Flavonoids and flavenoid synthesis affect white cotton fiber development.
Abstract
The cotton (Gossypium spp.) fiber is a unique elongated cell that is useful for investigating cell differentiation. Previous studies have demonstrated the importance of factors such as sugar metabolism, the cytoskeleton, and hormones, which are commonly known to be involved in plant cell development, while the secondary metabolites have been less regarded. By mining public data and comparing analyses of fiber from two cotton species (Gossypium hirsutum and Gossypium barbadense), we found that the flavonoid metabolism is active in early fiber cell development. Different flavonoids exhibited distinct effects on fiber development during ovule culture; among them, naringenin (NAR) could significantly retard fiber development. NAR is a substrate of flavanone 3-hydroxylase (F3H), and silencing the F3H gene significantly increased the NAR content of fiber cells. Fiber development was suppressed following F3H silencing, but the overexpression of F3H caused no obvious effects. Significant retardation of fiber growth was observed after the introduction of the F3H-RNA interference segment into the high-flavonoid brown fiber G. hirsutum T586 line by cross. A greater accumulation of NAR as well as much shorter fibers were also observed in the BC1 generation plants. These results suggest that NAR is negatively associated with fiber development and that the metabolism mediated by F3H is important in fiber development, thus highlighting that flavonoid metabolism represents a novel pathway with the potential for cotton fiber improvement.
Flavonoids are abundant and widely distributed plant secondary metabolites. They are the primary compounds of plant pigments, provide signals for pollinators and symbiotic bacteria (Taylor and Grotewold, 2005), protect plants from UV-B and environmentally induced oxidative stress (Pourcel et al., 2007), and are involved in pollen tube germination, seed dormancy, and auxin transport (Jacobs and Rubery, 1988; Debeaujon et al., 2000; Brown et al., 2001). The flavonoid pathway has been intensively studied in Arabidopsis (Arabidopsis thaliana) and petunia (Petunia hybrida) model plants, and numerous mutants in the pathway have furthered our understanding of the roles of flavonoids in plant development (Shirley et al., 1995; van Houwelingen et al., 1998; Wisman et al., 1998). The function and regulation of most flavonoid genes are conserved in plants (Uimari and Strommer, 1998; Dong et al., 2001), but the resulting flavonoids may have different functions in different species (Taylor and Grotewold, 2005). Flavanone 3-hydroxylase (F3H) as a core gene in the flavonoid pathway has been identified in more than 50 species. The enzyme activity was first characterized in petunia (Britsch and Grisebach, 1986), and the gene was first cloned in Antirrhinum majus (Martin et al., 1991). F3H is important to pigment biosynthesis in Arabidopsis. The F3H mutant or down-regulated plants exhibit petal color disruption and flavonoid content reduction (Britsch, 1990; Stephens et al., 1993; Peer et al., 2001; Flachowsky et al., 2012). Additionally, the development of seeds and seedlings is also affected in Arabidopsis and soybean (Glycine max) F3H mutants (Debeaujon et al., 2000; Zabala and Vodkin, 2005; Owens et al., 2008; Buer and Djordjevic, 2009). Previous work also showed that the transcript of F3H was abundant in cotton (Gossypium spp.; Udall et al., 2006; Tu et al., 2007).
Cotton is a major economic crop that is rich in secondary metabolites; however, few studies have examined the secondary metabolism in this plant. A cotton fiber is one of the longest single plant cells, and it undergoes four major developmental stages (initiation, elongation, secondary cell wall formation, and maturity; Kim and Triplett, 2001). In the past decade, large amounts of “omics” data about fiber development have been accumulated, and many critical developmental pathways were revealed (Ji et al., 2003; Ruan et al., 2003; Arpat et al., 2004; Shi et al., 2006; Wu et al., 2006; Hovav et al., 2008a). Flavonoid genes were widely detected in most of these data, which included all the studied cotton species (Arpat et al., 2004; Gou et al., 2007; Hovav et al., 2008b; Al-Ghazi et al., 2009; Rapp et al., 2010). In a comparison of the fiber development of Xuzhou142 and its fiberless mutant, flavonoid genes were found to be preferentially expressed in fiber cells over ovules (Gou et al., 2007). In fiber cells, the flavonoid genes were dominantly expressed in the fiber elongation stage (Gou et al., 2007; Hovav et al., 2008b; Rapp et al., 2010). Flavonoid genes were also abundant and showed much higher expression levels in brown fiber, which tends to be shorter than white fiber (Xiao et al., 2007). Proanthocyanidin (PA), a kind of flavonoid derivative, was detected in both white and brown fiber cells (Li et al., 2011). Recently, flavonoid genes were suggested to be correlated with specific fiber properties (Al-Ghazi et al., 2009).
Gossypium hirsutum and Gossypium barbadense are cultivated tetraploid cotton species. G. barbadense has good fiber quality (i.e. longer fiber, better micronaire property, etc.), but its cultivation areas are limited because of its low yield. G. hirsutum is widely cultivated, but its fiber quality is inferior to that of G. barbadense. Comparison of fiber development between these two cotton species is an effective way to analyze the mechanism of fiber development and to identify candidates to improve G. hirsutum quality (Al-Ghazi et al., 2009; Chaudhary et al., 2009). In this study, fiber development was compared between these two cotton species, and it was found that the flavonoid metabolism was differently regulated and might be associated with fiber quality. Further biological and metabolic analysis demonstrated the process whereby F3H catalyzed the metabolism of naringenin (NAR) and played an important role in fiber development. These results highlight the impact of secondary metabolites in fiber cell development and develop an alternative target for fiber quality improvement.
RESULTS
Flavonoid Metabolism during Fiber Elongation Exhibits Significantly Different Patterns in G. hirsutum Compared with G. barbadense
By mining publicly available data, we found that flavonoid genes were widely expressed during fiber development, not only in wild cotton but also in the cultivated cotton species Gossypium arboreum, G. hirsutum, and G. barbadense (Supplemental Table S1). The expression patterns of flavonoid genes were varied among different cotton species, and several of them were more highly expressed in the fibers of wild cotton than in those of cultivated cotton (Supplemental Table S2). Additionally, in fiber cells, most of these genes were more highly expressed during fiber elongation than in secondary cell wall formation (Supplemental Tables S1 and S2). These results indicated that the flavonoid pathway existed in cotton and might be involved in fiber development. To verify this possibility, flavonoid gene expression profiling was accomplished by real-time PCR during fiber development in G. hirsutum YZ1 and G. barbadense 3-79 (Fig. 1A). The flavonoid genes chalcone synthase (CHS), chalcone isomerase (CHI), F3H, flavonoid 3′,5′-hydroxylase (F3′5′H), dihydroflavonol reductase (DFR), and anthocyanidin reductase (ANR) all showed relatively high expression levels that were comparable with that of the internal control gene, ubiquitin7 (UBQ7), and with F3H and ANR, demonstrating the highest levels of transcription. The transcripts of flavonoid genes were obviously different between the two cotton species from 0 to 5 DPA, which is the early fiber elongation stage, whereas less difference was detected in the secondary cell wall developmental stage (15–20 DPA). The biggest difference was apparent in the upstream genes of the flavonoid pathway, including CHS, CHI, flavonoid 3′-hydroxylase (F3′H), and F3′5′H. These genes showed higher expression levels in YZ1. In contrast, F3H, DFR, and ANR, which lay downstream of NAR in the pathway, were more highly expressed in 3-79. Furthermore, these genes showed highest expression at 5 DPA. Interestingly, most genes showed peak expression at 5 DPA in these two cotton species, whereas CHS, CHI, F3′5′H, and ANS exhibited maximal expression at 0 DPA in 3-79.
