Metabolite profiles support a robust prediction of biomass across a range of conditions and accounts for environmental influences on metabolic networks.
Abstract
Natural genetic diversity provides a powerful tool to study the complex interrelationship between metabolism and growth. Profiling of metabolic traits combined with network-based and statistical analyses allow the comparison of conditions and identification of sets of traits that predict biomass. However, it often remains unclear why a particular set of metabolites is linked with biomass and to what extent the predictive model is applicable beyond a particular growth condition. A panel of 97 genetically diverse Arabidopsis (Arabidopsis thaliana) accessions was grown in near-optimal carbon and nitrogen supply, restricted carbon supply, and restricted nitrogen supply and analyzed for biomass and 54 metabolic traits. Correlation-based metabolic networks were generated from the genotype-dependent variation in each condition to reveal sets of metabolites that show coordinated changes across accessions. The networks were largely specific for a single growth condition. Partial least squares regression from metabolic traits allowed prediction of biomass within and, slightly more weakly, across conditions (cross-validated Pearson correlations in the range of 0.27–0.58 and 0.21–0.51 and P values in the range of <0.001–<0.13 and <0.001–<0.023, respectively). Metabolic traits that correlate with growth or have a high weighting in the partial least squares regression were mainly condition specific and often related to the resource that restricts growth under that condition. Linear mixed-model analysis using the combined metabolic traits from all growth conditions as an input indicated that inclusion of random effects for the conditions improves predictions of biomass. Thus, robust prediction of biomass across a range of conditions requires condition-specific measurement of metabolic traits to take account of environment-dependent changes of the underlying networks.
Plant biomass is the ultimate output of the interplay between metabolism and the cellular and developmental programs that control allocation (Poorter and Nagel, 2000; Hermans et al., 2006; Poorter et al., 2011) and cell and organ growth (Gonzalez et al., 2009; Krizek, 2009). A predictive understanding of these complex relationships would open up new perspectives in crop improvement. Given that an increase in the rate of growth must be underpinned by changes in metabolism, it should be possible to identify metabolic states that are associated with higher growth rates. One way to characterize metabolic states would be to measure fluxes. However, most flux measurements are in fact estimates based on fitting labeling patterns of metabolites to a selected metabolic model. This is technically challenging in multicellular life forms such as higher plants (Zamboni, 2011). Furthermore, such estimates would need to be very precise because small changes in flux can result in large changes in biomass; plant growth is exponential, with a typical increase in biomass of 10% to 25% per day, so a relatively small difference in fluxes and the momentary rate of growth will lead, within 1 to 2 weeks, to a large difference in biomass (Poorter, 1989; Stitt and Zeeman, 2012). A complementary approach is to identify metabolic traits, such as the levels of metabolites, which are associated with higher rates of growth and biomass formation. The attractiveness of this approach has been enhanced by the development of increasingly powerful platforms to measure metabolite levels and sophisticated tools to analyze the resulting data sets (Lisec et al., 2006; Saito and Matsuda, 2010; Fernie et al., 2011).
Metabolite profiling of large populations of Arabidopsis (Arabidopsis thaliana) natural accessions or inbred lines and the application of multivariate analysis tools, such as canonical correlation analysis and partial least squares (PLS) regression, has allowed the identification of descriptor sets of metabolites that are predictive of biomass (Meyer et al., 2007; Sulpice et al., 2009; Cuadros-Inostroza et al., 2010; Steinfath et al., 2010a; Carreno-Quintero et al., 2012) and physiological traits such as freezing tolerance (Korn et al., 2010) and herbivore resistance (Kliebenstein, 2012; Züst et al., 2012). The advantage of surveying a wide range of metabolites is underlined by the fact that multivariate analysis allows predictions to be made from data matrices in which no individual metabolite significantly correlates with biomass (Meyer et al., 2007). This approach was recently extended to hybrid vigor. The relative density of networks based on correlations extracted from metabolite profiles in Arabidopsis is modified in plants that show a strong degree of heterosis (Meyer et al., 2012). Furthermore, metabolite profiles measured in parents allow prediction of hybrid vigor in their progeny both in Arabidopsis (Steinfath et al., 2010b) and maize (Zea mays; Riedelsheimer et al., 2012). In an analogous approach, robotized platforms can be used to profile large numbers of enzymes and search for relationships between their maximum activities and growth (Sulpice et al., 2010).
Nevertheless, this top-down approach suffers from two major weaknesses. First, a statistical relationship between a set of metabolites and growth does not provide functional insights into how metabolism determines the rate of growth. Functional interpretation is compromised because the complexity of metabolic networks in primary metabolism makes it difficult to draw inferences about fluxes from changes in metabolite levels (Stitt et al., 2010; Sulpice et al., 2010; Fernie and Stitt, 2012) by the fact that current metabolite profiles only cover a small fraction of the total metabolome (Saito and Matsuda, 2010) and by the likelihood that many connections between metabolism and growth may be mediated by signaling pathways that impinge on physiological or developmental processes (LeClere et al., 2010; Lilley et al., 2012). The occurrence of a correlation between biomass and individual metabolites or linear combinations of metabolites also does not imply causality. Such correlations might arise if a given combination of metabolic traits supports increased biomass formation, but also if increased biomass formation resulted in a corresponding change in metabolite levels. Second, levels of metabolites in primary metabolism are dramatically influenced by the environment (Hannah et al., 2010; Caldana et al., 2011; Obata and Fernie, 2012), including the irradiance regime (Gibon et al., 2006, 2009; Bräutigam et al., 2009; Jänkänpää et al., 2012) and the nitrogen regime (Tschoep et al., 2009; Kusano et al., 2011; Amiour et al., 2012). It is not yet clear if the same sets of metabolites are predictive of biomass across different growth conditions.
Under short-day conditions, biomass is strongly and negatively correlated with starch content at dusk and with the total protein content per unit of fresh weight (FW) in a panel of Arabidopsis accessions (Sulpice et al., 2009). Multivariate data analysis using PLS revealed that biomass, starch, and protein are predicted by overlapping sets of metabolites, indicating that starch and protein concentration are integrative metabolic traits that capture information about the levels of many low-Mr metabolites and are closely linked to biomass formation. Starch is a transient carbon store, which accumulates in leaves during the day and is remobilized to support metabolism and growth at night (Smith and Stitt, 2007; Stitt and Zeeman, 2012). A negative correlation between biomass and starch levels at dusk implies that faster-growing accessions convert carbon more efficiently to biomass, at least during the night. A negative correlation between protein concentration and biomass will result in a larger leaf area per unit of invested protein and hence absorption of more light per plant. This finding is consistent with comparative studies of different species, where there is often a negative correlation between leaf area mass (dry weight per unit leaf area) and the growth rate, especially in limiting irradiance (Poorter and Nagel, 2000; Poorter et al., 2009). Furthermore, because protein synthesis is an energetically costly process (Piques et al., 2009; Raven, 2012), it is possible that the lower protein concentration might contribute to the observed increased efficiency of carbon use. Subsequently, Sulpice et al. (2010) showed that large accessions invested a large proportion of their protein in enzymes of photosynthesis. This will allow photosynthetic capacity per unit leaf area to be maintained irrespective of the fact that the total leaf protein concentration decreases.
The studies of Sulpice et al. (2009, 2010) were carried out in short-day conditions, where growth is limited by carbon (Gibon et al., 2009). Plant growth is also often restricted by the supply of nutrients, especially nitrogen (Poorter and Nagel, 2000; Krapp et al., 2005; Hirel et al., 2007; Xu et al., 2012). In the following experiments, we profiled metabolites and enzyme activities in the same set of accessions in conditions where nitrogen was limiting for growth (Tschoep et al., 2009) and in conditions where nitrogen was saturating and carbon was close to saturating for growth. These data were combined with our previously published data for short-day conditions and analyzed to identify which, if any, features of the relationships and connectivity between metabolic traits and growth are shared across different growth conditions.
RESULTS
Experimental Design
In earlier studies, we established three growth protocols for the reference accession Columbia-0 (Col-0) in which a decreased nitrogen or carbon supply leads to compensatory changes in metabolism and a mild and sustained decrease in growth rate. (1) Growth with a full nutrient supply in a 12-h photoperiod (12hHN) allows near-to-maximal growth rates of Col-0, and increasing the photoperiod does not lead to a major further stimulation of growth (Supplemental Fig. S1; Gibon et al., 2009). Longer photoperiods were avoided because they lead to early induction of flowering. (2) A low-nitrogen growth regime (12hLN) was established, in which there was a 20% to 25% decrease in the relative growth rate and an approximately 50% decrease in biomass after 29 to 35 d, compared with 12hHN (Tschoep et al., 2009). Protein levels were hardly altered, and some amino acids even increased, revealing that metabolism and growth have adjusted in a coordinated manner to the decreased nitrogen supply. (3) Similarly, in the 8-h photoperiod used by Sulpice et al. (2009, 2010), there was an approximately 30% decrease in the relative growth rate, compared with a 12-h photoperiod (Supplemental Table S2). Starch turnover was adjusted such that starch was almost but not completely exhausted at the end of the night, carbon was available throughout the 24-h cycle, and carbon starvation marker genes were not induced until after a short extension of the night (Usadel et al., 2008; Gibon et al., 2009; for review, see Stitt and Zeeman, 2012).
A panel of 97 Arabidopsis accessions selected to maximize genotypic and geographic variation and biomass variation (Sulpice et al., 2009, 2010; see Supplemental Table S1 for a list of accessions, abbreviations, and passport data) was grown with an optimal supply of nitrogen in a 12-h/12-h light/dark regime and a suboptimal supply of nitrogen in a 12-h/12-h light/dark regime (Tschoep et al., 2009). Sets of plants from both growth conditions were analyzed for rosette biomass and the levels of metabolites and enzyme activities at dusk. The resulting data were combined with published data for the same accessions grown in an 8-h/16-h light/dark regime (8hHN) to more strongly limit growth by the carbon supply (Sulpice et al., 2009, 2010). The combined data set included information about rosette biomass and 54 metabolic traits in three growth conditions. The metabolic traits included the three structural components (protein and chlorophylls a and b), the major transitory carbon store starch, 43 low-Mr metabolites, including a range of sugars, amino acids, organics acids, and other metabolites, and maximum activities of eight enzymes from central carbon and nitrogen metabolism (for a list of the metabolic traits and abbreviations, see Supplemental Table S1). As measurements of nitrate, Orn and spermidine were not available for the published 8hHN data set; random numbers were introduced for these traits in the calculations of condition-specific correlation matrices. However, these traits were not used in the PLS and mixed-model analyses.
