Acetylation of the cell wall affects plant resistance to pathogens.
Abstract
The plant cell wall has many significant structural and physiological roles, but the contributions of the various components to these roles remain unclear. Modification of cell wall properties can affect key agronomic traits such as disease resistance and plant growth. The plant cell wall is composed of diverse polysaccharides often decorated with methyl, acetyl, and feruloyl groups linked to the sugar subunits. In this study, we examined the effect of perturbing cell wall acetylation by making transgenic Arabidopsis (Arabidopsis thaliana) and Brachypodium (Brachypodium distachyon) plants expressing hemicellulose- and pectin-specific fungal acetylesterases. All transgenic plants carried highly expressed active Aspergillus nidulans acetylesterases localized to the apoplast and had significant reduction of cell wall acetylation compared with wild-type plants. Partial deacetylation of polysaccharides caused compensatory up-regulation of three known acetyltransferases and increased polysaccharide accessibility to glycosyl hydrolases. Transgenic plants showed increased resistance to the fungal pathogens Botrytis cinerea and Bipolaris sorokiniana but not to the bacterial pathogens Pseudomonas syringae and Xanthomonas oryzae. These results demonstrate a role, in both monocot and dicot plants, of hemicellulose and pectin acetylation in plant defense against fungal pathogens.
The cell wall is one of the most important compartments of the plant cell. This external cell skeleton plays an important role in cell and tissue shape determination. Besides its structural role, this extracellular complex is involved in the control of important functions such as cell-cell interactions, whole-plant growth, development, and interaction with the environment. The plant cell wall is mainly composed of highly dynamic heteropolysaccharides assembled in macromolecular networks (Albersheim et al., 2010). Cellulose is the major component of most plant cell walls, and it consists of unsubstituted (1,4)-β-d-glucan chains, which form microfibrils. Hemicelluloses include xyloglucan, arabino(glucurono)xylans, different mannans, and β-(1-3,1-4)-glucan (Scheller and Ulvskov, 2010). The pectic polysaccharides are a class of α-1,4-linked galacturonic acid (GalA)-containing polysaccharides and include homogalacturonan, xylogalacturonan, apiogalacturonan, rhamnogalacturonan II, and rhamnogalacturonan I (Caffall and Mohnen, 2009). Different plants have somewhat different cell wall compositions; dicotyledonous and non-Poales monocotyledonous plants (type I cell walls) have cellulose, pectin, and hemicelluloses, whereas grasses have cellulose with relatively small amounts of pectin and commensurately higher levels of hemicelluloses (type II cell walls), and their endosperm often contains high levels of mannans, galactomannans, galactoglucomannans, and β-(1-3,1-4)-glucans (Burton et al., 2010).
Besides the diversity of monosaccharide composition, cell wall polysaccharides are also modified with methyl, acetyl, and feruloyl groups, which are mainly O-linked to sugars. These functional groups are believed to protect polysaccharides from the action of specific glycosyl hydrolases and to cross link cell wall constituents controlling cell extensibility (Perrone et al., 2002; Gou et al., 2012). For example, methylesterification/deesterification acts in the control of cell growth, fruit ripening, and plant protection against biotic stresses (Koch and Nevins, 1989; Wolf et al., 2009; Lionetti et al., 2012). Demethylesterification of homogalacturonan allows the aggregation of polyuronides into calcium-linked structures, resulting in less porous, stiffer cell walls (Willats et al., 2001).
The complexity of cell wall polysaccharides and the formation of polysaccharide networks determine the strength, flexibility, and functionality of the primary cell wall. Recently, plant cell wall polysaccharides have attracted significant attention because they are a major source of reduced carbon for bioethanol production, which has the potential to supply part of our fossil fuel demand (Jordan et al., 2012). Direct modification of cell wall polysaccharides can increase the efficiency of biomass digestibility and improve saccharification during ethanol production (Lionetti et al., 2010; Pogorelko et al., 2011; Fursova et al., 2012; Jung et al., 2012; Song et al., 2012). However, modification of cell wall polysaccharides may also affect important cell wall functions, and the plant cell must maintain the functional integrity of the wall during morphogenesis and exposure to environmental cues (Hamman, 2012). Indeed, the existence of a dedicated cell wall integrity maintenance mechanism has been proposed (Nühse, 2012; Wolf et al., 2012a).
The cell wall acts as a mechanical barrier and also participates in signal transduction (Somerville et al., 2004; Underwood, 2012; Wolf et al., 2012a) that allows plants to sense and respond to changes in environmental conditions. Therefore, plants are very sensitive and responsive to changes in their cell walls. The mechanism of signal transduction through the cell wall during pathogen invasion is not well understood, but some evidence indicates that the signal originates from cytoplasmic membrane deformation caused by changes in wall structure and internal cell turgor pressure mediated by mechanosensitive ion channels (Haswell et al., 2008). Considering this, specific cell wall modifications could induce plant defense reactions even prior to pathogen attack, and these defenses may be able to reduce pathogen penetration and growth (Underwood, 2012). Indeed, increasing evidence demonstrates that changes in cell wall composition can induce reactions similar to the stress response in plants (Hamann, 2012; Nühse, 2012). For example, suppression of cellulose synthesis causes the activation of stress-related gene expression (Ellis et al., 2002; Hamann et al., 2009). The expression of fungal polygalacturonase (PG) and consequent reduction of homogalacturonan content in the cell wall increases resistance to pathogens in both tobacco (Nicotiana tabacum) and Arabidopsis (Arabidopsis thaliana; Ferrari et al., 2007).
Genetic and molecular evidence indicates the critical role of pectin methylesterification in plant defense; for example, in Arabidopsis and wheat (Triticum aestivum), high methylesterification of pectins decreased plant susceptibility to fungal and bacterial necrotrophs (Raiola et al., 2011; Volpi et al., 2011), and in wild strawberry plants (Fragaria spp.), overexpression of a fruit-specific pectin methylesterase (PME) increased plant resistance to Botrytis cinerea (Osorio et al., 2008). Demethylesterification makes homogalacturonans more susceptible to degradation by fungal pectic enzymes like PG) and pectate lyase (Limberg et al., 2000). Also, deesterification can be induced by pathogen action and is required for colonization by fungal and bacterial cells (Raiola et al., 2011). Similarly, the establishment of ferulic acid cross linking results in cell wall stiffening and growth deceleration (Buanafina, 2009), and negative correlations between the level of cell wall feruloyl esterification and pathogen invasion were observed for some plant species (Bily et al., 2003).
In contrast to other modifications, the biological function of O-acetyl substituents is not known, although there is some evidence that the degree of acetylation is modified during plant growth and development (Pauly et al., 2001; Obel et al., 2009). For example, it was proposed that the acetylation of cell wall polysaccharides impacts their noncovalent interactions, affecting their accessibility to invading pathogens (Gille and Pauly, 2012). Also, adding a small acetyl group instead of a bigger sugar residue seems to be an energetically favorable strategy that is broadly used in grass xyloglucan, where one xylosyl residue is regularly replaced with an acetyl group (Gibeaut et al., 2005; Gille and Pauly, 2012). The Arabidopsis reduced wall acetylation2 (rwa2) mutant, which exhibits a 20% reduction in cell wall acetylation, has increased resistance to the fungal pathogen B. cinerea (Manabe et al., 2011), although the authors did not consider a direct relationship between a decreased degree of cell wall acetylation and increased resistance. Mutations in other pectin-related proteins, powdery mildew-resistant PMR5 and PMR6, caused pectin alterations and increased resistance to powdery mildew (Vogel et al., 2002, 2004).
To investigate the effect of the deacetylation of different polysaccharides on type I and type II cell wall integrity and, consequently, on plant responses to biotic stresses, we generated transgenic Arabidopsis and Brachypodium (Brachypodium distachyon) plants expressing fungal acetyl esterases, which are proposed to act specifically on hemicellulose or pectin polysaccharides. The results of this study provide new insights into the relationship between cell wall acetylation, polysaccharide digestibility, and plant resistance to pathogens and demonstrate that postsynthetic polysaccharide modification is a powerful approach to investigate the contribution of the cell wall to plant fitness.
RESULTS
Generation and Characterization of Arabidopsis and Brachypodium Transgenic Plants Expressing Fungal Acetylesterases
To alter the acetylation of cell wall polysaccharides, we used two different enzymes from Aspergillus nidulans, xylan acetylesterase (AnAXE; AN6093.2) and rhamnogalacturonan acetylesterase (AnRAE; AN2528.2). AnAXE and AnRAE are annotated as specific for xylan deacetylation and pectin deacetylation, respectively. To target the enzymes to the plant apoplast, the 5′ end of the full-length complementary DNA (cDNA) of each gene was fused with the expansin-B signal peptide coding sequence from Arabidopsis (for expression in Arabidopsis) or from maize (Zea mays; for expression in Brachypodium), and the 3′ end of each gene was fused with the GFP coding sequence. DNA constructs harboring hydrolase-encoding genes were created to produce a single transcription unit containing the three fused parts (signal peptide, hydrolase cDNA, and GFP) in the same open reading frame (Fig. 1A).
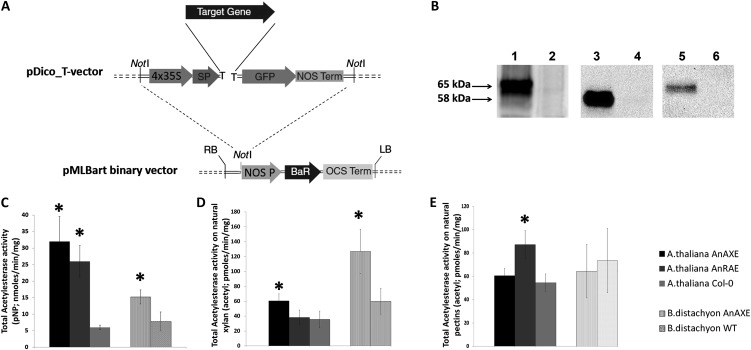
Figure 1.
A, The expression cassette of the vector developed for Arabidopsis transformation. BaR, Gene for herbicide resistance (Basta; dl-phosphinothricin); LB, left border; NOS Prom, promoter from nopaline synthase; NOS Term, terminator from nopaline synthase; OCS Term, octopine synthase terminator; RB, right border; SP, Arabidopsis expansin-B signal peptide; Target Gene, gene of interest; 4x35S, tetramer of the CaMV 35S RNA promoter. B, Western-blot analysis of total proteins from apoplast of transgenic and wild-type plants. On the left, the arrows show the size of proteins in kD. Lane 1, Arabidopsis plant expressing AnAXE fused with GFP (65 kD); lane 3, Arabidopsis plant expressing AnRAE fused with GFP (58 kD); lane 5, Brachypodium plant expressing AnAXE fused with GFP (65 kD); lanes 2, 4, and 6 represent total protein from wild-type plants prepared in parallel with transgenic plant protein extracts. Equal amounts of total protein were loaded for each line, and expressed proteins were detected using GFP monoclonal antibodies (1:5,000 dilution). C, Esterase activities in apoplast from transgenic and wild-type (WT) plants (pNP-acetyl was used as a substrate). D, Esterase activities in apoplast from transgenic and wild-type plants estimated using birch wood xylan. E, Esterase activities in apoplast from transgenic and wild-type plants determined using citrus pectin. In all assays, apoplast fluids were prepared from the combined three plants for each of three independent transgenic homozygous lines, of three lines expressing empty vector, and of three Col-0 plants, all of the same age. Bars represent mean ± sd of total acetylesterase activity averaged for three independent transgenic lines. Data represent average values obtained for three independent transgenic lines. Asterisks indicate data sets significantly different between transgenic plants and wild-type plants (Student’s t test, P < 0.05; n = 3).
