Oxalate decarboxylase-regulated metabolic networks may provide a widely applicable strategy for enhancing crop nutritional value.
Abstract
The plant metabolite oxalic acid is increasingly recognized as a food toxin with negative effects on human nutrition. Decarboxylative degradation of oxalic acid is catalyzed, in a substrate-specific reaction, by oxalate decarboxylase (OXDC), forming formic acid and carbon dioxide. Attempts to date to reduce oxalic acid levels and to understand the biological significance of OXDC in crop plants have met with little success. To investigate the role of OXDC and the metabolic consequences of oxalate down-regulation in a heterotrophic, oxalic acid-accumulating fruit, we generated transgenic tomato (Solanum lycopersicum) plants expressing an OXDC (FvOXDC) from the fungus Flammulina velutipes specifically in the fruit. These E8.2-OXDC fruit showed up to a 90% reduction in oxalate content, which correlated with concomitant increases in calcium, iron, and citrate. Expression of OXDC affected neither carbon dioxide assimilation rates nor resulted in any detectable morphological differences in the transgenic plants. Comparative proteomic analysis suggested that metabolic remodeling was associated with the decrease in oxalate content in transgenic fruit. Examination of the E8.2-OXDC fruit proteome revealed that OXDC-responsive proteins involved in metabolism and stress responses represented the most substantially up- and down-regulated categories, respectively, in the transgenic fruit, compared with those of wild-type plants. Collectively, our study provides insights into OXDC-regulated metabolic networks and may provide a widely applicable strategy for enhancing crop nutritional value.
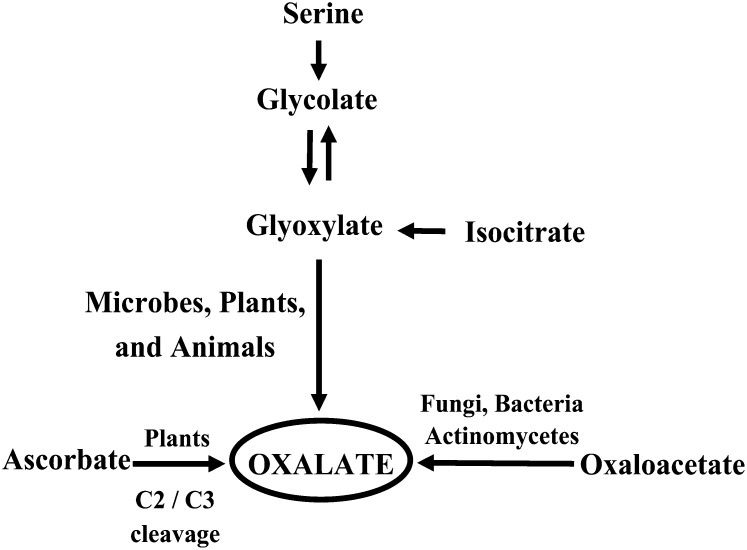
Oxalic acid, a simple low-Mr two-carbon dicarboxylic acid, is ubiquitous among algae, fungi, lichens, and plants as an inert end product of carbon metabolism arising from various metabolic pathways (Fig. 1; Millerd et al., 1963; Chang and Beevers, 1968; Loewus, 1980; Mehta and Datta, 1991). Historically, the presence of oxalate in plants was thought to be confined to the genus Oxalis. However, research conducted during the past decade suggests that oxalate metabolism is ubiquitous in plants and oxalate can be found in any organ, depending on the species (Webb, 1999). The strongly acidic nature and powerful chelating capacity of oxalic acid likely contribute to its action as a nutritional stress factor, and because it is present in almost all food crops (Libert and Franceschi, 1987; Holmes et al., 1998; Holmes and Kennedy, 2000), oxalic acid is a common antinutrient in the human diet. Oxalates in animals, including humans, mostly originate from the diet, especially through the ingestion of fruits, leafy vegetables, cereals, and legumes. Although a pathway for oxalate catabolism is present in bacteria, fungi, and plants, there is no such pathway in vertebrates, including humans. Once oxalate is ingested, it must be excreted through the kidneys, as no gastrointestinal route of oxalate excretion is known (Williams and Wilson, 1990). When oxalate-rich food crops are consumed in large quantities, they can cause primary and secondary hyperoxaluria in humans, which results in impaired renal function, disturbances in Gly metabolism, and reduced blood coagulability (de Castro, 1988; Conyers et al., 1990). Thus, excess ingestion of oxalate results in a variety of kidney-related disorders (Hodgkinson, 1970; Anderson et al., 1971; Suvachittanont et al., 1973; Finch et al., 1981; Curhan, 1997), in addition to neurolathyrism and coronary disease (Singh and Saxena, 1972). The precipitation of oxalic acid as calcium oxalate leads to kidney stones and hypocalcemia (Williams and Wandzilak, 1989) due to poor intestinal absorption of calcium ions in the presence of oxalate (Heaney et al., 1988; Kelsay et al., 1988). Furthermore, a high oxalate intake presumably impairs the utilization of iron, magnesium, and a number of trace metals because of the formation of insoluble salts (Peters et al., 1971). Thus, a reduction in the levels of oxalic acid in food crops would represent a potential attempt to prevent oxalate toxicity, as well as aid in increasing the levels of available minerals and micronutrients.
Figure 1.
Oxalic acid biosynthesis from its biochemical precursors in various organisms. While glycolic, glyoxalic, isocitric, oxaloacetic, and ascorbic acid are the main precursors of oxalic acid synthesis in most organisms, the primary substrate for its synthesis in plants is ascorbic acid.
Enhancing the nutritional composition of food crops represents an urgent worldwide health goal, with an estimated 925 million people currently suffering from food and nutrition insecurity (FAO, 2010). Over the last few years, there have been attempts to develop value-added food crops with the view that they will be more beneficial to human health than dietary supplements (Cooper, 2004). Bioengineering has been employed via many strategies to increase the levels of desirable phytonutrients, while also decreasing the concentrations of deleterious metabolites in both major and minor crops (for review, see Mattoo et al., 2010). Fruits and vegetables contain an array of nutritionally desirable phytochemicals, and those that are particularly noted for containing high levels of such health-promoting compounds are often termed functional foods (Grusak and DellaPenna, 1999). An example is tomato (Solanum lycopersicum), the principal model for fleshy fruit biology (Orzaez et al., 2006; Giovannoni, 2007; Orzaez and Granell, 2009; Centeno et al., 2011; Matas et al., 2011), whose fresh fruits and derived products (e.g. ketchup, juice, soups, and sauces) are rich sources of carotenoids, vitamins, and minerals (Römer et al., 2000). Furthermore, tomato has been associated, through epidemiological studies, with a reduced incidence of degenerative diseases, including heart disease and cancer (Hwang and Bowen, 2002; Murtaugh et al., 2004; Stacewicz-Sapuntzakis and Bowen, 2005). The economic demand and health-promoting constituents of the tomato therefore make it an important target for increasing its nutritional quality. Unfortunately, as with many green leafy vegetables, cereals, and legumes, tomato fruit accumulate considerable amounts of oxalate (U.S. Department of Agriculture, 1984; Noonan and Savage, 1999). In quality improvement programs targeting tomato, the priorities have been to increase carotenoids (Liu et al., 2003) and provitamin A (Römer et al., 2000), whereas reduction in oxalic acid has remained unexplored.
Efficient catabolism of oxalic acid is accomplished via two major pathways, oxidation and decarboxylation. While oxalate oxidase converts oxalate to CO2 and H2O2, decarboxylases catabolize oxalate directly to formate and CO2 (Dunwell et al., 2001). We propose that the introduction of oxalate decarboxylase (OXDC), a low-pH-inducible cupin superfamily protein (Mehta and Datta, 1991; Azam et al., 2002; Chakraborty et al., 2002) that is otherwise absent in plants, from a closely related species (Flammulina velutipes) is a potentially more effective strategy to reduce oxalate levels, as most of the oxalate resides in plant vacuole, where the pH is low. Studies involving FvOXDC overexpression in Arabidopsis (Arabidopsis thaliana; L’Haridon et al., 2011), tobacco (Nicotiana tabacum; Kesarwani et al., 2000), lettuce (Lactuca sativa; Dias et al., 2006), and soybean (Glycine max; Cunha et al., 2010) have suggested a role for this enzyme in fungal protection. However, despite evidence linking OXDC with increased stress tolerance and studies describing the enzymatic activity, catalytic properties, and spatial and temporal regulation of OXDC, our current understanding of the functional significance of OXDC in scavenging oxalic acid and its metabolic consequences in plants is still limited.
Following these observations and given that we have a long-standing interest in understanding oxalate management and consequence of oxalate down-regulation in plants, we attempted to elucidate the consequences of expressing FvOXDC in tomato fruit on oxalic acid degradation and cellular ion homeostasis. Here, we describe efficient decarboxylation of oxalate and removal of oxalic acid in tomato fruits via fruit-specific transgenic expression of OXDC. We demonstrate that genetic engineering of OXDC in tomato is a potentially useful strategy for oxalate management and modulation of the acid pool, thereby increasing bioavailable minerals and micronutrients. We also present a comparative proteomic analysis that provides insights into the dynamic molecular events and metabolic reprogramming leading to the E8.2-OXDC fruit chemotype. These results are discussed in the context of current theories of the cellular and metabolic cues underlying organic acid accumulation and homeostasis. The differences in protein expression pattern and protein function appeared to encompass diverse metabolic and signaling pathways that we propose contribute to the metabolic remodeling and decreased oxalic acid levels.
RESULTS
Expression and Stable Integration of F. velutipes OXDC in Transgenic Tomato
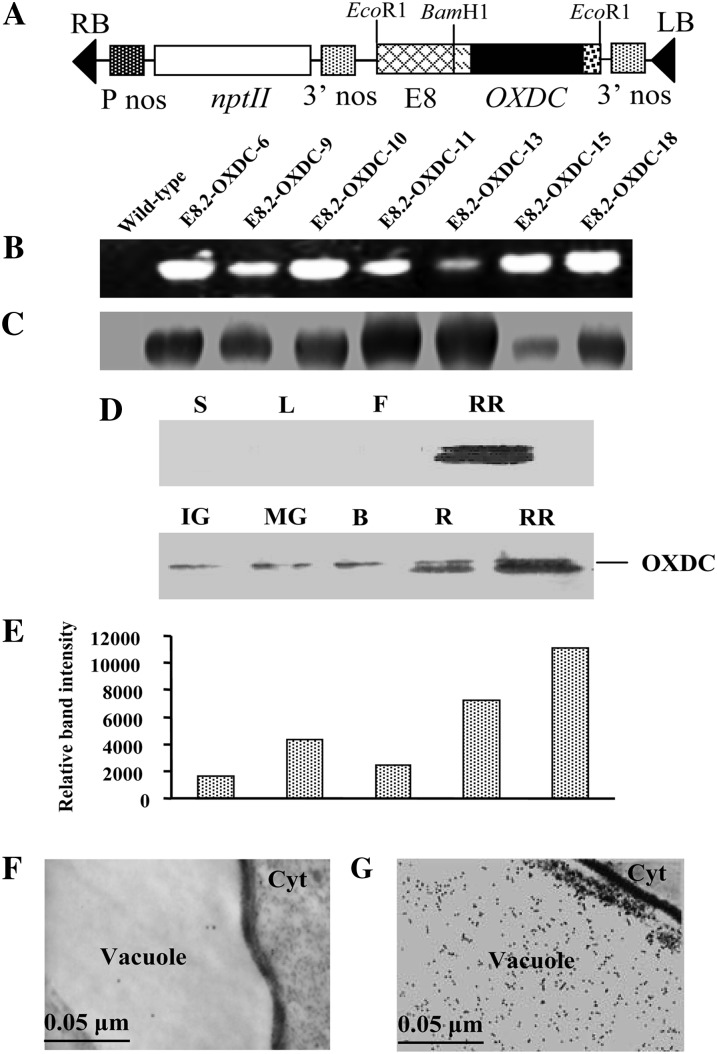
To reduce the oxalic acid content specifically in fruits of transgenic tomato plants without any deleterious developmental effects, a construct was generated to express the FvOXDC gene under the control of a fruit-specific promoter. The pS8.2 expression plasmid contained the OXDC coding sequence, a vacuolar targeting sequence, and a secretion signal under the control of the ethylene-inducible fruit-specific E8 promoter and a kanamycin resistance selectable marker (Fig. 2A). The plasmid was introduced into a commercial tomato cultivar via Agrobacterium tumefaciens-mediated transformation. We developed a robust, genotype-independent, reproducible transformation system using cotyledonary leaves as explants. The regenerated primary transgenic (E8.2-OXDC) plants were selected based on growth on medium containing kanamycin. Because copy number and site of integration greatly influence transgene expression, dozens of primary independent transformants were screened to obtain the desired phenotype. To select successful transformation events, the transgenic plants were analyzed via genomic PCR using a forward primer designed from the secretion signal sequence with a reverse primer based on the vacuolar targeting sequence. A PCR-amplified band of 1.6 kb was observed in all transgenic plants, confirming the intactness of the transgene (Fig. 2B). Real-time PCR analysis using TaqMan chemistry demonstrated the presence of one to three copies of the transgene in PCR-positive transgenic events. All of the kanamycin-selected, PCR-positive, and copy number-detected transgenic events across the population were transferred to an experimental field to generate fruits. E8.2-OXDC homozygous T2 plants were selected for further characterization.
