A thioredoxin reductase influences chlorophyll biosynthesis by reducing target cysteines in its biosynthesis enzymes.
Abstract
The NADPH-dependent thioredoxin reductase C (NTRC) is involved in redox-related regulatory processes in chloroplasts and nonphotosynthetic active plastids. Together with 2-cysteine peroxiredoxin, it forms a two-component peroxide-detoxifying system that acts as a reductant under stress conditions. NTRC stimulates in vitro activity of magnesium protoporphyrin IX monomethylester (MgPMME) cyclase, most likely by scavenging peroxides. Reexamination of tetrapyrrole intermediate levels of the Arabidopsis (Arabidopsis thaliana) knockout ntrc reveals lower magnesium protoporphyrin IX (MgP) and MgPMME steady-state levels, the substrate and the product of MgP methyltransferase (CHLM) preceding MgPMME cyclase, while MgP strongly accumulates in mutant leaves after 5-aminolevulinic acid feeding. The ntrc mutant has a reduced capacity to synthesize 5-aminolevulinic acid and reduced CHLM activity compared with the wild type. Although transcript levels of genes involved in chlorophyll biosynthesis are not significantly altered in 2-week-old ntrc seedlings, the contents of glutamyl-transfer RNA reductase1 (GluTR1) and CHLM are reduced. Bimolecular fluorescence complementation assay confirms a physical interaction of NTRC with GluTR1 and CHLM. While ntrc contains partly oxidized CHLM, the wild type has only reduced CHLM. As NTRC also stimulates CHLM activity in vitro, it is proposed that NTRC has a regulatory impact on the redox status of conserved cysteine residues of CHLM. It is hypothesized that a deficiency of NTRC leads to a lower capacity to reduce cysteine residues of GluTR1 and CHLM, affecting the stability and, thereby, altering the activity in the entire tetrapyrrole synthesis pathway.
During the last decades, almost all enzymes of tetrapyrrole biosynthesis and their complex network of transcriptional regulation have been comprehensively studied (Tanaka et al., 2011). These studies revealed a complex control of the expression of genes encoding enzymes in the light-regulated chlorophyll (Chl)-synthesizing branch of tetrapyrrole metabolism. In brief, 5-aminolevulinic acid (ALA) is synthesized in a transfer RNA (tRNA)GLU-mediated pathway, and eight molecules of ALA are ultimately converted in a series of enzymatic steps to protoporphyrin IX. The polymeric magnesium (Mg) chelatase complex consisting of the three different subunits CHLH, CHLI, and CHLD directs protoporphyrin IX into the Mg branch of tetrapyrrole biosynthesis. Methylation of magnesium protoporphyrin (MgP) by MgP methyltransferase (CHLM) at the C13 of pyrrole ring C initiates the formation of the typical fifth ring. The product of this step, magnesium protoporphyrin monomethylester (MgPMME), is then converted to divinyl protochlorophyllide (PChlide) by an oxidative cyclase complex. NADPH:protochlorophyllide oxidoreductase (POR) synthesizes chlorophyllide (Chlide). PChlide and Chlide are most likely the main substrates of a divinyl reductase that reduces the C7-C8 double bond, forming a monovinyl product. The two final steps of Chl a and b synthesis are likely to be consecutively performed in alternating order. Chl(ide) a oxygenase replaces the C3 methyl group at ring B with an aldehyde to form Chlide b, and Chl synthase esterifies Chlide a and b to the final pigments.
These biochemical and genetic studies pave the way for the exploitation of posttranslational regulatory mechanisms in the tetrapyrrole biosynthetic pathway. Feedback and feed-forward regulation between early and late enzymatic steps enables balancing the metabolic flux and preventing the accumulation of photoreactive tetrapyrrole intermediates (Czarnecki and Grimm, 2012). Two important posttranslational regulators, FLU and Genome Uncoupled4 (GUN4), have been identified that modulate the enzyme activities of glutamyl-tRNA reductase (GluTR) and Mg chelatase, respectively. In response to accumulating PChlide, the FLU protein represses ALA synthesis in darkness by interaction with GluTR1 (Meskauskiene et al., 2001; Richter et al., 2010; Kauss et al., 2012), which is one of the two analyzed isoforms. GUN4 has been identified in a mutant screen for modifications of retrograde signaling between chloroplasts and the nucleus (Larkin et al., 2003). In addition to a stimulatory role for Mg chelatase activity, GUN4 ultimately affects overall tetrapyrrole biosynthesis by stimulating ALA synthesis (Larkin et al., 2003; Peter and Grimm, 2009; Zhou et al., 2012).
Apart from these two posttranslational regulators that directly interact with metabolic activities of tetrapyrrole biosynthesis, additional factors are required to modulate the enzyme activities, stabilize proteins, preserve activity in certain plastid substructures, and facilitate substrate channeling. A GluTR-binding protein (GBP) was recently identified that is suggested to organize ALA synthesis in two subcompartments of the chloroplast. It is proposed that GBP subdivides a small portion of GluTR for ALA synthesis, which is not controlled by the FLU-mediated repression of ALA synthesis in darkness (Czarnecki et al., 2011). Mutual interaction of enzymes, such as the Mg chelatase subunit CHLH and MgP methyltransferase, have been reported to stimulate the enzymatic activities (Hinchigeri et al., 1997; Alawady et al., 2005; Shepherd et al., 2005). Apart from these regulatory interactions of proteins, attachments of chemical-modifying groups and the posttranslational processing of amino acid side chains control the functions of proteins.
Plastidic metabolism is basically also controlled by the organellar redox poise. Among the redox systems, the ferredoxin-thioredoxin-reductase catalyzes the transfer of electrons from the photosynthetic electron transport chain to target proteins to reduce their disulfide bridges. Among these proteins, the CHLI subunit of Mg chelatase is reduced by thioredoxin (TRX), leading to stimulation of its ATPase activity (Ikegami et al., 2007; Luo et al., 2012). This redox-dependent mechanism stimulates the Mg chelatase reaction. In addition, Glu-1-semialdehyde aminotransferase, porphobilinogen synthase, and uroporphyrinogen decarboxylase have been identified as potential TRX targets (Buchanan and Balmer, 2005).
A second system involves the transfer of electrons of NADPH to target proteins via NADPH-dependent-thioredoxin reductase (NTR). This enzyme contains a flavin-adenine-diphosphate-binding domain and a double Cys-forming peptide motif in the catalytic center. Three isoforms of NTR (NTRA, NTRB, and NTRC) are reported in Arabidopsis (Arabidopsis thaliana) that link TRXs to multiple target proteins of metabolic processes. While NTRA and NTRB are dually targeted to cytosol or mitochondria, NTRC is plastid localized and one of the main redox regulators in photosynthetic and nonphotosynthetic active plastids (Serrato et al., 2004; Pérez-Ruiz et al., 2006; Spínola et al., 2008; Kirchsteiger et al., 2009, 2012; Lepistö et al., 2009; Pérez-Ruiz and Cejudo, 2009; Pulido et al., 2010). NTRC contains a C-terminal TRX domain and acts as a bifunctional enzyme with NTR/TRX activity for the reduction of specific target proteins (Serrato et al., 2004). This initial report emphasizes NTRC function during abiotic, oxidative stress. NTRC reduces 2-Cys peroxiredoxins (2-CysPrx), which are small hydrogen peroxide (H2O2) peroxidases, indicating a key role of NTRC in the detoxification of reactive oxygen species (ROS; Pérez-Ruiz et al., 2006). Subsequently, NTRC has been shown to reduce heterotetrameric ADP-Glc pyrophosphorylase (AGP), a key enzyme in starch synthesis (Michalska et al., 2009). However, it is also reported that the activity of AGP is not influenced by the light-induced and redox-dependent switch between monomeric and dimeric forms of AGP (Li et al., 2012).
Interestingly, an ntrc mutant shows a chlorotic phenotype under normal growth conditions, which is not explained only by the accumulation of ROS. Loss of Chl in NTRC-deficient plants increases with shorter photoperiods and increasing light intensities, in comparison with wild-type seedlings (Lepistö et al., 2009). These phenotypical changes of the Chl content in the ntrc mutant resemble mutants with defects in Chl biosynthesis (Peter and Grimm, 2009; Peter et al., 2010). It was recently shown that NTRC stimulates the reactions of the MgPMME cyclase reaction by scavenging peroxides in vitro (Stenbaek et al., 2008). As the ntrc mutant accumulates MgPMME after ALA feeding, the authors proposed an inhibited activity of the MgPMME oxidative cyclase as a result of a compromised NTRC/2-CysPrx action, leading to impaired ROS depletion.
