A common histone modification code is associated with light induction and cell type-specific gene expression.
Abstract
C4 photosynthesis evolved more than 60 times independently in different plant lineages. Each time, multiple genes were recruited into C4 metabolism. The corresponding promoters acquired new regulatory features such as high expression, light induction, or cell type-specific expression in mesophyll or bundle sheath cells. We have previously shown that histone modifications contribute to the regulation of the model C4 phosphoenolpyruvate carboxylase (C4-Pepc) promoter in maize (Zea mays). We here tested the light- and cell type-specific responses of three selected histone acetylations and two histone methylations on five additional C4 genes (C4-Ca, C4-Ppdk, C4-Me, C4-Pepck, and C4-RbcS2) in maize. Histone acetylation and nucleosome occupancy assays indicated extended promoter regions with regulatory upstream regions more than 1,000 bp from the transcription initiation site for most of these genes. Despite any detectable homology of the promoters on the primary sequence level, histone modification patterns were highly coregulated. Specifically, H3K9ac was regulated by illumination, whereas H3K4me3 was regulated in a cell type-specific manner. We further compared histone modifications on the C4-Pepc and C4-Me genes from maize and the homologous genes from sorghum (Sorghum bicolor) and Setaria italica. Whereas sorghum and maize share a common C4 origin, C4 metabolism evolved independently in S. italica. The distribution of histone modifications over the promoters differed between the species, but differential regulation of light-induced histone acetylation and cell type-specific histone methylation were evident in all three species. We propose that a preexisting histone code was recruited into C4 promoter control during the evolution of C4 metabolism.
The current best estimate for the minimal number of independent evolutionary origins of C4 photosynthesis is 62. Thus, C4 photosynthesis belongs to the most prominent examples of parallel or convergent evolution in nature (Sage et al., 2012). C4 plants established a carbon pump that transports CO2 from mesophyll (M) to bundle sheath (B) cells, where Rubisco and the Calvin cycle are active (von Caemmerer and Furbank, 2003). One major aspect of C4 evolution was the recruitment of preexisting genes to encode the enzymes of the C4 pathway (Hibberd and Covshoff, 2010). Key enzymes for C4 photosynthesis in M cells are carbonic anhydrase (C4-Ca), phosphoenolpyruvate carboxylase (C4-Pepc), and pyruvate phosphate dikinase (C4-Ppdk). Conversely, in B cells, a decarboxylase such as NAD(P)-malic enzyme (C4-Me) and/or phosphoenolpyruvate carboxykinase (C4-Pepck) and Rubisco (RbcS for the genes encoding the small subunit) are required at high levels (Langdale, 2011). Maize (Zea mays) makes use of both a chloroplastic NADP-Me and a cytosolic Pepck for C4 acid decarboxylation (Furumoto et al., 1999; Wingler et al., 1999). During the recruitment process, C4 genes acquired new regulatory features: the genes show much higher expression than their C3 counterparts (Ku et al., 1996), they are activated in response to light (Sheen and Bogorad, 1987; Sheen, 1999), and their activity is often modulated by additional metabolic stimuli such as nitrate availability (Sugiharto et al., 1992) or sugar accumulation (Sheen, 1990). Much of this regulation takes place on the level of promoter activity (Sheen, 1999; Hibberd and Covshoff, 2010). Furthermore, C4 proteins show selective accumulation in M or B cells. Information for cell type specificity is encoded in promoter sequences (Gowik et al., 2004; Akyildiz et al., 2007), in untranslated transcript regions (Patel and Berry, 2008; Kajala et al., 2012), and/or in the coding sequence (Brown et al., 2011), depending on the C4 gene and the species. There is evidence that some genes in C3 plants were predisposed to their recruitment into the C4 pathway, because their C3 orthologs show aspects of C4 regulation already in the C3 plant (Brown et al., 2010; Kajala et al., 2012). Also, C4 genes are often correctly regulated when overexpressed in a C3 host, indicating that the relevant trans-acting factors are available in C3 plants (Ku et al., 1996; Engelmann et al., 2003).
Research in recent years has highlighted the role of chromatin structure and modification in the control of transcription. The primary repeat unit of chromatin is the nucleosome particle that is formed by DNA wound around a protein body. This protein body consists of two copies each of the histone proteins H2A, H2B, H3, and H4 (Kouzarides, 2007). Chromatin is a passive barrier for transcription and other DNA-associated biochemical processes (Kingston and Narlikar, 1999). Two major mechanisms that facilitate the transcription of chromatin have been identified. On the one hand, chromatin-remodeling complexes are recruited to specific chromatin domains and alter the mobility of nucleosomes in an ATP-dependent manner (Mellor, 2005). This results in differences in nucleosome occupancy (NO) at specific DNA positions. In both yeast and humans, active gene promoters are characterized by low NO (Lee et al., 2004; Nishida et al., 2006). On the other hand, histones can be posttranslationally modified in multiple ways (Bannister and Kouzarides, 2011; Tan et al., 2011). Using genome-wide correlation analyses, some of these modifications have been associated with transcriptionally active or inactive chromatin domains (Bernstein et al., 2007; Zhang, 2008). The best studied modification of histones is probably Lys acetylation. Multiple residues on the N-terminal tails of histones H3 and H4 can be acetylated. Histone acetylation is tightly correlated with gene transcription (Pokholok et al., 2005; Wang et al., 2009a). The activating properties of histone acetylation may be explained by two nonmutually exclusive hypotheses: either they are due to neutralization of the interaction of the positively charged Lys side chains with the negatively charged DNA, resulting in the mitigation of histone-DNA interaction (charge neutralization model; Dion et al., 2005; Henikoff and Shilatifard, 2011), or they are due to the provision of binding sites for transcription factors and other proteins that specifically bind to acetylated histones (histone code model; Berger, 2007; Hassan et al., 2007; Nelissen et al., 2007). The latter model also implies that histone modifications can be used for the storage of information about developmental and environmental cues on the promoters.
A second prominent modification of histones is Lys methylation. Again, multiple Lys residues on histones are prone to methylation (Sims et al., 2003). Depending on the Lys that is methylated, these modifications are either found in actively transcribed chromatin regions or in heterochromatic nontranscribed regions (Sims et al., 2003; Martin and Zhang, 2005). Furthermore, the terminal amino group of the Lys side chain can be monomethylated, dimethylated, or trimethylated, and this also affects the interpretation of the modification. Methylated Lys residues are recognized by specific protein domains (Kim et al., 2006) and, thus, can recruit proteins that, in turn, activate or repress transcription. The most prominent example of histone methylation associated with active genes is the trimethylation of Lys-4 on histone H3 (H3K4me3) at the start of the transcribed gene region (Santos-Rosa et al., 2002; Heintzman et al., 2007; Wang et al., 2009a).
We have previously studied information storage by histone modifications on the promoter of C4-Pepc in maize. Our studies revealed that the acetylation of Lys-9 on histone H3 (H3K9ac) and Lys-5 on histone H4 (H4K5ac) on the promoter is controlled by illumination (Offermann et al., 2006, 2008). The modifications were even set on inactive gene copies as long as the light stimulus was received, but they were removed in the dark. Other Lys residues such as H3K18 were constitutively acetylated on C4-Pepc (Offermann et al., 2008). Conversely, H3K4me3 on C4-Pepc was not affected by the light stimulus but was controlled by a cell type-specific signal specifically in M cells. In B cells, H3K4me2 was found instead (Danker et al., 2008). Here, we tested whether this control of specific modifications is conserved on other C4 genes in maize and on the orthologous C4-Pepc and C4-Me genes of the C4 grasses sorghum (Sorghum bicolor) and Setaria italica. Whereas maize and sorghum share a common C4 origin, S. italica evolved C4 metabolism independently (Christin et al., 2009a; Sage et al., 2012) and separated from the maize/sorghum lineage approximately 25 million years ago (Vicentini et al., 2008). Our data reveal a close similarity of the histone code within maize C4 genes and, for at least two key genes, across the species. This suggests that this code was recruited into C4 in two independent C4 lineages.
RESULTS
Chromatin Modification Profiles of C4 Genes in Maize
We used chromatin immunoprecipitation (ChIP) from illuminated leaves harvested 4 h after the onset of illumination to identify regions on C4 gene promoters in maize that show high H3K9ac or H3K4me3. C4 genes were identified from the genome sequence based on their homology to complementary DNAs (cDNAs) that had been shown before to encode C4-specific transcripts (see “Materials and Methods”; Supplemental Fig. S1). All modification data were standardized for the number of nucleosomes in the tested chromatin region (NO). NO was determined by precipitating chromatin with an antibody directed to an invariant domain of histone H3 and is shown in Supplemental Table S1. Y-scales always show the relative enrichment of acetylation or methylation at the respective site compared with the Actin1 housekeeping gene promoter (Haring et al., 2007; i.e. a value of 10 means that the modification is found 10 times more often on nucleosomes on the tested C4 gene promoter region than on nucleosomes on the Actin1 promoter [see “Materials and Methods”]).
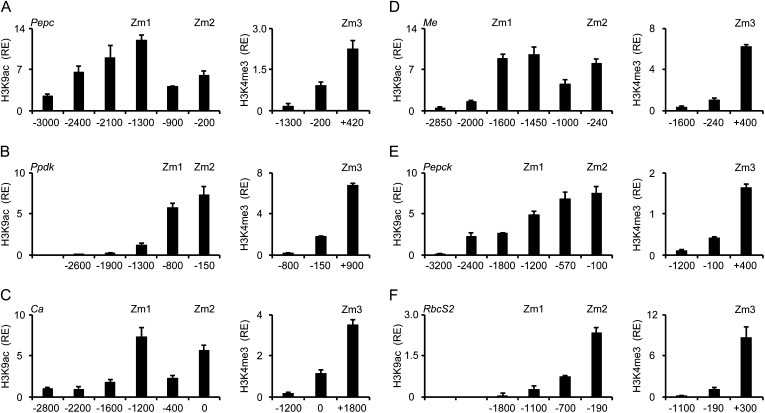
In the core promoter region (−200 relative to the transcription initiation site [TIS]) of the C4-Pepc promoter (Fig. 1A), we detected a 6-fold enrichment in acetylation compared with the Actin1 promoter. This number slightly decreased at position −900 and then increased to 12-fold enrichment at position −1,300. Further upstream, acetylation constantly decreased. For C4-Me (Fig. 1D), the situation was similar, with a peak of acetylation on the core promoter (−240) and a second peak further upstream (−1,450). Also, relative enrichment compared with Actin1 was similar on C4-Pepc and C4-Me. Therefore, for both genes, we selected a position on the upstream promoter (Zm1) and a position on the core promoter (Zm2) with high H3K9ac for the following analysis of light regulation (Fig. 2). These selected positions on the C4-Pepc and C4-Me promoters had already been shown to contain regulated chromatin modifications in previous studies (Offermann et al., 2006, 2008; Danker et al., 2008).
