Golgi-localized putative transporter proteins are involved in the intracellular transport of paraquat.
Abstract
Paraquat is one of the most widely used herbicides worldwide. In green plants, paraquat targets the chloroplast by transferring electrons from photosystem I to molecular oxygen to generate toxic reactive oxygen species, which efficiently induce membrane damage and cell death. A number of paraquat-resistant biotypes of weeds and Arabidopsis (Arabidopsis thaliana) mutants have been identified. The herbicide resistance in Arabidopsis is partly attributed to a reduced uptake of paraquat through plasma membrane-localized transporters. However, the biochemical mechanism of paraquat resistance remains poorly understood. Here, we report the identification and characterization of an Arabidopsis paraquat resistant1 (par1) mutant that shows strong resistance to the herbicide without detectable developmental abnormalities. PAR1 encodes a putative l-type amino acid transporter protein localized to the Golgi apparatus. Compared with the wild-type plants, the par1 mutant plants show similar efficiency of paraquat uptake, suggesting that PAR1 is not directly responsible for the intercellular uptake of paraquat. However, the par1 mutation caused a reduction in the accumulation of paraquat in the chloroplast, suggesting that PAR1 is involved in the intracellular transport of paraquat into the chloroplast. We identified a PAR1-like gene, OsPAR1, in rice (Oryza sativa). Whereas the overexpression of OsPAR1 resulted in hypersensitivity to paraquat, the knockdown of its expression using RNA interference conferred paraquat resistance on the transgenic rice plants. These findings reveal a unique mechanism by which paraquat is actively transported into the chloroplast and also provide a practical approach for genetic manipulations of paraquat resistance in crops.
Paraquat (or methyl viologen; N,N′-dimethyl-4,4′-bipyridinium dichloride) is a rapid-acting, nonselective herbicide that has been widely used for weed control (Haley, 1979). In green plants, paraquat causes rapid membrane damage by accepting electrons from PSI and subsequently transferring them to molecular oxygen, resulting in the production of toxic reactive oxygen species (ROS), which efficiently induce cell death (Dodge, 1971; Haley, 1979; Babbs et al., 1989; Fujii et al., 1990; Suntres, 2002; Bonneh-Barkay et al., 2005). As a fast-acting and nonselective herbicide for green plant tissues, paraquat rapidly kills a wide range of annual grasses as well as broad-leafed and perennial weeds upon contact. When entering the soil, paraquat becomes biologically inactive and has minimal or no toxicity toward roots and rhizomes. In addition, paraquat has no effects on mature bark (Dodge, 1971; Suntres, 2002). Because of these characteristics, paraquat is widely used in orchards, plantation crops, conservation tillage systems, and other applications (Bromilow, 2004).
During the course of commercial paraquat applications for decades, many paraquat-resistant biotypes, ecotypes, and mutants, including weeds and cultivated plants, have been characterized. To date, nearly 30 species of paraquat-resistant weeds have been reported worldwide (http://www.weedscience.org), and the best-studied examples are Conyza bonariensis (Fuerst et al., 1985; Amsellem et al., 1993), Lolium rigidum (Yu et al., 2004, 2007), Arctotheca calendula (Powles et al., 1989; Preston et al., 1994), and Rehmannia glutinosa (Chun et al., 1997a, 1997b). In the model plant species Arabidopsis (Arabidopsis thaliana), several paraquat-resistant mutants have been identified based on various genetic screens, including photoautotrophic salt tolerance1 (pst1), radical-induced cell death1 (rcd1), paraquat resistant2 (par2), pleiotropic drug resistance11 (atpdr11), and resistant to methyl viologen1 (rmv1; Tsugane et al., 1999; Ahlfors et al., 2004; Chen et al., 2009; Fujita et al., 2012; Xi et al., 2012).
Several mechanisms have been proposed for paraquat resistance in higher plants (Fuerst and Vaughn, 1990). In the early studies on paraquat-resistant weeds, a sequestration mechanism was proposed, in which paraquat is prevented from diffusing into PSI, the active site of the herbicide (Fuerst et al., 1985; Fuerst and Vaughn, 1990; Preston et al., 1992; Norman et al., 1993; Chun et al., 1997a). In addition, detoxification through reduction of the ROS level or the reduced uptake of paraquat have been suggested as alternative mechanisms for paraquat resistance in weeds (Shaaltiel and Gressel, 1986; Fuerst and Vaughn, 1990; Donahue et al., 1997; Jóri et al., 2007). Consistent with the observations that paraquat resistance is correlated with the increased activity of the ROS-scavenging enzymes superoxide dismutase, ascorbate peroxidase, and glutathione reductase (Shaaltiel and Gressel, 1986; Tsugane et al., 1999; Ye and Gressel, 2000), the expression level of these genes has been associated with the paraquat-resistant phenotype in several transgenic studies and in the Arabidopsis mutants rcd1 and pst1 (Bowler et al., 1991; Gupta et al., 1993; Arisi et al., 1998; Tsugane et al., 1999; Ye and Gressel, 2000; Ahlfors et al., 2004; Fujibe et al., 2004; Murgia et al., 2004). The characterization of the Arabidopsis par2-1 mutant has presented an alternative detoxification mechanism of paraquat resistance in plants. PAR2 encodes an S-nitrosoglutathione reductase that catalyzes the irreversible degradation of S-nitrosoglutathione, a major biologically active species of nitric oxide (NO), and mutations in PAR2 cause an increased level of NO species (Feechan et al., 2005; Lee et al., 2008; Chen et al., 2009). It has been proposed that an elevated NO level might exert an antagonistic effect against paraquat-induced oxidative stress (Chen et al., 2009), a mechanism similar to the reciprocal scavenging of NO and superoxide observed in the hypersensitive response (Delledonne et al., 2001).
Recent studies on two Arabidopsis paraquat-resistant mutants, atpdr11 and rmv1, have provided compelling evidence supporting the model that the uptake of paraquat is a critical factor responsible for resistance to paraquat (Fujita et al., 2012; Xi et al., 2012). AtPDR11 and RMV1 encode an ATP-binding cassette transporter and an l-type amino acid (LAT) transporter, respectively, both of which are localized to the plasma membrane and are involved in the uptake of paraquat (Fujita et al., 2012; Xi et al., 2012). In the cyanobacterium Synechocystis sp. PCC 6803, a heteromeric ATP-binding cassette-type transporter has been shown to function as a paraquat exporter for the exclusion of the herbicide from the photosynthetic machinery of the cells (Prosecka et al., 2009) and may act via a similar mechanism as AtPDR11 (Xi et al., 2012).
Despite this progress, it remains unknown how paraquat is transported into its major target site, the chloroplast, upon entry of a plant cell. Here, we report the identification and characterization of an Arabidopsis mutant, par1. We found that PAR1 encodes a putative LAT transporter that is localized to the Golgi apparatus and is required for the accumulation of paraquat in the chloroplast.
RESULTS
Identification and Characterization of the Paraquat-Resistant Mutant par1
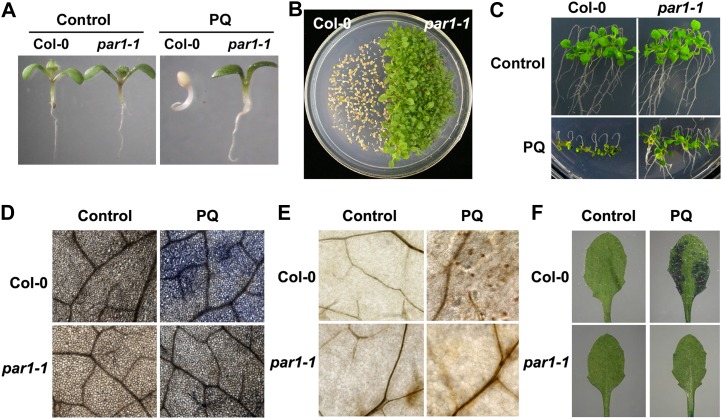
In a genetic screen for par mutants from an ethyl methanesulfonate-mutagenized M2 population in the Columbia-0 (Col-0) background (Chen et al., 2009), we identified four allelic mutants: par1-1 through par1-4 (see below for the genetic analysis). Because these four mutant alleles showed a similar phenotype under all tested conditions, we present only the data for the par1-1 mutant allele unless otherwise indicated. Under normal growth conditions, the par1 mutants were indistinguishable from wild-type plants throughout all of the developmental stages (Fig. 1A; Supplemental Fig. S1A). However, when germinated and grown in the presence of various concentrations of paraquat, the par1-1 mutant showed a phenotype that was more resistant than wild-type plants (Fig. 1, A and B; Supplemental Fig. S2). To determine whether the paraquat-resistant phenotype of par1-1 is also persistent during postgerminative growth, we transferred 5-d-old seedlings germinated and grown on paraquat-free medium onto paraquat-containing medium, and continued culturing for an additional 7 d. Under the assay conditions, par1-1 displayed substantial resistance to paraquat, whereas wild-type seedlings showed an inhibition of leaf and root growth (Fig. 1C). These results indicate that par1 mutations confer paraquat tolerance at both the germination and postgermination developmental stages.
Figure 1.
The par1-1 mutant phenotype. A, Wild type (Col-0) and par1-1 seedlings (10 d old) germinated and grown on MS medium supplemented with 0 and 1 μm paraquat (PQ). B, Wild type (Col-0) and par1-1 seedlings (20 d old) germinated and grown on MS medium in the presence of 1 μm paraquat. C, Wild type (Col-0) and par1-1 seedlings (5 d old) grown on MS medium were transferred to MS medium supplemented with 0 μm (top panels) or 1 μm paraquat (bottom panels) for an additional 7 d. D, Accumulation of superoxide induced by paraquat. Wild type (Col-0) and par1-1 seedlings (4 weeks old) were sprayed with water or 5 μm paraquat and then incubated for 24 h. The leaves were subsequently detached from the treated plants and stained with NBT. E, Accumulation of hydrogen peroxide induced by paraquat. Samples were treated as described in D and then stained with DAB. F, Cell death induced by paraquat. Samples were treated as described in D and then stained with Evans blue. [See online article for color version of this figure.]