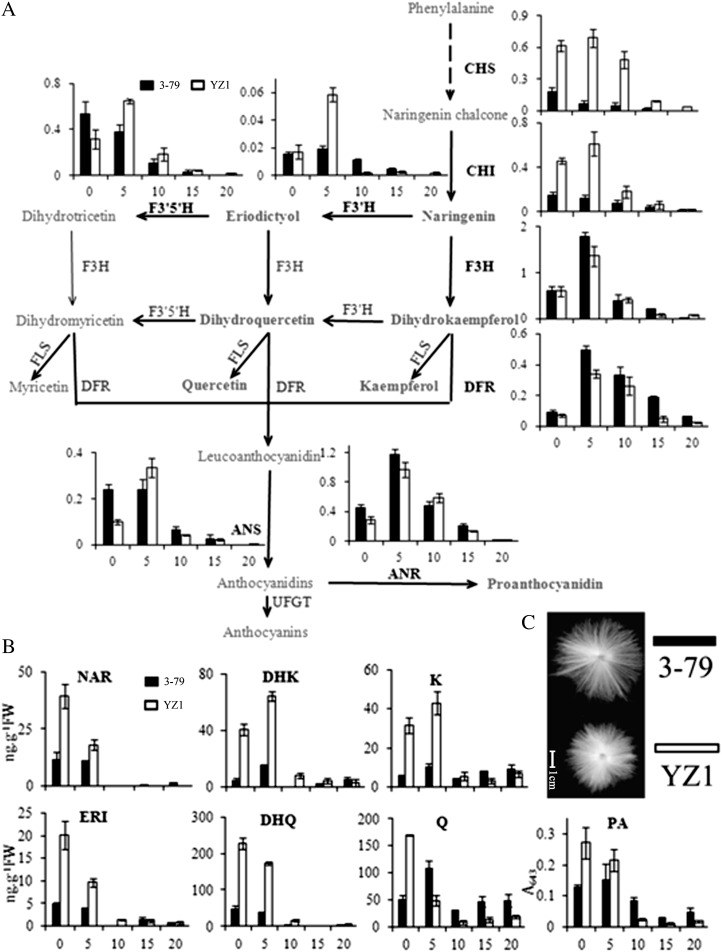
Figure 1.
Expression and metabolic analysis of flavonoids in G. barbadense and G. hirsutum. A, Expression of flavonoid genes was analyzed in developing fiber, and the data were normalized by the cotton ubiquitin gene UBQ7 (referred to as 1). The expression profiles are shown following the flavonoid pathway from Kyoto Encyclopedia of Genes and Genomes and are placed next to their corresponding enzymes. FLS, Flavonol synthase; UFGT, UDP-Glc:flavonoid 3-O-glucosyltransferase. B, Metabolite profiling of flavonoids in developing fibers exhibited high flavonoid levels in early-development-stage fibers corresponding to the gene expression pattern. FW, Fresh weight. Ovules from 0 and 5 DPA and fiber from 10, 15, and 20 DPA were analyzed and are exhibited as 0, 5, 10, 15, and 20, respectively. Error bars represent the sd of three biological replicates. Black columns represent G. barbadense 3-79, and white column represent G. hirsutum cv YZ1. C, The mature fiber of the two cotton species. Bar = 1 cm.
The flavonoid concentration patterns corresponded to flavonoid gene expression profiles during fiber development (Fig. 1B). The flavonoid concentration was higher in YZ1 than in 3-79 at the early stage of fiber development (0–5 DPA). The amounts of NAR, eriodictyol (ERI), dihydroquercetin (DHQ), quercetin (Q), and PA were highest in 0-DPA ovules, whereas dihydrokaempferol (DHK) and kaempferol (K) were highest at 5 DPA. The fibers of 3-79 had lower levels of flavonoids during the elongation stage, whereas during the secondary cell wall developmental stage (15–20 DPA), their flavonoid concentrations were comparable to that of YZ1. An active flavonoid metabolic pathway was present in both G. hirsutum and G. barbadense, whereas the species with higher quality fibers (G. barbadense; Fig. 1C) exhibited lower flavonoid content.
Exogenous NAR and DHK Retarded Fiber Development
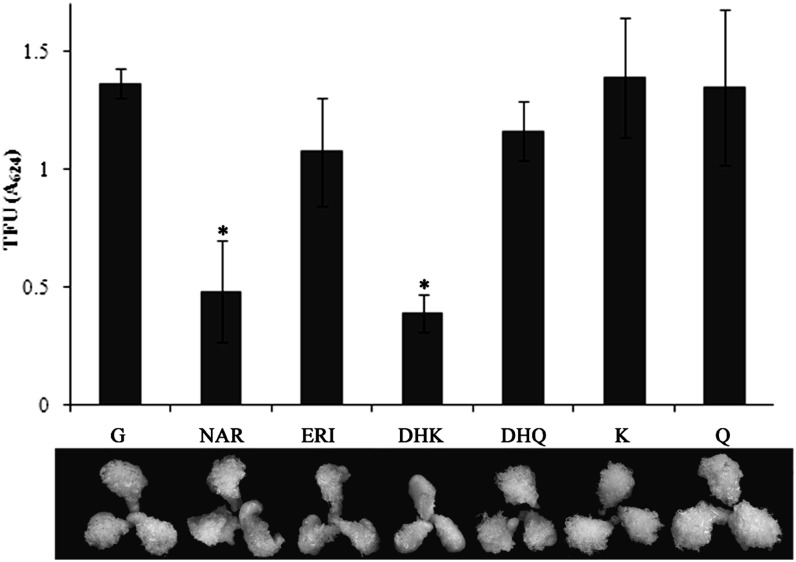
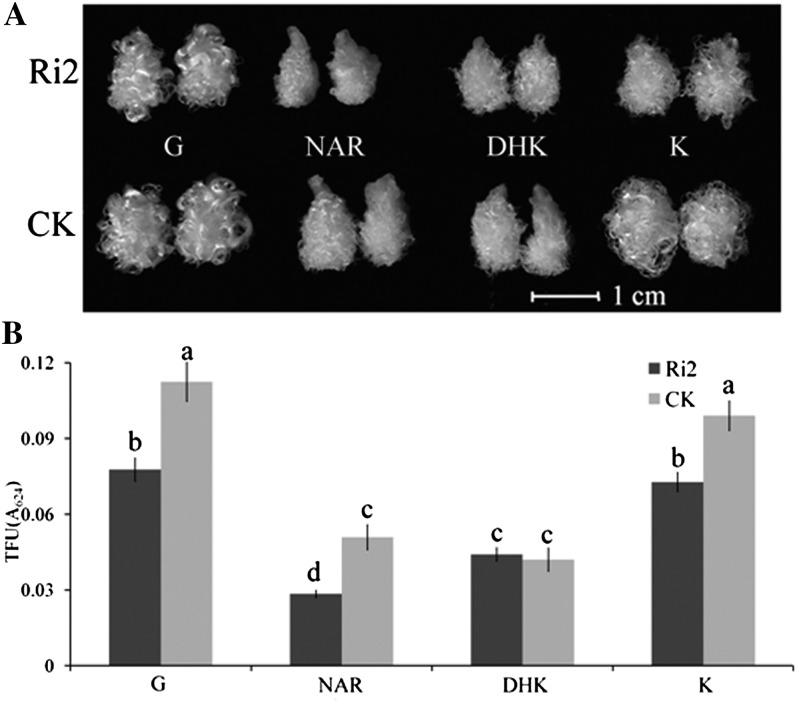
To verify whether flavonoids play a role in fiber development, an in vitro ovule culture assay was performed. Different kinds of flavonoids were applied to test their effect on fiber development in 0-DPA ovules of YZ1. The results showed that NAR and DHK strongly retarded fiber development after 20 d of culture, whereas ERI, DHQ, K, and Q had little effect (Fig. 2). Both flavonoids that exhibited a negative effect were found in lower amounts in G. barbadense than in G. hirsutum (Fig. 1B), suggesting that these two flavonoids might have a role in the difference of fiber quality between these two cotton species.
Figure 2.
