YFP (for yellow fluorescent protein) gene under the control of the same GAPC1 promoter sequence. For each expression cassette, several independent transgenic lines were isolated (Supplemental Fig. S2, A and B). The lines showing a similar expression pattern were used for further analyses, and these selected lines did not show changes in growth and morphology when compared with wild-type plants (data not shown). The western blot performed with total proteins extracted from the three selected translational reporter lines showed the presence of a band at the expected size recognized by the GFP antibody (Supplemental Fig. S2B). The GAPC1 promoter activity was detected throughout plant development, from seedlings (Supplemental Fig. S3A) to mature plants (Supplemental Fig. S3B), and was found to be expressed in root, rosette and cauline leaves, flowers, and silique abscission zone (Supplemental Fig. S3B).
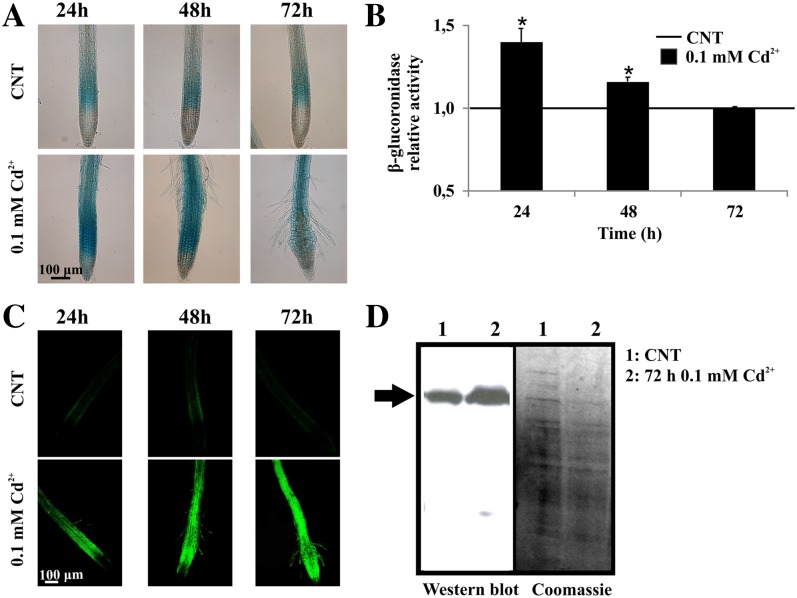
Transgenic Arabidopsis seedlings transformed with the transcriptional reporter pGAPC1::GUS treated with 0.1 mm cadmium showed an increased promoter activity in root tips at 24 h (Fig. 3, A and B), in agreement with quantitative real-time RT-PCR analysis of whole seedlings (Fig. 2). Quantitative determination of GUS activity showed a progressive decrease after 24-h treatment and reached control levels at 72 h (Fig. 3B), although GAPC1 promoter activation at this time point was still revealed by histochemical staining (Fig. 3A). At the cellular level, cadmium induced GAPC1 promoter activity both in elongating and differentiating cells, whereas the activity of the promoter appeared restricted to elongating cells in control roots (Fig. 3A).
Figure 3.
Effects of cadmium treatment on GAPC1 promoter activity and GAPC1-YFP level. A and B, GUS histochemical and quantitative enzymatic activity analyses performed on 7-d-old Arabidopsis pGAPC1::GUS transgenic seedlings treated with 0.1 mm cadmium for 24, 48, and 72 h. The GUS activity assay was performed using total protein extracts from 600 roots in three independent experiments. The relative activity values of GUS are related to the activity in control roots (set to 1). Asterisks indicate GUS activity levels that are significantly different from those found in control roots as calculated by Student’s t test (*P < 0.001). C, Confocal images of 7-d-old Arabidopsis pGAPC1::GAPC1-YFP transgenic seedlings treated with 0.1 mm cadmium for 24, 48, and 72 h. D, Two and one-half micrograms of total protein extracts from roots of 7-d-old Arabidopsis pGAPC1::GAPC1-YFP transgenic seedlings in control conditions or treated with 0.1 mm cadmium for 72 h were assayed by immunoblot analysis using a GFP antibody. Coomassie Blue was used as loading control. The blot was repeated twice.
Same experiments were performed on seedlings expressing the translational reporter GAPC1-YFP under the control of the GAPC1 promoter. Confocal microscope analyses (Fig. 3C) revealed the accumulation of GAPC1-YFP in elongating and differentiating cells, starting from 24-h cadmium treatments. After 48 and 72 h, GAPC1-YFP was accumulated also in the apical meristem and in differentiated root cells (Fig. 3C), pairing with the spread of the GUS histochemical signal of Figure 3A. Western-blot analyses of total proteins extracted from roots of control and 72-h-cadmium-treated seedlings confirmed that the increased fluorescence of Figure 3C was reflecting the accumulation of the GAPC1-YFP chimeric protein (Fig. 3D).
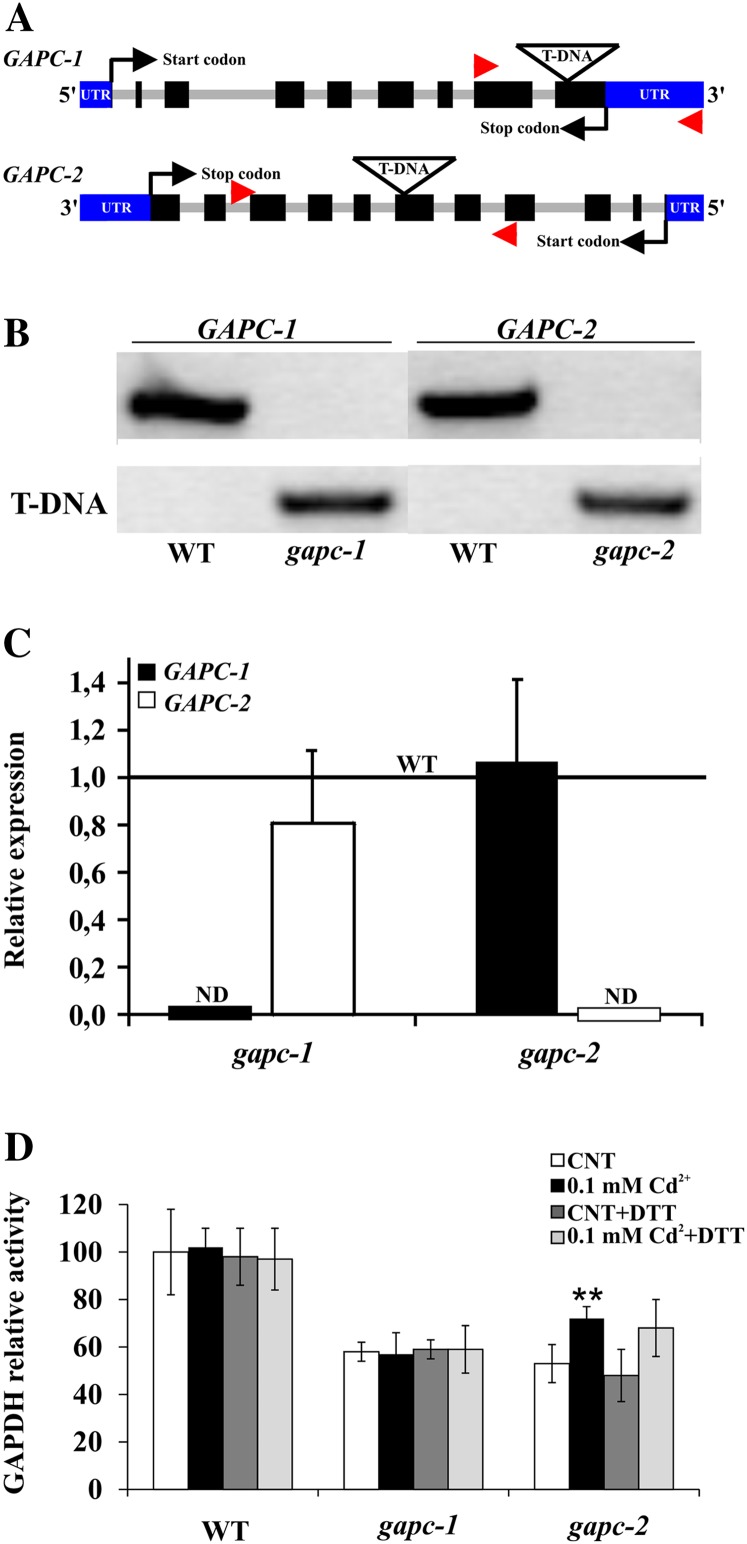
In spite of the induction of GAPC1 protein accumulation, 72-h cadmium treatments had no stimulating effects on the extractable NADH-specific GAPDH activity of whole seedlings (Fig. 4D). However, the interpretation of this result was complicated by the fact that the Arabidopsis genome codes for four different NADH-specific GAPDH isoforms (i.e. two cytosolic GAPC and two plastidial GAPCp; Rius et al., 2008; Muñoz-Bertomeu et al., 2010; Guo et al., 2012). To simplify the picture, knockout homozygous mutants for either GAPC1 or GAPC2 genes were thus isolated (see “Materials and Methods”; Fig. 4, A and B), and real-time PCR analysis revealed the complete absence of either GAPC1 or GAPC2 transcripts in their respective insertional mutant lines (Fig. 4C). Interestingly, no compensatory effects were observed, as neither GAPC2 nor GAPC1 transcript abundance was significantly affected in gapc1 and gapc2 mutants, respectively. Real-time PCR analysis also revealed a higher level of GAPC2 than GAPC1 transcripts in wild-type seedlings (Supplemental Fig. S4), consistent with published microarray data (Winter et al., 2007). In seedlings of each mutant line, the catalytic activity associated to NADH-specific cytosolic GAPDH isoforms (see “Materials and Methods”) was reduced to about 50% of the wild-type level (Fig. 4D), suggesting that both GAPC isoforms may similarly contribute to glycolysis in wild-type plants, with little or no contribution by GAPCp isoforms. Given the different expression of GAPC2 and GAPC1 transcripts (Supplemental Fig. S4), this result also suggests that regulatory mechanisms acting downstream of transcription would reequilibrate the GAPC1/GAPC2 ratio in terms of catalytic activity.