Paraquat is known to induce the generation of ROS and consequent cell death (Babbs et al., 1989; Fujii et al., 1990). When treated with paraquat, par1-1 showed a reduced accumulation of superoxide and hydrogen peroxide compared with wild-type plants, as revealed in leaves stained with nitroblue tetrazolium (NBT) and 3,3′-diaminobenzidine (DAB), respectively (Fig. 1, D and E). When stained with Evans blue, the reduced cell death was observed in par1-1 leaves (Fig. 1F). These results indicate that the paraquat-induced generation of ROS and resulting cell death are inhibited by the par1 mutation.
Because the generation of ROS is associated with the responses to abiotic stress and defense, we examined the responses of par1 to various stresses. The par1-1 mutant responded to salt, abscisic acid, salicylic acid, pathogens, and hydrogen peroxide in a similar pattern to the wild-type plants (Supplemental Fig. S1, B–F), suggesting that the responses to abiotic stress and defense remain relatively unaltered in par1 mutants. Collectively, these data suggest that the par1-1 mutation specifically confers paraquat resistance without detectable effects on the growth, development, and stress responses of the plants.
Molecular Characterization of the PAR1 Gene
To investigate the nature of the par1 mutation, we reciprocally crossed par1-1 with Col-0 plants. All examined F1 progeny (107 seedlings) showed a paraquat-sensitive phenotype similar to that of wild-type plants. In the F2 population, the par1-1 phenotype segregated at an approximately 1:3 ratio (par1:wild type = 66:219; χ2 = 0.42), indicating that par1-1 is a recessive mutation at a single nuclear locus.
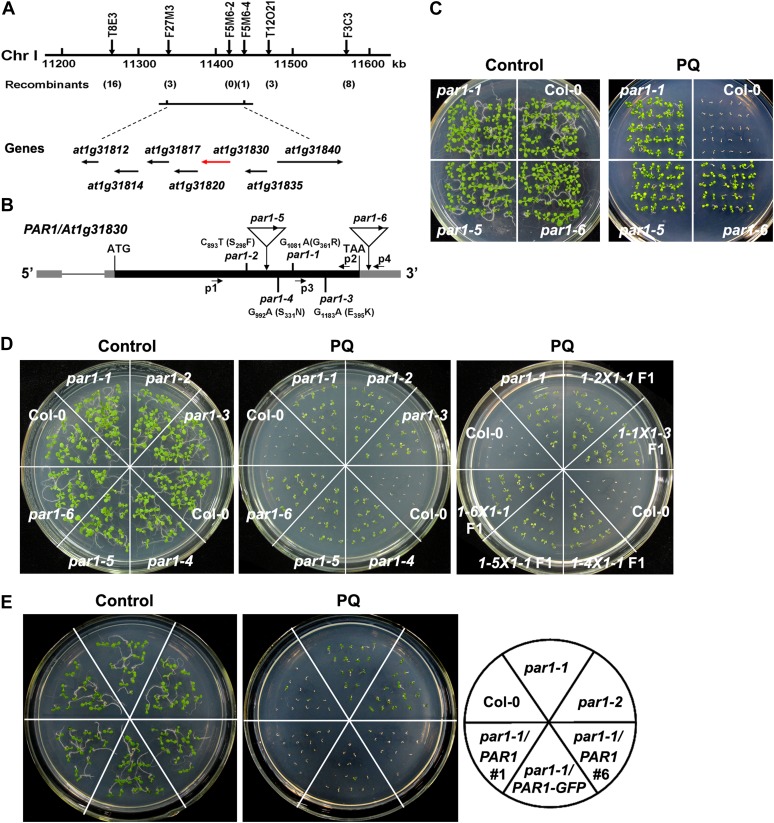
To identify the PAR1 gene, the par1-1 mutant was crossed with Landsberg erecta (Ler), and F2 progeny exhibiting paraquat resistance were used for genetic mapping. The par1 mutation was mapped to a 93-kb region on chromosome I between the markers F27M3 and F5M6-4 (Fig. 2A). DNA sequencing analysis of candidate genes in this region revealed a G-to-A transition at nucleotide 1,081 in the AT1G31830 gene, which resulted in the substitution of Gly-361 with an Arg residue (Fig. 2B). AT1G31830 encodes a polypeptide of 495 amino acid residues containing 12 predicted transmembrane domains (Supplemental Fig. S3A). The amino acid substitution in par1-1 occurs within the ninth transmembrane domain. In par1-2, a C-to-T transition at nucleotide 893 resulted in the substitution of Ser-298 with a Phe residue in the region between the seventh and eighth transmembrane domains in the same gene. In par1-3 and par1-4 of AT1G31830, G-to-A mutations were identified at nucleotides 1,183 and 992, which resulted in the substitution of Glu-395 with Lys and Ser-331 with Asn, respectively (Fig. 2B; Supplemental Fig. S3B). The mutated amino acid residues in par1-1 (Gly-361), par1-3 (Glu-395), and par1-4 (Ser-331) are highly conserved (Supplemental Fig. S3B). Notably, all four mutations occurred at the region between the seventh and 10th transmembrane domains, suggesting that this region is functionally important. Reverse transcription (RT)-PCR analysis revealed that these mutations had no obvious effects on the transcription of AT1G31830 (Supplemental Fig. S4).
Figure 2.
Map-based cloning of PAR1. A, Genetic mapping of PAR1. Markers used for the genetic mapping are shown on the top, and the number of recombinants for each marker is given below the map. Predicted genes are shown at bottom, and the arrows indicate the direction of transcription. The PAR1 candidate gene is shown in red. B, Genome structure of the PAR1 gene. The black boxes, gray boxes, and lines indicate exons, untranslated regions, and introns, respectively. The positions and the nature of the par1 mutant alleles are shown. The positions and orientations of PCR primers (for genotyping and RT-PCR analyses; P1–P4) are shown. C, Two-week-old seedlings of the wild type (Col-0) and par1 allelic mutants germinated and grown on MS medium in the presence or absence of 1 μm paraquat (PQ). D, Ten-day-old Col-0, par1, and F1 seedlings derived from the crosses between different combinations of par1 allelic mutants germinated and grown in the presence of 1 μm paraquat. E, Genetic complementation of the par1 mutant phenotype. Seven-day-old seedlings with the indicated genotypes were germinated and grown on MS medium in the presence or absence of paraquat. par1-1/PAR1 and par1-1/PAR1-GFP refer to par1-1 seedlings carrying a PAR1 and a PAR1-GFP transgene, respectively, under the control of the PAR1 promoter. [See online article for color version of this figure.]
We also identified two additional mutants that contained transfer DNA (T-DNA) insertions in AT1G31830, par1-5 (SALK_119707C) and par1-6 (SALK_129045; Alonso et al., 2003; Fig. 2B). PAR1 transcripts were not detected in par1-5, and a reduced expression of PAR1 was observed in par1-6, as revealed by RT-PCR (Supplemental Fig. S4). These two mutants showed a paraquat-resistant phenotype similar to par1-1 (Fig. 2C). To test the possible allelism among these mutants, we crossed par1-1 with other par1 mutants and found that all F1 progeny showed a paraquat-resistant phenotype (Fig. 2D), thus demonstrating that all par1 mutants are allelic.
To further verify the identity of the PAR1 gene, we transformed the par1-1 mutant with a 3.9-kb genomic DNA fragment containing the putative promoter and coding sequences of AT1G31830 fused with or without a GFP gene. Both transgenes fully restored the paraquat-resistant phenotype of par1-1 to the paraquat-sensitive phenotype, similar to that of wild-type plants (Fig. 2E), thereby confirming that the par1-1 mutant phenotype is caused by mutations in AT1G31830. Taken together, these results demonstrate that AT1G31830 represents PAR1.
PAR1 Encodes a Putative LAT Transporter or Amino Acid Permease
A sequence comparison revealed that PAR1 encodes a putative amino acid permease belonging to a small family of highly conserved proteins in eukaryotic organisms. In mammals, this class of proteins has been functionally characterized as LAT transporters, involved in the transport of LATs, polyamines, and organocations (Jack et al., 2000). In Arabidopsis, PAR1 belongs to a small gene family with four additional members, of which the encoded proteins share 43% to 75% identity (Supplemental Figs. S3B and S5). Among these members, LAT1 was characterized as RMV1 (Fujita et al., 2012). LAT3 (AT1G31820) is immediately adjacent to PAR1 (LAT4; AT1G31830). LAT3 shares the highest homology with PAR1 (75% identity). However, two T-DNA insertional mutants in LAT3 (lat3-1 and lat3-2) showed a paraquat-sensitive phenotype similar to that of wild-type plants (Supplemental Fig. S6), indicating that the paraquat-resistant phenotype is specific to mutations in PAR1/LAT4.