Figure 2.
Molecular analyses of transgenic tomato lines expressing OXDC. A, Schematic representation of OXDC expression plasmid pS8.2, containing the OXDC coding sequence under the control of the E8 promoter. RB, Right border; LB, left border; P nos, nopaline synthase promoter; nptII, neomycin phosphotransferase; 3′ nos, nopaline synthase terminator. B, PCR amplification of genomic DNA from pS8.2-transformed plantlets. C, Immunoblot analysis of OXDC protein expression in ripe fruits from multiple transgenic plants. Numbers are representative of the individual E8.2-OXDC transgenic lines, while the wild type corresponds to an untransformed plant. D, Immunoblot analysis of various plant organs showing the expression of OXDC. L, Leaf; S, stem; F, flower; RR, red ripe fruit. E, Differential expression of OXDC protein during different stages of fruit development. Fruit were from the E8.2-OXDC line. The OXDC immunoreactive band intensities were quantified by densitometry, and the values were plotted against each fruit stage. IG, Immature green; MG, mature green; B, breaker; R, ripe; RR, red ripe. F, Transmission electron micrograph of wild-type fruit vacuole probed with anti-OXDC antibody. G, Transmission electron micrograph showing immunolocalization of OXDC protein in the transgenic fruit vacuole.
Spatiotemporal Expression of OXDC and Its Subcellular Localization and Decarboxylation Activity
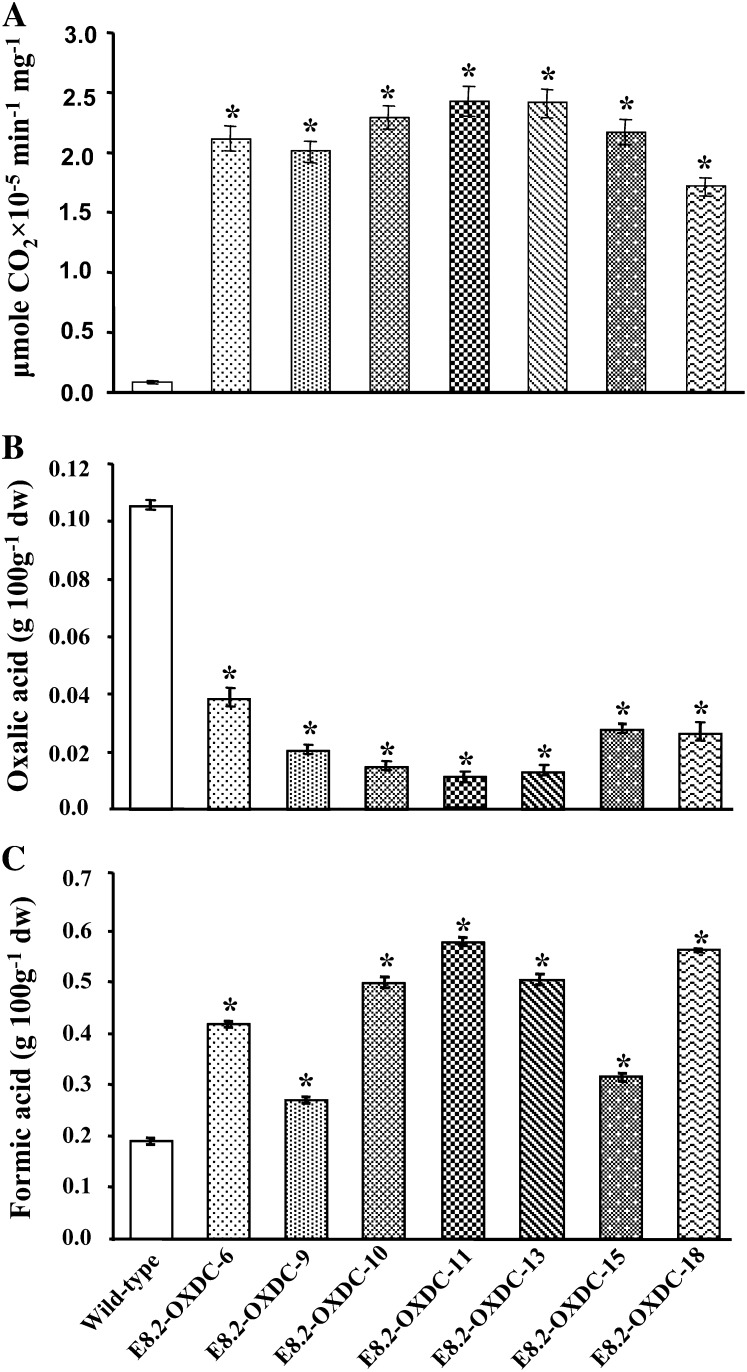
Immunoblot analysis of soluble proteins extracted from the E8.2-OXDC tomato fruits using a polyclonal anti-OXDC antibody showed an immunoreactive band of 55 kD, corresponding to the predicted size of the recombinant FvOXDC (Fig. 2C), confirming that the transgene was producing the desired protein, and to varying degrees among different transgenic lines. As expected, analysis of various plant organs, such as stems, leaves, flowers, and fruits, suggested that OXDC was specifically translated in tomato fruits (Fig. 2D). During fruit development and ripening, OXDC was first detected at low levels in the immature green fruit and peaked in the red ripe stage, with a 6-fold increase in expression being revealed through densitometric analysis (Fig. 2E). This is consistent with the E8.2 promoter being active upon ethylene induction only in ripening fruits. Immunolocalization analysis demonstrated that OXDC was targeted to the cell vacuole in the transgenic fruit, while no signal was detected in the wild-type fruit (Fig. 2, E and F). Furthermore, the expressed enzyme was found to be functionally active with 7- to 10-fold increase in CO2 production in transgenic fruits, as revealed via an in vitro decarboxylation assay. Fruits from E8.2-OXDC-11 showed the highest OXDC activity while those from E8.2-OXDC-18 showed the least, consistent with the reduction in oxalic acid content (Fig. 3A).
Figure 3.
Biochemical analyses of ripe tomato fruits expressing OXDC. A, Decarboxylase activity shown as micromoles of CO2 liberated per minute per milligram of protein at 37°C. B and C, Quantitation of oxalic acid and formic acid in tomato fruits. Data represent the values obtained from the means (± sd) of four plants, with two fruits per plant for each independent transgenic event and the wild type in triplicate represented by error bars. Asterisks indicate significant difference (P < 0.05) analyzed by Wilcoxon rank sum test.
Fruit-Specific Expression of OXDC Results in Decreased Levels of Oxalic Acid and Modulation of the Intracellular Acid Pool
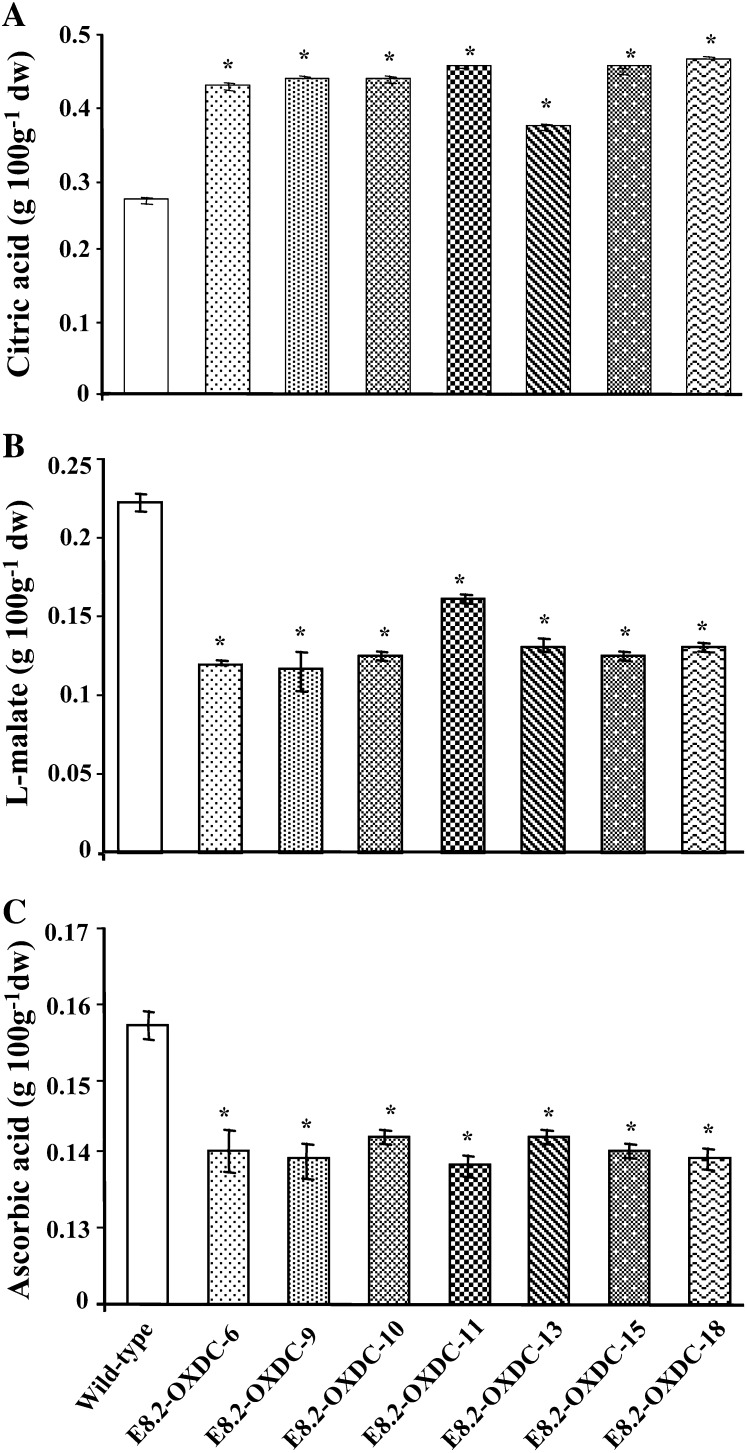
To assess whether transgene expression affected the oxalic acid content, oxalic acid levels were measured in E8.2-OXDC and wild-type fruits. Biochemical analysis revealed that the fruit-specific expression of FvOXDC in transgenic plants was accompanied by a substantial reduction, up to 90% in oxalic acid levels in ripe fruits (Fig. 3B). To investigate whether this decrease in oxalic acid levels led to corresponding changes in the abundance of formic acid, which is the product of oxalate catabolism by OXDC, the formic acid content was measured, and a concomitant increase (30%–67%) was observed (Fig. 3C). We hypothesized that this decrease in levels of oxalic acid could have a direct bearing on the fruit intracellular acid pool, and we detected changes in the levels of citric, malic, and ascorbic acid. Specifically, while the citric acid content was 27% to 42% higher (Fig. 4A), malic acid levels decreased by 24% to 52% (Fig. 4B), along with a 10% to 12% reduction in ascorbic acid (Fig. 4C).
Figure 4.
Organic acid levels in red ripe stage transgenic tomato fruits. A, Citric acid content. B, Malic acid content. C, Ascorbic acid content. Numbers are representative of the individual E8.2-OXDC lines, whereas the wild type corresponds to an untransformed plant. Data represent the values obtained from the means (± sd) of four plants, with two fruits per plant for each independent transgenic event and the wild type in triplicate represented by error bars. Asterisks indicate significant difference (P < 0.05) analyzed by Wilcoxon rank sum test.