Biomass Involves an Interaction between Accession and Growth Condition
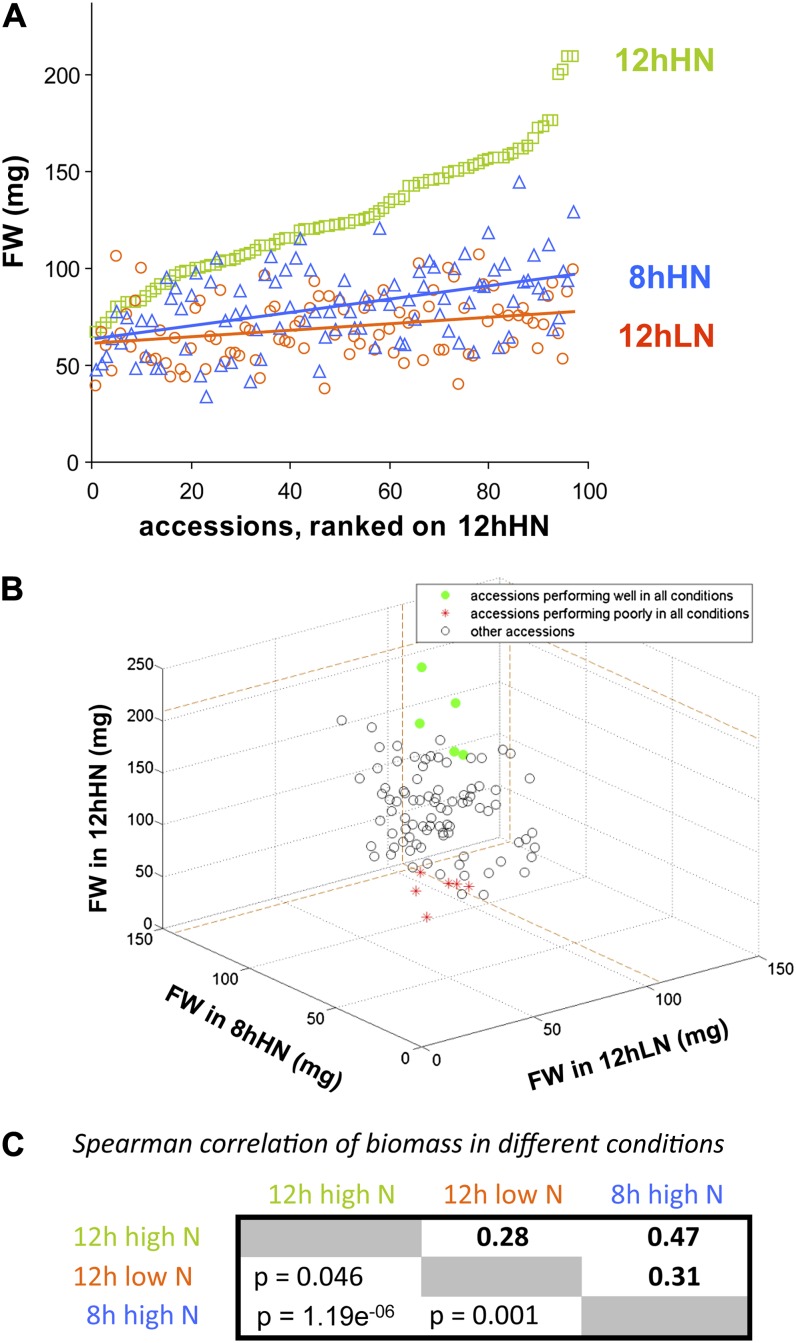
Biomass differed between the 97 accessions by 3.1-fold, 2.8-fold, and 4.2-fold in 12hHN, 12hLN, and 8hHN, respectively, relative to the accession with the lowest biomass in that growth regime (Fig. 1, A and B; Supplemental Table S1). The impact of low nitrogen and low carbon differed between accessions, with some accessions showing a greater than 70% decrease in biomass and others showing no decrease (Fig. 1, A and B). Accessions that had a large biomass in 12hHN tended to show a marked decrease in biomass in carbon-limiting or nitrogen-limiting conditions, while many of the accessions that had a small biomass in 12hHN showed little or no further decrease in carbon-limiting or nitrogen-limiting conditions. When individual accessions are inspected, some show a marked decrease in biomass in low-carbon and low-nitrogen conditions, some maintain biomass in low-carbon and low-nitrogen conditions, and others are especially sensitive to low carbon (Mh1, Nok2, and Lov5) or low nitrogen (Bur0, Dijon5, and Old1; Supplemental Table S1). Small sets of accessions ranked high (Bsch2, Da112, Dra0, Mt0, and Wei1) or low (Ang0, Bla11, Je54, Pyl1, RRS10, and TAMM2) for biomass in all three conditions (Fig. 1B). Overall, pairwise scatter plots revealed significant positive correlation (Spearman rank correlation coefficient [r] = 0.47, P = 1.19e–06) between biomass in 8hHN and 12hHN and weaker relationships between biomass at 8hHN and 12hLN (r = 0.31, P = 0.001) and biomass in 12hHN and 12hLN (r = 0.28, P = 0.0046; Fig. 1C).
Figure 1.
Biomass of 97 accessions in the three growth conditions. A panel of 97 Arabidopsis accessions was grown in 12hHN, 12hLN, and 8hHN. A, Biomass in each condition; the accessions are ordered on the x axis according to their biomass in the control treatment (12hHN). B, Three-dimensional plot of the biomass of each accession in the three growth conditions; accessions that have a high and low ranking for biomass in all three conditions are indicated by green and red symbols, respectively. C, Spearman correlation between biomass in the three growth conditions. The original data are provided in Supplemental Table S1.
Thus, the main trends are 3-fold: (1) subsets of accessions produce higher or lower biomass than others in all three conditions, (2) many accessions that produce high biomass in high-nitrogen and high-carbon conditions tend to show a larger decrease in biomass when nitrogen or carbon is decreased, and (3) many individual accessions respond differently to low nitrogen and low carbon.
Metabolic Traits Are Subject to Environmental and Genotypic Variation
The absolute levels of structural components, metabolites, enzymes, and a list of abbreviations are provided in Supplemental Table S1. ANOVA showed that all structural and metabolic traits, except succinate, showed highly significant changes (P < 0.0001) in the growth condition term (Supplemental Table S2). Significant traits in the accession term included biomass, starch, protein (all P < 0.0001), and many metabolites, including Fru, Glc, malate, myoinositol, Pro, Thr, and nicotinic acid at P < 0.0001 and Suc, raffinose, total amino acids, and many individual amino acids at P < 0.05, and enzyme activities, including nitrate reductase, phosphoenolpyruvate carboxylase (PEPCx), ADP Glc pyrophosphorylase (AGPase), and NAD-dependent Glu dehydrogenase (GlDH) at P < 0.0001 and Glu synthetase (GS) and NAD-dependent malate dehydrogenase (NAD-MDH) at P < 0.05).
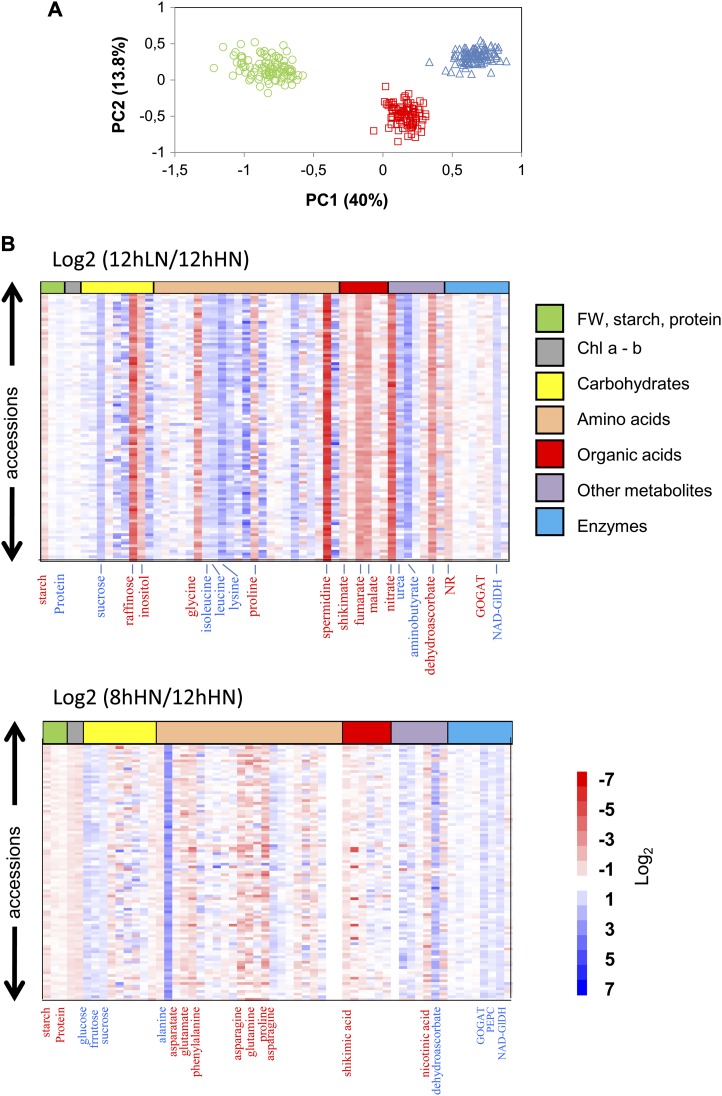
Principle components analysis generated three distinct groups corresponding to the three growth conditions (Fig. 2A; Supplemental Table S3). Principal component 1 (41.3% of total variance) separated carbon-limited (8hHN) and nitrogen-limited (12hLN) conditions, with near-optimal conditions (12hHN) in an intermediate position. Principle component 2 (14.3%) separated near-optimal conditions from the two limiting conditions. When axes are chosen that reflect the variance captured by each principal component, the 97 accessions formed a fairly compact group in near-optimal conditions and a slightly more spread-out group, especially in principal component 1, in low-carbon and low-nitrogen conditions. In principal component 1, positive weightings were found for myoinositol and traits related to nitrate assimilation (nitrate reductase activity and nitrate), ammonium assimilation (Gln:oxoglutarate aminotransferase [GOGAT]), and organic acid metabolism (PEPCx, malate, and fumarate), and negative weightings were found for protein, Suc, total amino acids, and several minor amino acids. In principal component 2, positive weightings were found for GlDH activity, several sugars (Suc, Glc, and Fru), spermidine, and Orn and negative weightings for shikimic acid, chlorophyll, starch, Glu, and nicotinic acid.
Figure 2.
Structural and metabolic traits. A, Principle components analysis based on z-score values for 97 accessions grown in three contrasting conditions (12hHN, green circles; 12hLN, red squares; and 8hHN, blue triangles). In total, 58 traits were determined per accession. The full data set for each growth condition are provided in Supplemental Table S1. VIP scores for the metabolic trait inputs are provided in Supplemental Table S7. B, Heat maps of the metabolite changes under nitrogen deficiency (12hLN) or short days (8hHN) compared with control conditions (12hHN). Each square represents the log2 ratio of the metabolite level using false color scale. Regions of red or blue indicate the metabolite level is decreased or increased, respectively. The full data set for each growth condition as provided in Supplemental Table S4.
Figure 2B (see Supplemental Table S4 for details) summarizes the changes of individual metabolic traits in 12hLN and 8hHN compared with 12hHN. Some metabolic traits showed consistent changes across all 97 accessions in low nitrogen. This included an increase of Suc, several amino acids (e.g. Leu, Ile, and Lys), urea, 4-aminobutyrate, and NAD-GlDH activity and a decrease in raffinose, myoinositol, Gly, Pro, spermidine, shikimate, malate, fumarate, dehydroascorbate, nitrate, and nitrate reductase activity. Nevertheless, the extent of the change varied. Some traits showed large variation between accessions, with an increase in some and a decrease in others (e.g. maltose, trehalose, Ala, Glu, Asn, and threonate). As previously reported for the reference accession Col-0 (Tschoep et al., 2009), there was, perhaps against expectations, a slight but consistent increase in the protein concentration in 12hLN compared with 12hHN. This, and the maintenance or increase in most amino acids, shows that all accessions adjust to compensate for the decrease in nitrogen supply.
A different set of metabolic traits showed consistent changes across all accessions in low carbon. This included an increase in Suc, Glc, Fru, Ala, and dehydroascorbate levels and GOGAT, PEPCx, and NAD-GlDH activities and a decrease of chlorophyll, several amino acids (including asparate, Glu, Phe, Asn, Gln, and Arg), shikimate, and nicotinic acid. As previously reported for the reference accession Col-0 (Gibon et al., 2009; Hannemann et al., 2009), there was a slight but consistent decrease in protein in all accessions in 8hHN compared with 12hHN. The decrease in the protein concentration and the levels of many amino acids reflects the strong dependence of nitrogen metabolism on the carbon supply (Nunes-Nesi et al., 2010). As in the previous comparison, many metabolites showed quite varied changes between 12hHN and 8hHN, again pointing to genotypic variation in the response to the growth condition.
