Cellulose synthase complexes may optimize cellulose properties and wall mechanical strength by selective exclusion of faulty CesA1 subunits.
Abstract
Multiple cellulose synthase (CesA) subunits assemble into plasma membrane complexes responsible for cellulose production. In the Arabidopsis (Arabidopsis thaliana) model system, we identified a novel D604N missense mutation, designated anisotropy1 (any1), in the essential primary cell wall CesA1. Most previously identified CesA1 mutants show severe constitutive or conditional phenotypes such as embryo lethality or arrest of cellulose production but any1 plants are viable and produce seeds, thus permitting the study of CesA1 function. The dwarf mutants have reduced anisotropic growth of roots, aerial organs, and trichomes. Interestingly, cellulose microfibrils were disordered only in the epidermal cells of the any1 inflorescence stem, whereas they were transverse to the growth axis in other tissues of the stem and in all elongated cell types of roots and dark-grown hypocotyls. Overall cellulose content was not altered but both cell wall crystallinity and the velocity of cellulose synthase complexes were reduced in any1. We crossed any1 with the temperature-sensitive radial swelling1-1 (rsw1-1) CesA1 mutant and observed partial complementation of the any1 phenotype in the transheterozygotes at rsw1-1’s permissive temperature (21°C) and full complementation by any1 of the conditional rsw1-1 root swelling phenotype at the restrictive temperature (29°C). In rsw1-1 homozygotes at restrictive temperature, a striking dissociation of cellulose synthase complexes from the plasma membrane was accompanied by greatly diminished motility of intracellular cellulose synthase-containing compartments. Neither phenomenon was observed in the any1 rsw1-1 transheterozygotes, suggesting that the proteins encoded by the any1 allele replace those encoded by rsw1-1 at restrictive temperature.
Cellulose microfibrils are major tension-bearing components of the cell walls of vascular plants. They consist of multiple β-1,4-linked glucan (cellulose) chains, which are synthesized at the plasma membrane from multi-CesA enzyme complexes, known as cellulose synthase complexes (CSCs) or rosettes (Brown et al., 1996; Kimura et al., 1999). Fluorescently tagged CesAs enable CSCs to be observed at the plasma membrane and their activity measured as a function of the velocity at which they move (Paredez et al., 2008). Recently a direct correlation between temperature and CSC velocity was demonstrated, which reinforces the concept that the glycosyltransferase activity is responsible for the movement of the CSCs, via displacement of the complex from the rigid, paracrystalline cellulose microfibril product (Fujita et al., 2011). The major objective of current research on CesAs is to identify the key structural features that define their glycosyltransferase activity as well as the interactions between CesAs that generate a functional enzyme complex. It is predicted that at least three different CesAs contribute to the synthetic activity of each CSC (Persson et al., 2007), the combinations of which change according to the tissue, developmental stage, or growth conditions. For example, of the 10 CesAs identified in Arabidopsis (Arabidopsis thaliana), CesA1, CesA2, CesA3, CesA5, CesA6, and CesA9 function during primary cell wall formation (Desprez et al., 2007; Persson et al., 2007), whereas CesA4, CesA7, and CesA8 are involved in secondary cell wall formation (Taylor et al., 2003).
Several conserved domains of the CesA protein orthologs have been characterized (illustrated schematically in Fig. 1B). The first cytoplasmic domain at the N terminus contains a Cys-rich region, which is suggested to form zinc- or RING-finger motifs to facilitate interaction with other CesAs (Kurek et al., 2002), along with a highly variable region that is not present in bacterial CesAs (Pear et al., 1996). After a cluster of two predicted transmembrane domains (TMDs), the central cytoplasmic domain contains the D, D, D, QxxRW motif, which is catalytically active and conserved in processive β-glycosyltransferases (Pear et al., 1996). The central cytoplasmic domain also contains a region that is relatively conserved between orthologs, called the class-specific region (Vergara and Carpita, 2001). Then, after a cluster of six predicted TMDs, there is a small cytoplasmic domain at the C-terminal region, which has been shown to be important for cellulose production, as demonstrated in analysis of the CesA3 mutant allele rsw5 (Wang et al., 2006).
Figure 1.
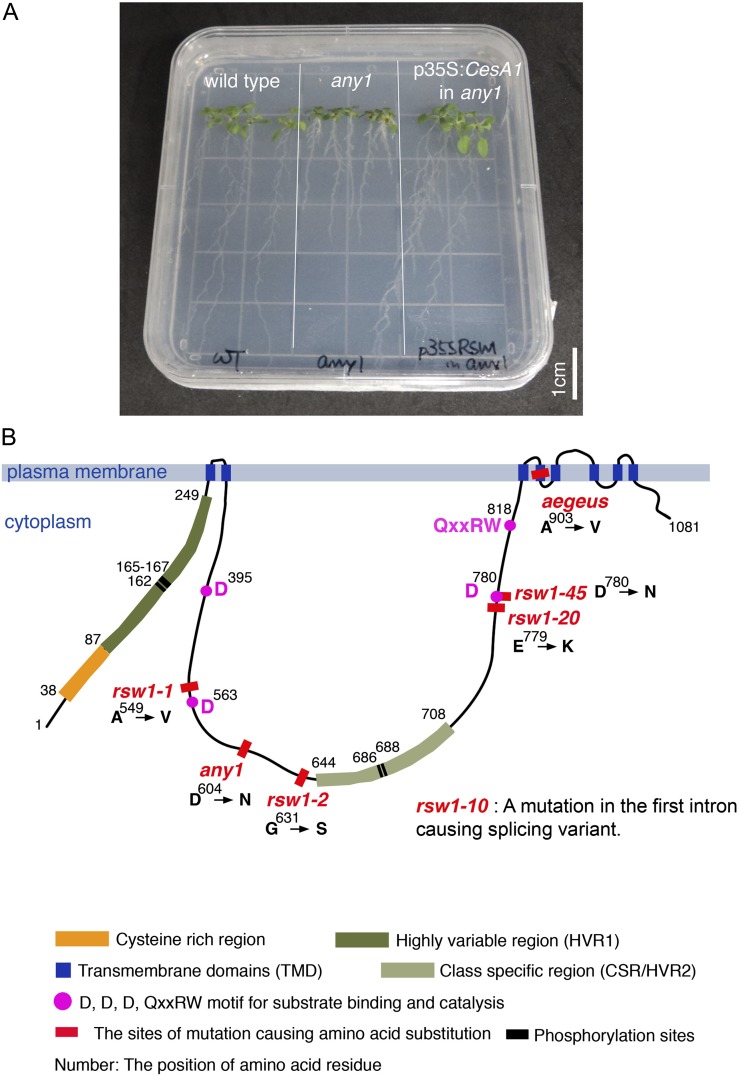
Identification of the any1 as an allele of CesA1. A, 14-d-old seedlings of the wild type, any1, and the any1 plants rescued by p35S:CesA1 cDNA. Scale bar = 1 cm. B, Schematic diagram of domains and the location of mutations in CesA1. CesA1 protein contains a cluster of two predicted TMDs, a Cys-rich domain suggested to form zinc- or RING-finger domains to facilitate interaction with other CesAs (Kurek et al., 2002), and a highly variable region (HVR1) at the N terminus that is not present in bacterial CesAs (Pear et al., 1996). The central domain contains the D, D, D, QxxRW motif found to be conserved in processive β-glycosyltransferases (Coutinho et al., 2003), and also contains a region that is relatively conserved between orthologs, called the class-specific region (CSR; Vergara and Carpita, 2001). After a cluster of six predicted TMDs, there is a small C-terminal region. The any1 mutation is indicated at residue 604. Four other previously described missense CesA1 mutant alleles are also located in the central cytosolic domain where the catalytic site of the β-glycosyltransferases is located.
During primary cell wall formation CesA1 and CesA3 are essential, while one or more of CesA2, CesA5, CesA6, or CesA9 must contribute to cellulose production (Robert et al., 2004; Desprez et al., 2007; Persson et al., 2007). CesA1 null mutations are gametophytic lethal (Persson et al., 2007). Another transfer DNA insertion line, radial swelling1-10 (rsw1-10), is a leaky allele that manifests in reduced CesA1 expression, causing a weak constitutive phenotype of postembryonic radial swelling (Fagard et al., 2000). In the cytoplasmic domain of CesA1, there are six phosphorylation sites, which are suggested to be involved in the directionality of CSC movement (Chen et al., 2010). To date, five missense mutant alleles of CesA1 have been identified. Four of these have altered amino acid sequence in the central cytosolic domain of the CesA1 protein, where the actual catalytic site of the β-glycosyltransferase enzyme is proposed to be located. The rsw1-1 mutant is temperature sensitive, and shows reduced cellulose production at 31°C, causing profound defects in growth and morphogenesis (Arioli et al., 1998; Williamson et al., 2001). The embryo-lethal rsw1-2 mutant was later identified based on its radially swollen phenotype during embryogenesis and was found to have a greatly reduced level of crystalline cellulose at the cotyledon stage of embryo development (Gillmor et al., 2002). The rsw1-20 and rsw1-45 mutations also cause defects in embryogenesis, including reduced cellulose production and the formation of thin and incomplete cell walls (Beeckman et al., 2002). The severe embryo-lethal phenotypes of rsw1-2, rsw1-20, and rsw1-45 suggest that the domains in which their missense mutations occur are essential for CesA1 function. Recently, another CesA1 mutant, aegeus, was identified in a screen for resistance to the cellulose biosynthesis inhibitor quinoxyphen (Harris et al., 2012). The aegeus mutant has a semidominant missense mutation in the C-terminal TMD (Harris et al., 2012).
