Nonhomologous gene exchange by zinc finger nucleases is a novel method for transgene replacement and gene stacking in plants.
Abstract
Stimulation of the homologous recombination DNA-repair pathway via the induction of genomic double-strand breaks (DSBs) by zinc finger nucleases (ZFNs) has been deployed for gene replacement in plant cells. Nonhomologous end joining (NHEJ)-mediated repair of DSBs, on the other hand, has been utilized for the induction of site-specific mutagenesis in plants. Since NHEJ is the dominant DSB repair pathway and can also lead to the capture of foreign DNA molecules, we suggest that it can also be deployed for gene replacement. An acceptor DNA molecule in which a green fluorescent protein (GFP) coding sequence (gfp) was flanked by ZFN recognition sequences was used to produce transgenic target plants. A donor DNA molecule in which a promoterless hygromycin B phosphotransferase-encoding gene (hpt) was flanked by ZFN recognition sequences was constructed. The donor DNA molecule and ZFN expression cassette were delivered into target plants. ZFN-mediated site-specific mutagenesis and complete removal of the GFP coding sequence resulted in the recovery of hygromycin-resistant plants that no longer expressed GFP and in which the hpt gene was unlinked to the acceptor DNA. More importantly, ZFN-mediated digestion of both donor and acceptor DNA molecules resulted in NHEJ-mediated replacement of the gfp with hpt and recovery of hygromycin-resistant plants that no longer expressed GFP and in which the hpt gene was physically linked to the acceptor DNA. Sequence and phenotypical analyses, and transmission of the replacement events to the next generation, confirmed the stability of the NHEJ-induced gene exchange, suggesting its use as a novel method for transgene replacement and gene stacking in plants.
Genome editing is a powerful tool for functional gene analysis and the genetic improvement of living cells. Developing methods for genome editing in plants will foster gene functional analysis and the introduction of novel traits into agriculturally important species (for review, see Puchta, 2002; Hanin and Paszkowski, 2003; Weinthal et al., 2010; Tzfira et al., 2012). Methods for genome editing have been developed for several model organisms, such as yeast (Saccharomyces cerevisiae), mouse embryonic stem cells, and Drosophila spp. (Scherer and Davis, 1979; Baribault and Kemler, 1989; Venken and Bellen, 2005; Hall et al., 2009; Laible and Alonso-González, 2009; Tenzen et al., 2010). These methods rely on homologous recombination (HR) between foreign donor DNA molecules and the target acceptor sequence in the genome. In plant species, however, domination of the nonhomologous end joining (NHEJ) DNA-repair machinery over that of HR (Ray and Langer, 2002; Britt and May, 2003) often leads to random integration of foreign DNA molecules, which in plants are often delivered by Agrobacterium tumefaciens-mediated gene transfer (Banta and Montenegro, 2008).
Several reports have described strategies to enhance the rate of HR DNA-repair events in plant cells or to select for rare HR-mediated donor DNA-integration events (for review, see Puchta, 2002; Hanin and Paszkowski, 2003; Weinthal et al., 2010; Tzfira et al., 2012). Thus, for example, HR-mediated gene targeting has been enhanced in Arabidopsis (Arabidopsis thaliana) plants by overexpression of RAD54, a yeast chromatin-remodeling protein (Shaked et al., 2005). Other examples include the use of strong positive- and negative-selection schemes or PCR-based screening for the selection of rare HR-mediated gene-targeting events in rice (Oryza sativa; Terada et al., 2002, 2007) and in Arabidopsis plants and tissues (Kempin et al., 1997; Hanin et al., 2001), respectively. While these approaches and others have proven useful, relying on the cell’s natural low rate of HR DNA repair has shown only limited success in the targeting of native and transgenic sequences in plant cells (for review, see Puchta, 2002; Hanin and Paszkowski, 2003; Reiss, 2003; Porteus, 2009; Weinthal et al., 2010; Tzfira et al., 2012).
The HR DNA-repair machinery can also be enhanced by the induction of genomic double-strand breaks (DSBs). DSBs can be induced at specific genomic locations using zinc finger nucleases (ZFNs). ZFNs are artificial hybrid restriction enzymes that are composed of a fusion between a designed zinc-finger protein DNA-binding domain and the cleavage domain of the FokI endonuclease (for review, see Porteus, 2009; Urnov et al., 2010; Weinthal et al., 2010; Tzfira et al., 2012). ZFNs have been designed to target a wide variety of native and artificial sequences in human, animal, and plant cells (Kumar et al., 2006; Porteus, 2006; Lombardo et al., 2007; Moehle et al., 2007; Doyon et al., 2008; Cai et al., 2009; Shukla et al., 2009; Liu et al., 2010; Zhang et al., 2010; de Pater et al., 2013). In many cases, ZFN-mediated DSBs have been used to stimulate the HR DNA-repair machinery for HR-mediated gene replacement and gene addition. This approach has been successfully implemented in animals (Beumer et al., 2006; Meng et al., 2008), human cell lines (Urnov et al., 2005; Lombardo et al., 2007), and plants (Wright et al., 2005; Cai et al., 2009; Shukla et al., 2009; Townsend et al., 2009; de Pater et al., 2013). However, it is important to note that while ZFN-induced DSBs can indeed induce the HR repair machinery, most of these breaks are repaired by the cell’s NHEJ DNA-repair machinery in plant and other species. Thus, ZFNs have also been used for the induction of site-specific mutagenesis in many eukaryotic cells, including plant cells (Lloyd et al., 2005; Tovkach et al., 2009; Marton et al., 2010; Osakabe et al., 2010; Zhang et al., 2010; Curtin et al., 2011; Even-Faitelson et al., 2011). More recently, ZFNs have been used for NHEJ-mediated targeted chromosomal deletions (Lee et al., 2010; Söllü et al., 2010), transgene removal (Petolino et al., 2010), and even NHEJ-mediated targeted DNA integration in mammalian genomes (Orlando et al., 2010). Since genomic DSBs can function as traps for the integration of A. tumefaciens transferred DNA (T-DNA) molecules via NHEJ (Salomon and Puchta, 1998; Chilton and Que, 2003; Tzfira et al., 2003), we decided to explore the possible use of the NHEJ DNA-repair pathway not only for site-specific mutagenesis and targeted gene insertion but also for gene replacement.
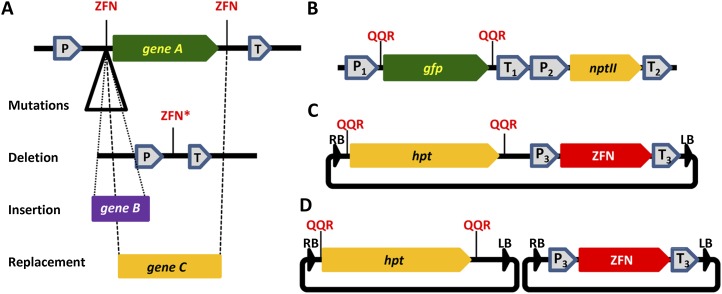
During plant transformation, A. tumefaciens delivers its T-DNA as a single-stranded molecule that, inside the plant cell, can be complemented into a double-stranded transferred DNA (dsT-DNA) intermediate by an as yet unknown mechanism (Tzfira et al., 2004; Ziemienowicz et al., 2008). Induction of DSBs by the transient expression of naturally occurring rare-cutting restriction enzymes results in the incorporation of the T-DNA molecules into a predetermined integration site in the plant cell (Salomon and Puchta, 1998; Chilton and Que, 2003; Tzfira et al., 2003). More importantly, T-DNA molecules can be digested by rare-cutting restriction enzymes prior to their final integration into the plant genome (Chilton and Que, 2003; Tzfira et al., 2003). These observations indicate that it is the dsT-DNA intermediates that function as substrates for the NHEJ integration machinery (Chilton and Que, 2003; Tzfira et al., 2003). Furthermore, sequencing analysis indicates that the digested dsT-DNA molecules may be integrated into the rare-cutter-induced genomic DSBs by a simple NHEJ ligation-like mechanism (Chilton and Que, 2003; Tzfira et al., 2003). These observations led us to suggest that NHEJ-mediated gene replacement might be achieved by coupling the release of a target DNA portion (by the expression of ZFN enzymes) with the delivery of donor T-DNA molecules. Our strategy, which relies on the induction of quadruple DSBs and on NHEJ-mediated incorporation of a T-DNA molecule into the broken target DNA (Fig. 1A), is substantially different from HR-mediated gene-replacement strategies, which rely on the induction of a single genomic DSB and stimulation of the HR repair machinery (Weinthal et al., 2010; Tzfira et al., 2012). Our strategy may thus provide an alternative not only for native gene replacement but also for editing and stacking a number of genes in the same chromosomal locus, several of which may carry similar regulatory sequences (Lyznik and Dress, 2008; Naqvi et al., 2010; Que et al., 2010), which could hinder the use of HR for their successive engineering.