PAR1 Is Constitutively Expressed throughout Arabidopsis Development
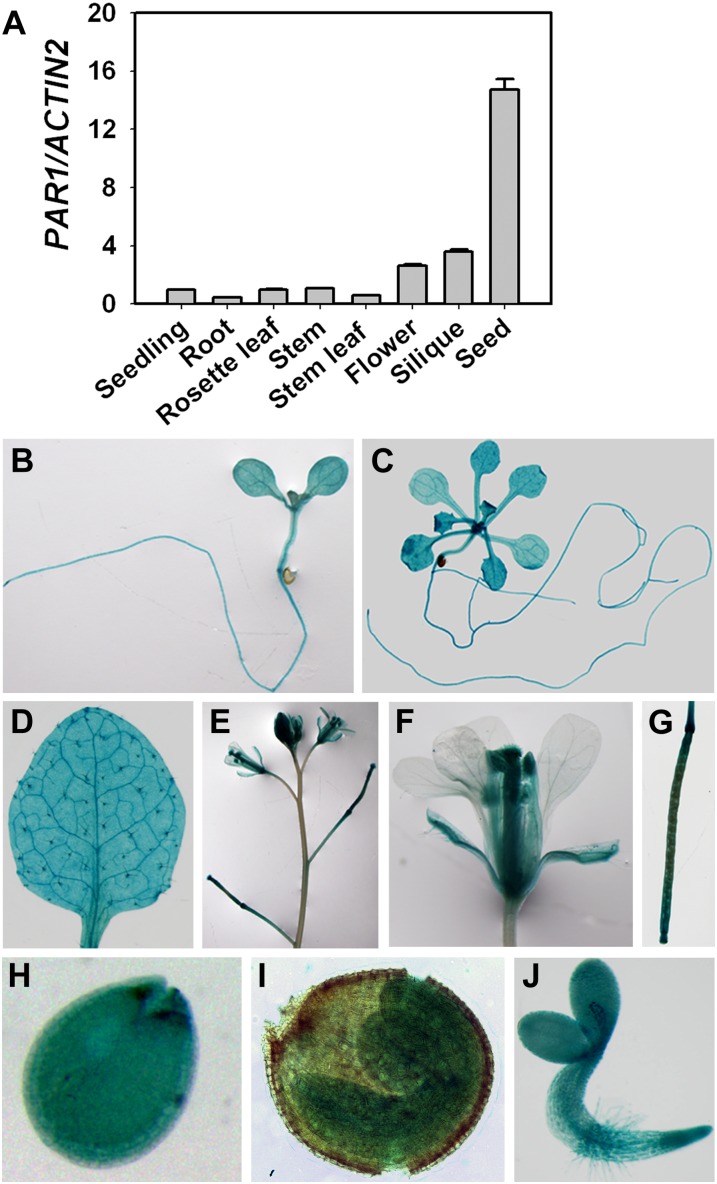
To examine the expression pattern of PAR1, quantitative reverse transcription (qRT)-PCR was performed using total RNA extracted from various tissues. PAR1 was ubiquitously expressed in all of the examined tissues, and the highest expression level was observed in seeds (Fig. 3A). To monitor the tissue-specific expression of PAR1, we generated a PAR1:GUS reporter construct, in which a 2.1-kb genomic fragment upstream of the translation start codon of PAR1 was used. Of the 18 independent lines that were examined, 14 lines showed similar GUS staining patterns. One representative line was selected for further detailed analysis of PAR1 expression patterns. PAR1:GUS showed strong GUS staining in almost all examined tissues, including roots, rosette leaves, cauline leaves, inflorescences, the floral organs (with the exception of petals), siliques, and seeds (Fig. 3, B–J). Consistent with the qRT-PCR analysis result, a higher level of PAR1:GUS expression was observed in the reproductive organs than in other organs (Fig. 3).
Figure 3.
Expression pattern of the PAR1 gene. A, Analysis of the expression of PAR1 in the roots, stems, rosette leaves, flowers, siliques, and seeds using qRT-PCR. The means of three replicates ± sd are shown. Similar results were obtained in three independent experiments. B to J, GUS expression in PAR1:GUS transgenic lines. GUS expression is shown in seedlings at the cotyledon stage (B) and eight-leaf stage (C), cauline leaf (D), inflorescence (E), flower (F), silique (G), immature seed (H), mature seed (I), and 2-d-old seedling (J). [See online article for color version of this figure.]
PAR1 Overexpression Confers Paraquat Hypersensitivity
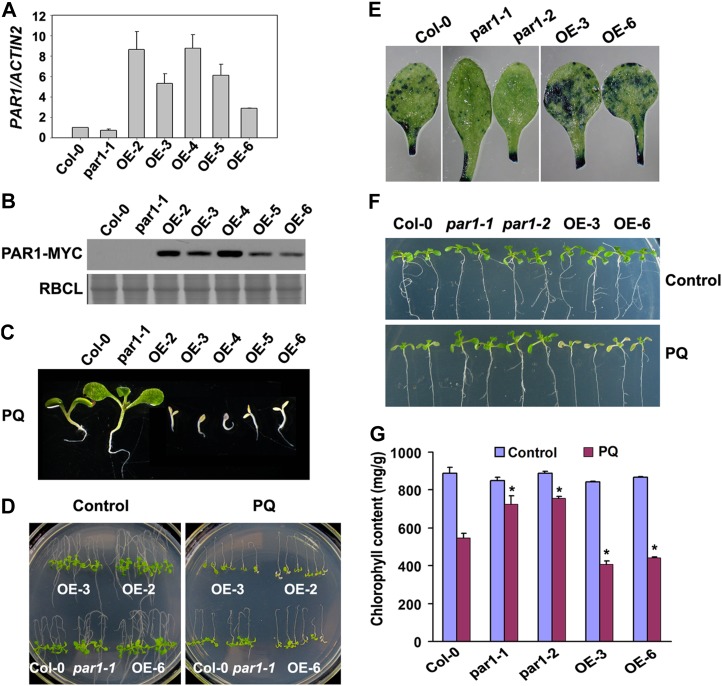
The data presented above indicate that loss-of-function mutations in PAR1 result in resistance to paraquat. To explore the correlation of PAR1 expression with paraquat sensitivity, we generated transgenic plants overexpressing a PAR1-MYC transgene under the control of a Super promoter (Li et al., 2001; Super:PAR1-MYC). These Super:PAR1-MYC transgenic lines showed a substantially higher level of PAR1 expression than the wild-type plants (Fig. 4A). Moreover, the accumulation of PAR1-MYC protein was readily detected (Fig. 4B). Under normal growth conditions, the Super:PAR1-MYC transgenic plants did not show any detectable abnormalities (Supplemental Fig. S7A). However, the overexpression of PAR1-MYC caused the transgenic plants to be hypersensitive to paraquat at both the seed germination and postgerminative growth stages (Fig. 4, C and D). The paraquat sensitivity of these transgenic plants was correlated with the expression levels of the transgene. When treated with paraquat, Super:PAR1-MYC transgenic plants showed increased cell death (Fig. 4E) and reduced chlorophyll content compared with wild-type plants (Fig. 4, F and G). Similar results were obtained in transgenic plants overexpressing PAR1 via an inducible promoter (Zuo et al., 2000; Supplemental Fig. S7, B–E). These results indicate that the overexpression of PAR1-MYC renders the transgenic plants hypersensitive to paraquat, and the paraquat sensitivity is correlated with the PAR1 expression level.
Figure 4.
Overexpression of PAR1 confers hypersensitivity to paraquat. A, Analysis of PAR1 overexpression in Col-0, par1-1, and Super:PAR1-MYC (OE lines) using qRT-PCR. RNA prepared from 2-week-old seedlings was used for this assay, and the means of three replicates ± sd are shown. Similar results were obtained in three independent experiments. B, Immunoblot analysis of PAR1 protein in the plants described in A using an anti-MYC antibody. Equal loading was verified by Coomassie blue staining. RBCL, Rubisco large subunit. C, Ten-day-old seedlings with the indicated genotypes germinated and grown in the presence of 0.5 μm paraquat (PQ). D, Phenotypes of Col-0, par1-1, and three independent overexpression seedlings (5 d old) grown on MS medium were transferred to MS medium supplemented with water (control) or 1 μm paraquat for an additional 7 d. E, Paraquat-induced cell death in leaves of wild-type (Col-0), par1, and PAR1-OE plants (4 weeks old) treated with water or 5 μm paraquat for 24 h by spraying. Leaves were then detached from treated plants and stained with Evans blue. F, Phenotypes of wild-type (Col-0), par1, and PAR1-OE seedlings (10 d old) transferred onto MS medium supplemented with 10 μm paraquat for 48 h. G, Chlorophyll content in the seedlings shown in F. Asterisks indicate P < 0.05 (Student’s t test) when compared with the paraquat-treated Col-0. [See online article for color version of this figure.]
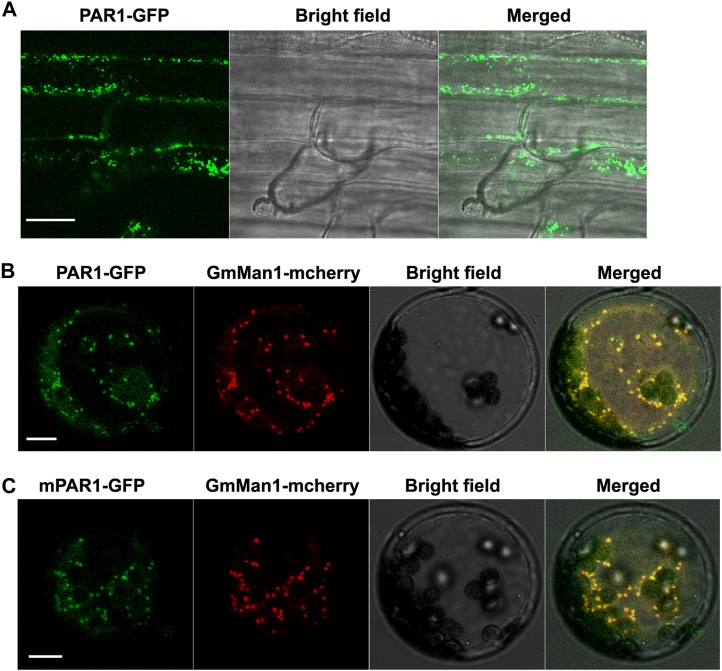
PAR1 Is Localized to the Golgi Apparatus
To gain insight into the molecular mechanism of PAR1-mediated paraquat sensitivity, we examined the subcellular localization of the PAR1-GFP fusion protein in PAR1:PAR1-GFP transgenic plants. This transgene fully rescued the par1-1 mutant phenotype (Fig. 2E). PAR1 is predicted to contain 12 transmembrane domains, implying possible localization to the plasma membrane. Unexpectedly, the fluorescent signal of PAR1-GFP was not detected in the plasma membrane but instead was detected in the cytoplasm with a punctate pattern (Fig. 5A). A similar pattern was observed in Arabidopsis protoplasts transformed with the Super:PAR1-GFP transgene (Fig. 5B). To determine the nature of the punctate structures containing PAR1-GFP, the Super:PAR1-GFP transgene was transiently coexpressed in protoplasts with various marker genes for the plant cell organelles, including the Golgi apparatus (GmMan1-mCherry), endoplasmic reticulum (ER-mCherry), and peroxisome (px-mCherry; Nelson et al., 2007). PAR1-GFP fluorescence colocalized with the Golgi marker GmMan1-mCherry, indicating that these PAR1-GFP-containing punctate structures were Golgi stacks (Fig. 5B). Notably, the mutated form of the par1-1-GFP (mPAR1-GFP) protein showed a similar subcellular localization pattern to that of PAR1-GFP (Fig. 5C), suggesting that the par1-1 mutation does not affect the subcellular localization of PAR1 but may affect its biochemical activity. In contrast, PAR1-GFP did not colocalize with ER-mCherry, px-mCherry, or the mitochondria dye Mitotracker Red (Supplemental Fig. S8). These results indicate that PAR1-GFP is localized to the Golgi apparatus.