After transformation, three independent transgenic lines were selected for each construct, and homozygous plants were obtained using herbicide resistance and confirmed to carry the complete construct by PCR. No visible phenotypes were observed during the development of all transgenic plants, which were grown side by side with wild-type plants.
Expression of Fungal Acetylesterases Increased Acetylesterase Activity in Arabidopsis and Brachypodium Apoplastic Fluids
Transgenic plants were first tested for the production of the fungal enzymes in the extracellular matrix. Apoplastic fluids were isolated from leaves of 4-week-old transgenic plants and analyzed by western blot using anti-GFP antibodies. Bands at molecular masses of 65 and 58 kD, corresponding to the expressed A. nidulans xylan acetylesterase and rhamnogalacturonan acetylesterase fused with GFP, respectively, were found (Fig. 1B).
The apoplastic fluids extracted from three independent homozygous lines for each gene were tested for acetylesterase activity using 4-nitrophenyl (pNP)-acetyl as a substrate. Apoplast extracts from the Arabidopsis plants expressing AnAXE and AnRAE showed 4.3- and 5.3-fold higher activity compared with apoplast from wild-type Columbia (Col-0) plants, whereas Brachypodium AnAXE lines showed 2-fold higher activity compared with wild-type plants (Fig. 1C). To confirm the annotated substrate specificity of AnAXE and AnRAE, the acetylesterase activity in the apoplast from transgenic plants was assayed using commercially available pure acetylated polysaccharides. The apoplastic fluids obtained from both Arabidopsis and Brachypodium transgenic plants expressing AnAXE have 1.7- to 2.1-fold higher activity deacetylating beech wood (Fagus spp.) xylan compared with wild-type plants, whereas there was no difference between apoplastic extracts from AnAXE and wild-type plants in the ability to deacetylate pectin (Fig. 1, D and E). By contrast, apoplastic fluid prepared from Arabidopsis AnRAE plants showed higher activity only when pectin was used as a substrate (1.6-fold), and no differences with wild-type plants were observed when beech xylan was used as a substrate (Fig. 1, D and 1E). Brachypodium plants expressing AnRAE (Fursova et al., 2012) did not show a difference in cell wall acetylation in comparison with wild-type plants, most likely due to the low amounts of pectin present in the Brachypodium cell wall; therefore, we did not extend our studies on these plants in this work.
Fungal Acetylesterases Deacetylate Specific Polysaccharides and Affect Their Accessibility to Glycosyl Hydrolases
The total contents of acetyl groups of the cell walls extracted from aerial parts of transgenic and wild-type plants were quantified. Brachypodium wild-type plants showed five times more acetyls in their cell wall in comparison with Arabidopsis wild-type plants (Fig. 2, A and B). AnAXE Arabidopsis and Brachypodium plants showed 1.8- and 1.3-fold reductions of acetyl levels in their cell walls, respectively, consistent with the different levels of AnAXE activity observed in this study. Expression of AnRAE in Arabidopsis plants reduced cell wall acetylation by 1.3-fold. Analysis of monosaccharide composition did not reveal significant differences between transgenic and wild-type plants (Supplemental Table S1).
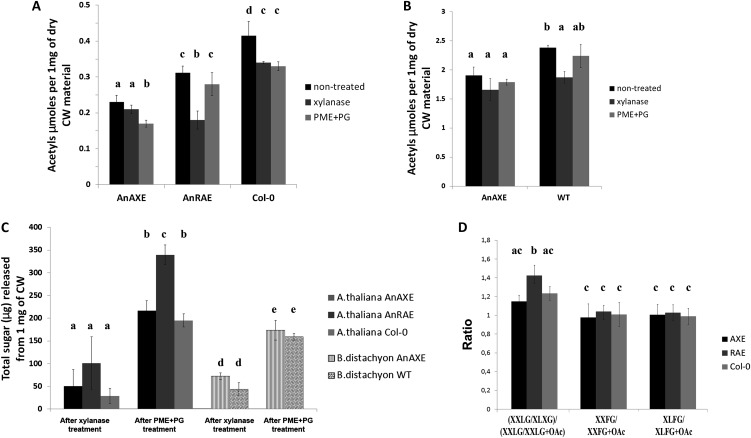
Figure 2.
A and B, Cell wall (CW) acetylation of transgenic Arabidopsis (A) and Brachypodium (B) lines and wild-type plants (WT). Shown mean values are averages for three independent transgenic lines for each construct and three wild-type plants grown simultaneously. Acetyl content is shown in µmol mg−1 dry cell wall material. Bars show means ± sd averaged for three independent transgenic lines. C, Amount of reducing sugars released after incubation of dry cell wall material from transgenic and wild-type control plants with xylanase and PME+PG during 24 h. D, Ratios of nonacetylated to acetylated xyloglucan oligosaccharides obtained after xyloglucan endoglucanase digestion of cell walls from Arabidopsis AnAXE, AnRAE, and wild-type plants analyzed by MALDI-TOF. Nomenclature of subunits is according to Fry et al. (1993). Data represent average values obtained for three independent transgenic lines. Letters (a–e) indicate significant differences among the combinations of genotypes and treatments (ANOVA, P ≤ 0.05).
To verify the substrate specificity of the expressed enzymes in vivo, we treated cell walls of transgenic and wild-type plants with xylanase that hydrolyzes xylan backbone between two consecutive unsubstituted xylose residues or with a mixture of PG and PME. If xylan is the substrate of AnAXE in vivo, treatment of AnAXE plants with xylanase should not affect the total polysaccharide acetyl content, as the degraded xylans would not have substantial acetyl content. By contrast, treatment of AnAXE plants with pectinase should substantially reduce the polysaccharide acetyl content. Conversely, treatment of AnRAE plants with xylanase should have the opposite effect, due to a substantial decrease in polysaccharide acetyl content. Treatments of cell walls with either xylanase or a mixture of PME and PG reduced the amount of acetylated polysaccharides to approximately the same level (1.25-fold) in the wild-type plants (Fig. 2A). In AnAXE plants, PME+PG treatment reduced the amount of acetylated polysaccharides, but no significant differences were observed after xylanase treatment. By contrast, in AnRAE plants, treatment with xylanases significantly reduced the amount of acetylated polysaccharides and treatment with PME+PG did not affect the amount of acetylated polysaccharides. In Brachypodium cell walls, due to the low amount of pectin, treatment with PME+PG did not change the amount of acetylated polysaccharides in wild-type plants or in AnAXE-expressing plants (Fig. 2B). However, treatment with xylanase reduced the amount of acetylated polysaccharides in wild-type Brachypodium but not in AnAXE plants (Fig. 2B), which is similar to results obtained for Arabidopsis AnAXE plants.
Oligo Mass Profiling analysis was used to examine whether expressed acetylesterases affect xyloglucan acetylation in Arabidopsis plants (Fig. 2D). Cell walls prepared from AnAXE, AnRAE, and Col-0 plants were treated with xyloglucan endoglucanase, and acetylated and nonacetylated xyloglucan subunits were measured using matrix-assisted laser-desorption ionization time of flight (MALDI-TOF). The nonacetylated XXLG-acetylated XXLG ratio was significantly higher in AnRAE in comparison with AnAXE and Col-0 plants, indicating a reduction of xyloglucan acetylation in AnRAE-expressing plants.
All these results, together with the activity assays using natural substrates, suggest that AnRAE deesterifies pectins and to some extent xyloglucan, but AnAXE is more specific toward xylan but not xyloglucan and pectin.
To detect whether deacetylation can alter polysaccharide digestibility, the amount of total sugars released during xylanase and PME+PG treatment of total plant cell walls was measured. Significantly higher amounts of sugars were released from the cell walls of AnRAE plants when they were treated with PME+PG (Fig. 2C), indicating their increased digestibility.
The monosaccharide composition of the cell wall residue left after xylanase and PME+PG treatments was analyzed. After xylanase treatment, higher amounts of pectin-related monosaccharides (Fuc, Rha, Ara, Gal, and a slight but not significant increase of GalA) were observed in Arabidopsis AnAXE cell walls in comparison with the wild type (Table I). Additionally, significant reduction of GlcA but not of Xyl was observed in cell wall residue that remained after xylanase treatment of AnAXE plants. On the contrary, cell wall residue from Arabidopsis AnRAE plants after treatment with xylanase contained higher amounts of Xyl, mainly present in hemicellulosic polysaccharides. These results indicate that the partial deacetylation of xylan does not significantly affect its xylanase digestibility, but pectin deacetylation seems to decrease xylan accessibility to xylanase.
Table I. Monosaccharide composition (mol %) of cell walls from transgenic and wild-type plants after xylanase treatment.
Asterisks indicate data sets significantly different between transgenic plants and wild-type plants, according to Student’s t test (P < 0.05; n = 3 plants from independent transformant progeny).
| Monosaccharide | Arabidopsis AnAXE | Arabidopsis AnRAE | Arabidopsis Col-0 | Brachypodium AnAXE | Brachypodium Wild Type |
|---|---|---|---|---|---|
| Fuc | 2.23 ± 0.16* | 1.78 ± 0.08 | 1.87 ± 0.25 | 0.24 ± 0.03 | 0.39 ± 0.01 |
| Rha | 8.32 ± 0.85* | 5.68 ± 1.34 | 7.20 ± 0.39 | 5.78 ± 0.27 | 5.56 ± 0.17 |
| Ara | 9.01 ± 0.97* | 6.16 ± 1.13 | 7.81 ± 0.39 | 6.27 ± 0.29 | 6.02 ± 0.15 |
| Gal | 18.4 ± 1.33* | 14.53 ± 0.42 | 15.77 ± 0.07 | 2.51 ± 0.12 | 2.69 ± 0.06 |
| Glc | 11.88 ± 1.49 | 14.48 ± 0.41 | 16.05 ± 3.14 | 27.75 ± 1.24 | 30.69 ± 0.76 |
| Xyl | 39.20 ± 2.28 | 46.38 ± 1.16* | 41.44 ± 1.51 | 47.96 ± 1.11 | 45.04 ± 4.34 |
| Man | 8.03 ± 0.89 | 9.58 ± 0.76 | 8.49 ± 0.91 | 9.62 ± 0.46 | 9.23 ± 0.59 |
| GalA | 2.81 ± 0.82 | 1.18 ± 0.63 | 0.9 ± 0.11 | 0.14 ± 0.02 | 0.12 ± 0.01 |
| GlcA | 0.15 ± 0.06* | 0.32 ± 0.08* | 0.54 ± 0.07 | 0.76 ± 0.09 | 0.32 ± 0.19 |
After PME+PG treatment, the cell wall residue of AnAXE Arabidopsis plants showed higher amounts of Gal but AnRAE plants showed a reduced amount of GalA. Interestingly, Fuc and GlcA were completely eliminated in all plants after treatment with PME+PG (Table II). This suggests that partial deacetylation of both pectin and xyloglucan increases pectin digestibility with PG.