Figure 1.
Experimental approach and constructs for analyzing NHEJ-mediated genome modification in plants. A, A target DNA molecule was engineered to carry a functional expression cassette in which the target gene (gene A) is flanked by ZFN recognition sites. Gene A loss of function can facilitate the detection of site-specific mutagenesis, gene deletion, donor gene insertion (gene B), and target gene replacement (gene C). B, A GFP- and kanamycin-expressing target DNA molecule in which the GFP coding sequence is flanked by two quasipalindromic QQR ZFN recognition sites. This construct was used to produce GFP-overexpressing transgenic plants. C, A donor T-DNA molecule that was engineered to carry a promoterless hygromycin resistance-encoding gene flanked by two quasipalindromic QQR ZFN recognition sites and a constitutive QQR ZFN expression cassette. D, A dual-donor T-DNA molecule in which the promoterless hygromycin resistance-encoding gene and the QQR ZFN-expressing cassette are launched from two separate T-DNA molecules. gfp, GFP-encoding gene; hpt, hygromycin resistance-encoding gene; LB, left border; nptII, kanamycin resistance-encoding gene; P, generic promoter; P1 and P3, dual CaMV 35S promoter; P2, octopine synthase promoter; QQR, QQR ZFN recognition site; RB, right border; T, generic terminator; T1 and T3, CaMV 35S terminators; T2, octopine synthase terminator; ZFN, ZFN recognition site; ZFN*, altered ZFN recognition site. [See online article for color version of this figure.]
Here, we describe a proof-of-concept report that demonstrates that the NHEJ DNA-repair pathway, when combined with the use of ZFNs, can serve for gene replacement in plant cells. Our approach is based on the use of a donor T-DNA molecule that carries ZFN recognition sites that are compatible with sites on the acceptor DNA sequence. We demonstrate that codelivery of the T-DNA molecule with ZFN expression cassettes leads to a variety of outcomes, including gene replacement, in addition to site-specific mutagenesis and gene deletion. Phenotypical and molecular analyses as well as transmission of the replacement events to the next generation in tobacco (Nicotiana tabacum) and Arabidopsis plants confirmed the nature and stability of the NHEJ-induced gene exchange.
RESULTS
Experimental System
We developed an experimental approach in which a variety of NHEJ-mediated genome modifications could be studied. Shown in Figure 1A is an outline of the target sequence (gene A), which is flanked by ZFN recognition sites. Targeting of gene A’s coding sequence at the 5′ end is expected to lead to gene A’s loss of function, while simultaneous digestion at both the 3′ and 5′ ends of gene A’s coding sequence may result in complete gene deletion. DSB induction at gene A’s 5′ end (or 3′ end) can also result in trapping of a foreign DNA molecule (i.e. gene B). More importantly, coupling the release of gene A by simultaneous digestion at both ends with the delivery of a foreign DNA molecule (i.e. gene C) can potentially result in NHEJ-mediated gene replacement.
We used a single-monomer ZFN approach to minimize the complexity that might be associated with variability in the expression, binding, and activity of two or more different monomers in plant cells. We used the QQR ZFN, which is a three-finger ZFN capable of binding to the sequence 5′-GGGGAAGAA-3′ (Smith et al., 2000). QQR ZFN has been successfully used for targeting experiments in Arabidopsis, tobacco, and petunia (Petunia hybrida) plants (Lloyd et al., 2005; Tovkach et al., 2009; Marton et al., 2010; Even-Faitelson et al., 2011). We selected gfp as the target DNA molecule to facilitate the phenotypical monitoring of putative targeting events by loss of function. The GFP coding sequence, which was driven by the cauliflower mosaic virus (CaMV) dual 35S constitutive promoter, was flanked by two quasipalindromic QQR ZFN recognition sites (Fig. 1B). A promoterless hygromycin resistance-encoding gene (hpt) was flanked by two quasipalindromic QQR ZFN recognition sites and used as donor DNA. The hpt gene was selected to facilitate the recovery of putative replacement events by positive selection. ZFN expression was driven by the CaMV dual 35S constitutive promoter (Tovkach et al., 2010). The hpt and ZFN expression cassettes were delivered either as a single T-DNA molecule (Fig. 1C) or from two independent T-DNA molecules (Fig. 1D). We produced several tobacco and Arabidopsis kanamycin-resistant and GFP-expressing target transgenic plants. Two Arabidopsis lines and two tobacco lines carrying a single and simple T-DNA target molecule and strong uniform GFP expression were selected for gene replacement and mutagenesis experiments.
QQR ZFN-Mediated Site-Specific Mutagenesis and Gene Deletion
Target transgenic tobacco and Arabidopsis plants were retransformed with the donor DNA and ZFN-expressing cassette (Fig. 1). Transgenic plantlets and seedlings were selected on hygromycin. Random integration of the donor DNA molecule can potentially lead to promoter trapping (Lindsey et al., 1998), which can result in nontargeted hygromycin-resistant plants. Therefore, we used a high concentration of hygromycin in an attempt to minimize the number of hygromycin-resistant nontarget plants. We recovered 30 tobacco and 21 Arabidopsis hygromycin-resistant plants that were further analyzed for GFP expression and integrity of the GFP coding sequence by PCR amplification and sequencing of the GFP coding locus. Table I summarizes the various types and numbers of targeting events we obtained. The percentages derived from the number of molecularly confirmed specific events (i.e. mutation, deletion, and replacement) from hygromycin-resistant plants that were recovered for each plant species.
Table I. Summary of targeting events in Arabidopsis and tobacco plants.
| Species | Classification | Events | Percentagea |
|---|---|---|---|
| Arabidopsis | Mutation | A17-5, A17-6 | 9.5 (2/21) |
| Deletion | A3-2, A3-3b, A7-1, A7-6, A7-10, A20 | 28.6 (6/21) | |
| Replacement | A15 | 4.8 (1/21) | |
| Tobacco | Mutation | T23, T8-21, T10-3, T6-23 | 13.3 (4/30) |
| Deletion | T8 | 3.3 (1/30) | |
| Replacements | T19, T14 | 6.7 (2/30) |
The number of events is shown in parentheses.
Figure 2 shows the sequencing data of several hygromycin-resistant lines that were found to carry the full or partial GFP coding sequence. As expected, the expression of ZFNs in target tissues led to site-specific mutagenesis at the QQR ZFN recognition sites flanking the GFP coding region. Nucleotide replacement and small deletions and/or additions were observed. While in most cases these changes did not result in the elimination of GFP expression, as observed by confocal microscopy, line T10-3 did not express GFP, most likely due to a truncation of its 3′ end (Fig. 2). Interestingly, while both QQR ZFN recognition sites were mutated in these lines (except for T23-6, which was mutated in only one QQR ZFN recognition site), the GFP coding sequence was not released. These observations indicated that the QQR ZFN recognition sites are most likely sequentially digested by QQR ZFN and repaired by the plant NHEJ machinery in a manner similar to that previously described by Gong and Golic (2003).
Figure 2.

Molecular analysis of ZFN-mediated mutagenesis events in tobacco (T23, T8-21, T10-3, and T23-6) and Arabidopsis (A17-5 and A17-6) plants. The QQR ZFN-binding sites, on the top strand of the parent target sequences, are in blue, and nucleotide changes to the parent target sequences are in red. gfp, GFP coding sequence; P, CaMV dual 35S promoter; T, CaMV 35S terminator. Dashes indicate base deletions. [See online article for color version of this figure.]