Figure 1.
Histone modification profiles of six C4 genes from maize. Amounts of chromatin precipitated with an antibody specific for H3K9ac or H3K4me3 in illuminated leaves (4L) are shown. Numbers on the x axis indicate bp positions relative to TIS. Positions chosen for further analyses are designated Zm1, Zm2, and Zm3. Values are presented as the relative enrichment (RE) of modifications per nucleosome over modifications per nucleosome found on maize Actin1. All data points are based on at least three independent experiments. Vertical lines indicate se.
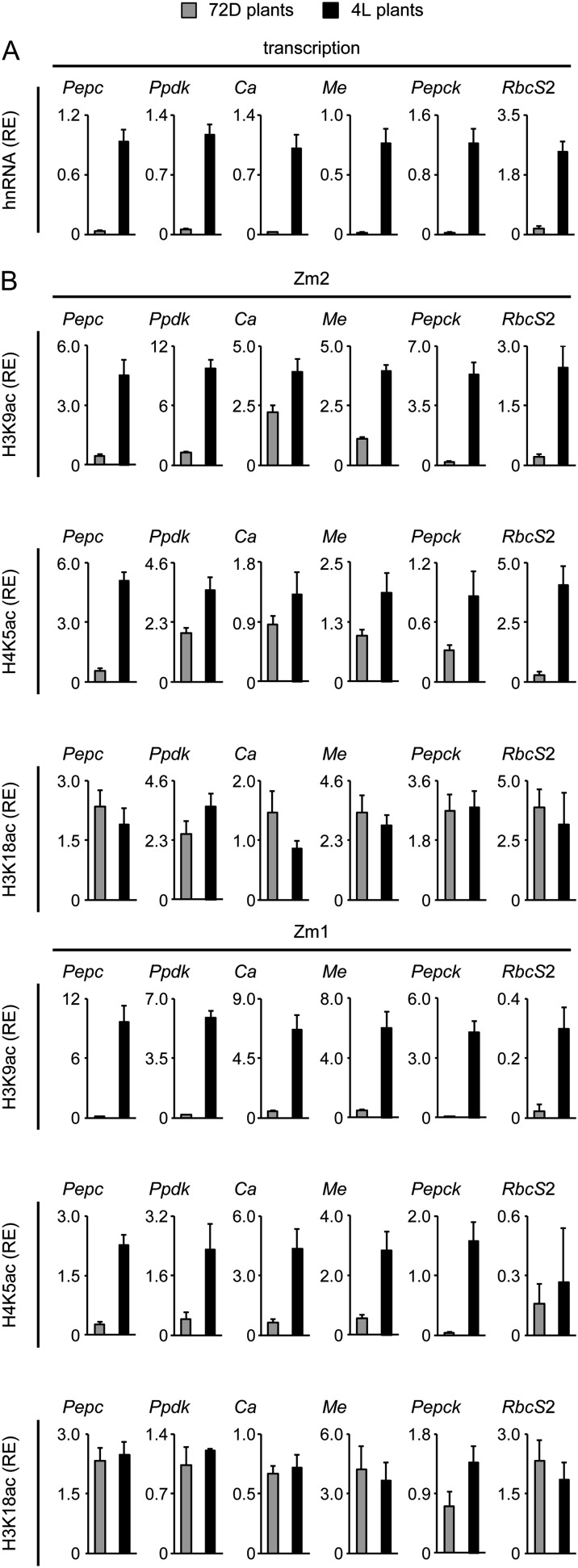
Figure 2.
Light regulation of transcription and histone acetylation on six C4 genes in maize. A, Relative quantification of hnRNA expression levels of the six C4 genes in leaves from plants that were exposed to 72 h of darkness (72D; gray columns) and in plants that were illuminated for 4 h (4L; black columns). Transcription is standardized for maize Actin1 expression (relative enrichment [RE]). hnRNA expression levels were determined by quantitative reverse transcription (RT)-PCR with a primer system specific for an intron (Supplemental Fig. S1). B, Light-dependent H3K9ac, H4K5ac, and H3K18ac on positions Zm1 and Zm2 of the six C4 genes. Values are presented as the relative enrichment of modifications per nucleosome over modifications per nucleosome found on the Actin1 promoter. All data points are based on at least four independent experiments. Vertical lines indicate se.
We further screened the promoters of the C4 genes C4-Ca, C4-Ppdk, C4-Pepck, and C4-RbcS2 with the aim to define comparable chromatin positions (i.e. a core promoter position and an upstream promoter position with high H3K9ac). On C4-Ppdk (Fig. 1B), we observed high H3K9ac on the core promoter (position −150) and at position −800 but a rapid decline at more upstream positions. Accordingly, Zm1 was positioned at −800 and Zm2 at −150. On C4-Ca (Fig. 1C), we detected high H3K9ac signals at position −1,200 (Zm1) and directly at the TIS (Zm2). We were unable to establish a functioning PCR system directly in the core promoter region of C4-Ca because of the very high GC content (more than 75% between −300 and 0) and multiple repetitive sequence elements; therefore, we had to use a PCR system centered on the TIS instead. On C4-Pepck (Fig. 1E), H3K9ac was highest on the core promoter (−100) and constantly declined toward the upstream promoter. Zm1 was defined at the most upstream position with at least 5-fold higher acetylation compared with Actin1 (−1,200), and Zm2 again was placed on the core promoter. On C4-RbcS2 (Fig. 1F), H3K9ac levels were generally lower. Maximum levels on the core promoter were only 2.3-fold higher compared with the Actin1 promoter. Moreover, H3K9ac rapidly declined toward more upstream promoter regions. In order to allow direct comparison with the other C4 genes, we decided to place positions Zm1 and Zm2 on C4-RbcS2 at −1,100 (the most upstream position with detectable H3K9ac) and −190 (highest acetylation), respectively.
H3K4 trimethylation (H3K4me3) is another chromatin mark of active genes that usually peaks in the 5′ region of the transcribed sequence (Santos-Rosa et al., 2002; Heintzman et al., 2007; Wang et al., 2009a). Therefore, we tested an additional position in the 5′ region of the transcribed sequence. This position is designated as Zm3 henceforth (for exact positions, see Supplemental Fig. S1). On all genes, H3K4me3 levels were clearly increased at Zm3 compared with the promoter positions (Fig. 1, right graphs). On C4-Pepc, C4-Ca, and C4-Pepck, significant relative enrichments of H3K4me3 were also detected at Zm2, whereas H3K4me3 levels were near background at the Zm2 position on the other genes. At Zm1, H3K4me3 signals were undetectable (C4-Ppdk, C4-Ca, C4-Me, C4-RbcS2) or very low (C4-Pepc, C4-Pepck). We selected the Zm3 position for further studies of histone methylation on the maize C4 genes.
Environmental and Developmental Regulation of C4 Gene Chromatin in Maize
A key feature of C4 gene regulation is gene induction by light. We compared promoter activity and histone acetylation at the Zm1 and Zm2 positions on the six maize C4 genes in response to illumination (Fig. 2). Beside H3K9ac, H4K5ac and H3K18ac were included, because these modifications showed specific responses to illumination on the C4-Pepc promoter before (Offermann et al., 2008). The amounts of heterogeneous nuclear RNA (hnRNA) derived from the genes were used as an approximation for promoter activity, because these primary transcripts are rapidly spliced after synthesis and do not accumulate (Elferink and Reiners, 1996; Delany, 2001; Wu et al., 2009). For all six C4 genes, hnRNA levels were low in plants that were exposed to 72 h of darkness (72D plants) and clearly increased in plants that were illuminated for 4 h (4L plants), indicating light-induced promoter activity (Fig. 2A).
For chromatin analyses, we used the same standardization methods as described for Figure 1. On the Zm2 site proximal to the TIS, H3K9ac was strongly induced (3- to 5-fold) on all genes after illumination, with the exception of C4-Ca (only 1.5-fold induction; Fig. 2B). H4K5ac at position Zm2 also responded positively to illumination on all tested genes, albeit to different extents. Light induction was 10-fold or more for C4-Pepc and C4-Rbcs2, approximately 2-fold for C4-Ppdk, C4-Me, and C4-Pepck, and less than 2-fold for C4-Ca. In contrast, H3K18ac at Zm2 remained largely unchanged in 72D plants compared with 4L plants on all C4 genes. At the more upstream-located Zm1 position, H3K9ac was strongly induced by illumination on all C4 genes. Likewise, H4K5ac showed a clear induction at Zm1 on all genes with the exception of RbcS2, where H4K5ac levels remained low in 4L plants. H3K18ac at the Zm1 position was unaffected by the light treatment on five of the six tested genes. A slight increase was only observed on C4-Pepck. Thus, with few exceptions, H3K9ac and H4K5ac responded positively to illumination at both the Zm1 and Zm2 promoter positions of the C4 genes, whereas H3K18ac remained unaffected by light.