Table II. Monosaccharide composition (mol %) of cell walls from transgenic and wild-type plants after PME+PG treatment.
Asterisks indicate data sets significantly different between transgenic plants and wild-type plants, according to Student’s t test (P < 0.05; n = 3 plants from independent transformant progeny).
| Monosaccharide | Arabidopsis AnAXE | Arabidopsis AnRAE | Arabidopsis Col-0 | Brachypodium AnAXE | Brachypodium Wild Type |
|---|---|---|---|---|---|
| Rha | 8.11 ± 1.02 | 5.44 ± 2.56 | 7.03 ± 0.34 | 1.91 ± 0.16 | 1.63 ± 0.32 |
| Ara | 8.79 ± 0.78 | 5.84 ± 2.14 | 7.55 ± 0.23 | 2.07 ± 0.11 | 1.72 ± 0.23 |
| Gal | 21.01 ± 0.42* | 18.52 ± 2.26 | 19.63 ± 0.58 | 6.25 ± 0.60 | 6.73 ± 0.27 |
| Glc | 15.64 ± 1.27 | 21.94 ± 2.9 | 17.98 ± 0.87 | 42.68 ± 4.61 | 39.38 ± 2.85 |
| Xyl | 44.39 ± 1.16 | 46.91 ± 3.03 | 45.68 ± 0.26 | 46.85 ± 4.32 | 50.35 ± 2.52 |
| Man | 7.99 ± 0.91 | 8.44 ± 1.31 | 8.22 ± 0.38 | 8.43 ± 0.29 | 9.06 ± 0.76 |
| GalA | 2.05 ± 0.72 | 1.4 ± 0.25* | 2.19 ± 0.06 | 0.24 ± 0.15 | 0.25 ± 0.05 |
Deacetylation of Different Cell Wall Polysaccharides Causes the Up-Regulation of Different Acetyltransferases
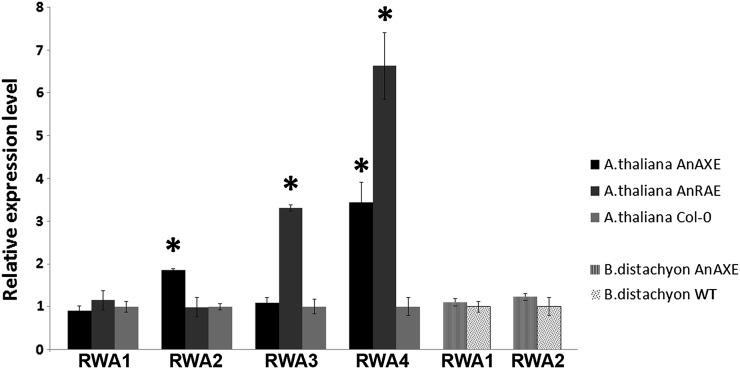
We next investigated the possible intensification of biosynthetic acetylation of polysaccharides as a plant compensatory reaction to postsynthetic deacetylation by overexpressed fungal acetylesterases. Quantitative reverse transcription (RT)-PCR was used to measure the transcript levels of four previously discovered acetyltransferase-encoding genes. Arabidopsis RWA1, RWA2, RWA3, and RWA4 were demonstrated to control xylan acetylation during secondary cell wall formation and to have high identity with each other (Lee et al., 2011). RT-quantitative PCR (qPCR) analysis showed higher expression of RWA2 and RWA4 (1.9- and 3.4-fold, respectively) in Arabidopsis AnAXE plants in comparison with wild-type and AnRAE plants and higher expression of RWA3 and RWA4 (3.3- and 6.6-fold, respectively) in AnRAE plants (Fig. 3). Using in silico analysis of primary DNA and protein sequences, two Brachypodium genes were identified and found to be 70% identical to the Arabidopsis RWA genes and to their protein products (Supplemental Fig. S1). No difference in expression of the two RWA-like genes was detected in Brachypodium AnAXE plants in comparison with wild-type plants.
Figure 3.
Real-time RT-PCR analysis of RWA genes in transgenic lines and wild-type plants (WT). Relative expression levels were calculated in comparison with an appropriate control gene from wild-type plants. ACTIN2 (At3g18780; for Arabidopsis) and GADPH (for Brachypodium), whose expression levels were not affected, were used as reference genes. The comparative threshold cycle method was used for determining the difference (asterisks indicate significant differences) between transcript copy numbers in wild-type and transgenic plants. Data represent average values ± se obtained for three independent transgenic lines.
Acetyltransferase-Expressing Plants Have Increased Resistance to Fungal Pathogens
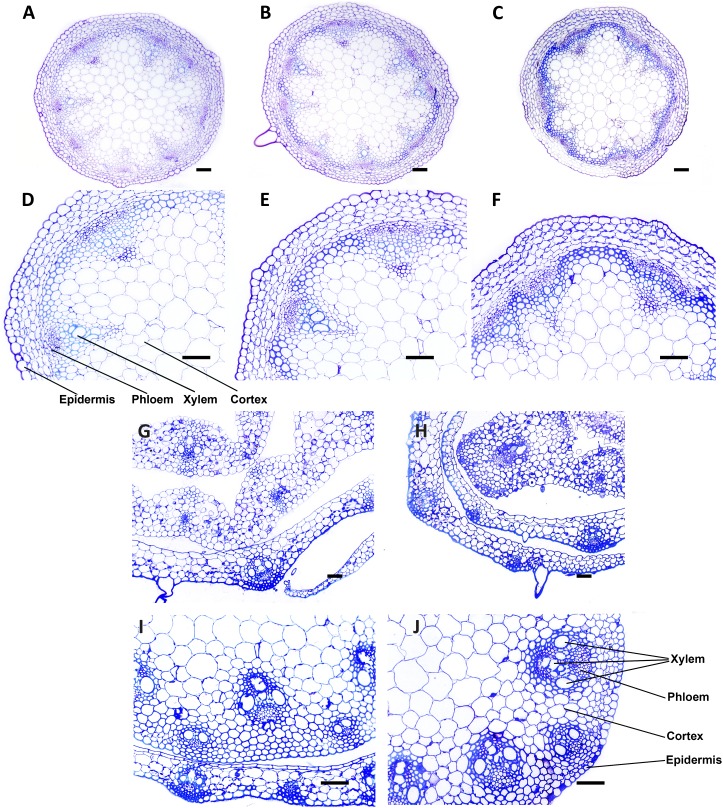
To examine whether the expression of acetylesterases in the plant cell wall causes any changes in cell morphology, thick cross sections were prepared from 5-week-old transgenic plant stems and stained with toluidine blue (Fig. 4). Examination of stained sections by light microscopy did not reveal differences in cell morphology or cell wall thickness between transgenic lines and wild-type plants.
Figure 4.
Toluidine blue-stained cross sections and enlargements of specific areas of stem from transgenic and wild-type plants. A to F, Arabidopsis AnAXE (A and D), AnRAE (B and E), and Col-0 (C and F) plants. G to J, Brachypodium AnAXE (G and I) and wild-type (H and J) plants. Bars = 1 mm in cross sections and 25 μm in enlargements. [See online article for color version of this figure.]
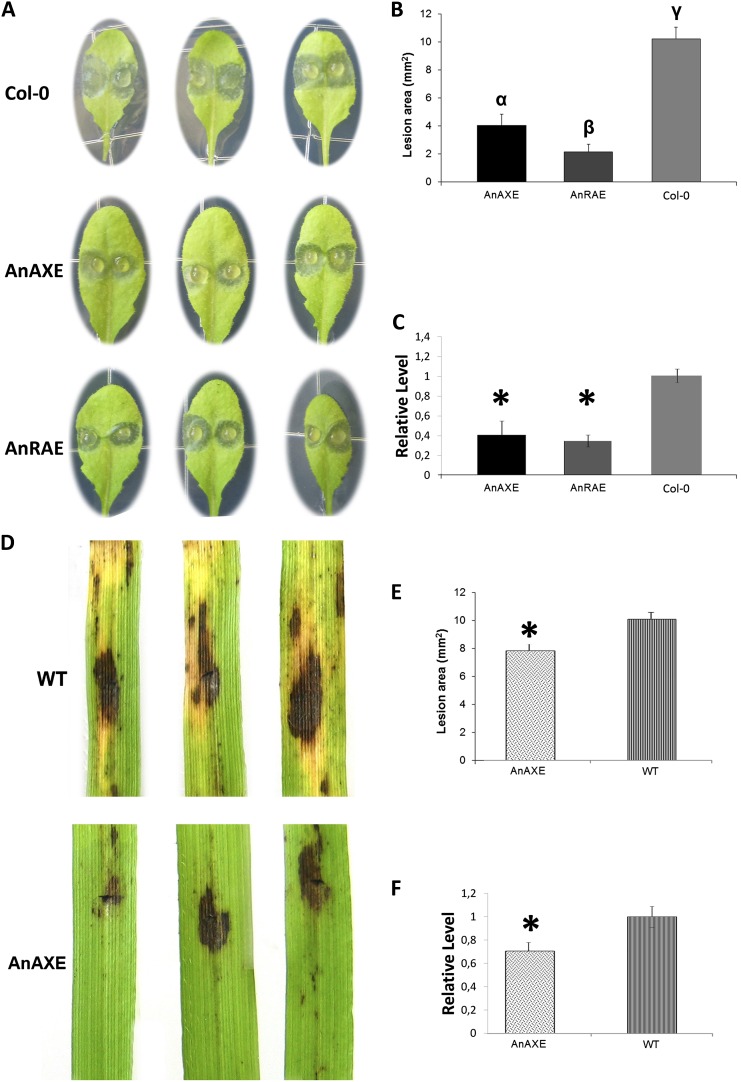
Since modification of plant cell wall structure may be critical for plant-pathogen interactions (Vorwerk et al., 2004; Lionetti et al., 2007; Ferrari et al., 2008), transgenic plants obtained in this study were challenged with pathogenic microorganisms. The susceptibility of Arabidopsis plants ectopically expressing AnAXE or AnRAE was assessed by inoculating leaves with B. cinerea conidia and subsequently evaluating symptoms 48 h post inoculation (hpi). AnAXE- and AnRAE-expressing plants displayed reduced lesion areas compared with Col-0 (Fig. 5, A and B), but no significant differences were observed in the number of expanding lesions per inoculated area between transformed lines and wild-type plants. The reduced symptoms detected in transgenic plants were related to reduced fungal growth, as determined by RT-qPCR analysis of transcript accumulation of the B. cinerea housekeeping gene Actin after 48 hpi in transgenic plants in comparison with wild-type plants (Fig. 5C).
Figure 5.