Reduction of Oxalic Acid Results in Increased Levels of Some Minerals and Micronutrients
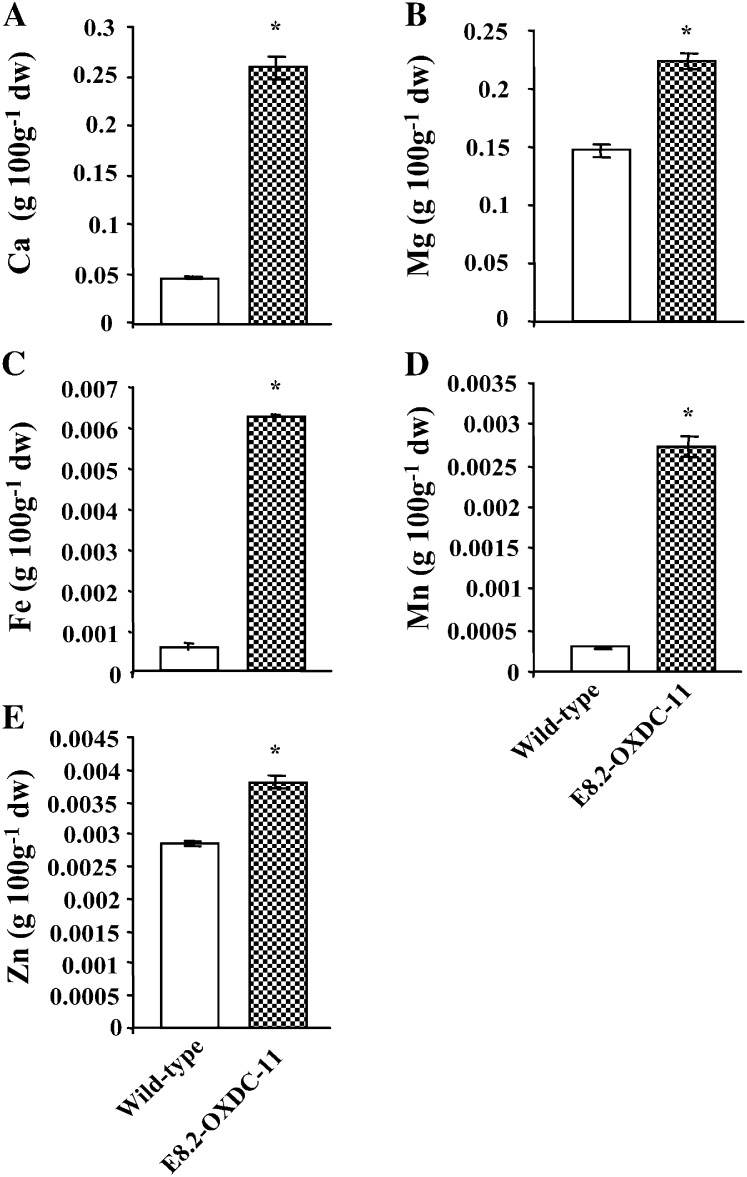
Oxalic acid is a strong chelator of divalent cations, especially calcium. The decrease of oxalic acid in E8.2-OXDC tomato fruits led us to investigate the effect on the inorganic pool of intracellular minerals and micronutrients. Wild-type and transgenic fruits were sampled at the red ripe stage for quantitative analysis, which revealed a marked increase in calcium, magnesium, iron, manganese, and zinc in the E8.2-OXDC fruits (Fig. 5, A–E). For example, calcium levels were increased by more than 5-fold in fruit from the E8.2-OXDC-11 line, while the concentrations of manganese and iron were 9- and 10-fold higher, respectively, than in wild-type fruits. Marginal increases in magnesium and zinc concentrations were also detected in the transgenic fruits. Taken together, these data suggest an increased concentration of minerals and micronutrients in the low-oxalate transgenic tomato.
Figure 5.
Quantification of calcium, other minerals, and micronutrients in red ripe stage fruit from transgenic line E8.2-OXDC-11. Levels of calcium (A), magnesium (B), and micronutrients iron (C), manganese (D), and zinc (E) were determined using atomic absorption spectrophotometry. Data represent the values obtained from the means (± sd) of four plants, two fruits per plant for each independent transgenic event and the wild type in triplicate represented by error bars. Asterisks indicate significant difference (P < 0.05) analyzed by Wilcoxon rank sum test.
Transgenic Plants Display Wild-Type Growth, Developmental Phenotypes, and Physiology
There were no observable differences in phenotype, development, or fertility in any of 24 independent E8.2-OXDC transgenic lines, compared with wild-type plants, either in the primary transgenic populations or in the subsequent generations. In addition, to assess the field performance of the E8.2-OXDC plants, 14 independent transgenic lines were selected for further characterization, transplanted in the experimental field alongside wild-type plants, and randomly replicated three times in two seasons. Despite the high levels of expression of FvOXDC in the transgenic tomato fruits, the plants exhibited a normal phenotype with similar heights and numbers of nodes per main stem. Determination of crop yields was carried out in fully senescent plants. There were no significant changes in the flowering time, vigor, or the total yield observed in any of the E8.2-OXDC plants. Moreover, expression of OXDC did not affect the net photosynthetic rates or other physiological parameters, including stomatal conductance and transpiration rate (data not shown).
Proteomic Changes in Low-Oxalate Transgenic Fruit
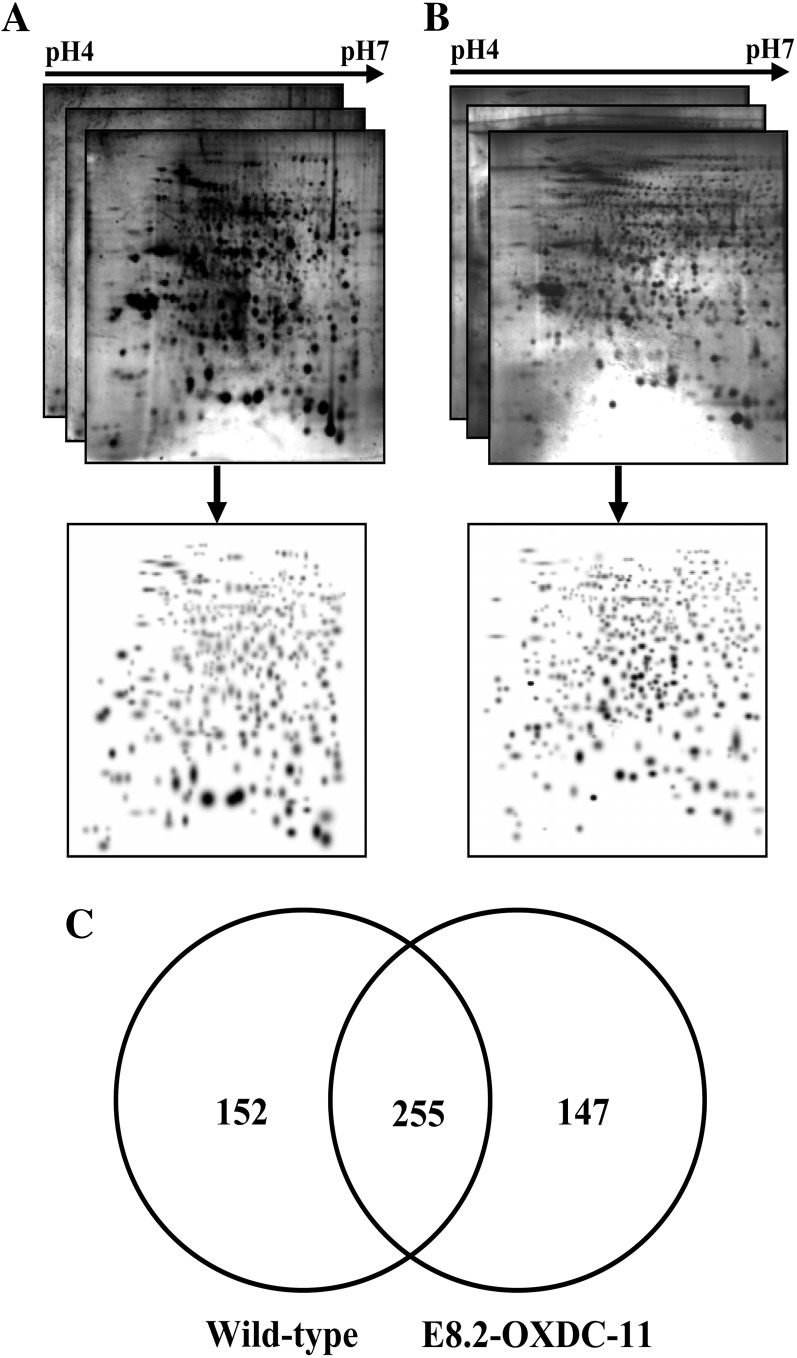
To identify additional processes that might be affected by fruit-specific overexpression of FvOXDC, we conducted a comparative proteomic analysis of E8.2-OXDC and wild-type fruits using two-dimensional gel electrophoresis (2-DE) analysis (Fig. 6, A and B). A flow chart of the experimental design, including the biological replicates used, is summarized in Supplemental Figure S1. Total protein samples were prepared from frozen mature fruit powder (see “Materials and Methods”), and proteome profiles were obtained. The proteome was resolved over a pH range of 4 to 7 and a molecular mass range of 10 to 120 kD. Data were analyzed using gels that showed more than 90% high-quality protein spots with a correlation coefficient of variation (CV) greater than 0.8 and P < 0.05, suggesting high reproducibility among the replicate 2-DE gels (Supplemental Table S1).
Figure 6.
Comparative proteomic analysis of wild-type and E8.2-OXDC tomato fruits. Three replicate 2-DE gels from wild-type (A) and E8.2-OXDC-11 (B) tomato were combined computationally using Bio-Rad PDQuest software version 7.2.0 to generate standard gels. C, Venn diagram showing the specific and overlapping protein spots from wild-type and E8.2-OXDC-11 extracts.
On average, 407 and 402 protein spots were detected in wild-type and E8.2-OXDC fruits, respectively. The degree of spot quantity variation inherent to the 2-DE process was assessed using spots matched to all six gels representing a quality score more than 30. A total of 300 spots (74% of the spots present in all six gels) met these requirements. The CV values ranged from 3% to 129% (Supplemental Fig. S2A). More than 52% of all spots were found to have CVs less than 30%, and more than 82% of the spots had CVs less than 50%. Only 53 spots (17% of the total number) exhibited a CV of greater than 50%. Closer visual inspection of the 53 spots with CVs greater than 50% revealed that they were affected by background, horizontal and vertical streaking, edge effects, and/or neighboring spots and thus were inaccurately quantified. To evaluate potential correlation between the variations in spot quantity and spot position, the average spot quantity CV from spots matched among all gels with a quality score of greater than 30 was related to their positions in the gel. It was apparent that the degree of quantitative variation was evenly distributed in the dimensions of both pI (Supplemental Fig. S2B) and molecular mass (Supplemental Fig. S2C), demonstrating the reproducibility of the 2-DE gels. A total of 255 spots were shared between the wild-type and E8.2-OXDC fruits (Fig. 6C). Changes in protein expression from E8.2-OXDC fruits, compared with wild-type fruits, were far larger, with 43 spots consistently differentially expressed (Supplemental Table S2). Some protein spots showed more than a 5-fold difference in abundance in extracts from OXDC fruits compared with those from the wild type, in addition to a few of the proteins that show only marginal differences. In total, 76 proteins (Supplemental Table S2) were identified through either liquid chromatography-tandem mass spectrometry (LC-MS/MS) or matrix-assisted laser-desorption ionization time-of-flight (MALDI-TOF/TOF) analysis.
Many seemingly well-resolved 2-DE spots were found to contain more than a single protein. In an attempt to minimize comigration of multiple protein species, we also used a relatively narrow pH range (4–7) for isoelectric focusing because the use of narrow-range immobilized pH gradient strips facilitates higher resolution (McGregor and Dunn, 2006; Sghaier-Hammami et al., 2009). However, MALDI-MS/MS is known to identify the most prevalent protein present in a gel spot sample (Jun et al., 2012), and the top-ranked hit resulting from LC-MS/MS analysis has also been shown typically to correspond to the most abundant protein among multiple proteins present in a spot (Yang et al., 2007). The spot intensities of different protein constituents were determined using the protein abundance index and the exponentially modified protein abundance index (emPAI; Supplemental Table S3), which have been routinely applied in proteomics workflows (Perkins et al., 1999; Ishihama et al., 2005; Zhang et al., 2006; Yang et al., 2007; Cilia et al., 2011; Taylor et al., 2011). In this study, if more than one protein was identified in a spot, the relative abundance of each protein was determined by calculating the emPAI from the MS/MS data, and the assumption was made that the most abundant protein would account for the observed regulation (Ishihama et al., 2005).
Taken together, the effects of comigrating proteins on the protein expression ratios observed in 2-DE analyses were deemed negligible. In cases where more than one protein was indicated with a significant score for the MS/MS-derived peptide sequence, the match was considered in terms of the highest ranked hit, molecular mass matches, and emPAI, an approach that has been routinely used for proteomics studies (Perkins et al., 1999; Poetsch and Wolters, 2008; Yoon et al., 2009). The details of the MS/MS analyses, including the protein identification, score of the identified protein, threshold score, number of matched peptides, percentage of sequence coverage, Sol Genomics Network (SGN) database accession number, theoretical and experimental Mr, and pI values for each protein, peptide sequence, and corresponding peptide score are provided in Supplemental Table S4. Any protein identified with a single peptide and the respective MS/MS spectrum is provided in Supplemental Table S5. In a number of cases, we identified the same protein in multiple spots. This observation is common in 2-DE studies for several reasons, such as proteolytic processing, the existence of multiple isoforms, and posttranslational modification(s) leading to changes in the pI, Mr, or both for identical proteins. If the same protein was identified in multiple spots that contained several shared peptides, a differential accumulation pattern was observed for each of the protein species, such as for alcohol dehydrogenase2 (Slt 797, 801, 904, and 1149), chaperone DnaK (Slt 5, 156, 162, and 321), and 14-3-3 proteins (Slt 41, 116, 117, 120, and 131). In such cases, these are listed as independent entities.