The coefficient of variation (CV, the sd divided by the mean) was estimated to provide insights into which metabolic traits show the largest genetic variation in a given growth condition (Supplemental Fig. S2A). The average CV of all metabolic traits in 8hHN, 12hHN, and 12hLN was 34%, 33%, and 31%, respectively. CV was generally low for structural components and higher for low-Mr metabolites. Protein, chlorophyll a, chlorophyll b, starch (CV < 10%), Suc, total amino acids, shikimate, and most enzymes (CV < 20%) had a low CV in all three conditions, while maltose, trehalose, raffinose, Gln, Asn, Arg, and Pro had a high CV in all conditions. Some metabolic traits showed a high CV in one condition; for example, nitrate had a high CV in 12hLN. This may be because nitrate accumulates to high levels in nitrogen-replete conditions but is used for growth in low nitrogen (see below).
Correlations between Individual Metabolites and Biomass in Each Growth Condition
We next investigated if the same or different individual metabolic traits correlate to biomass in the three growth conditions. Biomass-metabolite trait correlations that were significant at a false discovery rate (FDR) of less than 5% are listed in Table I (for a full list, see Supplemental Table 5).
Table I. Spearman rank correlation coefficient between biomass and metabolic traits in different growth conditions.
Adjusted P values were calculated using the Benjamini-Hochberg correction. This display summarizes metabolites that correlated at P < 0.05 in at least one growth condition. A full set of correlations is provided in Supplemental Table S5. NA, Not available; AA, total amino acids; NR_Vmax, maximum activity of nitrate reductase.
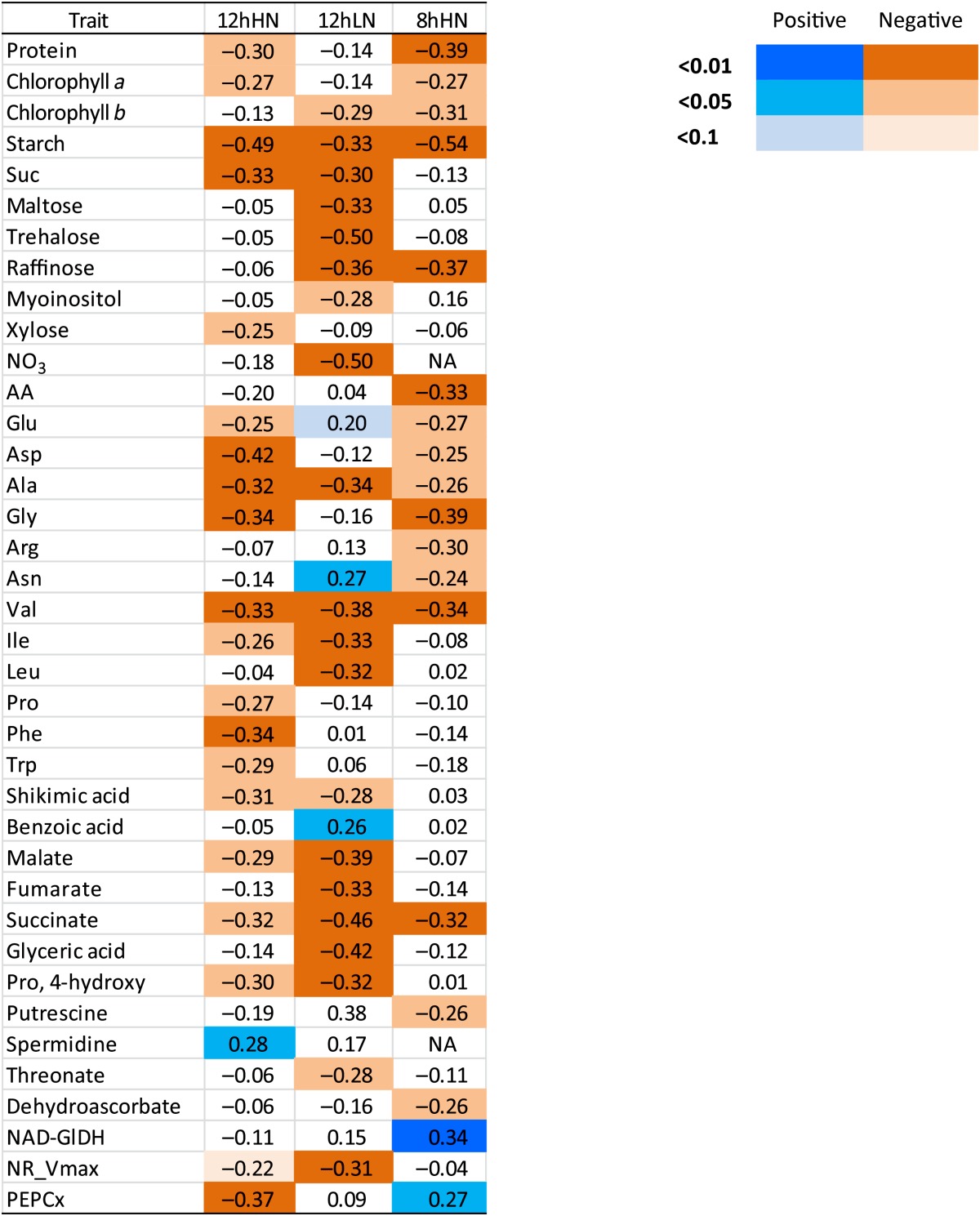
There was a highly significant negative correlation of starch with biomass in 8hHN (r = –0.54; Sulpice et al., 2009) and 12hHN (r = –0.49) and a weaker nonsignificant negative correlation in 12hLN (r = –0.33). The weakening of the negative correlation between biomass and starch in 12hLN is consistent with the hypothesis that allocation of carbon to transitory starch plays an especially important role when carbon limits growth. Ala, Val, and succinate were also negatively and significantly correlated with growth in all three conditions.
Protein showed a highly significant correlation with biomass in 8hHN (r = –0.39; Sulpice et al., 2009) that became weaker in 12hHN (r = –0.30) and was not significant in 12hLN (r = –0.014); Table I; see Supplemental Fig. S3 for scatter plots of biomass against starch and protein). This is in agreement with earlier reports that the negative relationship between biomass and protein observed in short-photoperiod conditions is lost when the photoperiod is longer than 12 h (Hannemann et al., 2009). This negative correlation may be related to a possible link in low-carbon conditions between efficient use of carbon and increased biomass formation, possibly because nitrogen assimilation and protein synthesis represent a major cost for growth (Piques et al., 2009; Amthor, 2010).
Suc, Ile, shikimic acid, malate, and 4-Hyp were negatively and significantly correlated with growth in both 12hHN and 12hLN, while Asp, Glu, and Gly were negatively correlated with growth in both 12hHN and 8hHN and raffinose was negatively and significantly correlated with growth in both 12hLN and 8hHN.
Other correlations were restricted to one growth condition. In 12hHN, biomass was negatively correlated with Xyl, Trp, and PEPCx activity and positively with spermidine. In 12hLN, biomass was negatively correlated to maltose, trehalose, myoinositol, nitrate, Leu, threonate, and nitrate reductase activity and positively correlated to Glu and Asn. Nitrate is the major source of inorganic nitrogen and is assimilated via nitrate reductase, while Asn is a major store for nitrogen. In 8hHN, biomass was negatively correlated with total amino acids and several individual amino acids, including Asn, dehydroascorbate, and putrescine and positively correlated with PEPCx and NAD-GlDH activity. Asn accumulates and NAD-GlDH activity is induced in carbon starvation (Melo-Oliveira et al., 1996; Gibon et al., 2004; Mayashita and Good, 2008; Gibon et al., 2009). Some metabolic traits were negatively correlated with growth in one condition and positively in another (Glu, Asn, and PEPCx activity).
Comparison of Metabolic Networks in the Three Growth Conditions
We next analyzed connectivity between metabolic traits. To this end, matrices were generated from the variation in metabolic traits across 97 accessions in each growth condition to reveal which traits are subject to coordinated changes between accessions in a given growth condition (Supplemental Table S6).
Of a total of 1,683 trait pairs, significant correlations at 10%, 5%, and 1% FDR were found for 493, 303, and 293 trait pairs in 12hHN, for 493, 261, and 129 trait pairs in 12hLN, and for 347, 261, and 129 trait pairs in 8hHN, respectively. The vast majority of the correlations were positive. This resembles earlier reports of high connectivity between metabolic traits in panels of cultivars, accessions, or inbred lines (Carrari et al., 2006; Meyer et al., 2007, 2012; Keurentjes et al., 2008; Schauer et al., 2008; Sulpice et al., 2009, 2010).
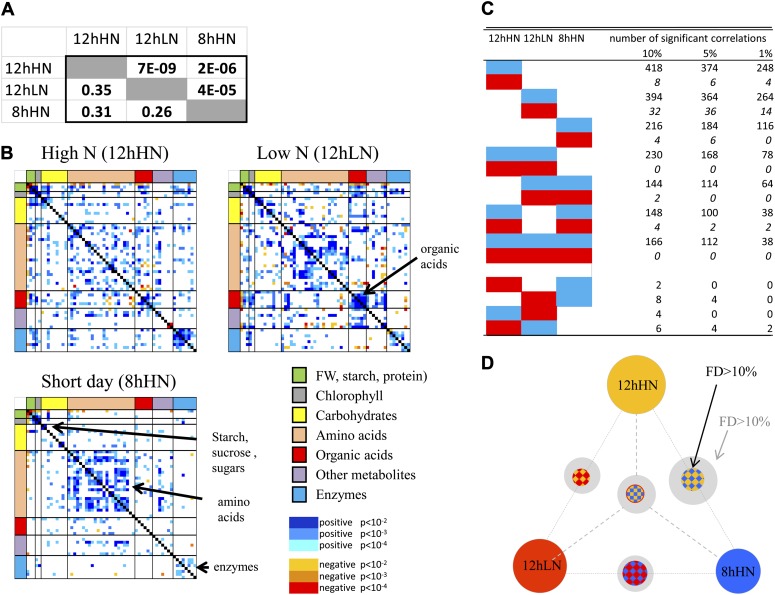
A total of 893, 737, and 434 trait pairs showed a significant correlation in at least one condition at 10%, 5%, and 1% FDR, respectively. These numbers are much higher than those for any single condition, indicating that there is considerable nonoverlap between the correlation matrices in the three growth conditions. The RV coefficient can be used to compare matrices in high-dimensional data analysis studies (Robert and Escoufier, 1976; Abdi, 2007). It is a measure of the similarity between two matrices and varies between +1 (if the two compared matrices are identical) and zero (if the two matrices are completely different). The RV coefficients (Fig. 3A) were between 0.35 and 0.26, which are rather low values, confirming that the metabolic networks are condition dependent. The P values were nonetheless significant, indicating there are some robustly shared features.
Figure 3.
Comparison of the correlation matrices for structural and metabolic traits in three growth conditions. Three separate networks were generated from the values of 50 metabolic traits, starch, protein, and biomass in 97 Arabidopsis accessions grown in the three different growth conditions 12hHN, 12hLN, and 8hHN. A, Analysis of the RV coefficient between the matrices. The lower left-hand segment shows the RV coefficients and the upper right-hand segment the P values. The RV coefficient values vary between +1 (two identical matrices) and zero (no similarity). B, Condensed heat map of correlation coefficients (r, FW basis) in the networks for 12hHN, 12hLN, and 8hHN. The individual values are provided in Supplemental Table S1. Positive (blue) and negative (red) correlations are indicated at a level of P < 0.01, P < 0.001, and P < 0.0001 (light, medium, and dark, respectively). The order of traits is as in Supplemental Table S2. C, Numbers of pairwise correlations between traits in one, two, or all three growth conditions. The original matrices are provided in Supplemental Table S5. D, Visualization of the numbers of shared and nonshared correlations (FDR [FD] < 0.01). The area of the circle depicts the number of shared correlations. The gray shell shows how overlap increases if FDR is relaxed to FDR less than 0.1 in all except one of the shared growth conditions.