Analysis of the susceptibility of Arabidopsis and Brachypodium transgenic plants to fungal pathogens. A and B, B. cinerea symptoms on Arabidopsis transgenic and wild-type leaves. α, β, and γ indicate data sets significantly different according to one-way ANOVA followed by Tukey’s tests (n = 44; P < 0.001). D and E, B. sorokiniana symptoms on Brachypodium transgenic and wild-type plants (WT). The asterisk indicates a significant difference between transgenic plants and wild-type plants, according to Student’s t test (n = 24; P < 0.001). All lesion areas were measured 48 h after inoculation. Data points represent average lesion areas ± se. Similar results were obtained from three independent experiments. C and F, Real-time RT-PCR analysis of housekeeping pathogen gene transcript accumulation in infected transgenic lines and wild-type plants. Relative expression levels of B. cinerea ACTIN (C) and B. sorokiniana GDP (F) genes were calculated as a comparison with the appropriate reference plant housekeeping gene ACTIN2 (for Arabidopsis) or GADPH (for Brachypodium), expression of which was not affected. The comparative threshold cycle method was used for determining differences between transcript copy numbers in wild-type and transgenic plants. Data represent average values obtained for three independent transgenic lines. Asterisks indicate significant differences between transgenic plants and wild-type plants (Student’s t test, P < 0.05; n = 3).
In contrast to fungal infection, when Arabidopsis transgenic and wild-type plants were treated with the bacterial pathogen Pseudomonas syringae pv tomato strain DC3000, no difference in bacterial growth was observed in the infected leaves (Supplemental Fig. S2A). However, in both AnAXE and AnRAE plants, the appearance of yellowish spots on the leaves, typical of bacterial infection, was delayed in comparison with the wild type (Supplemental Fig. S2B).
To verify whether the expression of AnAXE in grass species, which contain high levels of xylans in their cell walls, can affect resistance to fungal pathogens, Brachypodium plants expressing AnAXE were infected with Bipolaris sorokiniana. Water-soaked necrotic lesions developed between 48 and 72 hpi with B. sorokiniana on wild-type leaves and were followed at later stages by the development of a surrounding chlorotic area and the appearance of more widespread necrosis. After 48 hpi, the lesion areas on AnAXE plants were significantly reduced compared with wild-type plants (Fig. 5E). Also in this case, quantitative RT-PCR analysis of transcripts of the B. sorokiniana housekeeping gene GLYCERALDEYDE-3-PHOSPHATE DEHYDROGENASE-LIKE indicated that the reduced symptoms detected in transgenic plants were related to reduced B. sorokiniana growth (Fig. 5F). In addition, at a later stage of infection (at 170 hpi), leaves from transformed plants exhibited reduced chlorosis in comparison with wild-type plants (Fig. 5D).
No differences were observed between Brachypodium AnAXE and wild-type plants after treatment with Xanthomonas oryzae, a pathogenic bacterium of rice (Oryza sativa) and some weedy grasses (Supplemental Fig. S2C).
Expression of Acetylesterases in the Plant Cell Wall Leads to Changes in Pathogen-Responsive Gene Expression
Increased resistance to pathogens can be caused by the release of molecules that are toxic to microbial cells from the modified plant cell wall or by plant-specific responses to cell wall modification. Measurement of acetyls in apoplastic fluids of transgenic plants did not reveal any differences in comparison with wild-type plants (data not shown). In addition, incubation of B. cinerea cells with apoplastic fluids prepared from Arabidopsis AnRAE, AnAXE, or wild-type plants did not reveal differences in fungal growth (Supplemental Fig. S3), suggesting that any apoplastic compositional changes due to the presence of fungal acetylesterases do not have a direct effect on fungal growth.
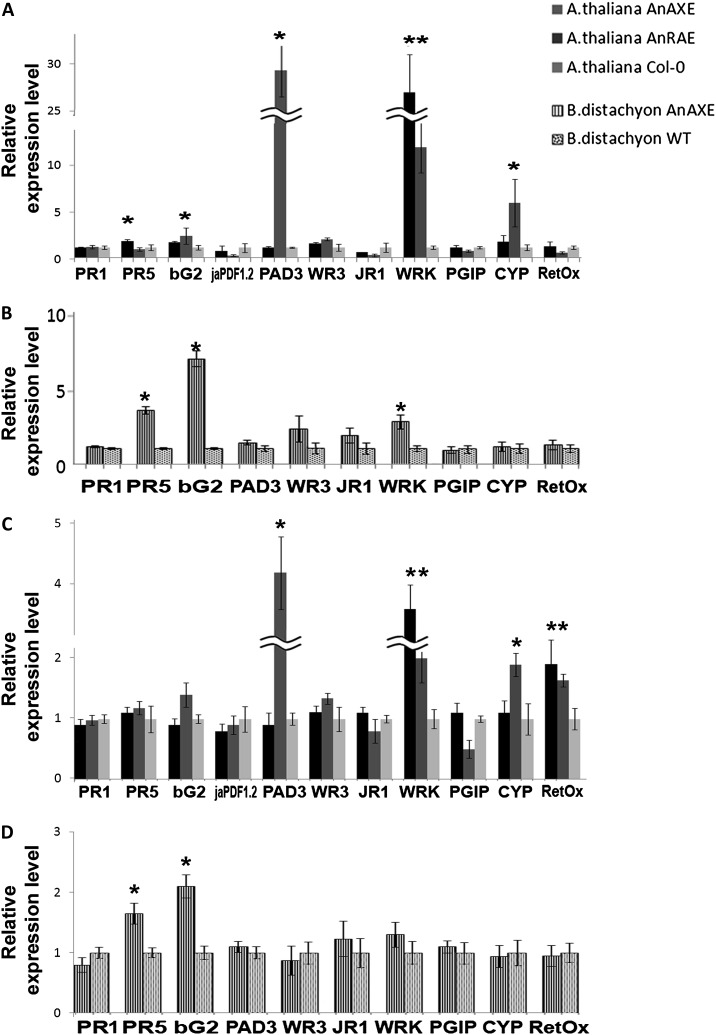
To investigate whether the expression of fungal acetylesterases and the consequent decrease of cell wall acetylation affect the pathways involved in plant defense responses to pathogens, the transcript levels of several defense genes were measured using RT-qPCR in healthy Arabidopsis and Brachypodium plants and upon fungal infection (Fig. 6). In Arabidopsis plants, the expression levels of genes encoding defense proteins (PR1 and PR5), β-glucanase2 (bG2), plant defensin1.2 (PDF1.2), phytoalexin deficient3 (PAD3), wound responsive3 (WR3), jasmonic acid responsive gene1 (JR1), pathogen-induced W box-containing transcription factor (WRKY40), and polygalacturonase-inhibiting protein1 (PGI1) involved in the defense response, CYP81F2 involved in the response to bacterium, fungus, and insects by initiating callose deposition in the cell wall, and oxidoreductase (RetOx) were analyzed. In noninfected AnAXE plants, the expression of PR5 (2-fold) and WRKY40 (27-fold) was higher than in wild-type plants. Increased levels of expression were also observed in PAD3 (29-fold), WRKY40 (11-fold), and CYP81F2 (5-fold) in Arabidopsis AnRAE plants in comparison with wild-type plants (Fig. 6A). Using BLAST searches of the Brachypodium genome, pathogen-related genes with high identity to Arabidopsis genes were identified. In noninfected Brachypodium AnAXE plants, the expression of PR5 (3.3-fold), bG2 (6.4-fold), and WRKY40 (2.6-fold) was higher in comparison with wild-type plants (Fig. 6B).
Figure 6.
Real-time RT-PCR analysis of selected pathogen-related genes in uninfected and infected transgenic lines and wild-type plants (WT). A and B, Expression levels of defense-related genes in Arabidopsis (A) and Brachypodium (B) transgenic plants. C and D, Gene expression in Arabidopsis (C) and Brachypodium (D) transgenic plants upon 48 h of infection with B. cinerea and B. sorokiniana, respectively. Gene expression levels in transgenic plants normalized to the expression of the same gene detected in wild-type plants (for which gene expression was set to 1), ACTIN2 (for Arabidopsis) or GADPH (for Brachypodium), were used as reference genes. The comparative threshold cycle method was used for determining differences between transcript copy numbers in wild-type and transgenic plants. Data represent average obtained for three independent transgenic lines. Asterisks indicate significant differences between transgenic plants and wild-type plants (Student’s t test, P < 0.05; n = 3).
After pathogen inoculation (48 hpi), expression levels of all analyzed defense-related genes were approximately 100-fold higher in comparison with noninfected plants (data not shown). While the difference in expression of PAD3, WRKY40, and CYP81F2 between transgenic and wild-type plants was lower than that in uninfected plants, the expression of RetOx was 2-fold higher in both Arabidopsis transgenic lines, AnAXE and AnRAE, in comparison with wild-type plants (Fig. 6C). In Brachypodium, the difference in defense gene expression was lowered to 1.6- and 2-fold for PR5 and bG2, respectively. No difference in WRKY40 between transgenic and wild-type plants after 48 hpi was detected (Fig. 6D). At 72 hpi for Arabidopsis and 170 hpi for Brachypodium plants, the difference in the level of defense gene expression disappeared completely, except for RetOx, which showed a 2- to 2.5-fold up-regulation in both Arabidopsis lines (Supplemental Fig. S4).
Arabidopsis AnAXE and AnRAE Plants Have Enhanced Defense Responses
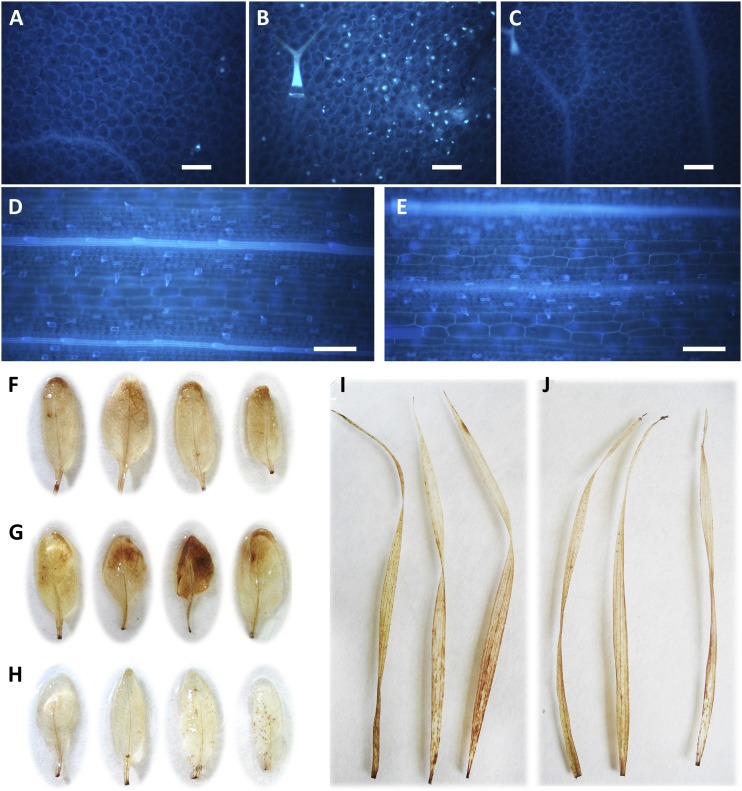
To verify if the reduced susceptibility to pathogens observed in Arabidopsis and Brachypodium transgenic plants is due to the constitutive activation of defense responses, we quantified callose deposition and hydrogen peroxide (H2O2) accumulation in leaves. Arabidopsis AnAXE and wild-type plants did not show callose deposition, but nontreated AnRAE plants showed a significant number of callose deposits (Fig. 7, A–C). This result is consistent with the induction of CYP81F2 detected in these plants. Brachypodium AnAXE and wild-type plants did not show any callose deposition (Fig. 7, D and E).