These findings encourage us to examine the impact of NTRC deficiency on the entire metabolic pathway of tetrapyrrole biosynthesis. We reinvestigated the role of NTRC on the regulation of tetrapyrrole synthesis by analyzing the steady-state levels of Chl intermediates and enzyme activities of chlorophyll biosynthesis in the ntrc mutant. A bimolecular fluorescence complementation (BiFC) assay was applied to study the interaction of NTRC with tetrapyrrole synthesis enzymes in vivo.
RESULTS
The ntrc Knockout Mutation Leads to Reduced Chl Content
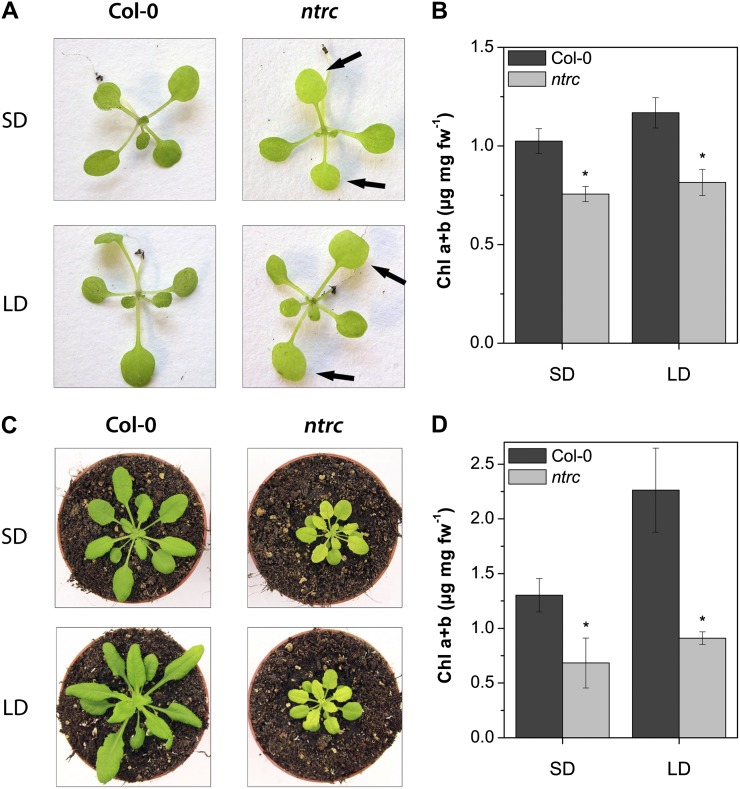
The chlorotic phenotype of ntrc is visible after development of the first true leaves (Fig. 1A, arrows). Mutant plants accumulate up to 60% less Chl than the wild type under both short-day (SD) and long-day (LD) growth conditions (Fig. 1, B and D). The Chl deficiency remains visible in 40- to 50-d-old ntrc plants and is even more pronounced in older than in younger ntrc leaves in comparison with older wild-type seedlings. The reduced Chl content of ntrc plants correlates with growth retardation in comparison with the wild type (Fig. 1C). SD- and LD-grown ntrc seedlings show reduced ALA-synthesizing capacity compared with the wild type (Table I). Compared with the wild type, ntrc ALA-synthesizing capacities in SD are diminished by 40% and in LD by 50%. These results indicate the close interdependence of strongly impaired ALA synthesis rate and low Chl content of ntrc in relation to the wild-type values and suggest an NTRC-dependent ALA biosynthesis.
Figure 1.
A and C, Phenotypes of Arabidopsis wild-type (Col-0) and ntrc seedlings growing in SD or LD conditions for 14 d (A) and 30 d (C). B and D, Chl a and b contents of 14- and 30-d-old plants, respectively. The ntrc seedlings show a pale-green phenotype of rosette leaves (arrows) and reduced Chl content. Data are given as means ± sd of three biological replicates. Asterisks denote significant differences from Col-0 with P < 0.05. fw, Fresh weight.
Table I. Enzyme activities in wild-type (Col-0) and ntrc mutant plants.
ALA synthesis capacity was determined with 21-d-old plants growing in SD and LD conditions. Plastid CHLM activity was measured with isolated plastids from 4-week-old wild-type and ntrc plants. For assay under oxidizing conditions, 10 mm H2O2 was added to the assay mixture of wild-type chloroplasts. CHLM activity was calculated from different time points and related to the amount of protein used. Data are given as means ± sd of three biological replicates. Asterisks denote significant differences from Col-0 with P < 0.05.
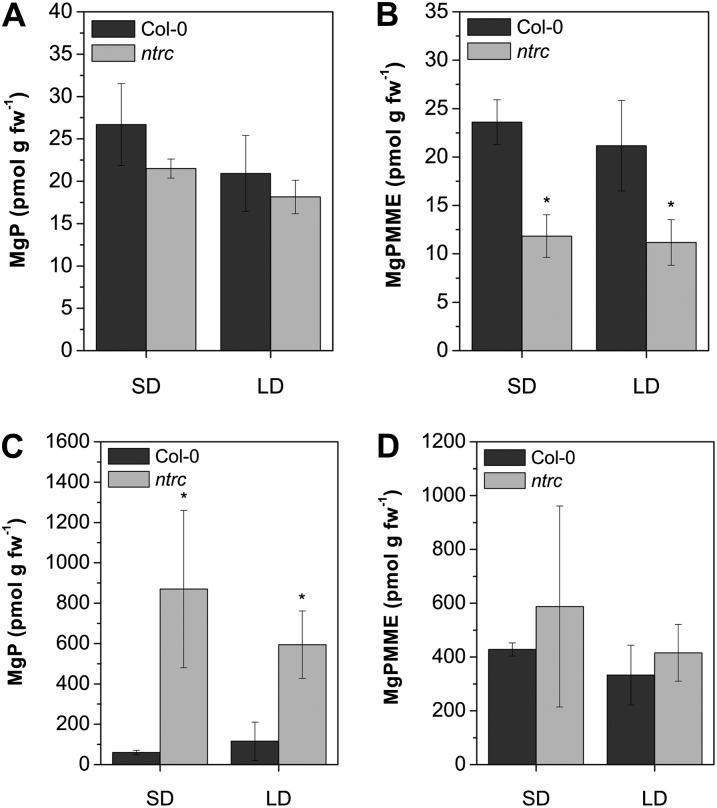
Analysis of the Mg porphyrin steady-state levels of 14- to 21-d-old plants grown under SD and LD conditions revealed that levels of MgPMME were reduced in ntrc seedlings (Fig. 2B). The reduced flow of intermediates can be attributed to the reduced activity of enzymes synthesizing ALA. In addition, when leaves of ntrc were incubated with ALA, 10 times more MgP, the substrate of the CHLM reaction, accumulated than in wild-type leaves (Fig. 2C). In contrast, the ALA-fed mutant leaves accumulate only slightly more MgPMME than the wild type (Fig. 2D). This latter result is consistent with a previous report (Stenbaek et al., 2008). These measurements of accumulating porphyrins of ntrc prompted us to assume that the loss of NTRC hampers the MgP methyltransferase reaction. To assess a possible interdependency between NTRC function and MgP methyltransferase, intact chloroplasts from mutant and wild-type plants were isolated and assayed for CHLM activity. Plastids isolated from 4- to 5-week-old ntrc plants display a 30% to 40% reduced CHLM activity in comparison with wild-type plastids (Table I).
Figure 2.
MgP and MgPMME contents of 14-d-old wild-type (Col-0) and ntrc plants growing in SD or LD conditions. A and B, Steady-state levels of Mg porphyrins. C and D, Accumulation of MgP and MgPMME after ALA feeding of plants for 16 h in darkness. The ntrc mutant shows a reduction in steady-state level of Mg porphyrins and a strong accumulation of MgP after ALA feeding. Data are given as means ± sd of three biological replicates. Asterisks denote significant differences from Col-0 with P < 0.05. fw, Fresh weight.