Exogenous flavonoids affected fiber development. The 0-DPA ovules of YZ1 were cultured for 20 d with several kinds of flavonoids. NAR and DHK significantly inhibited fiber development, as measured by total fiber units (TFU). G, Control, basic BT medium with 0.5 μm GA3; NAR, ERI, DHK, DHQ, K, and Q, an additional 10 μm NAR, ERI, DHK, DHQ, K, or Q was added to G, respectively. Five biological replicates were performed, and error bars represent sd (Student’s t test, *P < 0.05).
Characterization of F3H in Cotton
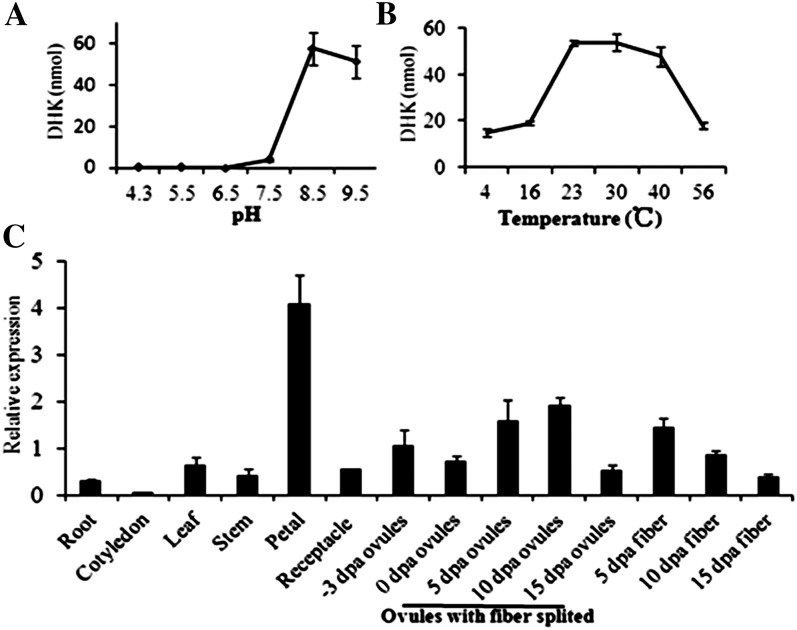
The transcript of F3H was most abundant in the publicly available data of fiber development (Supplemental Table S1; Udall et al., 2006). Additionally, NAR and DHK, the direct substrate and product of F3H in the flavonoid pathway (Fig. 1A), exhibited dramatic effects on fiber development. Therefore, we further analyzed the F3H gene to uncover the function of the flavonoid pathway in fiber development. Four highly homologous, full-length F3H cotton genes were found in the National Center for Biotechnology Information (NCBI) database (Supplemental Fig. S1A). A phylogenetic analysis indicated that F3H in cotton was more closely related to the F3H in petunia but less to rice (Oryza sativa) F3H (Supplemental Fig. S1B). A 42-kD recombinant protein was produced on the basis of the F3H sequence (DQ122181) from G. hirsutum (Supplemental Fig. S1C). Furthermore, an in vitro enzyme assay showed that the cotton F3H enzyme had optimum activity at pH 8.5 and 30°C (Fig. 3, A and B), which was similar to that of its homolog in Arabidopsis (Owens et al., 2008). Previous results have indicated that F3H is highly expressed in fiber cells (Supplemental Table S1). Our results showed that F3H is broadly expressed in G. hirsutum YZ1, and it was especially abundant in petals, while a relatively high transcript level was also seen in 5-DPA ovules and fibers and 10-DPA ovules (Fig. 3C).
Figure 3.
Recombinant protein characterization and expression patterns of F3H. A, pH assay of the recombinant protein through pH 4.3 to 9.5 at 30°C. B, Temperature assay from 4°C to 56°C at pH 8.5. DHK, the product of F3H, was measured to represent the enzyme activity with the same dose of NAR as the substrate. C, Real-time PCR analysis of F3H in different cotton tissues. All the expressions were normalized with UBQ7, and the expression of F3H in −3-DPA ovules was referred to as 1. Error bars represent sd of three biological replicates.
Suppression of F3H Significantly Disrupted Cotton Pigment Biosynthesis and Fiber Development
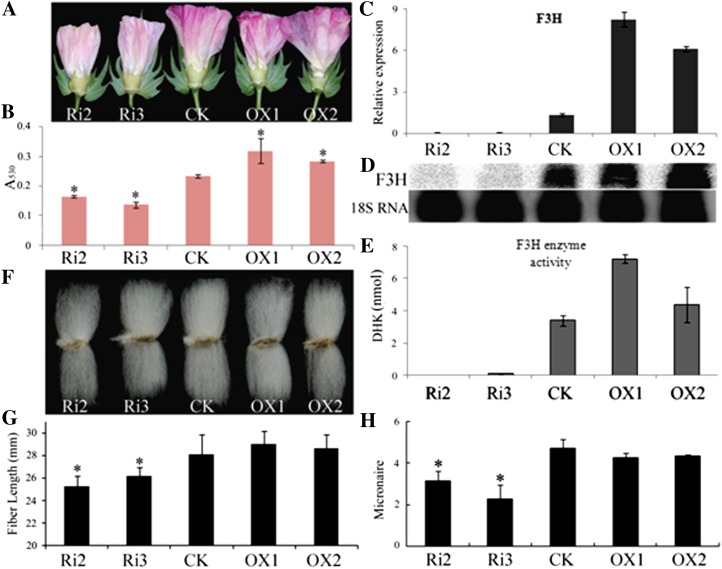
To further understand the role of F3H in cotton development, the RNA interference (RNAi) silencing and overexpression vectors pHG4-F3H and 35S:F3H were constructed to transform YZ1. Low-copy-number insertion RNAi plants Ri2 and Ri3 and overexpression plants OX1 and OX2, which were from different independent transgenic events, were further characterized (Supplemental Fig. S2). Homozygous T2 plants were analyzed in detail. Phenotypic analysis showed an obvious change in the petal color of transgenic plants (Fig. 4A). Total anthocyanin contents decreased in the flowers of RNAi plants and increased in the flowers of overexpression plants (Fig. 4B). Gene expression analysis of 5-DPA ovules from the transgenic plants showed that F3H was significantly suppressed in RNAi plants and up-regulated in overexpression plants (Fig. 4C). These results were confirmed by northern blotting and F3H enzyme assay (Fig. 4, D and E). Suppression of F3H significantly affected fiber quality (Fig. 4, F and G). The fiber length of RNAi plants was shorter than that of the control. The lengths of Ri2 and Ri3 were 25.3 and 26.2 mm, 10% and 7% decreases in comparison with the control (28.1 mm), respectively (Fig. 4G). RNAi plants showed a lower micronaire, at 3.2 and 2.3 for Ri2 and Ri3, representing decreases of 32% and 51%, respectively, compared with the control value of 4.7 (Fig. 4H). Overexpressing F3H had no effect on the fiber length or micronaire. These results were verified in T3 plants (Table I). The fiber of RNAi plants also showed a decrease in uniformity, but there was a less dramatic effect on fiber strength. The fiber length and micronaire decrease indicated that F3H suppression not only inhibited fiber elongation but also retarded fiber maturation.
Figure 4.
Suppression of F3H disrupted petal color and fiber development. Flowers at 1 DPA (A), total anthocyanins of the petals (B), and real-time PCR (C), northern blotting (18S RNA as a reference; D), and enzyme assay (E) of F3H in 5-DPA ovules were analyzed to characterize the transgenic plants. Analysis of homozygous T2 mature plant fibers showed that the fiber length (F and G) and micronaire (H) were significantly decreased in F3H-RNAi plants. Error bars represent the sd of three biological replicates. Asterisks in B, G, and H indicate significant differences by Student’s t test (P < 0.05). Ri, RNAi plants; CK, the wild-type plant; OX, overexpression plants.
Table I. Fiber quality analyses of field- and greenhouse-grown T3 plants.