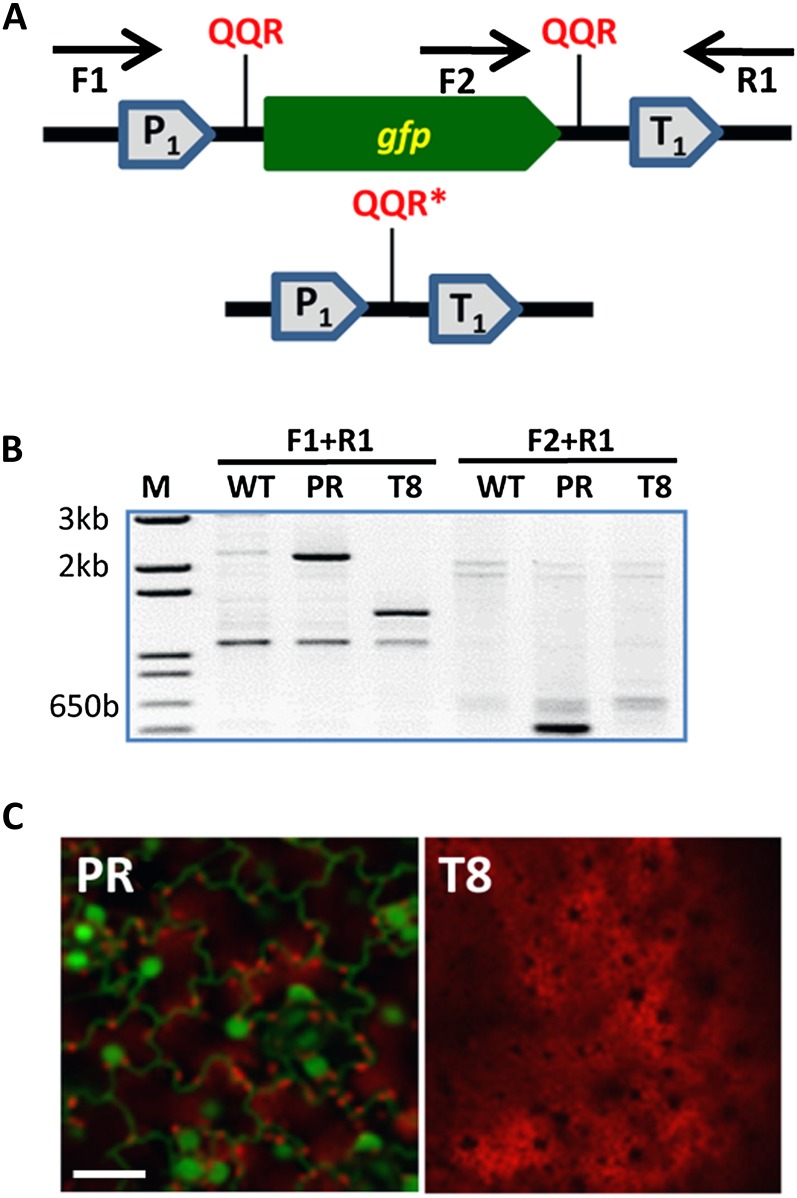
Complete release and deletion of the GFP coding region were observed in several tobacco and Arabidopsis lines (Fig. 3). We molecularly characterized these events by PCR amplification of total plant DNA using sets of primers capable of amplifying distinct regions of the GFP expression cassette (Fig. 3A). Exemplified by tobacco plant T8, amplification of the entire GFP expression cassette (primers F1 and R1; Fig. 3, A and B) resulted in a shorter product (approximately 1.4 kb) than that of the parental line (approximately 2.1 kb; Fig. 3B). Furthermore, no specific DNA band was observed in plant T8 when the GFP 3′ end and part of the terminator region were subjected to amplification by PCR (primers F2 and R1; Fig. 3, A and B). Sequencing analysis of short PCR products derived from the amplification of GFP expression cassettes in T8 and several Arabidopsis lines verified that the GFP coding sequence indeed had been removed from the parental genomic DNA (Fig. 4). Furthermore, confocal microscopy analysis indicated that deletion of the GFP coding sequence led to the elimination of GFP fluorescence in the targeted plants, as exemplified by tobacco plant T8 (Fig. 3C). In addition to T8, several transgene-deletion events were also characterized in Arabidopsis, all of which were verified by sequence analysis (Fig. 4). Subsequent characterization of lines T23, A6-23, T8, A3-2, A7-1, and A7-10 (which derived from dual T-DNA transformation) failed to produce a specific PCR product for the ZFN expression cassette. Next, we let T23, T8, A7-1, and A7-10 mature and collected seeds that were able to germinate in the presence of kanamycin. Subsequent characterization of these seedlings showed that about 16% of them did not produce an hpt-specific band. Our data thus showed that simultaneous induction of two DSBs is possible via transient expression of ZFN in the target plants and that the hpt gene used to detect gfp-free plants can be bred out in successive generations. This notion was further supported by the molecular characterization of randomly selected hygromycin-resistant T8 and T23 seedlings, which showed that these plants carry an independent nontargeted hpt-specific band (Fig. 5). This analysis further suggested that the gfp fragment that had been released from the genome of plant T8 did not reincorporate somewhere else inside the plant genome.
Figure 3.
NHEJ-mediated gene deletion in plants. A, Structure of the target gene region and outcome of gfp deletion events. The locations and names of primers used for PCR amplification of the targeting events are shown. gfp, GFP-encoding gene; P1, CaMV dual 35S promoter; QQR, QQR ZFN recognition site; QQR*, altered QQR ZFN recognition site; T1, CaMV 35S terminator. B, PCR analysis of wild-type (WT), parental (PR), and gfp-deleted (T8) tobacco genomes. M, DNA ladder. C, Confocal microscopy analysis of leaf tissue from parent transgenic (PR) and gfp-deleted targeted (T8) plants. GFP expression is in green, and plastid autofluorescence is in red. Images are single confocal sections. Bars = 40 μm. [See online article for color version of this figure.]
Figure 4.

Sequencing data of gfp deletion events in tobacco (T8) and Arabidopsis (A3-2, A3-2b, A7-1, A7-6, A7-10, and A20). The QQR ZFN-binding sites, on the top strand of the parent target sequences, are in blue. gfp, GFP coding sequence; P, CaMV dual 35S promoter; T, CaMV 35S terminator. [See online article for color version of this figure.]
Figure 5.
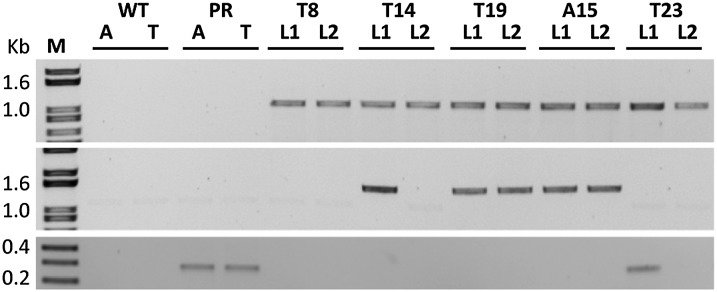
Molecular analysis of hygromycin-resistant offspring. Randomly integrated (top panel) and targeted (center panel) donor hpt and acceptor gfp (bottom panel) were amplified in offspring of mutated (T23), deleted (T8), and replaced (T14, T19, and A15) plants. A, Arabidopsis; L1 and L2, two randomly selected plants; M, DNA ladder; PR, parent plants; T, tobacco; WT, wild-type plants.