Posttranslational Reduction of GluTR1 and CHLM
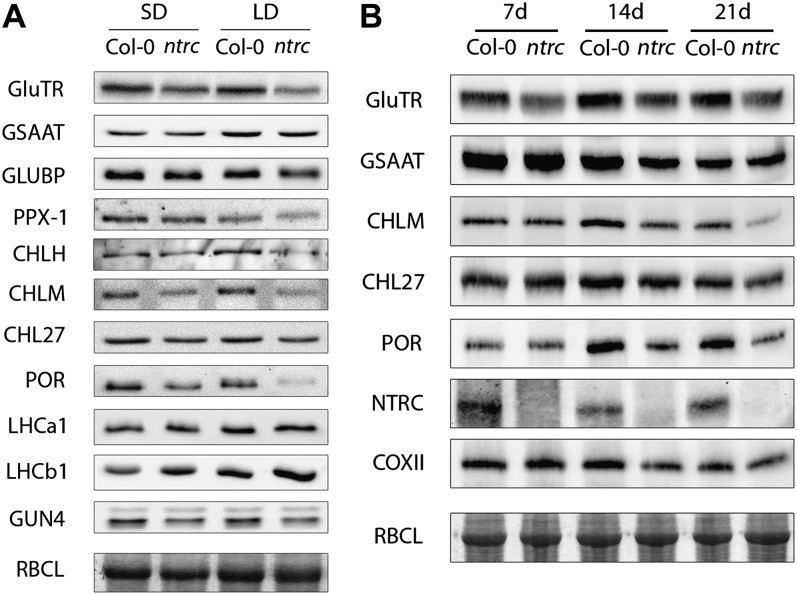
The diminished ALA synthesis rate and the lower CHLM activity in ntrc can be attributed to a posttranslational effect of NTRC on the stability and/or the catalytic properties of GluTR and CHLM in Chl biosynthesis. Protein and transcript analysis was performed for numerous enzymatic steps of the tetrapyrrole biosynthesis pathway. Western-blot analysis of total protein extracts of 14-d-old wild-type and ntrc plants indicates significantly reduced contents of GluTR, CHLM, and POR (Fig. 3A). The levels of other proteins, including Glu-1-semialdehyde aminotransferase, GluTR-binding protein (Czarnecki et al., 2011), protoporphyrinogen oxidase I, and the aerobic MgPMME cyclase subunit CHL27, were either not or hardly affected in ntrc compared with the wild type (Fig. 3A). Comparing the results of western blots from more than three biological replicates hints at a 50% reduction of GluTR and CHLM contents in ntrc seedlings grown under SD conditions relative to the wild-type protein levels (Fig. 3A). It is noteworthy that monitoring the protein contents during a growth kinetic revealed that the GluTR content was already considerably reduced in 7-d-old seedlings, whereas CHLM and POR levels were significantly diminished in late stages of seedling development (Fig. 3B).
Figure 3.
Contents of tetrapyrrole synthesis enzymes in wild-type (Col-0) and ntrc plants growing in SD or LD conditions. A, Enzyme levels of 14-d-old plants. B, Time-course analysis of enzyme levels during growth in SD of 7-, 14-, and 21-d-old wild-type and ntrc seedlings. GSAAT, Glu-1-semialdehyde aminotransferase; GLUBP, GluTR-binding protein; PPX-1, protoporphyrinogen IX oxidase; LHCa1 and LHCb1, light-harvesting complex a1 and b1; COXII: cytochrome c oxidase II subunit; RBCL: Rubisco large subunit (Coomassie stain).
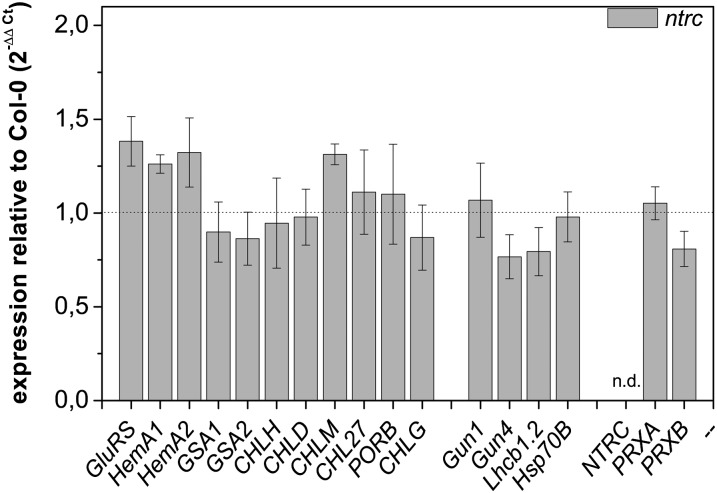
Although the contents of GluTR, CHLM, and POR are substantially reduced in the mutant seedlings, the contents of transcripts encoding these proteins were not significantly altered compared with the wild type (Fig. 4). In contrast, some levels of transcripts encoding enzymes of the tetrapyrrole biosynthesis pathway are slightly elevated, including those of glutamyl-tRNA synthetase (GluRS), HEMA1/HEMA2, and CHLM. The reduced protein contents do not result from a regulatory adaptation of transcriptional activity in ntrc seedlings but argue for a deficit in the posttranscriptional stability of these enzymes (Fig. 4). These results are consistent with previous microarray data published by Lepistö et al. (2009). An elevated transcript level of genes encoding enzymes of Chl synthesis was only detected in older SD-grown ntrc seedlings.
Figure 4.
Transcript analysis of ntrc mutant plants growing for 14 d in SD conditions. Transcript levels were calculated according to the comparative threshold cycle method with wild-type (Col-0) plants as a control and Actin2 (At3g18780) as a reference gene. The transcript for NTRC was not detectable (n.d.) in the ntrc mutant, confirming the knockout of NTRC. HEMA1 and HEMA2, GluTR1 and GluTR2; GSA1 and GSA2, Glu-1-semialdehyde aminotransferase1 and Glu-1-semialdehyde aminotransferase2; PORB, POR; LHCB1.2, light-harvesting complex b1.2; HSP70B, heat shock protein70B; PRXA and PRXB, 2-CysPrx A and B. Data are given as means ± sd of three biological replicates.
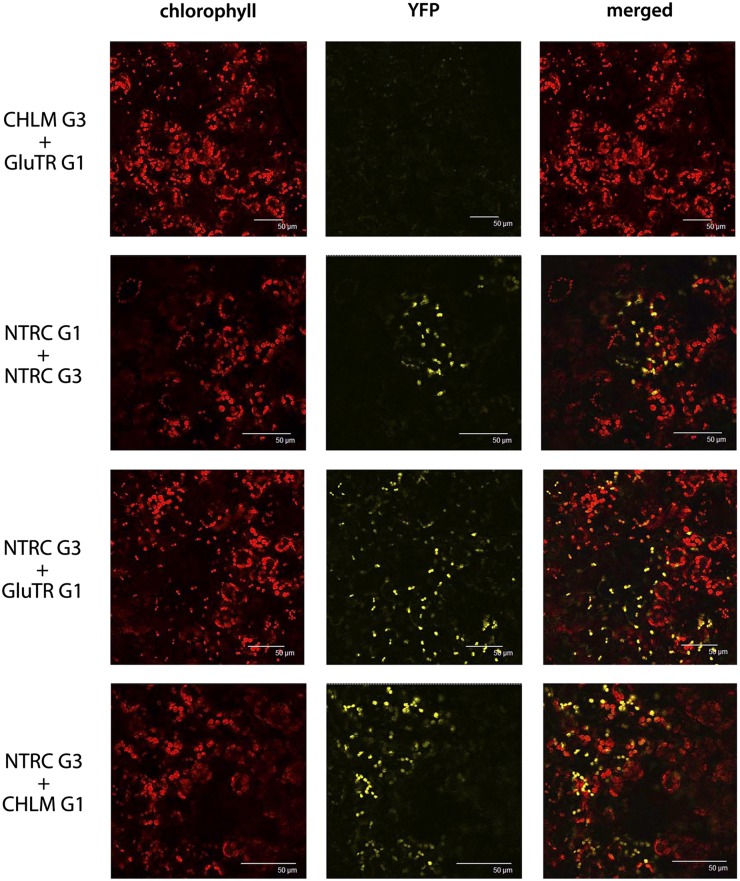
NTRC Interacts with GluTR1 and CHLM
As the reduced activities for ALA synthesis and CHLM in ntrc correlate with the reduced levels of the corresponding proteins, NTRC is a potential candidate for redox-controlled modulation of GluTR and CHLM activity and stability. Following the hypothesis that both proteins are enzymatic targets of NTRC, we examined the putative interactions of NTRC with GluTR1 (HEMA1) and CHLM using the in vivo BiFC assay. Tobacco (Nicotiana benthamiana) leaves were coinfiltrated with Agrobacterium tumefaciens cells carrying binary vector VyNE (G1) or VyCE (G3) with gene fusion constructs that encode NTRC, GluTR, or CHLM combined at their C termini with either the N- or the C-terminal part of the Venus protein (yellow fluorescent protein [YFP] derivative). In the two combinations of NTRC-GluTR and NTRC-CHLM interaction, a strong fluorescence signal was observed by confocal laser scanning microscopy, indicating the in planta interaction of NTRC with GluTR and CHLM, respectively (Fig. 5). Cotransformation of tobacco cells with NTRC-G1 or NTRC-G3 (NTRC fused to either the N- or C-terminal part of the Venus protein) was used as a positive BiFC control. This dimerization of NTRC confirmed previous findings (Pérez-Ruiz et al., 2009; Wulff et al., 2011). Coinfiltration of A. tumefaciens with vectors for the expression of CHLM-G3 and GluTR-G1 (and reverse combinations) did not result in yellow fluorescence and was used as a negative control (Fig. 5). As demonstrated by BiFC, the physical contact of NTRC with GluTR and CHLM is the prerequisite for NTRC-catalyzed posttranslational modifications of conserved Cys residues in both GluTR1 and CHLM. The protein sequences of both enzymes, GluTR and CHLM, from a broad range of plant species contain four and three highly conserved Cys residues, respectively (Supplemental Figs. S1 and S2).