Figure 4.
Isolation and characterization of Arabidopsis gapc1 and gapc2 mutants. A, Schemes of GAPC1 and GAPC2 gene structures of gapc1 (SALK_010839) and gapc2 (SALK_016539) insertional mutants. Arrowheads indicate the position of the primers used to screen homozygous mutants. B, PCR analysis of GAPC1 and GAPC2 genes performed on genomic DNA from the wild type and homozygous gapc1 and gapc2 mutants. The upper bands correspond to the amplification of wild-type alleles, and the lower bands correspond to mutated alleles. C, Quantitative real-time RT-PCR expression analysis of GAPC1 and GAPC2 genes in Arabidopsis wild-type and homozygous gapc1 and gapc2 mutant seedlings. The relative expression values are related to the expression levels of the same genes in the wild-type background, which were set to 1. Note that in control conditions the GAPC2 is expressed 100 times more than GAPC1 (Supplemental Fig. S4). Values represent mean ± sd of relative quantification value of three experiments performed by using templates from three independent biological samples. ND, Not detected. D, NADH-specific cytosolic GAPDH catalytic activity analyses performed with total protein extracts from 7-d-old Arabidopsis wild-type, gapc1, and gapc2 mutant seedlings treated with 0.1 mm cadmium for 72 h. Protein extracts were also treated with 10 mm DTT for 15 min and reported in the same graph. The relative activity values of control mutant seedlings and wild-type treated seedlings are related to the activity in control wild-type seedlings, which was set to 100 (corresponding to 418 ± 42 nmol min–1 mg–1). Asterisks indicate activity levels that are significantly different from those found in control seedlings as calculated by Student’s t test (**P < 0.01). [See online article for color version of this figure.]
Cadmium treatments of gapc1 and gapc2 mutant seedlings induced a moderate increase of NADH-specific GAPDH activity in gapc2 but no effects in gapc1 mutant, implying specific activation of GAPC1. The effect, however, was small (+20%), undetectable in wild-type seedlings (Fig. 4D) and hard to compare with the increase of GAPC1-YFP protein observed under identical experimental conditions (Fig. 3, C and D). These results seem thus to suggest that cadmium induced GAPC1 accumulation in an inactive form.
The reducing agent dithiothreitol (DTT) could not restore the activity of cadmium-overexpressed GAPC1 (Fig. 4D), indicating that reversible redox modifications of GAPC1 catalytic Cys (i.e. sulfonation, nitrosylation, or glutathionylation; Bedhomme et al., 2012) were not the cause of the apparent inactivation. Interestingly, Fourrat et al. (2007) showed that treatments of the protozoan Tetrahymena pyriformis with either sodium nitroprusside (NO donor) or H2O2 led to a dramatic increase of GAPDH protein level without affecting total enzymatic activity.
Catalytic Activity of Recombinant GAPC1 Is Sensitive to NO and H2O2, But Not to Cadmium
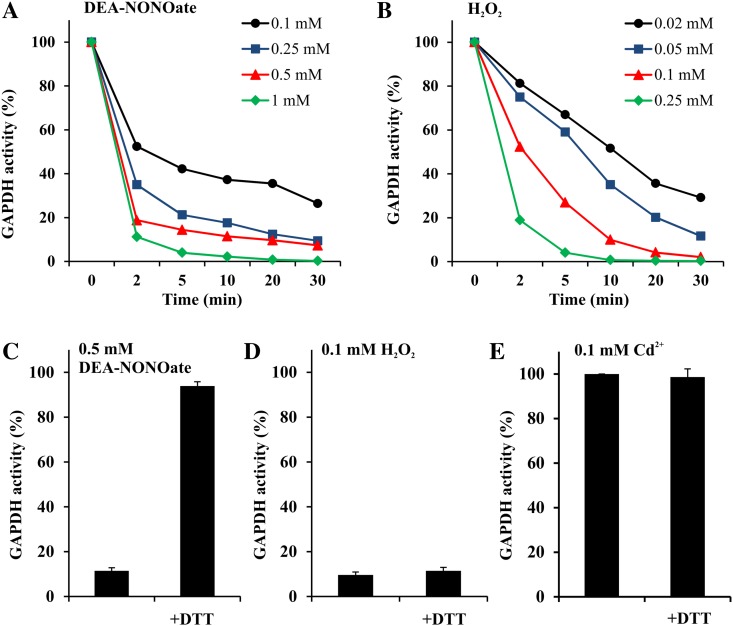
The direct effect of NO, H2O2, and cadmium on GAPC1 catalytic activity was investigated with the recombinant enzyme. Recombinant GAPC1 was inhibited by both NO (delivered by diethylamine [DEA]-NONOate) and H2O2 in a time- and concentration-dependent manner (Fig. 5, A and B). The reducing agent DTT could restore the activity of the NO-treated enzyme (Fig. 5C). In vitro inactivation by H2O2 was instead irreversible (Fig. 5D), although GSH was previously demonstrated to protect GAPC1 from H2O2 inactivation via glutathionylation of transiently oxidized GAPC1 sulfonate and recovery by glutaredoxins or thioredoxins (Bedhomme et al., 2012). It is thus concluded that both NO and H2O2 treatments of GAPC1 might result in a reversible inhibition of the enzyme in vivo, provided that GSH is available for recovery. On the other side, the activity of recombinant GAPC1 proved to be insensitive to cadmium treatment (0.1 mm; Fig. 5E).
Figure 5.
Oxidant treatments of recombinant GAPC1. A, Time- and concentration-dependent inactivation of recombinant GAPC1 by the NO donor DEA-NONOate. B, Time- and concentration-dependent inactivation of recombinant GAPC1 by H2O2. C, Reversibility of DEA-NONOate treatment. Recombinant GAPC1 was incubated for 10 min with 0.5 mm DEA-NONOate, and the reversibility of GAPC1 inactivation was then assessed by incubation for 10 min in the presence of 20 mm DTT. D, Reversibility of H2O2 treatment. Recombinant GAPC1 was incubated for 10 min with 0.1 mm H2O2, and the reversibility of GAPC1 inactivation was then assessed by incubation for 10 min in the presence of 20 mm DTT. E, Effect of cadmium treatment on GAPC1 activity. Recombinant GAPC1 was incubated with 0.1 mm cadmium for 10 min. The reversibility of the treatment was assessed by incubating the protein with 20 mm DTT after exposure to 0.1 mm cadmium for 10 min. Aliquots of the incubation mixtures were withdrawn at the indicated time points, and the NADH-dependent activity was determined. Data are represented as the mean percentage of maximal control activity ± sd. [See online article for color version of this figure.]
Evidence That GAPC1 Expression Is Affected by Glutathione Status
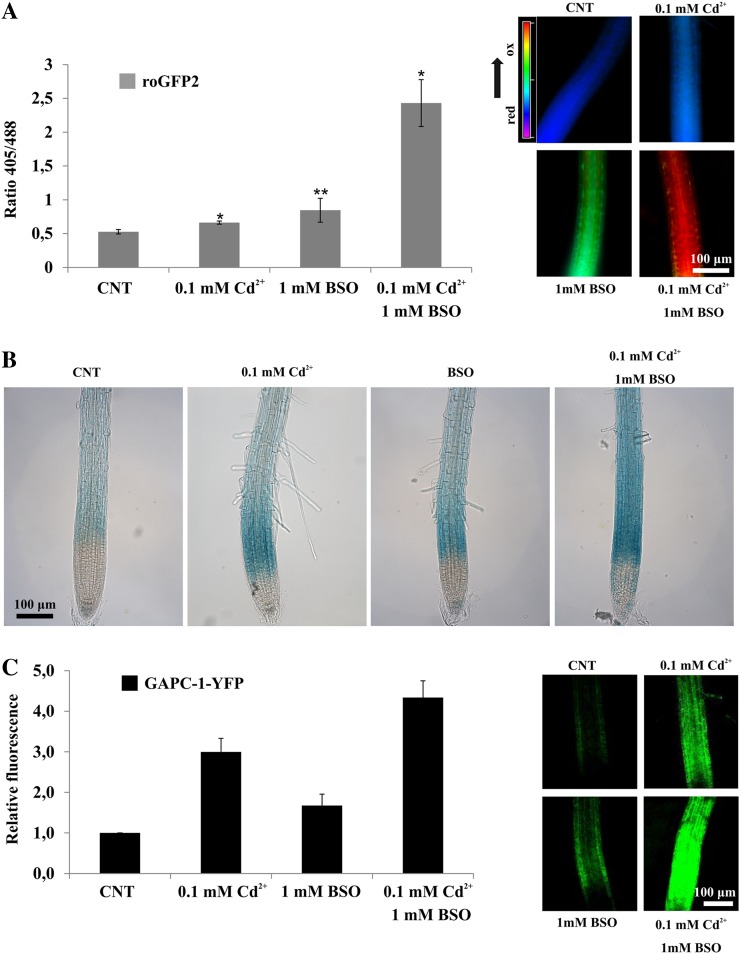
In further investigations, cytosolic oxidation was alternatively obtained by treating seedlings with l-buthionine-sulfoximine (BSO), an inhibitor of glutathione biosynthesis (Griffith, 1982), for 24 h (Fig. 6A). The oxidizing effect of BSO (1 mm), monitored by analyzing roGFP2 seedlings’ response, was comparable to that of cadmium (0.1 mm), and oxidation of the roGFP2 probe was severe when the seedlings were cotreated with both compounds. Same treatments applied to pGAPC1::GUS and pGAPC1::GAPC1-YFP seedlings showed that BSO itself induced both GAPC1 promoter activity and protein accumulation in the seedling root tips, as compared with control condition (Fig. 6, B and C). The effect was again amplified in seedlings treated with both BSO and cadmium together (Fig. 6, B and C). By contrast, 24-h treatment with GSH (1 mm) had opposite effects on both GAPC1 promoter activity and protein accumulation in root tips (data not shown). The BSO effect strongly suggests a link between the increased GAPC1-YFP accumulation and the cellular redox status, independently from the presence of cadmium. This observation was also strengthened by the fact that treatments with the catalase inhibitor 3-AT did also induce GAPC1-YFP accumulation (data not shown). Taking together the data shown in Figure 6, it can be observed that BSO affects the roGFP2 signal more than cadmium, but cadmium affects GAPC1 expression more than BSO, and that BSO and cadmium act synergistically or additively on both factors. This could reflect the fact that cadmium depletes the glutathione pool (for phytochelatin synthesis; De Michele et al., 2009) and generates ROS or other oxidants, whereas the effect of BSO is largely confined to depletion of the glutathione pool.