QQR ZFN-Mediated Gene Replacement
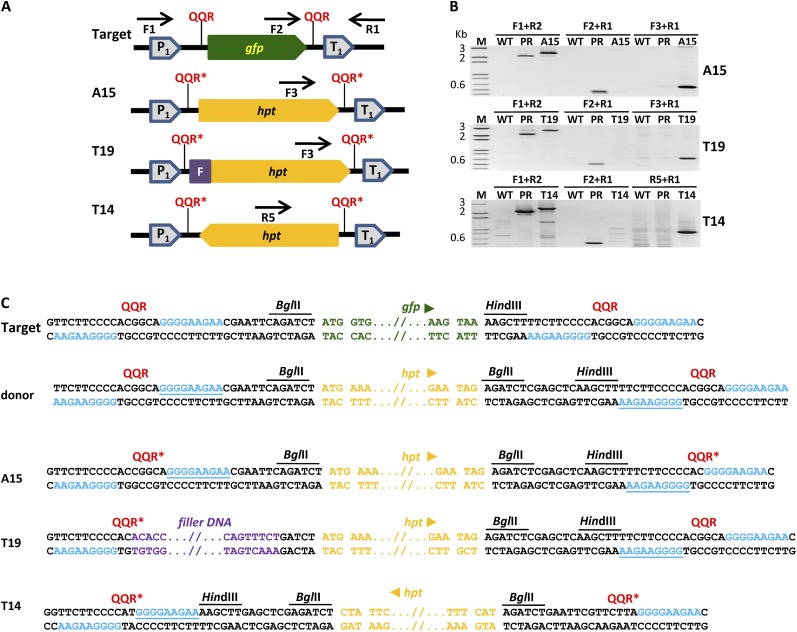
Our ultimate goal was to achieve NHEJ-mediated gene replacement in plant cells. Indeed, we were able to recover plants in which the GFP coding sequence had been replaced by hpt. The general structure of the gene-replacement events in Arabidopsis line A15 and tobacco lines T19 and T14 is shown in Figure 6A. Also shown are the pairs of primers used for the amplification of whole or distinct regions of the gfp (or hpt) expression cassettes. PCR amplification of total DNA from line A15, using primers that flanked the replacement gene (i.e. primers F1 and R1), produced a distinct 2.5-kb-long DNA fragment that was longer than the 2.1-kb DNA fragment in the parental line (Fig. 6B, top panel). Our data indicated that the GFP coding sequence had been replaced by a DNA fragment of a larger size, presumably hpt. Further analyses revealed a gfp-specific band in the parental line but not the A15 line (Fig. 6B, top panel, primers F2 and R1) and an hpt-specific band in A15 but not in the parental line (Fig. 6B, top panel, primers F3 and R1). Sequence analysis confirmed the nature of the replacement event and demonstrated that, indeed, the GFP coding sequence had been replaced by a complete hygromycin resistance-encoding gene (Fig. 6C). A similar replacement event was also observed in tobacco line T19, as detected by PCR analysis (Fig. 6B, center panel). However, the Mr of the PCR-amplified band obtained by using primers F1 and R1 or F3 and R1 was slightly higher than that obtained in Arabidopsis line A15. This was due to the insertion of a filler DNA of unknown origin at the hpt 5′ end, as detected by sequencing analysis (Fig. 6C). Tobacco regenerant T14 exhibited a poor growth rate and slow rooting under hygromycin-selection conditions. Further analysis revealed that T14 contained a replacement event in which the hpt gene was inserted in reverse orientation with respect to the target cassette’s constitutive promoter (Fig. 6A). The molecular structure of this replacement event was confirmed by PCR analysis (Fig. 6B, bottom panel) and DNA sequencing (Fig. 6C).
Figure 6.
NHEJ-mediated gene replacement in plants. A, Structure of the target gene region and outcome of gfp replacement events in Arabidopsis (A15) and tobacco (T19 and T14) plants. The locations and names of the primers used for PCR amplification of the replacement events are shown. F, Filler DNA; gfp, GFP-encoding gene; hpt, hygromycin resistance-encoding gene; P, dual CaMV 35S promoter; QQR, QQR ZFN recognition site; QQR*, altered QQR ZFN recognition site; T, CaMV 35S terminator. B, PCR analysis of wild-type (WT), parental (PR), and gfp replacement events in A15, T19, and T14 plants. M, DNA ladder. C, Sequencing data of gfp replacement events. QQR ZFN-binding sites are in blue, QQR ZFN-binding sites derived from donor DNA are underlined, gfp and hpt sequences are in green and orange, respectively, and filler DNA sequence is in purple. [See online article for color version of this figure.]
Phenotypical Analyses and Sexual Transmission of Replacement Events
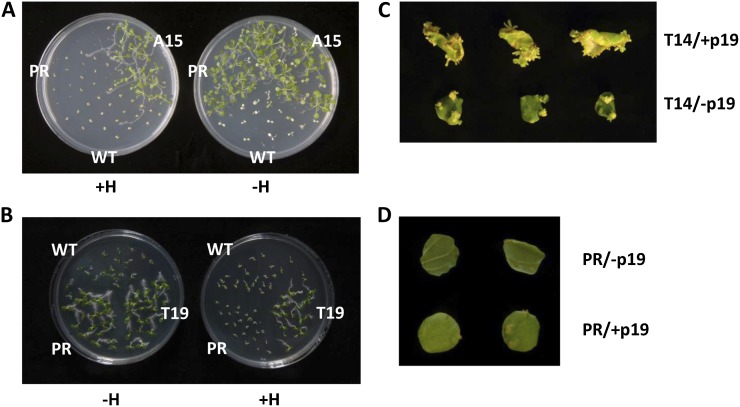
We further characterized the A15 and T19 replacement events, and as expected, neither the Arabidopsis seedling A15 nor the tobacco regenerant T19 displayed GFP fluorescence (data not shown). To characterize the stability of the gene-replacement events, we let A15 and T19 mature and set seed and tested the seedlings’ ability to germinate in the presence of hygromycin or kanamycin. Seeds derived from the parental lines germinated well on kanamycin but not on dual-selection germination medium (i.e. kanamycin and hygromycin selection media), while seeds of A15 and T19 germinated well on both selection media (Fig. 7, A and B). A15 and T19 seedlings segregated at a ratio of 3:1 when cultured on kanamycin, hygromycin, or dual kanamycin and hygromycin germination medium. These observations further support the notion that A15 and T19 were derived from a single replacement event and not from putative trapping of the hygromycin resistance gene promoter.
Figure 7.
Phenotypical analyses and sexual transmission of replacement events. A, Phenotypical analysis of a gfp replacement event in Arabidopsis. The wild type (WT), a parental line (PR), and offspring of targeted line A15 were grown on germination medium supplemented with kanamycin and with (+H) or without (−H) hygromycin. B, Phenotypical analysis of a gfp replacement event in tobacco. The wild type, a parental line, and offspring of targeted line T19 were grown on germination medium supplemented with kanamycin and with or without hygromycin. C, Shoot formation from leaf discs of targeted tobacco line T14 (with or without inoculation with p19-encoding T-DNA) on hygromycin-containing selection medium. D, Shoot formation from leaf discs of a parental tobacco line (with or without inoculation with p19-encoding T-DNA) on hygromycin-containing selection medium. [See online article for color version of this figure.]
Tobacco regenerant T14, which contained a reversed hpt gene insertion, exhibited a poor growth rate and slow rooting under hygromycin selection conditions. It is clear that the reverse orientation of the hpt gene did not promote the regeneration of T14. We thus suggested that a second hpt gene was activated by random integration into the T14 genome and that this plant’s poor growth rate might be linked to the silencing of this second, potentially functional, gene copy. To test this hypothesis, we determined whether application of the RNA-silencing suppressor p19 (Voinnet et al., 2003; Chapman et al., 2004) would reverse the putative silencing of hpt in T14. Indeed, as shown in Figure 7C, shoot regeneration under hygromycin-selective conditions was suppressed in T14 leaf discs, while inoculation of T14 leaf discs with a T-DNA molecule overexpressing p19 abolished the suppression, resulting in the efficient regeneration of hygromycin-resistant shoots. No shoot regeneration was observed when parental hygromycin-sensitive leaves were inoculated with p19-overexpressing T-DNA molecules (Fig. 7D).