Figure 7.
A to E, Determination of callose deposition. Leaves from Arabidopsis AnAXE (A), AnRAE (B), and Col-0 (C) and Brachypodium AnAXE (D) and wild-type (E) plants were stained with aniline blue for callose visualization. Calculation of callose deposit number in three microscopic fields of 0.5 mm2 for each leaf (six leaves were analyzed for each transgenic line) resulted in 89 ± 7 deposits in AnRAE against 9 ± 1 and 6 ± 1 deposits in Arabidopsis AnAXE and Col-0, respectively. The experiment was repeated twice with similar results. Bars = 100 µm. F to J, Accumulation of H2O2 in transgenic and wild-type plants. Staining of detached leaves with 3,3′-diaminobenzidine revealed an accumulation of H2O2 in leaves from Arabidopsis AnAXE (F) and AnRAE (G) plants and no accumulation in leaves from Col-0 plants (H). No difference between Brachypodium AnAXE (I) and wild-type plants (J) was observed.
Increased accumulation of H2O2 was observed in AnRAE and AnAXE Arabidopsis plants in comparison with wild-type plants (Fig. 7, F–H). No difference in the accumulation of H2O2 was observed between Brachypodium transgenic and wild-type leaves (Fig. 7, I and J).
DISCUSSION
Postsynthetic modification of the plant cell wall by polysaccharide-modifying enzymes overexpressed in the apoplast represents a promising strategy to examine the contributions of structural polysaccharides to plant fitness. Transgenic expression of microbial hydrolases with well-characterized enzymatic activities provides a useful approach for a targeted modification of the plant cell wall to study their physiological effects in planta.
Earlier, we reported the preparation of Arabidopsis plants expressing AnAXE and demonstrated that a signal peptide fused to the N terminus of the hydrolase ensures its delivery to the apoplast, and a fluorescent protein fused to the C terminus of the hydrolase does not affect its activity (Pogorelko et al., 2011). In this study, we constructed new versatile transformation vectors and expressed AnAXE and AnRAE esterases in the apoplast of Arabidopsis and Brachypodium plants characterized by type I and type II cell walls, respectively. Using Arabidopsis AnAXE created earlier and new Arabidopsis AnRAE and Brachypodium AnAXE transgenic plants, we have investigated the effect of deacetylation on plant cell wall integrity and obtained new insights into plant responses to cell wall modifications, specifically the potential role of polysaccharide acetylation in the defense against fungal infection.
Up-regulation of three putative acetyltransferases (RWA2, RWA3, and RWA4; Lee et al., 2011; Manabe et al., 2011) in Arabidopsis AnAXE and AnRAE plants suggested that plants respond to these postsynthetic modifications using compensatory mechanisms. Moreover, deacetylation of xylan in AnAXE plants promotes different acetyltransferases, most likely with different polysaccharide specificity, than deacetylation of pectin and xyloglucan in AnRAE plants. Similarly, a compensatory effect was observed when increased methylesterification of pectins achieved by the inhibition of PME in Arabidopsis plants caused the up-regulation of several PME-encoding genes via an unknown feedback mechanism for the BRI1 receptor (Wolf et al., 2012b). The lack of up-regulation of two RWA-like genes in Brachypodium AnAXE plants can be due to the involvement of other acetyltransferases in such compensatory reactions in Brachypodium. It is also possible that the low level of Brachypodium cell wall deacetylation in the transgenic plants is insufficient to induce compensatory responses.
Deacetylation of polysaccharides can lead to changes in cell wall integrity and, hence, can affect plant defense against environmental stresses. There is substantial evidence that esterification/deesterification and subsequent changes in the polysaccharide cross-linking network affect plant resistance to pathogenic microorganisms (Vogel et al., 2002, 2004; Lionetti et al., 2007; Manabe et al., 2011; Gille and Pauly, 2012). Here, we demonstrate that the reduction of polysaccharide acetylation in both Arabidopsis and Brachypodium transgenic plants significantly increases plant resistance to necrotrophic fungi, B. cinerea, and B. sorokiniana but not to bacterial pathogens, which suggests the importance of polysaccharide acetylation for the cell wall’s protective function against fungal invasion.
The observed lack of B. cinerea growth inhibition in the presence of apoplastic fluids prepared from either Arabidopsis transgenic or wild-type plants (Supplemental Fig. S3) suggests that the inhibition of fungal growth in the leaves of transgenic plants is not due to compositional changes in the apoplast of transgenic plants. Transcript analysis of the known plant defense-related genes PR1, PR5, bG2, jaPDF1.2, PAD3, WR3, JR1, WRKY40, PGIP1, CYP81F2, and RetOx (Galletti et al., 2008; Raiola et al., 2011) demonstrated that particular plant defense responses are constitutively initiated in our transgenic plants. Thus, overexpression of AnRAE in Arabidopsis and consequent deacetylation of pectins and xyloglucan in this plant induce the expression of PAD3, involved in camalexin biosynthesis and known to mainly affect plant defense against fungal pathogens (Schuhegger et al., 2006), and of CYP81F2, involved in glucosinolate metabolism and callose deposition in the cell wall (Po-Wen et al., 2013). However, we were unable to detect camalexin in Arabidopsis AnRAE plants (data not shown), most likely because other rate-limiting proteins involved in its biosynthesis were not induced in uninfected plants. Higher levels of PAD3 and CYP81F2 expression, together with callose accumulation, suggest a possible contribution of these specific defense pathways to the reduced susceptibility of AnRAE plants to B. cinerea. In addition, WRKY40 is known to act as a transcriptional suppressor in the mitogen-activated protein kinase cascade during pathogen invasion (Chen et al., 2010). The fact that this gene is up-regulated in all our transgenic lines, Arabidopsis AnAXE, AnRAE, and Brachypodium AnAXE, suggests that this regulatory pathway also responds to cell wall deacetylation.
Overexpression of AnAXE and deacetylation of xylan in both Arabidopsis and Brachypodium plants increased transcript levels of PR5 and bG2, two other defense-related genes. PR5 is normally induced by salicylic acid and secreted into the extracellular spaces (Linthorst, 1991). Its expression was associated with drought (Jakab et al., 2005) and salt stress responses and seed germination (Seo et al., 2008). bG2 is a β-glucanase-encoding gene that was shown to be up-regulated during the Arabidopsis response to biotic stress (Dong et al., 1991). β-Glucanase purified from pea (Pisum sativum) protein extract was shown to inhibit, together with chitinase, the growth of Fusarium solani f. sp. phaseoli (Mauch et al., 1988), Sclerotinia homoeocarpa, and Rhizoctonia solani (Wang et al., 2003).
Normally, upon infection with necrotrophic fungi, wild-type plants respond by the regulation of a large subset of signaling pathways involving a broad spectrum of defense-related genes (Denby et al., 2004). In our study, all analyzed defense-related genes, including those that had higher transcript levels in uninfected transgenic plants, PAD3, WRK, CYP, PR5, and bG2 in Arabidopsis and PR5 and bG2 in Brachypodium, were up-regulated in wild-type plants after infection. qPCR analysis performed for different postinfection time points (Supplemental Fig. S4) demonstrated that the expression of these genes in wild-type plants gradually became even with their expression levels in transgenic plants, suggesting that in uninfected transgenic plants, some defense-related pathways, which are induced in the wild type only in the response to fungal treatment, are induced prior to infection. Significantly higher up-regulation of the oxidoreductase-encoding gene RetOx (Carter and Thornburg, 2004) and the higher level of H2O2 observed in Arabidopsis AnAXE and AnRAE plants, but not in Brachypodium AnAXE plants, in comparison with wild-type plants indicate that dicots and monocots have different mechanisms to respond to pathogens, although both are sensitive to cell wall deacetylation. Our results also suggest that different polysaccharides and their acetylation contribute differently to plant defense reactions against fungal pathogens, most likely through the involvement in different specific signaling pathways.
We demonstrated that deacetylation of pectin and xyloglucan in the cell wall in transgenic plants makes pectic polysaccharides more accessible to pectinases. This can cause the production of small oligosaccharides and monosaccharides, which can act as elicitors of plant defense through signal transduction and transcription activation (Gille and Pauly, 2012). Ferrari et al. (2007, 2008) showed that transgenic plants expressing microbial PG were less susceptible to B. cinerea and had constitutively activated defense responses, possibly due to an increased accumulation of bioactive oligogalacturonides (OGs), elicitors perceived by the plant cell as danger-associated molecular patterns (DAMPs). It was demonstrated that OG-induced activation of basal resistance against fungal pathogens is mediated by molecular patterns independently of jasmonate-, salicylic acid-, and ethylene-mediated signaling but dependent on the expression of PAD3 (Ferrari et al., 2007). OGs also increase the levels of H2O2 and callose deposition. Considering that our Arabidopsis AnRAE plants have higher levels of PAD3 expression, H2O2 accumulation, and callose deposition, we hypothesize that the deacetylation of pectic polysaccharides increases their susceptibility to plant PG. This leads to the accumulation of elicitor OGs and the initiation of defense-related pathways in noninfected plants, which reduces their susceptibility to pathogen. Similarly, deacetylation of xylan might result in the production of elicitor-like oligosaccharides, which would act as DAMP-inducing defense reactions in the plant. It was reported that the expression of xylanase can increase resistance in rice (Weng et al., 2012); thus, it is possible that in AnAXE-expressing plants, xylan deacetylation results in the liberation of yet-unknown DAMPs. It is worth noting that some up-regulated genes were the same in both Brachypodium and Arabidopsis AnAXE-expressing plants, indicating that xylan deacetylation induces similar responses in dicot and monocot plants. It was reported earlier that reduction of polysaccharide acetylation by knockout of the acetyltransferase RWA2 results in Arabidopsis with increased resistance to B. cinerea (Manabe et al., 2011). Although the authors did not observe significant constitutive activation of PDF1.2 and PAD3, they proposed that rwa2 mutant plants have either other defense-related genes up-regulated or faster activation of defenses upon infection. It is also possible that rwa2 plants and our transgenic plants have different patterns of deacetylated polysaccharides in their cell walls, which might differently affect DAMP formation. Nevertheless, the results of Manabe et al. (2011) support our notion that it is polysaccharide deacetylation that affects the plant’s susceptibility to pathogens and not the presence of fungal acetylesterases in the apoplast. Further investigations will be needed to confirm the accumulation of oligosaccharide DAMP in the apoplast of transgenic plants.
In conclusion, the results obtained in this study point to a potential utility of the postsynthetic modification of cell wall properties. First, specific modifications of cell wall polysaccharides by the overexpression of polysaccharide-modifying enzymes can initiate a plant compensatory response including the up-regulation of biosynthetic genes. This may help to reveal key genes involved in polysaccharide biosynthesis and to elucidate the pathways involved in cell wall integrity signaling. Second, specific polysaccharide modifications can change plant fitness or induce responses to environmental stresses, which can be beneficial to plants. These results have key implications for strategies that seek to modify the plant cell wall for biofuels or other applications. Finally, we also propose that deacetylation of the plant cell wall activates plant defense mechanisms against pathogens and can be used in the future as a tool to increase the resistance of valuable crops to biotic stresses.