Fibers collected from T3 plants from the field and greenhouse were analyzed at the Center for Cotton Fiber Quality Inspection and Testing of the Chinese Ministry of Agriculture. Upper half mean length refers to the mean length of the top half of the fibers; mean length is the average length of fibers. Ri, RNAi plants; CK, the wild-type plant; OX, overexpression plants. The data presented are from three independent experiments. Asterisks indicate mean significance at P < 0.05 (Student’s t test).
| Fiber Samples | Upper Half Mean Length | Mean Length | Uniformity Index | Micronaire | Strength |
|---|---|---|---|---|---|
| mm | % | g tex−1 | |||
| T3 fiber harvested from the field | |||||
| Ri2 | 27.84 ± 0.52* | 23.55 ± 0.51* | 84.60 ± 0.64* | 4.28 ± 0.28* | 28.58 ± 0.90 |
| Ri3 | 26.62 ± 0.31* | 22.16 ± 0.29* | 83.23 ± 0.23* | 4.17 ± 0.38* | 26.20 ± 0.35* |
| CK | 29.15 ± 0.73 | 25.04 ± 0.68 | 85.90 ± 0.35 | 5.17 ± 0.06 | 29.17 ± 0.35 |
| OX1 | 28.12 ± 0.42 | 24.15 ± 0.43 | 85.87 ± 0.31 | 5.07 ± 0.06 | 27.60 ± 0.52 |
| OX2 | 28.82 ± 0.76 | 24.74 ± 1.04 | 85.87 ± 1.42 | 5.17 ± 0.06 | 28.63 ± 0.40 |
| T3 fiber harvested from the greenhouse | |||||
| Ri2 | 27.58 ± 0.2* | 22.97 ± 0.16* | 83.27 ± 0.15* | 3.97 ± 0.29* | 28.23 ± 0.55 |
| Ri3 | 27.78 ± 0.36* | 22.94 ± 0.26* | 82.60 ± 0.26* | 4.50 ± 0.10* | 27.40 ± 0.66 |
| CK | 29.89 ± 0.56 | 25.59 ± 0.35 | 85.63 ± 0.40 | 5.00 ± 0.20 | 28.77 ± 0.67 |
| OX1 | 28.69 ± 0.39 | 24.79 ± 0.34 | 86.43 ± 0.15 | 5.00 ± 0.00 | 27.70 ± 0.78 |
NAR Accumulated in F3H-RNAi Plants
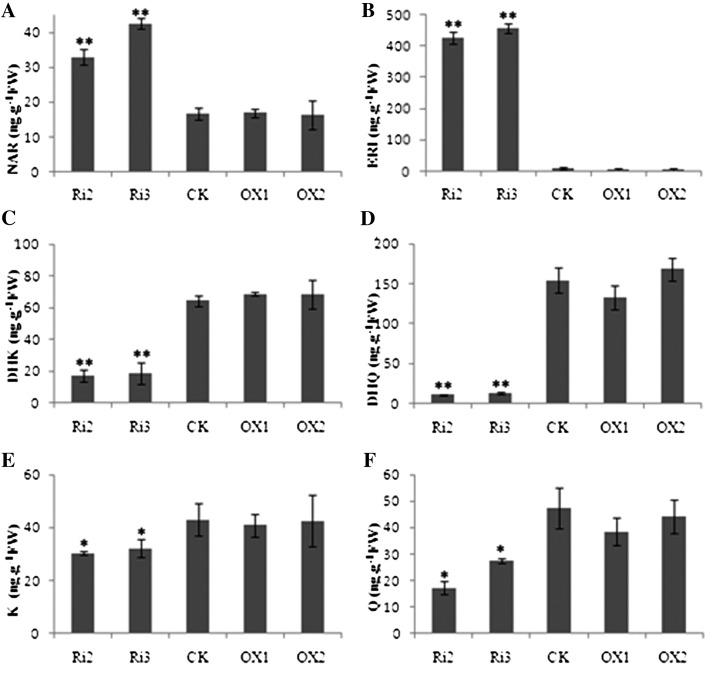
To determine how fiber development was retarded in F3H-suppressed plants, flavonoid metabolism was analyzed in the transgenic lines. Total flavonoid content was significantly different in the RNAi plants but not in the overexpression plants when compared with the control (Fig. 5). The suppression of F3H caused NAR and ERI to accumulate (Fig. 5, A and B), but it decreased the levels of DHK, DHQ, K, and Q (Fig. 5, C–F). Overexpression of F3H showed little influence on metabolite concentrations (Fig. 5). Although high ERI content was also detected in F3H-RNAi plants, ERI had less effect on fiber development than NAR in vitro (Fig. 2). All these results imply the NAR accumulation in F3H-RNAi plants was associated with retarded fiber development.
Figure 5.
Endogenous flavonoid analysis of F3H transgenic plants revealed a dramatic effect in RNAi plants but not in overexpression plants. The contents of NAR (A) and ERI (B), which were upstream of F3H in the metabolic pathway, were obviously accumulated, but DHK (C), DHQ (D), K (E), and Q (F), which were downstream of F3H, were decreased in F3H-RNAi plants. FW, Fresh weight; Ri, RNAi plants; CK, the wild-type plant; OX, overexpression plants. Ovules of 5 DPA were used for analysis. Three biological replicates were performed, and error bars represent sd (Student’s t test: *P < 0.05, **P < 0.01).
F3H Is More Important to Brown Fiber Development Than It Is in White Fiber
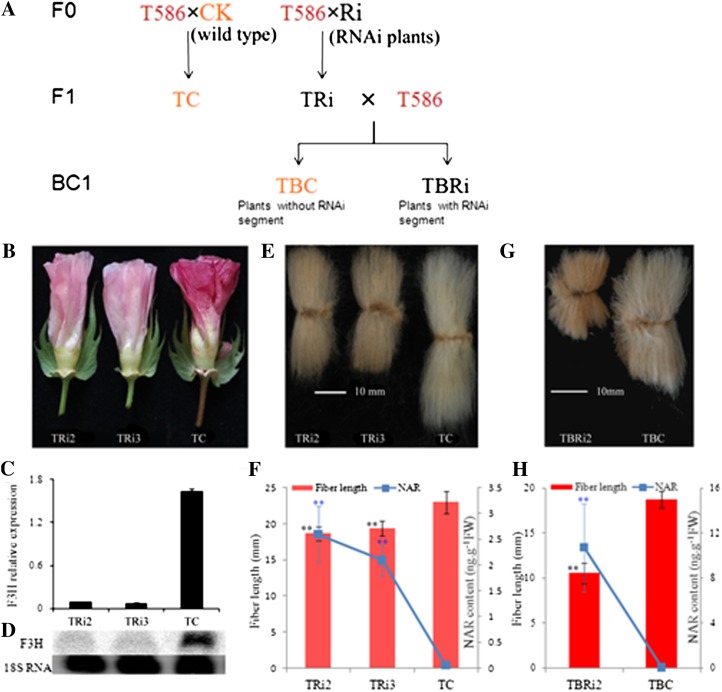
Previous studies demonstrated that the transcription of flavonoid genes and PA was more abundant in brown fiber than in white fiber (Xiao et al., 2007; Li et al., 2011). Our further analysis indicated that the other flavonoids were also abundant in brown fiber (Supplemental Table S3). The results exhibited that relative higher concentrations of NAR and DHK were present in 5-DPA ovules and 10-DPA fibers in comparison with the white fiber cotton. The high flavonoid levels might be the key factor for the short fiber development in brown fiber G. hirsutum. If this is the case, the suppression of F3H in brown fiber may cause a greater disruption of fiber growth as a consequence of its accumulation of more NAR during fiber development. To test this, we transferred the F3H-RNAi segment into the brown fiber G. hirsutum T586 via hybridization (Fig. 6A). Petal pigment in the F1 plants TRi2 and TRi3 changed significantly from red (the control TC) to pink (Fig. 6B). F3H enzyme assays and measurements of anthocyanin and flavonoids in TRi2 and TRi3 confirmed that the RNAi segment was effectively transferred to T586 (Supplemental Fig. S3). Gene expression analysis showed that F3H was obviously suppressed in TRi2 and TRi3 plants (Fig. 6, C and D). An analysis of F1 progeny fiber quality revealed that the fiber length and other fiber qualities were significantly decreased. The fiber lengths of TRi2 and TRi3 were only 81% and 84% of the control, respectively (Fig. 6, E and F), which was parallel with the high accumulation of NAR (Fig. 6F). The micronaire was only 35% of the control, and the fiber uniformity, fiber elongation, and yellowness were also decreased in the two RNAi segment-containing plants (Supplemental Table S4). All these results were caused by the down-regulation of the F3H gene in fiber, indicating that flavonoids were not only involved in fiber cell elongation and secondary cell wall deposition but also affect the fiber color in brown fibers. Comparing with the white fiber parent lines of Ri2 and Ri3 plants, the fiber quality of TRi2 and TRi3, which was brown and had much more NAR content, was much worse than the control (Figs. 4G and 6F). The BC1 plants were generated by backcrossing F1 plants of TRi2 with T586 (Fig. 6A). Then, an extreme short-fiber phenotype and a much higher accumulation of NAR were observed (Fig. 6, G and H). The fiber length of the F3H-RNAi segment-containing BC1 plant (TBRi2) was only 10.55 mm, 56.4% that of the control (TBC) and parent (TRi2) plants (Fig. 6G). TBRi2 had a much more significant accumulation of NAR than the control and TRi2 plants (Fig. 6, F and H). These results indicated that F3H played a more important role in brown fiber development than in white fibers, and these results also indicated that fiber development is significantly stunted in plants with higher NAR contents, consistent with the in vitro results (Fig. 2).