Figure 5.
Demonstration of the interaction of NTRC with GluTR1 and CHLM by BiFC assay. Proteins were either cloned to the N-terminal (G1) or C-terminal (G3) part of the Venus protein and cotransformed in leaves of tobacco. The autofluorescence of Chl, the reconstituted YFP fluorescence as a signal for a positive interaction, and an overlay (merge) image of the Chl and YFP fluorescence are shown.
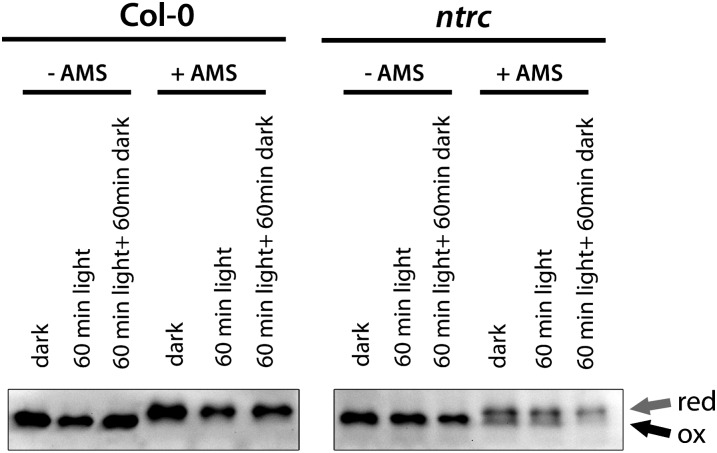
NTRC Controls the in Vivo Redox State of CHLM and GluTR
We assayed the reducing activity of NTRC on CHLM. The CHLM protein contains three highly conserved Cys residues at positions 111, 115, and 177 (Supplemental Fig. S1). Using 4-acetoamido-4-maleimidylstilbene-2,2-disulfonic acid (AMS), the in vivo redox status of CHLM in the wild type and ntrc was visualized. AMS binds to reduced Cys residues (thiol group/sulfhydryl group), thereby increasing the Mr of the protein. When AMS-labeled proteins are separated under nonreducing conditions, the labeled protein (reduced form) migrates slower on an SDS-polyacrylamide gel than the unlabeled protein (oxidized form). Oxidized proteins that are characterized by disulfide bonds or sulfenic acids cannot bind AMS and migrate like the nontreated proteins. In wild-type protein extracts, only one immunoreacting CHLM band for either AMS-treated or untreated samples can be observed. The immunoreactive band for CHLM of AMS-treated samples migrates slower compared with the untreated samples, indicating that CHLM is completely reduced in wild-type plants in all investigated conditions (Fig. 6). The three different dark/light regimes do not cause any effect on the content of reduced CHLM proteins. The change in gel mobility of CHLM in the wild type upon binding of AMS is explained with the AMS accessibility to CHLM. In contrast, AMS-treated protein extracts from ntrc mutant plants resulted in two immunoreactive CHLM bands (Fig. 6). The lower protein band migrates like the untreated CHLM and the upper one like the AMS-treated CHLM from the wild-type extracts, reflecting an oxidized and a reduced portion of CHLM. These results conclusively indicate that the lack of NTRC retains part of CHLM in the oxidized form.
Figure 6.
Visualization of the redox status of CHLM in 21-d-old wild-type (Col-0) and ntrc mutant plants growing under SD conditions. Protein extracts from different light treatments were extracted under nonreducing conditions and were either treated with (+AMS) or without (−AMS) AMS. Samples were separated on a nonreducing SDS-polyacrylamide gel, blotted, and probed with anti-CHLM antibody.
In a preexperiment to AMS treatment of GluTR, supply of dithiothreitol (DTT) to the protein extract led to the formation of distinct GluTR monomers, while oxidized GluTR was represented in an aggregation of oligomeric complexes (Supplemental Fig. S3). In addition, the similar Mr of the large subunit of Rubisco and GluTR affects the mobility of GluTR on SDS-polyacrylamide gels. Instead of a sharp and distinct band, GluTR migrates as a fuzzy band on the polyacrylamide gel. AMS-dependent shifts of GluTR are impaired under nonreducing conditions during SDS gel electrophoresis. Only a small portion of polymeric GluTR is accessible for AMS. However, a much larger fraction of GluTR in the wild-type sample shows the AMS-dependent mobility shift on the SDS gel than the GluTR fraction of the ntrc sample. In the ntrc samples, the bulk GluTR appears to be assembled in the high-Mr oligomeric aggregates (Supplemental Fig. S3).
NTRC Stimulates MgP Methyltransferase Activity
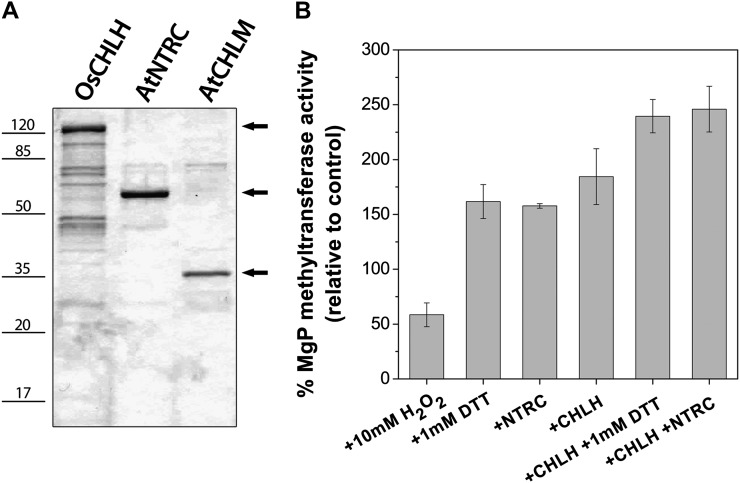
Purified recombinant CHLM was finally assayed for enzyme activity in the presence of NTRC. The sensitivity of CHLM to the redox state was assessed by the supply of DTT or H2O2 to the enzyme assay (Fig. 7). The series of measurements revealed that pretreatment with DTT elevated the CHLM activity almost 2-fold, while the oxidizing condition lowered the catalytic capacity of MgP methyltransferase. The loss of CHLM activity under the oxidizing condition was also determined in assays with purified chloroplasts of Arabidopsis wild-type plants, which were incubated with H2O2 (Table I). In comparison with the fully oxidized CHLM, the addition of DTT or NTRC to the assay mixture stimulated CHLM activity by approximately 3-fold (Fig. 7).
Figure 7.
Purification of 6×His-tagged OsCHLH, AtNTRC, and AtCHLM and recombinant CHLM activity assay. A, SDS-PAGE separation of proteins after purification and concentration. One microgram (OsCHLH and AtNTRC) or 0.5 µg (AtCHLM) of protein was loaded on the gel. B, CHLM activity assay with recombinant proteins. All assay mixtures contained 0.6 µm CHLM, 500 µm S-adenosyl-l-Met, and 500 µm NADPH. In addition, CHLM activity was analyzed after preincubation of AtCHLM with 10 mm H2O2, 1 mm DTT, 0.8 µm NTRC, 0.5 µm CHLH, or in combination with chemicals or proteins. All treatments result in a significant change of in vitro CHLM activity relative to the control assay. Data are given as means ± sd relative to control assays (without any treatment) from two experiments performed with proteins from independent purifications.
The presence of NTRC increased CHLM activity and substituted completely for DTT in our assay conditions. In combination with recombinant CHLH, the CHLM activity was also stimulated. A stimulatory effect of the recombinant CHLH-CHLM interaction has been demonstrated earlier (Shepherd and Hunter, 2004; Shepherd et al., 2005) and was confirmed in our enzyme assays, although MgP was applied in the CHLM assay and not the substrate of Mg chelatase, protoporphyrin IX. For a combined enzyme assay, the two times increased CHLM activity in the presence of CHLH was additionally enhanced when the reducing activity of NTRC or DTT was supplied (Fig. 7).