A second hallmark of C4 gene regulation is cell type-specific expression of C4 genes in M or B cells. We compared hnRNA levels of the tested C4 genes in leaves and isolated B strands (Fig. 3A). A rapid mechanical isolation protocol at low temperatures was used for B cell preparation in order to minimize hnRNA turnover during the preparation (Jenkins and Boag, 1985; Bassett et al., 1988). We did not isolate M cells for this assay, because the preparation of M cell protoplasts from leaves is a lengthy procedure during which C4 genes are often strongly suppressed. If promoter activity would contribute to gene regulation, a clear depletion of M-specific transcripts in isolated B cells compared with total leaves would be expected in our assay. In contrast, B-specific transcripts are expected to be more abundant in B preparations than in total leaves (assuming an even number of B cells and M cells in leaves, a 2-fold increase in B cells compared with total leaves would be expected). Accordingly, hnRNA levels were clearly lower in B cells compared with total leaves for C4-Pepc, C4-Ppdk, and C4-Ca, whereas C4-Me, C4-Pepck, and RbcS2 transcripts were 1.8- to 2.6-fold enriched in B preparations.
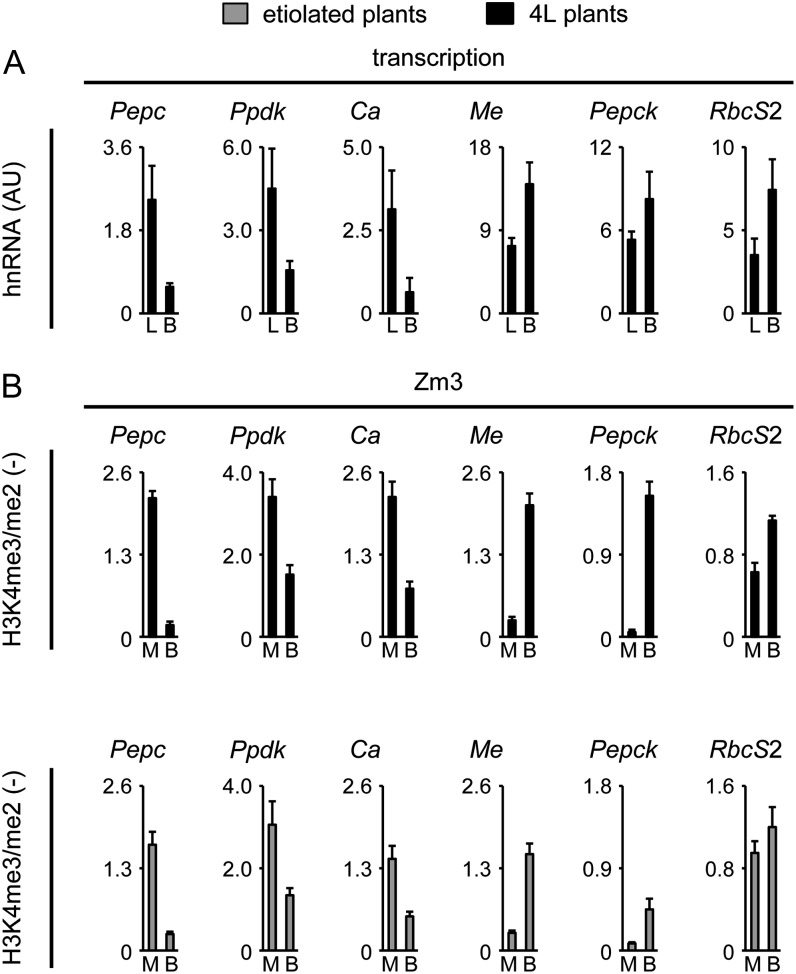
Figure 3.
Cell type-specific transcription and histone methylation on six C4 genes in maize. A, Quantification of hnRNA expression levels of the six C4 genes in leaves (L) and B cells (B) isolated from plants that were illuminated for 4 h (4L plants). Values are arbitrary units (AU) derived from a cDNA standard dilution series. hnRNA expression levels were determined by quantitative RT-PCR with a primer system specific for an intron (Supplemental Fig. S1). B, Ratio of the amount of chromatin precipitated with an antibody specific for H3K4me3 and H3K4me2 in isolated M or B cells. Black bars indicate data from illuminated plants (4L), whereas gray bars indicate data from etiolated plants. Ratios are without dimension. All data points are based on at least four independent experiments. Vertical lines indicate se.
We used chromatin isolated from M and B cells to test cell type-specific histone methylation on the C4 genes at position Zm3. Previous analyses indicated that methylation marks remain stable on M cell promoters during the preparation of protoplasts, even though C4 gene expression is suppressed (Danker et al., 2008). Figure 3B shows the ratio of H3K4me3 to H3K4me2 signals (H3K4me3/me2) at the Zm3 positions of the tested C4 genes in M and B cell chromatin. For C4-Pepc and C4-Me, we had shown before that high H3K4me3 signals and low H3K4me2 signals were established in the cell type where the particular gene can be activated by other stimuli (i.e. in M cells for C4-Pepc and in B cells for C4-Me). In each respective other cell type, we had observed the opposite pattern, with high H3K4me2 and low H3K4me3 instead. Consequently, an H3K4me3/me2 ratio of 1.9 was determined for C4-Pepc in chromatin from M cells, and this ratio dropped to 0.2 in chromatin from B cells. However, on C4-Me, the H3K4me3/me2 ratio was much higher in B cells than in M cells. For the other genes that showed preferential expression in either M cells (C4-Ca, C4-Ppdk) or B cells (C4-Pepck, C4-RbcS2), similar patterns were observed. C4-Ca and C4-Ppdk showed clearly higher H3K4me3/me2 ratios in chromatin from M cells than in chromatin from B cells, and C4-Pepck and C4-RbcS2 showed an opposite distribution of these histone modifications.
In order to determine whether this H3K4 methylation pattern was established independently of the transcription of C4 genes, we also measured H3K4me3/me2 levels in M and B chromatin isolated from etiolated leaves. In etiolated leaves, C4 genes are inactive because they never received a light stimulus (compare with Fig. 2). As shown in Figure 3B, H3K4me3/me2 ratios in M and B cells derived from etiolated leaves were very near those obtained from illuminated leaves. For M cell-specific genes, signals were higher in M cells than in B cells. B cell-specific genes showed the opposite pattern, although absolute levels were somewhat reduced. Only on C4-RbcS2 were ratios near 1 observed in chromatin from both M and B cells. Thus, cell type-specific histone methylation of H3K4 is found in maize on all tested C4 genes except C4-RbcS2. The specific role of the latter gene is revisited below in “Discussion.”
Histone Code at the C4-Pepc and C4-Me Genes in Independent C4 Lineages
The data described so far were obtained to study whether different C4 genes in maize showed similar chromatin modification profiles. We further analyzed whether homologous C4 genes in different species also showed similar chromatin modifications. To this end, we compared light-induced and cell type-specific chromatin marks on the C4-Pepc and C4-Me genes from the C4 model grasses sorghum and S. italica with those obtained from maize (Figs. 1–3). Identification of nearest gene homologs is described in “Materials and Methods” and Supplemental Figure S1. The three C4-Pepc genes and the three C4-Me genes showed very similar intron-exon organization in the coding regions (Supplemental Fig. S1) but no detectable sequence homology on the putative promoters. In order to delimit the maximum length of the promoters, we analyzed the distance to the next upstream gene. The next gene was predicted 30 kb upstream of the maize C4-Pepc gene, 100 kb upstream of the sorghum C4-Pepc gene, but only 7 kb upstream of the S. italica C4-Pepc gene. For C4-Me, distance to the next upstream gene was 13 kb (maize), 11 kb (sorghum), and 15 kb (S. italica).
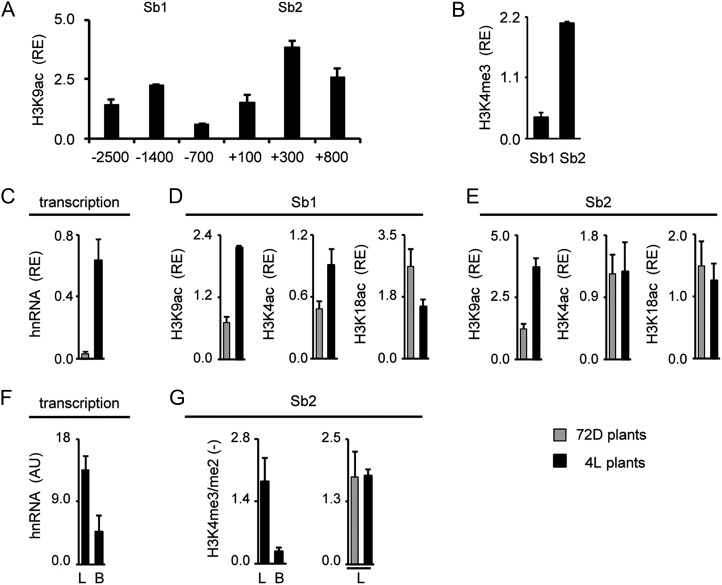
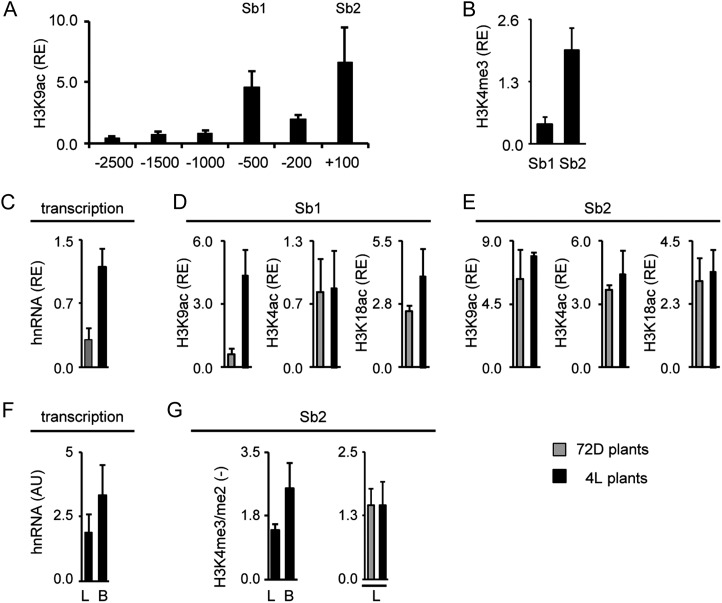
In order to define suitable gene positions for chromatin analyses, we again profiled H3K9ac over the putative promoter regions. Furthermore, we tested H3K4me3 levels at selected positions. Figure 4A shows the profile obtained for sorghum C4-Pepc. H3K9ac peaked at an upstream promoter position (−1,400; Sb1). Unlike in maize, a second peak was not observed in the core promoter (−100) but rather at the start of the transcribed region (+300; Sb2). At this position, also high H3K4me3 levels were observed (Fig. 4B). Positions Sb1 and Sb2 were chosen for the analysis of light-dependent histone acetylation (Fig. 4, C–E); in addition, Sb2 was chosen for the analysis of cell type-specific histone methylation (Fig. 4, F and G).