Figure 6.
Effects of inhibition of glutathione biosynthesis on redox status, GAPC1 promoter activity, and GAPC1-YFP level in control and cadmium-treated roots. A, Ratio values (405/488 nm) of 7-d-old Arabidopsis roGFP2 transgenic seedling root tips in control conditions or treated with 0.1 mm cadmium, 1 mm BSO, and 0.1 mm cadmium plus 1 mm BSO for 24 h. False-colored pictures are representative ratiometric images (405/488-nm ratios) of transition and elongation root tip zones considered for the data analyses reported in the graph. B, Histochemical analysis of GUS activity in 7-d-old Arabidopsis pGAPC-1::GUS transgenic seedling root tips in control conditions or treated with 0.1 mm cadmium, 1 mm BSO, and 0.1 mm cadmium plus 1 mm BSO for 24 h. C, YFP fluorescence of 7-d-old Arabidopsis pGAPC-1::GAPC1-YFP transgenic seedling root tips in control conditions or treated with 0.1 mm cadmium, 1 mm BSO, and 0.1 mm cadmium plus 1 mm BSO for 24 h. The graph reports the semiquantitative pixel intensity analysis. The relative fluorescence values are related to the fluorescence level in control roots (set to 1). Pictures are representative images of transition and elongation root tip zones considered for the data analyses reported in the graph. Asterisks indicate fluorescence levels that are significantly different from those found in control roots as calculated by Student’s t test (*P < 0.005 and **P < 0.05).
Cadmium Induces Nuclear Relocalization of Inactive GAPC1-YFP Chimeric Protein
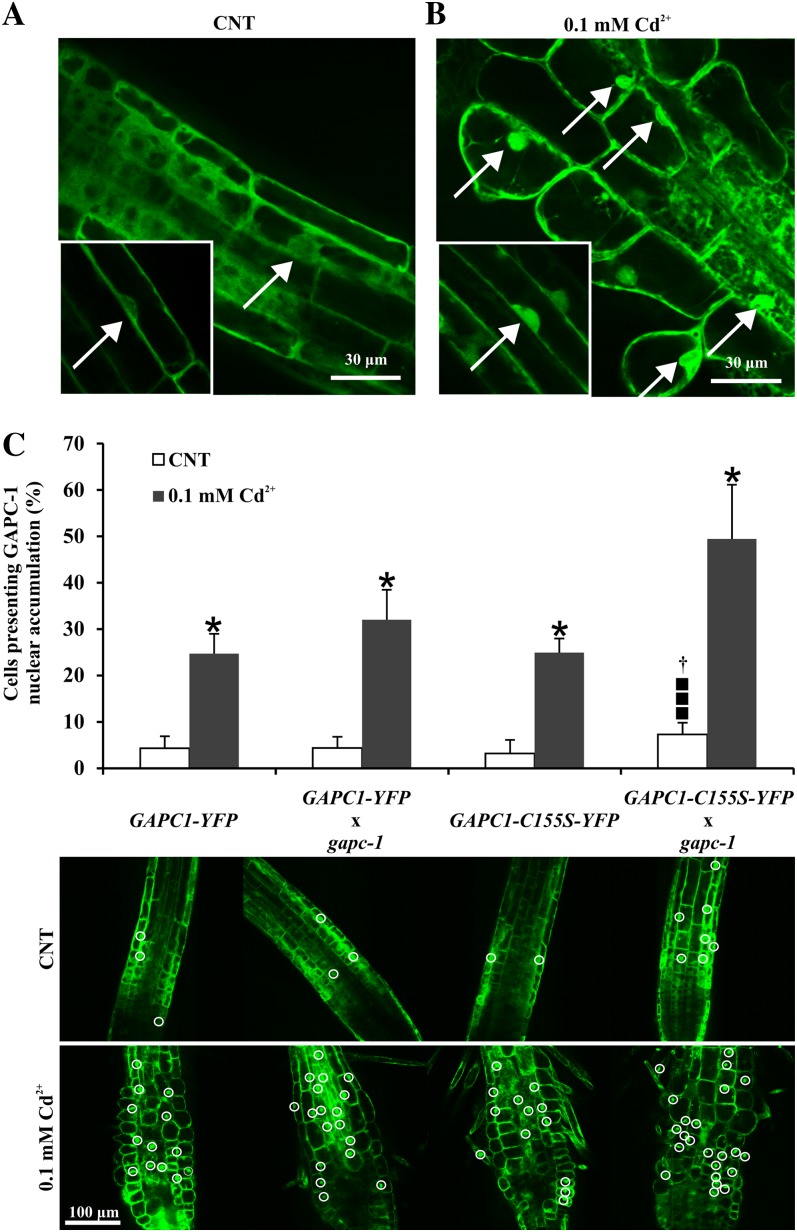
A subcellular localization analysis of root tips of pGAPC1::GAPC1-YFP seedlings treated for 72 h with cadmium showed a nuclear accumulation of GAPC1-YFP chimeric proteins in cells of the transition/elongation zone (Fig. 7B). Interestingly, green fluorescent nuclei were observed also in root epidermal cells induced to differentiate into root hairs under the effect of cadmium (Fig. 7B).
Figure 7.
GAPC1-YFP chimeric protein accumulation in the nucleus of cadmium-treated root cells. A, Cells from the elongation zone of a typical root tip of 7-d-old Arabidopsis showing the cytosolic localization of GAPC1-YFP chimeric protein. Inset, representative cell where the YFP signal clearly surrounds the nucleus but is not inside the nucleus. B, Cells from the elongation zone of a typical root tip of 7-d-old Arabidopsis challenged with 0.1 mm cadmium for 72 h showing the characteristic morphological alteration and the GAPC1-YFP accumulation in the nuclei of the responding cells. Inset, representative cell (same focal plane of the inset shown in A) where the YFP signal is inside the nucleus. Note the presence of ectopic root hairs as typical feature of cadmium treatment. C, Effect of mutation of Cys-155 on movement of the protein into the nucleus. Seven-day-old Arabidopsis pGAPC1::GAPC1-YFP, pGAPC1::GAPC1-YFP × gapc1, pGAPC1::GAPC1-C155S-YFP, and pGAPC1::GAPC1-C155S-YFP × gapc1 transgenic seedlings were treated with 0.1 mm cadmium for 72 H, and root tips were analyzed by means of confocal microscope. White circles indicate cells that present GAPC1-YFP nuclear accumulation. In the graph, values represent mean ± sd of the percentages of cells presenting GAPC1-YFP nuclear accumulation. Asterisks indicate values that are significantly different from those found in control roots as calculated by Student’s t test (*P < 0.001). Black squares indicate values that are significantly different from those found in the pGAPC1::GAPC1-YFP × gapc1 transgenic line as calculated by Student’s t test (P < 0.05). Dagger indicates values that are significantly different from those found in pGAPC1::GAPC1-C155S-YFP transgenic line as calculated by Student’s t test (P < 0.001).
Hara et al. (2005) described the nuclear relocalization of GAPDH in rat cells under oxidative stress conditions and showed it to be a consequence of (1) nitrosylation of its catalytic Cys-150 (C150) and (2) interaction with the nuclear carrier Siah1, with Lys-225 (K225) of GAPDH being required for such binary complex formation. Based on these findings, the following point mutations were introduced in the pGAPC1::GAPC1-YFP expression cassette: C155S (catalytic Cys, also known as Cys-149, based on the numbering of crystallized GAPDH from Bacillus stearothermophilus; Biesecker et al., 1977; Bedhomme et al., 2012), C159S, C155/C159S, and K230A (K230 is homologous to the Lys involved in GAPDH-Siah1 binding in animal GAPDH). Arabidopsis protoplasts were transiently transformed, and subcellular localization analyses demonstrated that all mutated GAPC1 forms were localized mainly in the cytosol, because we failed to detect any signal in the nucleus (Supplemental Fig. S5). Generation of stable transgenic lines by transforming wild-type plants with the four different constructs allowed testing nuclear relocalization upon cadmium treatments. Accumulation of GAPC1-YFP in cadmium-treated plants was confirmed, but also nuclear relocalization was maintained in each mutant, at a similar extent to that observed in the original pGAPC1::GAPC1-YFP plants (Supplemental Fig. S6).