Figure 6.
Phenotypic analysis of F1 and BC1 plants. A, Schematic diagram of the generation of brown fiber transgenic plant material. B to E, Characterization of F1 plants. B, Flower of 1 DPA. C and D, Real-time PCR (C) and northern-blot analysis (D) of F3H in F1 plants. E and G, Mature fiber of F1 and BC1 plants. F and H, Fiber length and NAR content in F1 and BC1 mature fibers. TRi2, TRi3, and TC were F1 progeny of F3H-RNAi plants Ri2 and Ri3 and the wild type crossed to the brown fiber plant T586, respectively. Error bars represent the sd of three biological replicates by Student’s t test (**P < 0.01).
DHK and K Could Not Recover the Inhibition of Fiber Development in YZ1 F3H-RNAi Plants
Down-regulation of F3H significantly increased NAR content and decreased the levels of DHK and K (Fig. 5). Therefore, we tested the effects of DHK and K in ovule culture to verify that the retarded fiber growth was caused by the decline in the concentration of downstream metabolites (Fig. 7A). The fiber yield was decreased in ovules collected from the YZ1 F3H-RNAi plant Ri2 in comparison with the ovule culture control, in keeping with the phenotype from the field (Fig. 4F). Neither DHK nor K could recover the inhibition of F3H silencing, but both exhibited an impact on fiber growth similar to the control (Fig. 7B). A less significant effect was noticed in the DHK treatment, which showed no difference between ovules of Ri2 and the wild type, although the fiber growth was suppressed in both in comparison with the G treatment (control). This might be because the ovules from the RNAi plants contain less DHK, and when both ovules were cultured with the same level of DHK, the ovules from Ri2 could suffer a relatively lower inhibition of fiber growth compared with that from wild-type plants. A more significant fiber growth inhibition was observed in F3H-silenced ovules compared with the NAR treatment control (Fig. 7). These results further confirmed that the effect of retarded fiber development in F3H-silenced plants is probably due to NAR accumulation.
Figure 7.
The downstream flavonoids of F3H could not recover retarded fiber development in F3H-RNAi plants. A, The 1-DPA ovules from RNAi plant Ri2 and the control plant (CK) were cultured with different media (see Fig. 2) for 10 d. B, Fiber products were measured by total fiber units (TFU). Error bars represent the sd of five biological replicates; different letters indicate statistically significant differences at P < 0.05 based on ANOVA (Tukey’s honestly significant difference).
DISCUSSION
Over decades of work, many important fiber development factors have been identified (Ruan et al., 2003; Li et al., 2005; Shi et al., 2006; Luo et al., 2007; Zhang et al., 2011). However, the exact mechanism involved is still largely unknown. Large-scale transcriptome analysis and other omics studies now provide a basis upon which to build an in-depth understanding of this mechanism (Supplemental Table S1). By mining publicly available data and combining gene transcription with a metabolic analysis of flavonoid biosynthesis during G. hirsutum YZ1 and G. barbadense 3-79 fiber development (Fig. 1), flavonoid metabolism was confirmed to be an active participant in fiber development. These data and our results also imply that flavonoids might play a negative or inhibitory role in fiber cell development, because cotton with higher fiber quality bears a relatively lower level of flavonoid content, particularly for NAR (Fig. 1; Supplemental Table S1–S3). The transcript levels of flavonoid genes F3H and ANR were as high as the levels of the housekeeping gene UBQ7. These genes might be the key factors for mediating this pathway in the fiber cell. F3H was thoroughly studied for decades (Britsch, 1990; Owens et al., 2008), but most research focused on its function in pigment biosynthesis (Wisman et al., 1998; Peer et al., 2001). A previous study showed that F3H was predominantly expressed at the fiber elongation stage in G. barbadense, a process that has no relationship with pigment formation (Tu et al., 2007). Transgenic analysis showed that the suppression of F3H significantly disrupted petal pigment biosynthesis and fiber development (Figs. 4 and 5). Surprisingly, F3H is not only important for common fiber quality (such as fiber length and micronaire) but also for fiber color development in brown fiber (Supplemental Table S4).
Exogenous NAR, which is the substrate of F3H, is negatively associated with fiber development (Fig. 2). In vivo suppression of F3H increased NAR accumulation in transgenic plants and subsequently inhibited fiber development (Fig. 6, F and H). The metabolites of F3H, DHK, and K could not recover the inhibition of fiber development in F3H-RNAi plants, and exogenous NAR caused a more significant inhibition in RNAi plants during ovule culture (Fig. 7B). Additionally, a peak of NAR content appeared at 0 DPA, but the highest transcript level of F3H occurred at 5 DPA (Fig. 1). Given that NAR could retard fiber development, it is reasonable to infer that there is a mechanism in fiber cells to modulate NAR content in vivo. F3H appears to be the most important regulator, and F3′H, which catalyzed NAR to ERI, was an alternative.
The fiber quality of G. barbadense 3-79 is better than that of G. hirsutum YZ1. The flavonoid content was low in 3-79, but the transcripts of F3H, DFR, and ANR were higher than those in YZ1, which conferred lower levels of NAR and DHK (Fig. 1B) and subsequently promoted fiber development. The quality of brown fiber in T586 was worse than that in YZ1 (Xiao et al., 2007), and flavonoids were more abundant in the brown fiber (Supplemental Table S3). When the F3H-RNAi segment was transferred into brown fiber plants, they accumulated to a much higher level of NAR and yielded more stunted fiber (Fig. 6). In addition, the transcription of flavonoid genes was higher in wild cotton than in the cultivated cottons (Supplemental Table S2), and higher levels of transcription of these genes were associated with the worst fiber quality in wild cottons (Hovav et al., 2008a; Rapp et al., 2010). All these results confirm that NAR has a negative impact on fiber development and that F3H plays an important role in NAR metabolism. Because DHK, the product of F3H, also played a negative role in fiber development, it is possible that F3H may not be the only mediator to decrease the high level of flavonoids in vivo (Fig. 2). The high transcription of ANR in developing fiber cells also implied that other genes might have similar roles in fiber cell development to F3H (Fig. 1). Ongoing research will advance our understanding of flavonoid mechanisms during cotton fiber development. The results presented here demonstrate that the transcription of flavonoid genes and enhanced flavonoid content are negatively correlated with fiber quality. This finding was supported by the finding that the down-regulation of F3H significantly altered fiber length and micronaire.