DISCUSSION
NTRC Is a Posttranslational Regulator of Chl Biosynthesis
The redox state in chloroplasts is mainly determined through the photosynthetic activities but differs not only between day and night but also in response to varying environmental light conditions. The adaptation to these changes requires a rapid posttranslational regulation of primary metabolism and organellar gene expression. The metabolic flux and the enzyme activities of Chl biosynthesis are also posttranslationally modulated in response to varying light conditions (Ikegami et al., 2007; Stenbaek and Jensen, 2010; Czarnecki and Grimm, 2012).
It has been reported that several enzymes of Chl biosynthesis are linked to two redox systems, the ferredoxin-dependent TRX reductase and the NTRC. The ATPase activity of the Mg chelatase CHLI subunit is activated by TRXs (Ikegami et al., 2007). The TRX-dependent reduction of CHLI was also shown by AMS treatment. The presence of TRX f stimulates the in vitro Mg chelatase activity of the recombinant subunits of rice (Oryza sativa), while the deficiency of TRX f and TRX m by virus-inducible gene silencing of pea (Pisum sativum) plants leads to reduced activity of Mg chelatase and ALA biosynthesis (Luo et al., 2012). The MgPMME oxidative cyclase activity was stimulated by NTRC and 2-CysPrx (Stenbaek et al., 2008). It was proposed that NTRC and 2-CysPrx act protectively on the oxygenase reaction of the cyclase by the detoxification of peroxides. An indirect impact on MgP methyltransferase was suggested by a cyclase-triggered negative feedback inhibition (Stenbaek et al., 2008).
Here, we demonstrate a direct reducing activity of NTRC on MgP methyltransferase and GluTR, modifying their stability and activity. The following experiments demonstrate the direct role of NTRC as the interacting partner and reductant of methyltransferase and GluTR. First, BIFC assays display NTRC interaction with GluTR and CHLM in plastids (Fig. 5). NTRC and GluTR also form homodimers. The BIFC-mediated dimerization of NTRC corroborates the previous report on the dimeric structure of the NTRC (Wulff et al., 2011). Second, the addition of NTRC stimulates the activity of purified MgP methyltransferase and substitutes DTT in the in vitro enzyme assay (Fig. 7). In contrast, the CHLM activity decreases under oxidizing conditions in vitro and in vivo (Fig. 7; Table I). Third, the lack of NTRC in plant extracts prevents the complete reduction of CHLM. In ntrc protein extracts, only partial AMS conjugation with CHLM was detected, indicating a substantial amount of oxidized CHLM in the absence of NTRC (Fig. 6). Although CHLM was not identified as a target of TRX-dependent reduction, it could be speculated that the loss of a specific redox regulator can at least partially be compensated by other redox modulators (Bohrer et al., 2012; Luo et al., 2012; Thormählen et al., 2013). In principle, similar AMS experiments with GluTR indicated a larger amount of reduced GluTR in wild-type extracts in comparison with ntrc, but GluTR in nonreducing conditions migrates in high-Mr molecular polymeric aggregates, which disappeared only when protein extracts were reduced with DTT (Supplemental Fig. S3). Moreover, the ntrc mutant contains less GluTR and CHLM than the wild type (Fig. 3). Lack of NTRC affects the stability and redox state of these two enzymes and fosters the structure of both proteins for optimized enzyme activity, conformational integrity, and stability. We suggest that the reduction of the target Cys residues is essential for an enhanced stability of CHLM in light and darkness. It was previously shown that the MgP methyltransferase is present and active in both light- and dark-incubated tissues of tobacco plants (Alawady et al., 2005). Keeping CHLM in a reduced status in darkness (Fig. 6) guarantees the availability of sufficient active CHLM upon the shift from dark to light under photoperiodic growth conditions.
Previous reports showed that the loss of NTRC can be more tolerated under growth conditions with a prolonged light period. Lepistö et al. (2009) reported that ntrc plants grown at 100 µmol photons m−2 s−1 under 8 h of light/16 h of dark (SD) showed a more drastic phenotype than plants growing under 16 h of light/8 h of dark (LD). Those growth conditions, however, are not directly comparable to those used in this paper. Here, the plants were grown at 120 µmol photons m−2 s−1 under 10 h of light/14 h of dark (SD) and 14 h of light/10 h of dark (LD). Besides the difference in the total amount of light, the photoperiods used here were different. This might explain the phenotypical differences between the data published here and the plants analyzed by Lepistö et al. (2009). However, the difference in the strength of the phenotype of ntrc plants with varying photoperiods phenocopies the growth behavior of other Chl synthesis mutants, like CHL27 antisense tobacco plants (Peter et al., 2010) or GUN4 knockdown/knockout mutants from Arabidopsis (Peter and Grimm, 2009). Plants with reduced expression or even knockout of proteins that are involved in tetrapyrrole synthesis show the strongest phenotype under SD growth conditions. When these plants are transferred to continuous light or low-light conditions, the loss of the proteins can better be tolerated and the phenotype becomes less conspicuous. In terms of the ntrc mutant, the reduced amount of CHLM has a stronger effect the shorter the photoperiod and the higher the light intensity. This observation again underlines the need for the delicate control of the reduction of CHLM during photoperiodic growth.
Finally, the ntrc mutant also possesses reduced CHLM activity and ALA synthesis rates (Table I). Both reduced enzyme activities can be assigned to the reduced content of both proteins, CHLM and GluTR. CHLM activity was measured only in plants grown under the SD photoperiod, but because the CHLM content of ntrc is equally reduced in LD conditions (Fig. 3), we assume that the corresponding activity is also diminished in LD. As the levels of transcripts encoding Chl synthesis enzymes are not modified in ntrc compared with wild-type seedlings (Fig. 4), the reduced contents of GluTR and CHLM argue for posttranslational redox-dependent regulation ultimately affecting protein stability.
It is proposed that the lower redox-dependent activities of these two enzymes contribute in the ntrc mutant to the reduced formation of Chl and, ultimately, to lower Chl content (Fig. 1). Then, lower ALA synthesis rates minimize the metabolic flow and are also responsible for lower steady-state levels of tetrapyrrole intermediates (Fig. 2; Table I). When ALA feeding bypasses the repression of the tightly controlled ALA biosynthesis in ntrc, then those enzymatic steps, which are redox controlled by NTRC, show strong accumulation of the catalytic substrate of the MgP methyltransferase in the absence of NTRC in comparison with the wild type (Fig. 2, C and D). Thus, the highest levels of MgP are observed upon ALA feeding in ntrc.
The described posttranslational modifications in tetrapyrrole biosynthesis in response to impaired redox-controlled CHLM phenotypically resemble CHLM antisense tobacco plants. CHLM antisense plants are not only impaired in CHLM synthesis and, ultimately, in MgP methyltransferase activity, but they also have lower ALA synthesis rate and Mg chelatase activity. The feedback control of ALA biosynthesis has been explained by posttranslational feedback control and the down-regulation of genes involved in ALA and Chl biosynthesis in response to the lower plastid-localized activities of Mg chelatase and MgP methyltransferase (Alawady and Grimm, 2005). In NTRC-deficient Arabidopsis seedlings, the reduced MgP methyltransferase activity and ALA-synthesizing capacity are not explained by reduced transcript levels (Fig. 4) but by redox-dependent changes in protein stability and activity. While the reducing activity of NTRC modulates CHLM and GluTR stability, the CHLM antisense expression in tobacco induces a complex regulatory response, which also includes plastid-derived retrograde signaling for the modulation of transcriptional activity. It is not excluded that different regulatory mechanisms exist in tobacco and Arabidopsis seedlings, as both species are characterized by different growth rates that also require different metabolic activities. Accelerated Chl biosynthesis rates in tobacco seedlings are connected with elevated steady-state levels of porphyrins and Mg porphyrins. In order to prevent photooxidation by accumulating tetrapyrroles, ROS-induced signaling alters nuclear gene expression (Apel and Hirt, 2004). In contrast, in the absence of NTRC, a higher level of oxidized forms of CHLM and GluTR reduces their stability and activity, which attenuates metabolic flow in tetrapyrrole synthesis without an obvious direct effect on transcript levels for the corresponding enzymes.