In a screen for defective microtubule patterns in leaf epidermal cells, we identified a missense cesA1 allele, anisotropy1 (any1). Unlike previously described cesA1 alleles with point mutations in the central cytosolic domain, any1 has neither a lethal nor conditional phenotype, yet it does have dwarf stature and defective cell morphology, in particular, a reduction in growth anisotropy. Here we report that the any1 mutant’s defective anisotropy is attributed to reduced wall crystallinity and that this is correlated with slower displacement velocities of the CSCs in expanding cells. Using live-cell imaging of CSCs in transheterozygotes of any1 and the temperature-sensitive rsw1-1 allele, we demonstrate that the any1-encoded enzyme complements the rsw1-1 phenotype by restoring CSCs to the plasma membrane.
RESULTS
any1 Is a CesA1 Missense Allele
The any1 mutant was isolated from M2 lines of an ethyl methanesulfonate-treated population. The mutation is completely recessive. The any1 heterozygotes cannot be distinguished from wild-type plants. The F2 segregation ratio for wild type:any1 is therefore 3:1. Coarse mapping (see “Materials and Methods”) linked the any1 mutation to the CesA1 locus (At4g32410). The any1 mutant phenotype was rescued by transforming the mutants to express the CesA1 complementary DNA (cDNA) under the constitutive 35S promoter element (Fig. 1A). Sequencing the CESA1 gene in the any1 mutant background identified a single G to A nucleotide substitution (1810 bp of cDNA) that results in an Asp to Asn substitution at residue 604 (Fig. 1B). Thus, any1 is a missense allele of CESA1.
any1 Has a Constitutive Phenotype throughout Development
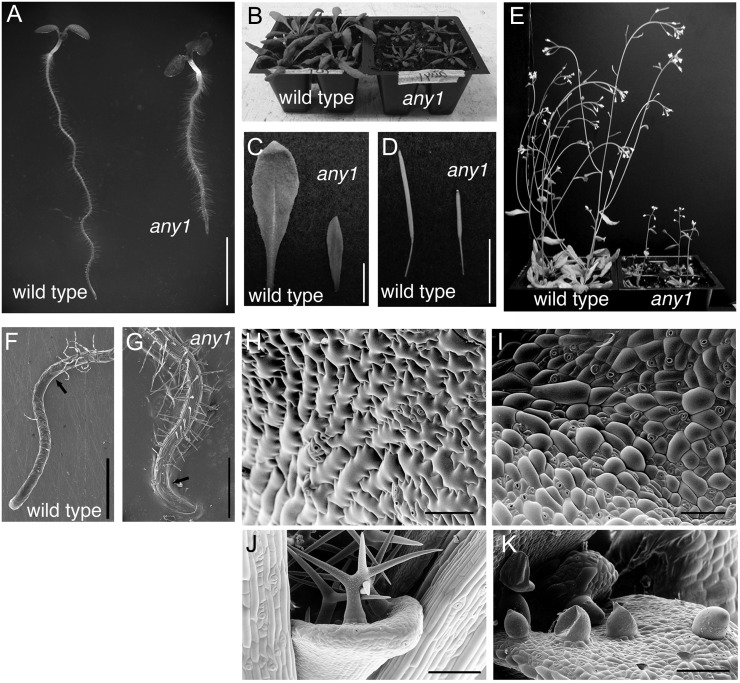
The any1 mutant has a dwarf phenotype throughout development. any1 seedlings have short, thick roots and hypocotyls (Fig. 2A). Aerial organs, including rosette leaves (Fig. 2, B and C), siliques (Fig. 2D), and inflorescence stems (Fig. 2E) are smaller than wild-type counterparts. Whereas bolting occurs 4 weeks after germination in the wild type, under identical conditions, bolting is delayed by 1 week in any1.
Figure 2.
The any1 mutant is a dwarf and has altered cell morphology. A, 6-d-old seedlings of the wild type and any1. Note that any1 has a shorter, thicker root compared with the wild type. Scale bar = 5 mm. B, 3-week-old wild-type and any1 plants. C, Rosette leaves from 4-week-old wild-type and any1 plants. D, Siliques. Scale bars = 1 cm (B–D). E, 6-week-old wild-type and any1 plants. F to K, Cryoscanning electron micrographs. F, Wild-type root. G, any1 root (note shorter elongation zone). Arrows show the end of elongation zone. Scale bars = 500 µm. H, Epidermal cells of cotyledons in the wild type showing typical interdigitation unlike any1 cotyledon epidermal cells (I). Scale bars = 200 µm (H and I). J, Wild-type trichomes from first leaves have two or more tapering branches. K, any1 trichomes are fragile, resulting in rupture of some trichomes. Scale bars = 100 µm (J and K).
Severe epidermal defects were observed in any1 mutants, especially in aerial organs. In the roots, scanning electron microscopy (SEM) analysis demonstrated a normal differentiation of the epidermis into trichoblast and atrichoblast files (Fig. 2, F and G). Root epidermal cell production rate (calculated by dividing root growth rate by cell length) was similar for the wild type (1.6 cell h–1) and any1 (1.5 cell h–1), but cells did not elongate normally, resulting in very short epidermal cells and increased root hair density (Fig. 2A), although root hairs were of normal length (Fig. 2A) and shape (Fig. 2G). In leaves, comparison of wild-type (Fig. 2H) and any1 (Fig. 2I) epidermis by SEM demonstrated that the any1 mutation impaired the ability of pavement cells to elongate and interdigitate (Fig. 2I). Compared with the slender and branched trichomes of wild-type leaves (Fig. 2J), any1 trichomes initiate but do not develop beyond round protrusions, which rarely branch and frequently rupture (Fig. 2K). The any1 trichomes and leaves were easily detached when manipulated by forceps, suggesting that any1 has defects in cell-to-cell adhesion.
Despite the altered cellular morphology of any1 mutants, microtubules were abundant and well organized in epidermal cells of leaves (Supplemental Fig. S1A), hypocotyls (Supplemental Fig. S1B), and roots (Supplemental Fig. S1C).