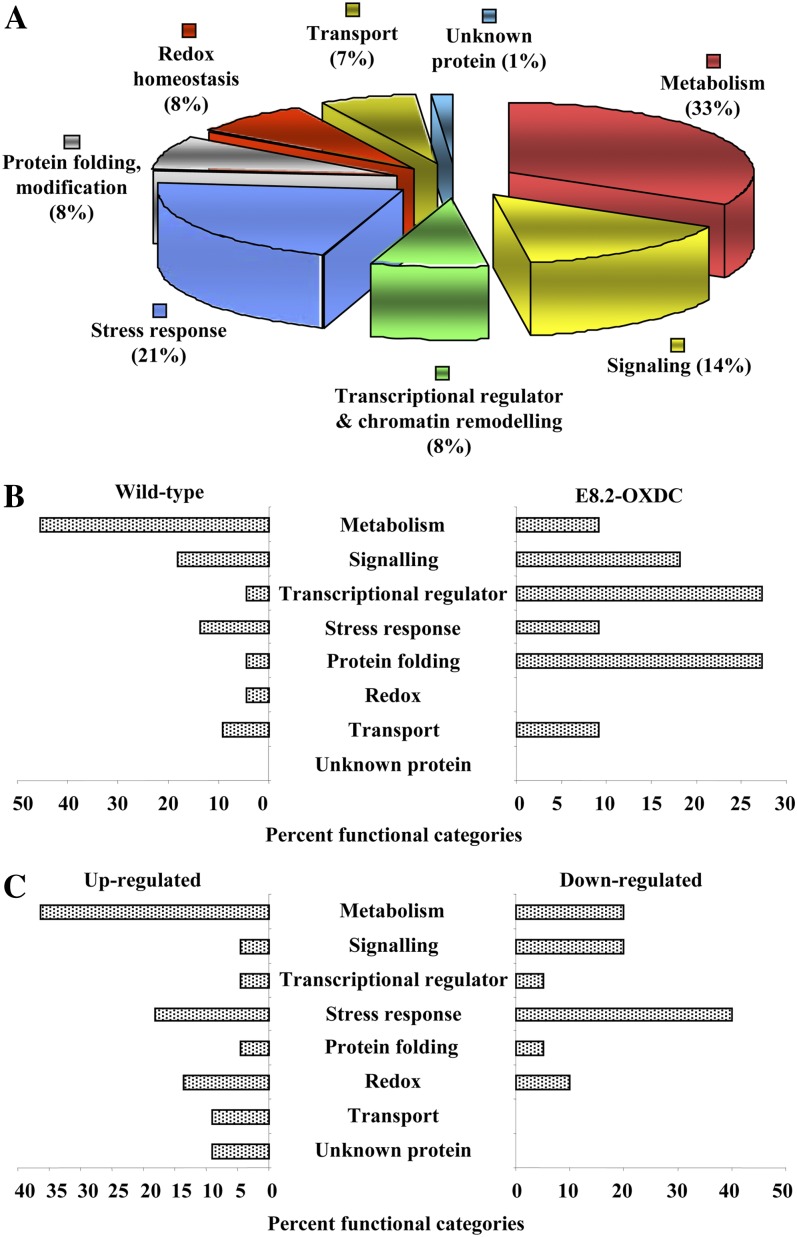
To gain a more comprehensive understanding of the changes in protein expression in the fruits as a consequence of expression of the transgene, differentially expressed proteins were grouped into seven functional categories (Fig. 7A; Supplemental Table S2), with one group containing proteins of unknown function and proteins that could not be assigned to any other category, accounting for approximately 1% of the proteins. Of the identified proteins, 32% were implicated in metabolism, with stress responses accounting for the second largest category (21%). Significant fractions corresponded to functions involved in signaling (14%) and transcription regulators (7%). Another category included proteins involved in protein folding (7%), and a further 6% were related with transport (6%), such as well-known molecular transporters or cargo protein carriers. A total of 7% of the proteins were involved in redox functions associated with maintaining cellular homeostasis. Further details and features of the identified proteins are described in Supplemental Table S2. Twenty-two spots were absent from E8.2-OXDC proteins, whereas 11 spots were present exclusively in the corresponding gels. In addition, 23 spots were clearly up-regulated in E8.2-OXDC, compared with wild-type fruits, while 20 spots showed down-regulation (Fig. 7, B and C).
Figure 7.
Functional assignment of protein identified from wild-type and E8.2-OXDC-11 fruits. A, The proteins were assigned a putative function using Pfam and InterPro databases and functionally categorized as represented in the pie chart. B, Bars illustrate the percentage of protein spots within various categories based on the total numbers identified from wild-type and E8.2-OXDC-11 fruit. C, Bars represent the percentage of up-regulated and down-regulated proteins within various functional categories in E8.2-OXDC-11 fruit.
To determine the putative functions of the unknown protein, the protein sequences were subjected to functional domain analysis using the InterPro database and queried for domains in the Simple Modular Architecture Research Tool, Panther, and Pfam databases. These analyses identified conserved domains in the proteins, some of which corresponded to known activities, as reported in Supplemental Table S6. A few of the proteins showed discrepancies with their theoretical Mr or pI values. In general, the apparent Mr value predicted by SDS-PAGE has been reported to exhibit an error deviation of approximately 10% compared with the theoretical value (Liu et al., 2007). From our data, among the 76 identified proteins, a set of 17 were found to show observed Mr values that were smaller than the theoretical values (Supplemental Table S2), suggesting that these proteins might represent degradation products. Of these, seven belonged to the metabolism category, one was a transcriptional regulator, three were associated with transport, and two each were assigned to the stress response, protein folding, and redox homeostasis categories.
Comparative Proteome Analyses Show Metabolic Remodeling in E8.2-OXDC Tomato
To examine the impact of OXDC in decreasing oxalic acid contents, changes in the protein profiles were analyzed, suggesting functionally diverse proteins at the interface of several cellular pathways that may be related to metabolic remodeling. For example, identification of some of the proteins present in either E8.2-OXDC or wild-type fruits provided a plausible mechanism for OXDC-mediated biochemical changes. These included proteins belonging to the transcriptional regulator and chromatin remodeling categories, namely γ-glutamyl transferase-like protein (Slt 20), elongation factor-like protein (Slt 49), and translation elongation factor-Tu precursor (Slt 927), which were present only in E8.2-OXDC. This finding might indicate the existence of a compensatory response to adjust the expression of oxalic acid-metabolizing proteins to oxalic acid levels. One of the identified protein spots present exclusively in E8.2-OXDC 2-DE gels was a variant of class I heat shock protein (Slt 265) belonging to the stress response category that might contribute to protein homeostasis. Importin subunit β (Slt 70) from the transport category, another protein only detected in in E8.2-OXDC extracts, is known to function at the interface of 14-3-3 proteins (Slt 41, 116, 117, 120, and 131) and chaperone proteins (Slt 239 and 685) to modulate the nuclear trafficking of EF-Tu (Harvey, 2009). To identify proteins that might be involved in mediating the regulatory changes observed in E8.2-OXDC fruits, we examined the set of proteins present in the wild type and absent from E8.2-OXDC. Malate dehydrogenase (Slt 791), alcohol dehydrogenase2 (Slt 797), and NAD-dependent malic enzyme2 (Slt 326) in the metabolic category, V-type ATP synthase β chain (Slt 53 and 272) from the transport category, and GTP-binding nuclear protein Ran-A1 (Slt 1031) from the signaling category were only detected in the wild-type fruits.
The protein spots up-regulated in E8.2-OXDC fruits included, as expected, a large percentage (36%) presumably associated with primary metabolism, suggesting associations between core metabolic pathways and oxalic acid degradation. Of the nine features in this category, four, glyceraldehyde-3-P dehydrogenase (Slt 907, 1070, and 1148) and alcohol dehydrogenase (Slt 801), are predicted to be glycolytic, suggesting primary metabolism of carbohydrates as a positive regulator of OXDC-regulated processes in E8.2-OXDC fruits, while adenosylhomocysteinase (Slt 323) plays a role in amino acid metabolism (Textor et al., 2004). Inspection of the energy metabolism and redox homeostasis category revealed the involvement of cell wall-localized proteins in oxalic acid degradation, specifically, monodehydroascorbate reductase (Slt 816) and peroxiredoxin (Slt 73 and 106), both of which were up-regulated.
Functional categories in which there was a higher percentage of down-regulated than up-regulated proteins in E8.2-OXDC fruits were more diverse. A total of 14 spots (56%) involved in metabolism were found to be down-regulated or absent in E8.2-OXDC fruits, represented by NAD-dependent malic enzyme2 (Slt 326), malate dehydrogenase (Slt 791), and adenosylhomocysteinase (Slt 534), which are known to affect the electron transport chain (Zhou et al., 2011). A total of 50% of the proteins in the stress response category were down-regulated, comprising a class II heat shock protein (Slt 969) and heat shock protein 90 (Slt 163). Another set of down-regulated protein spots associated with signaling (36%) included members of the 14-3-3 proteins (Slt 41, 117, and 120) and actin (Slt 312). Three of the categories mentioned above specifically control highly connected nodes in the pathways regulating processes as diverse as metabolic remodeling. Notably, 33% of the proteins involved in redox homeostasis, such as thioredoxin/protein disulfide isomerase (Slt 304 and 305) were also down-regulated. This protein is known to play a role in modulating chaperone activity (Saccoccia et al., 2012). No proteins belonging to the transport category were down-regulated.
DISCUSSION
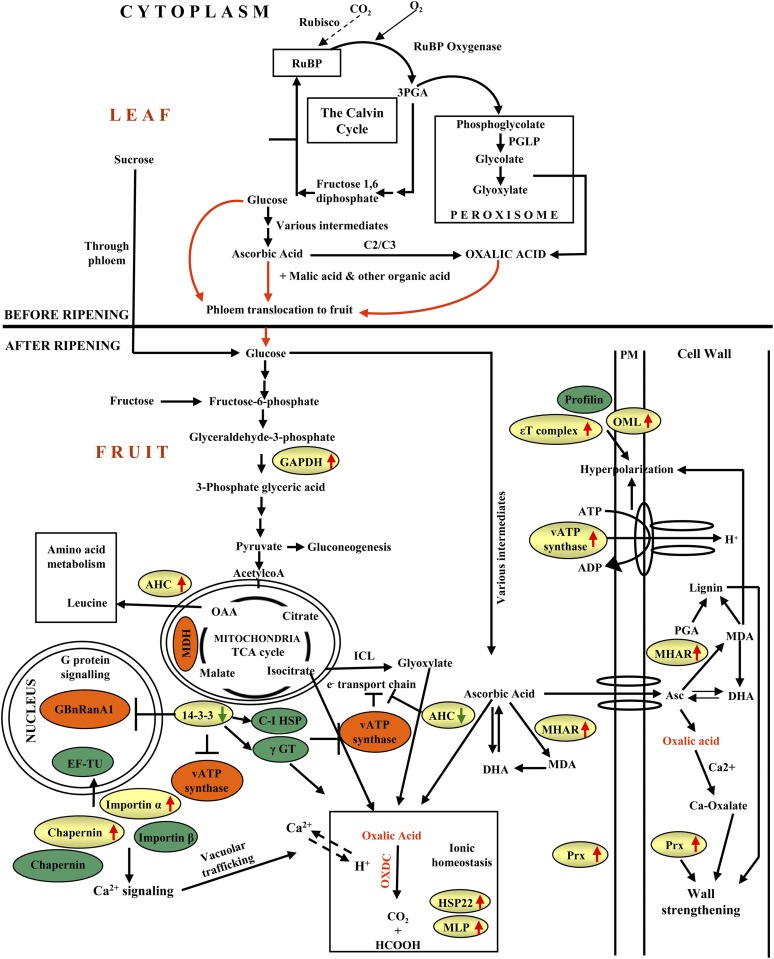
While there has been considerable success in enhancing the nutritional value of food crops in terms of macronutrients (protein, oils, and carbohydrates) and efforts to increase the abundance of micronutrients, vitamins, and antioxidants, there has been less progress in the management of antinutrients and the bioavailability of micronutrients and minerals. Furthermore, the consequence of such modulation by metabolic engineering on the cellular metabolism is poorly understood. In this study, we performed molecular, physiological, and proteomic analyses of transgenic tomato expressing FvOXDC to reduce oxalic acid contents in a fruit-specific manner. Our results provide evidence that OXDC levels have a substantial effect on the metabolic status of tomato fruits and reveal a strategy for degradation of an antinutrient with parallel enhancement of micronutrients and health-promoting acids, with essentially no negative collateral effects on crop yields by expressing a single transgene. We also investigated the regulatory and functional protein network operating in response to reducing oxalate levels by the action of OXDC, which resulted in metabolic remodeling. A model representing the functional network of OXDC-responsive proteins involved in metabolic remodeling is depicted in Figure 8.
Figure 8.
A model summarizing the cellular pathways associated with OXDC overexpression. Green circles represent proteins exclusively detected from E8.2-OXDC extracts and orange circles indicate proteins apparently absent in E8.2-OXDC, while differentially expressed proteins in E8.2-OXDC fruit identified in this study are shown in the yellow-colored boxes. Red arrows indicate up-regulation, while green arrows represent down-regulation. GAPDH, Glyceraldehyde-3-P dehydrogenase; MDH, malate dehydrogenase; Asc, ascorbic acid; MHAR, monodehydroascorbate reductase; DHA, dehydroascorbic acid; MDA, monodehydroascorbic acid; PM, plasma membrane; OML, outer membrane lipoprotein; MLP, major latex protein; AHC, adenosylhomocysteinase; γGT, γ-glutamyl transferase-like protein; Prx, peroxiredoxin; C-I HSP, class I heat shock protein; GBnRanA1, GTP-binding nuclear protein Ran-A1; PGA, polygalacturonic acid.