MATERIALS AND METHODS
Generation of Transgenic Plants
The pDico_T-vector was constructed using the pXcmkn12 (AB107950·1) vector obtained from Dr. S. Yasuda (National Institute of Genetics, Shizuoka, Japan). The cauliflower mosaic virus (CaMV) promoter of the 35S RNA (CaMV 35S promoter) was amplified using Encyclo DNA polymerase (Evrogen; PK001) from the pEnLox vector (Pogorelko et al., 2008) with the use of primers CaMV_F and CaMV_R harboring EcoRI+NotI and EcoRI restriction sites, respectively. The CaMV 35S promoter PCR fragment was cloned into pGEM-T Easy vector (Promega; A1360) and moved by NotI and EcoRI restriction enzymes into NotI-EcoRI-pretreated pXcmkn12 plasmid, resulting in the pXcmkn12-CaMV vector. The Arabidopsis (Arabidopsis thaliana) expansin gene signal peptide-coding sequence was amplified by PCR with primers AtSP_F and AtSP_R containing EcoRI and KpnI restriction sites, respectively (for primer sequences, see Supplemental Data S1), using Encyclo DNA polymerase from the pBAt plasmid (obtained from Dr. M.G. Hahn, The Complex Carbohydrate Research Center), and the product was cloned into the pGEM-T Easy vector. After the digestion of recombinant pGEM-T_Easy_AtSP vector, the EcoRI-KpnI fragment was moved into the pXcmkn12-CaMV plasmid treated with the same enzymes to obtain the pXcmkn12-CaMV-AtSP vector. The GFP coding sequence, together with the transcription terminator, was amplified with Encyclo DNA polymerase from the psmGFP plasmid (www.arabidopsis.org; CD3-326) using GFP_F and GFP_R primers containing XbaI and PstI restriction sites, respectively. The product was cloned into a pGEM-T Easy vector. After digestion of the recombinant pGEM-T_Easy_GFP vector, the XbaI-PstI fragment was moved into the pXcmkn12-CaMV-AtSP plasmid cleaved with the same enzymes. To obtain the final pDico_T-vector, a PCR fragment amplified with CaMV_F and GFP_R primers from the pXcmkn12-AtSP-GFP plasmid was cloned into the pGEM-T Easy vector containing two NotI restriction sites flanking the cloning site (for complete sequence, see Supplemental Data S2).
Aspergillus nidulans AN2528.2 (AnRAE) and AN6093.2 (AnAXE) cDNAs extracted from Pichia pastoris recombinant strains (Bauer et al., 2006) obtained from the Fungal Genetics Stock Center (www.fgsc.net) were amplified with Encyclo DNA polymerase with appropriate forward and reverse primers (Supplemental Data S1). All forward primers started from the second nucleotide of the ATG start codon to fit into the same open reading frame with the signal peptide and GFP sequences from pDico_T-vector pretreated with XcmI (New England Biolabs) restriction enzyme to provide single T-overhanging ends. The AnRAE-coding sequence was cloned into the pDico_T-vector and treated with NotI to move the CaMV-AtSP-AnRAE-GFP cassette into the dicot-specific pMLBart binary vector (Gleave, 1992) treated with the same enzyme. For Brachypodium (Brachypodium distachyon) transformation to express AnAXE, the pMono_T/pUbiP2 vector system described previously (Fursova et al., 2012) was used. The binary vectors were transformed into Agrobacterium tumefaciens EHA101 cells by electroporation, and Arabidopsis plants were transfected with these cells by the floral-dip method (Clough and Bent, 1998). Brachypodium transgenic plants were created via A. tumefaciens-mediated transformation of calluses (Vogel and Hill, 2008) at the Plant Transformation Facility of Iowa State University (http://www.agron.iastate.edu/ptf/).
Apoplastic Fluid Preparation and Western-Blot Analysis
Apoplastic fluids from aerial plant parts were extracted as described previously (Pogorelko et al., 2011). Immunoblot analyses were performed according to Magi and Liberatori (2005). Apoplastic proteins after precipitation with 10% (v/v) TCA were resuspended in 1× Laemmli’s buffer (125 mm Tris-HCl [pH 6.8], 1% SDS, 37.7% glycerol, 1% β-mercaptoethanol, and 0.01% bromphenol blue), boiled for 5 min, and used immediately for SDS-PAGE. Fifteen micrograms of total protein was loaded per well for 10% SDS-PAGE. After separation, the proteins were electrophoretically transferred to a Trans-Blot Transfer Medium membrane (Bio-Rad; 162-0112) using the Trans-Blot Cell (Bio-Rad) with standard transfer buffer. Primary anti-GFP monoclonal antibody (Covance; MMS-118P) at a dilution of 1:5,000 and secondary anti-mouse IgG (whole molecule) peroxidase antibody (Sigma; A9044) at a dilution of 1:40,000 were used for the detection of GFP fusion proteins. Membranes were treated with the HyGlo Quick Spray Reagent B for peroxidase activity (Denville; E2400) and immediately visualized by the ChemiDoc XRS+ System (Bio-Rad; 170-8265).
Assays for Acetyl Esterase Activity
Acetylxylan esterase and rhamnogalacturonan acetylesterase activities were measured in 100 µL of reaction mixture containing 2 mm pNP-acetyl (Sigma; N8130) in 0.1 m assay buffer (25 mm Tris-HCl, 50 mm EDTA, and 150 mm MgCl2, pH 7.4). Apoplastic fluids were added to obtain 1.28 × 10−1 mg of protein in each reaction mixture. As a negative control, reaction mixtures with boiled apoplastic extract were used. The reactions were carried out at 37°C and terminated at different time points. Absorbance was measured at 460 nm on a microplate reader. Specific activity was expressed as the amount of pNP (mmol) released per 1 min by 1 mg of protein.
Acetylxylan esterase and rhamnogalacturonan acetylesterase activities in apoplastic fluids were also assayed using xylan from beech wood (Fagus spp.; Sigma; X4252) and pectin from citrus (Citrus aurantium) fruit (Sigma; P9436). Apoplastic fluid (1.28 × 10−1 mg of total protein) was incubated with 20 mg of xylan or pectin in 1 mL of 0.1 m sodium phosphate buffer (pH 7.0) at 37°C for 24 h. At each time point, 100 µL of sample was boiled for 15 min at 100°C to terminate the reaction, dried, and used for acetyl amount determination according to the protocol of McComb and McCready (1957; see below).
Cell Wall Composition Analysis and Treatments
Cell walls were isolated from the plant tissue residues after apoplastic fluid extraction as described by Zabotina et al. (2008). To determine monosaccharide composition, 1 mg of dry cell wall was hydrolyzed with 2 n trifluoroacetic acid at 120°C for 2 h. After the acid was evaporated, the hydrolysate was redissolved in water and analyzed by high-performance anion-exchange chromatography-pulsed-amperometric detection using a CarboPac PA-20 column (Dionex) as described earlier (Zabotina et al., 2008). Monosaccharide standards included l-Fuc, l-Rha, l-Ara, d-Gal, d-Glc, d-Xyl, d-Man, d-GalA, and d-GlcA (all from Sigma). To determine response factors, standard curves were created using mixtures of all standard monosaccharides at different concentrations.
Acetyl content in cell walls was estimated using an assay adapted from McComb and McCready (1957). Briefly, 10 mg of dry cell wall material was mixed with 2.5 mL of 0.5 m hydroxylamine hydrochloride solution, followed by the gradual addition of 2.5 mL of 2 n sodium hydroxide solution. A total of 0.5 mL of the resulting mixture was mixed with 0.2 mL of water and 0.5 mL of acid methanol solution (35.2 mL of 70% perchloric acid in 500 mL of absolute methanol). A total of 1.3 mL of ferric perchlorate solution was added in small increments, mixing after each addition. After 15 min of reaction, the absorbance of the formed ferric acetohydroxamic complex was determined at 510 nm. The amount of acetyls was calculated using a standard curve created using Glc pentaacetate as a standard.
Partial removal of hemicelluloses was done using 30 mg of dry cell wall material incubated with a mixture of 150 units of endo-1,4-β-xylanase M6 (rumen microorganism; Megazyme) and 15 units of endo-1,4-β-xylanase M1 (Trichoderma viride; Megazyme) in a 1-mL total volume of sodium phosphate buffer (pH 6.0) for 24 h at 37°C. For the partial removal of pectins, 30 mg of dry cell wall material was incubated with a mixture of 150 units of endo-polygalacturonase (Megazyme) and 50 units of PME (PROZOMIX) in a 1-mL total volume of sodium phosphate buffer (pH 6.0) for 24 h at 37°C.
Reducing sugars were measured using the PAHBAH assay (Lever, 1972) with minor modifications. Briefly, 15 µL of supernatant was mixed with 135 µL of freshly prepared PAHBAH reagent (1 volume of 5% p-hydroxybenzoic acid hydrazide in 5% HCl mixed with 9 volumes of 1.25% trisodium citrate, 0.11% calcium chloride, and 2% sodium hydroxide) and heated at 95°C for exactly 6 min. Absorbance was measured at 410 nm. Calculations were done using a standard curve prepared using different concentrations of Glc.
The cell walls were analyzed by Oligo Mass Profiling according to previously published methods (Lerouxel et al., 2002; Obel et al., 2009). The dry cell wall (2 mg) was suspended in 50 mL of 100 mm ammonium formate buffer, pH 4.5, containing 0.02 units of purified recombinant xyloglucan endoglucanase (EC 3.2.1.151; Pauly et al., 1999) and incubated at 37°C for 18 h. The reactions were centrifuged to pellet undigested material, and the supernatant containing soluble xyloglucan oligosaccharides was removed and dried in a Speed-Vac. The released xyloglucan oligosaccharides were dissolved in 10 μL of water containing approximately 10 beads of Bio-Rex MSZ 501(D) resin (Bio-Rad) to remove buffer salts. One microliter of the oligosaccharide solution was mixed with 2,5-dihydroxybenzoic acid, spotted onto a MALDI-TOF sample plate, and dried under vacuum. Spectra from five independent randomly distributed spots for each sample were collected and analyzed on a Voyager DE-Pro MALDI-TOF-mass spectrometry instrument in positive reflection mode with an acceleration voltage of 20 kV and an extraction delay time of 350 ns. Ratios of corresponding peak intensities for each xyloglucan subunit and acetylated subunit were determined, and Student’s t test was performed to compare transgenic and wild-type plants.
Sample Fixation, Sectioning, and Light Microscopy
Four-week-old Arabidopsis and 6-week-old Brachypodium stems were fixed as described previously (Pattathil et al., 2010) with minor modifications. Stem segments 3 to 5 mm long were incubated for 2.5 h in 1.6% (w/v) paraformaldehyde and 0.2% (w/v) glutaraldehyde in 25 mm sodium phosphate buffer, pH 7.1, washed with water twice for 15 min, and dehydrated using a graded ethanol series (35%, 50%, 75%, 95%, 100%, 100%, and 100% [v/v] ethanol) for 30 min at each step. The dehydrated samples were moved to 4°C and then gradually infiltrated with cold LR White embedding resin (Ted Pella) using 33% (v/v) and 66% (v/v) resin in 100% ethanol for 24 h for each concentration, followed by 100% resin for 24 h, three times. The infiltrated samples were transferred to gelatin capsules containing 100% resin for embedding, and resin was polymerized by incubation at 65°C for 24 h.