Cellulose Content Is Not Altered But Cell Wall Crystallinity Is Reduced in the any1 Inflorescence Stems
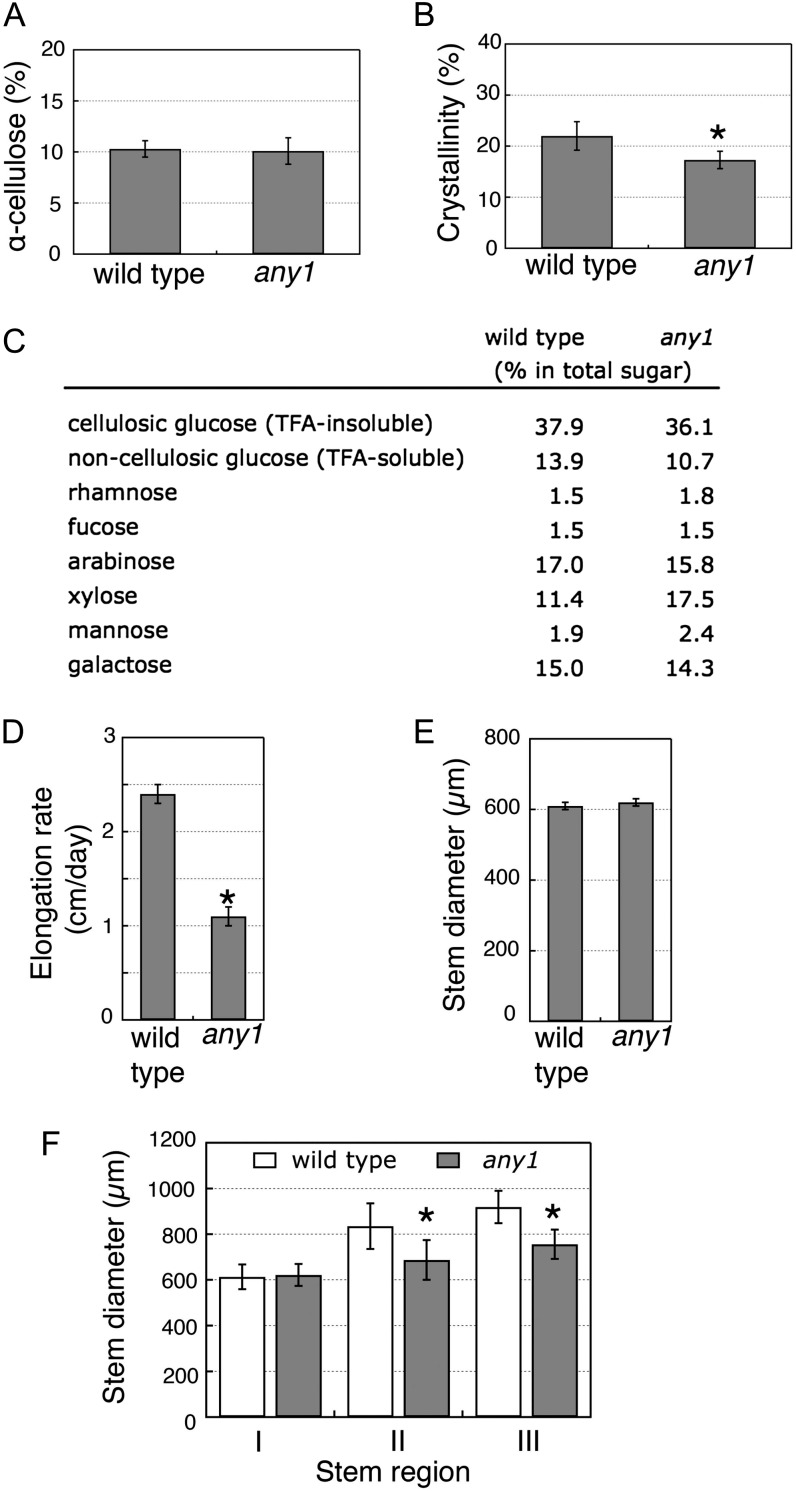
Because cellulose production is greatly reduced in severe CesA1 mutant alleles (Supplemental Table S1 and references therein), we sought to determine if cellulose content and/or wall crystallinity are affected in the any1 mutant using the growing region of the inflorescence stems because these organs provide the most tissue for this purpose. However, we found no significant reduction in cellulose content in the any1 mutant. Analysis of wall material collected from the growing regions of inflorescence stems determined that the α-cellulose content was 10.3% ± 0.8% in the wild type and 10.1% ± 1.3% in any1 (Fig. 3A). In contrast, cell wall crystallinity (the proportion of crystalline cellulose in the cell wall) was significantly reduced. X-ray diffraction analysis estimated a cell wall crystallinity of 22.0% ± 2.8% for the wild type and 17.3% ± 1.7% for any1 (Fig. 3B). This analysis suggests that the any1 mutation primarily affects cellulose structure rather than its production. Analysis of the neutral sugars from the cellulosic (trifluoroacetic acid-insoluble) and noncellulosic (trifluoroacetic acid-soluble) cell wall fraction by gas chromatography-mass spectrometry (GC-MS) showed that the any1 mutant has a higher proportion of Xyl (17.5%) in the wall compared with that in the wild type (11.4%), whereas other sugars were present in relatively normal proportions (Fig. 3C). These changes in cell wall structure and composition are associated with defective cell elongation, as indicated by the reduced ratio of cell length to width in the epidermis and cortex layers of the mutant stem (Table I). The any1 inflorescence stems grow more slowly (Fig. 3D), but are also thinner at maturity compared with wild-type stems (Fig. 3, E and F).
Figure 3.
Growth, cellulose content, and wall crystallinity analysis from inflorescence stems grown at 21°C. A, α-Cellulose content measured as Glc after removal of noncellulosic components from the growing region of inflorescence stems. Values are means ± sd (n = 3). B, Cell wall crystallinity measured from growing regions of wild-type and any1 inflorescence stems grown at 21°C. Asterisk indicates significant difference (Student’s t test: P < 0.05). Values are means ± sd (n = 10). C, GC-MS analysis of cell wall fraction showing the proportions of monosaccharides in total sugar (note higher proportion of Xyl in any1 compared with the wild type). Values are means from duplicate samples. D, Stem elongation rates were measured after marking the top 2 cm once stem heights reached 5 to 8 cm, then measuring the length increases after 1 d. E, Diameters measured from upper growing regions of inflorescence stems. Values are means ± se (n = 20). F, Stem diameters measured 1 d after marking points 1 (I), 2 (II), and 3 (III) cm from the top of stem. Asterisks in I–III indicate statistically significant differences relative to the wild type (Student’s t test: P < 0.05). Values are means ± se (n = 15).
Table I. Degree of growth anisotropy is reduced in epidermal cells of the any1 inflorescence stems.
Cell length and width were measured and the cell dimension ratio was calculated as length divided by width. The higher the value of the ratio, the higher degree of growth anisotropy. More than three inflorescence stems for each genotype were used. Asterisks indicate differences that are statistically significant (Student’s t test: P < 0.05). n, Number of cells that were measured.
| Cell Length | Cell Dimension Ratio (Length/Width) |
|
|---|---|---|
| Wild Type (n) | any1 (n) | |
| Epidermal cells | ||
| ≤20 μm | 2.4 ± 0.7 (56) | 2.4 ± 0.7 (81) |
| 20 to 40 μm | 4.2 ± 1.4 (121) | 3.7 ± 1.2 (108)* |
| 40 to 60 μm | 7.4 ± 2.4 (29) | 4.7 ± 1.3 (24)* |
| >60 μm | 9.9 ± 1.5 (6) | 7.6 ± 1.7 (9)* |
| Cortex cells | ||
| ≤5 μm | 0.4 ± 0.1 (69) | 0.5 ± 0.1 (85)* |
| 5 to 10 μm | 0.7 ± 0.2 (142) | 0.6 ± 0.2 (138)* |
Cellulose Microfibrils Are Disorganized in the Epidermis of the any1 Inflorescence Stems But Otherwise Align Transverse to the Growth Axis of Elongating Organs
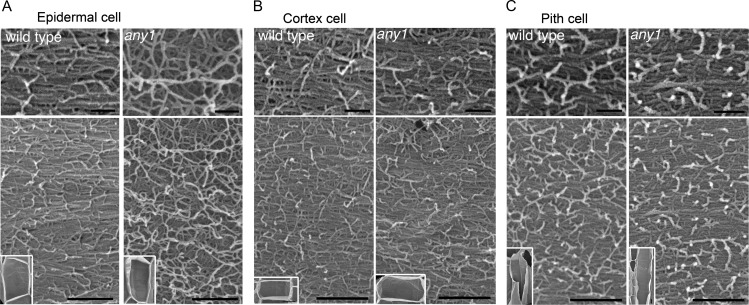
Previously it was reported that reduced cellulose synthesis caused by 2,6-dichlorobenzonitrile treatments of wild-type seedlings or caused by the rsw1-1 mutation at the restrictive temperature profoundly disrupt the appearance and orientation of cellulose microfibrils (Sugimoto et al., 2001). Since our chemical analysis of cellulose was carried out on the growing regions of inflorescence stems, we used field emission scanning electron microscopy (FESEM) on cryoplaned inflorescence stems to analyze cellulose microfibrillar patterns in several tissue layers. FESEM analysis showed that compared with the predominantly transverse cellulose microfibrils found in wild-type cells, microfibrils appeared to be less densely packed and lacked any preferred orientation in the any1 epidermal cells (Fig. 4A). In the cortex (Fig. 4B) and pith (Fig. 4C) layers, however, any1 cellulose microfibril patterns were indistinguishable from those in equivalent wild-type cells, with a predominant orientation transverse to the growth axis. FESEM analyses on the any1 root and dark-grown hypocotyl epidermal cells demonstrated that any1 cellulose microfibrils also aligned transversely to the cell growth axis as in wild-type plants (Supplemental Fig. S2).
Figure 4.
FESEM cellulose microfibril analysis. A to C, FESEM micrographs show cellulose microfibrils in representative cells from different tissues of the inflorescence stems of wild type and any1. Small insets show the cell in which cellulose microfibrils were observed. Upper panel shows the high magnification image of cellulose microfibrils shown in the lower panel. A, Cellulose microfibrils are aligned transversely to the cell growth axis in wild-type epidermal cells, but are random in any1 epidermal cells. B and C, Both cortex cells and pith cells have transverse cellulose microfibrils in the wild type and any1. Scale bars = 100 nm (Top), 300 nm (Bottom).