However, because GAPC1 is a tetrameric protein, we speculated that wild-type GAPC1 subunits might build spurious GAPDH tetramers with mutated GAPC1-YFP subunits in transgenic plants, thus preventing a clear-cut interpretation of the relocalization experiments. To overcome this problem, plants expressing the chimeric proteins (GAPC1-YFP and GAPC1-C155S-YFP) were crossed with gapc1 plants. F2 lines with no wild-type GAPC1 alleles were then selected (GAPC1::GAPC1-YFP-gapc1–/– and GAPC1::GAPC1-C155S-YFP-gapc1–/–). Reevaluation of the effect of cadmium treatments on these plants revealed that nuclear accumulation of GAPC1-YFP was higher in gapc1 than in the wild-type background (Fig. 7C). Interestingly, the absence of wild-type GAPC1 (gapc1 background) allowed to detect a clear positive effect of C155S mutation on the extent of nuclear accumulation (Fig. 7C), i.e. exactly the opposite of what previously suggested by works on different systems (Hara et al., 2005; Kornberg et al., 2010), and this effect was observed both in the absence or in the presence of cadmium.
DISCUSSION
Increasing attention has been recently paid to the study of “new roles” played by “old enzymes.” An emerging idea is that certain enzymes, so far only considered as key players in fundamental metabolic pathways, may play different roles in signal transduction or other stress-related responses. To recognize their versatility, these have been named moonlighting proteins (Jeffery, 1999; Huberts and van der Klei, 2010; Sirover, 2011) and different mechanisms were proposed for the control of their moonlighting activity, chiefly posttranslational modifications and protein-protein interactions (Moore, 2004). Several reviews, largely inspired by studies on the animal field, have recently proposed plant GAPDH as an example of a moonlighting protein and S-nitrosylation of its catalytic Cys as a possible means to control GAPDH moonlighting activities (Astier et al., 2011; Gaupels et al., 2011). However, despite a general interest in nonglycolytic roles of plant GAPDHs, few reports have experimentally addressed the topic.
Cadmium, a well-known inducer of oxidative stress (De Michele et al., 2009), is shown here to induce a production of NO in root tip cells of Arabidopsis seedlings (DAF-FM diacetate probe) and to cause cytosolic oxidation as determined by the redox probe roGFP2. Arabidopsis plants express two different cytosolic GAPDHs, GAPC1 and GAPC2, each accounting for nearly 50% of total NADH-specific GAPDH activity in whole seedlings. Both GAPC1 and GAPC2 transcripts were up-regulated by cadmium, but GAPC1 was the more responsive between the two genes. Further studies on GAPC1 gene expression and protein accumulation were thus conducted with the help of Arabidopsis transgenic lines expressing either the GUS transcriptional reporter or the GAPC1-YFP translational reporter. With these plants, we could show that cadmium induced the activation of the GAPC1 promoter, and the accumulation of the GAPC1-YFP chimeric protein, in the same root cells where NO and cytosolic oxidation also increased. Cytosolic oxidation could also be induced by the catalase inhibitor 3-AT or BSO, an inhibitor of glutathione biosynthesis, and again this condition was associated to GAPC1 accumulation in root tips.
However, the relationship between cadmium and GAPC1 in Arabidopsis plants is made complicated by the fact that cadmium, in vivo, seems to cause GAPC1 inactivation, concomitantly with the induction of GAPC1 protein accumulation. In vitro experiments showed that GAPC1 enzyme activity is insensitive to cadmium but rapidly inactivated by oxidants such as NO or H2O2. In principle, GAPC1 oxidative inactivation should be countered in vivo by glutathione, either directly or through glutaredoxins. However, in root tips of cadmium-treated seedlings, the glutathione pool is probably not reduced enough to keep active this recovery system, as suggested by the progressive oxidation of the roGFP2 probe. This conclusion fits with our observations that cadmium induced in vivo an accumulation over time of chimeric GAPC1-YFP (Fig. 3C), without significantly increasing GAPC1 activity (Fig. 4D). Perhaps the oxidative stress conditions imposed by cadmium activate a signaling cascade that enhances GAPC1 expression to compensate its ongoing inactivation. Interestingly, similarly to our results with cadmium, salt stress has been shown to induce, in Arabidopsis root cells, the accumulation of GAPC1 and proteins involved in ROS scavenging (Jiang et al., 2007).
Though normally considered cytosolic, GAPC was proposed to be localized in the nucleus of plant cells, and nuclear localization of a glycolytic enzyme is taken as a hint of moonlighting activity. Anderson et al. (2004) have shown by immunogold localization the presence of GAPC both in the cytosol and the nucleus of pea leaf cells. Holtgrefe et al. (2008) have proposed that, in Arabidopsis protoplasts, both GAPC1 and GAPC2 could localize also in the nucleus, in addition to the cytosol, and bind DNA. In the same Arabidopsis system, tobacco (Nicotiana tabacum) cytosolic GAPDH was shown to interact, both in the cytosol and in the nucleus, with an osmotic stress-activated protein kinase (NtOSAK) from tobacco too (Wawer et al., 2010). Although complex formation in the nucleus was not unaffected by stress conditions or NO donors, the interaction was abolished when both active-site cysteines of GAPDH were mutated into serines (Wawer et al., 2010). These studies were performed with Arabidopsis protoplasts expressing GAPDH under the control of the p35S promoter (Holtgrefe et al., 2008; Wawer et al., 2010), and in similar experiments, we could nicely confirm the nuclear localization of GAPC1-YFP expressed under the control of p35S (data not shown). However, replacing this constitutive strong promoter with the GAPC1 endogenous promoter largely prevented, in control conditions, the nuclear accumulation of GAPC1 (Supplemental Fig. S5). We therefore suspect that, in transiently transformed protoplasts with a strong promoter such as p35S, nuclear GAPDH accumulation could be a mere consequence of its overexpression. To overcome this problem, we generated stable transgenic lines expressing GAPC1-YFP under the control of the endogenous GAPC1 promoter. These plants proved instrumental to investigate GAPDH subcellular localization under both control and stress conditions. Nuclear localization of GAPC1-YFP in pGAPC1::GAPC1-YFP Arabidopsis plants was monitored in root tips and found to be very rare in seedlings grown under control conditions, but strongly stimulated by cadmium treatments.
Cadmium-induced nuclear relocalization of GAPC1 in Arabidopsis root tips is reminiscent of the nuclear relocalization of GAPDH described by Hara et al. (2005) in mammalian cells challenged with different types of stimuli. In the mammalian system, GAPDH needs to be nitrosylated on its catalytic Cys before forming a binary complex with Siah1 that can move into the nucleus, where the ubiquitinating activity of Siah1 is exerted. In spite of superficial analogies, our results on the Arabidopsis system challenged with cadmium underpin some crucial differences. In the search for unambiguous results, the GAPC1-YFP chimera was introduced into a gapc1 background and it was demonstrated that cadmium-induced nuclear localization of GAPC1 was not inhibited, but rather stimulated, by the substitution of the catalytic Cys C155 with a Ser. Based on this unexpected result, we could rule out that S-nitrosylation or any other type of redox modification of the catalytic Cys is an essential prerequisite for binding with a putative partner protein that allows relocalization of GAPC1 into the nucleus. It is possible, however, that inactivation of the enzyme, either by redox modification or by mutagenesis, may be a sufficient condition to drive GAPDH into the nucleus. In animal cells, mechanisms independent on S-nitrosylation of catalytic Cys were also proposed for nuclear localization of GAPDH under several conditions (Sirover, 2011). Interestingly, Wawer et al. (2010) also found that nuclear localization of the GAPDH-NtOSAK complex was not stimulated by NO donors, though complex formation did depend, somehow, on GAPDH catalytic Cys. Interestingly, in mammalian cells, several nonmetabolic functions have been reported for GAPDH in the nucleus, including posttranscriptional regulation and apoptosis (Sirover, 2011; Tristan et al., 2011). The demonstration that mammalian and also chloroplastic GAPDHs have uracil glycosylase activity (Wang et al., 1999; Mazzola and Sirover, 2003) opens an interesting scenario on a possible role played by the GAPC1 in the nucleus. The uracil glycosylase activity of GAPDH is associated with its monomeric form present in the nucleus (Mazzola and Sirover, 2003), whereas the glycolytic cytosolic form is tetrameric. Bearing in mind that the uracil glycosylase activity is important for the maintenance of DNA integrity and is necessary to prevent mutagenesis by eliminating uracil from DNA molecules, it is possible that the nuclear accumulation we observed for GAPC1 may fit with the cell requirement of preventing possible DNA damaging due to oxidative stress.
Whatever the mechanism, we show in this work that GAPC1, in addition to its glycolytic function, may well behave as a moonlighting protein involved in an oxidative stress-signaling pathway. Oxidative stress conditions, in our case promoted in vivo by cadmium treatments, induce the expression of GAPC1, but the protein accumulates as inactive form and is relocalized into the nucleus. The complex relationships between GAPC1 and the cellular redox environment suggest that it may act as an oxidative stress sensor.
The understanding of the mechanisms involved in GAPC1 nuclear relocalization and the role played by GAPC1 in the nucleus, along with the study of the role played by redox posttranslational modifications, will be important topics to investigate in the future. Future research will be also addressed to discovery, by means of proteomic approaches, which are the proteins that might interact with GAPC1 during oxidative stress and are potentially responsible for its nuclear accumulation. The transgenic lines generated would be helpful in such analyses.