Balancing Calcium and Associated Processes
The fruit-specific expression of OXDC in tomato led to a significant reduction in oxalate levels and a concomitant substantial increase in calcium. The decrease in oxalic acid might reduce the content of calcium oxalate, possibly by limiting the availability of oxalic acid for chelating calcium. Most analytical studies to date that have addressed the occurrence and distribution of oxalates in land plants have focused on their possible calcium-sequestering influence in the human diet (Nakata, 2003; Nishizawa, 2006; Nakata and McConn, 2007). Calcium in its ionic form, Ca2+, performs critical functions in metabolism (White and Broadley, 2003), and Ca2+ deficiency is the most common nutritional problem affecting tomato, the second largest vegetable crop worldwide (Park et al., 2005).
Calcium is required for the activation and/or stabilization of certain enzymes; for example, plant cells require calcium to release peroxidases, which are involved in the control of cell elongation because they can rigidify walls through their cross-linking activity and their ability to participate in the formation of lignin. Peroxiredoxin, an antioxidant that detoxifies H2O2 (Mika et al., 2010) along with a major stress protein, Heat shock protein22 (Slt 183), might maintain the ionic homeostasis in E8.2-OXDC tomato. This might represent an adaptive mechanism in E8.2-OXDC tomato to protect the mitochondria from oxidative damage. Furthermore, it can be presumed that while oxalic acid is being continually catabolized, free calcium is transported to the cytoplasm from the vacuole by tonoplast antiporters. It is also possible that plants may have taken up more calcium and other minerals from the soil due to the high turnover of oxalic acid.
Among the other micronutrients tested, there were also increases in manganese, magnesium, and zinc in the transgenic tomato fruits along with a substantial increase in iron (Fig. 5, A–E). Activation of Cation exchanger1, the Ca2+/H+ antiporter, leads to increases in divalent cations, such as iron, zinc, magnesium, and manganese (Park et al., 2005). It is therefore conceivable that a similar regulatory mechanism might lead to the increased accumulation of these minerals.
Engineering of OXDC Balances Organic Acid by Affecting Carbon Utilization and Carbohydrate Metabolism
The metabolic status of tomato fruits is dependent to a large extent on the intermediates of the photosynthetic carbon reduction/oxidation (PCR/PCO) cycle (Liu et al., 2003). Long-distance phloem translocation of organic acids, such as malic acid, citric acid, and oxalic acid, provides a supply of metabolites before ripening; while after ripening, the metabolic pool is maintained through a feedback mechanism associated with various cellular pathways (Carrari et al., 2006; van der Merwe et al., 2009; Osorio et al., 2011). The proteomic analysis of E8.2-OXDC fruits provided insights into an effective mechanism for continual degradation of oxalic acid and maintenance of the metabolite pool, possibly through the combinatorial action of OXDC-responsive proteins. In addition, the organic acid pools (primarily citrate and malate levels) might play a significant role in decreasing oxalic acid contents.
To relate the information obtained from the analysis of organic acid levels to metabolic changes, we next assessed the processes that might regulate organic acids by affecting carbohydrate metabolism. Our analysis revealed increased levels of citric acid and a marginal decrease in malic acid contents in E8.2-OXDC tomato. The increase in citric acid, a central metabolic hub, is important because the citric acid cycle contributes to the formation of important biochemical intermediates in addition to energy-rich molecules, such as NADPH and flavin adenine dinucleotide. Moreover, increasing evidence suggests that citric acid is one of the dietary substances that promote the bioavailability of the micronutrients iron and zinc (Welch, 1995, 2002). Thus, it is possible that the increase in citrate levels observed in E8.2-OXDC fruit would promote the absorption of iron and zinc.
Glyceraldehyde-3-P dehydrogenase (Slt 907, 1070, and 1148), a key intermediate in the PCR/PCO cycle, is a primary enzyme in the glycolytic pathway that is known to synthesize the two-carbon compounds glycolate and glyoxylate, which ultimately lead to the production of oxalic acid. We hypothesize that up-regulation of glyceraldehyde-3-P dehydrogenase might redirect the pathway toward gluconeogenesis, the tricarboxylic acid (TCA) cycle, and amino acid metabolism in E8.2-OXDC fruits. A pool of hexose coordinately regulates various organic acids, including ascorbate, a precursor of oxalic acid in the fruit (Loewus, 1999). Phloem translocation of l-ascorbic acid thus appears to be crucial for its supply to sink organs. Ascorbic acid largely provides the substrate for oxalic acid biosynthesis in plants and is used for calcium oxalate formation (Horner et al., 2000). In this study, the concentration of endogenous ascorbic acid (14 mg 100 g–1 fresh weight) detected in E8.2-OXDC fruit, even after a 10% to 12% reduction, was within the normal range reported for ripe tomato fruits (Zhang et al., 2011; Cronje et al., 2012) and so is not expected to have any negative nutritional effect in the E8.2-OXDC fruit. Rather, the decrease in ascorbic acid levels may reflect its increased conversion to oxalic acid to compensate for the reduction in oxalate levels following OXDC expression. This is congruent with the observation that the Medicago truncatula calcium oxalate defective4 mutants have lower ascorbate levels due to the utilization of more oxalate for the formation of oxalate crystals (Macrae, 1971).
Enzymes involved in ascorbate metabolism, including monodehydroascorbate reductase, functioning in both the cytosol and cell wall, have been suggested to play a dual role in the production of oxalic acid and cell expansion/tightening (Smirnoff, 1996). Moreover, ascorbic acid metabolism and cross-linking of the cell wall with calcium is known to influence lignin biosynthesis (Delless et al., 2011). It is plausible that the transport of ascorbic acid into the wall via a carrier may promote the enzymatic oxidation of this organic acid by monodehydroascorbate reductase to form monodehydroascorbate, which might stimulate membrane depolarization in the presence of V-type ATP synthase β chain (Slt 146). Depolarized membrane in conjunction with profilin (Slt 76) and outer membrane lipoprotein (Slt 601) and T-complex protein η (Slt 1123) promote cell wall strengthening (Smirnoff, 1996). These proteins were found to be either exclusively present or up-regulated in E8.2-OXDC protein extracts, indicating that they might affect wall architecture in transgenic fruits. Moreover, the hydrophobic outer membrane lipoprotein, a key enzyme in the xanthophyll cycle that plays a role in fruit metabolism, was highly expressed in E8.2-OXDC tomato. Amino acid metabolism through the isocitrate-malate pathway is well characterized (Textor et al., 2004). In E8.2-OXDC fruits, the expression of adenosylhomocysteinase, one of the isocitrate-malate pathway enzymes known to produce Leu (Textor et al., 2004), may shift the intermediates toward amino acid metabolism. V-type ATP synthase β chain (Slt 53 and 272) also appeared to be absent in E8.2-OXDC fruits, indicating that TCA cycle and electron transport chain proteins are affected, whereas the abundance of glycolytic proteins, such as glyceraldehyde-3-P dehydrogenase, appears to be induced due to the expression of OXDC.
Taken together, these data indicate that proteins associated with the electron transport chain and the TCA cycle are down-regulated in E8.2-OXDC fruits, while glycolytic proteins are induced. In addition, our data suggest that proteins associated with oxalic acid degradation are induced in E8.2-OXDC fruits.
Proteomic Changes in E8.2-OXDC Fruit Are Indicative of OXDC-Mediated G-Protein and Ca2+ Signaling
The comparative proteome data indicated that OXDC-mediated regulation is influenced by various signaling pathways, such as G-protein signaling and Ca2+ signaling. A previous study provided strong evidence that 14-3-3 isoforms require Ca2+ for the activation of various kinases to phosphorylate many important enzymes (Rutschmann et al., 2002). Furthermore, the interplay between 14-3-3 proteins, Ca2+, Ca2+-binding ligands, and nuclear import proteins such as importin α/β (Slt 70 and 238), as well as the GTP-binding nuclear protein Ran-A1, has been postulated to affect G-protein signaling in animals (Agassandian et al., 2010). This highlights the complexity involved in the regulation of nuclear trafficking and ion channels in cells. We found reduced expression of 14-3-3 proteins, which likely adversely affected G-protein signaling, leading to the observed reduction in the integrator protein Ran-A1. The absence of Ran-A1 compels importin α/β and chaperonins (Slt 239 and 685) to transport EF-Tu from the cytosol to the nucleus (Agassandian et al., 2010), which might promote protein synthesis by adjusting the expression of oxalic acid-metabolizing proteins to maintain oxalic acid levels through a compensatory response. Up-regulation of importin α/β and chaperonins due to the down-regulation of 14-3-3 proteins might lead to Ca2+ signaling to regulate vacuolar trafficking. This highly regulated interplay could fulfill fundamental roles in maintaining the OXDC-mediated responses by modulating G-protein and Ca2+ signaling.
Furthermore, 14-3-3 proteins interact with various chaperones, such as chaperonins, class I heat shock protein (Slt 265), and γ-glutamyl transferase-like protein to promote vacuolar trafficking. This regulation of nuclear and vacuolar trafficking most likely reflects the effect of OXDC on the biosynthesis of enzymes that produce the intermediates of the PCR/PCO cycle and the time required to accumulate the necessary levels of PCR/PCO cycle intermediates. This hypothesis is in accord with our data showing that 14-3-3 proteins, the GTP-binding nuclear protein Ran-A1, thioredoxin/protein disulfide isomerase, and phosphomanomutase2 (Slt 768) exhibited either reduced expression or no expression in E8.2-OXDC, reflecting the fact that vacuolar trafficking, rather than nuclear trafficking, predominates in E8.2-OXDC fruits. Our findings also point toward feedback activation or repression of vacuolar trafficking. The aforementioned interplay activates this trafficking, whereas inhibition of V-ATP synthase (Slt 53 and 272) represses the trafficking. This mechanism maintains ion homeostasis in the vacuole and may regulate OXDC-responsive processes.
The data presented here reveal a spectrum of OXDC-regulated proteins and pathways previously unknown as OXDC targets. We also propose a working model of OXDC-mediated regulation involving metabolic pathways and cross talk between various cellular processes in feedback-dependent activation or repression of vacuolar trafficking and cellular homeostasis.
MATERIALS AND METHODS
Construction of the OXDC Transformation Vector
To overexpress OXDC specifically in tomato (Solanum lycopersicum) fruits, the coding sequence of OXDC was cloned under the regulation of the ethylene-inducible fruit-specific promoter E8, producing the plasmid pS8.2. pS8.2 was constructed by replacing the Cauliflower mosaic virus 35S promoter of pSOVA (Kesarwani et al., 2000) with the E8 promoter (Deikman and Fischer, 1988; Davuluri et al., 2005). The EcoRI-filled in/BamHI-digested fragment of pE8.2 containing the E8 promoter was ligated to the HindIII-filled in/BamHI-digested backbone of pSOVA to yield pS8.2. In this construct, OXDC was fused to a 50-bp vacuolar targeting sequence from the tobacco (Nicotiana tabacum) chitinase gene at the 3′-terminus and a secretion signal at the 5′-terminus (Neuhaus et al., 1991). pS8.2 was mobilized into Agrobacterium tumefaciens strain EHA105 using the helper strain HB101::pRK2013 via the triparental mating technique (Van Haute et al., 1983).
Transformation and Selection of Transgenic Events
Seeds of tomato ‘Pusa Ruby’ were grown in vitro in Murashige and Skoog (MS) basal medium (Sigma) containing 3.0% (w/v) Suc and 0.6% (w/v) agar at 23° ± 2°C under a 16-h/8-h photoperiod. The cotyledonary leaves of 2-to-3-week-old germinated tomato seedlings were transformed using A. tumefaciens containing the pS8.2 plasmid. The explants were incubated for 30 min in a 0.7 to 0.9 optical density mL–1 A. tumefaciens culture, resuspended in MS liquid medium, soaked on sterile Whatman paper, and cocultivated for 48 h. The explants were then transferred to MS medium containing 250 mg L–1 cefotaxime for 1 week, followed by being transferred into shoot induction medium (MS basal medium with 3.0% [w/v] Suc, 0.8% [w/v] agar, and 1 mg L–1 zeatin) containing 250 mg L–1 cefotaxime and 50 mg L–1 kanamycin. Kanamycin-selected shoots were transferred to rooting medium (MS basal medium with 3.0% [w/v] Suc, 0.6% [w/v] agar, 0.1 mg L–1 indole-3-acetic acid, 250 mg L–1 cefotaxime, and 50 mg L–1 kanamycin). The kanamycin-selected transformed plants were maintained in the growth room before being transferred to the experimental field.
Nucleic Acid Isolation and Analysis
Genomic DNA was extracted from leaf tissue (100 mg) using the DNeasy Plant Mini Kit (Qiagen). The intactness of the OXDC gene in the E8.2-OXDC plants was determined via PCR using the forward (5′-CGTTTGCATTTCACCAG-3′) and reverse (5′-CGATTATAGTCGTGATCCC-3′) primers, respectively. The OXDC gene copy number in E8.2-OXDC plants was estimated via real-time PCR using TaqMan chemistry with a 7500 Real-Time PCR System (Applied Biosystems), according to the manufacturer’s protocol.
Plant Material and Experimental Design
Wild-type and E8.2-OXDC tomato plants were grown in an experimental field and equivalent-sized red ripe fruits were harvested at maturity. Fruits were collected from three randomized plots and pooled to normalize the effect of variations in the biological replicates. Each biological replicate consisted of nine fruits at the same ripening stage obtained from three different plants (Supplemental Fig. S1). Fruits were immediately frozen in liquid nitrogen, ground to a fine powder, and stored at −80°C until being used for proteomic analysis.