CSC Velocity Is Reduced in any1
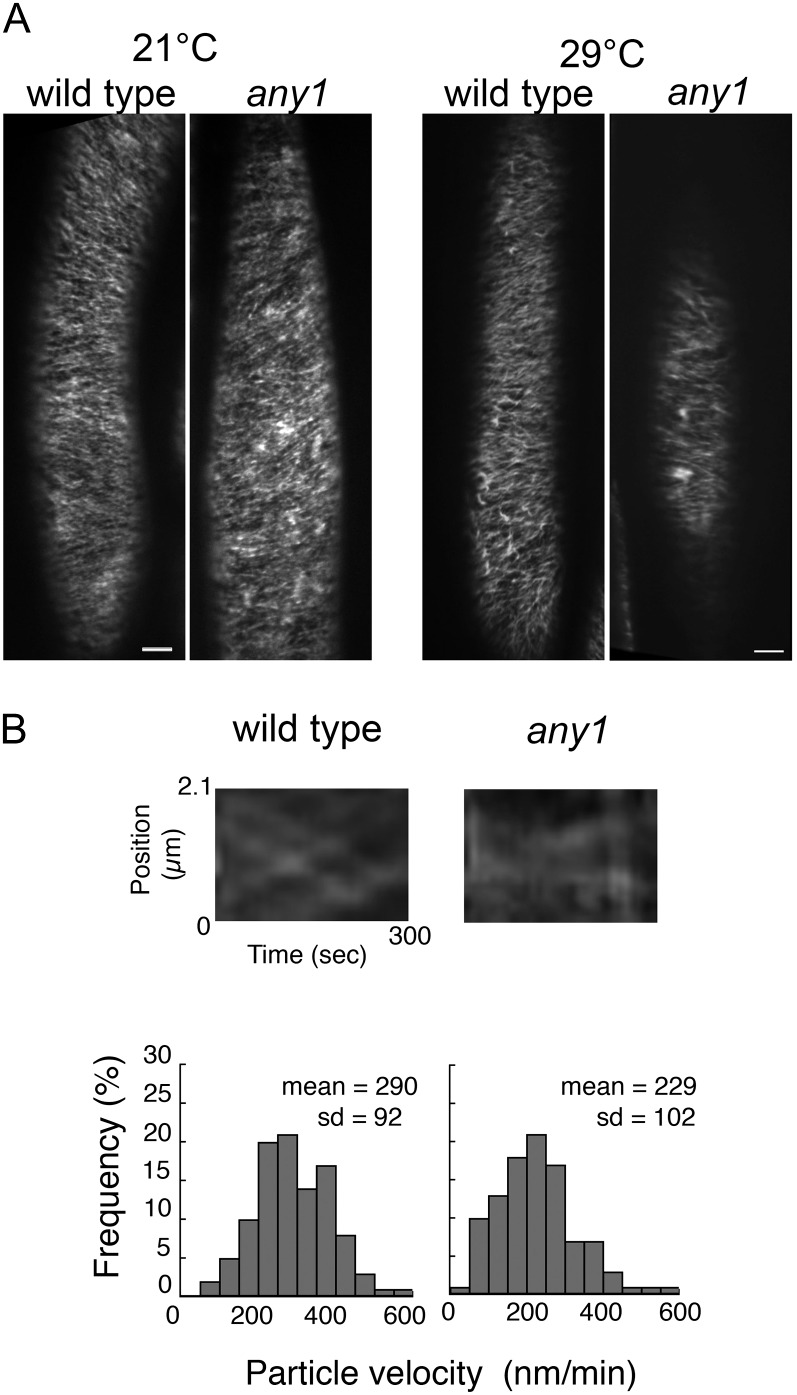
To determine if the any1 mutation affects the assembly and behavior of the CSCs at the plasma membrane, we crossed any1 with transgenic plants expressing yellow fluorescent protein-tagged CesA6 (YFP-CesA6) in the prc1-1 null CesA6 mutant background and isolated any1 prc1-1 double homozygotes expressing YFP-CesA6. The CSCs were tracked in the hypocotyl epidermal cells of dark-grown seedlings using a spinning disc confocal microscope. Previously, we found that at 29°C the CSC velocities are greatly increased and tracking the CSCs is more accurate (Fujita et al., 2011). We therefore measured the CSC velocities either in seedlings grown at 21°C for 3 d, or at 21°C for 2 d and 29°C for 1 d. At both temperatures, we noted that 3-d-old dark-grown seedlings expressing YFP-CesA6 in any1 had shorter hypocotyls compared with the wild type (Supplemental Fig. S3), indicating that mutant dwarf phenotypes are consistent in the hypocotyls at both temperatures. The fluorescence intensity of YFP-CesA6 in any1 was consistently slightly lower than that of the wild type. Combined images of 31 frames collected over 5 min showed linear trajectories of YFP-CesA6 in both the wild type and any1 at 21°C (Fig. 5A) and at 29°C after 1 d (Fig. 5A; Supplemental Movie S1 for wild type and Supplemental Movie S2 for any1). Kymograph analysis revealed that movement of CSCs is bidirectional in both the wild type and any1, but that the CSC velocity was significantly reduced in any1 (229 ± 102 nm min–1) compared with that of the wild type (290 ± 92 nm min–1; Wilcoxon rank sum test: P < 0.05; Fig. 5B).
Figure 5.
The CSC velocity is reduced in any1. A, Projection of 5-min time lapse images of YFP-CesA6 showing similar CSC trajectories in the wild type and any1 growing at both 21°C and 29°C. Scale bars = 5 µm. B, Kymographs show bidirectional movement of CSCs in the wild type and any1 after 1 d at 29°C. Frequency distribution of YFP-CesA6 velocity indicates that the CSCs move significantly more slowly in any1 than in the wild type (Wilcoxon rank sum test: P < 0.05). The velocity of 400 particles was measured in nine cells from six seedlings after 1 d at 29°C.
any1 Rescues the rsw1-1 Temperature-Sensitive Phenotype and rsw1-1 Partially Rescues any1
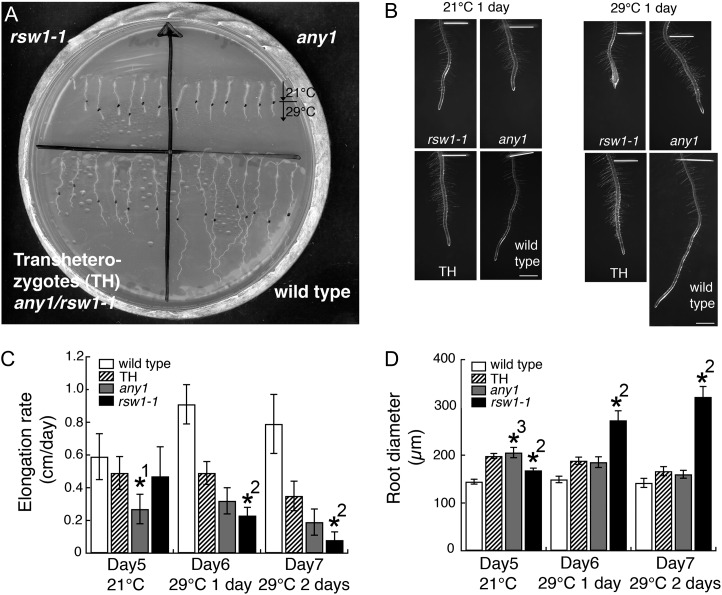
When any1 CesA mutants (D604N allele) were crossed with temperature-sensitive rsw1-1 CesA mutants (A549V allele), we observed partial allelic complementation (Fig. 6). To examine the extent of this allelic complementation, we compared root elongation rates (Fig. 6C) and diameters (Fig. 6D) between any1, rsw1-1, the any1/rsw1-1 transheterozygotes, and wild-type controls. Plants were grown at 21°C (permissive temperature) for 5 d and then grown at 29°C (restrictive temperature) for 2 d. At 21°C, any1 roots had a reduced elongation rate (0.27 ± 0.02 cm d–1) and an increased root diameter (205 ± 11 µm) compared with the wild type (elongation rate: 0.59 ± 0.02 cm d–1; diameter: 145 ± 5 µm) and rsw1-1 (elongation rate: 0.47 ± 0.03 cm d–1, diameter: 168 ± 5 µm). Root elongation rates for any1/rsw1-1 transheterozygotes (0.49 ± 0.02 cm d–1) were greater than those of any1 (0.27 ± 0.02 cm d–1), and root diameters of the transheterozygotes (198 ± 5 µm) and any1 (205 ± 11 µm) were similar. These results indicate that there is a partial allelic complementation of the any1 phenotype by rsw1-1 at 21°C. At the restrictive temperature (29°C) for rsw1-1, the root elongation rate was significantly reduced in rsw1-1 (0.23 ± 0.01 cm d–1) compared with the wild type (0.91 ± 0.02 cm d–1) and any1 (0.32 ± 0.02 cm d–1). Transheterozygotes showed a root elongation rate (0.49 ± 0.02 cm d–1) that was intermediate between the wild type and any1. Although root diameter was significantly increased in rsw1-1 (273 ± 20 µm) compared with the wild type (149 ± 7 µm) and any1 (185 ± 11 µm), the transheterozygote roots had a similar diameter (188 ± 8 µm) to that of any1 roots and did not swell (Fig. 6B), indicating that any1 fully rescued the rsw1-1 phenotype at 29°C.