MATERIALS AND METHODS
Plant Material and Growth Conditions
All the Arabidopsis (Arabidopsis thaliana) plants used in this study were of the Columbia ecotype. Plants were grown on Jiffy-Pot (http://www.jiffypot.com/) under 16-h/8-h cycles of white light (approximately 75 µE) at 22°C. Seedlings were grown in vitro for the selection of transformants and experiments. Seeds were surface-sterilized and, after cold treatment for 2 to 3 d, were exposed to 16-h/8-h cycles of white light approximately 75 μE) in growth chambers. Seeds were grown at 23°C on vertical plates containing one-half-strength Murashige and Skoog medium (Murashige and Skoog, 1962) including 0.8% (w/v) agar (Duchefa). The medium was enriched with 0.1% (w/v) Suc and 0.05% (w/v) MES. The pH of the media was adjusted to 6.0 ± 0.1 with 0.5 n KOH before autoclaving at 121°C for 20 min. Seeds of the transgenic roGFP2 Arabidopsis line (Meyer et al., 2007) were provided by Markus Schwarzländer. Seeds of gapc1 and gapc2 Arabidopsis transfer DNA (T-DNA) insertional mutant lines were from the SALK Institute (SALK_010839 and SALK_016539, respectively).
Genotyping of Insertional gapc1 and gapc2 Mutants
T-DNA insertion mutant lines gapc1 (SALK_010839) and gapc2 (SALK_016539) were selected from the Salk collection (Alonso et al., 2003). GAPC1 gene (At3g04120) is composed of nine exons, and the gapc1 insertion line SALK_010839 presents the T-DNA insertion in the ninth exon (Fig. 4A). This line is a single T-DNA insertion line and was previously isolated and characterized (Rius et al., 2008). The GAPC2 gene (At1g13440) is composed of 11 exons, and the T-DNA insertion in the SALK_016539 line is located in the sixth exon (Fig. 4A) and was previously isolated and characterized (Guo et al., 2012).
The wild-type GAPC1, GAPC2, and T-DNA insertion gapc1 and gapc2 alleles were identified using PCR with the following primers: LP-GAPC1 (5′-CCGCACATCTGTTAATGAATTTC-3′) and RP-GAPC1 (5′-CTCAGAAGACTGTTGATGGGC-3′) to identify GAPC1 wild-type alleles, LP-GAPC2 (5′-GGTTAGGACTGAGGGTCCTTG-3′) and RP-GAPC2 (5′-GGCATCAGGTACATAATCATGG-3′) to identify GAPC2 wild-type alleles, and LBa1-SALK (5′-TGGTTCACGTAGTGGGCCATCG-3′) with RP-GAPC1 (5′-CTCAGAAGACTGTTGATGGGC-3′) or RP-GAPC2 (5′-GGCATCAGGTACATAATCATGG-3′) to identify, respectively, gapc1 and gapc2 mutated alleles.
Enzyme Analyses
For the analyses of GAPDH activities, 400 7-d-old Arabidopsis seedlings were pooled and homogenized in 50 mm Tris-HCl (pH 7.4) and 0.05% (w/v) Cys at 4°C. After centrifugation at 16,100g for 20 min at 4°C, the protein content was quantified according to the method of Bradford (1976). GAPDH activity was monitored spectrophotometrically at 340 nm and 25°C in a reaction mixture containing 50 mm Tris-HCl (pH 7.5), 5 mm MgCl2, 3 mm 3-phosphoglycerate, 1 mm EDTA, 5 units mL–1 of 3-phosphoglycerate kinase (from rabbit muscle; Sigma), 2 mm ATP, and 0.2 mm NADH. For calculations, an extinction coefficient of 340 for NADH of 6.23 mm–1 was used. The NADH-dependent GAPDH activity of crude extracts may depend on both NADH-specific GAPDH (i.e. glycolytic isoforms) and NAD(P)H-GAPDH isoforms of the Calvin-Benson cycle. The maximal NADPH-dependent GAPDH activity was assayed as above, except that 0.2 mm NADPH substituted NADH and the extracts were incubated with a 1,3-bisphosphoglycerate generating system for 10 min before assaying. Because the NADH activity of fully activated NAD(P)H-GAPDH is approximately one-third of its NADPH activity (Sparla et al., 2005), the activity of glycolytic GAPDH isoforms was estimated by subtracting one-third of the fully activated NADPH activity from the total NADH activity of the crude extract.
Detection of NO Production
For detection of NO production, 7-d-old Arabidopsis seedlings were incubated with the cell-permeable fluorescent probe DAF-FM diacetate (Alexis Biochemicals), with a final concentration of 5 μm in the loading buffer solution (0.25 mm KCl, 1 mm CaCl2, and 5 mm MES-KOH, pH 5.7) for 15 min. Seedlings were then washed three times with fresh buffer and examined by confocal microscope as previously reported (Zottini et al., 2007).
RNA Isolation and cDNA Synthesis
Seven-day-old Arabidopsis seedlings were harvested, frozen in liquid nitrogren, and stored at –80°C. The frozen samples were powdered in liquid nitrogen, and RNA was isolated with the TRIzol method. Then, the total RNA was purified using an RNeasy kit, including DNase digestion (Qiagen). Complementary DNA (cDNA) was synthesized by ImProm-II Reverse Transcriptase (Promega) from 1 μg of purified RNA.
Quantitative Real-Time RT-PCR Primer
The quantitative real-time RT-PCR expression analysis of GAPC1 and GAPC2 genes was performed using the following primers: GAPC1-F (GAGGTGATGGGAGTTTGTAGAC) and GAPC1-R (TACGTCATCATCAACGGG) for the expression analysis of the GAPC1 gene, GAPC2-F (TGCGCAGTCATGAGAGTT) and GAPC2-R (AGTTGCCAGTTGGGTTTG) for the expression analysis of the GAPC2 gene, and EF-1α-F (TGAGCACGCTCTTCTTGCTTTCA) and EF1α-R (GGTGGTGGCATCCATCTTGTTACA) for the expression analysis of the elongation factor1α (EF1α) gene.
RNA Analysis
Quantitative real-time RT-PCR using FAST SYBR Green I technology was performed on an ABI PRISM 7500 sequence detection system (Applied Biosystems) using the following cycling conditions: initial denaturation at 95°C for 10 min, 40 cycles of 15 s at 95°C, and 1 min at 60°C, followed by melt-curve stage analysis to check for specificity of the amplification.
The reactions contained Power SYBR Green PCR Master Mix (Applied Biosystems), 300 nm of gene-specific forward and reverse primers, and 1 μL of the diluted cDNA in a 20-μL reaction. The negative controls contained 1 μL RNase-free water instead of the cDNA. The primer efficiencies were calculated as E = 10–1/slope on a standard curve generated using a 4- or 2-fold dilution series over at least five dilution points of cDNA. The expression analysis of GAPC genes was performed by the Pfaffl method, using EF1α as the reference gene (Pfaffl, 2001; Remans et al., 2008).
Genetic Materials
The Arabidopsis GAPC1 coding sequence was amplified by PCR from Arabidopsis cDNA using the following primers where the NcoI sites were introduced: forward 5′-CATGCCATGGCTGACAAGAAGATTAGG-3′ and reverse 5′-CATGCCATGGCGGAGGCCTTTGACATGTGGACGATCAA-3′. The Arabidopsis GAPC1 promoter sequence (–633 bp to –1 bp) was amplified by PCR from Arabidopsis genomic DNA using the following primers where the EcoRI site was introduced: forward 5′-CATGGAATTCCGAGTTTTTGATAGGGACTTTTGCT-3′ and reverse 5′-CATGGAATTCTGTAGAATCGAAAACGAGAGTTAGA-3′.
DNA Constructs
For the expression of the GUS transcriptional reporter and the GAPC1-YFP translational reporter genes, the pGreen0029 (Hellens et al., 2000) binary vector was used.
To isolate the GAPC1 promoter, we amplified by PCR 633 bp upstream of the GAPC1 ATG start codon. The amplicon was digested with EcoRI and ligated into a modified pGreen0029 binary vector upstream of the GUS coding sequence fused with the nopaline synthase terminator (Valerio et al., 2011). The obtained vector was named pGAPC1-GUS.
To obtain the pGreen0029-p35S::GAPC1-YFP binary vector, the GAPC1 coding sequence was amplified by PCR using as a template the cDNA obtained by the retrotranscription of total RNA extracted by 4-week-old Arabidopsis leaves. The GAPC1 amplicon was digested with the NcoI enzyme and ligated upstream of the YFP coding sequence in the pGreen-p35S::YFP plasmid (Valerio et al., 2011). The pGreen0029-pGAPC1::GAPC1-YFP binary vector was constructed introducing the GAPC1 promoter sequence upstream of the GAPC1-YFP coding sequence by replacing the 35S promoter using the EcoRI restriction sites.
To study the role of specific GAPC1 residues, several transgenic Arabidopsis lines carrying the pGAPC1::GAPC1-YFP expression cassette mutated in the GAPC1 sequence were produced. The introduction of specific mutations in the GAPC1 sequence was carried out as described by Makarova et al. (2000) with some modifications. The mutagenesis was performed by PCR reaction using 0.5 µg of pGreen0029-pGAPC1::GAPC1-YFP binary vector as template in a reaction volume of 25 µL containing 0.5 units Phusion High-Fidelity DNA Polymerase (Finnzymes), 0.2 mm deoxynucleotide triphosphates, and 0.2 µm of primer harboring the specific mutation(s). To introduce the mutations C155S, C159S, and K230A and the double mutation C155S-C159S, the following primers were used: 5′-CATTGTCTCCAACGCTAGCTCCACCACTAACTGCCTTGCTC-3′, 5′-CGCTAGCTGCACCACTAACTCCCTTGCTCCCCTTGCCAAGG-3′, 5′-CTCCAACGCTAGCTCCACCACTAACTCCCTTGCTCCCCTTG-3′, and 5′-GCTTCCAGCTCTTAACGGAGCATTGACTGGAATGTCTTTCC-3′, respectively. The PCR reaction was performed using the following cycling conditions: initial denaturation at 95°C for 3 min, 18 cycles of 15 s at 95°C, 1 min of annealing at 61°C, 5 min of extension at 72°C, and a final step of 10 min at 72°C. Then, the PCR product was treated with DpnI endonuclease for 2 h at 37°C. Finally, 2 µL of digested DNA was transformed into Escherichia coli XL1 Blue supercompetent cells.