Figure 3B provides a visual overview of the correlation matrix in each condition (for original data and the full matrices, see Supplemental Table S1). Color coding is used to distinguish positive and negative correlations and to denote significance at P < 0.01, P < 0.001, and P < 0.0001. Some general features were conserved across all growth conditions. First, there were many more positive correlations than negative correlations, and second, while there were many correlations between metabolites and many correlations between enzymes, there were relatively few correlations between enzymes and metabolites (Sulpice et al., 2010). However, closer inspection reveals that many correlations were condition dependent. The 12hHN data set showed a relatively low connectivity between metabolites, while enzymes were strongly correlated. In 12hLN, correlations between metabolites were stronger, especially between amino acids and between organic acids. Nitrate assimilation is closely linked with organic acid synthesis because organic acids act as counteranions for nitrate and provide carbon skeletons for the synthesis of amino acids (Nunes-Nesi et al., 2010; Xu et al., 2012). In 8hHN, the matrix is dominated by positive correlations between amino acids and positive correlations between enzymes. The positive correlations between amino acids reveals that the decrease in the levels of different amino acid levels noted above (Fig. 2B) occurs in a coordinated manner and is larger in some accessions than in others.
The extent of overlap of individual links (correlations) in the three correlation matrices is further explored in Figure 3, C and D. Of the links that are significant at FDR of less than 0.01, only 19 were shared across all three growth conditions. These were restricted to metabolites that are immediately adjacent to each other in metabolic pathways or have very similar functionalities (Glc and Fru; Asp and Glu; the three basic amino acids Lys, Asn, and Arg, the three aliphatic amino acids Val, Leu, and Ile, and aminobutyric acid; and Asp, Arg, Pro, and 4-Hyp), two closely adjacent enzymes that are involved in malate formation (PEPCx and NADH-MDH), and the three structural components (protein and chlorophylls a and b; Supplemental Table S6).
Testing for shared links at FDR of less than 1% may result in false negatives because traits pairs that are significant in one growth condition may lie slightly below this stringent threshold in another. We therefore investigated how many conserved links are found for trait pairs that show a correlation at FDR of less than 0.01 in at least one growth condition and a more relaxed significance level of FDR of less than 10% for the other two conditions (Fig. 3D). This analysis revealed that up to 83 (4.9% of all possible) links are conserved in all three conditions. The additional shared links include Suc with protein, amino acids with chlorophylls a and b, further pairs of amino acids, and a set of enzymes involved in starch and nitrogen metabolism (AGPase, GS, PEPCx, and NAD-MDH).
We also tested for links that were conserved in two of the three growth conditions. At FDR of less than 0.01, another 39, 19, and 32 pairwise correlations were significant in the 12hHN versus 12hLN, 12hHN versus 8hHN, and 12hLN versus 8hHN comparisons, respectively, rising to 115, 74, and 72 when the criteria were relaxed, as discussed above. We also asked for selected metabolic traits if the variation between accessions was conserved across different growth conditions. Pairwise plots revealed a weak but significant agreement for starch (r = 0.30, P = 0.05) and protein (r = 0.35, P = 0.001) when 8hHN was compared with 12hHN and a nonsignificant correlation for starch (r = 0.16, P = 0.12) and protein (r = 0.15, P = 0.15) when 12hHN was compared with 12hLN (Supplemental Fig. S4).
Altogether, these results point to a strong impact of the growth condition on the links in networks extracted from metabolic profiles. While there are a small proportion of conserved links, these are mainly for metabolites or enzymes that are closely related with respect to pathway topology or trait function.
PLS Regression of Biomass, Starch, and Protein on Other Metabolic Traits
As already noted, some individual metabolic traits correlate with biomass (Table I). Predictive power can be increased by using multivariate analysis to predict biomass from a linear combination of a set of low-Mr metabolites (Meyer et al., 2007; Sulpice et al., 2009). Therefore, we investigated whether multivariate analysis reveals shared features in the network linking metabolic traits and biomass formation that are not apparent at the level of pairwise comparisons.
In data sets such as ours, where the number of predictors (52) is lower than the number of accessions (97), the predictive power of linear models can be improved by dimensionality reduction methods such as PLS regression. PLS identifies combinations of the original input traits (also termed variables or predictors) that have the maximum covariance with the output trait of interest. These orthogonal combinations of input traits, referred to as latent variables, are then used to predict the output trait. Sulpice et al. (2009) previously found, for Arabidopsis accessions growing in short-day conditions, that biomass, starch levels, and protein concentration are correlated and that each is predicted by a similar combination of low-Mr metabolites. We repeated this analysis for all three growth conditions. The idea of PLS is to provide models of high predictive power while selecting a small number of latent variables, which might correspond to a reduced number of metabolic traits in the model. Selection of the number of latent variables was performed based on minimization of the residual mean-squared prediction error after leave-one-out (LOO) cross validation. The predictive power of the model was evaluated in a second round of cross validation by determining the correlation between the predicted values and the respective response variable with the previously determined fixed number of latent variables; significance was evaluated by permutation of the data 5,000 times (see “Materials and Methods” for details). Shuffling was conducted to allow consideration of permutations specific for a metabolic trait. The use of two separate rounds of cross validation for robust estimation of the number of latent variables and for evaluation of the predicting power of the model reduces the possibility of overfitting.
In each growth condition, PLS regression using metabolite levels as an input allowed a significant prediction of biomass (Pearson correlation of 0.36, 0.58, and 0.27 for 12hHN, 12hLN, and 8hHN, respectively, P < 0.05) and starch (Pearson correlation of 0.67, 0.39, and 0.23 for 12hHN, 12hLN, and 8hHN, respectively, P < 0.05). It also allowed a significant prediction of protein concentration in 12hHN and 12hLN (Pearson correlation of 0.46 and 0.57, respectively, P > 0.05) but not in 8hHN (Table II; Supplemental Table S7). The predictive power was improved compared with individual metabolites (Table I).
Table II. PLS regression analysis of the relation between low molecular weight metabolites and biomass, starch level, or protein concentration.
PLS was performed using 52 metabolic traits from condition A as input and FW, starch, or protein as output traits from condition B, for each of the nine combinations of conditions A and B. The correlation between the predicted values and the response variables, denoted by R, was determined after cross-validated selection of the number of latent variables (n). The number of latent variables (n) and the P values were determined in two successive and independent rounds of cross validation, and significance was then determined by permutation test (see “Materials and Methods”). PLS was also performed on the means of traits across the three growth conditions, and on a combined data set containing all individual values from the three growth conditions; in these regressions, growth conditions are not given because the output traits were averaged or combined, respectively, across all three growth conditions. Italic font indicates regressions in which the input and output traits are from the same growth condition. Significant regressions are indicated by boldface.
| Growth Condition | Output Trait |
12hHN |
12hLN |
8hHN |
Mean |
Combination of All Individual Values |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| R/n | P | R/n | P | R/n | P | R/n | P | R/n | P | ||
| 12hHN | FW | 0.36/1 | 0.002 | 0.26/1 | 0.006 | 0.28/1 | 0.002 | ||||
| Starch | 0.67/1 | 0.001 | 0.47/1 | 0.001 | 0.68/1 | 0.001 | |||||
| Protein | 0.46/1 | 0.001 | 0.34/1 | 0.001 | 0.46/1 | 0.001 | |||||
| 12hLN | FW | 0.51/1 | 0.001 | 0.58/1 | 0.001 | 0.51/1 | 0.001 | ||||
| Starch | 0.35/1 | 0.001 | 0.39/1 | 0.001 | 0.36/3 | 0.001 | |||||
| Protein | 0.45/1 | 0.001 | 0.57/2 | 0.001 | 0.34/1 | 0.002 | |||||
| 8hHN | FW | 0.27/1 | 0.003 | 0.21/1 | 0.023 | 0.27/1 | 0.013 | ||||
| Starch | 0.26/1 | 0.006 | 0.05/2 | 0.302 | 0.23/1 | 0.023 | |||||
| Protein | 0.13/1 | 0.102 | 0.17/1 | 0.057 | 0.17/1 | 0.072 | |||||
| Mean or All | FW | 0.45/1 | 0.001 | 0.73/3 | 0.001 | ||||||
| Starch | 0.61/2 | 0.001 | 0.77/31 | 0.001 | |||||||
| Protein | 0.57/3 | 0.001 | 0.94/32 | 0.001 | |||||||
We also asked whether metabolite profiles measured in one growth condition allow prediction of biomass, starch, or protein in a different growth condition. While almost all PLS regressions were significant (except for the prediction of protein in 8hHN by metabolic traits from any condition), the cross-validated correlations were generally smaller (Table II). For significant regressions, the range and average of P values was 0.001 to 0.003 and 0.013 for within-growth condition comparisons and 0.001 to 0.030 and 0.023 for cross-growth condition comparisons, respectively.
We also analyzed two additional scenarios. In the first, we conducted PLS on the means of the traits across all three conditions, while in the second, we employed PLS on a combination of the three data sets (“Mean” and “All,” respectively, in Table II). More specifically, in the “Mean” scenario, we used the mean input traits across the three conditions to build a PLS regression on the mean output traits across the three conditions; in the “All” scenario, we conducted PLS analysis on the concatenated data matrices from the three conditions. Using the mean of the traits over the three conditions in the PLS analysis, we found that all regressions are significant and of greater predictive power than the average power of condition-specific PLS. This gain was even greater when we used all three data sets (“All”). However, although similar observations with much greater correlation values were obtained using the combined data sets, these regressions were based on a much larger number of latent variables, especially when we combined the data sets (as many as 32 from 52 available). Altogether, these findings suggest that PLS regression on metabolites to predict output traits such as biomass and starch may be specific and more robust for comparisons within a given growth condition than for comparison across conditions.
The importance of individual input traits (variables) in the linear combination is provided by the variable importance in the projection (VIP; Chon and Jung, 2005). The VIP for each individual metabolic trait as input in each growth condition is provided in Supplemental Table S7. We next investigated the correlation between the VIP of the input traits from the PLS regressions on the pairs of output traits. This analysis led to two main conclusions. First, there was close agreement between the VIP of metabolic input traits in the prediction of the three output traits in 12hHN (P < 0.001 in all pairwise comparisons of biomass, starch, and protein; Table III). The agreement was lower under 12hLN (especially for the comparison protein versus biomass, P = 0.17) and still weaker in 8hHN, when there was good agreement between biomass and protein (P < 0.0001) but not between biomass and starch (P = 0.059) or starch and protein (P = 0.28). The latter differs from a previous report (Sulpice et al., 2009; see “Discussion”). Second, different metabolic traits were important in the PLS regression in different conditions (assessments are based on VIP > 1). Taking the PLS regressions on biomass as an example, in 12hHN, Xyl, several central amino acids (Gln, Ala, Asp, and Gly), all three aromatic amino acids (Phe, Trp, and Tyr), Pro, 4-Hyp, malate, and succinate had a high VIP, while in 8hHN, raffinose, a similar set of central amino acids (Gln, Glu, Ala, Asp, and Gly), all nitrogen-rich amino acids (Lys, Asn, and Arg), fumarate, threonate, and putrescine had a high VIP and in 12hLN, raffinose, maltose, trehalose, erythritol, a different set of amino acids (Ala, Asn, and Arg), succinate, glycerate, 4-Hyp, dehydroascorbate, threonate, putrescine, and nitrate reductase activity had a high VIP for biomass. Some metabolic traits (e.g. Ala and 4-Hyp) were represented in all three conditions, while others (e.g. Pro, Asn, and Arg) were represented in two conditions and many in only one condition. With the exception of nitrate reductase, enzyme activities did not show high VIP. By comparing Table I with Supplemental Table S5, we observed that many of the metabolic traits with a high VIP show significant correlations with biomass in that condition. The correlation between the VIP in the regression for biomass and the Spearman rank correlation coefficients of individual metabolic traits and biomass, with values of 0.65, 0.72, and 0.42 in 12hHN, 12hLN, and 8hHN, respectively, provide statistical support for this observation.
Table III. Correlation (R) between the VIP of metabolic traits in the PLS regression on biomass, starch, and protein.