Although it is obvious that NADPH for dark-dependent activity of NTRC originates from the oxidative pentose phosphate cycle, it has also been reported that NTRC is also involved in redox-controlled starch synthesis in light by ADP-Glc pyrophosphorylase (Michalska et al., 2009). Chl formation is also a light-dependent process in photosynthetic cells of angiosperms. Consequently, NTRC-dependent activation of enzymes in the Chl-synthesizing pathway is facilitated by the use of NADPH derived from functional photosynthetic electron transfer in light. However, we also observed in light- and dark-grown wild-type seedlings a permanent reduced state of CHLM. Thus, our experiments confirm previous conclusions that NTRC not only has regulatory functions in plants in darkness or under prolonged dark treatments but also is essential for their growth in normal daily light/dark conditions. Consequently, NTRC complies with a housekeeping function in the stabilization and activation of CHLM. This observation indicates the need of posttranslational redox control of important enzymatic steps of Chl synthesis in light/dark growth periods and temporally light-limiting conditions and requires further study to verify the redox control of other enzymes of tetrapyrrole biosynthesis.
CHLM Activity Is Redox Dependent and Can Be Stimulated by NTRC
The in vitro methyltransferase activity is higher in the presence of NTRC. The NTRC function on CHLM is suggested to be a direct reduction of oxidized Cys residues. These results do not entirely exclude the observed stimulatory effects of NTRC and 2-CysPrx in in vitro assays of MgPMME cyclase (Stenbaek et al., 2008). The oxygenase reaction with molecular oxygen at the catalytic diiron cluster of the aerobic MgPMME cyclase (Tottey et al., 2003) can fail by the generation of ROS. Stenbaek et al. (2008) proposed that ROS can directly inhibit the cyclase reaction. Therefore, it has been assumed that the cyclase reaction is promoted by NTRC/2-CysPrx catalyzing H2O2 detoxification in in vitro assays. However, we consider an active participation of 2-CysPrx to be unlikely for the stimulatory effect of NTRC on the CHLM activity.
The wild-type-like phenotype of the double knockout of two PRX genes (PRXA and PRXB), which interact with NTRC, also supports the direct interaction of CHLM with NTRC. Although the prxAprxB double mutant accumulates higher levels of H2O2 compared with the wild type, the enzymatic steps of Chl biosynthesis are not affected (Pulido et al., 2010). The green phenotype of the prxAprxB double mutant differs from that of the ntrc mutant, indicating that the lack of 2-CysPrx does not primarily compromise Chl content. Thereby, the ROS-induced alteration of Chl biosynthesis in the ntrc mutant can be excluded under our experimental conditions. In conclusion, the lack of NTRC-dependent redox regulation of GluTR and CHLM sufficiently explains the phenotype of the ntrc mutant, whereas the combined activity of the two redox modulators NTRC and 2-CysPrx seems to have a minor effect on the reduction of Chl content in mutant leaves.
In the in vitro activity assay, the stimulatory effect of the reductant NTRC on CHLM is additive with the positive action of CHLH (Fig. 7). The stimulatory interaction of CHLM and CHLH on the enzymatic activity of MgP methyltransferase corroborates previous reports (Shepherd et al., 2005) and may likely release regulatory feedback signals on the early enzymes of tetrapyrrole biosynthesis. But only future experiments can elucidate the molecular mechanisms of the CHLH-mediated stimulation of CHLM. Which mechanism supports the enzymatic conversion from MgP to MgPMME and enables the regulatory interdependency between late and early enzymes of the pathway? Either CHLH activity contributes to the methyltransferase activity, as proposed by Shepherd et al. (2005), or CHLH is needed for the stability of a multienzymatic complex including CHLM. In this context, a protein complex consisting of CHL27, POR, GluTR, and other proteins has been recently reported for the tight repression of ALA synthesis in darkness by the negative regulator FLU in angiosperms (Meskauskiene et al., 2001; Kauss et al., 2012). It remains open whether other proteins of Chl synthesis, including the CHLH subunit and CHLM, are part of this protein complex. And it cannot be excluded that the redox control of GluTR and other proteins in Chl biosynthesis contributes to the difference in metabolic activities during light irradiation and darkness.
At present, the redox mechanism controlling the activity and stability of GluTR and CHLM in planta is not known. We suggest that the reduction of target Cys residues induces conformational changes in the GluTR and CHLM proteins, which affects the stability of these proteins and is essential for the high activity of the enzymes and/or for the interaction of the enzymes with other proteins of the Chl biosynthesis pathway. Therefore, in future experiments, we are going to identify the Cys residues in CHLM and GluTR1 that are targeted to NTRC-dependent redox regulation.
MATERIALS AND METHODS
Plant Growth
Arabidopsis (Arabidopsis thaliana) wild-type (ecotype Columbia [Col-0]) and ntrc knockout (At2g41680/SALK 096776) plants were routinely grown on soil at 110 to 120 µmol photons m−2 s−1 in either SD (10 h of light) or LD (14 h of light) conditions at 22°C to 24°C.
Porphyrin Extraction and HPLC Analysis
Approximately 50 to 100 mg of frozen leaf material was homogenized and resuspended in porphyrin extraction buffer (10 volumes of acetone, 9 volumes of methanol, and 1 volume of NH4OH). After centrifugation for 10 min at 16,000g, the pellet was extracted three times and the supernatants were combined in a new reaction tube. The samples were separated on an HPLC reverse-phase column (Nova-Pak C18 [Waters]; 3.9 × 150 mm, 4 μm, 60Å, HPLC 1100 system [Agilent Technologies]) to determine the contents of MgP and MgPMME by using the following program: 7 min of 100% solvent A (80% water, 10% methanol, 10% ammonium acetate [1 m; pH 7.0]) followed by a linear gradient to 100% solvent B (90% methanol, 10% ammonium acetate [1 m; pH 7.0]) for 16 min. After 1 min with 100% solvent B, a linear gradient to 100% solvent A was continued for 5 min. The Mg porphyrins were detected by fluorescence (excitation wavelength, 420 nm; emission wavelength, 595 nm), then confirmed and quantified by using authentic standards.
ALA Synthesis Capacity Measurement and ALA Feeding
ALA synthesis capacity was measured using the method of Mauzerall and Granick (1956). Frozen leaf material was homogenized under liquid nitrogen and resuspended in 20 mm potassium phosphate buffer (pH 6.8). After centrifugation for 10 min at 16,000g, 400 μL of the supernatant was mixed with 100 μL of ethyl acetoacetate and boiled for 10 min at 100°C. Chilled samples were mixed with 500 μL of modified Ehrlich’s reagent (373 mL of acetic acid, 90 mL of 70% [v/v] perchloric acid, 1.55 g of HgCl2, 9.10 g of 4-dimethylamino benzaldehyde, and 500 mL of water) and centrifuged for 5 min at 16,000g. The absorption of the ALA pyrrole was measured at 526, 553, and 720 nm. The ALA content was calculated using a dilution curve with an ALA solution (Sigma-Aldrich) and was related to the incubation time and fresh weight of the samples. One millimolar ALA feeding of leaf discs was performed in 20 mm Tris-HCl buffer (pH 7.2) for 14 to 16 h in darkness. After incubation, samples were dried on a tissue, then subsequently frozen in liquid nitrogen and extracted for Mg porphyrin analyses.
Protein Extraction, AMS Treatment, and Western Blot
Leaf material was ground in liquid nitrogen and resuspended in protein extraction buffer (2% [w/v] SDS, 56 mm NaCO3, 12% [w/v] Suc, 56 mm DTT, and 2 mm EDTA, pH 8.0) and heated for 20 min at 70°C. After centrifugation for 10 min, protein concentration of the supernatant was determined using the BCA Protein Assay Kit (Thermo Fisher Scientific). Ten to 15 µg of the extracts of reduced proteins was separated by 12% SDS-PAGE and blotted onto Hybond-C membranes (GE Healthcare). Membranes were probed with protein-specific antibodies according to Sambrook (2001).
The in vivo redox state of proteins was indicated when proteins were extracted without reducing agents in the extraction buffer, as described previously (Hendriks et al., 2003), with a single modification. Proteins were precipitated with 16% (w/v) TCA and washed twice with 100% acetone, before the protein pellet was resuspended in 1 volume of 0.1 m NaOH and 1 volume protein gel-loading buffer (without DTT). One part of the samples was incubated with AMS (Molecular Probes Life Technologies) for 20 min at room temperature. Both AMS-treated and untreated samples were separated, blotted, and probed with antibodies (see above).