For biochemical and molecular analyses, fruits were harvested at red ripe stage from the wild type and individual E8.2-OXDC transgenic lines. The samples were either used immediately after harvest or frozen in liquid nitrogen and stored at −80°C until analyses. The experiments were performed in at least three biological replicates. Each replicate consisted of a pool of six fruits per genotype.
Expression and Immunodetection of OXDC
Soluble proteins were extracted from 10 mg of lyophilized fruit tissues in 0.25 mL of extraction buffer (50 mm Tris-HCl, pH 7.8, 0.25 m NaCl, 1 mm phenylmethylsulfonyl fluoride, and 1 mm EDTA). The suspension was homogenized for three cycles of 30 s each using a PRO200 homogenizer (PRO Scientific). The homogenate was then centrifuged at 12,000g for 10 min, and the protein concentration in the supernatant was determined using the Bradford protein assay kit (Bio-Rad). An equal amount of protein (50 µg) from each sample was fractionated on a 12.5% (w/v) SDS-PAGE gel and immobilized onto a Hybond-C membrane (GE Biosciences). OXDC was detected with a rabbit polyclonal anti-OXDC antibody (Mehta and Datta, 1991) in combination with alkaline phosphatase-conjugated goat anti-rabbit IgG, using nitroblue tetrazolium 5-bromo-4-chloro-3-indolyl phosphate reaction. Representative blots are shown in Figure 2. OXDC protein quantification was performed with a Fluor-S MultiImager (Bio-Rad) and Quantity 1-D Analysis software (Bio-Rad) using the volume analysis function, and the relative signal strengths were calculated.
Decarboxylation Assay
The decarboxylation activity of OXDC was determined in 1 mL of reaction mixture containing [14C]oxalic acid (GE Biosciences) as a substrate using a method described previously (Mehta and Datta, 1991). An aliquot of 100 µg of soluble protein from each biological replicate, as noted above in “Plant Material and Experimental Design,” was used for the decarboxylation assay. Each sample was analyzed in triplicate, and the mean value was calculated. One unit of decarboxylase activity is equivalent to 1 μmol m–1 [14C]CO2 produced at 37°C. The activity data were expressed as the means ± sd of at least three independent experiments.
Organ-Specific Expression of OXDC
The organ-specific and fruit stage-specific expression patterns of OXDC were determined by western-blot analysis. Aliquots of 100 µg of protein extracted from flowers, leaves, stems, and red ripe fruits, together with the same amounts of protein from five different stages during fruit development, were fractionated on 12.5% (w/v) SDS-PAGE gels. These proteins were then subjected to immunoblot analysis using an anti-OXDC antibody as described previously (Mehta and Datta, 1991).
Immunolocalization Analysis
Immunolocalization of OXDC in fruit was performed as described in Jones et al. (1997) and Chakraborty et al. (2000). Fruit pericarp was fixed in 2% (w/v) glutaraldehyde in 0.1 m phosphate-buffered saline (PBS; pH 7.2) dehydrated in a graded ethanol-water series (70%–100%, v/v) and embedded in London Resin White (hard). Thin sections (60–90 nm) lifted on 400-mesh nickel grids were blocked with 1% (w/v) bovine serum albumin in PBS for 2 h at room temperature. Immunolabeling was conducted with an anti-OXDC antibody diluted 1:100 in PBS containing 1% (w/v) bovine serum albumin and 0.02% (w/v) sodium azide overnight at 4°C. After washing, the grids were incubated with 1:100 (v/v) 10 nm colloidal-gold-conjugated goat anti-rabbit IgG (Auroprobe EM, GAR 10, Amersham Biosciences) containing 0.5% (v/v) Tween 20 for 1 h at room temperature. The grids were postfixed in 2.5% (w/v) glutaraldehyde in 0.1 m PBS (pH 7.2) for 10 min and stained with 2% (w/v) uranyl acetate and lead citrate at room temperature. Electron micrographs were obtained using a Philips CM10 transmission electron microscope.
Extraction and Determination of Oxalic Acid and Formic Acid
Oxalic acid and formic acid contents were determined using an analysis kit according to a procedure recommended by the manufacturer (Boehringer Mannheim/R-Biopharm). Each sample was crushed to a fine powder using a mortar and pestle and suspended in 300 μL of Milli Q water. The suspension was homogenized using a PRO 200 homogenizer for five cycles of 30 s each. The pH of the homogenates was adjusted according to the manufacturer’s instructions. The homogenate was then centrifuged at 12,000g for 15 min to recover the supernatant. The activity was assayed using 100 µL of the supernatant in triplicates, and the absorbance was read at 340 nm.
Analysis of Ascorbic Acid, Citric Acid, and l-Malic Acid
The contents of ascorbic acid, citric acid, and malic acid were determined using analysis kits (l-ascorbic acid [product code 10 409 677 035], citric acid [product code 10 139 076 035], and l-malic acid [product code 10 139 068 035], respectively) according to a procedure recommended by the manufacturer (Boehringer Mannheim/R-Biopharm). The samples used for these assays were prepared as described above. The assays were performed using a 100 µL of tomato extract in triplicates, and the absorbance was read at 340 nm for citric acid and malic acid and at 540 nm for ascorbic acid.
Determination of Calcium and Other Micronutrients
The concentrations of calcium, magnesium, and other micronutrients (iron, manganese, and zinc) were analyzed by subjecting the lyophilized samples to graphite chamber atomic absorption spectrophotometry (Varian AA-880 with GTA-100 atomizer). The concentration of each metal ion was determined using 10 mg of dried fruit tissue with reference to previously constructed standard curves for each of the cations tested.
Morphological Characterization and Photosynthetic Activity
The field experiment was laid out in a randomized block design with three replicates. Plant height and the number of nodes per main stem were measured at 65 d after planting, and photosynthetic rates were quantified with a GFS3000 portable photosynthesis measurement system (Waltz). The photosynthetic capacity of the plants on the basis of single-leaf measurements conducted on three different leaves in each plant was recorded under standard atmospheric (360 μL L–1 CO2) and light conditions (750 μmol m–2 s–1). Relative humidity was maintained at 70%, and leaf temperature was set at 25°C in the leaf chamber. The leaves were held in the chamber for 2 to 3 min until the photosynthetic rates reached steady-state levels.
Fruit Protein Isolation and 2-DE
Briefly, 2.5 g of frozen and powdered fruit (whole fruit) was homogenized in three volumes of extraction buffer (700 mm Suc, 500 mm Tris-HCl, pH 7.5, 100 mm potassium chloride, 50 mm EDTA, 2% [v/v] β-mercaptoethanol, and 1 mm phenylmethanesulfonyl fluoride) by vortexing for 15 min on ice. Total proteins were recovered by the addition of an equal volume of phenol-saturated Tris-HCl (pH 7.5). The upper phenol phase was removed, and the protein was extracted twice with the same extraction buffer. The mixture was vortexed for 10 min and then centrifuged at 10,000g at 4°C, and the soluble proteins were recovered as the supernatant. The proteins were then precipitated by the addition of five volume of 100 mm ammonium acetate in methanol overnight at –20°C. The precipitated proteins were pelleted by centrifugation at 10,000g for 30 min, and the pellets were washed once with ice-cold methanol and three times with ice-cold acetone, air-dried, and resuspended in 2-D rehydration buffer (9 m urea, 2 m thiourea, 4% [w/v] CHAPS, 20 mm dithiothreitol, 0.5% [v/v] Pharmalyte [pH 4–7], and 0.05% [w/v] bromophenol blue). Protein concentrations were determined using the 2-D Quant kit (GE Healthcare). Isoelectric focusing was carried out with 300 μg of protein. Protein samples were loaded by an in-gel rehydration method onto 13 cm isoelectric focusing strips (pH 4–7) and electrofocused using the IPGphor system (GE Healthcare) at 20°C for 35,000 volt hours. The focused strips were subjected to reduction with 1% (w/v) dithiothreitol in 10 mL of equilibration buffer (6 m urea, 50 mm Tris-HCl [pH 8.8], 30% [v/v] glycerol, and 2% [w/v] SDS), followed by alkylation with 2.5% (w/v) iodoacetamide in the same buffer. The strips were then loaded onto 12.5% (w/v) polyacrylamide gels for SDS-PAGE. The gels were stained with Silver Stain Plus (Bio-Rad).
Image Acquisition and Data Analysis
After 2-DE and gel staining, gel images were scanned using the Bio-Rad FluorS system equipped with a 12-bit camera. For each data set from both wild-type and E8.2-OXDC samples, at least three 2-DE gels representing three biological replicates were included in the data analysis. The scanned gel images were processed and analyzed with PDQuest gel analysis software version 7.2.0 (Bio-Rad). The following preprocessing steps were included in the two-dimensional gel analysis: (1) cropping of the gel images to a uniform size, (2) normalizing intensities to remove the effects of differential loading and staining, (3) transformation of outcome variables to normally distributed variables, and (4) inputting values for missing spot intensities. All images were subsequently processed using the software settings for spot detection, background subtraction, spot quantitation, and gel-to-gel matching of spot patterns as described previously (Ruebelt et al., 2006). These spot detection parameters were chosen because they allowed detection of the majority of spots. To compare spots across gels, a match set representing a standard image of three replicates was created from both the wild-type and E8.2-OXDC samples. Each spot included in the standard gel met the criteria of being qualitatively consistent in size and shape in the replicate gels and being within the linear range of detection. Before the software could detect and document the correspondence between different spots, a few landmarks in the gel series were manually defined to improve the automated matching results (Ruebelt et al., 2006; Kannan et al., 2012). All of the spots detected by the software program were verified manually to eliminate any possible artifacts, such as gel background or streaks. False-positive spots (e.g. artifacts and multiple spots in a cluster) were manually removed. In addition to quantity scores (based on spot density and area), the PDQuest software was used to assign quality scores to each gel spot (Lonosky et al., 2004). Low-quality spots (quality score <30) were removed from further analysis. Spots with a quality score greater than 30 that met the above-mentioned stringent criteria were thereafter referred to as high-quality spots. The high-quality quantities were used to calculate the median value for a given spot, and this value was used as the spot quantity on the standard gel. Here, the correlation coefficient is a measure of the association between the spot intensities on replicates, which was maintained at a minimum of 0.8 between gel images (Supplemental Fig S3; Supplemental Table S1). Next, for comparison, the protein spots observed in wild-type and E8.2-OXDC samples were normalized to the “total density in gel image” mode, and spots were manually annotated. These normalized density values were employed to calculate the sd and CV. The spot volumes were further normalized using three unaltered protein spots across all of the gels. Mr and pI values were also assigned to the master image using the MrpI parameter in the software. A report was extracted from the software in a Microsoft Excel-compatible format, and the normalized spot volume data were taken for further analysis. All statistical analyses were performed as explained below.
Protein Identification
A total of 34 spots were analyzed using LC-MS/MS and 42 spots by MALDI-TOF/TOF. Protein samples were excised mechanically using pipette tips, destained in 30 mm potassium ferricyanide and 100 mm sodium thiosulfate solution, and in-gel digested with trypsin, and the peptides were extracted according to standard techniques (Meywald et al., 1996; Casey et al., 2005). For LC-MS/MS analysis, trypsin-digested peptides were loaded onto a C18 PepMap100 column (3 μm, 100 Å, 75 µm ID_15 cm; LCPackings) at 300 nL min–1, separated with a linear gradient of water/acetonitrile/0.1% formic acid (v/v), and analyzed by electrospray ionization using the Ultimate 3000 Nano HPLC system (Dionex) coupled to a 4000 QTRAP mass spectrometer (Applied Biosystems). The peptides were eluted with a gradient of 10% to 40% acetonitrile with 0.1% formic acid (v/v) over 60 min. The MS/MS data were extracted using Analyst software version 1.5.1 (Applied Biosystems). Peptide analysis was performed through data-dependent acquisition of mass spectrometry scans (mass-to-charge ratio from 400 to 1800), followed by MS/MS scans. Peptides were identified by searching the peak list against the SGN tomato database ITAG release 2.3 (34,727 sequences and 11,956,401 residues) available by the International Tomato Genome Annotation team (http://www.solgenomics.net/organism/Solanum_lycopersicum/genome) using the Mascot version 2.1 search engine (http://www.matrixsciences.com). The database search criteria were as follows: taxonomy, all entries; peptide tolerance, ±1.2 D; MS/MS tolerance, ±0.6 D; peptide charge, +1, +2, or +3; maximum allowed missed cleavage, 1; fixed modification, Cys carbamidomethylation; variable modification, Met oxidation; and instrument type, electrospray ionization (ESI)-ion trap. Protein scores were derived from ion scores as a nonprobabilistic basis for ranking protein hits and as the sum of a series of peptide scores. The score threshold to achieve P < 0.05 was set by the Mascot algorithm and was based on the size of the database used in the search. We considered only those protein spots whose molecular weight search score was above the significant threshold level determined by Mascot. For MALDI-TOF/TOF analysis, after in-gel tryptic digestion, the peptides were extracted according to standard techniques (Casey et al., 2005). The α-cyano-4-hydroxycinnamic acid matrix was prepared at one-half saturation in acetonitrile/water (1:1, v/v) acidified with 0.1% (v/v) trifluoroacetic acid. A 1-μL aliquot of each sample was mixed with an equal volume of matrix solution. The mixture was immediately spotted onto the MALDI target plate and allowed to dry at room temperature. The analysis was performed on a 4800 MALDI-TOF/TOF analyzer (Applied Biosystems). The reflected spectra were obtained over a mass range of 850 to 4,000 D. The spectra of 100 laser shots were summed to generate a peptide mass fingerprinting for each protein digest. Suitable precursors for MS/MS sequencing analyses were selected, and fragmentation was carried out using collision-induced dissociation (atmospheric gas was used) in the 1-kV ion reflector mode and precursor mass windows of ±5 D. The plate model and default calibration were optimized for processing the MS/MS spectra. Searches for peptides were performed in batch mode using GPS Explorer version 3.6 software (Applied Biosystems) with a licensed version of Mascot 2.1 against the SGN tomato database ITAG release 2.3. The search parameters were set as follows: taxonomy, all entries; digestion enzyme, trypsin with one missed cleavage; fixed modification, Cys carbamidomethylation; variable modification, Met oxidation; mass spectrometry (precursor ion) peak filtering: monoisotopic, minimum signal-to-noise ratio of 10, and mass tolerance of ±100 ppm; and MS/MS (fragment ion) peak filtering: monoisotopic, minimum signal-to-noise ratio of 3, and MS/MS fragment tolerance of ±0.4 D. Proteins with a confidence interval percentage of greater than 95% were required to represent a positive identification and were also evaluated on the basis of various parameters, such as the number of peptides matched, MOWSE score, quality of the peptide maps, and percent coverage of the matched protein. For proteins identified based on only one peptide with a score greater than 40, the peptide sequence was systematically checked manually. In all of the protein identifications, the probability scores were greater than the score fixed by Mascot as significant, with a P value < 0.05. The abundance of each identified protein was estimated by determining the protein abundance index (Liu et al., 2004) and the emPAI (Ishihama et al., 2005). The corresponding protein content in mole percentage was calculated as described previously (Ishihama et al., 2005). For the total number of observed peptides per protein, the unique sequences were counted and were imported into Microsoft Excel (Supplemental Table S3. When there was more than one accession number for the same peptide, the match was considered in terms of the putative function. The protein functions were assigned using the Pfam (http://pfam.sanger.ac.uk) and InterPro (http://www.ebi.ac.uk/interpro) protein function database.