Figure 5.
PAR1 is localized to the Golgi apparatus. A, Subcellular localization of PAR1 in the roots of PAR1:PAR1-GFP transgenic plants. Confocal laser scanning microscopy revealed punctate PAR1-GFP fluorescence signals. Bar = 40 μm. B and C, Colocalization of PAR1-GFP (B) and mutated PAR1-GFP (C; mPAR1-GFP) with the Golgi marker in protoplasts. The PAR1-GFP or mPAR1-GFP transgene was cotransformed with a Golgi marker gene (GmMan1-mCherry) into protoplasts prepared from mature leaves of 5-week-old Arabidopsis plants. mPAR1 harbors a mutation identical to the par1-1 mutant allele (see Fig. 2B). Bars = 10 μm. [See online article for color version of this figure.]
We observed that LAT3 shares greater than 75% identity with PAR1 and is also predicted to contain 12 transmembrane domains (Supplemental Fig. S9A). However, mutations in LAT3 did not alter the sensitivity to paraquat (Supplemental Fig. S6). In contrast to the plasma membrane-localized RMV1-GFP (Fujita et al., 2012) and the Golgi-localized PAR1-GFP, the LAT3-GFP protein was localized to the endoplasmic reticulum when transiently expressed in protoplasts (Supplemental Fig. S9, B and C). The different sensitivities of the rmv1, par1, and lat3 mutants to paraquat may be partly attributed to their different subcellular localization patterns.
PAR1 Is Involved in the Transport of Paraquat into Chloroplasts
To characterize the biochemical basis of the paraquat-resistant phenotype of par1, we examined the possible transporter activity of PAR1 on paraquat. We first tested the capability of PAR1 for paraquat uptake. Wild-type, par1-1, and PAR1-overexpressing seedlings were treated with 14C-labeled paraquat, and the radioactivity was measured in the seedlings. No obvious difference in paraquat uptake was observed in the wild-type, par1-1 mutant, and PAR1-overexpressing plants (Supplemental Fig. S10). Because PAR1 is localized to the Golgi, it is expected that PAR1 is not directly involved in the uptake of paraquat.
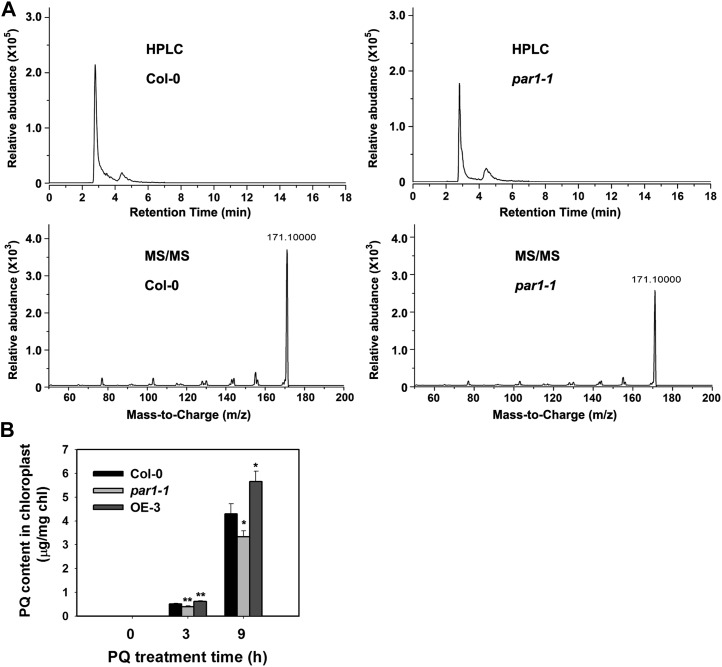
Because the paraquat resistance of a number of weeds has been attributed to a reduced level of intracellular paraquat transport to chloroplasts (Fuerst et al., 1985; Fuerst and Vaughn, 1990; Preston et al., 1992; Norman et al., 1993), it is reasonable to assume that PAR1 may function in the transport of paraquat into the chloroplast. To test this possibility, we analyzed the accumulation of paraquat in the chloroplast of wild-type and par1-1 mutant plants using HPLC coupled with tandem mass spectrometry (MS/MS). We obtained high-quality chloroplast preparations, which did not have detectable contamination with other cellular components (Supplemental Fig. S11). When incubated with 10 μm paraquat for 9 h, the accumulation of paraquat in the chloroplasts of par1-1 was approximately 67% of that in wild-type plants (Fig. 6; Supplemental Fig. S12). In contrast, the accumulation of paraquat is markedly increased in chloroplasts prepared from PAR1-overexpressing plants compared with wild-type plants (Fig. 6B). These results suggest that PAR1 is required for the transport of paraquat to the chloroplast, and the paraquat-resistant phenotype of par1 is likely attributed to the reduced transport of paraquat into the chloroplast.
Figure 6.
Measurement of the paraquat contents in chloroplasts. A, Analysis of the paraquat levels in the chloroplasts of wild-type (Col-0) and par1-1 seedlings. Chloroplasts used for paraquat measurements were prepared from 2-week-old plants treated with 10 μm paraquat for 9 h. HPLC-MS/MS was used to measure the paraquat levels. The HPLC chromatograms (top) and the MS/MS spectra (bottom) are shown. B, Quantitative analysis of the paraquat (PQ) contents in chloroplasts prepared from seedlings of the indicated genotypes assayed using HPLC-MS/MS as shown in A. Seedlings were treated with 10 μm paraquat for 0, 3, or 9 h. The means of three replicates ± sd are shown. Asterisks indicate P < 0.05 (*) and P < 0.01 (**) by Student’s t test when compared with Col-0. Similar results were obtained in three independent experiments.
The Inhibition of Vesicle Trafficking Antagonizes the Cellular Toxicity of Paraquat
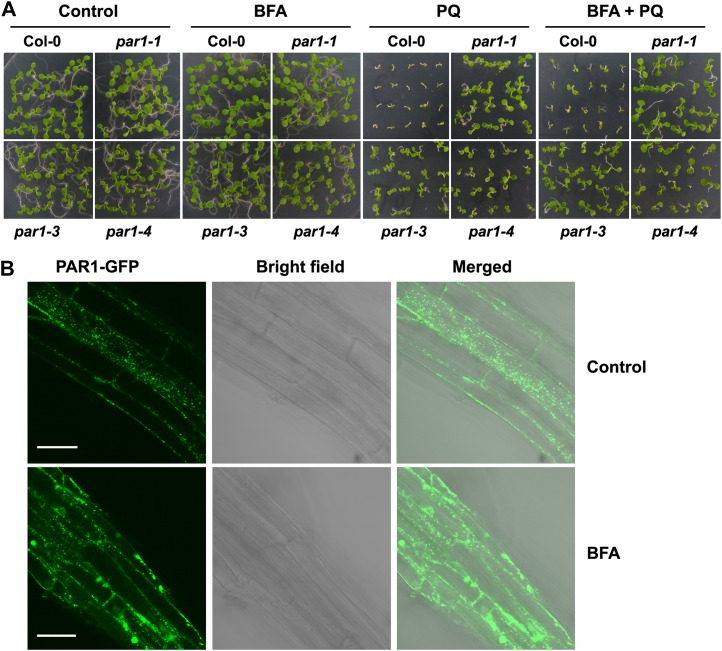
Given that PAR1 is involved in the transport of paraquat into the chloroplast, it is reasonable to expect that the inhibition of intracellular trafficking may reduce the cellular toxicity of paraquat. To test this possibility, we treated wild-type and par1-1 mutant plants with brefeldin A (BFA), an inhibitor that blocks the intracellular trafficking of proteins and causes the formation of visible aggregates (BFA compartments) in the Golgi apparatus and the trans-Golgi network (Nebenführ et al., 2002). Under the assay conditions, BFA did not have detectable effects on plant growth and development (Fig. 7A). However, BFA reduced the sensitivity of wild-type seedlings to paraquat and slightly enhanced the paraquat-resistant phenotype of par1 mutants (Fig. 7A), suggesting that BFA is capable of antagonizing the cellular toxicity of paraquat. Consistent with these observations, when the PAR1:PAR1-GFP transgenic plants were treated with BFA, apparent aggregates of PAR1-GFP fluorescence were observed in the root cells (Fig. 7B). These results provide additional evidence supporting that PAR1-GFP is localized to the Golgi apparatus and that PAR1 is involved in the intracellular transport of paraquat, which is partially inhibited by BFA.
Figure 7.
BFA affects the subcellular localization of PAR1-GFP. A, BFA causes a reduced sensitivity to paraquat. Ten-day-old seedlings of the wild type (Col-0) and par1-1 were germinated and grown on MS medium supplemented with different combinations of BFA (4 μm) and paraquat (1 μm). B, Subcellular localization of PAR1-GFP in the roots of PAR1:PAR1-GFP transgenic seedlings treated with 30 μm BFA for 2.5 h. The GFP fluorescence signal was analyzed using confocal laser scanning microcopy. BFA induces the formation of aggregates of PAR1-GFP. Bars = 40 μm. [See online article for color version of this figure.]