The VIP of the metabolite traits in the PLS regressions on biomass, starch, and protein concentration are provided in Supplemental Table S7). Significant correlations are indicated by boldface.
Relationship between Biomass and Nitrate and Total Nitrogen Content in Plants Growing with a Restricted Nitrogen Supply
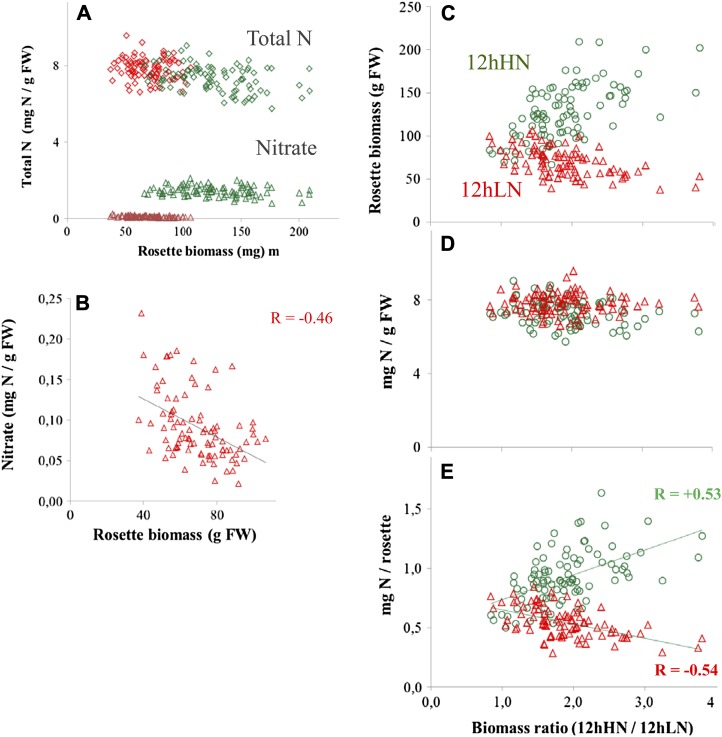
We next asked whether nitrate or other metabolic traits related to nitrogen adopt a more important role as a predictor for biomass in low-nitrogen conditions. These analyses were limited to 12hHN and 12hLN because values for nitrate were not available for the published 8hHN study. Nitrate levels were higher in 12hHN, where they typically accounted for about 20% of the nitrogen in the rosette, than in 12hLN (Fig. 4A). Nitrate levels were negatively correlated to biomass in 12hLN but unrelated to biomass in 12hHN (r = –0.5, P = 8e–06 and r = –0.18, P = 0.18, respectively; Table I; Fig. 4, A and B). Total nitrogen content was estimated by summing nitrogen in nitrate, protein, amino acids, and chlorophyll. Total nitrogen content was similar in both 12hLN and 12hHN and was unrelated to biomass in both conditions (Fig. 4A).
Figure 4.
Nitrogen concentration and nitrogen content in accessions growing in low and optimal nitrogen supply. A, Rosette biomass and total nitrogen in nitrate, protein, amino acids, and chlorophyll (red and green diamonds) and against nitrogen in nitrate (red and green triangles). Total nitrogen contents are calculated in Supplemental Table S8. B, Relationship between nitrate concentration and rosette biomass in 12hLN; data in A is replotted with an expanded y-axis scale. C to E, The x axis ranks accessions according to their gain in biomass between low-nitrogen and high-nitrogen growth conditions. The x axis shows the values for a given accession for 12hHN (green circles) and 12hLN (red triangles). C, Rosette biomass. D, Summed nitrogen content in protein, amino acids, chlorophyll, and nitrate. E, Summed nitrogen content per rosette.
As already noted, accessions that maintained a relatively high biomass in low nitrogen tended to show only a small increase in biomass in high nitrogen, whereas accessions that showed a relatively small biomass in low nitrogen showed a large (greater than 3-fold) increase in biomass in high nitrogen (Fig. 4C). The ability of plants to grow with a low nitrogen supply, sometimes termed nitrogen use efficiency, can be divided into two components: the ability to produce more biomass per unit nitrogen in the plant and the ability to obtain nitrogen from the soil (Moll et al., 1982). The total nitrogen concentration in the rosette was unrelated to the biomass difference between low and high nitrogen (Fig. 4D). The nitrogen content (milligrams of nitrogen per rosette) was strongly related to the response of an accession to nitrogen; accessions that maintained biomass in low nitrogen contained more nitrogen in the rosette than accessions that showed a large gain in biomass in high nitrogen (Fig. 4E). These results imply that accessions differ in the extent to which they can acquire nitrogen from low-nitrogen soil and that this is far more important for the response of biomass to nitrogen supply than changes in the nitrogen content of the rosette.
Mixed-Model Analysis
The results presented in the previous sections were mainly based on per-condition PLS regressions without controlling for the effects of environmental conditions. This is usually referred to as a by-group approach, where each group corresponds to a condition. A by-group approach does not detect relationships that are conserved across conditions and may highlight very specific effects for the individual conditions. We therefore asked if a more generalizable model for each of the three output traits (i.e. biomass, starch, and protein) can be obtained by combining the data sets from the three conditions, using an approach based on linear mixed models.
Linear mixed models are a type of generalized linear mixed model (Breslow and Clayton, 1993) that offers a parsimonious way to account for group level structure in data while simultaneously assessing effects within and across groups (i.e. conditions). In addition to individual level noise, linear mixed models allow for normally distributed group level differences centered on the individual level parameters. Our analysis is based on a linear mixed model with random intercepts by condition (defined as a grouping factor), formulated as:
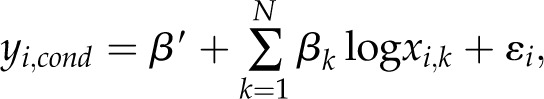 |
where ε follows normal distribution N(0, σε), βcond follows N(0, σcond), and βcond is perpendicular to ε.
In this model, the intercept (β′) is the sum of the ordinary intercept (i.e. the global mean [β0]) and the adjustment based on the group (i.e. the condition [βcond]) for each of the three output traits. The adjustment is assumed to be normally distributed, centered on zero, and orthogonal to the individual level noise (ε). This adjustment is termed the random intercept because it adjusts the overall intercept to reflect a randomly distributed condition-specific intercept.
Here, we first ask whether we can remove the random intercept without sacrificing the power of the model. This is achieved by 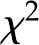 test (with 1 degree of freedom) over the difference in deviance (defined as twice the log likelihood) between the model with a random intercept against the same model without a random per-condition intercept. This test aims to determine whether the added number of parameters (due to the random intercepts) significantly improves the model quality. While inclusion of random intercepts increases the quality for the model of biomass (P < 0.05), this is not the case for starch and protein concentration (Supplemental Table S9). The random intercepts in the case of a linear mixed model for biomass were 35.75, 2.21, and –37.96 for 12hHN, 12hLN, and 8hHN, respectively, indicating the condition specificity. Moreover, analysis of the deviance table (Supplemental Table S9) reported that chlorophyll a, Suc, myoinositol, Asp, Gly, Ser, and nicotinic acid had significant coefficients in the regression for biomass at a significance level of 0.05. While some of these metabolites (Suc and Gly) had a significant correlation (Table I) or a high VIP in the PLS regression on biomass (Supplemental Table S5) in two of the growth conditions, others (e.g. Ser and nicotinic acid) had not been uncovered in the previous analyses. This is due to the more important role of these metabolic traits in the generalized (cross-condition) model. Nevertheless, as for the PLS regressions, enzymes make only a weak contribution (none significant at P < 0.05 and only one, GS, at P < 0.1).
test (with 1 degree of freedom) over the difference in deviance (defined as twice the log likelihood) between the model with a random intercept against the same model without a random per-condition intercept. This test aims to determine whether the added number of parameters (due to the random intercepts) significantly improves the model quality. While inclusion of random intercepts increases the quality for the model of biomass (P < 0.05), this is not the case for starch and protein concentration (Supplemental Table S9). The random intercepts in the case of a linear mixed model for biomass were 35.75, 2.21, and –37.96 for 12hHN, 12hLN, and 8hHN, respectively, indicating the condition specificity. Moreover, analysis of the deviance table (Supplemental Table S9) reported that chlorophyll a, Suc, myoinositol, Asp, Gly, Ser, and nicotinic acid had significant coefficients in the regression for biomass at a significance level of 0.05. While some of these metabolites (Suc and Gly) had a significant correlation (Table I) or a high VIP in the PLS regression on biomass (Supplemental Table S5) in two of the growth conditions, others (e.g. Ser and nicotinic acid) had not been uncovered in the previous analyses. This is due to the more important role of these metabolic traits in the generalized (cross-condition) model. Nevertheless, as for the PLS regressions, enzymes make only a weak contribution (none significant at P < 0.05 and only one, GS, at P < 0.1).
We next tested if random effects for the slope of these seven significant metabolic traits improve the quality of the model for biomass. 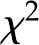 test (7 degrees of freedom) indicated that adding random slopes, presented in Supplemental Table S9, improves the predictive power of the model. Subsequent analysis of the deviance table indicated that the combined effects (i.e. fixed and random) for Suc and Gly are significant, while Ala has a significant fixed effect (P < 0.05; Supplemental Table S9). Ala was one of the very few metabolites that in all three growth conditions correlated significantly (P < 0.05) with biomass (Table I) and had a high VIP in the PLS regression on biomass (Supplemental Table S5).
test (7 degrees of freedom) indicated that adding random slopes, presented in Supplemental Table S9, improves the predictive power of the model. Subsequent analysis of the deviance table indicated that the combined effects (i.e. fixed and random) for Suc and Gly are significant, while Ala has a significant fixed effect (P < 0.05; Supplemental Table S9). Ala was one of the very few metabolites that in all three growth conditions correlated significantly (P < 0.05) with biomass (Table I) and had a high VIP in the PLS regression on biomass (Supplemental Table S5).
DISCUSSION
While it can be anticipated that metabolism will affect growth and that this dependence should be reflected in the values of metabolic traits, this connection is often masked due to the complexity of the network that links metabolism with growth (Fernie and Stitt, 2012). Natural genetic diversity provides a powerful tool to analyze complex networks because it allows the study of thousands of genetic perturbations that vary independently between different genotypes. Profiling of populations of Arabidopsis natural accessions or inbred lines and application of multivariate analysis tools has allowed sets of metabolites to be identified that are predictive of biomass (Meyer et al., 2007; Sulpice et al., 2009; Steinfath et al., 2010a, 2010b; Cuadros-Inostroza et al., 2010; Carreno-Quintero et al., 2012) and in some cases has allowed hypotheses to be formulated with respect to which aspects of metabolism play a key role in the determination of growth (Sulpice et al., 2009, 2010). However, metabolite levels depend on the growth condition (Caldana et al., 2011; Obata and Fernie, 2012; see the introduction for further references). We have investigated (1) whether metabolite profiles provide information that is predictive for biomass in three different growth conditions and (2) whether the network connectivity is conserved or changes between growth conditions. To do this, a panel of 97 genetically diverse Arabidopsis accessions was grown in three growth conditions: near-optimal carbon and nitrogen supply, restricted carbon supply, and restricted nitrogen supply. The growth protocols used to restrict carbon and nitrogen decreased biomass by, on average, about 2-fold compared with near-optimal carbon and nitrogen. This represents a small decrease in the rate of growth. Previous work in the reference accession Col-0 has shown that Arabidopsis adjusts to these regimes to avoid an acute carbon limitation (Gibon et al., 2009; Stitt and Zeeman, 2012) or nitrogen limitation of metabolism and growth (Tschoep et al., 2009).
The large genetic diversity in Arabidopsis for biomass is apparent, with approximately 3-fold differences in biomass between the smallest and largest accessions in a given growth condition. Accessions vary in their response to the growth condition (Fig. 1). While there is a trend for accessions that are large in one condition to also be large in other conditions, this is modified by two further trends; first, accessions that develop a high biomass in near-optimal conditions show a larger decrease of biomass in limiting conditions and second, many individual accessions show differing responses to low carbon and low nitrogen.