Statistical Analyses
Analyses of the statistical significance of the data set of normalized spot volumes comparing the two sets of proteomic data (wild-type and E8.2-OXDC fruit) were performed by unpaired Student’s t tests using MultiExperiment Viewer version 4.8 (Saeed et al., 2003) as described previously (Chaiyarit and Thongboonkerd, 2012; Sanchez-Bel et al., 2012). The statistically significant differences with regard to the results of the biochemical (enzyme activity and organic acid quantification) and micronutrient analyses were determined with the nonparametric Welch’s t test using GraphPad PRISM 5.0 and the nonparametric Wilcoxon rank sum test from the Statistics Online Computational Resource (http://www.SOCR.ucla.edu). The levels of significant differences are indicated with an asterisk in the graphs. P values < 0.05 were considered statistically significant. The values of all parameters analyzed are from three biological replicates per sample.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Plant material and experimental design.
Supplemental Figure S2. 2-DE pattern reproducibility.
Supplemental Figure S3. Scatter plots displaying a CV above 0.8 among the three replicates of E8.2-OXDC-11 and the corresponding wild-type fruit proteome.
Supplemental Table S1. Reproducibility of 2-DE gels.
Supplemental Table S2. List of proteins identified by MS/MS analysis.
Supplemental Table S3. The probable candidate proteins of the identified spots by LC-ESI-MS/MS and MALDI-TOF/TOF.
Supplemental Table S4. The MS/MS detail of the identified protein spots by LC-ESI-MS/MS and MALDI-TOF/TOF.
Supplemental Table S5. Fragment spectra of the proteins identified with a single peptide.
Supplemental Table S6. Domain analysis of proteins with unknown function.
Acknowledgments
We thank Shankar Acharya and C. Ravi Shankar for assistance with the tissue culture and field experiments and Jasbeer Singh for illustrations and graphical representation in the manuscript.
Glossary
- 2-DE
two-dimensional gel electrophoresis
- CV
correlation coefficient of variation
- LC
liquid chromatography
- MS/MS
tandem mass spectrometry
- MALDI-TOF
matrix-assisted laser-desorption ionization time-of-flight
- MALDI
matrix-assisted laser-desorption ionization
- emPAI
exponentially modified protein abundance index
- PCR/PCO
photosynthetic carbon reduction/oxidation
- TCA
tricarboxylic acid
- MS
Murashige and Skoog
- PBS
phosphate-buffered saline
- SGN
Sol Genomics Network
- ESI
electrospray ionization
- EF
elongation factor
References
- Agassandian M, Chen BB, Schuster CC, Houtman JCD, Mallampalli RK. (2010) 14-3-3ζ escorts CCTα for calcium-activated nuclear import in lung epithelia. FASEB J 24: 1271–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson W, Hollins JG, Bond PS. (1971) The composition of tea infusions examined in relation to the association between mortality and water hardness. J Hyg (Lond) 69: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam M, Kesarwani M, Chakraborty S, Natarajan K, Datta A. (2002) Cloning and characterization of the 5′-flanking region of the oxalate decarboxylase gene from Flammulina velutipes. Biochem J 367: 67–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrari F, Baxter C, Usadel B, Urbanczyk-Wochniak E, Zanor M-I, Nunes-Nesi A, Nikiforova V, Centero D, Ratzka A, Pauly M, et al. (2006) Integrated analysis of metabolite and transcript levels reveals the metabolic shifts that underlie tomato fruit development and highlight regulatory aspects of metabolic network behavior. Plant Physiol 142: 1380–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey TM, Arthur PG, Bogoyevitch MA. (2005) Proteomic analysis reveals different protein changes during endothelin-1- or leukemic inhibitory factor-induced hypertrophy of cardiomyocytes in vitro. Mol Cell Proteomics 4: 651–661 [DOI] [PubMed] [Google Scholar]
- Centeno DC, Osorio S, Nunes-Nesi A, Bertolo AL, Carneiro RT, Araújo WL, Steinhauser MC, Michalska J, Rohrmann J, Geigenberger P, et al. (2011) Malate plays a crucial role in starch metabolism, ripening, and soluble solid content of tomato fruit and affects postharvest softening. Plant Cell 23: 162–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaiyarit S, Thongboonkerd V. (2012) Changes in mitochondrial proteome of renal tubular cells induced by calcium oxalate monohydrate crystal adhesion and internalization are related to mitochondrial dysfunction. J Proteome Res 11: 3269–3280 [DOI] [PubMed] [Google Scholar]
- Chakraborty S, Chakraborty N, Datta A. (2000) Increased nutritive value of transgenic potato by expressing a nonallergenic seed albumin gene from Amaranthus hypochondriacus. Proc Natl Acad Sci USA 97: 3724–3729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S, Chakraborty N, Jain D, Salunke DM, Datta A. (2002) Active site geometry of oxalate decarboxylase from Flammulina velutipes: role of histidine-coordinated manganese in substrate recognition. Protein Sci 11: 2138–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Beevers H. (1968) Biogenesis of oxalate in plant tissues. Plant Physiol 43: 1821–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cilia M, Tamborindeguy C, Rolland M, Howe K, Thannhauser TW, Gray S. (2011) Tangible benefits of the aphid Acyrthosiphon pisum genome sequencing for aphid proteomics: enhancements in protein identification and data validation for homology-based proteomics. J Insect Physiol 57: 179–190 [DOI] [PubMed] [Google Scholar]
- Conyers RAJ, Bais R, Rofe AM. (1990) The relation of clinical catastrophes, endogenous oxalate production, and urolithiasis. Clin Chem 36: 1717–1730 [PubMed] [Google Scholar]
- Cooper DA. (2004) Carotenoids in health and disease: recent scientific evaluations, research recommendations and the consumer. J Nutr 134: 221S–224S [DOI] [PubMed] [Google Scholar]
- Cronje C, George GM, Fernie AR, Bekker J, Kossmann J, Bauer R. (2012) Manipulation of l-ascorbic acid biosynthesis pathways in Solanum lycopersicum: elevated GDP-mannose pyrophosphorylase activity enhances l-ascorbate levels in red fruit. Planta 235: 553–564 [DOI] [PubMed] [Google Scholar]
- Cunha WG, Tinocoab MLP, Pancotia HL, Ribeiroa RE, Araga FJL. (2010) High resistance to Sclerotinia sclerotiorum in transgenic soybean plants transformed to express an oxalate decarboxylase gene. Plant Pathol 59: 654–660 [Google Scholar]
- Curhan GC. (1997) Dietary calcium, dietary protein, and kidney stone formation. Miner Electrolyte Metab 23: 261–264 [PubMed] [Google Scholar]
- Davuluri GR, van Tuinen A, Fraser PD, Manfredonia A, Newman R, Burgess D, Brummell DA, King SR, Palys J, Uhlig J, et al. (2005) Fruit-specific RNAi-mediated suppression of DET1 enhances carotenoid and flavonoid content in tomatoes. Nat Biotechnol 23: 890–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Castro MD. (1988) Determination of oxalic acid in urine: a review. J Pharm Biomed Anal 6: 1–13 [DOI] [PubMed] [Google Scholar]
- Deikman J, Fischer RL. (1988) Interaction of a DNA binding factor with the 5′-flanking region of an ethylene-responsive fruit ripening gene from tomato. EMBO J 7: 3315–3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delless G, Boyer N, Gaspar T. (2011) Thigmomorphogenesis in Bryonia dioica: change in soluble and wall peroxidase, phenylalanine ammonia-lyase activity, cellulose, lignin content and monomeric constituents. Plant Growth Regul 3: 133–148 [Google Scholar]
- Dias BBA, Cunha WG, Morais LS, Vianna GR, Rech EL, de Capdeville G, Aragao FJL. (2006) Expression of an oxalate decarboxylase gene from Flammulina sp. in transgenic lettuce (Lactuca sativa) plants and resistance to Sclerotinia. Plant Pathol 55: 187–193 [Google Scholar]
- Dunwell JM, Moya-León MA, Herrera R. (2001) Transcriptome analysis and crop improvement (a review). Biol Res 34: 153–164 [DOI] [PubMed] [Google Scholar]
- Finch AM, Kasidas GP, Rose GA. (1981) Urine composition in normal subjects after oral ingestion of oxalate-rich foods. Clin Sci (Lond) 60: 411–418 [DOI] [PubMed] [Google Scholar]
- FAO (2010) The State of Food Insecurity in the World. Food and Agricultural Organization of the United Nations, Rome [Google Scholar]
- Giovannoni JJ. (2007) Fruit ripening mutants yield insights into ripening control. Curr Opin Plant Biol 10: 283–289 [DOI] [PubMed] [Google Scholar]
- Grusak MA, DellaPenna D. (1999) Improving the nutrient composition of plants to enhance human nutrition and health. Annu Rev Plant Physiol Plant Mol Biol 50: 133–161 [DOI] [PubMed] [Google Scholar]
- Harvey WR. (2009) Voltage coupling of primary H+ V-ATPases to secondary Na+- or K+-dependent transporters. J Exp Biol 212: 1620–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaney RP, Weaver CM, Recker RR. (1988) Calcium absorbability from spinach. Am J Clin Nutr 47: 707–709 [DOI] [PubMed] [Google Scholar]
- Hodgkinson A. (1970) Determination of oxalic acid in biological material. Clin Chem 16: 547–557 [Google Scholar]
- Holmes RP, Assimos DG, Goodman HO. (1998) Genetic and dietary influences on urinary oxalate excretion. Urol Res 26: 195–200 [DOI] [PubMed] [Google Scholar]
- Holmes RP, Kennedy M. (2000) Estimation of the oxalate content of foods and daily oxalate intake. Kidney Int 57: 1662–1667 [DOI] [PubMed] [Google Scholar]
- Horner HT, Kausch AP, Wagner BL. (2000) Ascorbic acid: a precursor of oxalate in crystal idioblasts of Yucca torreyi in liquid root culture. Int J Plant Sci 1: 861–868 [Google Scholar]
- Hwang ES, Bowen PE. (2002) Can the consumption of tomatoes or lycopene reduce cancer risk? Integr Cancer Ther 1: 121–132 [DOI] [PubMed] [Google Scholar]
- Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M. (2005) Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics 4: 1265–1272 [DOI] [PubMed] [Google Scholar]
- Jones L, Seymour GB, Knox JP. (1997) Localization of pectic galactan in tomato cell walls using a monoclonal antibody specific to (1→4)-β-d-galactan. Plant Physiol 113: 1405–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun H, Kieselbach T, Jönsson LJ. (2012) Comparative proteome analysis of Saccharomyces cerevisiae: a global overview of in vivo targets of the yeast activator protein 1. BMC Genomics 13: 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan S, Sujitha MV, Sundarraj S, Thirumurugan R. (2012) Two dimensional gel electrophoresis in cancer proteomics. In Magdeldin S, ed, Gel Electrophoresis–Advanced Techniques. In Tech, Rijeka, Croatia, pp 359–390 [Google Scholar]
- Kelsay JL, Prather ES, Clark WM, Canary JJ. (1988) Mineral balances of men fed a diet containing fiber in fruits and vegetables and oxalic acid in spinach for six weeks. J Nutr 118: 1197–1204 [DOI] [PubMed] [Google Scholar]
- Kesarwani M, Azam M, Natarajan K, Mehta A, Datta A. (2000) Oxalate decarboxylase from Collybia velutipes. Molecular cloning and its overexpression to confer resistance to fungal infection in transgenic tobacco and tomato. J Biol Chem 275: 7230–7238 [DOI] [PubMed] [Google Scholar]
- L’Haridon F, Besson-Bard A, Binda M, Serrano M, Abou-Mansour E, Balet F, Schoonbeek HJ, Hess S, Mir R, Léon J, et al. (2011) A permeable cuticle is associated with the release of reactive oxygen species and induction of innate immunity. PLoS Pathog 7: e1002148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libert B, Franceschi VR. (1987) Oxalate in crop plants. J Agric Food Chem 35: 926–938 [Google Scholar]