Figure 4.
Histone modification profile of the C4-Pepc gene from sorghum. A and B, Amounts of chromatin precipitated with an antibody specific for H3K9ac and H3K4me3 in illuminated leaves (4L). Positions chosen for further analyses are designated Sb1 and Sb2. Values are presented as the relative enrichment (RE) of modifications per nucleosome over modifications per nucleosome found on sorghum Actin1. Numbers on the x axis indicate bp positions relative to TIS. C, Relative quantification of C4-Pepc hnRNA expression levels in sorghum leaves from plants that were exposed to 72 h of darkness (72D; gray columns) and in plants that were illuminated for 4 h (4L; black columns). Transcription is standardized for Actin1 expression (RE). hnRNA expression levels were determined by quantitative RT-PCR with a primer system specific for an intron (Supplemental Fig. S1). D and E, Light-dependent H3K9ac, H4K5ac, and H3K18ac on positions Sb1 and Sb2. Values are presented as the relative enrichment of modifications per nucleosome over modifications per nucleosome found on sorghum Actin1. F, Quantification of C4-Pepc hnRNA expression levels in sorghum leaves (L) and B cells (B) isolated from plants that were illuminated for 4 h (4L plants). Values are arbitrary units (AU) derived from a cDNA standard dilution series. hnRNA expression levels were determined by quantitative RT-PCR with a primer system specific for an intron (Supplemental Fig. S1). G, Ratio of the amount of chromatin precipitated with an antibody specific for H3K4me3 and H3K4me2 in leaves and B cells. Black bars indicate data from illuminated plants (4L), whereas gray bars indicate data from plants that were exposed to 72 h of darkness (72D). Ratios are without dimension. All data points are based on at least three independent experiments. Vertical lines indicate se.
As expected, sorghum C4-Pepc hnRNA levels were much higher in 4L than in 72D plants (Fig. 4C). On position Sb1, acetylation of H3K9 (3-fold) and H4K5 (2-fold) was enhanced by light, whereas H3K18ac was even slightly reduced in 4L compared with 72D plants (Fig. 4D). On Sb2, H3K9ac was 3-fold induced, but H4K5ac and H3K18ac remained unaffected by the light stimulus (Fig. 4E).
C4-Pepc hnRNA amounts in total leaves were about 3.5-fold higher than in isolated B strands (Fig. 4F). This is typical for genes that are preferentially transcribed in M cells (compare with Fig. 3). Unlike in maize, we analyzed cell type-specific histone methylation in sorghum by comparing methylation levels in total leaves and B cells, because we failed to prepare intact M cell protoplasts from sorghum leaves. H3K4me3/me2 ratios were more than 2-fold higher in total leaves compared with isolated B cells. In order to provide evidence that this H3K4 methylation pattern is established independent from the actual rate of transcription, we tested whether the H3K4me3/me2 ratio is different between chromatin from 4L and 72D leaves (Fig. 4H). No differences were detected, indicating that H3K4 methylations do not respond to the light stimulus or the rate of transcription.
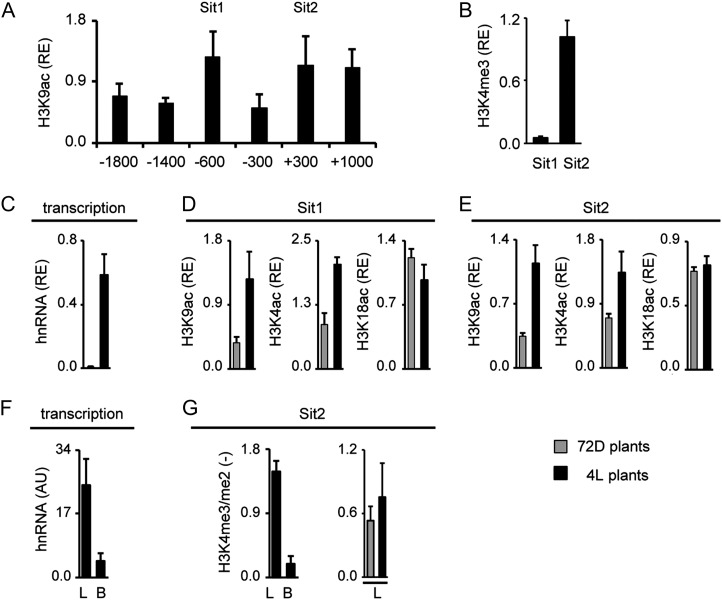
An analogous data set for S. italica C4-Pepc is shown in Figure 5. On the S. italica C4-Pepc promoter, acetylation peaked at positions −600 (Sit1) and +300 (Sit2; Fig. 5A). Again, core promoter acetylation (−300) was relatively low. At Sit2, but not Sit1, also high H3K4me3 levels were detected. hnRNA levels of S. italica C4-Pepc were strongly induced by light (Fig. 5C). This correlated with light-induced H3K9ac and H4K5ac at both tested gene positions. Again, H3K18ac remained unaffected by the light stimulus (Fig. 5, D and E). When comparing hnRNA levels in total leaves and B cells, 4-fold higher levels were observed in leaves, indicating cell type-specific transcription (Fig. 5F). Consistently, H3K4me3/me2 ratio was much higher in total leaves compared with B cells but was unaffected by light and, therefore, transcription rates (Fig. 5G). Thus, the sorghum and S. italica C4-Pepc genes showed clearly different distributions of histone acetylation over the promoters compared with maize, but regulation of transcription, histone acetylation, and H3K4 methylation were highly similar to maize C4-Pepc.
Figure 5.
Histone modification profile of the C4-Pepc gene from S. italica. A and B, Amount of chromatin precipitated with an antibody specific for H3K9ac and H3K4me3 in illuminated leaves (4L). Positions chosen for further analyses are designated Sit1 and Sit2. Values are presented as the relative enrichment (RE) of modifications per nucleosome over modifications per nucleosome found on S. italica Actin1. Numbers on the x axis indicate bp positions relative to TIS. C, Relative quantification of C4-Pepc hnRNA expression levels in S. italica leaves from plants that were exposed to 72 h of darkness (72D; gray columns) and in plants that were illuminated for 4 h (4L; black columns). Transcription is standardized for Actin1 expression (RE). hnRNA expression levels were determined by quantitative RT-PCR with a primer system specific for an intron (Supplemental Fig. S1). D and E, Light-dependent H3K9ac, H4K5ac, and H3K18ac on positions Sit1 and Sit2. Values are presented as the relative enrichment of modifications per nucleosome over modifications per nucleosome found on S. italica Actin1. F, Quantification of C4-Pepc hnRNA expression levels in S. italica leaves (L) and B cells (B) isolated from plants that were illuminated for 4 h (4L plants). Values are arbitrary units (AU) derived from a cDNA standard dilution series. hnRNA expression levels were determined by quantitative RT-PCR with a primer system specific for an intron (Supplemental Fig. S1). G, Ratio of the amount of chromatin precipitated with an antibody specific for H3K4me3 and H3K4me2 in leaves and B cells. Black bars indicate data from illuminated plants (4L), whereas gray bars indicate data from plants that were exposed to 72 h of darkness (72D). Ratios are without dimension. All data points are based on at least three independent experiments. Vertical lines indicate se.
In order to substantiate these observations, we also compared the transcription and chromatin regulation of the C4-Me genes from the three species. Data sets are organized identically to the C4-Pepc data in Figures 4 and 5. On sorghum C4-Me, H3K9ac peaked at positions −500 (Sb1) and +100 (Sb2; Fig. 6A). High H3K4me3 levels were again found at Sb2 (Fig. 6B). Promoter activity was induced 3-fold by light. On position Sb1, H3K9ac increased 6-fold after illumination, whereas H4K5ac did not change. On position Sb2, all tested histone acetylations were unaffected by illumination. Figure 6, F and G, shows the results regarding cell type specificity of this gene. The hnRNA for C4-Me accumulated to 1.8-fold higher levels in B cells compared with total leaves (Fig. 6F). A theoretical increase of 2-fold would be expected in these assays for genes that show B cell-specific transcription or modification, assuming even numbers of M and B cells in a leaf (compare with Fig. 3). The H3K4me3/me2 ratio was also 1.8-fold higher in B cells (Fig. 6G). Very similar H3K4me3/me2 ratios were recorded from leaves of 4L and 72D plants (Fig. 6G). Thus, H3K4 methylation levels were unaffected by light.
Figure 6.