We next tested the heredity and stability of the gene replacement in eight randomly selected hygromycin-resistant seedlings from A15 and T19 plants. As expected, no GFP expression could be detected in these seedlings, and sequencing analysis of these plants confirmed that they retained the replacement DNA structure and sequences of the parents from which they were derived (Fig. 8). Molecular characterization of randomly selected hygromycin-resistant seedlings of T19 and A15 demonstrated that the hpt coding sequence is linked to the target site and that the gfp fragment that was released from the genome is no longer present in these lines (Fig. 5). The existence of a second hpt fragment in the genome of T14 lines was evident from the recovery of hpt-resistant, but nontargeted, plants (i.e. line T14-L2; Fig. 5), which were also gfp free. Furthermore, PCR analysis of some of these seedlings failed to detect traces of the QQR ZFN expression cassette, indicating that the latter did not integrate into the genomes of A15 and T19 plants.
Figure 8.
Molecular analysis of randomly selected hygromycin-resistant Arabidopsis (A15) and tobacco (T19) seedlings. QQR ZFN-binding sites are in blue, QQR ZFN-binding sites derived from donor DNA are underlined, hpt (hygromycin resistance-encoding gene) sequences are in orange, and filler DNA sequence is in purple. [See online article for color version of this figure.]
DISCUSSION
In previous studies, HR-mediated gene-replacement strategies have been the preferred mode of gene replacement in plants (Wright et al., 2005; Shukla et al., 2009; Townsend et al., 2009; de Pater et al., 2013), while NHEJ-mediated repair of ZFN-induced DSBs has been limited to targeted mutagenesis and transgene removal (Lloyd et al., 2005; Morton et al., 2006; Maeder et al., 2008; de Pater et al., 2009; Tovkach et al., 2009; Osakabe et al., 2010; Petolino et al., 2010; Zhang et al., 2010). Our findings show that the NHEJ DNA-repair pathway can be harnessed for gene replacement in plant species. Our study may thus provide an alternative to HR-mediated gene-replacement strategies in plant cells.
There can be several advantages to using NHEJ-mediated gene replacement in plant cells. First, data show that T-DNA molecules preferentially integrate in plant cells via NHEJ and not HR (Tzfira et al., 2004, 2012; Ziemienowicz et al., 2008; Weinthal et al., 2010). Since NHEJ is most likely the major DNA-repair pathway in a wide range of somatic plant cells (which are primary targets in many existing plant transformation protocols), relying on the plant’s natural DNA-repair pathway may be preferable to stimulating the HR DNA-repair machinery.
Second, studies have shown that dsT-DNAs integrate into genomic DSBs (Salomon and Puchta, 1998; Chilton and Que, 2003; Tzfira et al., 2003) and that DSBs may act as “hot spots” for T-DNA integration (Tzfira et al., 2003). It has thus been suggested that dsT-DNA, and not T-strands, is the preferred substrate for the integration machinery (Tzfira et al., 2003, 2004; Ziemienowicz et al., 2008). Since ZFNs digest double-stranded DNA, our finding shows that, indeed, it is the dsT-DNA that integrates into the plant genome. While the possibility of T-strands integrating into undamaged plant genome regions cannot be ruled out, the use of dsT-DNA as the substrate for integration, in particular into DSBs, may enhance the efficiency of gene replacement in plants.
Third, studies in yeast have revealed that the host DNA-repair machinery, and not just the T-DNA sequence, greatly determines the route by which T-DNA molecules integrate into the transformed cell’s genome (van Attikum et al., 2001; van Attikum and Hooykaas, 2003). More specifically, it was shown that KU70, a key determinant of the NHEJ repair pathway, is essential for NHEJ-mediated T-DNA integration (van Attikum et al., 2001), while Rad52, a determinant of the HR machinery, is required for HR-mediated T-DNA integration (van Attikum and Hooykaas, 2003). Studies have also shown that KU80 is an essential protein for T-DNA integration in plants (Li et al., 2005). KU80, which may function together with KU70 (Tamura et al., 2002), is likely to act by recognizing dsT-DNA molecules, mostly likely as broken genomic fragments, and directing them to integration into naturally occurring DSBs via NEHJ and not HR. These observations suggest that, while under specific conditions, DSBs may stimulate the HR machinery, most of the T-DNA molecules may still be destined to integration via NHEJ and not HR. In this study, we used a single, well-characterized QQR ZFN. Naturally, the application of our strategy for native sequences will require the design and assembly of two ZFNs, or some other type of artificially designed restriction enzyme, such as transcription activator-like effector-based nucleases (Cermak et al., 2011; Li et al., 2012), and designed endonucleases (Gao et al., 2010).
Fourth, NHEJ-mediated gene replacement is particularly powerful for gene stacking and for genome editing of multitransgene arrays in transgenic plants, which are some of the major challenges today for plant biotechnologists (Halpin, 2005; Lyznik and Dress, 2008; Naqvi et al., 2010; Que et al., 2010). More specifically, the assembly of multigene arrays by traditional cloning or recombination systems typically leaves repetitive sequences within the DNA molecule (Dafny-Yelin and Tzfira, 2007), which can include recombination sites, traces of multicloning sites, and even reparative promoters and terminator regions: these may hinder the use of HR-mediated recombination methods for their targeting. Our approach may facilitate the engineering of multigene arrays by using pairs of ZFNs, which are designed to flank the target genes and to be used in conjunction with matching donor DNA molecules for NHEJ-mediated replacement. Alternatively, multitransgene arrays can be designed to carry unique sets of quasipalindromic ZFN recognition sites at key locations across the transforming vector. The latter can then be used for NHEJ-mediated gene replacement and deletion via the transient expression of a single ZFN monomer and may facilitate the stacking of several genes in the same chromosomal locus. Indeed, we have previously shown that ZFNs can be used for the construction of multigene plant transformation vectors (Zeevi et al., 2008, 2010, 2012). In these vectors, each ZFN expression cassette is flanked by quasipalindromic ZFN recognition sites, and the ZFNs that were used for in vivo assembly of the transformation vector (Zeevi et al., 2008, 2010, 2012) can potentially be used for the replacement and deletion of specific expression cassettes in transgenic plants. Thus, the NHEJ DNA-repair pathway provides an important complementary, and sometimes even superior, strategy to HR-mediated gene replacement in plant cells.
de Pater et al. (2009) reported that the rate of site-specific mutagenesis in ZFN-overexpressing transgenic plants is much lower than expected, and Osakabe et al. (2010) discovered that the ZFN-mediated site-specific mutagenesis rate is similar in wild-type and KU80-deficient plants. However, Osakabe et al. (2010), showed that the lack of KU80, which functions in protecting exposed DNA ends in DSBs, leads to larger deletions at the ZFN-induced break site as compared with shorter mutations in the wild-type plants. It was thus suggested that an alternative NHEJ pathway might exist in Arabidopsis (Osakabe et al., 2010). Our site-specific mutagenesis rate (19% and 26% for Arabidopsis and tobacco, respectively, when calculated for two individual mutations per event; Table I) was similar to those reported by others (Lloyd et al., 2005; de Pater et al., 2009; Tovkach et al., 2009). We observed that in several cases, both target sites had been mutated, yet the target sequence was not released (Fig. 2). While we cannot rule out the possibility that both sites were sequentially (as described previously by Gong and Golic [2003]), and not simultaneously, targeted, it is also possible that the GFP coding region was physically released and quickly reintegrated by the NHEJ machinery into the break site. Interestingly, one of the sites in line T6-23 was true to type. Although it could be that this site was simply not targeted, reconstruction of the QQR ZFN site in line T19 (Fig. 6) clearly indicated that NHEJ-mediated gene targeting can occur via simple ligation between the donor and acceptor DNA. Indeed, it was previously shown that T-DNA molecules engineered to carry a site for the rare-cutting restriction enzyme I-SceI are digested and integrated, via a ligation-like mechanism, into I-SceI-induced genomic DSBs (Tzfira et al., 2003).