Sections of 300 nm were prepared using a Leica VT1000S vibratome, stained with 0.05% toluidine blue, and visualized with a Zeiss Axioskop 2 plus (Carl Zeiss) microscope. Images were captured with a Nikon DXM 1200 digital camera.
RNA Extraction, cDNA Synthesis, and Real-Time qPCR
Total RNA was extracted from the uninfected and infected leaves of 5-week-old plants using the SV Total RNA Isolation kit (Promega), and cDNA synthesis was performed with the SuperScript III First Strand Synthesis system (Invitrogen) following the manufacturer’s recommendations. The Maxima SYBR Green qPCR Master Mix (2X; Fermentas) with appropriate primers (Supplemental Data S1) and the Roche LightCycler 480 qPCR apparatus (Plant Science Institute at Iowa State University) were used to determine relative expression of the genes. Relative expression levels were calculated in comparison with an appropriate control gene (for primer sequences, see Supplemental Data S1) from wild-type plants, and the ACTIN2 gene (At3g18780; for Arabidopsis) and the GADPH gene (for Brachypodium), whose expression levels were not affected, were used as reference genes. Gene expression levels in transgenic plants are presented relative to the expression of the same gene detected in wild-type plants (for which gene expression was set to 1). The comparative threshold cycle method (Schmittgen and Livak, 2008) was used for determining differences between transcript copy numbers in wild-type and transgenic plants.
Arabidopsis Infection with Botrytis cinerea
B. cinerea strain SF1 (Lionetti et al., 2007) was grown for 15 d on potato dextrose agar at 39 g L–1 at 23°C with a 12-h photoperiod before spore collection. The spores were harvested by washing the culture surface of the agar and suspended in 5 mL of sterile distilled water. Spore suspensions were filtered through glass wool to remove residual mycelia, and the concentration was determined using a Thoma chamber. To synchronize germination, the conidia, at the concentration of 5 × 105 conidia mL–1, were germinated in potato dextrose broth at 24 g L–1 at room temperature for 3 h. The infection of Arabidopsis leaves was performed as described previously (Ferrari et al., 2008; Manabe et al., 2011). Fully developed leaves were detached from four 6-week-old Arabidopsis plants (three leaves per plant) and grown in a growth chamber maintained at 22°C and 70% relative humidity with a 12-h/12-h light/dark photoperiod. The detached leaves were placed in square petri dishes with petioles embedded in 0.8% agar. Two droplets of spore suspension (5 µL each) were placed on the surface of each leaf. Mock inoculation was performed using potato dextrose broth. Leaves were incubated at 24°C with a 12-h photoperiod, and lesion diameter was measured 48 hpi. Lesion sizes were measured using ImageJ software (Abramoff and Magalhães, 2004).
Brachypodium Infection with Bipolaris sorokiniana
Macroconidia of B. sorokiniana strain DSMZ 62608 (kindly provided by Prof. F. Favaron, University of Padua) were produced by culturing the fungus on potato dextrose agar medium before spore collection. The conidia were collected by washing the culture surface with 3 mL of sterile water, and conidia concentrations were estimated by using a Thoma chamber. The infection of Brachypodium leaves was performed as described previously (Peraldi et al., 2011). Twenty-four fully expanded leaves were detached from 60-d-old Brachypodium plants (three leaves per plant) grown under a 16-h/8-h light/dark cycle in a climatically controlled chamber with a relative humidity of 70% at 22°C. The leaves were cut to 5-cm lengths and placed in square petri dishes containing 0.8% agar. Two droplets (10 μL) of conidial suspension (1 × 106 conidia mL−1), with 0.05% Tween 20, were deposited onto each leaf at a distance of about 2 cm from each other. Mock inoculation was performed using sterile distilled water with 0.05% Tween 20. The plates were incubated at 22°C under a 16-h/8-h light/dark cycle. Disease symptoms were recorded every 24 h, and lesion sizes were measured using ImageJ software (Abramoff and Magalhães, 2004).
Determination of H2O2 Accumulation and Callose Deposition
For H2O2 visualization, leaves were cut from adult plants and dipped for 16 h in a solution containing 1 mg mL−1 3,3′-diaminobenzidine, pH 5.0. Chlorophyll was extracted with hot ethanol, and the leaves were photographed. The two more expanded leaves from six independent 4-week-old plants were cleared with 100% ethanol. Leaves were fixed in an acetic acid:ethanol (1:3) solution for 2 h, sequentially incubated for 15 min in 75% ethanol, in 50% ethanol, and in 150 mm phosphate buffer, pH 8.0, and then stained for 2 h at 25°C in 150 mm phosphate buffer, pH 8.0, containing 0.01% (w/v) aniline blue. After staining, leaves were mounted in 50% glycerol and examined by UV2A filter (excitation wavelength = 330–380) using the epifluorescence microscope Nikon Eclipse E200. Photographs were taken with a Nikon Digital Sight DS-Fi1c camera.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. In silico identification of Brachypodium genes similar to Arabidopsis RWA genes.
Supplemental Figure S2. Infection of transgenic Arabidopsis lines with P. syringe and Brachypodium transgenic lines with X. oryzae.
Supplemental Figure S3. Analysis of apoplast from Arabidopsis transgenic and wild-type plant inhibition of B. cinerea growth.
Supplemental Figure S4. Real-time RT-PCR analysis of selected pathogen-related genes in infected transgenic lines and wild-type plants.
Supplemental Table S1. Monosaccharide composition (mol %) of total cell wall extracted from aerial parts of transgenic plants expressing AnAXE and AnRAE and wild-type plants.
Supplemental Data S1. Primer sequences used in this work.
Supplemental Data S2. pDico_T-vector sequence.
Glossary
- PG
polygalacturonase
- Col-0
Columbia
- PME
pectin methylesterase
- PME+PG
pectin methylesterase plus polygalacturonase
- MALDI-TOF
matrix-assisted laser-desorption ionization time of flight
- RT
reverse transcription
- qPCR
quantitative PCR
- hpi
h post inoculation
- H2O2
hydrogen peroxide
- OG
oligogalacturonide
- DAMP
danger-associated molecular pattern
- CaMV
cauliflower mosaic virus
- cDNA
complementary DNA
- GalA
α-1,4-linked galacturonic acid
- pNP
4-nitrophenyl
References
- Abramoff MD, Magalhães PJ. (2004) Image processing with ImageJ. Biophotonics Int 11: 36–42 [Google Scholar]
- Albersheim P, Darvill A, Roberts K, Sederoff R, Staehelin A (2010) Plant Cell Walls. Garland Science, New York [Google Scholar]
- Bauer S, Vasu P, Persson S, Mort AJ, Somerville CR. (2006) Development and application of a suite of polysaccharide-degrading enzymes for analyzing plant cell walls. Proc Natl Acad Sci USA 10: 11417–11422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bily AC, Reid LM, Taylor JH, Johnston D, Malouin C, Burt AJ, Bakan B, Regnault-Roger C, Pauls KP, Arnason JT, et al. (2003) Dehydrodimers of ferulic acid in maize grain pericarp and aleurone: resistance factors to Fusarium graminearum. Phytopathology 93: 712–719 [DOI] [PubMed] [Google Scholar]
- Buanafina MMdO. (2009) Feruloylation in grasses: current and future perspectives. Mol Plant 2: 861–872 [DOI] [PubMed] [Google Scholar]
- Burton RA, Gidley MJ, Fincher GB. (2010) Heterogeneity in the chemistry, structure and function of plant cell walls. Nat Chem Biol 6: 724–732 [DOI] [PubMed] [Google Scholar]
- Caffall KH, Mohnen D. (2009) The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr Res 344: 1879–1900 [DOI] [PubMed] [Google Scholar]
- Carter CJ, Thornburg RW. (2004) Tobacco nectarin V is a flavin-containing berberine bridge enzyme-like protein with glucose oxidase activity. Plant Physiol 134: 460–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Lai Z, Shi J, Xiao Y, Chen Z, Xu X. (2010) Roles of Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol 10: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Denby KJ, Kumar P, Kliebenstein DJ. (2004) Identification of Botrytis cinerea susceptibility loci in Arabidopsis thaliana. Plant J 38: 473–486 [DOI] [PubMed] [Google Scholar]
- Dong X, Mindrinos M, Davis KR, Ausubel FM. (1991) Induction of Arabidopsis defense genes by virulent and avirulent Pseudomonas syringae strains and by a cloned avirulence gene. Plant Cell 3: 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C, Karafyllidis I, Wasternack C, Turner JG. (2002) The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14: 1557–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S, Galletti R, Denoux C, De Lorenzo G, Ausubel FM, Dewdney J. (2007) Resistance to Botrytis cinerea induced in Arabidopsis by elicitors is independent of salicylic acid, ethylene, or jasmonate signaling but requires PHYTOALEXIN DEFICIENT3. Plant Physiol 144: 367–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S, Galletti R, Pontiggia D, Manfredini C, Lionetti V, Bellincampi D, Cervone F, De Lorenzo G. (2008) Transgenic expression of a fungal endo-polygalacturonase increases plant resistance to pathogens and reduces auxin sensitivity. Plant Physiol 146: 669–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC, York WS, Albersheim P, Darvill A, Hayashi T, Joseleau J-P, Lorences EP, Maclachlan GA, McNeil M, Mort AJ, et al. (1993) An unambiguous nomenclature for xyloglucan-derived oligosaccharides. Physiol Plant 89: 1–3 [Google Scholar]
- Fursova O, Pogorelko G, Zabotina OA. (2012) An efficient method for transient gene expression in monocots applied to modify the Brachypodium distachyon cell wall. Ann Bot (Lond) 110: 47–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletti R, Denoux C, Gambetta S, Dewdney J, Ausubel FM, De Lorenzo G, Ferrari S. (2008) The AtrbohD-mediated oxidative burst elicited by oligogalacturonides in Arabidopsis is dispensable for the activation of defense responses effective against Botrytis cinerea. Plant Physiol 148: 1695–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille S, Pauly M. (2012) O-Acetylation of plant cell wall polysaccharides. Front Plant Sci 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave AP. (1992) A versatile binary vector system with a T-DNA organizational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 6: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Gou JY, Miller LM, Hou G, Yu XH, Chen XY, Liu CJ. (2012) Acetylesterase-mediated deacetylation of pectin impairs cell elongation, pollen germination, and plant reproduction. Plant Cell 24: 50–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann T. (2012) Plant cell wall integrity maintenance as an essential component of biotic stress response mechanisms. Front Plant Sci 3: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann T, Bennett M, Mansfield J, Somerville C. (2009) Identification of cell-wall stress as a hexose-dependent and osmosensitive regulator of plant responses. Plant J 57: 1015–1026 [DOI] [PubMed] [Google Scholar]