Histone modification profile of the C4-Me gene from sorghum. A and B, Amount of chromatin precipitated with an antibody specific for H3K9ac and H3K4me3 in illuminated leaves (4L). Positions chosen for further analyses are designated Sb1 and Sb2. Values are presented as the relative enrichment (RE) of modifications per nucleosome over modifications per nucleosome found on sorghum Actin1. Numbers on the x axis indicate bp positions relative to TIS. C, Relative quantification of C4-Me hnRNA expression levels in sorghum leaves from plants that were exposed to 72 h of darkness (72D; gray columns) and in plants that were illuminated for 4 h (4L; black columns). Transcription is standardized for Actin1 expression (RE). hnRNA expression levels were determined by quantitative RT-PCR with a primer system specific for an intron (Supplemental Fig. S1). D and E, Light-dependent acetylation of H3K9ac, H4K5ac, and H3K18ac on positions Sb1 and Sb2. Values are presented as the relative enrichment of modifications per nucleosome over modifications per nucleosome found on sorghum Actin1. F, Quantification of C4-Me hnRNA expression levels in sorghum leaves (L) and B cells (B) isolated from plants that were illuminated for 4 h (4L plants). Values are arbitrary units (AU) derived from a cDNA standard dilution series. hnRNA expression levels were determined by quantitative RT-PCR with a primer system specific for an intron (Supplemental Fig. S1). G, Ratio of the amount of chromatin precipitated with an antibody specific for H3K4me3 and H3K4me2 in leaves and B cells. Black bars indicate data from illuminated plants (4L), whereas gray bars indicate data from plants that were exposed to 72 h darkness (72D). Ratios are without dimension. All data points are based on at least three independent experiments. Vertical lines indicate se.
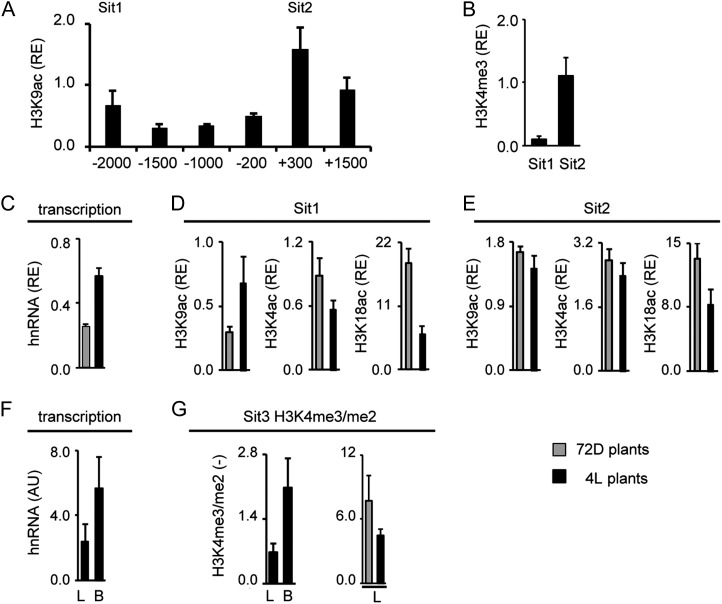
On S. italica C4-Me, the highest H3K9ac signals were observed at position +300 at the start of the transcribed sequence (Sit2). Acetylation declined toward the upstream promoter, but a second peak was detected at position −2,000 (Sit1; Fig. 7A). Comparable H3K9ac levels were also found at an even more upstream position (−2,500; data not shown). As for the other genes, high H3K4me3 signals were only detected at position Sit2 (Fig. 7B). A 2-fold increase in S. italica C4-Me gene transcription was induced by light. This was accompanied by a more than 2-fold increase in H3K9 acetylation at the more upstream Sit1 position. H4K5ac remained largely unaffected and H3K18ac was down-regulated by light at this position. At position Sit2 at the start of the transcribed sequence, H3K9ac and H4K5ac remained unaffected by the light stimulus, whereas H3K18ac was again down-regulated after illumination. When comparing B cells with total leaves, C4-Me hnRNA levels were 2.3-fold higher in B cells (Fig. 7F) and the H3H4me3/me2 ratio was 2.9-fold higher (Fig. 7G). Again, the H3K4me3/me2 ratio remained largely unaffected by light (Fig. 7G).
Figure 7.
Histone modification profile of the C4-Me gene from S. italica. A and B, Amount of chromatin precipitated with an antibody specific for H3K9ac and H3K4me3 in illuminated leaves (4L). Positions chosen for further analyses are designated Sit1 and Sit2. Values are presented as the relative enrichment (RE) of modifications per nucleosome over modifications per nucleosome found on S. italica Actin1. Numbers on the x axis indicate bp positions relative to TIS. C, Relative quantification of C4-Me hnRNA expression levels in S. italica leaves from plants that were exposed to 72 h of darkness (72D; gray columns) and in plants that were illuminated for 4 h (4L; black columns). Transcription is standardized for Actin1 expression (RE). hnRNA expression levels were determined by quantitative RT-PCR with a primer system specific for an intron (Supplemental Fig. S1). D and E, Light-dependent acetylation of H3K9ac, H4K5ac, and H3K18ac on positions Sit1 and Sit2. Values are presented as the relative enrichment of modifications per nucleosome over modifications per nucleosome found on S. italica Actin1. F, Quantification of C4-Me hnRNA expression levels in S. italica leaves (L) and B cells (B) isolated from plants that were illuminated for 4 h (4L plants). Values are arbitrary units (AU) derived from a cDNA standard dilution series. hnRNA expression levels were determined by quantitative RT-PCR with a primer system specific for an intron (Supplemental Fig. S1). G, Ratio of the amount of chromatin precipitated with an antibody specific for H3K4me3 and H3K4me2 in leaves and B cells. Black bars indicate data from illuminated plants (4L), whereas gray bars indicate data from plants that were exposed to 72 h of darkness (72D). Ratios are without dimension. All data points are based on at least three independent experiments. Vertical lines indicate se.
DISCUSSION
We wanted to analyze promoter histone modifications on C4 genes in maize. The definition of promoters in eukaryotic genomes is complicated, because of the large genome size and the resulting long distance to the next upstream gene that can be used to define maximal promoter size. Indeed, the next upstream gene was annotated between 5 kb and more than 100 kb distant from the predicted TIS of the maize genes analyzed (Goodstein et al., 2012; Supplemental Fig. S1). Promoter elements can act over a long distance, as described before for the element controlling M-specific expression of C4-Pepc in Flaveria spp. (Gowik et al., 2004) or the upstream promoter region of Flowering Locus T in Arabidopsis (Adrian et al., 2010). Promoter-deletion studies were instrumental in identifying such functional elements. However, these deletion studies require the use of transgenes that randomly integrate into plant genomes (Francis and Spiker, 2005; Kim et al., 2007). Epigenetic traits such as NO and histone modification can be strongly affected by the transgene integration site (Yan and Boyd, 2006; Yamasaki et al., 2011; Yin et al., 2012). Transgenic promoter studies, therefore, are of limited value for the analysis of epigenetic mechanisms controlling promoters. Chromatin signatures have been used instead to identify functional promoter elements in humans (Heintzman et al., 2007; Müller-Tidow et al., 2010). Our analyses revealed that all maize C4 genes but RbcS2 had extended promoter regions enriched in acetylation more than 1 kb upstream of the TIS (Fig. 1). This often coincided with reduced NO (Supplemental Table S1). This pattern is unexpected, as H3K9ac peaks around the TIS on the average maize gene and acetylation in upstream promoter regions is usually low (Wang et al., 2009a). However, the functional significance of these regions is suggested by the strong reaction of upstream promoter histone acetylation to illumination and, thus, gene transcription (Fig. 2). Moreover, the upstream promoter region of C4-Pepc also contains DNA methylation sites that are regulated by light, further supporting the involvement of these regions in gene regulation (Tolley et al., 2012). We cannot discriminate in this assay whether the increase in acetylation on the upstream promoter is necessary for transcriptional activation or just accompanies this process. However, we have shown before for C4-Pepc that upstream promoter acetylation can be induced even when gene activation is suppressed, supporting the autonomous regulation of histone acetylation in this promoter region (Offermann et al., 2006, 2008).
The distribution of acetylation over the C4-Pepc and C4-Me promoters was strikingly different in S. italica and sorghum compared with maize. The highest acetylation was detected in the 5′ part of the transcribed region in these species. Localization of the second acetylation peak was highly variable, with positions between −500 (sorghum C4-Me) and −2,000 (S. italica C4-Me) relative to the TIS (Figs. 4A–7A). One obvious explanation for the lack of core promoter acetylation on some of the genes is the clearly smaller genome size of S. italica (490 Mb; Doust et al., 2009) and sorghum (730 Mb; Paterson et al., 2009) compared with maize (2,300 Mb; Schnable et al., 2009), which might limit promoter sizes. Indeed, on S. italica C4-Pepc, the gene with the shortest distance to the next upstream gene, the upstream acetylation peak was already found at position −600.
Independent of the variable positions of acetylation peaks, light regulation of acetylation at these peak positions was highly similar when comparing the three C4 grasses. All C4-Pepc and all C4-Me genes showed clear light regulation of H3K9ac at least at one of the tested positions, mostly at the more upstream position. Light regulation of H4K5ac was evident on the C4-Pepc genes but not on C4-Me genes in sorghum and S. italica. The lack of H4K5ac regulation correlates with a rather weak light induction of C4-Me transcription in these species (only 2- to 3-fold light induction for C4-Me compared with 20- to 60-fold for C4-Pepc), suggesting that transcription levels might contribute to the degree of modification of this site. Remarkably, H3K18ac was never induced by light on any of the C4 genes but remained unchanged or even declined after illumination. Thus, the positive regulation of selected acetylation sites, most notably H3K9ac, by light is a common feature of the histone code on C4 genes in maize, sorghum, and S. italica. These results are in line with other observations in plants. Earley et al. (2007) reported that histone H4 is progressively acetylated from the inner modification Lys-16 to the N-terminal modification Lys-5. When yeast Lys residues on the N-terminal tail of H4 were replaced by Arg residues, mimicking unacetylated Lys residues, the inner Lys→Arg-16 mutation affected gene expression differently from all the other mutations, which rather showed additive effects on gene expression (Dion et al., 2005). Thus, acetylations on more C-terminal residues of the histone tails might play different roles than on N-terminal residues. However, in a genome-wide analysis of changes in histone modifications during deetiolation in Arabidopsis, the outer H3K9ac and the inner H3K27ac showed a high degree of coregulation (Charron et al., 2009). More chromatin analyses in dynamic, and not static, configurations (Roudier et al., 2009) are required to analyze whether the light-induced acetylation pattern observed on C4 genes can be generalized for all genes that respond to illumination.