A PAR1-Like Gene, OsPAR1, Is Involved in Paraquat Resistance in Rice
PAR1-like genes are highly conserved (Supplemental Fig. S5), implying a similar function in both monocots and dicots. To explore the possible applications of PAR1-like genes, we investigated the function of a rice (Oryza sativa) PAR1-like gene in paraquat resistance. The rice genome contains four PAR1-like genes, of which Os03g0576900 shows the highest similarity to PAR1 (62%; Supplemental Figs. S3B and S5). We designated Os03g0576900 as OsPAR1.
When expressed in protoplast cells of Arabidopsis and rice, an OsPAR1-GFP fusion protein colocalized with the Golgi marker GmMan1-mCherry (Supplemental Fig. S13), suggesting that OsPAR1 may function similarly to PAR1. To further test this possibility, we generated transgenic rice plants (in the Nipponbare background) overexpressing OsPAR1 driven by a Ubi1 promoter (OsPAR1-OE) or knocked down OsPAR1 expression using an RNA interference (RNAi) construct. In the OsPAR1-overexpressing transgenic lines, the expression of the PAR1 transgene was over 30-fold higher than the expression of the endogenous gene. In contrast, the expression of the endogenous OsPAR1 gene was substantially reduced in the RNAi transgenic lines (Fig. 8A). We have followed the complete life cycle of these transgenic lines and did not observe detectable phenotypes under field growth conditions. In addition, the grain yield of the transgenic plants was similar to that of wild-type plants (Supplemental Fig. S14). These results suggest that the overexpression or knockdown of OsPAR1 has no detrimental effects on the growth and development of rice.
Figure 8.
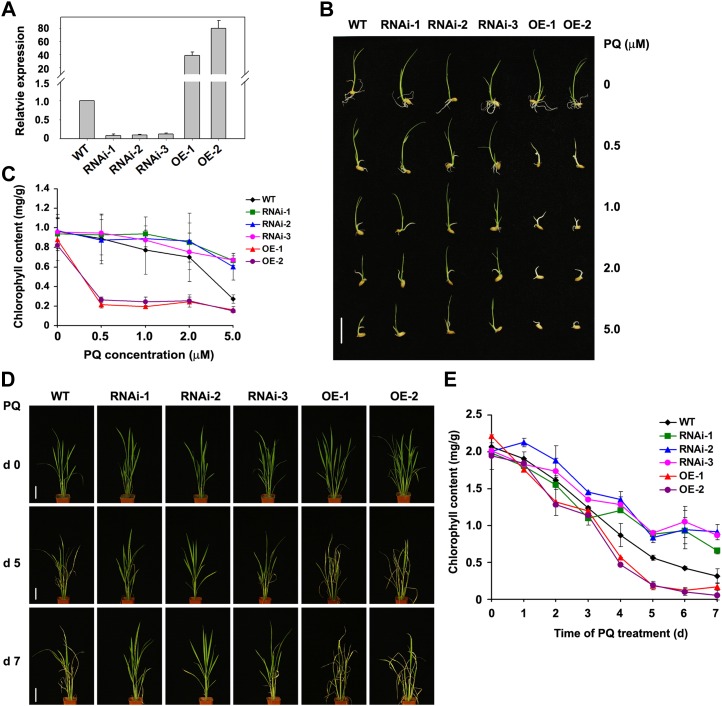
Genetic manipulation of OsPAR1 alters paraquat resistance in transgenic rice plants. A, Analysis of OsPAR1 expression in wild-type (WT; Nipponbare), RNA interference (RNAi), and PAR1-overexpressing (OE) transgenic plants using qRT-PCR. Both RNAi and OE transgenes were expressed under the control of the Ubi1 promoter. The numbers refer to transgenic lines. RNA prepared from 6-d-old seedlings was used for the qRT-PCR analysis. B, Six-day-old wild-type, RNA interference, and PAR1-overexpressing transgenic seedlings germinated and grown in the presence of different concentrations of paraquat (PQ) as indicated. Bar = 2 cm. C, Chlorophyll content of the seedlings shown in B. The means of three replicates (biological repeats) ± sd are shown. D, Field-grown plants (8 weeks old) sprayed with 140 μm paraquat followed by continued growth in the field for different durations. Representative plants are shown. Bars = 10 cm. E, Analysis of chlorophyll contents in the leaves of wild-type and transgenic plants treated with 140 μm paraquat for the indicated durations. The means of three replicates (biological repeats) ± sd are shown. [See online article for color version of this figure.]
We examined the sensitivity of these transgenic plants to paraquat during germination. Wild-type rice seedlings were slightly more resistant to paraquat than Arabidopsis. Under our assay conditions, rice was tolerant to approximately 1 μm paraquat (Fig. 8, B and C). In the germination assay, the RNAi knockdown transgenic lines were more resistant to paraquat than the wild-type plants, especially when treated with the relatively high concentrations of paraquat (Fig. 8, B and C). In contrast, the OsPAR1-overexpressing transgenic seedlings were hypersensitive to paraquat (Fig. 8, B and C). When treated with 0.5 μm paraquat, the chlorophyll level in the OsPAR1-overexpressing transgenic seedlings was reduced by greater than 70% (Fig. 8C). Under field growth conditions, we sprayed the transgenic plants with paraquat using a concentration equivalent to that used by farmers (approximately 140 μm). Similar to the results of the germination assay, the overexpression of OsPAR1 dramatically increased sensitivity to paraquat, leading to the death of the transgenic plants at 4 to 5 d after spraying (Fig. 8, D and E; Supplemental Fig. S15). However, knockdown of the OsPAR1 expression level conferred the transgenic plants significant resistance to paraquat (Fig. 8, D and E). These results indicate that OsPAR1 functions similar to PAR1 in Arabidopsis in the regulation of paraquat sensitivity.
DISCUSSION
In this study, we report the identification and characterization of a paraquat-resistant mutant, par1. We present multiple lines of evidence obtained from studies of molecular genetics, biochemistry, and cell biology, demonstrating that PAR1 encodes a Golgi-localized putative transporter protein involved in the intracellular transport of paraquat to the chloroplast. Because PSI is the main target site of paraquat in higher plants, the paraquat-resistant phenotype of par1 is likely caused by the reduced transport of paraquat to the chloroplast.
Since paraquat was used as an herbicide worldwide half a century ago, many paraquat-resistant biotypes, mostly as weeds, have been identified. The biochemical mechanisms of paraquat resistance in these weeds have been extensively investigated, from which several models have been proposed (Fuerst and Vaughn, 1990; Norman et al., 1993). Paraquat is presumed to enter plant cells through plasma membrane-localized transporters (Fujita et al., 2012; Xi et al., 2012) and to be subsequently transported into the chloroplast by an unknown mechanism. Paraquat resistance in several weeds is associated with the reduced accumulation of paraquat in the chloroplast of resistant biotypes compared with that of susceptible biotypes, which was proposed as excluding paraquat from the chloroplast (Fuerst et al., 1985; Fuerst and Vaughn, 1990; Preston et al., 1992; Norman et al., 1993). However, it has also been argued that reduced paraquat translocation is a consequence, but not a cause, of paraquat resistance (Chun et al., 1997a; Soar et al., 2003). The discovery that Golgi-localized PAR1 is involved in the accumulation of paraquat in the chloroplast suggests the involvement of an intracellular transport mechanism that actively translocates paraquat into the chloroplast. Although one cannot exclude the possibility that the reduced accumulation of paraquat is caused by the exclusion of the herbicide from the chloroplast, as proposed in the sequestration model (Fuerst and Vaughn, 1990; Norman et al., 1993), our results indicate that active transport of paraquat into the chloroplast is an important mechanism that causes the toxicity to plant cells. This view is supported by the observation made in studies on the paraquat-resistant weed R. glutinosa, in which paraquat tolerance is attributed to paraquat metabolism outside of the chloroplast (Chun et al., 1997a). Additionally, we notice that the accumulation of paraquat in the chloroplast is only partially blocked by the par1 mutation, implying the presence of functionally redundant genes or PAR1-independent routes to mediate the transport of paraquat into the chloroplast. Consistent with this notion, a considerable fraction of paraquat is distributed in the cytosol and the vacuole of paraquat-treated maize (Zea mays) seedlings, implying that other organelles may also be targets of paraquat in plant cells (Hart et al., 1992).
Resistance to paraquat has also been demonstrated by the reduced uptake of paraquat by two Arabidopsis mutants, atpdr11 and rmv1 (Fujita et al., 2012; Xi et al., 2012). Both AtPDR11 and RMV1 are plasma membrane-localized transporters. RMV1 (LAT1) and PAR1 (LAT4) belong to the same LAT family and have considerable structural similarities. However, these two proteins are localized to different subcellular foci, implying that they are involved in distinctive cellular activities. Consistent with this notion, whereas RMV1 plays an important role in the intercellular uptake of paraquat, PAR1 is involved in the accumulation of paraquat in the chloroplast. These two processes are presumed to be major factors responsible for the toxicity of paraquat in plant cells. Notably, RMV1/LAT1 is a functional transporter for polyamines that are structurally similar to paraquat (Fujita et al., 2012). A recent study showed OsPUT1 (for POLYAMINE UPTAKE TRANSPORTER1), which shares approximately 60% identity with OsPAR1, is a high-affinity transporter of polyamines when expressed in yeast (Saccharomyces cerevisiae) cells (Mulangi et al., 2012). Likewise, PAR1 and OsPAR1 may have a similar function in the transport of polyamines. Again, the different subcellular localization patterns of RMV1/LAT1, PAR1, and OsPAR1 imply that these proteins are involved in different processes of polyamine transport.