To conclude, our results show that F3H is an important gene mediating the metabolism of NAR in fiber cells and that NAR is negatively associated with fiber development. These data may provide a novel alternative way to improve fiber quality by decreasing the endogenous NAR content and engineering the flavonoid pathway.
MATERIALS AND METHODS
Plant Materials
The cotton plants Gossypium hirsutum YZ1, brown cotton G. hirsutum T586, and Gossypium barbadense 3-79 used in this study were cultivated in Wuhan, China, with standard farming practices and management. Bolls were tagged on the day of anthesis (0 DPA). Ovules and fiber were harvested at different developmental stages and were either immediately ground into powder in liquid nitrogen or immersed in liquid nitrogen and then stored at −70°C until use. Roots, cotyledons, and leaves were harvested from 20-d-old seedlings.
Data Assembly
Data from transcriptome, proteome, and deep RNA-seq studies of fiber cells were obtained from previously published papers, and all flavonoid-related data are assembled and reviewed in Supplemental Table S1. Data from Rapp et al. (2010) were redefined. More detailed information is presented in Supplemental Table S2.
Ovule Culture
The bolls at 0 and 1 DPA were harvested, disinfested in 0.1% (w/v) HgCl2 for 15 min, and washed three times in sterilized distilled, deionized water. Ovules were carefully dissected from the ovaries under sterile conditions, immediately placed in liquid BT medium supplemented with various chemicals in 50-mL flasks, and incubated at 30°C in the dark without agitation (Beasley and Ting, 1973; Tan et al., 2012). The control contained basic BT medium with 0.5 μm GA3. The additional flavonoids were added to the medium as a final concentration of 10 μm according to a previous study (Brown et al., 2001). All chemicals were filter sterilized. The 10- and 20-d cultured ovules were harvested for photography with a Nikon D40 camera and fiber yield analysis. The fiber yield was expressed in terms of total fiber units, as described previously (Beasley et al., 1974). The cultured ovules were immersed in hot water to disperse the fibers, and after drying, they were stained for 30 s in 0.02% toluidine blue O and then washed immediately with running water for 2 min. The ovules were destained in glacial acetic acid:ethanol:water (10:95:5) for 2 h. The solvent absorbance was measured with an Infinite 200 PRO multimode reader (Tecan) at 624 nm. Five independent experiments were conducted, and more than 10 ovules were analyzed for each assay. DHK was obtained from TransMIT; the other flavonoids and phytohormones were obtained from Sigma-Aldrich.
Quantitative Real-Time Reverse Transcription-PCR
Ovules at 0 and 5 DPA and fibers at 10, 15, and 20 DPA were harvested for transcript analysis of their flavonoid genes from the cotton species under study. RNA of cotton petals, receptacles, and ovules was obtained for expression profiling of F3H at −3 and 0 DPA; ovules and fibers were examined at 5, 10, and 15 DPA; and roots, cotyledons, leaves, and stems were collected from seedlings. RNA extraction, complementary DNA synthesis, and quantitative reverse transcription-PCR were performed as described previously (Zhu et al., 2005; Tu et al., 2007), with cotton UBQ7 as the reference gene (Tan et al., 2012). The expression of all genes was normalized by referring to UBQ7 as “1.” Sequences of flavonoid genes were obtained from the public NCBI UniGene data bank (Supplemental Table S5). Three biological repeats and at least two technical repeats for each were performed. Error bars represent the sd. All primers are listed in Supplemental Table S5. SuperScript III reverse transcription kits were obtained from Invitrogen. Reagents (iTaq SYBR Green supermix with ROX) for real-time PCR were obtained from Bio-Rad.
Gene Cloning, Recombinant Protein, and Enzyme Assay
There are four full-length F3H genes in the NCBI public data bank (DQ122181, EF187440, GU434116, and DQ912945). The full-length coding sequence of F3H was inserted into pET-28a and transformed into strain Escherichia coli BL21(DE3) for recombinant protein analysis. The recombinant protein was purified by nickel-nitrilotriacetic acid agarose resin (Qiagen). The crude protein of 5-DPA ovules was extracted and quantified as described previously (Deng et al., 2012). The F3H enzyme assay was performed according to a previously published method (Britsch and Grisebach, 1986). Each 100-μL reaction contained 5 μL of α-ketoglutaric acid (20 mm), 5 μL of ascorbic acid (200 mm), 10 μL of ferrous sulfate (2 mm), 50 μL of Gly (160 mm), 2 μL of NAR (100 μm), 1 μg of purified recombinant protein or 10 μg of ovule crude protein, and distilled, deionized water. The optimum pH for the enzyme was determined with Gly over a wide pH range (4.3–9.5 at increments of 1) at 30°C for 30 min. The temperature assay was performed at pH 8.5 at a range of temperatures (4°C–56°C) for 30 min. The F3H activity in ovules was determined at pH 8.5 and 30°C for 1 h with 10 mg of crude protein. Reactions were initiated by the addition of substrate and terminated by extraction with ethyl acetate (1:1, v/v). The extraction was repeated twice. The supernatant was combined, dried in nitrogen, and redissolved in methanol. The products DHK were quantified with a 4000 Q-TRAP liquid chromatography-mass spectrometry device (Applied Biosystems).
Plasmid Construction and Plant Transformation
Full-length F3H was amplified from fiber complementary DNA from YZ1 and inserted into the BamHI and SalI sites of pCAMBIA 2300S to generate vector 35S:F3H for overexpression (Munis et al., 2010). The conserved region of the four published F3H genes was selected as the RNAi target and cloned into the RNAi vector pHellsgate 4 through the Gateway system to produce pHG4-F3H (Helliwell et al., 2002). Transformation was performed via Agrobacterium tumefaciens (EHA105) according to a previously described method (Jin et al., 2006b). Because several regenerated transgenic seedlings failed in healthy root growth, a grafting method was performed to rescue these seedlings (Jin et al., 2006a). Plantlets from the same T0 transgenic event by grafting were treated as one transgenic line for further analysis. Positive transgenic T0 plants were verified by PCR (the sense primer 5′-TTCATTTGGAGAGGACACGCTG-3′, which was designed from the cauliflower mosaic virus 35S promoter, was paired with the corresponding antisense primers of RNAi or overexpression in Supplemental Table S5 to check the positive transgenic plants), and Southern blotting and homozygous T2 and T3 plants were used for further analysis. The Gateway recombination kit was obtained from Invitrogen. Restriction enzymes were obtained from New England Biolabs.
As brown fiber G. hirsutum T586 transgenic plants are difficult to produce, the F3H-RNAi segment was transferred from the transgenic T2 plants, Ri2 and Ri3, derived from YZ1 into T586 via hybridization. The F1 plants from T586 × Ri2 and T586 × Ri3 were recorded as TRi2 and TRi3, respectively. The F1 of wild-type YZ1 crossed with T586 was referred to as the control (TC). The F1 plant TRi2 was further backcrossed with T586 to generate BC1 plants. In total, five BC1 plants were obtained, and transgenic segment-containing plants were determined by both PCR detection and the changing petal color. Three of the BC1 plants with the F3H-RNAi segment were recorded as TBRi2, and the other two without the segment were referred to as the control (TBC).
Southern and Northern Blotting
Cotton DNA and RNA were extracted and transferred to membranes as described previously (Zhu et al., 2005; Li et al., 2010). A fragment of NPTII was used as a probe for Southern blotting for transgene verification. A conserved region of F3H was used as the probe for northern-blot analysis of F3H in transgenic plants. 18S RNA served as the reference.