Figure 6.
Root growth analysis shows that rsw1-1 partially complements any1, and any1 complements the radial swelling of rsw1-1 in transheterozygotes. A, Wild type, any1, rsw1-1, and transheterozygotes (any1/rsw1-1) were grown for 5 d at 21°C, then moved to 29°C. Black dots indicate the location of the root tip when the plants were moved to 29°C. B, Images of roots grown for 6 d at 21°C and for 5 d at 21°C and 1 d at 29°C. The distance between the white line and the root tip show the root growth for 1 d at either 21°C or 29°C. Scale bars = 1 mm. C, Root elongation rates showing that transheterozygote (TH) has an elongation rate intermediate between wild type and any1. Values are means ± sd (n = 20). D, Root diameter of rsw1-1 increases when plants are grown at 29°C, while transheterozygotes have similar root thickness to any1 at both 21°C and 29°C. Values are means ± sd (n = 20). Asterisks show significant differences (P < 0.0001). *1, Significant difference between any1 and wild type, TH and rsw1-1; *2, Significant difference between rsw1-1 and wild type, any1 and TH; *3, Significant difference between any1 and wild type and rsw1-1.
From the root growth analyses, we also concluded that the any1 root phenotype, including slow elongation rate and thick roots, is constitutive at both temperatures.
Reduced Density of the CSCs in rsw1-1 at Restrictive Temperature Is Accompanied by Less Motile CesA6-Containing Endomembrane Compartments
The apparent functional rescue of the rsw1-1 temperature-conditional phenotype by the presence of the any1 allele prompted us to examine the behavior of the CSCs in hypocotyl epidermal cells of rsw1-1 homozygotes and rsw1-1/any1 transheterozygotes. Analysis of hypocotyl length and diameter in dark-grown wild-type, any1, rsw1-1, and any1/rsw1-1 transheterozygotes expressing YFP-CesA6 confirmed that there is partial allelic complementation in the any1 rsw1-1 transheterozygote similar to that observed in the roots (Supplemental Fig. S3).
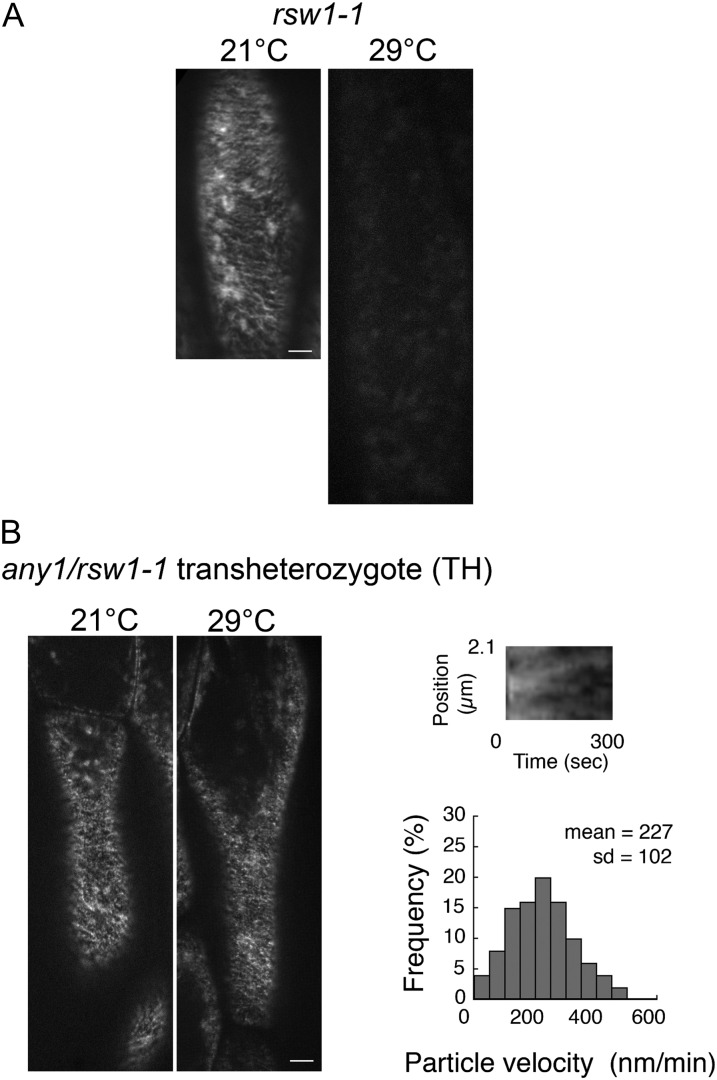
In rsw1-1 hypocotyl epidermal cells observed at 21°C using a temperature-controlled stage, YFP-CesA6 at the plasma membrane exhibited typical movements and distribution (Fig. 7A), but YFP-CesA6 particles were not detected at the plasma membrane after 1 d at 29°C (Fig. 7A). We also examined the distribution of YFP-CesA6 in the endomembrane system, which is largely associated with ring-shaped structures that have previously been identified as Golgi bodies (Paredez et al., 2006). In comparison with the very rapid movement in the wild type after 1 d at 29°C (Supplemental Movie S1), in rsw1-1, compartments carrying YFP-CesA6 moved very slowly (Supplemental Movie S3) and, in addition, the overall rate of cytoplasmic streaming was greatly reduced, as observed by light microscopy.
Figure 7.
The CSC velocity is reduced in any1 rsw1-1 transheterozygotes, and the CSCs are lost from the plasma membrane in rsw1-1 at 29°C. A, Projection of 5-min time lapse images of YFP-CesA6 in rsw1-1 showing the trajectories of YFP-CesA6-containing CSCs at 21°C; such CSCs are absent at 29°C after 1 d. More than 10 seedlings were measured. B, Projection of 5-min time lapse images of YFP-CesA6 in an any1/rsw1-1 transheterozygote showing the trajectories of YFP-CesA6-containing CSCs, which are present both at 21°C and after 1 d at 29°C. The loss of the CSCs in rsw1-1 after 1 d at 29°C is rescued by the presence of CesA1any1. Scale bars = 5 µm. A kymograph showing bidirectional movement of the CSCs in any1/rsw1-1 after 1 d at 29°C. The velocity of 400 particles was measured in 12 cells from seven seedlings. The velocity of the CSCs in any1/rsw1-1 transheterozygotes, as shown in the bar graph, was similar to those in any1 after 1 d at 29°C (see Fig. 5B; Wilcoxon rank sum test: P > 0.05).
We next sought to determine how quickly the CSCs are lost from the plasma membrane in the rsw1-1 mutant at restrictive temperature and whether they can recover when the permissive temperature is restored. Live-cell imaging of rsw1-1 confirmed that YFP-CesA6 particles become less numerous within 30 min at 29°C (Supplemental Movie S4). This loss coincided with reduced motility of YFP-CesA6-containing compartments in the cytoplasm as well as reduced cytoplasmic streaming. After 3 h at 29°C, very few YFP-CesA6 particles could be detected at the plasma membrane and these were only present in some cells. After 1 d at 29°C, YFP-CesA6 particles were no longer detected at the plasma membrane (Supplemental Movie S3).
We also examined how quickly YFP-CesA6 particles reappear at the plasma membrane when seedlings were transferred to 21°C after 1 d at 29°C. We observed a gradual recovery of the motility of YFP-CesA6-containing compartments in the cytoplasm over the first hour at 21°C, but particles were first detected at the plasma membrane after 2 h in 10 cells from more than 4 seedlings.
Finally, we examined the CSC behavior in hypocotyl epidermal cells of rsw1-1/any1 transheterozygotes. After 1 d at 29°C, in which rsw1-1 shows no detectable YFP-CesA6 particles at the plasma membrane (Fig. 7A), we observed YFP-CesA6 particles at the plasma membrane and rapid movement of the CesA6-containing cytoplasmic compartments in transheterozygotes (Fig. 7B; Supplemental Movie S5). The CSC catalytic velocity at the plasma membrane in transheterozygotes was 227 ± 102 nm min–1 (Fig. 7B), similar to that of any1 at 29°C.