In animal models, paraquat has been associated with Parkinson’s disease via the induction of dopaminergic toxicity in the brain (Berry et al., 2010). To execute its neurotoxicity, paraquat must cross the blood-brain barrier to enter the brain. How paraquat penetrates the blood-brain barrier and whether paraquat is a cause of Parkinson’s disease in humans remain controversial (Bartlett et al., 2009; Berry et al., 2010). Nonetheless, the results of a competition experiment using various amino acids suggest that paraquat is taken up into the brain through a neutral amino acid transport system (Shimizu et al., 2001). Therefore, the utilization of the transporter system for structural analogs, such as amino acids or polyamines, for the transport of paraquat appears to be a conserved mechanism in both animals and plants.
In mammalian cells, the transport of polyamines has been proposed to be mediated by a vesicular sequestration mechanism or an endocytosis-based mechanism (Soulet et al., 2004; Poulin et al., 2012). In both models, polyamines are actively transported by LAT transporters into the cytosol or various cellular organelles such as lysosomes (Poulin et al., 2012). The observation that PAR1 is localized in the Golgi apparatus and that BFA partially relieves the paraquat toxicity suggests that paraquat should be, at least partly, imported into the chloroplast through a BFA-sensitive transport system. However, this has not been reported for the presence of a vesicle trafficking-based transport route from the Golgi apparatus to the chloroplast. Therefore, it remains questionable that paraquat is directly transported from the Golgi apparatus into the chloroplast. We speculate that PAR1 may transport paraquat (or polyamines) into the cytosol or an unidentified organelle by endocytosis, and paraquat is then transported into the chloroplast by unknown transporters. An alternative explanation is that a small fraction of PAR1 is also localized at the chloroplast, which is under the detection limit in our assay. In this case, paraquat is presumably to be directly transported from the cytosol into the chloroplast.
Paraquat is one of the most widely used herbicides in the world. In the agricultural application of herbicides, a major concern is to minimize the toxicity to crops and the ecological system. Moreover, an increasing problem is the generation of herbicide-resistant weed biotypes. Currently, more than 350 herbicide-resistant biotypes have been identified, including several biotypes with multiple herbicide resistance (Vaughn, 2003; Powles and Preston, 2006; Yu et al., 2007; http://www.weedscience.org). An efficient approach to killing herbicide-resistant weeds is to use herbicides with different modes of action or with different targets in plant cells. Paraquat targets PSI, which is mechanistically distinct from other commonly used herbicides, such as glyphosate, phosphinothricin, and sulfonylurea. Moreover, paraquat exhibits nonselective and rapid action and is biologically inactive in soil. Despite these advantageous features, the application of paraquat in agriculture is relatively limited, and this herbicide is primarily used in orchards, plantation crops, and conservation tillage systems (Bromilow, 2004). This limitation is partly attributed to the lack of a molecular target that can be genetically modified to confer paraquat resistance to crops, in contrast to several other herbicides, such as glyphosate, whose utilization has been successfully manipulated by genetic engineering (Comai et al., 1983, 1985). Therefore, it will be of paramount importance to identify key targets that can be used to genetically manipulate paraquat-resistant crops. The identification and characterization of AtPDR11, RMV1, PAR1, and, in particular, OsPAR1 have made genetic manipulations of paraquat-resistant crops feasible.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) accessions Col-0 and Ler and the rice (Oryza sativa japonica) Nipponbare variety were used in this study. The Arabidopsis plants were grown at 22°C under a long-day (16-h-light/8-h-dark cycle) photoperiod at 100 μmol m−2 s−1 light intensity with 50% to 70% relative humidity in soil or on solid Murashige and Skoog (MS) medium (Sigma-Aldrich) containing 1.5% Suc and 0.8% agar. The rice plants were grown in a greenhouse or in the field in Beijing and Hainan.
The T-DNA insertional mutants par1-5 (SALK_119707C), par1-6 (SALK_129045), and lat3-1 (SAIL_270_G10) were obtained from the Arabidopsis Biological Resource Center, and lat3-2 (GABI_890C10) was obtained from the European Arabidopsis Stock Center. Homozygous T-DNA insertional mutants were identified using PCR-based genotyping with a T-DNA-specific primer and the gene-specific primers PAR1-p5 and PAR1-p6 (for par1-5 and par1-6) or LAT3-p1 and LAT3-p2 (for lat3-1 and lat3-2), respectively. All primers used in this study are listed in Supplemental Tables S1 and S2.
Detection of Cell Death, Superoxide, and Hydrogen Peroxide
The leaves were stained with Evans blue, NBT, and DAB to detect cell death, superoxide, and hydrogen peroxide, respectively, as described previously (Chen et al., 2009).
Genetic Mapping and Cloning of the PAR1 Gene
To map the par1-1 mutation, the par1-1 mutant (Col-0 background) was crossed with wild-type Ler plants. A total of 1,014 F2 plants exhibiting the paraquat-resistant phenotype were selected as a mapping population. Genomic DNA from these F2 plants was extracted and used for PCR-based mapping using simple sequence length polymorphism, cleaved-amplified polymorphic sequence, and derived cleaved-amplified polymorphic sequence markers. Additional mapping markers were developed based on insertions/deletions identified from the Cereon Arabidopsis polymorphism and Ler sequence collection (www.arabidopsis.org). These markers are listed in Supplemental Table S2. Genomic DNA fragments corresponding to candidate genes were PCR amplified from par1 mutants and used in the DNA sequencing analysis to identify the mutations.
Plasmid Construction and Plant Transformation
To generate the PAR1:GUS fusion, a 2.1-kb genomic fragment upstream of the PAR1 ATG start codon was PCR amplified using PAR1-p7 and PAR1-p8 primers. The amplified fragment was fused with the GUS reporter gene in the binary vector pZPGUS2 (Diener et al., 2000).
The PAR1 complementary DNA (cDNA) fragment lacking a stop codon was PCR amplified using primers PAR1-p9 and PAR1-p10 and fused with GFP in frame in pUC-GFP (Walter et al., 2004) to generate the transient expression vector 35S:PAR1-GFP. The fragment was also fused upstream of GFP or MYC under the control of a Super promoter in the pCAMBIA1300 vector (CAMBIA) to generate the Super:PAR1-GFP or Super:PAR1-MYC construct, respectively.
For the molecular complementation assay, a 3.9-kb genomic fragment comprising the PAR1 promoter, coding region, and 3′ untranslated region was PCR amplified from the genomic DNA of Col-0 plants using PAR1-p3 and PAR1-p2 primers. The PCR product was subsequently cloned into pCAMBIA1300 to generate the PAR1:PAR1 construct. To generate the PAR1:PAR1-GFP construct, an XbaI/ClaI fragment of PAR1:PAR1 containing the promoter region and the 5′ end region of the PAR1 genomic DNA was ligated with a ClaI/SacI fragment of Super:PAR1-GFP containing 3′ end sequences of PAR1 genomic DNA fused in frame with GFP, which was cloned into pCAMBIA1300-NOS to generate the PAR1:PAR1-GFP construct.
To generate pER10-PAR1-FLAG, a PAR1 genomic DNA fragment was PCR amplified using primers PAR-p11 and PAR-p12 and cloned into the XhoI and SmaI sites of pSK-c-FLAG vector, which contain a FLAG epitope followed by an in-frame stop codon. The PAR1-FLAG fusion fragment was subsequently cloned into the XhoI and SpeI sites of pER10 (Zuo et al., 2000).
A 5.3-kb rice genomic DNA fragment containing the OsPAR1 coding sequences was PCR amplified from the Nipponbare genomic DNA using the primer pairs OsPAR1F1 and OsPAR1B1. The PCR fragment was digested with BamHI and NheI and then cloned into the BamHI and SpeI sites of pTCK303 under the control of a Ubi1 promoter (Wang et al., 2004). To construct the RNAi vector, a 0.43-kb OsPAR1 cDNA fragment corresponding to part of exon 4 (the last exon) and the 3′ untranslated region was obtained by RT-PCR, using the primer pair OsPAR1F2 and OsPAR1B2. Two copies of this cDNA fragment were inserted into pTCK303 in a tail-to-tail configuration (Wang et al., 2004).
The Agrobacterium tumefaciens strain GV3101 carrying different constructs was used to transform wild-type or par1 mutant plants using the floral dip method (Clough and Bent, 1998). Genetic transformation of rice was performed as described (Hiei et al., 1994).
qRT-PCR
Total RNA was prepared with TriReagent (Sigma-Aldrich) followed by treatment with RNase-free DNase I (Promega) at 37°C for 1 h. The treated RNA samples (1 μg each) were used as templates for first-strand cDNA synthesis. Real-time PCR was performed using an Applied Biosystems 7500 real-time PCR system with SYBR Premix Ex Taq (Takara). The relative expression levels were calculated as described previously (Deng et al., 2010; Wang et al., 2011).
Histochemical GUS Staining and Analysis of Subcellular Localization
The histochemical detection of GUS activity was performed as described previously (Jefferson et al., 1987). For the transient expression analysis, the Super:PAR1-GFP and various organelle-specific marker genes (Nelson et al., 2007) were cotransformed into protoplasts prepared from mature leaves of 5-week-old Arabidopsis plants using a previously described protocol (Yoo et al., 2007). The preparation and transformation of rice protoplast cells were performed as described (Zhang et al., 2011). The roots of 7-d-old stable transgenic plants expressing PAR1:PAR1-GFP were treated without or with BFA (30 μm) for 2.5 h as described previously (Lam et al., 2009).
The fluorescence of GFP and mCherry in the protoplasts and transgenic plants was visualized using a confocal laser scanning microscope (LSM510; Carl Zeiss).
Preparation of Protein Extracts and Immunoblot Analysis
Total protein exacts were prepared from freshly collected or frozen materials as described previously (Huang et al., 2009). Immunoblot analysis was carried out as described previously (Huang et al., 2009).