Agrobacterium tumefaciens Strains
For the use of pGreen-derived binary vectors, the A. tumefaciens GV3101-pSoup strain was used (Hellens et al., 2000). Binary vectors were introduced in A. tumefaciens GV3101 strain by freeze-thaw method. One microgram of plasmid DNA was added to the competent cells, frozen in liquid nitrogen for 5 min, and shocked at 37°C for 5 min. The bacterial culture was incubated at 28°C for 3 h with gentle shaking in 1 mL yeast extract peptone medium (10 g L–1 bacto-tryptone, 10 g L–1 yeast [Saccharomyces cerevisiae] extract, and 5 g L–1 NaCl, pH 7.0) and then spread out on a yeast extract peptone agar plate containing the appropriate antibiotic selection (50 mg L–1 gentamycin, 50 mg L–1 rifampicin, 50 mg L–1 kanamycin, and 5 mg L–1 tetracycline).
Transgenic Plants
Transgenic Arabidopsis plants were generated by floral dip method (Clough and Bent, 1998) using transformed A. tumefaciens strain GV3101. Transformed seedlings were selected on medium supplemented with 50 μg mL–1 kanamycin. For each construct, different Arabidopsis independent transgenic lines were isolated. None of the transgenic lines selected, with the different constructs, showed phenotypic differences or abnormalities in our standard growth conditions.
Among 12 independently isolated lines transformed with the pGAPC1::GUS expression cassette, 11 of them showed a similar expression pattern (Supplemental Fig. S2A). Seventeen independent transgenic lines transformed with the pGAPC1::GAPC1-YFP expression cassette were isolated, and all of them showed a similar expression pattern based on the YFP fluorescence analyses (data not shown). Expression of GAPC1-YFP chimeric protein was confirmed by immunoblot analysis with anti-GFP antibodies of protein extracts from three independent lines (Supplemental Fig. S2B). The immune detection was revealed by alkaline phosphatase reaction.
Detection of GAPC1-YFP by Immunoblot Analysis
For the quantification of GAPC1-YFP induction in root tips, 500 roots of 7-d-old seedlings for each condition, control or treated for 72 h with 0.1 mm cadmium, were harvested and homogenized in 250 μL of protein extraction buffer (0.320 m Suc, 50 mm Tris, pH 7.4, 1 mm EDTA, 10 mm DTT, 1 mm phenylmethanesulfonylfluoride, and 5% [w/v] polyvinylpyrrolidone). The homogenate was centrifuged at 2,000 rpm for 2 min at 4°C to eliminate debris. The supernatant was centrifuged at 10,200 rpm for 20 min at 4°C, and the supernatant was collected. Protein concentration was determined by the Bradford (1976) method, using the Bio-Rad protein assay. Two and one-half micrograms of extracted proteins were separated by SDS-PAGE with precasted ProGel Tris-Gly Gradient Gels 4% (w/v) to 12% (w/v) Tris/Gly Gels (anamed Elektrophorese), transferred to polyvinylidene fluoride 0.2-μm membrane (Bio-Rad), and analyzed with antibodies raised against GFP (Invitrogen). Equal loading of proteins was checked by staining an equally loaded gel with Coomassie Blue.
The chemiluminescence analysis (Fig. 3D) was performed by using a horseradish peroxidase-conjugated secondary antibody detected with the ECL Western Blotting Detection System by Pierce (Thermo Fisher Scientific).
GUS Staining and GUS Activity Assay
Histochemical analyses of the transcriptional reporter GUS and measurement of GUS enzymatic activity were performed as described by Jefferson et al. (1987).
Samples for histochemical analysis of GUS activity were fixed by 30-min vacuum infiltration in the fixation solution (50 mm phosphate buffer, pH 7.2, 1.5% [v/v] formaldehyde, and 0.5% [w/v] Triton X-100), and then the samples were washed three times with 50 mm phosphate buffer (pH 7.2) and vacuum infiltrated for 15 min in the reaction mixture containing 50 mm phosphate buffer (pH 7.2), 1 mm 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid, 0.5 mm K3Fe(CN)6, 0.5 mm K4Fe(CN)6⋅3H2O, and 0.5% (w/v) Triton X-100. Then, the samples were incubated at 37°C overnight or for 20 min for qualitative or semiquantitative analyses, respectively. Samples were cleared by washing them in a methanol:acetic acid solution (3:1, v/v).
For GUS enzymatic activity assay, 300 roots of 7-d-old seedlings treated up to 72 h were homogenized in 2 mL of GUS extraction buffer (50 mm NaH2PO4, pH 7.0, 0.1% [w/v] sodium lauryl sarcosine, 20% [v/v] methanol, 10 mm β-mercaptoethanol, and 10 mm EDTA). The homogenate was centrifuged at 400g for 2 min at 4°C to eliminate debris. The supernatant was centrifuged at 14,000 rpm for 15 min at 4°C, and the supernatant was collected. Protein concentration was determined by the Bradford (1976) method, using the Bio-Rad protein assay.
An appropriate amount of sample was incubated in the reaction solution (GUS extraction buffer and 2 mm 4-methylumbelliferyl-β-d-glucuronide trihydrate) for 1 h at 37°C. Aliquots were collected and mixed with stop reaction buffer (0.2 m Na2CO3) every 15 min starting from time zero. Fluorescence of the samples collected was measured with excitation at 365 nm and emission at 455 nm on a Luminescence Spectrometer LS 55 (PerkinElmer).
Confocal Microscopy Analyses
Confocal microscopy analyses were performed using a Nikon PCM2000 (Bio-Rad) and an inverted SP5II laser scanning confocal imaging system (Leica). For DAF-FM detection, excitation was at 488 nm and emission between 515 and 530 nm. To extract quantitative data, pixel values were measured over root regions, which were located manually on confocal images and calculated using ImageJ bundle software (http://rsb.info.nih.gov/ij/). For NO and GAPC1-YFP detection, 15 roots from three independent experiments were observed. Then, quantitative fluorescence values of root pictures were determined by analyzing pixel intensity. Data were expressed as (fluorescence intensity of treated root)/(fluorescence intensity of control root) ratios.
roGFP2 Microscopy Analyses
For roGFP2 ratiometric analysis, the seedling roots were imaged in vivo by a Nikon Ti-E inverted fluorescence microscope with a Nikon CFI Plan Apo VC 20× Air 0.75-numerical aperture dry objective. Excitation light was produced by a Prior Lumen 200 PRO fluorescent lamp. The roots were excited at 470/40 nm and then at 405/40 nm, and for both excitation wavelengths, roGFP2 fluorescence was collected with a band-pass filter of 505 to 530 nm. Filters and dichroic mirrors were purchased from Chroma. The fluorescences emitted were collected with an Hamamatsu Dual CCD Camera ORCA-D2. Exposure time was 30 ms with a 2×2 CCD binning for the 470/40 nm excitation and 300 ms with a 2×2 CCD binning for the 405/40 nm excitation. The images where acquired every 5 s for a total time of 90 s. NIS-Elements (Nikon) was used as a platform to control microscope, illuminator, camera, and postacquisition analyses.
As regards time-course experiments, fluorescence intensity was determined in the transition/elongation zone of the seedlings’ root tip. The background was subtracted, and the two emissions of the analyzed roots were used for the ratio calculation and eventually normalized to the initial ratio.
Cloning, Expression, and Purification of Recombinant GAPC1
The recombinant GAPC1 protein was expressed and produced as described previously (Bedhomme et al., 2012).
The molecular mass and purity of the protein were analyzed by SDS-PAGE and Coomassie Blue staining after desalting on PD-10 columns equilibrated with 50 mm potassium phosphate buffer (pH 7.5). The protein concentration was determined spectrophotometrically using a molar extinction coefficient at 280 nm of 40,910 m–1 cm–1 of the monomer. The resulting homogeneous protein was stored at –20°C.
Inactivation Treatments of Recombinant GAPC1
Before any treatments, recombinant GAPC1 was incubated with 10 mm DTT for 30 min at room temperature for reduction. Thereafter, DTT was removed by desalting on NAP-5 columns equilibrated with 50 mm BisTris buffer (pH 7.0). Reduced GAPC1 (2.5 µm final concentration) was incubated in 50 mm BisTris buffer (pH 7.0) supplemented with 0.14 mm NAD+ in the presence of different concentrations of DEA-NONOate (diazeniumdiolate) diethylammonium salt or H2O2. At indicated time, an aliquot of the sample was withdrawn for the assay of enzyme activity as described above. The reversibility of GAPC1 inactivation (10-min incubation with 0.5 mm DEA-NONOate or 0.1 mm H2O2) was assessed by measuring GAPC1 activity after incubation for 10 min in the presence of 20 mm DTT. Reduced GAPC1 was also incubated in 50 mm BisTris buffer (pH 7.0) supplemented with 0.14 mm NAD+ in the presence of 0.1 mm cadmium. After 10-min incubation, an aliquot of the sample was withdrawn for the assay of enzyme activity as described above. The reversibility of the treatment (10-min incubation with 0.1 mm cadmium) was assessed by measuring GAPC1 activity after incubation for 10 min in the presence of 20 mm DTT.
Generation of Arabidopsis Transgenic Lines Expressing Transgenes in gapc1 Mutant Background
The transgenic Arabidopsis gapc1 mutant plants stably transformed with pGAPC1::GAPC1-YFP and pGAPC1::GAPC1(C155S)-YFP were obtained by crossing their mature flowers: ovary of pGAPC1::GAPC1-YFP and pGAPC1::GAPC1-C155S-YFP were pollinated by brushing them with stamens of fully mature flowers from Arabidopsis gapc1 mutant plants. Seeds obtained were sown, and seedlings were selected by using PCR analyses.