Chloroplast Isolation and CHLM Activity Assay
Four- to 5-week-old Arabidopsis plants grown on soil were homogenized in a modified Waring blender with homogenization buffer (0.45 m sorbitol, 20 mm Tricine-KOH, pH 8.4, 10 mm EDTA, 10 mm NaHCO3, and 0.1% bovine serum albumin). The crude homogenate was filtered through one layer of Miracloth (Merck Millipore), the filtrate was centrifuged at 500g for 8 min (acceleration maximum/deceleration maximum), and the pellet was resuspended in resuspension buffer (0.3 m sorbitol, 20 mm Tricine-KOH, pH 8.4, 2.5 mm EDTA, and 5 mm MgCl2). The crude extract was centrifuged through a 40% to 80% Percoll gradient in a swing-out rotor for 30 min at 6,500g. Intact chloroplasts were carefully washed in fresh homogenization buffer and pelleted (swing-out rotor, 3,800g, 6 min). The pellet of chloroplasts was thoroughly resuspended in resuspension buffer. For assays with the stroma fraction, isolated plastids were lysed in resuspension buffer without sorbitol, vortexed, and physically stressed by pipetting up and down several times. Separation of the stroma and thylakoid fractions was achieved by centrifugation of the lysed chloroplasts. Chloroplast isolation was performed in a cold chamber at 4°C, and the samples were kept on ice.
Cloning, Expression, and Purification of Recombinant Proteins
Arabidopsis CHLM (At4g25080) and NTRC (At2G41680) were cloned into pQE80L (Qiagen) for the expression of N-terminal 6×His-tagged proteins. Supplemental Table S1 shows the primers used in the cloning procedures. The complementary DNA (cDNA) with correct sequence inserted either in pQE80L_CHLM or pQE80L_NTRC was transformed into Escherichia coli Rosetta strain, and the bacterial clones were expressed as follows. The bacterial culture was inoculated with 1/100th of an overnight culture. Cultures were grown for 2.5 to 3 h at 37°C until the optical density at 600 nm was 0.6 to 0.8. Then, the expression of recombinant proteins was induced by adding 1 mm isopropylthio-β-galactoside. After 3 to 4 h at 30°C, cells were harvested and stored at −20°C until the pellets were used for purification. Proteins were purified under native conditions according to the QIAexpressionist protocol (Qiagen) using nickel-nitrilotriacetic acid agarose. After purification, protein-containing fractions were concentrated with Amicon Ultra-4 Centrifugal Filter Units (Merck-Millipore); thereby, the elution buffer with high concentration of imidazole was exchanged against the lysis buffer. For the overproduction of recombinant rice (Oryza sativa) CHLH, the CHLH cDNA sequence in the pet28a vector was used, which was kindly provided by Zhou et al. (2012).
CHLM Activity Assay with Isolated Plastids and Recombinant Proteins
The CHLM activity assay was performed according to Shepherd et al. (2005) with total chloroplast protein extracts, the stroma fraction of isolated plastids, or purified recombinant proteins. For assays with purified chloroplasts, prewarmed 2× assay buffer (600 mm sorbitol, 10 mm MgCl2, 1,000 µm S-adenosyl-Met, 40 µm MgP, and 40 mm Tricine, pH 8.0, with KOH) was mixed 1:1 with chloroplast extract and immediately incubated at 30°C. The activity of recombinant CHLM was assayed in assay buffer without sorbitol (500 µm S-adenosyl-Met, 500 µm NADPH, 20 µm MgP, and 50 mm Tris-HCl, pH 7.5) at 30°C. After preincubation of recombinant CHLM with or without the addition of reducing or oxidizing chemicals (1 mm DTT/10 mm H2O2) and NTRC and/or CHLH proteins for 10 min at 30°C, the reaction was started by adding 2× assay buffer. Samples were incubated at 30°C. A typical assay contained 0.6 µm CHLM per time point. The reaction was stopped in acetone:NH4OH (9:1), and the product formation was determined by HPLC, quantified, and related to the protein used in the assay. All assays were repeated at least two times with aliquots from independent chloroplast or protein purifications.
Transcript Analysis
Total RNA was extracted from 50 to 100 mg of frozen material of 14-d-old SD Arabidopsis using the TRIsure reagent (Bioline). After DNase (Thermo Fisher Scientific) treatment, cDNA was transcribed from 2 µg of RNA using Moloney murine leukemia virus reverse transcriptase (Thermo Fisher Scientific) and oligo(dT)18 primer following the instructions of the supplier. Quantitative reverse transcription PCR was performed with 2xSensiMix (Bioline) on 96-well plates in a BFX96 thermo cycler (Bio-Rad). ACT2 (AT3G18780) was used as a reference gene. The expression levels were calculated according to the normalized fold expression difference. Supplemental Table S1 shows the primer pairs used for expression analysis.
BiFC
Full-length cDNA fragments of target genes were cloned into pDONR221 (Life Technologies) using gene-specific primers (Supplemental Table S1) and the BP cloning kit (Life Technologies). The positive sequenced clones were used for the LR cloning reaction (Life Technologies) into the Gateway-compatible pVyNE and pVyCE plasmids (Gehl et al., 2009) carrying the N-terminal part and the C-terminal part of Venus protein (YFP derivative), respectively. For BiFC assays (Walter et al., 2004), the constructs were transformed into Agrobacterium tumefaciens GV2260. Transient coexpression of tagged putative interaction partners was achieved by transformation of tobacco (Nicotiana benthamiana) leaves with the infiltration technique. After 2 to 3 d of dark incubation, leaf discs were analyzed by confocal laser scanning microscopy using the Leica TCS SP2 apparatus at λex = 514 nm and λem = 530 to 555 nm for YFP and 600 to 700 nm for Chl emission.
Miscellaneous
All experiments were performed with three to four biological replicates and were repeated at least two times. Significant differences were tested using Student’s t test with P < 0.05.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of CHLM protein sequences.
Supplemental Figure S2. Alignment of GluTR protein sequences.
Supplemental Figure S3. Visualization of the redox status of GluTR.
Supplemental Table S1. Overview of primers used for quantitative reverse transcription PCR analysis and cloning procedure.
Glossary
- Chl
chlorophyll
- ALA
5-aminolevulinic acid
- Mg
magnesium
- MgP
magnesium protoporphyrin
- MgPMME
magnesium protoporphyrin monomethylester
- PChlide
protochlorophyllide
- POR
NADPH:protochlorophyllide oxidoreductase
- Chlide
chlorophyllide
- TRX
thioredoxin
- 2-CysPrx
2-Cys peroxiredoxin
- ROS
reactive oxygen species
- AGP
ADP-Glc pyrophosphorylase
- SD
short-day
- LD
long-day
- BiFC
bimolecular fluorescence complementation
- YFP
yellow fluorescent protein
- AMS
4-acetoamido-4-maleimidylstilbene-2,2-disulfonic acid
- DTT
dithiothreitol
- H2O2
hydrogen peroxide
- Col-0
ecotype Columbia
- cDNA
complementary DNA
- tRNA
transfer RNA
References
- Alawady A, Reski R, Yaronskaya E, Grimm B. (2005) Cloning and expression of the tobacco CHLM sequence encoding Mg protoporphyrin IX methyltransferase and its interaction with Mg chelatase. Plant Mol Biol 57: 679–691 [DOI] [PubMed] [Google Scholar]
- Alawady AE, Grimm B. (2005) Tobacco Mg protoporphyrin IX methyltransferase is involved in inverse activation of Mg porphyrin and protoheme synthesis. Plant J 41: 282–290 [DOI] [PubMed] [Google Scholar]
- Apel K, Hirt H. (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Bohrer AS, Massot V, Innocenti G, Reichheld JP, Issakidis-Bourguet E, Vanacker H. (2012) New insights into the reduction systems of plastidial thioredoxins point out the unique properties of thioredoxin z from Arabidopsis. J Exp Bot 63: 6315–6323 [DOI] [PubMed] [Google Scholar]
- Buchanan BB, Balmer Y. (2005) Redox regulation: a broadening horizon. Annu Rev Plant Biol 56: 187–220 [DOI] [PubMed] [Google Scholar]
- Czarnecki O, Grimm B. (2012) Post-translational control of tetrapyrrole biosynthesis in plants, algae, and cyanobacteria. J Exp Bot 63: 1675–1687 [DOI] [PubMed] [Google Scholar]