Chloroplast Preparations
Two-week-old Col-0, par1-1, and overexpressing transgenic plants were submerged in 10 μm paraquat solutions for 3 and 9 h in the dark. The preparation of chloroplasts was performed as described previously (Nishimura et al., 1976). After discontinuous Suc density gradient centrifugation, 0.5-mL fractions were collected, and aliquots were used for measuring the paraquat and chlorophyll contents.
Analysis of Paraquat Uptake
Seedlings from the wild type, the par1-1 mutant, and the PAR1-OE3 line were grown on MS medium for 7 d under dark conditions. The paraquat uptake experiment was performed as described previously (Xi et al., 2012) with modifications. Briefly, the seedlings were submerged in 3 mL of pretreatment buffer (5 mm MES-Tris buffer, pH 6.0, and 0.02 mm CaCl2) for 20 min followed by incubation in 3 mL of treatment buffer (5 mm MES-Tris buffer, pH 6.0, 0.02 mm CaCl2, and 2 μm [14C]paraquat [2 kilobecquerel mL−1]) for 0.5 to 9 h. After the incubation, the sample was washed four times with 10 mL of washing buffer (5 mm MES-Tris buffer, pH 6.0, 0.02 mm CaCl2, and 10 μm paraquat). The seedlings were blotted, weighed, and soaked in 1 mL of scintillation fluid. The radioactivity in the seedlings was measured using a Liquid Scintillation Spectrometer-1450 (Perkin Elmer Wallac).
Measurement of Paraquat Level by HPLC-MS/MS
Approximately 0.5 mL of the chloroplast preparations was lysed by adding 2 mL of milli-Q water, followed by centrifugation at 10,000g for 10 min. The supernatant was used for subsequent HPLC-MS/MS analysis.
The samples were purified using solid-phase extraction columns (Oasis Waters). The column was activated with 3 mL of methanol and 3 mL of water, and a 2-mL sample was loaded onto the column. A volume of 1 mL of a methanol:milli-Q water (1:1) solution was used to rinse the column, and paraquat was eluted with 1 mL of a trifluoroacetic acid:acetonitrile:milli-Q water (2:84:14) solution. The eluted solution was dried in a 50°C water bath under a stream of nitrogen and reconstituted with 500 μL of a milli-Q water:acetonitrile (1:1) solution for HPLC-MS/MS analysis.
HPLC-MS/MS analysis was performed using an Agilent 1260 HPLC device coupled with an Agilent 6460 triple quadrupole mass spectrometer (Agilent Technologies). A Syncronis HILIC (100 mm × 2.1 mm, 5 μm; ThermoFisher Scientific) column was used. Mobile phase A consisted of water with 10 mm ammonium formate and 0.1% acetonitrile, and mobile phase B contained acetonitrile. The samples were separated using gradient elution. The flow rate was 200 μL min−1, and the injection volume was 10 μL. The Agilent 6460 mass spectrometer was operated in the positive product ion scan mode. The following instrument parameters were used: spray voltage, 3,500 V; heater temperature, 350°C; sheath gas pressure, 35 ψ; auxiliary gas pressure, 10 ψ; capillary temperature, 300°C; fragmenter voltage, 135 V; and collision-induced dissociation energy, 30 eV. The peak area of the paraquat fragment at mass-to-charge ratio = 171 was used for quantitative measurements.
Measurement of Chlorophyll Level
Total chlorophyll was extracted from Arabidopsis or rice leaves and analyzed as described (Lichtenthaler, 1987). All leaves from randomly collected seedlings or plants were used for the extraction of total chlorophyll. All experiments were repeated at least three times with biological replicates, and mean values of these replicates are presented.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers PAR1/LAT4 (AT1G31830), LAT3 (AT1G31820), and OsPAR1 (Os03g0576900).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The par1 mutant phenotype under normal growth conditions and abiotic stresses.
Supplemental Figure S2. Dose response of wild type and par1-1 plants to paraquat.
Supplemental Figure S3. Analysis of the structures of PAR1 and PAR1-like proteins.
Supplemental Figure S4. Analysis of PAR1 expression in par1 mutants.
Supplemental Figure S5. A phylogenetic tree of PAR1 and PAR1-like proteins.
Supplemental Figure S6. Characterization of the lat3 mutants.
Supplemental Figure S7. Characterization of the pER10-PAR1 transgenic plants.
Supplemental Figure S8. Subcellular localization of PAR1 in Arabidopsis protoplasts.
Supplemental Figure S9. Subcellular localization of LAT3 in Arabidopsis protoplasts.
Supplemental Figure S10. Measurement of paraquat uptake.
Supplemental Figure S11. Immunoblot analysis of chloroplast proteins.
Supplemental Figure S12. Analysis of paraquat by HPLC-MS/MS.
Supplemental Figure S13. Analysis of subcellular localization of OsPAR1.
Supplemental Figure S14. Analysis of the grain phenotype of the transgenic rice.
Supplemental Figure S15. Analysis of the paraquat sensitivity of the transgenic rice.
Supplemental Table S1. PCR primers used in this study.
Supplemental Table S2. Markers used for genetic mapping of par1-1.
Acknowledgments
We thank the Arabidopsis Biological Resource Center, the European Arabidopsis Stock Center, Chengbin Xiang (University of Science and Technology of China), and Kazuo Shinozaki (RIKEN Plant Science Center) for providing mutant seeds and the cellular organelle markers, Kang Chong (Institute of Botany, Chinese Academy of Sciences) and Zhizhong Gong (China Agricultural University) for providing vectors, and Bo Ren (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for advice on the phylogenetic analysis.
Glossary
- ROS
reactive oxygen species
- NO
nitric oxide
- LAT
l-type amino acid
- Col-0
Columbia-0
- DAB
3,3′-diaminobenzidine
- NBT
nitroblue tetrazolium
- Ler
Landsberg erecta
- RT
reverse transcription
- qRT
quantitative reverse transcription
- MS/MS
tandem mass spectrometry
- BFA
brefeldin A
- RNAi
RNA interference
- MS
Murashige and Skoog
- T-DNA
transfer DNA
- cDNA
complementary DNA
References
- Ahlfors R, Lång S, Overmyer K, Jaspers P, Brosché M, Tauriainen A, Kollist H, Tuominen H, Belles-Boix E, Piippo M, et al. (2004) Arabidopsis RADICAL-INDUCED CELL DEATH1 belongs to the WWE protein-protein interaction domain protein family and modulates abscisic acid, ethylene, and methyl jasmonate responses. Plant Cell 16: 1925–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Amsellem Z, Jansen MAK, Driesenaar ARJ, Gressel J. (1993) Developmental variability of photooxidative stress tolerance in paraquat-resistant Conyza. Plant Physiol 103: 1097–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arisi A-CM, Cornic G, Jouanin L, Foyer CH. (1998) Overexpression of iron superoxide dismutase in transformed poplar modifies the regulation of photosynthesis at low CO2 partial pressures or following exposure to the prooxidant herbicide methyl viologen. Plant Physiol 117: 565–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babbs CF, Pham JA, Coolbaugh RC. (1989) Lethal hydroxyl radical production in paraquat-treated plants. Plant Physiol 90: 1267–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett RM, Holden JE, Nickles RJ, Murali D, Barbee DL, Barnhart TE, Christian BT, DeJesus OT. (2009) Paraquat is excluded by the blood brain barrier in rhesus macaque: an in vivo PET study. Brain Res 1259: 74–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry C, La Vecchia C, Nicotera P. (2010) Paraquat and Parkinson’s disease. Cell Death Differ 17: 1115–1125 [DOI] [PubMed] [Google Scholar]
- Bonneh-Barkay D, Reaney SH, Langston WJ, Di Monte DA. (2005) Redox cycling of the herbicide paraquat in microglial cultures. Brain Res Mol Brain Res 134: 52–56 [DOI] [PubMed] [Google Scholar]
- Bowler C, Slooten L, Vandenbranden S, De Rycke R, Botterman J, Sybesma C, Van Montagu M, Inzé D. (1991) Manganese superoxide dismutase can reduce cellular damage mediated by oxygen radicals in transgenic plants. EMBO J 10: 1723–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromilow RH. (2004) Paraquat and sustainable agriculture. Pest Manag Sci 60: 340–349 [DOI] [PubMed] [Google Scholar]
- Chen R, Sun S, Wang C, Li Y, Liang Y, An F, Li C, Dong H, Yang X, Zhang J, et al. (2009) The Arabidopsis PARAQUAT RESISTANT2 gene encodes an S-nitrosoglutathione reductase that is a key regulator of cell death. Cell Res 19: 1377–1387 [DOI] [PubMed] [Google Scholar]
- Chun J-C, Kim S-E, Ma S-Y. (1997a) Inactivation of paraquat by an aqueous extract of Rehmannia glutinosa. Pestic Sci 50: 5–10 [Google Scholar]
- Chun JC, Ma SY, Kim SE, Lee HJ. (1997b) Physiological responses of Rehmannia glutinosa to paraquat and its tolerance mechanisms. Pestic Biochem Physiol 59: 51–63 [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Comai L, Facciotti D, Hiatt WR, Thompson G, Rose RE, Stalker DM. (1985) Expression in plants of a mutant aroA gene from Salmonella typhimurium confers tolerance to glyphosate. Nature 317: 741–744 [Google Scholar]
- Comai L, Sen LC, Stalker DM. (1983) An altered aroA gene product confers resistance to the herbicide glyphosate. Science 221: 370–371 [DOI] [PubMed] [Google Scholar]
- Delledonne M, Zeier J, Marocco A, Lamb C. (2001) Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive disease resistance response. Proc Natl Acad Sci USA 98: 13454–13459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Dong H, Mu J, Ren B, Zheng B, Ji Z, Yang W-C, Liang Y, Zuo J. (2010) Arabidopsis histidine kinase CKI1 acts upstream of histidine phosphotransfer proteins to regulate female gametophyte development and vegetative growth. Plant Cell 22: 1232–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener AC, Li H, Zhou W, Whoriskey WJ, Nes WD, Fink GR. (2000) Sterol methyltransferase 1 controls the level of cholesterol in plants. Plant Cell 12: 853–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodge AD. (1971) The mode of action of the bipyridylium herbicides, paraquat and diquat. Endeavour 30: 130–135 [DOI] [PubMed] [Google Scholar]