Statistical Analysis
Pictures shown in the figures are representative of at least 15 roots from three independent experiments, and values reported in the graph represent mean ± sd. The statistical significance of differences was evaluated by Student’s t test.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Evans blue staining of Arabidopsis roots exposed to cadmium.
Supplemental Figure S2. Validation of the pGAPC1::GUS and pGAPC1::GAPC1-YFP transgenic lines produced.
Supplemental Figure S3. Characterization of GAPC1 promoter activity pattern.
Supplemental Figure S4. Quantitative real-time RT-PCR expression analysis of GAPC genes in Arabidopsis wild-type seedlings.
Supplemental Figure S5. Transient expression of mutated GAPC1-YFP chimeric proteins in Arabidopsis protoplasts.
Supplemental Figure S6. Wild-type and mutant GAPC1-YFP chimeric proteins accumulate in the nucleus of cadmium-treated root cells in a similar way.
Acknowledgments
We thank Markus Schwarzländer (Institute of Crop Science and Resource Conservation, University of Bonn) for providing the roGFP2 Arabidopsis seeds and Michael Riefler (Institute of Biology/Applied Genetics, Dahlem Centre of Plant Sciences, Free University of Berlin) for help with quantitative real-time RT-PCR expression analysis. M.V., A.C., M.Z., and P.T. designed research; M.V., A.C., M.Z., and M.F. performed the experiments; M.V., A.C., and M.Z. analyzed data; P.T. proposed the study; and P.T., A.C., and F.L.S. supervised the research and wrote the article.
Glossary
- ROS
reactive oxygen species
- RNS
reactive nitrogen species
- MAPK
mitogen-activated protein kinase
- H2O2
hydrogen peroxide
- NO
nitric oxide
- GSH
reduced glutathione
- DAF-FM
4-amino-5-methylamino-2,7-difluorofluorescein diacetate
- 3-AT
3-amino-1,2,4-triazole
- RT
reverse transcription
- DTT
dithiothreitol
- DEA
diethylamine
- BSO
l-buthionine-sulfoximine
- T-DNA
transfer DNA
- cDNA
complementary DNA
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Anderson LE, Ringenberg MR, Carol AA. (2004) Cytosolic glyceraldehyde-3-P dehydrogenase and the B subunit of the chloroplast enzyme are present in the pea leaf nucleus. Protoplasma 223: 33–43 [DOI] [PubMed] [Google Scholar]
- Apel K, Hirt H. (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Astier J, Rasul S, Koen E, Manzoor H, Besson-Bard A, Lamotte O, Jeandroz S, Durner J, Lindermayr C, Wendehenne D. (2011) S-nitrosylation: an emerging post-translational protein modification in plants. Plant Sci 181: 527–533 [DOI] [PubMed] [Google Scholar]
- Bedhomme M, Adamo M, Marchand CH, Couturier J, Rouhier N, Lemaire SD, Zaffagnini M, Trost P. (2012) Glutathionylation of cytosolic glyceraldehyde-3-phosphate dehydrogenase from the model plant Arabidopsis thaliana is reversed by both glutaredoxins and thioredoxins in vitro. Biochem J 445: 337–347 [DOI] [PubMed] [Google Scholar]
- Besson-Bard A, Gravot A, Richaud P, Auroy P, Duc C, Gaymard F, Taconnat L, Renou JP, Pugin A, Wendehenne D. (2009) Nitric oxide contributes to cadmium toxicity in Arabidopsis by promoting cadmium accumulation in roots and by up-regulating genes related to iron uptake. Plant Physiol 149: 1302–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besson-Bard A, Pugin A, Wendehenne D. (2008) New insights into nitric oxide signaling in plants. Annu Rev Plant Biol 59: 21–39 [DOI] [PubMed] [Google Scholar]
- Biesecker G, Harris JI, Thierry JC, Walker JE, Wonacott AJ. (1977) Sequence and structure of d-glyceraldehyde 3-phosphate dehydrogenase from Bacillus stearothermophilus. Nature 266: 328–333 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Chen IP, Haehnel U, Altschmied L, Schubert I, Puchta H. (2003) The transcriptional response of Arabidopsis to genotoxic stress - a high-density colony array study (HDCA). Plant J 35: 771–786 [DOI] [PubMed] [Google Scholar]
- Cho UH, Seo NH. (2005) Oxidative stress in Arabidopsis thaliana exposed to cadmium is due to hydrogen peroxide accumulation. Plant Sci 168: 113–120 [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Chaki M, Fernández-Ocaña A, Valderrama R, Palma JM, Carreras A, Begara-Morales JC, Airaki M, del Río LA, Barroso JB. (2008) Metabolism of reactive nitrogen species in pea plants under abiotic stress conditions. Plant Cell Physiol 49: 1711–1722 [DOI] [PubMed] [Google Scholar]
- Correa-Aragunde N, Graziano M, Lamattina L. (2004) Nitric oxide plays a central role in determining lateral root development in tomato. Planta 218: 900–905 [DOI] [PubMed] [Google Scholar]
- Cuypers A, Smeets K, Ruytinx J, Opdenakker K, Keunen E, Remans T, Horemans N, Vanhoudt N, Van Sanden S, Van Belleghem F, et al. (2011) The cellular redox state as a modulator in cadmium and copper responses in Arabidopsis thaliana seedlings. J Plant Physiol 168: 309–316 [DOI] [PubMed] [Google Scholar]
- De Michele R, Vurro E, Rigo C, Costa A, Elviri L, Di Valentin M, Careri M, Zottini M, Sanità di Toppi L, Lo Schiavo F. (2009) Nitric oxide is involved in cadmium-induced programmed cell death in Arabidopsis suspension cultures. Plant Physiol 150: 217–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delledonne M, Zeier J, Marocco A, Lamb C. (2001) Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive disease resistance response. Proc Natl Acad Sci USA 98: 13454–13459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DP, Fordham-Skelton AP, Edwards R. (2005) Redox regulation of a soybean tyrosine-specific protein phosphatase. Biochemistry 44: 7696–7703 [DOI] [PubMed] [Google Scholar]
- Foreman J, Demidchik V, Bothwell JH, Mylona P, Miedema H, Torres MA, Linstead P, Costa S, Brownlee C, Jones JD, et al. (2003) Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422: 442–446 [DOI] [PubMed] [Google Scholar]
- Fourrat L, Iddar A, Valverde F, Serrano A, Soukri A. (2007) Effects of oxidative and nitrosative stress on Tetrahymena pyriformis glyceraldehyde-3-phosphate dehydrogenase. J Eukaryot Microbiol 54: 338–346 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. (2005) Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 17: 1866–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. (2009) Redox regulation in photosynthetic organisms: signaling, acclimation, and practical implications. Antioxid Redox Signal 11: 861–905 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. (2011) Ascorbate and glutathione: the heart of the redox hub. Plant Physiol 155: 2–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaupels F, Kuruthukulangarakoola GT, Durner J. (2011) Upstream and downstream signals of nitric oxide in pathogen defence. Curr Opin Plant Biol 14: 707–714 [DOI] [PubMed] [Google Scholar]
- Gechev TS, Minkov IN, Hille J. (2005) Hydrogen peroxide-induced cell death in Arabidopsis: transcriptional and mutant analysis reveals a role of an oxoglutarate-dependent dioxygenase gene in the cell death process. IUBMB Life 57: 181–188 [DOI] [PubMed] [Google Scholar]
- Griffith OW. (1982) Mechanism of action, metabolism, and toxicity of buthionine sulfoximine and its higher homologs, potent inhibitors of glutathione synthesis. J Biol Chem 257: 13704–13712 [PubMed] [Google Scholar]
- Guo L, Devaiah SP, Narasimhan R, Pan X, Zhang Y, Zhang W, Wang X. (2012) Cytosolic glyceraldehyde-3-phosphate dehydrogenases interact with phospholipase Dδ to transduce hydrogen peroxide signals in the Arabidopsis response to stress. Plant Cell 24: 2200–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Luan S. (2003) Redox control of protein tyrosine phosphatases and mitogen-activated protein kinases in plants. Plant Physiol 132: 1149–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock J, Desikan R, Harrison J, Bright J, Hooley R, Neill S. (2006) Doing the unexpected: proteins involved in hydrogen peroxide perception. J Exp Bot 57: 1711–1718 [DOI] [PubMed] [Google Scholar]
- Hancock JT, Henson D, Nyirenda M, Desikan R, Harrison J, Lewis M, Hughes J, Neill SJ. (2005) Proteomic identification of glyceraldehyde 3-phosphate dehydrogenase as an inhibitory target of hydrogen peroxide in Arabidopsis. Plant Physiol Biochem 43: 828–835 [DOI] [PubMed] [Google Scholar]
- Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, Takahashi M, Cheah JH, Tankou SK, Hester LD, et al. (2005) S-Nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol 7: 665–674 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Herbette S, Taconnat L, Hugouvieux V, Piette L, Magniette ML, Cuine S, Auroy P, Richaud P, Forestier C, Bourguignon J, et al. (2006) Genome-wide transcriptome profiling of the early cadmium response of Arabidopsis roots and shoots. Biochimie 88: 1751–1765 [DOI] [PubMed] [Google Scholar]