Transgene removal from transgenic plants is an important technology for molecular breeding and for gene containment (Moon et al., 2010), and it is frequently achieved using novel vectors and site-specific recombination systems (Darbani et al., 2007). Transgene deletion by ZFNs was recently reported by Petolino et al. (2010), who flanked a transgene with the ZFN CCR5 (Perez et al., 2008). In their report, Petolino et al. (2010) used a transgenic strategy in which transgenic target plants were crossed with CCR5 ZFN-overexpressing plants, giving rise to targeted offspring. Here, we expand the use of transgene delivery to the important model plant Arabidopsis with an efficiency of 28% (calculated for actual deletion events; Table I). Furthermore, we demonstrate that deploying transient ZFN expression in standard transformation procedures (i.e. leaf disc transformation and flower dip) is sufficient for the recovery of deletion events. A clear advantage of our strategy is that it can potentially be implemented in existing transformation and regeneration procedures. In addition, it does not rely on crosses between target and ZFN-expressing plants, which may be difficult or even impossible to implement in clonally propagated plant species. It should be noted that while promoter trapping of the hpt selection gene under stringent conditions enabled the selection of GFP-free transgenic lines, this gene could be bred out in successive generations. Other, more direct screening and selection methods potentially can be deployed for the detection and selection of mutants with target gene deletions. No less important, our data support the vision of Moon et al. (2010) of using ZFNs for transgene biocontaminant strategies in somatic cells.
We recovered three NEHJ gene-replacement events (with rates of 4% and 6% for Arabidopsis and tobacco, respectively; Table I) and further characterized them both molecularly and phenotypically. An approximately 200-bp filler DNA was inserted at the hpt 5′ end in line T19. This filler DNA showed no similarity to any other known DNA. Scrambled filler DNA is often observed at the integration sites of plasmid and T-DNA molecules in transgenic plants (Gorbunova and Levy, 1997; Kumar and Fladung, 2000; Makarevitch et al., 2003; Windels et al., 2003). In their report, Gorbunova and Levy (1997) proposed that the origin of scrambled DNA may involve the invasion of ectopic templates and multiple template switches during DNA integration. Those authors also suggested that genomic DSB repair may involve extensive end degradation and suggested that capturing “floating” DNA may not be a major process by which filler DNA is formed. Our observations, however, suggest that while the formation of scrambled DNA at the hpt 5′ end may indeed derive from multiple template switches, the accurate ligation-like integration at the 3′ end indicates that the final substrate for integration may have been a double-stranded filler-DNA/T-DNA floating molecule that was captured by the DSB. A better understanding of the DNA-repair machinery and the mechanisms by which genomic DSBs are repaired is required to fully exploit the NHEJ pathway for genome editing in plant cells.
MATERIALS AND METHODS
DNA Constructs
To produce the acceptor DNA, the pSAT6-2xQQR plasmid, in which two quasipalindromic QQR ZFN recognition sites flank unique BglII and HindIII sites, was constructed as follows. The first QQR ZFN recognition site was cloned by annealing primers 5′-CCCAAGCTTTTCTTCCCCACGGCAGGGGAAGAACTCGAGCGG-3′ and 5′-CCGCTCGAGTTCTTCCCCTGCCGTGGGGAAGAAAAGCTTGGG-3′ and cloning the resultant DNA as a HindIII/XhoI fragment into HindIII/SalI sites of pSAT6-MCS (Tzfira et al., 2005), producing pSAT6-1xQQR. The second QQR ZFN recognition site was cloned by annealing the 5′-CATGCCATGGGTTCTTCCCCACGGCAGGGGAAGAACGAATTCAGATCTTC-3′ and 5′-GAAGATCTGAATTCGTTCTTCCCCTGCCGTGGGGAAGAACCCATGGCATG-3′ primers and cloning the resultant DNA as an NcoI/BglII fragment into the same sites of pSAT6-1xQQR, producing pSAT6-2xQQR. pSAT6[QQR-TS.GFP.QQR-TS], in which the GFP coding sequence is flanked by two quasipalindromic QQR ZFN recognition sites, was constructed by PCR amplification of the enhanced GFP coding sequence from pSAT6-enhanced GFP-N1 (Tzfira et al., 2005) using 5′-GAAGATCTATGAGCAAGGGCGAGGAGCTG-3′ and 5′-CCCAAGCTTTTACTTGTACAGCTCGTCCATG-3′ and cloning the DNA as a BglII/HindIII fragment into the same sites of pSAT6-2xQQR. The constitutive GFP expression cassette was transferred as a PI-PspI fragment from pSAT6[QQR-TS.GFP.QQR-TS] into pRCS2[ocs.nptII] (Chung et al., 2005), producing the acceptor plant transformation binary vector pRCS2[ocs.nptII][QQR-TS.GFP.QQR-TS].
The donor hygromycin-encoding gene was constructed by PCR amplification of the hpt-coding sequence from pRCS2[ocs-hpt] (Chung et al., 2005) using 5′-GGAAGATCTATGAAAAAGCCTGAACTCAC-3′ and 5′-GGAAGATCTCTATTCCTTTGCCCTCGGACG-3′ and cloning the resultant DNA fragment into the BglII site of pSAT6-2xQQR, producing pSAT6[QQR-TS.hpt.QQR-TS]. QQR-TS.hpt.QQR-TS was then transferred as an NcoI-NotI fragment into the same sites of pAUX3133, producing pAUX3133[QQR-TS.hpt.QQR-TS]. The promoterless hpt gene was next transferred as a PI-PspI fragment from pAUX3133[QQR-TS.hpt.QQR-TS] into pRCS2 (Tzfira et al., 2005) or into pRCS2[QQR-ZFN] (Tovkach et al., 2009), producing the donor plant transformation binary vectors pRCS2[QQR-TS.HYG.QQR-TS] and pRCS2[QQR-ZFN][QQR-TS.HYG.QQR-TS], respectively.
Transgenic Plants
The pRCS2[ocs.nptII][QQR-TS.GFP.QQR-TS] binary vector carrying a target GFP expression cassette and a functional plant kanamycin resistance gene was used for the transformation of tobacco (Nicotiana tabacum) ‘Turk’ and Arabidopsis (Arabidopsis thaliana) using the standard leaf disc (Guterman et al., 2006) and floral dip (Clough and Bent 1998) transformation methods, respectively. Plants were selected on kanamycin selection medium. The same methods were used for retransformation experiments, using pRCS2[QQR-ZFN][QQR-TS.HYG.QQR-TS] or a mixture of pRCS2[QQR-ZFN] and pRCS2[QQR-TS.HYG.QQR-TS]. Screening for putative targeting events was performed on dual kanamycin and hygromycin selection medium.
Analysis of Gene-Targeting Events
Gene-targeting events were detected by a combination of confocal laser-scanning microscopy to image GFP expression, PCR analysis, and DNA sequencing. For molecular analysis of targeting events, total DNA was isolated from hygromycin-resistant plants according to Bernatzky and Tanksley (1986) and was subjected to PCR amplification using a combination of primers (F1, 5′-GTCAGTGTCCGCATAAAGAACC-3′; R1, 5′-GTAGATGTTAACATCCAACGTCGC-3′; F2, 5′-CATGGTCCTGCTGGAGTTCGTG-3′; F3, 5′-GTATATGCTCCGCATTGGTCTTGACC-3′; R5, 5′-ATACACATGGGGATCAGCAATCG-3′).
Acknowledgments
We thank Dr. G.N. Drews for the gift of pHS::QQR-QEQ/2300.