DISCUSSION
In this study we have shown that the impaired growth anisotropy in the any1 mutant is severe in epidermal cells. Trichomes, for example, initiate normally but expand isotropically and frequently rupture, giving the leaves a glabrous appearance. Despite these detrimental effects on the epidermis, the any1 phenotype is mild enough to enable the mutants to complete their life cycle, in contrast to other constitutive, embryo-lethal or conditional CesA1 alleles (Arioli et al., 1998; Fagard et al., 2000; Gillmor et al., 2002; Beeckman et al., 2002; Persson et al., 2007). The any1 mutant is therefore an important experimental model for exploring cellulose synthesis throughout plant growth and development. Although the any1 mutation is located in the central, cytoplasmic domain of CesA1 in close proximity to the enzyme’s catalytic motifs, the FESEM observations showing relatively normal wall texture combined with the motility of YFP-CesA6 particles at the plasma membrane suggest that the CSCs in any1 can produce near normal amounts of cellulose. Nevertheless, the CSC velocity, as measured in hypocotyl epidermal cells, and cell wall crystallinity, as assessed in inflorescence stems, are both significantly reduced. In addition, cellulose microfibrils are disordered in the epidermal cells of growing inflorescence stems.
The reduced crystalline cellulose in the any1 mutant can be interpreted in two ways. Given that CSC displacement results from cellulose polymer formation, it is possible that the reduced CSC velocity in any1 could reflect a slow rate of Glc polymerization and/or UDP-Glc uptake. This might in turn generate intermittent synthesis and inconsistent cellulose chain lengths, impairing the ability of neighboring cellulose chains to form crystalline structures. Another explanation is that cellulose of altered ultrastructure triggers compensatory production of noncellulosic wall components, thus reducing the proportion of crystalline cellulose. It has been shown that pectin and hemicellulose content or composition is altered in cellulose-deficient mutants such as rsw1-1 (Peng et al., 2000), procuste1-1 (prc1-1; Fagard et al., 2000), korrigan (Sato et al., 2001), kobito (kob; Pagant et al., 2002), chitinase-like1 (ctl1)/pom-pom1 (Sánchez-Rodriguez et al., 2012), and aegeus (Harris et al., 2012), and in plants treated with cellulose biosynthesis inhibitors such as thaxtomin A (Bischoff et al., 2009). Our GC-MS results indicate that there is an increase in Xyl in any1, which could be due to increased xyloglucan synthesis. A similar increase in Xyl has also been observed in wall-defective rice (Oryza sativa) mutants lacking a kinesin-4 protein (Zhang et al., 2010).
To date, studies other than our own (this study and Fujita et al., 2011) have not monitored or controlled temperature while measuring the CSC velocity, making it impossible to compare absolute values across all studies. Nevertheless, velocities measured relative to wild type indicate that the direct correlation we found in this study between cell wall crystallinity and the CSC catalytic velocity is consistent with data from other studies. The CSC velocity is similarly reduced in crystalline cellulose-deficient mutants such as korrigan (Paredez et al., 2008), cellulose-interactive protein1 (Gu et al., 2010), prc1-1 (Bischoff et al., 2011), and ctl1-1 (Sánchez-Rodriguez et al., 2012). High crystallinity is correlated with increased YFP-CesA6 velocity in the microtubule organization1-1 mutant at restrictive temperature (Fujita et al., 2011). The exceptions to the crystallinity-CSC velocity correlation is the aegeus mutant, (Harris et al., 2012) and wild type grown at high temperature (Fujita et al., 2011), which were found to have increased CSC velocity but lower crystallinity.
It remains unclear whether the any1 mutation affects the catalytic function of CesA1 or whether it affects the integrity of the CSCs, or cellular components associated with the CSCs. The fact that the CSCs in any1 move bidirectionally with a constant speed along the linear trajectories suggests that the any1 protein only mildly interferes with catalytic function. The normal abundance of the CSCs at the plasma membrane in any1 implies that this mutation does not alter the assembly or delivery of the CSCs to the plasma membrane.
In contrast, the rsw1-1 mutation at restrictive temperature caused the disappearance of YFP-CesA6 particles from the plasma membrane (Fig. 7). This was also reported by live cell imaging by Chen et al. (2010), but they did not use a temperature-controlled stage, and the image showing apparent loss of CSCs in rsw1-1 is an optical section of the cytoplasm. Our current analysis therefore provides important confirmation of the study by Arioli et al. (1998), in which freeze-fracture transmission electron microscopy demonstrated the apparent disassembly of CSCs in rsw1-1 homozygotes at the restrictive temperature (Arioli et al., 1998). In that study, however, individual globular-shaped subunits persisted at the plasma membrane after loss of the rosette structure, suggesting that some fragments of the complex remain (Arioli et al., 1998). The fact that we do not see persistent YFP-CesA6 fluorescence at the plasma membrane in rsw1-1 at restrictive temperature can be interpreted in several ways. The particles observed by freeze fracture might lack the CesA6 enzyme, or they could be single enzyme particles that are undetectable with fluorescence microscopy. Alternatively, they could be freeze-fixation artifacts.
In rsw1-1, the loss of the CSCs from the plasma membrane coincided with a severe impairment of CSC-containing compartment motility. Our recovery experiments showed that the reappearance of the CSCs at the plasma membrane coincided with the recovery of CSC-containing compartment motility. These results suggest that there is a coordinated sensing mechanism between the cellulose synthesis machinery at the plasma membrane and the intracellular trafficking of the CSCs, and that the motility of CSC-containing compartments may be dependent on the correct assembly of CSCs. This sensing mechanism could be mediated by the tethering of CSC-containing compartments to cortical microtubules, which has been reported in cellulose synthesis inhibitor-treated and osmotically stressed cells (Crowell et al., 2009; Gutierrez et al., 2009).
The analysis of CSC behavior in any1/rsw1-1 transheterozygotes raises the question as to whether CSCs can selectively incorporate different CesA1 mutant proteins during their assembly. Clearly the rsw1-1 protein remains functional at 21°C, although its ability to only partially complement the any1 phenotype suggests that it is either not fully functional or that it is haploinsufficient. At 29°C, however, the rsw1-1 enzyme function and/or the structural integrity of the CSCs appear to be impaired to the extent that the CSC complexes disappear from the plasma membrane. In the transheterozygotes, delivery of the CSCs to the plasma membrane and the CSC velocity at the plasma membrane occur just as they do in any1 homozygotes. These results suggest that there is no measurable interference by the rsw1-1 protein in the transheterozygotes at the restrictive temperature, perhaps due to the more faulty rsw1-1 protein being absent from CSCs and the any1 protein produced in the haploid state being sufficient to maintain the integrity and delivery of the CSCs to the plasma membrane. A goal of future studies will be to determine the relative proportions of CesA1rsw1-1 and CesA1any1 in the CSCs in transheterozygotes at restrictive and permissive temperatures.
In relation to the above suggestion, domain swapping experiments between CesA1 and CesA3 in the rsw1-1 or rsw5 (CesA3) mutants led Wang et al. (2006) to conclude that the regions between the second transmembrane domain and the C terminus of the protein are important for CesA1 (or CesA3) to access its particular sites in the CSCs. It has been suggested that entry of a CesA to a particular site in the rosette will be restricted if another CesA preferentially occupies this site. The predicted order for CesA1 incorporation is CesA1wild type > 21°CCesA1rsw1-1 > 29°CCesA1rsw1-1 (Wang et al., 2006). In the case of the any1/rsw1-1 transheterozygotes, the incorporation hierarchy is predicted to be 21°CCesA1rsw1-1 > 21°CCesA1any1 and 29°CCesA1any1 > 29°CCesA1rsw1-1.
Although CesA1 is expressed in all growing tissues including stems, roots, leaves, flowers, young seedlings, and embryos (Beeckman et al., 2002; Burn et al., 2002; Hamann et al., 2004), this does not preclude variations in the contribution of CesA1 to different tissues. In addition, the contribution of cellulose to cell morphology may also vary from tissue to tissue. FESEM analyses showed that cellulose microfibrils in elongating any1 cells in the inflorescence stems were transversely aligned with the exception of the epidermal cells, which had obviously disorganized microfibril patterns. Our growth analyses of roots and hypocotyls showed that the extent of any1 complementation of the rsw1-1 phenotype varied slightly between roots and hypocotyls. These observations support a model in which the contribution of CesA1 to cellulose synthesis might vary from tissue to tissue, and from organ to organ (Williamson et al., 2001).
We found no overall decrease in cellulose content in the any1 mutant, although the any1 mutation results in a specific loss of transverse parallel microfibril orientation in the epidermal cells of inflorescence stems, which might be correlated with a reduction of cellulose content specifically in this tissue. Random cellulose microfibril orientation often correlates with reduced cellulose content and loss of growth anisotropy (Wasteneys, 2004), as shown in rsw1-1 mutants and 2,6-dichlorobenzonitrile-treated plants (Sugimoto et al., 2001; Himmelspach et al., 2003) and in kob1-1 mutants (Pagant et al., 2002).