While there are no other published studies of the response of biomass to low carbon in Arabidopsis, three earlier studies used a much smaller but partly overlapping panel of accessions to study the response to low nitrogen (http://dbsgap.versailles.inra.fr/vnat/; Supplemental Table S10; North et al., 2009; Chardon et al., 2010). The trend for accessions that produce a high biomass in high nitrogen to show a larger decrease in biomass in low nitrogen is visible in these earlier studies. However, detailed comparison is difficult because of differences in accession ranking for biomass. In high-nitrogen conditions, there is very good agreement between biomass in our study and the Natural Variation of Arabidopsis thaliana (VNAT) database (20 shared accessions, r = 0.69) and weak agreement with the study of Chardon et al. (2010; 18 shared accessions, r = 0.23, due to two accessions that show low biomass in our study and the VNAT database but have a high biomass in the study of Chardon et al., 2010). In low-nitrogen treatments, the agreement between our data and that of the VNAT database and Chardon et al. (2010) breaks down (r = –0.03 and 0.07, respectively). This poor agreement may be due to different protocols in the low-nitrogen treatments. Whereas we used large pots containing soil with a high or a very low nitrogen content from the beginning of the experiment, earlier studies grew plants in small pots with sand and watered regularly with nutrient solution containing different amounts of nitrogen. In our growth protocol, nitrogen-restricted plants contain less nitrate and show a slower rate of growth but maintain rosette levels of amino acids and protein (Figs. 2 and 4; Supplemental Table S1; Tschoep et al., 2009), whereas rosette nitrogen concentration decreased by 20% to 30% in North et al. (2009) and Chardon et al. (2010). Furthermore, while the variation in biomass is similar in high-nitrogen and low-nitrogen treatments in our growth protocol, in the other growth protocols, there is less variation in biomass formation in low-nitrogen than in high-nitrogen treatments. Despite this variation between studies, our analysis confirms previous reports (Chardon et al., 2010, 2012) that Bur0 shows a large response to nitrogen, reveals that this accession is relatively insensitive to low carbon, and identifies further accessions that show a similar response (Dijon5 and Old1). Our study also identifies accessions that show a reverse response, with a large decrease in biomass in low carbon, and maintained biomass in low-nitrogen conditions (Mh1, Nok2, and Lov5).
The response of metabolic traits is dominated by the growth condition (Fig. 2), with low carbon or low nitrogen leading to marked and differing changes in many metabolic traits across all the accessions. Nevertheless, there is genetic variation for metabolic traits. This can be captured in each growth condition as a correlation matrix (Fig. 3). These networks identify metabolic traits that are subject to coordinated changes between accessions in a given growth condition. The correlation matrices show some shared general features, in particular a predominance of positive correlations, and the presence of many correlations between metabolite levels, many correlations between enzyme activities, and few correlations between metabolite levels and enzyme activities. As previously discussed (Sulpice et al., 2010), this may reflect the complexity of the network that links enzyme activities with metabolite levels. A small number of links are found in all three growth conditions, mainly between topologically adjacent or functionally similar metabolic traits. Nonetheless, the main feature emerging from our large study is that both the metabolic traits and the correlation network depend strongly on growth conditions. First, low nitrogen and low carbon lead to characteristic changes in metabolite levels that affect all accessions (Fig. 2). In low nitrogen, this includes an increase in many amino acid levels, a decrease in organic acids, and a decrease in nitrate reductase activity. In low carbon, this includes an increase in Suc and reducing sugars, a decrease in many amino acids, with the exception of Ala, which increases, and an increase in NAD-GlDH activity. Second, most of the individual links in the metabolic network are condition specific (Fig. 3). In low nitrogen, the correlation network is dominated by strong connectivity between amino acids and between organic acids, in low carbon, the network is dominated by strong connectivity between amino acids, and in near-optimal conditions, the network is dominated by a less topologically defined response.
The growth condition modifies the relationship between metabolic traits and biomass. A different set of individual metabolic traits correlate to biomass in each growth condition (Table I). While PLS regression allows a highly significant prediction of biomass in each growth condition and often between growth conditions, the statistical significance for the predictive power tends to be stronger and a smaller number of latent variables is required when these analyses are made within a given growth condition (Table II). Application of linear mixed models highlighted that the inclusion of random (condition-dependent) effects for the intercept in the regression increases the quality of the model for biomass, but not for starch and protein concentrations. This further supports the condition specificity of biomass prediction that is suggested by the PLS regressions. Additional analysis suggested that the inclusion of random slopes for metabolic traits that have significant coefficients in the linear mixed models could further improve the quality of these models.
A small number of individual metabolic traits are linked to biomass in all three growth conditions. For example, Ala correlated with biomass in all three growth conditions (Table I), had high VIP in PLS regressions on biomass in all conditions (Supplemental Table S7), and, together with Suc and Gly, was highlighted as important in the mixed linear model (Supplemental Table S9). We previously reported that biomass is negatively correlated with starch and protein (Sulpice et al., 2009). This finding is confirmed for starch in all conditions used in this study and for protein in near-optimal and low-carbon conditions, but not in low nitrogen (Table I). We also reported that a similar set of metabolites has a high VIP in a PLS regression on all three traits and proposed that starch and protein concentration are integrative metabolic traits that capture information about the levels of many low-Mr metabolites and are closely linked to biomass formation (Sulpice et al., 2009). This relationship with biomass is confirmed in near-optimal (12hHN) conditions for starch and for protein (P < 0.001), in low nitrogen for starch (P < 0.05), but not for protein (P = 0.17), and in low-carbon conditions for protein (P < 0.001), but not, or only very weakly, for starch (P = 0.059; Table III). The analysis in Sulpice et al. (2009) was carried out in short-day (low-carbon) conditions; hence, there is a discrepancy in this particular condition. This may be due to use of a more stringent procedure for selection of the number of latent variables and validation of prediction in this study and because the published 8hHN data set was obtained using a weaker experimental design than that used to obtain the 12hHN data set in this study (see “Materials and Methods”).
The metabolic traits that adopt a major role in the network linking metabolism and growth in a given growth condition are often closely related to the metabolic resource that limits growth in that condition. In short-day (low-carbon) conditions, low starch is the most powerful single predictor of biomass (Table I). Protein is also negatively correlated to biomass, as are many amino acids (Table I). Furthermore, protein and many amino acids decrease in short-day conditions (Table II). As outlined in the introduction, in low-carbon conditions, low protein concentration may increase the efficiency with which resources are used to generate biomass, which in turn may explain why starch reserves can be decreased (Sulpice et al., 2009, 2010). The second most strongly correlating individual metabolic trait is a positive correlation with NADH-GlDH activity. NAD-GlDH activity is induced by carbon starvation (Melo-Oliveira et al., 1996; Gibon et al., 2004; Mayashita and Good, 2008). This prompts the hypothesis that large accessions, which contain less starch, operate with a lower margin of carbon than small accessions. When more carbon is available for growth in a 12-h photoperiod (Gibon et al., 2009), the negative correlation between biomass and starch is retained, but the links to protein concentration, amino acid metabolism, and NAD-GlDH activity are weakened or abolished. This is consistent with the idea that this link is driven by metabolic adjustment to low carbon and that there is variation between accessions for the way in which this interaction is regulated (Table I; Fig. 3).
By contrast, in low-nitrogen conditions, the metabolic traits that correlate strongly with biomass include nitrate reductase activity and nitrate, with the latter being the most strongly correlating individual metabolic trait (Table I). A trivial explanation for the negative relationship between biomass and nitrate would be that accessions with a larger biomass in low-nitrogen conditions exhaust nitrate; this, however, can be excluded because total nitrogen concentration was independent of accession biomass (Fig. 4). A similar observation has been made in earlier studies with a small panel of Arabidopsis accessions (Chardon et al., 2010, 2012). Nitrate is typically taken up in the day and the night but is mainly assimilated during the day, when nitrate reductase is posttranslationally activated and photosynthetic electron transport provides reducing equivalents of the reduction of nitrate and the subsequent reduction of nitrite (Lea et al., 2006; Lillo, 2008). This results in a diurnal rhythm in which nitrate levels decrease in the light and recover during the night (Stitt and Krapp, 1999; Matt et al., 2001). The lower level of nitrate and higher activity of nitrate reductase at dusk in accessions that maintain a large biomass in low-nitrogen conditions is consistent with them assimilating more of the incoming nitrate during the day. Furthermore, accessions that maintain a larger biomass in low-nitrogen conditions absorb far more nitrogen from the soil (Fig. 4). Earlier studies of small panels of Arabidopsis accessions indicate that differences in the root system may partly explain differences in nitrogen uptake (Loudet et al., 2005). It is possible that the lower rosette nitrate levels may promote root growth and nitrogen uptake, although more studies of root growth and transport activity will be needed to test this hypothesis.
In conclusion, while metabolic traits can be used to predict biomass in different growth conditions, this will require collection of data for the metabolic input traits in each growth condition. The growth condition has a large impact on the values of metabolic traits and on the connectivity between metabolic traits and influences the connectivity between metabolism and growth. While metabolic traits determined in one growth condition allow prediction of biomass in other conditions, the analysis is more robust when full input and output trait data are available for all conditions under study. Application of linear mixed models also reveals a marked condition effect on the biomass prediction and reveals that prediction can be improved when metabolic input data in all conditions is used as part of the model. Based on the growth conditions related to carbon and nitrogen availability analyzed in our study, in a given condition, metabolic traits related to the limiting resource can adopt a more central role in the network that connects metabolism and growth. This implies that there is substantial natural variation in Arabidopsis for adjustment of metabolism to improve growth in low-carbon and low-nitrogen conditions. This variation, however, means that environmental conditions must be taken into account when searching for individual metabolites or sets of metabolites that act as biomarkers and may compromise attempts to make predictions about genotype performance between different growth conditions.
MATERIALS AND METHODS
Selection of the Accessions and Growth Conditions
Arabidopsis (Arabidopsis thaliana) accessions used in this study were obtained from various sources as previously described (Sulpice et al., 2009, 2010). Geographical origin of the accessions is available at VNAT (http://dbsgap.versailles.inra.fr/vnat/). For the 8hHN treatment, plants were grown in multiple overlapping experiments as previously described (Sulpice et al., 2009, 2010). For the 12hHN and 12hLN treatments, plants were grown in large replicated experiments with all accessions. To eliminate effects due to seedling germination and establishment, in all growth regimes, seeds were germinated and grown for 7 d with a 16-h daylength (irradiance, 145 µmol m–2 s–1; temperature, 20°C in the light and 6°C at night; humidity, 75%) and then in an 8-h-light/16-h-dark regime for 7 d (irradiance, 145 µmol m–2 s–1; respective temperatures and humidities of 20°C and 60% during the day and 16°C and 75% at night). At 14 d, plants of average sizes were transferred to 6-cm-diameter pots (five plants per pot). In all experiments, the position of the pots containing individual accessions was randomized.
For the 8hHN treatment, the soil substrate was GS90 (composition: peat, clay, coconut fiber, 2 g L–1 salt, 160 mg L–1 nitrogen, 190 mg L–1 P2O5, 230 mg L–1 K2O, pH 6; Werner Tantau GmbH and Co.) and vermiculite (Gebrueder Patzer; Cross et al., 2006). For the 12hLN treatment, the soil substrate was 50% (v/v) white peat (Gramoflor GmbH) and 30% (v/v) fine- and 20% (v/v) coarse-grained vermiculite (AGRA-RHP, Kausek GmbH) fertilized with 260 mg K2HPO4, 396 mg GRANUKAL 85 (80% [w/v] CaCO3 and 5% [w/v] MgCO3, Kreidewerke Dammann KG), 1.6 mg Fetrilon-Combi micronutrient fertilizer (BASF AG), and 30 mL of tap water per 100-mL pot (Tschoep et al., 2009). For the 12hHN treatment, the soil substrate was identical to 12hLN, except that it was additionally supplemented with 90 mg solid NH4NO3 per 100-mL pot. Prior to use, soils were stored for 2 weeks at 10°C to allow homogenization of nutrients. At 21 d, plants were transferred to a controlled small growth chamber (145 μmol m–2 s–1, 20°C day and night) for 2 additional weeks in either an 8-h photoperiod (8hHN) or a 12-h photoperiod (12hHN and 12hLN). Plants were watered daily. Within each experiment, the position of the pots containing individual accessions was randomized. Harvests (five samples per accession, each consisting of three rosettes) were performed at the end of the light period. The entire sample was powdered under liquid nitrogen and stored at –80°C until its use.