H3K4me3 was always weak on the upstream promoter, but strong signals were obtained at the start of the transcribed region. Surprisingly, H3K4me3 did not respond to the light stimulus either on the six maize C4 genes or on C4-Pepc or C4-Me in sorghum and S. italica (Figs. 3–7). The observed pattern is unexpected, because C4 gene activity is very low in etiolated plants and plants exposed to prolonged darkness (Fig. 2) and H3K4me3 is frequently used as the key epigenetic indicator of active genes (Santos-Rosa et al., 2002; Heintzman et al., 2007; Wang et al., 2009a). Our previous analyses had shown that high H3K4me3 (and low H3K4me2) were found on C4-Pepc in M cells and on C4-Me in B cells, suggesting a function in the establishment of cell type specificity (Danker et al., 2008). The data presented here strongly support this hypothesis, because all the C4 genes in maize, sorghum, and S. italica showed cell type-specific but light-independent regulation of H3K4 methylation (Figs. 3–7). Developmentally regulated H3K4 trimethylation in one of the two photosynthetic cell types, therefore, constitutes a second element of the common histone code in leaves of C4 grasses.
Promoter histone modifications rather contribute to gene regulation on the transcriptional level than on the posttranscriptional level. In accordance with the histone methylation data described above, hnRNA accumulation patterns from leaves and isolated B cells recorded in this study suggested that the cell type specificity of C4 gene expression was controlled on the transcriptional level. However, different from our observation, it had been repeatedly shown for B cell-specific genes in maize such as RbcS and C4-Me that posttranscriptional mechanisms control the cell type specificity of gene expression (Viret et al., 1994; Sheen, 1999; Brown et al., 2011). On the other hand, a reporter construct containing the promoter and the 5′ untranslated region of maize C4-Me was exclusively expressed in B cells of transgenic maize plants, indicating an important role of the promoter in B cell specificity (Nomura et al., 2005). In this line, transient promoter-reporter assays with maize leaves suggested that an RbcS promoter element, together with sequence elements in the transcribed region, contributed to the repression of gene expression in M cells (Xu et al., 2001). The gene analyzed in the study by Xu et al. (2001) is identical to RbcS1 as defined by Ewing et al. (1998), whereas we studied RbcS2 here. However, both RbcS1 and RbcS2 showed B cell-specific expression in the latter study. Together, B-specific gene expression seems to be regulated simultaneously on multiple levels. H3K4 methylation might constitute a first level of this regulation that primes genes for possible activation by other stimuli.
Chromatin patterns on the maize RbcS2 gene investigated here differed in several respects from the other maize C4 genes analyzed. The acetylated promoter region was shorter than the acetylated region on the other promoters (Fig. 1), and the light response of acetylation was stronger at the core promoter position Zm2 than at Zm1 (Fig. 2). In addition, different from the other C4 genes, illumination contributed to the establishment of the histone methylation pattern on RbcS2 (Fig. 3). The latter observation is in accordance with in situ hybridization studies on RbcS expression in etiolated maize leaves that showed basal RbcS expression in both M and B cells. After illumination, M cell expression was suppressed and B cell expression was enhanced. Both processes together established a cell type-specific expression pattern (Langdale et al., 1988). RbcS expression in both M and B cells was also observed in very young Amaranthus spp. leaves (Wang et al., 1992). This type of RbcS gene regulation would explain the relatively high H3K4me3 levels on RbcS2 in etiolated M cells.
It is an unsolved question how the promoters of C4 genes acquired the regulatory elements necessary for efficient functioning of the C4 pathway. Within a single species, several genes must have evolved C4 expression patterns in parallel, and the demands for regulation of the new promoters such as high expression, light inducibility, and B or M specificity were highly overlapping. Thus, it is tempting to speculate that common regulatory elements were recruited by the different C4 genes. Such corecruitment was not detectable by an analysis of primary DNA sequences, although it might exist, taking the low conservation and short sequence lengths of transcription factor-binding sites (Sandelin et al., 2004) into consideration. Instead, we observed a high degree of similarity on the level of regulated histone modifications on the different C4 genes in maize. Thus, we propose that C4 promoters rather jointly acquired a histone code than a DNA code. This hypothesis is supported by the extensive conservation of this code on orthologous C4-Pepc and C4-Me genes from two separate C4 lineages, the maize/sorghum lineage and the S. italica lineage (Figs. 4–7). Because these two lineages evolved C4 metabolism independently (Brutnell et al., 2010), a preexisting epigenetic mechanism for promoter control was probably recruited into C4. This hypothesis is analogous to what has been proposed for regulatory DNA sequences in the transcribed region of C4 genes (Brown et al., 2010, 2011; Kajala et al., 2012). In this respect, it will be interesting to see whether light-induced and tissue-specific genes in C3 plants share the described histone code.
CONCLUSION
Analysis of histone modification profiles on C4 genes in maize revealed a common histone modification code associated with light induction and cell type-specific gene expression. Comparative modification profiling on two selected C4 genes in sorghum and S. italica suggested that this code is used in independent C4 lineages and, thus, was probably recruited into C4 from an ancient mechanism already existing in C3 plants.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Maize (Zea mays ‘Montello’), sorghum (Sorghum bicolor ‘BTx623’), and Setaria italica ‘Set20’ were cultivated in growth chambers with a 16-h photoperiod and a day/night temperature regime of 25°C/20°C. Seedlings were grown in soil (VM; Einheitserde) with a photon flux density of 120 to 180 µmol m−2 s−1 or in complete darkness (etiolated plants) until the third leaf was fully expanded. 72D plants were grown in the normal light rhythm but darkened for 3 d before harvest.
Sequence Identification
Coding sequences for maize C4 genes were derived from the literature. References for individual genes are given in Supplemental Figure S1. Nearest C4 gene homologs were identified for sorghum by Wang et al. (2009b) and for S. italica by Christin et al. (2007, 2009b) and Besnard et al. (2003). The corresponding genes including exon-intron predictions and surrounding genome sequences were derived from www.phytozome.net (Goodstein et al., 2012). Coordinates of the respective loci are also listed in Supplemental Figure S1.
Isolation of B Cells
For gene expression analyses in maize, B strands were isolated mechanically as described before by Hahnen et al. (2003), but without diethylether treatment. For the isolation of B strands from sorghum and S. italica, leaves were washed extensively in ice-cold water and homogenized in a Waring Blendor three times for 3 s each time. The mixture was sieved through a household sieve, and the homogenization step was repeated with the filter residue. The suspension was then filtered through Miracloth (VWR), and the residue was washed extensively with ice-cold water. The isolated B strands were shortly dried with paper and frozen in liquid nitrogen.
The material used for ChIP from isolated maize M and B cells was already described by Danker et al. (2008). Leaves were treated with formaldehyde as described below and afterward incubated in SMC buffer (0.5 m sorbitol, 5 mm MES, and 10 mm CaCl2, pH 5.8) containing 15% (w/v) Rohament CL (AB Enzymes), 10% (w/v) Rohament PL (AB Enzymes), and 0.6% (w/v) Macerozyme R-10 (Serva) for 2.5 h at 25°C.
For ChIP analysis from isolated sorghum and S. italica B strands, 4 g of leaves was cross linked as described below and afterward incubated in SMC buffer containing 3% (w/v) Cellulase Onozuka R-10 (Serva) and 0.6% (w/v) Macerozyme R-10 (Serva) for 20 h at 25°C under constant agitation. M protoplasts and remaining epidermal strips were separated manually in ice-cold water. The quality of each preparation was evaluated microscopically.
ChIP
As described previously by Horst et al. (2009), 6 g of leaves from 10- to 12-d-old maize seedlings was harvested and cross linked. For sorghum and S. italica, 4 g of leaves from 14- to 16-d-old seedlings was harvested and vacuum infiltrated with 1% (v/v) formaldehyde instead of 3% (v/v) for maize. ChIP was performed as described by Haring et al. (2007).
The material was ground, resuspended in extraction buffer (10 mm sodium-butyrate, 400 mm Suc, 10 mm Tris-HCl, pH 8.0, 5 mm β-mercaptoethanol, 0.1 mm phenylmethylsulfonyl fluoride [PMSF], and 1× Complete [Roche Applied Science]), and incubated for 15 min at 4°C. Afterward, the solution was filtered through four layers of Miracloth (VWR), and the residue was washed with purification buffer 1 (10 mm sodium-butyrate, 250 mm Suc, 10 mm Tris-HCl, pH 8.0, 5 mm β-mercaptoethanol, 0.1 mm PMSF, 10 mm MgCl2, 1% [w/v] Triton X-100, and 1× Complete) and afterward with purification buffer 2 (10 mm sodium-butyrate, 1.64 m Suc, 10 mm Tris-HCl, pH 8.0, 5 mm β-mercaptoethanol, 0.1 mm PMSF, 2 mm MgCl2, 0.15% [w/v] Triton X-100, and 1× Complete). After purification, nuclei were resuspended in nuclei lysis buffer (25 mm Tris-HCl, pH 8.0, 5 mm EDTA, 0.5% [w/v] SDS, 0.1 mm PMSF, and 1× Complete).
Chromatin was sheared with a Bioruptor (Diagenode) for 10 min (setting, high; interval, 30/30 s) under constant cooling. The sheared chromatin solution was diluted 2-fold with ChIP buffer (50 mm Tris-HCl, pH 8.0, 1 mm EDTA, 150 mm NaCl, and 0.1% [w/v] Triton X-100) and precleared with 40 µL of protein A agarose (Roche Applied Science). Precleared chromatin was split into aliquots of 400 µL for immunoprecipitation and one aliquot of 40 µL for determination of the amount of input. The chromatin aliquots were added to 30 µL of protein A agarose, and modified histones were detected with 5 µL of anti-acetyl H4K5 (07-327; Millipore), 5 µL of anti-acetyl H3K9 (07-352; Millipore), 1 µL of anti-acetyl H3K18 (07-354; Millipore), 5 µL of anti-dimethyl H3K4 (07-030; Millipore), 2.5 µL of anti-trimethyl H3K4 (04-745 [Millipore] and ab8580 [Abcam]), and 1 µL of anti-H3 C-term (ab1791; Abcam). The control serum for the determination of background precipitation was derived from rabbits immunized with an unrelated protein from potato (Solanum tuberosum).