From the perspective of organ expansion, inflorescence stems generally become thicker at the base. Radial expansion of organs generates hoop stress (Schopfer, 2006) and generally, the outermost epidermal layer of organs is structurally designed to resist this. It is therefore surprising that any1 with a random arrangement of cellulose microfibrils in the epidermal layer has thin stems. It is possible that inflorescence stem expansion is reduced in response to the weakened mechanical properties of the any1 epidermis, to protect the stem from hoop stress. Given that shoot epidermal cells need to coordinate their growth with the inner tissues, our results show that the any1 mutant inflorescence stem is a suitable model for studying the contribution of epidermis to organ growth.
MATERIALS AND METHODS
Plant Materials and Growth Condition
The anisotropy1 (any1) mutant was isolated from a genetic screen of an M2 ethyl methanesulfonate-mutagenized Arabidopsis (Arabidopsis thaliana) Columbia-0 population. The homozygous any1 mutant, which had been backcrossed eight times, and wild-type segregants from this backcrossing were used for all studies. Plants were grown on Hoagland media in agar plates and placed vertically in a 21°C growth cabinet after an initial cold treatment for 4 d. Ten-day-old seedlings were transferred onto soil and grown at 21°C for harvesting inflorescence stems.
Plants expressing p35S::GFP-tagged α-tubulin6 (GFP-TUA6; Ueda et al., 1999) were crossed with any1, and F2 segregants expressing GFP-TUA6/any1 were used for viewing microtubules.
Homozygous YFP-CesA6/prc1-1 was obtained from a T1 generation provided by Chris Somerville (Paradez et al., 2006) as described (Fujita et al., 2011), and was crossed with any1 and rsw1-1 (Arioli et al., 1998) Plants expressing YFP-CesA6 were screened from F2 lines. F3 and F4 lines were used for screening lines carrying homozygous YFP-CesA6 and homozygous prc1-1 mutation as described in Fujita et al. (2011). YFP-CesA6/any1 and YFP-CesA6/rsw1-1 were crossed to generate YFP-CesA6/any1/rsw1-1 transheterozygotes. These lines were grown in the dark for the observation of YFP-CesA6 for 3 d at 21°C, or for 2 d at 21°C and for 1 d at 29°C.
Map-Based Cloning and Genetic Analysis
The any1 mutant Columbia-0 ecotype was crossed with the Arabidopsis ecotype Landsberg erecta. Genomic DNA was isolated from F2 lines showing the any1 phenotype, and was used to map the ANY1 locus. Using cleaved amplified polymorphic sequence markers, the any1 mutation was mapped to the lower arm of chromosome 4. There was a tight linkage with marker F10N7H, which is close to the RSW1/CesA1 locus (Arioli et al., 1998). The any1 mutant was therefore transformed to express CESA1 under the constitutive Cauliflower mosaic virus 35S promoter (p35S:CESA1), the construct of which was kindly provided by Richard Williamson (Arioli et al., 1998). Transformants selected by antibiotic resistance were found to fully complement the any1 phenotype. Sequence analysis identified a G to A nucleotide substitution in the 10th exon, predicted to substitute the amino acid Asp 604 with Asn.
The any1 mutant was also crossed with the rsw1-1 temperature-sensitive allele to look for noncomplementation. The F1 progeny showed rescue of the rsw1-1 temperature-sensitive radial swelling and partial rescue of the any1 phenotype at 21°C, indicating partial allelic complementation. F2 seedlings segregated in a 1:2:1 (52:122:59; χ2 = 0.94) ratio for any1, partially rescued any1, and rsw1-1 phenotypes. There were no wild-type segregants in the F2 generation, confirming that any1 and rsw1-1 are allelic. The any1, rsw1-1, and any1/rsw1-1 seedling phenotypes are shown in Figure 6A.
Growth Analysis
The any1, rsw1-1, transheterozygous and wild-type seedlings were grown in continuous light for 5 d at 21°C, then for 1 d at 29°C for root growth analysis. Growth measurements of inflorescence stems were performed as described in Fujita et al. (2011). For hypocotyl growth analysis, YFP-CesA6 in prc1-1 and YFP-CesA6 in any1/prc1-1 seedlings were grown in foil-wrapped plates either for 3 d at 21°C or for 2 d at 21°C and 1 d at 29°C. Images were taken using a stereomicroscope (Leica MZ16FA, Leica) equipped with a digital camera (DC 350 FXR2, Leica), and the measurements were made using Leica Application Suite (Leica) and ImageJ software with the Neuron J plug-in. ses and deviations were calculated, and means were compared by the independent Student’s t test for samples with unequal variance at a significance level of 0.05.
Cell Wall Analysis
Growing region of inflorescence stems were harvested for cell wall analyses. Preparation of the materials for α-cellulose content and cell wall crystallinity analyses was as described in Fujita et al. (2011). Carbohydrate analysis was performed as described previously (Lane et al., 2001). Cell wall preparation for FESEM was performed as described previously (Fujita et al., 2011).
Cryo-SEM
Fresh samples were frozen and viewed as described previously (Kazama et al., 2004).
Microtubule Labeling
To examine microtubules in aerial organs, the GFP-TUA6 reporter obtained from Takashi Hashimoto (Ueda et al., 1999) was crossed into any1. To view microtubules in roots, immunofluorescence labeling was performed as described previously (Sugimoto et al., 2000; Fujita et al., 2011).
Live-Cell Imaging of YFP-CesA6
Hypocotyl epidermal cells of dark-grown 3-d-old YFP-CesA6/prc1-1 and YFP-CesA6/any1/prc1-1 seedlings were used for imaging the CSCs. These seedlings were grown for 3 d at 21°C or 2 d at 21°C and for 1 d at 29°C. Images were collected using the Perkin-Elmer Ultraview VoX spinning disk system mounted on a Leica DMI6000 microscope equipped with a 63× numerical aperture 1.3 glycerol objective lens as described previously using temperature control stage and objective lens heater to maintain the temperature at either 21°C or 29°C (Fujita et al., 2011). Citrine-YFP was excited with a 514-nm laser. Images were acquired every 10 s for 5 min using a Hamamatsu 9100-02 CCD camera controlled with VOLOCITY software, using emission band filters 540/30 for citrine-YFP.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Cortical microtubules are unaffected in the any1 mutant.
Supplemental Figure S2. Cellulose microfibrils are aligned transversely in the epidermal cells of the any1 root and dark-grown hypocotyls.
Supplemental Figure S3. Analysis of dark-grown hypocotyls of any1, rsw1-1, and any1 rsw1-1 transheterozygotes at 29°C. At 21°C rsw1-1 partially complements the any1 short hypocotyl phenotype, while at 29°C the any1 allele partially complements the rsw1-1 phenotype.
Supplemental Table S1. Summary of CesA1 mutant phenotypes.
Supplemental Movie S1. Distribution and movement of YFP-CesA6 in the wild type at 29°C after 1 d.
Supplemental Movie S3. Distribution and movement of YFP-CesA6 in rsw1-1 at 29°C after 1 d.
Supplemental Movie S4. Distribution and movement of YFP-CesA6 in rsw1-1 at 29°C after 21 min.
Supplemental Movie S5. Distribution and movement of YFP-CesA6 in any1/rsw1-1 transheterozygote at 29°C after 1 d.
Acknowledgments
We thank Ming Kalanon (Australian National University) for assistance with map-based cloning and Tobias Baskin (University of Massachusetts) for calculating cell production rates. We thank Roger Heady (Australian National University Electron Microscopy Unit) and Kevin Hodgson and Derrick Horne (University of British Columbia Bioimaging Facility) for microscopy assistance. We thank Alex Paredez (Carnegie Institution, Stanford University) and Chris Somerville (Energy Bioscience Institute, University of California at Berkeley) for the YFP-CesA6 line and Richard Williamson (Australian National University) for the rsw1-1 mutant and CESA1 cDNA.