- Czarnecki O, Hedtke B, Melzer M, Rothbart M, Richter A, Schröter Y, Pfannschmidt T, Grimm B. (2011) An Arabidopsis GluTR binding protein mediates spatial separation of 5-aminolevulinic acid synthesis in chloroplasts. Plant Cell 23: 4476–4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehl C, Waadt R, Kudla J, Mendel RR, Hänsch R. (2009) New Gateway vectors for high throughput analyses of protein-protein interactions by bimolecular fluorescence complementation. Mol Plant 2: 1051–1058 [DOI] [PubMed] [Google Scholar]
- Hendriks JH, Kolbe A, Gibon Y, Stitt M, Geigenberger P. (2003) ADP-glucose pyrophosphorylase is activated by posttranslational redox-modification in response to light and to sugars in leaves of Arabidopsis and other plant species. Plant Physiol 133: 838–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchigeri SB, Hundle B, Richards WR. (1997) Demonstration that the BchH protein of Rhodobacter capsulatus activates S-adenosyl-L-methionine:magnesium protoporphyrin IX methyltransferase. FEBS Lett 407: 337–342 [DOI] [PubMed] [Google Scholar]
- Ikegami A, Yoshimura N, Motohashi K, Takahashi S, Romano PG, Hisabori T, Takamiya K, Masuda T. (2007) The CHLI1 subunit of Arabidopsis thaliana magnesium chelatase is a target protein of the chloroplast thioredoxin. J Biol Chem 282: 19282–19291 [DOI] [PubMed] [Google Scholar]
- Kauss D, Bischof S, Steiner S, Apel K, Meskauskiene R. (2012) FLU, a negative feedback regulator of tetrapyrrole biosynthesis, is physically linked to the final steps of the Mg(++)-branch of this pathway. FEBS Lett 586: 211–216 [DOI] [PubMed] [Google Scholar]
- Kirchsteiger K, Ferrández J, Pascual MB, González M, Cejudo FJ. (2012) NADPH thioredoxin reductase C is localized in plastids of photosynthetic and nonphotosynthetic tissues and is involved in lateral root formation in Arabidopsis. Plant Cell 24: 1534–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchsteiger K, Pulido P, González M, Cejudo FJ. (2009) NADPH thioredoxin reductase C controls the redox status of chloroplast 2-Cys peroxiredoxins in Arabidopsis thaliana. Mol Plant 2: 298–307 [DOI] [PubMed] [Google Scholar]
- Larkin RM, Alonso JM, Ecker JR, Chory J. (2003) GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299: 902–906 [DOI] [PubMed] [Google Scholar]
- Lepistö A, Kangasjärvi S, Luomala EM, Brader G, Sipari N, Keränen M, Keinänen M, Rintamäki E. (2009) Chloroplast NADPH-thioredoxin reductase interacts with photoperiodic development in Arabidopsis. Plant Physiol 149: 1261–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Almagro G, Muñoz FJ, Baroja-Fernández E, Bahaji A, Montero M, Hidalgo M, Sánchez-López AM, Ezquer I, Sesma MT, et al. (2012) Post-translational redox modification of ADP-glucose pyrophosphorylase in response to light is not a major determinant of fine regulation of transitory starch accumulation in Arabidopsis leaves. Plant Cell Physiol 53: 433–444 [DOI] [PubMed] [Google Scholar]
- Luo T, Fan T, Liu Y, Rothbart M, Yu J, Zhou S, Grimm B, Luo M. (2012) Thioredoxin redox regulates ATPase activity of magnesium chelatase CHLI subunit and modulates redox-mediated signaling in tetrapyrrole biosynthesis and homeostasis of reactive oxygen species in pea plants. Plant Physiol 159: 118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauzerall D, Granick S. (1956) The occurrence and determination of delta-amino-levulinic acid and porphobilinogen in urine. J Biol Chem 219: 435–446 [PubMed] [Google Scholar]
- Meskauskiene R, Nater M, Goslings D, Kessler F, op den Camp R, Apel K. (2001) FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 98: 12826–12831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalska J, Zauber H, Buchanan BB, Cejudo FJ, Geigenberger P. (2009) NTRC links built-in thioredoxin to light and sucrose in regulating starch synthesis in chloroplasts and amyloplasts. Proc Natl Acad Sci USA 106: 9908–9913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Ruiz JM, Cejudo FJ. (2009) A proposed reaction mechanism for rice NADPH thioredoxin reductase C, an enzyme with protein disulfide reductase activity. FEBS Lett 583: 1399–1402 [DOI] [PubMed] [Google Scholar]
- Pérez-Ruiz JM, González M, Spínola MC, Sandalio LM, Cejudo FJ. (2009) The quaternary structure of NADPH thioredoxin reductase C is redox-sensitive. Mol Plant 2: 457–467 [DOI] [PubMed] [Google Scholar]
- Pérez-Ruiz JM, Spínola MC, Kirchsteiger K, Moreno J, Sahrawy M, Cejudo FJ. (2006) Rice NTRC is a high-efficiency redox system for chloroplast protection against oxidative damage. Plant Cell 18: 2356–2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter E, Grimm B. (2009) GUN4 is required for posttranslational control of plant tetrapyrrole biosynthesis. Mol Plant 2: 1198–1210 [DOI] [PubMed] [Google Scholar]
- Peter E, Rothbart M, Oelze ML, Shalygo N, Dietz KJ, Grimm B. (2010) Mg protoporphyrin monomethylester cyclase deficiency and effects on tetrapyrrole metabolism in different light conditions. Plant Cell Physiol 51: 1229–1241 [DOI] [PubMed] [Google Scholar]
- Pulido P, Spínola MC, Kirchsteiger K, Guinea M, Pascual MB, Sahrawy M, Sandalio LM, Dietz KJ, González M, Cejudo FJ. (2010) Functional analysis of the pathways for 2-Cys peroxiredoxin reduction in Arabidopsis thaliana chloroplasts. J Exp Bot 61: 4043–4054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter A, Peter E, Pörs Y, Lorenzen S, Grimm B, Czarnecki O. (2010) Rapid dark repression of 5-aminolevulinic acid synthesis in green barley leaves. Plant Cell Physiol 51: 670–681 [DOI] [PubMed] [Google Scholar]
- Sambrook DW., Jr (2001). Molecular Cloning: A Laboratory Manual, Ed 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Serrato AJ, Pérez-Ruiz JM, Spínola MC, Cejudo FJ. (2004) A novel NADPH thioredoxin reductase, localized in the chloroplast, which deficiency causes hypersensitivity to abiotic stress in Arabidopsis thaliana. J Biol Chem 279: 43821–43827 [DOI] [PubMed] [Google Scholar]
- Shepherd M, Hunter CN. (2004) Transient kinetics of the reaction catalysed by magnesium protoporphyrin IX methyltransferase. Biochem J 382: 1009–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd M, McLean S, Hunter CN. (2005) Kinetic basis for linking the first two enzymes of chlorophyll biosynthesis. FEBS J 272: 4532–4539 [DOI] [PubMed] [Google Scholar]
- Spínola MC, Pérez-Ruiz JM, Pulido P, Kirchsteiger K, Guinea M, González M, Cejudo FJ. (2008) NTRC new ways of using NADPH in the chloroplast. Physiol Plant 133: 516–524 [DOI] [PubMed] [Google Scholar]
- Stenbaek A, Hansson A, Wulff RP, Hansson M, Dietz KJ, Jensen PE. (2008) NADPH-dependent thioredoxin reductase and 2-Cys peroxiredoxins are needed for the protection of Mg-protoporphyrin monomethyl ester cyclase. FEBS Lett 582: 2773–2778 [DOI] [PubMed] [Google Scholar]
- Stenbaek A, Jensen PE. (2010) Redox regulation of chlorophyll biosynthesis. Phytochemistry 71: 853–859 [DOI] [PubMed] [Google Scholar]
- Tanaka R, Kobayashi K, Masuda T. (2011). Tetrapyrrole metabolism in Arabidopsis thaliana. The Arabidopsis Book 9: e0145, doi/10.1199/tab.0145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thormählen I, Ruber J, von Roepenack-Lahaye E, Ehrlich SM, Massot V, Hümmer C, Tezycka J, Issakidis-Bourguet E, Geigenberger P. (2013) Inactivation of thioredoxin f1 leads to decreased light activation of ADP-glucose pyrophosphorylase and altered diurnal starch turnover in leaves of Arabidopsis plants. Plant Cell Environ 36: 16–29 [DOI] [PubMed] [Google Scholar]
- Tottey S, Block MA, Allen M, Westergren T, Albrieux C, Scheller HV, Merchant S, Jensen PE. (2003) Arabidopsis CHL27, located in both envelope and thylakoid membranes, is required for the synthesis of protochlorophyllide. Proc Natl Acad Sci USA 100: 16119–16124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wulff RP, Lundqvist J, Rutsdottir G, Hansson A, Stenbaek A, Elmlund D, Elmlund H, Jensen PE, Hansson M. (2011) The activity of barley NADPH-dependent thioredoxin reductase C is independent of the oligomeric state of the protein: tetrameric structure determined by cryo-electron microscopy. Biochemistry 50: 3713–3723 [DOI] [PubMed] [Google Scholar]
- Zhou S, Sawicki A, Willows RD, Luo M. (2012) C-terminal residues of Oryza sativa GUN4 are required for the activation of the ChlH subunit of magnesium chelatase in chlorophyll synthesis. FEBS Lett 586: 205–210 [DOI] [PubMed] [Google Scholar]