Enzyme and Metabolite Assays
Chemicals were purchased as described (Gibon et al., 2004). Total protein, starch, Glc, Fru, Suc, and total amino acids were assayed as described (Cross et al., 2006). Malate and fumarate were assayed as described (Nunes-Nesi et al., 2007). For enzyme measurements, aliquots of 20 mg frozen FW were extracted by vigorous mixing with extraction buffer (Nunes-Nesi et al., 2007). AGPase, fumarase, GlDH, PEPCx, invertase, GOGAT, nitrate reductase, and GS were assayed as described (Gibon et al., 2004). NAD-MDH was assayed as described by Nunes-Nesi et al. (2007). Metabolite extraction for Gas chromatography-mass spectrometry was performed as described previously (Schauer et al., 2008).
Derivatization and gas chromatography-mass spectrometry analysis were performed as described previously (Lisec et al., 2006), starting from aliquots of 20 mg frozen FW. Because measurements of nitrate, Orn, and spermidine were not available for the published 8hHN data set, random numbers were introduced for these traits in the calculations of condition-specific correlation matrices. However, these were not used in the PLS and mixed-model analyses.
Statistical Analysis
PLS regression is a dimensionality reduction method that aims at determining predictor combinations with maximum covariance with the response variable (Wold, 1966; Eriksson et al., 2001). The identified combinations, called latent variables, are used to predict the response variable. Selection of the number of latent variables was performed based on minimization of the residual mean-squared prediction error after LOO cross validation. The predicted vector was correlated with the measured values to assess the predictive power of the predictor variables with the fixed number of latent variables. The significance of the prediction power was evaluated by permutation test with 5,000 permutations of the data. We note that in every permutation, each row of the data matrix, corresponding to the profile of a metabolic trait, was shuffled independently of the others. Such permutation strategy is intended to break correlations in pairs of metabolic traits, while maintaining the range that is specific for each metabolic trait. Then, for each permutation, a PLS model with the predetermined number of latent variables was built to predict the randomized response variable and a Pearson correlation between the permuted response variable and in LOO cross validation. The 5,000 random correlations are compared with the performance of the PLS model that was used to predict the true response variable. The predictors were ranked according to their VIP (Chon and Jung, 2005). The VIP measure of a predictor estimates its contribution in the PLS regression. The predictors having VIP values greater than one are considered important for the PLS prediction of the response variable. All procedures were applied after log scaling the metabolic profiles. Our computations were carried out using the R package PLS (Mevik and Wehrens, 2007). For the analysis based on linear mixed models, we used the lmer function from the R package lme4 (Bates and Maechler, 2010), while analysis of the deviance table was carried out with the ANOVA function from the R package car (Fox and Weisberg, 2011).
The RV coefficient is a multivariate generalization of the squared Pearson correlation coefficient, provides a measure of the similarity between two squared symmetric positive semidefinite matrices, and varies between +1 (two identical matrices) and zero (no similarity; Robert and Escoufier, 1976; Abdi, 2007). It was calculated using the R package located at http://CRAN.R-project.org/package=FactoMineR. CV is defined as the ratio of the sd to the mean of all mean values obtained for a trait/accession pair.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Relationship between photoperiod and biomass in the Col-0 wild type.
Supplemental Figure S2. CV for the metabolic traits analyzed between the accessions grown in a given growth condition or averaged CV of all the accessions for the variation in metabolite amounts between two growth conditions.
Supplemental Figure S3. Relationship between biomass and selected metabolic traits in different growth conditions.
Supplemental Figure S4. Relationship between selected metabolic traits (starch and protein) in different growth conditions.
Supplemental Table S1. Accessions, biomass, structural components, starch, low-Mr metabolites, enzyme activities, and abbreviations plus correlation matrices.
Supplemental Table S2. ANOVA analysis.
Supplemental Table S3. Principle components analysis.
Supplemental Table S4. Heat map of changes in metabolic traits under low-carbon and low-nitrogen conditions compared with near-optimal conditions.
Supplemental Table S5. Correlation of individual metabolic traits with biomass.
Supplemental Table S6. Correlation matrices for metabolic traits in each of the three growth conditions.
Supplemental Table S7. PLS regression for biomass, starch, or protein concentration on low-Mr metabolites in three growth conditions.
Supplemental Table S8. Calculation of nitrogen contents.
Supplemental Table S9. Linear mixed models for biomass, starch, and protein concentration: analysis of deviance tables.
Supplemental Table S10. Comparison with published data for biomass in Arabidopsis accessions.
Glossary
- PLS
partial least squares
- FW
fresh weight
- Col-0
Columbia-0
- 12hHN
12-h photoperiod with high nitrogen
- 12hLN
12-h photoperiod with low nitrogen
- 8hHN
8-h photoperiod with high nitrogen
- CV
coefficient of variation
- FDR
false discovery rate
- LOO
leave-one-out
- VIP
variable importance in the projection
- r
Spearman rank correlation coefficient
- VNAT
Natural Variation of Arabidopsis thaliana
- GOGAT
Gln:oxoglutarate aminotransferase
- PEPCx
phosphoenolpyruvate carboxylase
- AGPase
ADP Glc pyrophosphorylase
- GlDH
Glu dehydrogenase
- GS
Glu synthetase
- MDH
malate dehydrogenase
- VNAT
Natural Variation of Arabidopsis thaliana
References
- Abdi H. (2007) RV coefficient and congruence coefficient. In Salkind Neil, ed, Encyclopedia of Measurement and Statistics. Sage, Thousand Oaks, CA [Google Scholar]
- Amiour N, Imbaud S, Clément G, Agier N, Zivy M, Valot B, Balliau T, Armengaud P, Quilleré I, Cañas R, et al. (2012) The use of metabolomics integrated with transcriptomic and proteomic studies for identifying key steps involved in the control of nitrogen metabolism in crops such as maize. J Exp Bot 63: 5017–5033 [DOI] [PubMed] [Google Scholar]
- Amthor JS. (2010) From sunlight to phytomass: on the potential efficiency of converting solar radiation to phyto-energy. New Phytol 188: 939–959 [DOI] [PubMed] [Google Scholar]
- Bates D, Maechler M (2010) Lme4: linear mixed effects models using S4 classes. CRAN:R-project. http://CRAN.R-project.org/package=lme4 (April 4, 2013)
- Bräutigam K, Dietzel L, Kleine T, Ströher E, Wormuth D, Dietz KJ, Radke D, Wirtz M, Hell R, Dörmann P, et al. (2009) Dynamic plastid redox signals integrate gene expression and metabolism to induce distinct metabolic states in photosynthetic acclimation in Arabidopsis. Plant Cell 21: 2715–2732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow E, Clayton DG. (1993) Approximate inference in generalized linear mixed models. J Am Stat Assoc 88: 9–25 [Google Scholar]
- Caldana C, Degenkolbe T, Cuadros-Inostroza A, Klie S, Sulpice R, Leisse A, Steinhauser D, Fernie AR, Willmitzer L, Hannah MA. (2011) High-density kinetic analysis of the metabolomic and transcriptomic response of Arabidopsis to eight environmental conditions. Plant J 67: 869–884 [DOI] [PubMed] [Google Scholar]
- Carrari F, Baxter C, Usadel B, Urbanczyk-Wochniak E, Zanor MI, Nunes-Nesi A, Nikiforova V, Centero D, Ratzka A, Pauly M, et al. (2006) Integrated analysis of metabolite and transcript levels reveals the metabolic shifts that underlie tomato fruit development and highlight regulatory aspects of metabolic network behavior. Plant Physiol 142: 1380–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreno-Quintero N, Acharjee A, Maliepaard C, Bachem CWB, Mumm R, Bouwmeester H, Visser RGF, Keurentjes JJB. (2012) Untargeted metabolic quantitative trait loci analyses reveal a relationship between primary metabolism and potato tuber quality. Plant Physiol 158: 1306–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chardon F, Barthélémy J, Daniel-Vedele F, Masclaux-Daubresse C. (2010) Natural variation of nitrate uptake and nitrogen use efficiency in Arabidopsis thaliana cultivated with limiting and ample nitrogen supply. J Exp Bot 61: 2293–2302 [DOI] [PubMed] [Google Scholar]
- Chardon F, Noël V, Masclaux-Daubresse C. (2012) Exploring NUE in crops and in Arabidopsis ideotypes to improve yield and seed quality. J Exp Bot 63: 3401–3412 [DOI] [PubMed] [Google Scholar]
- Chon IG, Jung CH. (2005) Performance of some variable selection methods when multicollinearity is present. Chemom Intell Lab Syst 78: 103–112 [Google Scholar]
- Cross JM, von Korff M, Altmann T, Bartzetko L, Sulpice R, Gibon Y, Palacios N, Stitt M. (2006) Variation of enzyme activities and metabolite levels in 24 Arabidopsis accessions growing in carbon-limited conditions. Plant Physiol 142: 1574–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadros-Inostroza A, Giavalisco P, Hummel J, Eckardt A, Willmitzer L, Peña-Cortés H. (2010) Discrimination of wine attributes by metabolome analysis. Anal Chem 82: 3573–3580 [DOI] [PubMed] [Google Scholar]
- Eriksson L, Joahnsson N, Kettane-Wold N, Tyrgg J, Wikstom C, Wold S. (2001) Multi- and Megavariate Data Analysis: Principles and Applications. Umetrics Academy, Umea, Sweden [Google Scholar]
- Fernie AR, Aharoni A, Willmitzer L, Stitt M, Tohge T, Kopka J, Carroll AJ, Saito K, Fraser PD, DeLuca V. (2011) Recommendations for reporting metabolite data. Plant Cell 23: 2477–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Stitt M. (2012) On the discordance of metabolomics with proteomics and transcriptomics: coping with increasing complexity in logic, chemistry, and network interactions scientific correspondence. Plant Physiol 158: 1139–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J, Weisberg S (2011) An R Companion to Applied Regression, Ed 2. Sage Publications, Thousand Oaks, CA
- Gibon Y, Blaesing OE, Hannemann J, Carillo P, Höhne M, Hendriks JHM, Palacios N, Cross J, Selbig J, Stitt M. (2004) A robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell 16: 3304–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Pyl ET, Sulpice R, Lunn JE, Höhne M, Günther M, Stitt M. (2009) Adjustment of growth, starch turnover, protein content and central metabolism to a decrease of the carbon supply when Arabidopsis is grown in very short photoperiods. Plant Cell Environ 32: 859–874 [DOI] [PubMed] [Google Scholar]
- Gibon Y, Usadel B, Blaesing OE, Kamlage B, Hoehne M, Trethewey R, Stitt M. (2006) Integration of metabolite with transcript and enzyme activity profiling during diurnal cycles in Arabidopsis rosettes. Genome Biol 7: R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N, Beemster GTS, Inzé D. (2009) David and Goliath: what can the tiny weed Arabidopsis teach us to improve biomass production in crops? Curr Opin Plant Biol 12: 157–164 [DOI] [PubMed] [Google Scholar]