Glossary
- HR
homologous recombination
- NHEJ
nonhomologous end joining
- DSB
double-strand break
- ZFN
zinc finger nuclease
- T-DNA
transferred DNA
- dsT-DNA
double-stranded transferred DNA
- CaMV
cauliflower mosaic virus
References
- Banta LM, Montenegro M (2008) Agrobacterium and plant biotechnology. In T Tzfira, V Citovsky, eds, Agrobacterium Springer, New York, pp 73–147 [Google Scholar]
- Baribault H, Kemler R. (1989) Embryonic stem cell culture and gene targeting in transgenic mice. Mol Biol Med 6: 481–492 [PubMed] [Google Scholar]
- Bernatzky R, Tanksley SD. (1986) Genetics of actin-related sequences in tomato. Theor Appl Genet 72: 314–321 [DOI] [PubMed] [Google Scholar]
- Beumer K, Bhattacharyya G, Bibikova M, Trautman JK, Carroll D. (2006) Efficient gene targeting in Drosophila with zinc-finger nucleases. Genetics 172: 2391–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britt AB, May GD. (2003) Re-engineering plant gene targeting. Trends Plant Sci 8: 90–95 [DOI] [PubMed] [Google Scholar]
- Cai CQ, Doyon Y, Ainley WM, Miller JC, Dekelver RC, Moehle EA, Rock JM, Lee YL, Garrison R, Schulenberg L, et al. (2009) Targeted transgene integration in plant cells using designed zinc finger nucleases. Plant Mol Biol 69: 699–709 [DOI] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. (2011) Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 39: e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman EJ, Prokhnevsky AI, Gopinath K, Dolja VV, Carrington JC. (2004) Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev 18: 1179–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilton M-D, Que Q. (2003) Targeted integration of T-DNA into the tobacco genome at double-stranded breaks: new insights on the mechanism of T-DNA integration. Plant Physiol 133: 956–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung SM, Frankman EL, Tzfira T. (2005) A versatile vector system for multiple gene expression in plants. Trends Plant Sci 10: 357–361 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Curtin SJ, Zhang F, Sander JD, Haun WJ, Starker C, Baltes NJ, Reyon D, Dahlborg EJ, Goodwin MJ, Coffman AP, et al. (2011) Targeted mutagenesis of duplicated genes in soybean with zinc-finger nucleases. Plant Physiol 156: 466–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dafny-Yelin M, Tzfira T. (2007) Delivery of multiple transgenes to plant cells. Plant Physiol 145: 1118–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darbani B, Eimanifar A, Stewart CN, Jr, Camargo WN. (2007) Methods to produce marker-free transgenic plants. Biotechnol J 2: 83–90 [DOI] [PubMed] [Google Scholar]
- de Pater S, Neuteboom LW, Pinas JE, Hooykaas PJ, van der Zaal BJ. (2009) ZFN-induced mutagenesis and gene-targeting in Arabidopsis through Agrobacterium-mediated floral dip transformation. Plant Biotechnol J 7: 821–835 [DOI] [PubMed] [Google Scholar]
- de Pater S, Pinas JE, Hooykaas PJ, van der Zaal BJ. (2013) ZFN-mediated gene targeting of the Arabidopsis protoporphyrinogen oxidase gene through Agrobacterium-mediated floral dip transformation. Plant Biotechnol J (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ, et al. (2008) Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol 26: 702–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Even-Faitelson L, Samach A, Melamed-Bessudo C, Avivi-Ragolsky N, Levy AA. (2011) Localized egg-cell expression of effector proteins for targeted modification of the Arabidopsis genome. Plant J 68: 929–937 [DOI] [PubMed] [Google Scholar]
- Gao H, Smith J, Yang M, Jones S, Djukanovic V, Nicholson MG, West A, Bidney D, Falco SC, Jantz D, et al. (2010) Heritable targeted mutagenesis in maize using a designed endonuclease. Plant J 61: 176–187 [DOI] [PubMed] [Google Scholar]
- Gong WJ, Golic KG. (2003) Ends-out, or replacement, gene targeting in Drosophila. Proc Natl Acad Sci USA 100: 2556–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova V, Levy AA. (1997) Non-homologous DNA end joining in plant cells is associated with deletions and filler DNA insertions. Nucleic Acids Res 25: 4650–4657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guterman I, Masci T, Chen X, Negre F, Pichersky E, Dudareva N, Weiss D, Vainstein A. (2006) Generation of phenylpropanoid pathway-derived volatiles in transgenic plants: rose alcohol acetyltransferase produces phenylethyl acetate and benzyl acetate in petunia flowers. Plant Mol Biol 60: 555–563 [DOI] [PubMed] [Google Scholar]
- Hall B, Limaye A, Kulkarni AB (2009) Overview: generation of gene knockout mice. Curr Protoc Cell Biol 44: 19.12.1–19.12.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpin C. (2005) Gene stacking in transgenic plants: the challenge for 21st century plant biotechnology. Plant Biotechnol J 3: 141–155 [DOI] [PubMed] [Google Scholar]
- Hanin M, Paszkowski J. (2003) Plant genome modification by homologous recombination. Curr Opin Plant Biol 6: 157–162 [DOI] [PubMed] [Google Scholar]
- Hanin M, Volrath S, Bogucki A, Briker M, Ward E, Paszkowski J. (2001) Gene targeting in Arabidopsis. Plant J 28: 671–677 [DOI] [PubMed] [Google Scholar]
- Kempin SA, Liljegren SJ, Block LM, Rounsley SD, Yanofsky MF, Lam E. (1997) Targeted disruption in Arabidopsis. Nature 389: 802–803 [DOI] [PubMed] [Google Scholar]
- Kumar S, Allen GC, Thompson WF. (2006) Gene targeting in plants: fingers on the move. Trends Plant Sci 11: 159–161 [DOI] [PubMed] [Google Scholar]
- Kumar S, Fladung M. (2000) Transgene repeats in aspen: molecular characterisation suggests simultaneous integration of independent T-DNAs into receptive hotspots in the host genome. Mol Gen Genet 264: 20–28 [DOI] [PubMed] [Google Scholar]
- Laible G, Alonso-González L. (2009) Gene targeting from laboratory to livestock: current status and emerging concepts. Biotechnol J 4: 1278–1292 [DOI] [PubMed] [Google Scholar]
- Lee HJ, Kim E, Kim JS. (2010) Targeted chromosomal deletions in human cells using zinc finger nucleases. Genome Res 20: 81–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Vaidya M, White C, Vainstein A, Citovsky V, Tzfira T. (2005) Involvement of KU80 in T-DNA integration in plant cells. Proc Natl Acad Sci USA 102: 19231–19236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Liu B, Spalding MH, Weeks DP, Yang B. (2012) High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat Biotechnol 30: 390–392 [DOI] [PubMed] [Google Scholar]
- Lindsey K, Topping JF, Muskett PR, Wei W, Horne KL. (1998) Dissecting embryonic and seedling morphogenesis in Arabidopsis by promoter trap insertional mutagenesis. Symp Soc Exp Biol 51: 1–10 [PubMed] [Google Scholar]
- Liu PQ, Chan EM, Cost GJ, Zhang L, Wang J, Miller JC, Guschin DY, Reik A, Holmes MC, Mott JE, et al. (2010) Generation of a triple-gene knockout mammalian cell line using engineered zinc-finger nucleases. Biotechnol Bioeng 106: 97–105 [DOI] [PubMed] [Google Scholar]
- Lloyd A, Plaisier CL, Carroll D, Drews GN. (2005) Targeted mutagenesis using zinc-finger nucleases in Arabidopsis. Proc Natl Acad Sci USA 102: 2232–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, Ando D, Urnov FD, Galli C, Gregory PD, et al. (2007) Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol 25: 1298–1306 [DOI] [PubMed] [Google Scholar]
- Lyznik LA, Dress V. (2008) Gene targeting for chromosome engineering applications in eukaryotic cells. Recent Pat Biotechnol 2: 94–106 [DOI] [PubMed] [Google Scholar]
- Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, et al. (2008) Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell 31: 294–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevitch I, Svitashev SK, Somers DA. (2003) Complete sequence analysis of transgene loci from plants transformed via microprojectile bombardment. Plant Mol Biol 52: 421–432 [DOI] [PubMed] [Google Scholar]
- Marton I, Zuker A, Shklarman E, Zeevi V, Tovkach A, Roffe S, Ovadis M, Tzfira T, Vainstein A. (2010) Nontransgenic genome modification in plant cells. Plant Physiol 154: 1079–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. (2008) Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol 26: 695–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehle EA, Rock JM, Lee YL, Jouvenot Y, DeKelver RC, Gregory PD, Urnov FD, Holmes MC. (2007) Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc Natl Acad Sci USA 104: 3055–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon HS, Li Y, Stewart CN., Jr (2010) Keeping the genie in the bottle: transgene biocontainment by excision in pollen. Trends Biotechnol 28: 3–8 [DOI] [PubMed] [Google Scholar]