After washing, the antibody-bound complexes were released and decross linked by incubation in elution buffer (62.5 mm Tris-HCl, pH 6.8, 200 mm NaCl, 2% [w/v] SDS, and 10 mm dithiothreitol) at 65°C overnight. The coprecipitated DNA was purified using the MSB Spin PCRapace kit (Invitek). Typically, 2 µL of eluted DNA was used as a template for quantitative PCR analysis.
Data Normalization
Real-time PCR signals obtained from an immunoprecipitate with an antibody directed against a specific histone acetylation or methylation were first corrected for the real-time PCR signals precipitated using a negative control serum (see above). The negative control serum signal was never more than 10% of the signal obtained with a specific antibody. The signal obtained with the antibody against an invariant domain of histone H3 (anti-H3 C-term; see above) was defined as NO. The acetylation or methylation signal at a gene position was divided by NO at the same position to obtain the modification signal per nucleosome (MN). MN is always shown as a relative enrichment compared with the MN on the promoter of the Actin1 housekeeping gene. For H3K4me3/me2 ratios, the MN obtained with the antibody directed to H3K4me3 was divided by the MN obtained with the antibody directed to H3K4me2. The resulting ratio is dimensionless.
RNA Isolation and Reverse Transcription
Total RNA isolation was performed by phenol-chloroform extraction as described by Haring et al. (2007). About 25 to 30 mg of ground plant material was dissolved in 1 mL of Trizol and agitated for 15 min. After the addition of 0.2 volume of chloroform and agitation for 10 min, phases were separated by centrifugation (13,000 rpm, 4°C, 15 min). The aqueous phase was transferred to a new reaction tube and washed twice with 1 volume of chloroform. RNA was precipitated with 2 volumes of ice-cold ethanol (96%) for 20 min at −20°C and following centrifugation (13,000 rpm, 4°C, 15 min). After washing with 70% ethanol, the RNA was dissolved in 30 µL of water. The quality of the isolated RNA was controlled by electrophoresis, and the concentration was determined photometrically.
One unit of DNaseI (Fermentas) per microgram of RNA and MgCl2 to a final concentration of 2 mm were added, and reactions were incubated for 30 min at 37°C, followed by a denaturation step of 15 min at 70°C to remove traces of contaminating DNA. cDNA synthesis was performed with approximately 1 µg of total RNA and 50 pmol of random nonamer primer. Reactions were incubated for 5 min at 70°C and cooled down on ice before adding 200 units of Moloney murine leukemia virus reverse transcriptase (Promega) and 1 mm deoxyribonucleotide triphosphates in reaction buffer as specified by the manufacturer. hnRNAs were amplified from cDNA using primer systems specific for introns (Supplemental Fig. S1). A dilution series of cDNA from illuminated leaves was used as a standard.
Quantitative PCR
Quantitative PCR was performed on an ABI PRISM 7300 sequence detection system (Life Technologies) using SYBR Green fluorescence (Platinum SYBR Green QPCR Mix; Life Technologies) for detection. Oligonucleotides were purchased from Metabion. Oligonucleotide sequences are given in Supplemental Figure S1. Amplification conditions were 2 min of initial denaturation at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. Afterward, a melting curve was recorded. General reaction conditions were 3 mm MgCl2 and 200 nm of each oligonucleotide. Sizes of the amplified molecules were confirmed by gel electrophoresis.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NM_001111948, NM_001112268, U08401.1, NM_001111843, AB018744, Y092214.1, J01238, XM_002438476, XM_002454985, XM_002456645, AF495586, FN397881, and AF288226.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Nucleosome occupancy on the gene promoters investigated in this study.
Supplemental Table S1. Gene information and oligonucleotide sequences.
Glossary
- M
mesophyll
- B
bundle sheath
- NO
nucleosome occupancy
- TIS
transcription initiation site
- hnRNA
heterogeneous nuclear RNA
- ChIP
chromatin immunoprecipitation
- PMSF
phenylmethylsulfonyl fluoride
- MN
modification signal per nucleosome
- RT
reverse transcription
- cDNA
complementary DNA
References
- Adrian J, Farrona S, Reimer JJ, Albani MC, Coupland G, Turck F. (2010) cis-Regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell 22: 1425–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akyildiz M, Gowik U, Engelmann S, Koczor M, Streubel M, Westhoff P. (2007) Evolution and function of a cis-regulatory module for mesophyll-specific gene expression in the C4 dicot Flaveria trinervia. Plant Cell 19: 3391–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. (2011) Regulation of chromatin by histone modifications. Cell Res 21: 381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett CL, Rawson JR, Jernstedt JA. (1988) DNA and RNA levels in bundle sheath and mesophyll cells of pearl millet (Pennisetum americanum). Plant Physiol 87: 307–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL. (2007) The complex language of chromatin regulation during transcription. Nature 447: 407–412 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES. (2007) The mammalian epigenome. Cell 128: 669–681 [DOI] [PubMed] [Google Scholar]
- Besnard G, Pinçon G, D’Hont A, Hoarau JY, Cadet F, Offmann B. (2003) Characterisation of the phosphoenolpyruvate carboxylase gene family in sugarcane (Saccharum spp.). Theor Appl Genet 107: 470–478 [DOI] [PubMed] [Google Scholar]
- Brown NJ, Newell CA, Stanley S, Chen JE, Perrin AJ, Kajala K, Hibberd JM. (2011) Independent and parallel recruitment of preexisting mechanisms underlying C4 photosynthesis. Science 331: 1436–1439 [DOI] [PubMed] [Google Scholar]
- Brown NJ, Palmer BG, Stanley S, Hajaji H, Janacek SH, Astley HM, Parsley K, Kajala K, Quick WP, Trenkamp S, et al. (2010) C acid decarboxylases required for C photosynthesis are active in the mid-vein of the C species Arabidopsis thaliana, and are important in sugar and amino acid metabolism. Plant J 61: 122–133 [DOI] [PubMed] [Google Scholar]
- Brutnell TP, Wang L, Swartwood K, Goldschmidt A, Jackson D, Zhu XG, Kellogg E, Van Eck J. (2010) Setaria viridis: a model for C4 photosynthesis. Plant Cell 22: 2537–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron J-BF, He H, Elling AA, Deng XW. (2009) Dynamic landscapes of four histone modifications during deetiolation in Arabidopsis. Plant Cell 21: 3732–3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin P-A, Salamin N, Kellogg EA, Vicentini A, Besnard G. (2009a) Integrating phylogeny into studies of C4 variation in the grasses. Plant Physiol 149: 82–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin P-A, Salamin N, Savolainen V, Duvall MR, Besnard G. (2007) C4 photosynthesis evolved in grasses via parallel adaptive genetic changes. Curr Biol 17: 1241–1247 [DOI] [PubMed] [Google Scholar]
- Christin P-A, Samaritani E, Petitpierre B, Salamin N, Besnard G. (2009b) Evolutionary insights on C4 photosynthetic subtypes in grasses from genomics and phylogenetics. Genome Biol Evol 1: 221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danker T, Dreesen B, Offermann S, Horst I, Peterhänsel C. (2008) Developmental information but not promoter activity controls the methylation state of histone H3 lysine 4 on two photosynthetic genes in maize. Plant J 53: 465–474 [DOI] [PubMed] [Google Scholar]
- Delany AM. (2001) Measuring transcription of metalloproteinase genes: nuclear run-off assay vs analysis of hnRNA. Methods Mol Biol 151: 321–333 [DOI] [PubMed] [Google Scholar]
- Dion MF, Altschuler SJ, Wu LF, Rando OJ. (2005) Genomic characterization reveals a simple histone H4 acetylation code. Proc Natl Acad Sci USA 102: 5501–5506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doust AN, Kellogg EA, Devos KM, Bennetzen JL. (2009) Foxtail millet: a sequence-driven grass model system. Plant Physiol 149: 137–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Shook MS, Brower-Toland B, Hicks L, Pikaard CS. (2007) In vitro specificities of Arabidopsis co-activator histone acetyltransferases: implications for histone hyperacetylation in gene activation. Plant J 52: 615–626 [DOI] [PubMed] [Google Scholar]
- Elferink CJ, Reiners JJ., Jr (1996) Quantitative RT-PCR on CYP1A1 heterogeneous nuclear RNA: a surrogate for the in vitro transcription run-on assay. Biotechniques 20: 470–477 [DOI] [PubMed] [Google Scholar]
- Engelmann S, Bläsing OE, Gowik U, Svensson P, Westhoff P. (2003) Molecular evolution of C4 phosphoenolpyruvate carboxylase in the genus Flaveria: a gradual increase from C3 to C4 characteristics. Planta 217: 717–725 [DOI] [PubMed] [Google Scholar]
- Ewing RM, Jenkins GI, Langdale JA. (1998) Transcripts of maize RbcS genes accumulate differentially in C3 and C4 tissues. Plant Mol Biol 36: 593–599 [DOI] [PubMed] [Google Scholar]
- Francis KE, Spiker S. (2005) Identification of Arabidopsis thaliana transformants without selection reveals a high occurrence of silenced T-DNA integrations. Plant J 41: 464–477 [DOI] [PubMed] [Google Scholar]
- Furumoto T, Hata S, Izui K. (1999) cDNA cloning and characterization of maize phosphoenolpyruvate carboxykinase, a bundle sheath cell-specific enzyme. Plant Mol Biol 41: 301–311 [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowik U, Burscheidt J, Akyildiz M, Schlue U, Koczor M, Streubel M, Westhoff P. (2004) cis-Regulatory elements for mesophyll-specific gene expression in the C4 plant Flaveria trinervia, the promoter of the C4 phosphoenolpyruvate carboxylase gene. Plant Cell 16: 1077–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahnen S, Joeris T, Kreuzaler F, Peterhänsel C. (2003) Quantification of photosynthetic gene expression in maize C(3) and C(4) tissues by real-time PCR. Photosynth Res 75: 183–192 [DOI] [PubMed] [Google Scholar]