- Haswell ES, Peyronnet R, Barbier-Brygoo H, Meyerowitz EM, Frachisse JM. (2008) Two MscS homologs provide mechanosensitive channel activities in the Arabidopsis root. Curr Biol 18: 730–734 [DOI] [PubMed] [Google Scholar]
- Jakab G, Ton J, Flors V, Zimmerli L, Métraux JP, Mauch-Mani B. (2005) Enhancing Arabidopsis salt and drought stress tolerance by chemical priming for its abscisic acid responses. Plant Physiol 139: 267–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan DB, Bowman MJ, Braker JD, Dien BS, Hector RE, Lee CC, Mertens JA, Wagschal K. (2012) Plant cell walls to ethanol. Biochem J 442: 241–252 [DOI] [PubMed] [Google Scholar]
- Jung HJ, Samac DA, Sarath G. (2012) Modifying crops to increase cell wall digestibility. Plant Sci 185-186: 65–77 [DOI] [PubMed] [Google Scholar]
- Koch JL, Nevins DJ. (1989) Tomato fruit cell wall. I. Use of purified tomato polygalacturonase and pectinmethylesterase to identify developmental changes in pectins. Plant Physiol 91: 816–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Teng Q, Zhong R, Ye ZH. (2011) The four Arabidopsis reduced wall acetylation genes are expressed in secondary wall-containing cells and required for the acetylation of xylan. Plant Cell Physiol 52: 1289–1301 [DOI] [PubMed] [Google Scholar]
- Lerouxel O, Choo TS, Séveno M, Usadel B, Faye L, Lerouge P, Pauly M. (2002) Rapid structural phenotyping of plant cell wall mutants by enzymatic oligosaccharide fingerprinting. Plant Physiol 130: 1754–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever M. (1972) A new reaction for colorimetric determination of carbohydrates. Anal Biochem 47: 273–279 [DOI] [PubMed] [Google Scholar]
- Limberg G, Körner R, Buchholt HC, Christensen TM, Roepstorff P, Mikkelsen JD. (2000) Analysis of different de-esterification mechanisms for pectin by enzymatic fingerprinting using endopectin lyase and endopolygalacturonase II from A. niger. Carbohydr Res 327: 293–307 [DOI] [PubMed] [Google Scholar]
- Linthorst HJM. (1991) Pathogenesis-related proteins of plants. Crit Rev Plant Sci 10: 123–150 [Google Scholar]
- Lionetti V, Cervone F, Bellincampi D. (2012) Methyl esterification of pectin plays a role during plant-pathogen interactions and affects plant resistance to diseases. J Plant Physiol 169: 1623–1630 [DOI] [PubMed] [Google Scholar]
- Lionetti V, Francocci F, Ferrari S, Volpi C, Bellincampi D, Galletti R, D’Ovidio R, De Lorenzo G, Cervone F. (2010) Engineering the cell wall by reducing de-methyl-esterified homogalacturonan improves saccharification of plant tissues for bioconversion. Proc Natl Acad Sci USA 107: 616–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionetti V, Raiola A, Camardella L, Giovane A, Obel N, Pauly M, Favaron F, Cervone F, Bellincampi D. (2007) Overexpression of pectin methylesterase inhibitors in Arabidopsis restricts fungal infection by Botrytis cinerea. Plant Physiol 143: 1871–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magi B, Liberatori S. (2005) Immunoblotting techniques. Methods Mol Biol 295: 227–254 [DOI] [PubMed] [Google Scholar]
- Manabe Y, Nafisi M, Verhertbruggen Y, Orfila C, Gille S, Rautengarten C, Cherk C, Marcus SE, Somerville S, Pauly M, et al. (2011) Loss-of-function mutation of REDUCED WALL ACETYLATION2 in Arabidopsis leads to reduced cell wall acetylation and increased resistance to Botrytis cinerea. Plant Physiol 155: 1068–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauch F, Mauch-Mani B, Boller T. (1988) Antifungal hydrolases in pea tissue. II. Inhibition of fungal growth by combinations of chitinase and β-1,3-glucanase. Plant Physiol 88: 936–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McComb EA, McCready RM. (1957) Determination of acetyl in pectin and in acetylated carbohydrate polymers. Anal Chem 29: 819–821 [Google Scholar]
- Nühse TS. (2012) Cell wall integrity signaling and innate immunity in plants. Front Plant Sci 3: 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obel N, Erben V, Schwarz T, Kühnel S, Fodor A, Pauly M. (2009) Microanalysis of plant cell wall polysaccharides. Mol Plant 2: 922–932 [DOI] [PubMed] [Google Scholar]
- Osorio S, Castillejo C, Quesada MA, Medina-Escobar N, Brownsey GJ, Suau R, Heredia A, Botella MA, Valpuesta V. (2008) Partial demethylation of oligogalacturonides by pectin methyl esterase 1 is required for eliciting defence responses in wild strawberry (Fragaria vesca). Plant J 54: 43–55 [DOI] [PubMed] [Google Scholar]
- Pattathil S, Avci U, Baldwin D, Swennes AG, McGill JA, Popper Z, Bootten T, Albert A, Davis RH, Chennareddy C, et al. (2010) A comprehensive toolkit of plant cell wall glycan-directed monoclonal antibodies. Plant Physiol 153: 514–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly M, Andersen LN, Kauppinen S, Kofod LV, York WS, Albersheim P, Darvill A. (1999) A xyloglucan-specific endo-beta-1,4-glucanase from Aspergillus aculeatus: expression cloning in yeast, purification and characterization of the recombinant enzyme. Glycobiology 9: 93–100 [DOI] [PubMed] [Google Scholar]
- Pauly M, Qin Q, Greene H, Albersheim P, Darvill A, York WS. (2001) Changes in the structure of xyloglucan during cell elongation. Planta 212: 842–850 [DOI] [PubMed] [Google Scholar]
- Peraldi A, Beccari G, Steed A, Nicholson P. (2011) Brachypodium distachyon: a new pathosystem to study Fusarium head blight and other Fusarium diseases of wheat. BMC Plant Biol 11: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone P, Hewage CM, Thomson AR, Bailey K, Sadler IH, Fry SC. (2002) Patterns of methyl and O-acetyl esterification in spinach pectins: new complexity. Phytochemistry 60: 67–77 [DOI] [PubMed] [Google Scholar]
- Pogorelko G, Fursova O, Lin M, Pyle E, Jass J, Zabotina OA. (2011) Post-synthetic modification of plant cell walls by expression of microbial hydrolases in the apoplast. Plant Mol Biol 77: 433–445 [DOI] [PubMed] [Google Scholar]
- Pogorelko GV, Fursova OV, Ogarkova OA, Tarasov VA. (2008) A new technique for activation tagging in Arabidopsis. Gene 414: 67–75 [DOI] [PubMed] [Google Scholar]
- Po-Wen C, Singh P, Zimmerli L. (2013) Priming of the Arabidopsis pattern-triggered immunity response upon infection by necrotrophic Pectobacterium carotovorum bacteria. Mol Plant Pathol 14: 58–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiola A, Lionetti V, Elmaghraby I, Immerzeel P, Mellerowicz EJ, Salvi G, Cervone F, Bellincampi D. (2011) Pectin methylesterase is induced in Arabidopsis upon infection and is necessary for a successful colonization by necrotrophic pathogens. Mol Plant Microbe Interact 24: 432–440 [DOI] [PubMed] [Google Scholar]
- Scheller HV, Ulvskov P. (2010) Hemicelluloses. Annu Rev Plant Biol 61: 263–289 [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Schuhegger R, Nafisi M, Mansourova M, Petersen BL, Olsen CE, Svatos A, Halkier BA, Glawischnig E. (2006) CYP71B15 (PAD3) catalyzes the final step in camalexin biosynthesis. Plant Physiol 141: 1248–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo PJ, Lee AK, Xiang F, Park CM. (2008) Molecular and functional profiling of Arabidopsis pathogenesis-related genes: insights into their roles in salt response of seed germination. Plant Cell Physiol 49: 334–344 [DOI] [PubMed] [Google Scholar]
- Somerville C, Bauer S, Brininstool G, Facette M, Hamann T, Milne J, Osborne E, Paredez A, Persson S, Raab T, et al. (2004) Toward a systems approach to understanding plant cell walls. Science 306: 2206–2211 [DOI] [PubMed] [Google Scholar]
- Song L, Siguier B, Dumon C, Bozonnet S, O’Donohue MJ. (2012) Engineering better biomass-degrading ability into a GH11 xylanase using a directed evolution strategy. Biotechnol Biofuels 5: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood W. (2012) The plant cell wall: a dynamic barrier against pathogen invasion. Front Plant Sci 3: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Hill T. (2008) High-efficiency Agrobacterium-mediated transformation of Brachypodium distachyon inbred line Bd21-3. Plant Cell Rep 27: 471–478 [DOI] [PubMed] [Google Scholar]
- Vogel JP, Raab TK, Schiff C, Somerville SC. (2002) PMR6, a pectate lyase-like gene required for powdery mildew susceptibility in Arabidopsis. Plant Cell 14: 2095–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Raab TK, Somerville CR, Somerville SC. (2004) Mutations in PMR5 result in powdery mildew resistance and altered cell wall composition. Plant J 40: 968–978 [DOI] [PubMed] [Google Scholar]
- Volpi C, Janni M, Lionetti V, Bellincampi D, Favaron F, D’Ovidio R. (2011) The ectopic expression of a pectin methyl esterase inhibitor increases pectin methyl esterification and limits fungal diseases in wheat. Mol Plant Microbe Interact 24: 1012–1019 [DOI] [PubMed] [Google Scholar]
- Vorwerk S, Somerville S, Somerville C. (2004) The role of plant cell wall polysaccharide composition in disease resistance. Trends Plant Sci 9: 203–209 [DOI] [PubMed] [Google Scholar]
- Wang Y, Kausch AP, Chandlee JM, Luo H, Ruemmele BA, Browning M, Jackson N, Goldsmith MR. (2003) Co-transfer and expression of chitinase, glucanase, and bar genes in creeping bentgrass for conferring fungal disease resistance. Plant Sci 165: 497–506 [Google Scholar]
- Weng X, Huang Y, Hou C, Jiang D. (2012) Effects of an exogenous xylanase gene expression on the growth of transgenic rice and the expression level of endogenous xylanase inhibitor gene RIXI. J Sci Food Agric 93: 173–179 [DOI] [PubMed] [Google Scholar]
- Willats WG, Orfila C, Limberg G, Buchholt HC, van Alebeek GJ, Voragen AG, Marcus SE, Christensen TM, Mikkelsen JD, Murray BS, et al. (2001) Modulation of the degree and pattern of methyl-esterification of pectic homogalacturonan in plant cell walls. Implications for pectin methyl esterase action, matrix properties, and cell adhesion. J Biol Chem 276: 19404–19413 [DOI] [PubMed] [Google Scholar]
- Wolf S, Hématy K, Höfte H. (2012a) Growth control and cell wall signaling in plants. Annu Rev Plant Biol 63: 381–407 [DOI] [PubMed] [Google Scholar]
- Wolf S, Mouille G, Pelloux J. (2009) Homogalacturonan methyl-esterification and plant development. Mol Plant 2: 851–860 [DOI] [PubMed] [Google Scholar]
- Wolf S, Mravec J, Greiner S, Mouille G, Höfte H. (2012b) Plant cell wall homeostasis is mediated by brassinosteroid feedback signaling. Curr Biol 22: 1732–1737 [DOI] [PubMed] [Google Scholar]
- Zabotina O, Malm E, Drakakaki G, Bulone V, Raikhel N. (2008) Identification and preliminary characterization of a new chemical affecting glucosyltransferase activities involved in plant cell wall biosynthesis. Mol Plant 1: 977–989 [DOI] [PubMed] [Google Scholar]