Glossary
- FESEM
field emission scanning electron microscopy
- TMD
transmembrane domain
- CSC
cellulose synthase complex
- cDNA
complementary DNA
- GC-MS
gas chromatography-mass spectrometry
- YFP
yellow fluorescent protein
- SEM
scanning electron microscopy
References
- Arioli T, Peng LC, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Höfte H, Plazinski J, Birch R, et al. (1998) Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279: 717–720 [DOI] [PubMed] [Google Scholar]
- Beeckman T, Przemeck GKH, Stamatiou G, Lau R, Terryn N, De Rycke R, Inzé D, Berleth T. (2002) Genetic complexity of cellulose synthase a gene function in Arabidopsis embryogenesis. Plant Physiol 130: 1883–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff V, Cookson SJ, Wu S, Scheible WR. (2009) Thaxtomin A affects CESA-complex density, expression of cell wall genes, cell wall composition, and causes ectopic lignification in Arabidopsis thaliana seedlings. J Exp Bot 60: 955–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff V, Desprez T, Mouille G, Vernhettes S, Gonneau M, Höfte H. (2011) Phytochrome regulation of cellulose synthesis in Arabidopsis. Curr Biol 21: 1822–1827 [DOI] [PubMed] [Google Scholar]
- Brown RM, Jr, Saxena IM, Kudlicka K. (1996) Cellulose biosynthesis in higher plants. Trends Plant Sci 1: 149–156 [Google Scholar]
- Burn JE, Hocart CH, Birch RJ, Cork AC, Williamson RE. (2002) Functional analysis of the cellulose synthase genes CesA1, CesA2, and CesA3 in Arabidopsis. Plant Physiol 129: 797–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Ehrhardt DW, Somerville CR. (2010) Mutations of cellulose synthase (CESA1) phosphorylation sites modulate anisotropic cell expansion and bidirectional mobility of cellulose synthase. Proc Natl Acad Sci USA 107: 17188–17193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho PM, Deleury E, Davies GJ, Henrissat B. (2003) An evolving hierarchical family classification for glycosyltransferases. J Mol Biol 328: 307–317 [DOI] [PubMed] [Google Scholar]
- Crowell EF, Bischoff V, Desprez T, Rolland A, Stierhof YD, Schumacher K, Gonneau M, Höfte H, Vernhettes S. (2009) Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 21: 1141–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, Höfte H, Gonneau M, Vernhettes S. (2007) Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 15572–15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard M, Desnos T, Desprez T, Goubet F, Refregier G, Mouille G, McCann M, Rayon C, Vernhettes S, Höfte H. (2000) PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 12: 2409–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Himmelspach R, Hocart CH, Williamson RE, Mansfield SD, Wasteneys GO. (2011) Cortical microtubules optimize cell-wall crystallinity to drive unidirectional growth in Arabidopsis. Plant J 66: 915–928 [DOI] [PubMed] [Google Scholar]
- Gillmor CS, Poindexter P, Lorieau J, Palcic MM, Somerville C. (2002) Alpha-glucosidase I is required for cellulose biosynthesis and morphogenesis in Arabidopsis. J Cell Biol 156: 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Kaplinsky N, Bringmann M, Cobb A, Carroll A, Sampathkumar A, Baskin TI, Persson S, Somerville CR. (2010) Identification of a cellulose synthase-associated protein required for cellulose biosynthesis. Proc Natl Acad Sci USA 107: 12866–12871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez R, Lindeboom JJ, Paredez AR, Emons AMC, Ehrhardt DW. (2009) Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat Cell Biol 11: 797–806 [DOI] [PubMed] [Google Scholar]
- Hamann T, Osborne E, Youngs HL, Misson J, Nussaume L, Somerville C. (2004) Global expression analysis of CESA and CSL genes in Arabidopsis. Cellulose 11: 279–286 [Google Scholar]
- Harris DM, Corbin K, Wang T, Gutierrez R, Bertolo AL, Petti C, Smilgies DM, Estevez JM, Bonetta D, Urbanowicz BR, et al. (2012) Cellulose microfibril crystallinity is reduced by mutating C-terminal transmembrane region residues CESA1A903V and CESA3T942I of cellulose synthase. Proc Natl Acad Sci USA 109: 4098–4103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelspach R, Williamson RE, Wasteneys GO. (2003) Cellulose microfibril alignment recovers from DCB-induced disruption despite microtubule disorganization. Plant J 36: 565–575 [DOI] [PubMed] [Google Scholar]
- Kazama H, Dan H, Imaseki H, Wasteneys GO. (2004) Transient exposure to ethylene stimulates cell division and alters the fate and polarity of hypocotyl epidermal cells. Plant Physiol 134: 1614–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Laosinchai W, Itoh T, Cui XJ, Linder CR, Brown RM., Jr (1999) Immunogold labeling of rosette terminal cellulose-synthesizing complexes in the vascular plant vigna angularis. Plant Cell 11: 2075–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurek I, Kawagoe Y, Jacob-Wilk D, Doblin M, Delmer D. (2002) Dimerization of cotton fiber cellulose synthase catalytic subunits occurs via oxidation of the zinc-binding domains. Proc Natl Acad Sci USA 99: 11109–11114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DR, Wiedemeier A, Peng LC, Höfte H, Vernhettes S, Desprez T, Hocart CH, Birch RJ, Baskin TI, Burn JE, et al. (2001) Temperature-sensitive alleles of RSW2 link the KORRIGAN endo-1,4-β-glucanase to cellulose synthesis and cytokinesis in Arabidopsis. Plant Physiol 126: 278–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagant S, Bichet A, Sugimoto K, Lerouxel O, Desprez T, McCann M, Lerouge P, Vernhettes S, Höfte H. (2002) KOBITO1 encodes a novel plasma membrane protein necessary for normal synthesis of cellulose during cell expansion in Arabidopsis. Plant Cell 14: 2001–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez AR, Somerville CR, Ehrhardt DW. (2006) Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312: 1491–1495 [DOI] [PubMed] [Google Scholar]
- Paredez AR, Persson S, Ehrhardt DW, Somerville CR. (2008) Genetic evidence that cellulose synthase activity influences microtubule cortical array organization. Plant Physiol 147: 1723–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM. (1996) Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci USA 93: 12637–12642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng LC, Hocart CH, Redmond JW, Williamson RE. (2000) Fractionation of carbohydrates in Arabidopsis root cell walls shows that three radial swelling loci are specifically involved in cellulose production. Planta 211: 406–414 [DOI] [PubMed] [Google Scholar]
- Persson S, Paredez A, Carroll A, Palsdottir H, Doblin M, Poindexter P, Khitrov N, Auer M, Somerville CR. (2007) Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc Natl Acad Sci USA 104: 15566–15571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert S, Mouille G, Höfte H. (2004) The mechanism and regulation of cellulose synthesis in primary walls: lessons from cellulose-deficient Arabidopsis mutants. Cellulose 11: 351–364 [Google Scholar]
- Sánchez-Rodríguez C, Bauer S, Hématy K, Saxe F, Ibáñez AB, Vodermaier V, Konlechner C, Sampathkumar A, Rüggeberg M, Aichinger E, et al. (2012) CHITINASE-LIKE1/POM-POM1 and its homolog CTL2 are glucan-interacting proteins important for cellulose biosynthesis in Arabidopsis. Plant Cell 24: 589–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Kato T, Kakegawa K, Ishii T, Liu YG, Awano T, Takabe K, Nishiyama Y, Kuga S, Sato, et al. (2001) Role of the putative membrane-bound endo-1,4-beta-glucanase KORRIGAN in cell elongation and cellulose synthesis in Arabidopsis thaliana. Plant Cell Physiol 42: 251–263 [DOI] [PubMed] [Google Scholar]
- Schopfer P. (2006) Biomechanics of plant growth. Am J Bot 93: 1415–1425 [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Williamson RE, Wasteneys GO. (2000) New techniques enable comparative analysis of microtubule orientation, wall texture, and growth rate in intact roots of Arabidopsis. Plant Physiol 124: 1493–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K, Williamson RE, Wasteneys GO. (2001) Wall architecture in the cellulose-deficient rsw1 mutant of Arabidopsis thaliana: microfibrils but not microtubules lose their transverse alignment before microfibrils become unrecognizable in the mitotic and elongation zones of roots. Protoplasma 215: 172–183 [DOI] [PubMed] [Google Scholar]
- Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR. (2003) Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc Natl Acad Sci USA 100: 1450–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda K, Matsuyama T, Hashimoto T. (1999) Visualization of microtubules in living cells of transgenic Arabidopsis thaliana. Protoplasma 206: 201–206 [Google Scholar]
- Vergara CE, Carpita NC. (2001) Beta-D-glycan synthases and the CesA gene family: lessons to be learned from the mixed-linkage (1→3),(1→4)beta-D-glucan synthase. Plant Mol Biol 47: 145–160 [PubMed] [Google Scholar]
- Wang J, Howles PA, Cork AH, Birch RJ, Williamson RE. (2006) Chimeric proteins suggest that the catalytic and/or C-terminal domains give CesA1 and CesA3 access to their specific sites in the cellulose synthase of primary walls. Plant Physiol 142: 685–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasteneys GO. (2004) Progress in understanding the role of microtubules in plant cells. Curr Opin Plant Biol 7: 651–660 [DOI] [PubMed] [Google Scholar]
- Williamson RE, Burn JE, Birch R, Baskin TI, Arioli T, Betzner AS, Cork A. (2001) Morphology of rsw1, a cellulose-deficient mutant of Arabidopsis thaliana. Protoplasma 215: 116–127 [DOI] [PubMed] [Google Scholar]
- Zhang M, Zhang B, Qian Q, Yu Y, Li R, Zhang J, Liu X, Zeng D, Li J, Zhou Y. (2010) Brittle Culm 12, a dual-targeting kinesin-4 protein, controls cell-cycle progression and wall properties in rice. Plant J 63: 312–328 [DOI] [PMC free article] [PubMed] [Google Scholar]