- Haring M, Offermann S, Danker T, Horst I, Peterhänsel C, Stam M. (2007) Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods 3: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan AH, Awad S, Al-Natour Z, Othman S, Mustafa F, Rizvi TA. (2007) Selective recognition of acetylated histones by bromodomains in transcriptional co-activators. Biochem J 402: 125–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318 [DOI] [PubMed] [Google Scholar]
- Henikoff S, Shilatifard A. (2011) Histone modification: cause or cog? Trends Genet 27: 389–396 [DOI] [PubMed] [Google Scholar]
- Hibberd JM, Covshoff S. (2010) The regulation of gene expression required for C4 photosynthesis. Annu Rev Plant Biol 61: 181–207 [DOI] [PubMed] [Google Scholar]
- Horst I, Offermann S, Dreesen B, Niessen M, Peterhansel C. (2009) Core promoter acetylation is not required for high transcription from the phosphoenolpyruvate carboxylase promoter in maize. Epigenetics Chromatin 2: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins CL, Boag S. (1985) Isolation of bundle sheath cell chloroplasts from the NADP-ME type C4 plant Zea mays: capacities for CO2 assimilation and malate decarboxylation. Plant Physiol 79: 84–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajala K, Brown NJ, Williams BP, Borrill P, Taylor LE, Hibberd JM. (2012) Multiple Arabidopsis genes primed for recruitment into C4 photosynthesis. Plant J 69: 47–56 [DOI] [PubMed] [Google Scholar]
- Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, Zhang Y, Bedford MT. (2006) Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep 7: 397–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-I, Veena, Gelvin SB. (2007) Genome-wide analysis of Agrobacterium T-DNA integration sites in the Arabidopsis genome generated under non-selective conditions. Plant J 51: 779–791 [DOI] [PubMed] [Google Scholar]
- Kingston RE, Narlikar GJ. (1999) ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev 13: 2339–2352 [DOI] [PubMed] [Google Scholar]
- Kouzarides T. (2007) Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Ku MSB, Kano-Murakami Y, Matsuoka M. (1996) Evolution and expression of C4 photosynthesis genes. Plant Physiol 111: 949–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdale JA. (2011) C4 cycles: past, present, and future research on C4 photosynthesis. Plant Cell 23: 3879–3892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdale JA, Zelitch I, Miller E, Nelson T. (1988) Cell position and light influence C4 versus C3 patterns of photosynthetic gene expression in maize. EMBO J 7: 3643–3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD. (2004) Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet 36: 900–905 [DOI] [PubMed] [Google Scholar]
- Martin C, Zhang Y. (2005) The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 6: 838–849 [DOI] [PubMed] [Google Scholar]
- Mellor J. (2005) The dynamics of chromatin remodeling at promoters. Mol Cell 19: 147–157 [DOI] [PubMed] [Google Scholar]
- Müller-Tidow C, Klein HU, Hascher A, Isken F, Tickenbrock L, Thoennissen N, Agrawal-Singh S, Tschanter P, Disselhoff C, Wang Y, et al. (2010) Profiling of histone H3 lysine 9 trimethylation levels predicts transcription factor activity and survival in acute myeloid leukemia. Blood 116: 3564–3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen H, Boccardi TM, Himanen K, Van Lijsebettens M. (2007) Impact of core histone modifications on transcriptional regulation and plant growth. Crit Rev Plant Sci 26: 243–263 [Google Scholar]
- Nishida H, Suzuki T, Kondo S, Miura H, Fujimura Y, Hayashizaki Y. (2006) Histone H3 acetylated at lysine 9 in promoter is associated with low nucleosome density in the vicinity of transcription start site in human cell. Chromosome Res 14: 203–211 [DOI] [PubMed] [Google Scholar]
- Nomura M, Higuchi T, Ishida Y, Ohta S, Komari T, Imaizumi N, Miyao-Tokutomi M, Matsuoka M, Tajima S. (2005) Differential expression pattern of C4 bundle sheath expression genes in rice, a C3 plant. Plant Cell Physiol 46: 754–761 [DOI] [PubMed] [Google Scholar]
- Offermann S, Danker T, Dreymüller D, Kalamajka R, Töpsch S, Weyand K, Peterhänsel C. (2006) Illumination is necessary and sufficient to induce histone acetylation independent of transcriptional activity at the C4-specific phosphoenolpyruvate carboxylase promoter in maize. Plant Physiol 141: 1078–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offermann S, Dreesen B, Horst I, Danker T, Jaskiewicz M, Peterhansel C. (2008) Developmental and environmental signals induce distinct histone acetylation profiles on distal and proximal promoter elements of the C4-Pepc gene in maize. Genetics 179: 1891–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M, Berry JO. (2008) Rubisco gene expression in C4 plants. J Exp Bot 59: 1625–1634 [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, et al. (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556 [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. (2005) Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122: 517–527 [DOI] [PubMed] [Google Scholar]
- Roudier F, Teixeira FK, Colot V. (2009) Chromatin indexing in Arabidopsis: an epigenomic tale of tails and more. Trends Genet 25: 511–517 [DOI] [PubMed] [Google Scholar]
- Sage RF, Christin P-A, Edwards EJ. (2012) The C4 plant lineages of planet Earth. J Exp Bot 62: 3155–3169 [DOI] [PubMed] [Google Scholar]
- Sandelin A, Alkema W, Engström P, Wasserman WW, Lenhard B. (2004) JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res 32: D91–D94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. (2002) Active genes are tri-methylated at K4 of histone H3. Nature 419: 407–411 [DOI] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Sheen J. (1990) Metabolic repression of transcription in higher plants. Plant Cell 2: 1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. (1999) C4 gene expression. Annu Rev Plant Physiol Plant Mol Biol 50: 187–217 [DOI] [PubMed] [Google Scholar]
- Sheen JY, Bogorad L. (1987) Differential expression of C4 pathway genes in mesophyll and bundle sheath cells of greening maize leaves. J Biol Chem 262: 11726–11730 [PubMed] [Google Scholar]
- Sims RJ, III, Nishioka K, Reinberg D. (2003) Histone lysine methylation: a signature for chromatin function. Trends Genet 19: 629–639 [DOI] [PubMed] [Google Scholar]
- Sugiharto B, Burnell JN, Sugiyama T. (1992) Cytokinin is required to induce the nitrogen-dependent accumulation of mRNAs for phosphoenolpyruvate carboxylase and carbonic anhydrase in detached maize leaves. Plant Physiol 100: 153–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, et al. (2011) Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146: 1016–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolley BJ, Woodfield H, Wanchana S, Bruskiewich R, Hibberd JM. (2012) Light-regulated and cell-specific methylation of the maize PEPC promoter. J Exp Bot 63: 1381–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicentini A, Barber J, Aliscioni S, Ciussani L, Kellogg E. (2008) The age of the grasses and clusters of origins of C4 photosynthesis. Glob Change Biol 14: 2963–2977 [Google Scholar]
- Viret J-F, Mabrouk Y, Bogorad L. (1994) Transcriptional photoregulation of cell-type-preferred expression of maize rbcS-m3: 3′ and 5′ sequences are involved. Proc Natl Acad Sci USA 91: 8577–8581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Caemmerer S, Furbank RT. (2003) The C(4) pathway: an efficient CO(2) pump. Photosynth Res 77: 191–207 [DOI] [PubMed] [Google Scholar]
- Wang JL, Klessig DF, Berry JO. (1992) Regulation of C4 gene expression in developing Amaranth leaves. Plant Cell 4: 173–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Elling AA, Li X, Li N, Peng Z, He G, Sun H, Qi Y, Liu XS, Deng XW. (2009a) Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell 21: 1053–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Gowik U, Tang H, Bowers JE, Westhoff P, Paterson AH. (2009b) Comparative genomic analysis of C4 photosynthetic pathway evolution in grasses. Genome Biol 10: R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingler A, Walker RP, Chen ZH, Leegood RC. (1999) Phosphoenolpyruvate carboxykinase is involved in the decarboxylation of aspartate in the bundle sheath of maize. Plant Physiol 120: 539–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang X, Ling G, D’Agostino J, Ding X. (2009) Mechanisms of differential expression of the CYP2A13 7520C and 7520G alleles in human lung: allelic expression analysis for CYP2A13 heterogeneous nuclear RNA, and evidence for the involvement of multiple cis-regulatory single nucleotide polymorphisms. Pharmacogenet Genomics 19: 852–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Purcell M, Zucchi P, Helentjaris T, Bogorad L. (2001) TRM1, a YY1-like suppressor of rbcS-m3 expression in maize mesophyll cells. Proc Natl Acad Sci USA 98: 2295–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki S, Oda M, Daimon H, Mitsukuri K, Johkan M, Nakatsuka T, Nishihara M, Mishiba K. (2011) Epigenetic modifications of the 35S promoter in cultured gentian cells. Plant Sci 180: 612–619 [DOI] [PubMed] [Google Scholar]
- Yan C, Boyd DD. (2006) Histone H3 acetylation and H3 K4 methylation define distinct chromatin regions permissive for transgene expression. Mol Cell Biol 26: 6357–6371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Kong QR, Zhao ZP, Wu ML, Mu YS, Hu K, Liu ZH. (2012) Position effect variegation and epigenetic modification of a transgene in a pig model. Genet Mol Res 11: 355–369 [DOI] [PubMed] [Google Scholar]
- Zhang X. (2008) The epigenetic landscape of plants. Science 320: 489–492 [DOI] [PubMed] [Google Scholar]