- Holtgrefe S, Gohlke J, Starmann J, Druce S, Klocke S, Altmann B, Wojtera J, Lindermayr C, Scheibe R. (2008) Regulation of plant cytosolic glyceraldehyde 3-phosphate dehydrogenase isoforms by thiol modifications. Physiol Plant 133: 211–228 [DOI] [PubMed] [Google Scholar]
- Huberts DH, van der Klei IJ. (2010) Moonlighting proteins: an intriguing mode of multitasking. Biochim Biophys Acta 1803: 520–525 [DOI] [PubMed] [Google Scholar]
- Jaspers P, Kangasjärvi J. (2010) Reactive oxygen species in abiotic stress signaling. Physiol Plant 138: 405–413 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery CJ. (1999) Moonlighting proteins. Trends Biochem Sci 24: 8–11 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Yang B, Harris NS, Deyholos MK. (2007) Comparative proteomic analysis of NaCl stress-responsive proteins in Arabidopsis roots. J Exp Bot 58: 3591–3607 [DOI] [PubMed] [Google Scholar]
- Kojima H, Nakatsubo N, Kikuchi K, Kawahara S, Kirino Y, Nagoshi H, Hirata Y, Nagano T. (1998) Detection and imaging of nitric oxide with novel fluorescent indicators: diaminofluoresceins. Anal Chem 70: 2446–2453 [DOI] [PubMed] [Google Scholar]
- Kornberg MD, Sen N, Hara MR, Juluri KR, Nguyen JV, Snowman AM, Law L, Hester LD, Snyder SH. (2010) GAPDH mediates nitrosylation of nuclear proteins. Nat Cell Biol 12: 1094–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosová K, Vítámvás P, Prášil IT, Renaut J. (2011) Plant proteome changes under abiotic stress—contribution of proteomics studies to understanding plant stress response. J Proteomics 74: 1301–1322 [DOI] [PubMed] [Google Scholar]
- Kwak JM, Mori IC, Pei ZM, Leonhardt N, Torres MA, Dangl JL, Bloom RE, Bodde S, Jones JD, Schroeder JI. (2003) NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J 22: 2623–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindermayr C, Saalbach G, Durner J. (2005) Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol 137: 921–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova O, Kamberov E, Margolis B. (2000) Generation of deletion and point mutations with one primer in a single cloning step. Biotechniques 29: 970–972 [DOI] [PubMed] [Google Scholar]
- Mazzola JL, Sirover MA. (2003) Subcellular alteration of glyceraldehyde-3-phosphate dehydrogenase in Alzheimer’s disease fibroblasts. J Neurosci Res 71: 279–285 [DOI] [PubMed] [Google Scholar]
- Meyer AJ, Brach T, Marty L, Kreye S, Rouhier N, Jacquot JP, Hell R. (2007) Redox-sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. Plant J 52: 973–986 [DOI] [PubMed] [Google Scholar]
- Meyer Y, Verdoucq L, Vignols F. (1999) Plant thioredoxins and glutaredoxins: identity and putative roles. Trends Plant Sci 4: 388–394 [DOI] [PubMed] [Google Scholar]
- Møller IM, Jensen PE, Hansson A. (2007) Oxidative modifications to cellular components in plants. Annu Rev Plant Biol 58: 459–481 [DOI] [PubMed] [Google Scholar]
- Moore Bd. (2004) Bifunctional and moonlighting enzymes: lighting the way to regulatory control. Trends Plant Sci 9: 221–228 [DOI] [PubMed] [Google Scholar]
- Muñoz-Bertomeu J, Cascales-Miñana B, Irles-Segura A, Mateu I, Nunes-Nesi A, Fernie AR, Segura J, Ros R. (2010) The plastidial glyceraldehyde-3-phosphate dehydrogenase is critical for viable pollen development in Arabidopsis. Plant Physiol 152: 1830–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Noctor G, Queval G, Mhamdi A, Chaouch S, Foyer CH. (2011) Glutathione. The Arabidopsis Book 9: e0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parani M, Rudrabhatla S, Myers R, Weirich H, Smith B, Leaman DW, Goldman SL. (2004) Microarray analysis of nitric oxide responsive transcripts in Arabidopsis. Plant Biotechnol J 2: 359–366 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polverari A, Molesini B, Pezzotti M, Buonaurio R, Marte M, Delledonne M. (2003) Nitric oxide-mediated transcriptional changes in Arabidopsis thaliana. Mol Plant Microbe Interact 16: 1094–1105 [DOI] [PubMed] [Google Scholar]
- Poole LB, Karplus PA, Claiborne A. (2004) Protein sulfenic acids in redox signaling. Annu Rev Pharmacol Toxicol 44: 325–347 [DOI] [PubMed] [Google Scholar]
- Remans T, Smeets K, Opdenakker K, Mathijsen D, Vangronsveld J, Cuypers A. (2008) Normalisation of real-time RT-PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations. Planta 227: 1343–1349 [DOI] [PubMed] [Google Scholar]
- Rius SP, Casati P, Iglesias AA, Gomez-Casati DF. (2008) Characterization of Arabidopsis lines deficient in GAPC-1, a cytosolic NAD-dependent glyceraldehyde-3-phosphate dehydrogenase. Plant Physiol 148: 1655–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez MC, Petersen M, Mundy J. (2010) Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol 61: 621–649 [DOI] [PubMed] [Google Scholar]
- Romero-Puertas MC, Campostrini N, Mattè A, Righetti PG, Perazzolli M, Zolla L, Roepstorff P, Delledonne M. (2008) Proteomic analysis of S-nitrosylated proteins in Arabidopsis thaliana undergoing hypersensitive response. Proteomics 8: 1459–1469 [DOI] [PubMed] [Google Scholar]
- Roth U, von Roepenack-Lahaye E, Clemens S. (2006) Proteome changes in Arabidopsis thaliana roots upon exposure to Cd2+. J Exp Bot 57: 4003–4013 [DOI] [PubMed] [Google Scholar]
- Sanità di Toppi L, Gabbrielli R. (1999) Response to cadmium in higher plants. Environ Exp Bot 41: 105–130 [Google Scholar]
- Sarry JE, Kuhn L, Ducruix C, Lafaye A, Junot C, Hugouvieux V, Jourdain A, Bastien O, Fievet JB, Vailhen D, et al. (2006) The early responses of Arabidopsis thaliana cells to cadmium exposure explored by protein and metabolite profiling analyses. Proteomics 6: 2180–2198 [DOI] [PubMed] [Google Scholar]
- Sirover MA. (2011) On the functional diversity of glyceraldehyde-3-phosphate dehydrogenase: biochemical mechanisms and regulatory control. Biochim Biophys Acta 1810: 741–751 [DOI] [PubMed] [Google Scholar]
- Sparla F, Zaffagnini M, Wedel N, Scheibe R, Pupillo P, Trost P. (2005) Regulation of photosynthetic GAPDH dissected by mutants. Plant Physiol 138: 2210–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tristan C, Shahani N, Sedlak TW, Sawa A. (2011) The diverse functions of GAPDH: views from different subcellular compartments. Cell Signal 23: 317–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukagoshi H, Busch W, Benfey PN. (2010) Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143: 606–616 [DOI] [PubMed] [Google Scholar]
- Urano K, Kurihara Y, Seki M, Shinozaki K. (2010) ‘Omics’ analyses of regulatory networks in plant abiotic stress responses. Curr Opin Plant Biol 13: 132–138 [DOI] [PubMed] [Google Scholar]
- Valderrama R, Corpas FJ, Carreras A, Fernández-Ocaña A, Chaki M, Luque F, Gómez-Rodríguez MV, Colmenero-Varea P, Del Río LA, Barroso JB. (2007) Nitrosative stress in plants. FEBS Lett 581: 453–461 [DOI] [PubMed] [Google Scholar]
- Valerio C, Costa A, Marri L, Issakidis-Bourguet E, Pupillo P, Trost P, Sparla F. (2011) Thioredoxin-regulated beta-amylase (BAM1) triggers diurnal starch degradation in guard cells, and in mesophyll cells under osmotic stress. J Exp Bot 62: 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Du Y, Li Y, Ren D, Song CP. (2010) Hydrogen peroxide-mediated activation of MAP kinase 6 modulates nitric oxide biosynthesis and signal transduction in Arabidopsis. Plant Cell 22: 2981–2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Sirover MA, Anderson LE. (1999) Pea chloroplast glyceraldehyde-3-phosphate dehydrogenase has uracil glycosylase activity. Arch Biochem Biophys 367: 348–353 [DOI] [PubMed] [Google Scholar]
- Wawer I, Bucholc M, Astier J, Anielska-Mazur A, Dahan J, Kulik A, Wysłouch-Cieszynska A, Zareba-Kozioł M, Krzywinska E, Dadlez M, et al. (2010) Regulation of Nicotiana tabacum osmotic stress-activated protein kinase and its cellular partner GAPDH by nitric oxide in response to salinity. Biochem J 429: 73–83 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57: 781–803 [DOI] [PubMed] [Google Scholar]
- Yu M, Yun BW, Spoel SH, Loake GJ. (2012) A sleigh ride through the SNO: regulation of plant immune function by protein S-nitrosylation. Curr Opin Plant Biol 15: 424–430 [DOI] [PubMed] [Google Scholar]
- Zaffagnini M, Bedhomme M, Lemaire SD, Trost P. (2012a) The emerging roles of protein glutathionylation in chloroplasts. Plant Sci 185-186: 86–96 [DOI] [PubMed] [Google Scholar]
- Zaffagnini M, Bedhomme M, Marchand CH, Morisse S, Trost P, Lemaire SD. (2012b) Redox regulation in photosynthetic organisms: focus on glutathionylation. Antioxid Redox Signal 16: 567–586 [DOI] [PubMed] [Google Scholar]
- Zottini M, Costa A, De Michele R, Ruzzene M, Carimi F, Lo Schiavo F. (2007) Salicylic acid activates nitric oxide synthesis in Arabidopsis. J Exp Bot 58: 1397–1405 [DOI] [PubMed] [Google Scholar]