Fiber Quality Analysis of Transgenic Plants
Bolls of the T2, T3, and F1 generations from similar positions on each cotton plant were harvested simultaneously. The fiber length was measured manually with a comb. After ginning, the fiber was sent to the Center for Cotton Fiber Quality Inspection and Testing at the Chinese Ministry of Agriculture for quality assessment. Three biological replicates were performed. Data were analyzed using Student’s t test.
Quantification of Anthocyanins, PAs, and Flavonoids
Anthocyanin was extracted from petals at 1 DPA, and 0.5-g samples were ground and extracted for 48 h in acidic (1% HCl, w/v) methanol. Anthocyanin was quantified as a function of extract A530 (Mancinelli and Schwartz, 1984). For PA and flavonoid measurements, samples were ground to powder; 0.1 g was extracted with 500 μL of 80% (v/v) methanol at 4°C for 12 h; the residue was extracted with an additional 500 μL of 80% (v/v) methanol; and the supernatant was combined, dried with nitrogen, and resolved with 300 μL of methanol. Ten-microliter extracts were mixed with 150 μL of 3 m HCl/50% (v/v) methanol containing 0.1% (w/v) 4-dimethylaminocinnamaldehyde and stained for 20 min at room temperature; the absorbance of the supernatant was measured at 643 nm for PA quantification (Li et al., 1996). Ten-microliter extracts were injected into the ultrafast liquid chromatograph (UFLC) system (Shimadzu) for flavonoid measurement with a UFLC-electrospray ionization-tandem mass spectrometry system (ABI 4000 Q-Trap; Applied Biosystems; Liu et al., 2012) that was equipped with a C18 (Agilent) column. The UFLC parameters were as follows: column temperature of 35°C, water as solvent A, acetonitrile as solvent B, with both A and B containing 0.1% acetic acid, at a constant flow rate of 250 μL min−1. A linear gradient profile of solvent B was applied as follows: 0 min, 10% B; 0 to 15 min, 10% to 100% B; 15 to 20 min, 100% B; 20 to 25 min, 100% to 10% B; and 25 to 28 min, 10% B for reequilibration. Compounds were separated by reverse-phase UFLC and analyzed by electrospray ionization-tandem mass spectrometry in the negative ionization mode. The compounds were confirmed by analysis of the ion fragments obtained by the hybrid triple quadrupole/linear ion-trap mass spectrometer with a source voltage of 4.5 kV and source temperature of 500°C, and fragments in the range of 50 to 500 mass-to-charge ratio were detected. Reagents for UFLC-mass spectrometry were obtained from Fisher.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Characterization of F3H in cotton.
Supplemental Figure S2. Southern-blot analysis of T0 plants.
Supplemental Figure S3. Characterization of flavonoid metabolism in F1 plants.
Supplemental Table S1. Global assembly of flavonoid-related genes in published data.
Supplemental Table S2. Flavonoid genes differentially expressed in wild and domesticated cotton.
Supplemental Table S3. Flavonoid contents of the developing white and brown fibers.
Supplemental Table S4. Fiber quality analysis of F1 plants.
Supplemental Table S5. Primers used in this study.
Acknowledgments
We thank Dongqin Li (National Key Laboratory of Crop Genetic Improvement) for helping with the quantification of flavonoids and data analysis.
Glossary
- NAR
naringenin
- ERI
eriodictyol
- DHQ
dihydroquercetin
- Q
quercetin
- PA
proanthocyanidin
- DHK
dihydrokaempferol
- K
kaempferol
- NCBI
National Center for Biotechnology Information
- RNAi
RNA interference
- UFLC
ultrafast liquid chromatograph
References
- Al-Ghazi Y, Bourot S, Arioli T, Dennis ES, Llewellyn DJ. (2009) Transcript profiling during fiber development identifies pathways in secondary metabolism and cell wall structure that may contribute to cotton fiber quality. Plant Cell Physiol 50: 1364–1381 [DOI] [PubMed] [Google Scholar]
- Arpat AB, Waugh M, Sullivan JP, Gonzales M, Frisch D, Main D, Wood T, Leslie A, Wing RA, Wilkins TA. (2004) Functional genomics of cell elongation in developing cotton fibers. Plant Mol Biol 54: 911–929 [DOI] [PubMed] [Google Scholar]
- Beasley CA, Birnbaum EH, Dugger WM, Ting IP. (1974) A quantitative procedure for estimating cotton fiber growth. Stain Technol 49: 85–92 [DOI] [PubMed] [Google Scholar]
- Beasley CA, Ting IP. (1973) The effects of plant growth substances on in vitro fiber development from fertilized cotton ovules. Am J Bot 60: 130–139 [Google Scholar]
- Britsch L. (1990) Purification of flavanone 3β-hydroxylase from Petunia hybrida: antibody preparation and characterization of a chemogenetically defined mutant. Arch Biochem Biophys 276: 348–354 [DOI] [PubMed] [Google Scholar]
- Britsch L, Grisebach H. (1986) Purification and characterization of (2S)-flavanone 3-hydroxylase from Petunia hybrida. Eur J Biochem 156: 569–577 [DOI] [PubMed] [Google Scholar]
- Brown DE, Rashotte AM, Murphy AS, Normanly J, Tague BW, Peer WA, Taiz L, Muday GK. (2001) Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiol 126: 524–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer CS, Djordjevic MA. (2009) Architectural phenotypes in the transparent testa mutants of Arabidopsis thaliana. J Exp Bot 60: 751–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary B, Hovav R, Flagel L, Mittler R, Wendel JF. (2009) Parallel expression evolution of oxidative stress-related genes in fiber from wild and domesticated diploid and polyploid cotton (Gossypium). BMC Genomics 10: 378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I, Léon-Kloosterziel KM, Koornneef M. (2000) Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol 122: 403–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng F, Tu L, Tan J, Li Y, Nie Y, Zhang X. (2012) GbPDF1 is involved in cotton fiber initiation via the core cis-element HDZIP2ATATHB2. Plant Physiol 158: 890–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Braun EL, Grotewold E. (2001) Functional conservation of plant secondary metabolic enzymes revealed by complementation of Arabidopsis flavonoid mutants with maize genes. Plant Physiol 127: 46–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flachowsky H, Halbwirth H, Treutter D, Richter K, Hanke M-V, Szankowski I, Gosch C, Stich K, Fischer TC. (2012) Silencing of flavanone-3-hydroxylase in apple (Malus × domestica Borkh.) leads to accumulation of flavanones, but not to reduced fire blight susceptibility. Plant Physiol Biochem 51: 18–25 [DOI] [PubMed] [Google Scholar]
- Gou JY, Wang LJ, Chen SP, Hu WL, Chen XY. (2007) Gene expression and metabolite profiles of cotton fiber during cell elongation and secondary cell wall synthesis. Cell Res 17: 422–434 [DOI] [PubMed] [Google Scholar]
- Helliwell CA, Wesley SV, Wielopolska AJ, Waterhouse PM. (2002) High-throughput vectors for efficient gene silencing in plants. Funct Plant Biol 29: 1217–1225 [DOI] [PubMed] [Google Scholar]
- Hovav R, Udall JA, Chaudhary B, Hovav E, Flagel L, Hu G, Wendel JF. (2008a) The evolution of spinnable cotton fiber entailed prolonged development and a novel metabolism. PLoS Genet 4: e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovav R, Udall JA, Hovav E, Rapp R, Flagel L, Wendel JF. (2008b) A majority of cotton genes are expressed in single-celled fiber. Planta 227: 319–329 [DOI] [PubMed] [Google Scholar]