- Liu H, Sadygov RG, Yates JR., III (2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem 76: 4193–4201 [DOI] [PubMed] [Google Scholar]
- Liu Q, Tan G, Levenkova N, Li T, Pugh EN, Jr, Rux JJ, Speicher DW, Pierce EA. (2007) The proteome of the mouse photoreceptor sensory cilium complex. Mol Cell Proteomics 6: 1299–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YS, Gur A, Ronen G, Causse M, Damidaux R, Buret M, Hirschberg J, Zamir D. (2003) There is more to tomato fruit colour than candidate carotenoid genes. Plant Biotechnol J 1: 195–207 [DOI] [PubMed] [Google Scholar]
- Loewus FA. (1980) The Biochemistry of Plants, Vol 3 Academic Press, New York, pp 77–99 [Google Scholar]
- Loewus FA. (1999) Biosynthesis and metabolism of ascorbic acid in plants and of analogs of ascorbic acid in fungi. Phytochemistry 52: 193–210 [Google Scholar]
- Lonosky PM, Zhang X, Honavar VG, Dobbs DL, Fu A, Rodermel SR. (2004) A proteomic analysis of maize chloroplast biogenesis. Plant Physiol 134: 560–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrae AR. (1971) Malic enzyme activity in plant mitochondria. Phytochemistry 10: 2343–2347 [Google Scholar]
- Matas AJ, Yeats TH, Buda GJ, Zheng Y, Chatterjee S, Tohge T, Ponnala L, Adato A, Aharoni A, Stark R, et al. (2011) Tissue- and cell-type specific transcriptome profiling of expanding tomato fruit provides insights into metabolic and regulatory specialization and cuticle formation. Plant Cell 23: 3893–3910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattoo AK, Shukla V, Fatima T, Handa AK, Yachha SK. (2010) Genetic engineering to enhance crop-based phytonutrients (nutraceuticals) to alleviate diet-related diseases. Adv Exp Med Biol 698: 122–143 [DOI] [PubMed] [Google Scholar]
- Murtaugh MA, Khe-ni M, Benson J, Curtin K, Caan B, Slattery ML. (2004) Antioxidants, carotenoids, and risk of rectal cancer. Am J Epidemiol 1: 32–41 [DOI] [PubMed] [Google Scholar]
- McGregor E, Dunn MJ. (2006) Proteomics of the heart: unraveling disease. Circ Res 98: 309–321 [DOI] [PubMed] [Google Scholar]
- Mehta A, Datta A. (1991) Oxalate decarboxylase from Collybia velutipes. Purification, characterization, and cDNA cloning. J Biol Chem 266: 23548–23553 [PubMed] [Google Scholar]
- Meywald T, Scherthan H, Nagl W. (1996) Increased specificity of colloidal silver staining by means of chemical attenuation. Hereditas 124: 63–70 [DOI] [PubMed] [Google Scholar]
- Mika A, Boenisch MJ, Hopff D, Lüthje S. (2010) Membrane-bound guaiacol peroxidases from maize (Zea mays L.) roots are regulated by methyl jasmonate, salicylic acid, and pathogen elicitors. J Exp Bot 61: 831–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millerd A, Morton RK, Wells JR. (1963) Enzymic synthesis of oxalic acid in Oxalis pes-caprae. Biochem J 88: 281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata PA. (2003) Advances in our understanding of calcium oxalate crystal formation and function in plants. Plant Sci 164: 901–909 [Google Scholar]
- Nakata PA, McConn M. (2007) Isolated Medicago truncatula mutants with increased calcium oxalate crystal accumulation have decreased ascorbic acid levels. Plant Physiol Biochem 45: 216–220 [DOI] [PubMed] [Google Scholar]
- Neuhaus JM, Sticher L, Meins F, Jr, Boller T. (1991) A short C-terminal sequence is necessary and sufficient for the targeting of chitinases to the plant vacuole. Proc Natl Acad Sci USA 88: 10362–10366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa K. (2006) Bottle cultivation of culinary-medicinal Enokitake mushroom Flammulina velutipes (W. Curt.: Fr.) Singer (Agaricomycetidae) in Nagano prefecture (Japan). Int J Med Mushroom 2: 173–178 [Google Scholar]
- Noonan SC, Savage GP. (1999) Oxalate content of food and its effect on humans. Asia Pac J Clin Nutr 8: 64–74 [PubMed] [Google Scholar]
- Orzaez D, Granell A. (2009) Reverse genetics and transient gene expression in fleshy fruits: overcoming plant stable transformation. Plant Signal Behav 4: 864–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orzaez D, Mirabel S, Wieland WH, Granell A. (2006) Agroinjection of tomato fruits. A tool for rapid functional analysis of transgenes directly in fruit. Plant Physiol 140: 3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio S, Alba R, Damasceno CMB, Lopez-Casado G, Lohse M, Zanor MI, Tohge T, Usadel B, Rose JKC, Fei Z, et al. (2011) Systems biology of tomato fruit development: combined transcript, protein, and metabolite analysis of tomato transcription factor (nor, rin) and ethylene receptor (Nr) mutants reveals novel regulatory interactions. Plant Physiol 157: 405–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Cheng NH, Pittman JK, Yoo KS, Park J, Smith RH, Hirschi KD. (2005) Increased calcium levels and prolonged shelf life in tomatoes expressing Arabidopsis H+/Ca2+ transporters. Plant Physiol 139: 1194–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20: 3551–3567 [DOI] [PubMed] [Google Scholar]
- Peters TJ, Jr, Apt L, Ross JF. (1971) Effect of phosphates upon iron absorption studied in normal human subjects and in an experimental model using dialysis. Gastroenterology 61: 315–322 [PubMed] [Google Scholar]
- Poetsch A, Wolters D. (2008) Bacterial membrane proteomics. Proteomics 8: 4100–4122 [DOI] [PubMed] [Google Scholar]
- Römer S, Fraser PD, Kiano JW, Shipton CA, Misawa N, Schuch W, Bramley PM. (2000) Elevation of the provitamin A content of transgenic tomato plants. Nat Biotechnol 18: 666–669 [DOI] [PubMed] [Google Scholar]
- Ruebelt MC, Leimgruber NK, Lipp M, Reynolds TL, Nemeth MA, Astwood JD, Engel KH, Jany KD. (2006) Application of two-dimensional gel electrophoresis to interrogate alterations in the proteome of genetically modified crops. 1. Assessing analytical validation. J Agric Food Chem 54: 2154–2161 [DOI] [PubMed] [Google Scholar]
- Rutschmann F, Stalder U, Piotrowski M, Oecking C, Schaller A. (2002) LeCPK1, a calcium-dependent protein kinase from tomato. Plasma membrane targeting and biochemical characterization. Plant Physiol 129: 156–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccoccia F, Di Micco P, Boumis G, Brunori M, Koutris I, Miele AE, Morea V, Sriratana P, Williams DL, et al. (2012) Moonlighting by different stressors: crystal structure of the chaperone species of a 2-Cys peroxiredoxin. Structure 20: 429–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378 [DOI] [PubMed] [Google Scholar]
- Sanchez-Bel P, Egea I, Sanchez-Ballesta MT, Sevillano L, Del Carmen Bolarin M, Flores FB. (2012) Proteome changes in tomato fruits prior to visible symptoms of chilling injury are linked to defensive mechanisms, uncoupling of photosynthetic processes and protein degradation machinery. Plant Cell Physiol 53: 470–484 [DOI] [PubMed] [Google Scholar]
- Sghaier-Hammami B, Drira N, Jorrín-Novo JV. (2009) Comparative 2-DE proteomic analysis of date palm (Phoenix dactylifera L.) somatic and zygotic embryos. J Proteomics 73: 161–177 [DOI] [PubMed] [Google Scholar]
- Singh PP, Saxena SN. (1972) Effect of maturity on the oxalate and cation contents of six leafy vegetables. Indian J Nutr Diet 9: 269–276 [Google Scholar]
- Smirnoff N. (1996) The function and metabolism of ascorbic acid in plants. Ann Bot (Lond) 78: 661–669 [Google Scholar]
- Stacewicz-Sapuntzakis M, Bowen PE. (2005) Role of lycopene and tomato products in prostate health. Biochim Biophys Acta 1740: 202–205 [DOI] [PubMed] [Google Scholar]
- Suvachittanont O, Meksongsee LA, Dhanamitta S, Valyasevi A. (1973) The oxalic acid content of some vegetables in Thailand, its possible relationships with the bladder stone disease. J Med Assoc Thai 56: 645–653 [PubMed] [Google Scholar]
- Taylor NL, Heazlewood JL, Millar AH. (2011) The Arabidopsis thaliana 2-D gel mitochondrial proteome: refining the value of reference maps for assessing protein abundance, contaminants and post-translational modifications. Proteomics 11: 1720–1733 [DOI] [PubMed] [Google Scholar]
- Textor S, Bartram S, Kroymann J, Falk KL, Hick A, Pickett JA, Gershenzon J. (2004) Biosynthesis of methionine-derived glucosinolates in Arabidopsis thaliana: recombinant expression and characterization of methylthioalkylmalate synthase, the condensing enzyme of the chain-elongation cycle. Planta 218: 1026–1035 [DOI] [PubMed] [Google Scholar]
- U.S. Department of Agriculture (1984) Composition of Foods: Vegetables and Vegetable Products (Agriculture Handbook No 8–11). Government Printing Office, Washington, DC, pp 502 [Google Scholar]
- van der Merwe MJ, Osorio S, Moritz T, Nunes-Nesi A, Fernie AR. (2009) Decreased mitochondrial activities of malate dehydrogenase and fumarase in tomato lead to altered root growth and architecture via diverse mechanisms. Plant Physiol 149: 653–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Haute E, Joos H, Maes M, Warren G, Van Montagu M, Schell J. (1983) Intergeneric transfer and exchange recombination of restriction fragments cloned in pBR322: a novel strategy for the reversed genetics of the Ti plasmids of Agrobacterium tumefaciens. EMBO J 2: 411–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb MA. (1999) Cell-mediated crystallization of calcium oxalate in plants. Plant Cell 11: 751–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch RM. (1995) Micronutrient nutrition of plants. Crit Rev Plant Sci 14: 49–82 [Google Scholar]
- Welch RM. (2002) Breeding strategies for biofortified staple plant foods to reduce micronutrient malnutrition globally. J Nutr 132: 495S–499S [DOI] [PubMed] [Google Scholar]
- White PJ, Broadley MR. (2003) Calcium in plants. Ann Bot (Lond) 92: 487–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AW, Wilson DM. (1990) Dietary intake, absorption, metabolism, and excretion of oxalate. Semin Nephrol 10: 2–8 [PubMed] [Google Scholar]
- Williams HE, Wandzilak TR. (1989) Oxalate synthesis, transport and the hyperoxaluric syndromes. J Urol 141: 742–749 [DOI] [PubMed] [Google Scholar]
- Yang Y, Thannhauser TW, Li L, Zhang S. (2007) Development of an integrated approach for evaluation of 2-D gel image analysis: impact of multiple proteins in single spots on comparative proteomics in conventional 2-D gel/MALDI workflow. Electrophoresis 28: 2080–2094 [DOI] [PubMed] [Google Scholar]
- Yoon JH, Yea K, Kim J, Choi YS, Park S, Lee H, Lee CS, Suh P-G, Ryu SH. (2009) Comparative proteomic analysis of the insulin-induced L6 myotube secretome. Proteomics 9: 51–60 [DOI] [PubMed] [Google Scholar]
- Zhang C, Liu J, Zhang Y, Cai X, Gong P, Zhang J, Wang T, Li H, Ye Z. (2011) Overexpression of SlGMEs leads to ascorbate accumulation with enhanced oxidative stress, cold, and salt tolerance in tomato. Plant Cell Rep 30: 389–398 [DOI] [PubMed] [Google Scholar]
- Zhang LJ, Wang XE, Peng X, Wei YJ, Cao R, Liu Z, Xiong JX, Yin XF, Ping C, Liang S. (2006) Proteomic analysis of low-abundant integral plasma membrane proteins based on gels. Cell Mol Life Sci 63: 1790–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Benavente LM, Stepanova AN, Alonso JM. (2011) A recombineering-based gene tagging system for Arabidopsis. Plant J 66: 712–723 [DOI] [PubMed] [Google Scholar]