- Donahue JL, Okpodu CM, Cramer CL, Grabau EA, Alscher RG. (1997) Responses of antioxidants to paraquat in pea leaves (relationships to resistance). Plant Physiol 113: 249–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feechan A, Kwon E, Yun B-W, Wang Y, Pallas JA, Loake GJ. (2005) A central role for S-nitrosothiols in plant disease resistance. Proc Natl Acad Sci USA 102: 8054–8059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst EP, Nakatani HY, Dodge AD, Penner D, Arntzen CJ. (1985) Paraquat resistance in Conyza. Plant Physiol 77: 984–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst EP, Vaughn KC. (1990) Mechanisms of paraquat resistance. Weed Technol 4: 150–156 [Google Scholar]
- Fujibe T, Saji H, Arakawa K, Yabe N, Takeuchi Y, Yamamoto KT. (2004) A methyl viologen-resistant mutant of Arabidopsis, which is allelic to ozone-sensitive rcd1, is tolerant to supplemental ultraviolet-B irradiation. Plant Physiol 134: 275–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii T, Yokoyama E, Inoue K, Sakurai H. (1990) The sites of electron donation of photosystem I to methyl viologen. Biochim Biophys Acta 1015: 41–48 [Google Scholar]
- Fujita M, Fujita Y, Iuchi S, Yamada K, Kobayashi Y, Urano K, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K. (2012) Natural variation in a polyamine transporter determines paraquat tolerance in Arabidopsis. Proc Natl Acad Sci USA 109: 6343–6347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta AS, Heinen JL, Holaday AS, Burke JJ, Allen RD. (1993) Increased resistance to oxidative stress in transgenic plants that overexpress chloroplastic Cu/Zn superoxide dismutase. Proc Natl Acad Sci USA 90: 1629–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley TJ. (1979) Review of the toxicology of paraquat (1,1′-dimethyl-4,4′-bipyridinium chloride). Clin Toxicol 14: 1–46 [DOI] [PubMed] [Google Scholar]
- Hart JJ, Di Tomaso JM, Linscott DL, Kochian LV. (1992) Characterization of the transport and cellular compartmentation of paraquat in roots of intact maize seedlings. Pestic Biochem Physiol 43: 212–222 [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Huang X, Zhang X, Yang S. (2009) A novel chloroplast-localized protein EMB1303 is required for chloroplast development in Arabidopsis. Cell Res 19: 1205–1216 [DOI] [PubMed] [Google Scholar]
- Jack DL, Paulsen IT, Saier MH. (2000) The amino acid/polyamine/organocation (APC) superfamily of transporters specific for amino acids, polyamines and organocations. Microbiology 146: 1797–1814 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jóri B, Soós V, Szegő D, Páldi E, Szigeti Z, Rácz I, Lásztity D. (2007) Role of transporters in paraquat resistance of horseweed Conyza canadensis (L.) Cronq. Pestic Biochem Physiol 88: 57–65 [Google Scholar]
- Lam SK, Cai Y, Tse YC, Wang J, Law AH, Pimpl P, Chan HY, Xia J, Jiang L. (2009) BFA-induced compartments from the Golgi apparatus and trans-Golgi network/early endosome are distinct in plant cells. Plant J 60: 865–881 [DOI] [PubMed] [Google Scholar]
- Lee U, Wie C, Fernandez BO, Feelisch M, Vierling E. (2008) Modulation of nitrosative stress by S-nitrosoglutathione reductase is critical for thermotolerance and plant growth in Arabidopsis. Plant Cell 20: 786–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Gong Z, Koiwa H, Niu X, Espartero J, Zhu X, Veronese P, Ruggiero B, Bressan RA, Weller SC, et al. (2001) Bar-expressing peppermint (Mentha × piperita L. var. Black Mitcham) plants are highly resistant to the glufosinate herbicide Liberty. Mol Breed 8: 109–118 [Google Scholar]
- Lichtenthaler HK. (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148: 350–382 [Google Scholar]
- Mulangi V, Chibucos MC, Phuntumart V, Morris PF. (2012) Kinetic and phylogenetic analysis of plant polyamine uptake transporters. Planta 236: 1261–1273 [DOI] [PubMed] [Google Scholar]
- Murgia I, Tarantino D, Vannini C, Bracale M, Carravieri S, Soave C. (2004) Arabidopsis thaliana plants overexpressing thylakoidal ascorbate peroxidase show increased resistance to paraquat-induced photooxidative stress and to nitric oxide-induced cell death. Plant J 38: 940–953 [DOI] [PubMed] [Google Scholar]
- Nebenführ A, Ritzenthaler C, Robinson DG. (2002) Brefeldin A: deciphering an enigmatic inhibitor of secretion. Plant Physiol 130: 1102–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Nishimura M, Graham D, Akazawa T. (1976) Isolation of intact chloroplasts and other cell organelles from spinach leaf protoplasts. Plant Physiol 58: 309–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman MA, Fuerst EP, Smeda RJ, Vaughn KC. (1993) Evaluation of paraquat resistance mechanisms in Conyza. Pestic Biochem Physiol 46: 236–249 [Google Scholar]
- Poulin R, Casero RA, Soulet D. (2012) Recent advances in the molecular biology of metazoan polyamine transport. Amino Acids 42: 711–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powles SB, Preston C. (2006) Evolved glyphosate resistance in plants: biochemical and genetic basis of resistance. Weed Technol 20: 282–289 [Google Scholar]
- Powles SB, Tucker ES, Morgan TR. (1989) A capeweed (Arctotheca calendula) biotype in Australia resistant to bipyridyl herbicides. Weed Sci 37: 60–62 [Google Scholar]
- Preston C, Balachandran S, Powles SB. (1994) Investigations of mechanisms of paraquat resistance to bipyridyl herbicides in Arctotheca calendula (L.) Levyns. Plant Cell Environ 17: 1113–1123 [Google Scholar]
- Preston C, Holtum JAM, Powles SB. (1992) On the mechanism of resistance to paraquat in Hordeum glaucum and H. leporinum: delayed inhibition of photosynthetic O2 evolution after paraquat application. Plant Physiol 100: 630–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosecka J, Orlov AV, Fantin YS, Zinchenko VV, Babykin MM, Tichy M. (2009) A novel ATP-binding cassette transporter is responsible for resistance to viologen herbicides in the cyanobacterium Synechocystis sp. PCC 6803. FEBS J 276: 4001–4011 [DOI] [PubMed] [Google Scholar]
- Shaaltiel Y, Gressel J. (1986) Multienzyme oxygen radical detoxifying system correlated with paraquat resistance in Conyza bonariensis. Pestic Biochem Physiol 26: 22–28 [Google Scholar]
- Shimizu K, Ohtaki K, Matsubara K, Aoyama K, Uezono T, Saito O, Suno M, Ogawa K, Hayase N, Kimura K, et al. (2001) Carrier-mediated processes in blood-brain barrier penetration and neural uptake of paraquat. Brain Res 906: 135–142 [DOI] [PubMed] [Google Scholar]
- Soar CJ, Karotam J, Preston C, Powles SB. (2003) Reduced paraquat translocation in paraquat resistant Arctotheca calendula (L.) Levyns is a consequence of the primary resistance mechanism, not the cause. Pestic Biochem Physiol 76: 91–98 [Google Scholar]
- Soulet D, Gagnon B, Rivest S, Audette M, Poulin R. (2004) A fluorescent probe of polyamine transport accumulates into intracellular acidic vesicles via a two-step mechanism. J Biol Chem 279: 49355–49366 [DOI] [PubMed] [Google Scholar]
- Suntres ZE. (2002) Role of antioxidants in paraquat toxicity. Toxicology 180: 65–77 [DOI] [PubMed] [Google Scholar]
- Tsugane K, Kobayashi K, Niwa Y, Ohba Y, Wada K, Kobayashi H. (1999) A recessive Arabidopsis mutant that grows photoautotrophically under salt stress shows enhanced active oxygen detoxification. Plant Cell 11: 1195–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn KC. (2003) Herbicide resistance work in the United States Department of Agriculture-Agricultural Research Service. Pest Manag Sci 59: 764–769 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang Z, Chen C, Xu Y, Jiang R, Han Y, Xu Z, Chong K. (2004) A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L.). Plant Mol Biol Rep 22: 409–417 [Google Scholar]
- Wang Z, Meng P, Zhang X, Ren D, Yang S. (2011) BON1 interacts with the protein kinases BIR1 and BAK1 in modulation of temperature-dependent plant growth and cell death in Arabidopsis. Plant J 67: 1081–1093 [DOI] [PubMed] [Google Scholar]
- Xi J, Xu P, Xiang C-B. (2012) Loss of AtPDR11, a plasma membrane-localized ABC transporter, confers paraquat tolerance in Arabidopsis thaliana. Plant J 69: 782–791 [DOI] [PubMed] [Google Scholar]
- Ye B, Gressel J. (2000) Transient, oxidant-induced antioxidant transcript and enzyme levels correlate with greater oxidant-resistance in paraquat-resistant Conyza bonariensis. Planta 211: 50–61 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yu Q, Cairns A, Powles S. (2007) Glyphosate, paraquat and ACCase multiple herbicide resistance evolved in a Lolium rigidum biotype. Planta 225: 499–513 [DOI] [PubMed] [Google Scholar]
- Yu Q, Cairns A, Powles SB. (2004) Paraquat resistance in a population of Lolium rigidum. Funct Plant Biol 31: 247–254 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Su J, Duan S, Ao Y, Dai J, Liu J, Wang P, Li Y, Liu B, Feng D, et al. (2011) A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Chua NH. (2000) An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24: 265–273 [DOI] [PubMed] [Google Scholar]