- Jacobs M, Rubery PH. (1988) Naturally occurring auxin transport regulators. Science 241: 346–349 [DOI] [PubMed] [Google Scholar]
- Ji SJ, Lu YC, Feng JX, Wei G, Li J, Shi YH, Fu Q, Liu D, Luo JC, Zhu YX. (2003) Isolation and analyses of genes preferentially expressed during early cotton fiber development by subtractive PCR and cDNA array. Nucleic Acids Res 31: 2534–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Liang S, Zhang X, Nie Y, Guo X. (2006a) An efficient grafting system for transgenic plant recovery in cotton (Gossypium hirsutum L.). Plant Cell Tissue Organ Cult 85: 181–185 [Google Scholar]
- Jin S, Zhang X, Nie Y, Guo X, Liang S, Zhu H. (2006b) Identification of a novel elite genotype for in vitro culture and genetic transformation of cotton. Biol Plant 50: 519–524 [Google Scholar]
- Kim HJ, Triplett BA. (2001) Cotton fiber growth in planta and in vitro: models for plant cell elongation and cell wall biogenesis. Plant Physiol 127: 1361–1366 [PMC free article] [PubMed] [Google Scholar]
- Li T, Fan H, Li Z, Wei J, Lin Y, Cai Y. (2011) The accumulation of pigment in fiber related to proanthocyanidins synthesis for brown cotton. Acta Physiol Plant 34: 813–818 [Google Scholar]
- Li XB, Fan XP, Wang XL, Cai L, Yang WC. (2005) The cotton ACTIN1 gene is functionally expressed in fibers and participates in fiber elongation. Plant Cell 17: 859–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Liu D, Tu L, Zhang X, Wang L, Zhu L, Tan J, Deng F. (2010) Suppression of GhAGP4 gene expression repressed the initiation and elongation of cotton fiber. Plant Cell Rep 29: 193–202 [DOI] [PubMed] [Google Scholar]
- Li YG, Tanner G, Larkin P. (1996) The DMACA-HCl protocol and the threshold proanthocyanidin content for bloat safety in forage legumes. J Sci Food Agric 70: 89–101 [Google Scholar]
- Liu H, Li X, Xiao J, Wang S. (2012) A convenient method for simultaneous quantification of multiple phytohormones and metabolites: application in study of rice-bacterium interaction. Plant Methods 8: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Xiao Y, Li X, Lu X, Deng W, Li D, Hou L, Hu M, Li Y, Pei Y. (2007) GhDET2, a steroid 5α-reductase, plays an important role in cotton fiber cell initiation and elongation. Plant J 51: 419–430 [DOI] [PubMed] [Google Scholar]
- Mancinelli AL, Schwartz OM. (1984) The photoregulation of anthocyanin synthesis. IX. The photosensitivity of the response in dark and light-grown tomato seedlings. Plant Cell Physiol 25: 93–105 [Google Scholar]
- Martin C, Prescott A, Mackay S, Bartlett J, Vrijlandt E. (1991) Control of anthocyanin biosynthesis in flowers of Antirrhinum majus. Plant J 1: 37–49 [DOI] [PubMed] [Google Scholar]
- Munis MFH, Tu L, Deng F, Tan J, Xu L, Xu S, Long L, Zhang X. (2010) A thaumatin-like protein gene involved in cotton fiber secondary cell wall development enhances resistance against Verticillium dahliae and other stresses in transgenic tobacco. Biochem Biophys Res Commun 393: 38–44 [DOI] [PubMed] [Google Scholar]
- Owens DK, Crosby KC, Runac J, Howard BA, Winkel BS. (2008) Biochemical and genetic characterization of Arabidopsis flavanone 3beta-hydroxylase. Plant Physiol Biochem 46: 833–843 [DOI] [PubMed] [Google Scholar]
- Peer WA, Brown DE, Tague BW, Muday GK, Taiz L, Murphy AS. (2001) Flavonoid accumulation patterns of transparent testa mutants of Arabidopsis. Plant Physiol 126: 536–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcel L, Routaboul JM, Cheynier V, Lepiniec L, Debeaujon I. (2007) Flavonoid oxidation in plants: from biochemical properties to physiological functions. Trends Plant Sci 12: 29–36 [DOI] [PubMed] [Google Scholar]
- Rapp RA, Haigler CH, Flagel L, Hovav RH, Udall JA, Wendel JF. (2010) Gene expression in developing fibres of Upland cotton (Gossypium hirsutum L.) was massively altered by domestication. BMC Biol 8: 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan YL, Llewellyn DJ, Furbank RT. (2003) Suppression of sucrose synthase gene expression represses cotton fiber cell initiation, elongation, and seed development. Plant Cell 15: 952–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi YH, Zhu SW, Mao XZ, Feng JX, Qin YM, Zhang L, Cheng J, Wei LP, Wang ZY, Zhu YX. (2006) Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell 18: 651–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM. (1995) Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J 8: 659–671 [DOI] [PubMed] [Google Scholar]
- Stephens PA, Nickell CD, Vodkin LO. (1993) Pink flower color associated with increased protein and seed size in soybean. Crop Sci 33: 1135–1137 [Google Scholar]
- Tan J, Tu L, Deng F, Wu R, Zhang X. (2012) Exogenous jasmonic acid inhibits cotton fiber elongation. J Plant Growth Regul 31: 599–605 [Google Scholar]
- Taylor LP, Grotewold E. (2005) Flavonoids as developmental regulators. Curr Opin Plant Biol 8: 317–323 [DOI] [PubMed] [Google Scholar]
- Tu LL, Zhang XL, Liang SG, Liu DQ, Zhu LF, Zeng FC, Nie YC, Guo XP, Deng FL, Tan JF, et al (2007) Gene expression analyses of sea-island cotton (Gossypium barbadense L.) during fiber development. Plant Cell Rep 26: 1309–1320 [DOI] [PubMed] [Google Scholar]
- Udall JA, Swanson JM, Haller K, Rapp RA, Sparks ME, Hatfield J, Yu Y, Wu Y, Dowd C, Arpat AB, et al. (2006) A global assembly of cotton ESTs. Genome Res 16: 441–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uimari A, Strommer J. (1998) Anthocyanin regulatory mutations in pea: effects on gene expression and complementation by R-like genes of maize. Mol Gen Genet 257: 198–204 [DOI] [PubMed] [Google Scholar]
- van Houwelingen A, Souer E, Spelt K, Kloos D, Mol J, Koes R. (1998) Analysis of flower pigmentation mutants generated by random transposon mutagenesis in Petunia hybrida. Plant J 13: 39–50 [DOI] [PubMed] [Google Scholar]
- Wisman E, Hartmann U, Sagasser M, Baumann E, Palme K, Hahlbrock K, Saedler H, Weisshaar B. (1998) Knock-out mutants from an En-1 mutagenized Arabidopsis thaliana population generate phenylpropanoid biosynthesis phenotypes. Proc Natl Acad Sci USA 95: 12432–12437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Machado AC, White RG, Llewellyn DJ, Dennis ES. (2006) Expression profiling identifies genes expressed early during lint fibre initiation in cotton. Plant Cell Physiol 47: 107–127 [DOI] [PubMed] [Google Scholar]
- Xiao YH, Zhang ZS, Yin MH, Luo M, Li XB, Hou L, Pei Y. (2007) Cotton flavonoid structural genes related to the pigmentation in brown fibers. Biochem Biophys Res Commun 358: 73–78 [DOI] [PubMed] [Google Scholar]
- Zabala G, Vodkin LO. (2005) The wp mutation of Glycine max carries a gene-fragment-rich transposon of the CACTA superfamily. Plant Cell 17: 2619–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Zheng X, Song S, Zeng Q, Hou L, Li D, Zhao J, Wei Y, Li X, Luo M, et al. (2011) Spatiotemporal manipulation of auxin biosynthesis in cotton ovule epidermal cells enhances fiber yield and quality. Nat Biotechnol 29: 453–458 [DOI] [PubMed] [Google Scholar]
- Zhu L, Tu L, Zeng F, Liu D, Zhang X. (2005) An improved simple protocol for isolation of high quality RNA from Gossypium spp. suitable for cDNA library construction. Acta Agron Sin 31: 1657–1659 [Google Scholar]