- Morton J, Davis MW, Jorgensen EM, Carroll D. (2006) Induction and repair of zinc-finger nuclease-targeted double-strand breaks in Caenorhabditis elegans somatic cells. Proc Natl Acad Sci USA 103: 16370–16375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi S, Farré G, Sanahuja G, Capell T, Zhu C, Christou P. (2010) When more is better: multigene engineering in plants. Trends Plant Sci 15: 48–56 [DOI] [PubMed] [Google Scholar]
- Orlando SJ, Santiago Y, DeKelver RC, Freyvert Y, Boydston EA, Moehle EA, Choi VM, Gopalan SM, Lou JF, Li J, et al. (2010) Zinc-finger nuclease-driven targeted integration into mammalian genomes using donors with limited chromosomal homology. Nucleic Acids Res 38: e152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe K, Osakabe Y, Toki S. (2010) Site-directed mutagenesis in Arabidopsis using custom-designed zinc finger nucleases. Proc Natl Acad Sci USA 107: 12034–12039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL, et al. (2008) Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol 26: 808–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petolino JF, Worden A, Curlee K, Connell J, Strange Moynahan TL, Larsen C, Russell S. (2010) Zinc finger nuclease-mediated transgene deletion. Plant Mol Biol 73: 617–628 [DOI] [PubMed] [Google Scholar]
- Porteus MH. (2006) Mammalian gene targeting with designed zinc finger nucleases. Mol Ther 13: 438–446 [DOI] [PubMed] [Google Scholar]
- Porteus MH. (2009) Plant biotechnology: zinc fingers on target. Nature 459: 337–338 [DOI] [PubMed] [Google Scholar]
- Puchta H. (2002) Gene replacement by homologous recombination in plants. Plant Mol Biol 48: 173–182 [PubMed] [Google Scholar]
- Que Q, Chilton MD, de Fontes CM, He C, Nuccio M, Zhu T, Wu Y, Chen JS, Shi L. (2010) Trait stacking in transgenic crops: challenges and opportunities. GM Crops 1: 220–229 [DOI] [PubMed] [Google Scholar]
- Ray A, Langer M. (2002) Homologous recombination: ends as the means. Trends Plant Sci 7: 435–440 [DOI] [PubMed] [Google Scholar]
- Reiss B. (2003) Homologous recombination and gene targeting in plant cells. Int Rev Cytol 228: 85–139 [DOI] [PubMed] [Google Scholar]
- Salomon S, Puchta H. (1998) Capture of genomic and T-DNA sequences during double-strand break repair in somatic plant cells. EMBO J 17: 6086–6095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer S, Davis RW. (1979) Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci USA 76: 4951–4955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaked H, Melamed-Bessudo C, Levy AA. (2005) High-frequency gene targeting in Arabidopsis plants expressing the yeast RAD54 gene. Proc Natl Acad Sci USA 102: 12265–12269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla VK, Doyon Y, Miller JC, DeKelver RC, Moehle EA, Worden SE, Mitchell JC, Arnold NL, Gopalan S, Meng X, et al. (2009) Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature 459: 437–441 [DOI] [PubMed] [Google Scholar]
- Smith J, Bibikova M, Whitby FG, Reddy AR, Chandrasegaran S, Carroll D. (2000) Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic Acids Res 28: 3361–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söllü C, Pars K, Cornu TI, Thibodeau-Beganny S, Maeder ML, Joung JK, Heilbronn R, Cathomen T. (2010) Autonomous zinc-finger nuclease pairs for targeted chromosomal deletion. Nucleic Acids Res 38: 8269–8276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Adachi Y, Chiba K, Oguchi K, Takahashi H. (2002) Identification of Ku70 and Ku80 homologues in Arabidopsis thaliana: evidence for a role in the repair of DNA double-strand breaks. Plant J 29: 771–781 [DOI] [PubMed] [Google Scholar]
- Tenzen T, Zembowicz F, Cowan CA. (2010) Genome modification in human embryonic stem cells. J Cell Physiol 222: 278–281 [DOI] [PubMed] [Google Scholar]
- Terada R, Johzuka-Hisatomi Y, Saitoh M, Asao H, Iida S. (2007) Gene targeting by homologous recombination as a biotechnological tool for rice functional genomics. Plant Physiol 144: 846–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada R, Urawa H, Inagaki Y, Tsugane K, Iida S. (2002) Efficient gene targeting by homologous recombination in rice. Nat Biotechnol 20: 1030–1034 [DOI] [PubMed] [Google Scholar]
- Tovkach A, Zeevi V, Tzfira T. (2009) A toolbox and procedural notes for characterizing novel zinc finger nucleases for genome editing in plant cells. Plant J 57: 747–757 [DOI] [PubMed] [Google Scholar]
- Tovkach A, Zeevi V, Tzfira T. (2010) Validation and expression of ZFNs and in plant cells. Methods Mol Cell Biol 649: 315–336 [DOI] [PubMed] [Google Scholar]
- Townsend JA, Wright DA, Winfrey RJ, Fu F, Maeder ML, Joung JK, Voytas DF. (2009) High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature 459: 442–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T, Frankman LR, Vaidya M, Citovsky V. (2003) Site-specific integration of Agrobacterium tumefaciens T-DNA via double-stranded intermediates. Plant Physiol 133: 1011–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T, Li J, Lacroix B, Citovsky V. (2004) Agrobacterium T-DNA integration: molecules and models. Trends Genet 20: 375–383 [DOI] [PubMed] [Google Scholar]
- Tzfira T, Tian G-W, Lacroix B, Vyas S, Li J, Leitner-Dagan Y, Krichevsky A, Taylor T, Vainstein A, Citovsky V. (2005) pSAT vectors: a modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol Biol 57: 503–516 [DOI] [PubMed] [Google Scholar]
- Tzfira T, Weinthal D, Marton I, Zeevi V, Zuker A, Vainstein A. (2012) Genome modifications in plant cells by custom-made restriction enzymes. Plant Biotechnol J 10: 373–389 [DOI] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. (2005) Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435: 646–651 [DOI] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. (2010) Genome editing with engineered zinc finger nucleases. Nat Rev Genet 11: 636–646 [DOI] [PubMed] [Google Scholar]
- van Attikum H, Bundock P, Hooykaas PJJ. (2001) Non-homologous end-joining proteins are required for Agrobacterium T-DNA integration. EMBO J 20: 6550–6558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H, Hooykaas PJJ. (2003) Genetic requirements for the targeted integration of Agrobacterium T-DNA in Saccharomyces cerevisiae. Nucleic Acids Res 31: 826–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJ, Bellen HJ. (2005) Emerging technologies for gene manipulation in Drosophila melanogaster. Nat Rev Genet 6: 167–178 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D. (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Weinthal D, Tovkach A, Zeevi V, Tzfira T. (2010) Genome editing in plant cells by zinc finger nucleases. Trends Plant Sci 15: 308–321 [DOI] [PubMed] [Google Scholar]
- Windels P, De Buck S, Van Bockstaele E, De Loose M, Depicker A. (2003) T-DNA integration in Arabidopsis chromosomes: presence and origin of filler DNA sequences. Plant Physiol 133: 2061–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright DA, Townsend JA, Winfrey RJ, Jr, Irwin PA, Rajagopal J, Lonosky PM, Hall BD, Jondle MD, Voytas DF. (2005) High-frequency homologous recombination in plants mediated by zinc-finger nucleases. Plant J 44: 693–705 [DOI] [PubMed] [Google Scholar]
- Zeevi V, Liang Z, Arieli U, Tzfira T. (2012) Zinc finger nuclease and homing endonuclease-mediated assembly of multigene plant transformation vectors. Plant Physiol 158: 132–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevi V, Tovkach A, Tzfira T. (2008) Increasing cloning possibilities using artificial zinc finger nucleases. Proc Natl Acad Sci USA 105: 12785–12790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevi V, Tovkach A, Tzfira T. (2010) Artificial zinc finger nucleases for DNA cloning. Methods Mol Biol 649: 209–225 [DOI] [PubMed] [Google Scholar]
- Zhang F, Maeder ML, Unger-Wallace E, Hoshaw JP, Reyon D, Christian M, Li X, Pierick CJ, Dobbs D, Peterson T, et al. (2010) High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proc Natl Acad Sci USA 107: 12028–12033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemienowicz A, Tzfira T, Hohn B (2008) Mechanisms of T-DNA integration. In T Tzfira, V Citovsky, eds, Agrobacterium Springer, New York, pp 395–440 [Google Scholar]