- Hannah MA, Caldana C, Steinhauser D, Balbo I, Fernie AR, Willmitzer L. (2010) Combined transcript and metabolite profiling of Arabidopsis grown under widely variant growth conditions facilitates the identification of novel metabolite-mediated regulation of gene expression. Plant Physiol 152: 2120–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannemann J, Poorter H, Usadel B, Tardieu F, Atkin O, Pons T, Stitt M, Gibon Y. (2009) XEML Lab: a tool that supports the design of experiments at a graphical interface and generates computer-readable metadata files, which capture information about genotypes, growth conditions, environmental perturbations and sampling strategy. Plant Cell Environ 32: 1185–1200 [DOI] [PubMed] [Google Scholar]
- Hermans C, Hammond JP, White PJ, Verbruggen N. (2006) How do plants respond to nutrient shortage by biomass allocation? Trends Plant Sci 11: 610–617 [DOI] [PubMed] [Google Scholar]
- Hirel B, Le Gouis J, Ney B, Gallais A. (2007) The challenge of improving nitrogen use efficiency in crop plants: towards a more central role for genetic variability and quantitative genetics within integrated approaches. J Exp Bot 58: 2369–2387 [DOI] [PubMed] [Google Scholar]
- Jänkänpää HJ, Mishra Y, Schröder WP, Jansson S. (2012) Metabolic profiling reveals metabolic shifts in Arabidopsis plants grown under different light conditions. Plant Cell Environ 35: 1824–1836 [DOI] [PubMed] [Google Scholar]
- Keurentjes JJB, Sulpice R, Gibon Y, Steinhauser MC, Fu JY, Koornneef M, Stitt M, Vreugdenhil D. (2008) Integrative analyses of genetic variation in enzyme activities of primary carbohydrate metabolism reveal distinct modes of regulation in Arabidopsis thaliana. Genome Biol 9: R129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ. (2012) Plant defense compounds: systems approaches to metabolic analysis. Annu Rev Phytopathol 50: 155–173 [DOI] [PubMed] [Google Scholar]
- Korn M, Gärtner T, Erban A, Kopka J, Selbig J, Hincha DK. (2010) Predicting Arabidopsis freezing tolerance and heterosis in freezing tolerance from metabolite composition. Mol Plant 3: 224–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp A, Saliba-Colombani V, Daniel-Vedele F. (2005) Analysis of C and N metabolisms and of C/N interactions using quantitative genetics. Photosynth Res 83: 251–263 [DOI] [PubMed] [Google Scholar]
- Krizek BA. (2009) Making bigger plants: key regulators of final organ size. Curr Opin Plant Biol 12: 17–22 [DOI] [PubMed] [Google Scholar]
- Kusano M, Fukushima A, Redestig H, Saito K. (2011) Metabolomic approaches toward understanding nitrogen metabolism in plants. J Exp Bot 62: 1439–1453 [DOI] [PubMed] [Google Scholar]
- Lea US, Leydecker MT, Quilleré I, Meyer C, Lillo C. (2006) Posttranslational regulation of nitrate reductase strongly affects the levels of free amino acids and nitrate, whereas transcriptional regulation has only minor influence. Plant Physiol 140: 1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeClere S, Schmelz EA, Chourey PS. (2010) Sugar levels regulate tryptophan-dependent auxin biosynthesis in developing maize kernels. Plant Physiol 153: 306–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley JLS, Gee CW, Sairanen I, Ljung K, Nemhauser JL. (2012) An endogenous carbon-sensing pathway triggers increased auxin flux and hypocotyl elongation. Plant Physiol 160: 2261–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillo C. (2008) Signalling cascades integrating light-enhanced nitrate metabolism. Biochem J 415: 11–19 [DOI] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Loudet O, Gaudon V, Trubuil A, Daniel-Vedele F. (2005) Quantitative trait loci controlling root growth and architecture in Arabidopsis thaliana confirmed by heterogeneous inbred family. Theor Appl Genet 110: 742–753 [DOI] [PubMed] [Google Scholar]
- Matt P, Geiger M, Walch-Liu P, Engels C, Krapp A, Stitt M. (2001) Elevated carbon dioxide increases nitrate uptake and nitrate reductase activity when tobacco is growing on nitrate, but increases ammonium uptake and inhibits nitrate reductase activity when tobacco is growing on ammonium nitrate. Plant Cell Environ 24: 1119–1137 [Google Scholar]
- Mayashita Y, Good AC. (2008) NAD(H)-dependent glutamate dehydrogenase is essential for the survival of Arabidopsis thaliana during dark-induced carbon starvation. J Exp Bot 59: 667–680 [DOI] [PubMed] [Google Scholar]
- Melo-Oliveira R, Oliveira IC, Coruzzi GM. (1996) Arabidopsis mutant analysis and gene regulation define a nonredundant role for glutamate dehydrogenase in nitrogen assimilation. Proc Natl Acad Sci USA 93: 4718–4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RC, Steinfath M, Lisec J, Becher M, Witucka-Wall H, Törjék O, Fiehn O, Eckardt A, Willmitzer L, Selbig J, et al. (2007) The metabolic signature related to high plant growth rate in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 4759–4764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RC, Witucka-Wall H, Becher M, Blacha A, Boudichevskaia A, Dörmann P, Fiehn O, Friedel S, von Korff M, Lisec J, et al. (2012) Heterosis manifestation during early Arabidopsis seedling development is characterized by intermediate gene expression and enhanced metabolic activity in the hybrids. Plant J 71: 669–683 [DOI] [PubMed] [Google Scholar]
- Mevik BH, Wehrens R. (2007) The pls package: principal component and partial least squares regression in r. J Stat Softw 18: 1–24 [Google Scholar]
- Moll RH, Kamprath EJ, Jackson WA. (1982) Analysis and interpretation of factors which contribute to efficiency of nitrogen utilization. Agron J 74: 562–564 [Google Scholar]
- North KA, Ehlting B, Koprivova A, Rennenberg H, Kopriva S. (2009) Natural variation in Arabidopsis adaptation to growth at low nitrogen conditions. Plant Physiol Biochem 47: 912–918 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A, Carrari F, Gibon Y, Sulpice R, Lytovchenko A, Fisahn J, Graham J, Ratcliffe RG, Sweetlove LJ, Fernie AR. (2007) Deficiency of mitochondrial fumarase activity in tomato plants impairs photosynthesis via an effect on stomatal function. Plant J 50: 1093–1106 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A, Fernie AR, Stitt M. (2010) Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Mol Plant 3: 973–996 [DOI] [PubMed] [Google Scholar]
- Obata T, Fernie AR. (2012) The use of metabolomics to dissect plant responses to abiotic stresses. Cell Mol Life Sci 69: 3225–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piques M, Schulze WX, Höhne M, Usadel B, Gibon Y, Rohwer J, Stitt M. (2009) Ribosome and transcript copy numbers, polysome occupancy and enzyme dynamics in Arabidopsis. Mol Syst Biol 5: 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poorter H. (1989) Growth analysis: towards a synthesis of the classical and the functional approach. Physiol Plant 75: 237–244 [Google Scholar]
- Poorter H, Nagel O. (2000) The role of biomass allocation in the growth response of plants to different levels of light, CO2, nutrients and water: a quantitative review. Aust J Plant Physiol 27: 595–607 [Google Scholar]
- Poorter H, Niinemets U, Poorter L, Wright IJ, Villar R. (2009) Causes and consequences of variation in leaf mass per area (LMA): a meta-analysis. New Phytol 182: 565–588 [DOI] [PubMed] [Google Scholar]
- Poorter H, Niklas KJ, Reich PB, Oleksyn J, Poot P, Mommer L. (2011) Biomass allocation to leaves, stems and roots: meta-analyses of interspecific variation and environmental control. New Phytol 193: 30–50 [DOI] [PubMed] [Google Scholar]
- Raven JA. (2012) Protein turnover and plant RNA and phosphorus requirements in relation to nitrogen fixation. Plant Sci 190: 131. [DOI] [PubMed] [Google Scholar]
- Riedelsheimer C, Czedik-Eysenberg A, Grieder C, Lisec J, Technow F, Sulpice R, Altmann T, Stitt M, Willmitzer L, Melchinger AE. (2012) Genomic and metabolic prediction of complex heterotic traits in hybrid maize. Nat Genet 44: 217–220 [DOI] [PubMed] [Google Scholar]
- Robert P, Escoufier Y. (1976) A unifying tool for linear multivariate statistical methods: the RV-coefficient. J R Stat Soc Ser C Appl Stat 25: 257–265 [Google Scholar]
- Saito K, Matsuda F. (2010) Metabolomics for functional genomics, systems biology, and biotechnology. Annu Rev Plant Biol 61: 463–489 [DOI] [PubMed] [Google Scholar]
- Schauer N, Semel Y, Balbo I, Steinfath M, Repsilber D, Selbig J, Pleban T, Zamir D, Fernie AR. (2008) Mode of inheritance of primary metabolic traits in tomato. Plant Cell 20: 509–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Stitt M. (2007) Coordination of carbon supply and plant growth. Plant Cell Environ 30: 1126–1149 [DOI] [PubMed] [Google Scholar]
- Steinfath M, Gärtner T, Lisec J, Meyer RC, Altmann T, Willmitzer L, Selbig J. (2010b) Prediction of hybrid biomass in Arabidopsis thaliana by selected parental SNP and metabolic markers. Theor Appl Genet 120: 239–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinfath M, Strehmel N, Peters R, Schauer N, Groth D, Hummel J, Steup M, Selbig J, Kopka J, Geigenberger P, et al. (2010a) Discovering plant metabolic biomarkers for phenotype prediction using an untargeted approach. Plant Biotechnol J 8: 900–911 [DOI] [PubMed] [Google Scholar]
- Stitt M, Krapp A. (1999) The interaction between elevated carbon dioxide and nitrogen nutrition: the physiological and molecular background. Plant Cell Environ 22: 583–621 [Google Scholar]
- Stitt M, Sulpice R, Keurentjes J. (2010) Metabolic networks: how to identify key components in the regulation of metabolism and growth. Plant Physiol 152: 428–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M, Zeeman SC. (2012) Starch turnover: pathways, regulation and role in growth. Curr Opin Plant Biol 15: 282–292 [DOI] [PubMed] [Google Scholar]
- Sulpice R, Pyl ET, Ishihara H, Trenkamp S, Steinfath M, Witucka-Wall H, Gibon Y, Usadel B, Poree F, Piques MC, et al. (2009) Starch as a major integrator in the regulation of plant growth. Proc Natl Acad Sci USA 106: 10348–10353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulpice R, Trenkamp S, Steinfath M, Usadel B, Gibon Y, Witucka-Wall H, Pyl ET, Tschoep H, Steinhauser MC, Guenther M, et al. (2010) Network analysis of enzyme activities and metabolite levels and their relationship to biomass in a large panel of Arabidopsis accessions. Plant Cell 22: 2872–2893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschoep H, Gibon Y, Carillo P, Armengaud P, Szecowka M, Nunes-Nesi A, Fernie AR, Koehl K, Stitt M. (2009) Adjustment of growth and central metabolism to a mild but sustained nitrogen-limitation in Arabidopsis. Plant Cell Environ 32: 300–318 [DOI] [PubMed] [Google Scholar]
- Usadel B, Bläsing OE, Gibon Y, Retzlaff K, Höhne M, Günther M, Stitt M. (2008) Global transcript levels respond to small changes of the carbon status during progressive exhaustion of carbohydrates in Arabidopsis rosettes. Plant Physiol 146: 1834–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold H. (1966) Estimation of Principal Components and Related Models by Iterative Least Squares. Academic Press, New York [Google Scholar]
- Xu G, Fan X, Miller AJ. (2012) Plant nitrogen assimilation and use efficiency. Annu Rev Plant Biol 63: 153–182 [DOI] [PubMed] [Google Scholar]
- Zamboni N. (2011) 13C metabolic flux analysis in complex systems. Curr Opin Biotechnol 22: 103–108 [DOI] [PubMed] [Google Scholar]
- Züst T, Heichinger C, Grossniklaus U, Harrington R, Kliebenstein DJ, Turnbull LA. (2012) Natural enemies drive geographic variation in plant defenses. Science 338: 116–119 [DOI] [PubMed] [Google Scholar]