Abstract
Pichia pastoris is an efficient expression system for production of recombinant proteins. To understand its physiology for building novel applications it is important to understand and reconstruct its metabolic network. The metabolic reconstruction approach connects genotype with phenotype. Here, we have attempted to reconstruct carbohydrate metabolism pathways responsible for high biomass density and N-glycosylation pathways involved in the post translational modification of proteins of P. pastoris CBS7435. Both these metabolic pathways play a crucial role in heterologous protein production. We report novel, missing and unannotated enzymes involved in the target metabolic pathways. A strong possibility of cellulose and xylose metabolic processes in P. pastoris CBS7435 suggests its use in the area of biofuels. The reconstructed metabolic networks can be used for increased yields and improved product quality, for designing appropriate growth medium, for production of recombinant therapeutics and for making biofuels.
Keywords: Pichia, Reconstruction, Carbohydrate, System biology
Introduction
Yeasts are being continuously used for producing eukaryotic foreign proteins as they offer easy microbial growth and gene manipulations like those of bacteria, as well as possess eukaryotic characteristics (Balamurugan et al. 2007). The methylotrophic yeast Pichia pastoris has emerged as an efficient expression system for production of recombinant proteins for basic research and medical applications. It has a natural ability to do so, owing to its high cell density, a strong methanol inducible AOX promoter, post-translational modifications of the protein (glycosylation) and efficient secretion of these proteins into the culture medium itself (Schutter et al. 2009). A large number of heterologous proteins have been produced in P. pastoris so far. Escherichia coli, another prokaryotic expression system, can also be used for recombinant protein production but it does not carry out the post-translational modifications of proteins like folding, disulphide bond formation and glycosylation (Yadava and Ockenhouse 2003). As a result P. pastoris, as an expression system, is given preference over other systems. After E. coli, Saccharomyces cerevisiae emerged as an efficient expression system and is still being used as a model organism in laboratories for expression of recombinant proteins. However, the glycosylation pattern of S. cerevisiae gave rise to hyperglycosylated proteins which were deemed to be inactive. On the other hand, P. pastoris glycosylation pathways have been re-engineered to produce mammalian-type glycosylated proteins and availability of such engineered strains has increased its use in industrial production of therapeutic proteins (Hu et al. 2011). Another major advantage of P. pastoris over other expression vectors is that it can secrete the produced recombinant protein into the growth medium. This makes isolation of the expressed protein easier because the endogenous proteins of P. pastoris are not secreted (Weidner et al. 2010).
With the increase in genome sequencing projects, understanding of the biological capabilities of the organisms has also increased. On the basis of availability of annotated genomes all the relevant information about the metabolic capacity of the organisms can be deciphered, which can further be used to give rise to genome scale metabolic networks (Notebaart et al. 2006). The number of organisms for which metabolic networks have been reconstructed is constantly increasing, since it gives us an insight into the systems biology of metabolism of a given organism. The process of metabolic reconstruction involves identifying, characterizing and interconnecting the genes, proteins, reactions and metabolites participating in the metabolic activity of an organism (Feist et al. 2006). The metabolic networks provide information about numerous relationships between gene and gene products. It’s an organism specific process since it uses genomic information to associate genes with enzymatic activities (Kharchenko et al. 2004). The genome scale metabolic network reconstruction is a manual and an iterative process that may take up to one-man year to collect all organism specific reaction information (Forster et al. 2003). To overcome this problem of time consumption automation at the level of annotation using similarity searches has cropped up as the most obvious and convenient option, but this technique leaves behind a lot of gaps, which need to be filled during reconstruction process (DeJongh et al. 2007).
The use of reconstructed metabolic networks has increased since their advent nearly a decade ago. Reconstructed networks highlight the differences and similarities between intracellular mechanisms of various species and are hence important for comparative studies (Mazurie et al. 2010). Identification of potential drug targets and the causative reactions of a disease is also possible due to availability of metabolic network models (Folger et al. 2011). One of the most significant applications of metabolic network reconstruction is metabolic engineering which involves altering the metabolism to improve a desired cellular function. The reconstructed networks help in choosing a target, most commonly enzymes, for metabolic engineering (Oberhardt et al. 2009). Pichia pastoris can be used for production of therapeutic glycoproteins and thus, is a promising host for industrial production of such proteins. Therefore it becomes extremely important to understand its physiology and work towards its improvement. Metabolic networks can help in identifying the potential pathway targets of P. pastoris that can be metabolically engineered so as to give rise to an improved strain, and hence improved yields of the recombinant proteins produced by it (Chung et al. 2010).
In the present study we reconstruct the metabolic network of the pathways of the carbohydrate metabolism and the N-glycosylation pathways of P. pastoris CBS7435. Since, the high biomass density of P. pastoris, obtained as a result of carbon and energy metabolism, gives it an edge over other competent expression systems, if we choose to target and reconstruct this particular metabolism of P. pastoris and elucidate its unique and important features to identify the growth requirements of the organism, we are able to design an appropriate growth medium that would favor higher biomass yields. Since production of biofuels depends largely on the carbohydrate metabolism we try to ascertain if P. pastoris can be employed for production of biofuels by exploring its genome to locate genes and gene products (enzymes) that may use the carbohydrates required for biofuel production. P. pastoris carries the machinery for mammalian type post-translational modifications like glycosylation, which makes it a preferred choice for production of therapeutic proteins. As a result the N-glycosylation pathway of other strains of P. pastoris has been engineered earlier to produce humanized glycoproteins for therapeutic purposes. In the present study, we reconstruct the metabolic network of this pathway on similar lines for P. pastoris CBS7435 so that it can be metabolically engineered to produce human like proteins in future, as and when required.
Materials and methods
The process of metabolic reconstruction is labor-intensive and time consuming, and ranges from 6 months for a small bacterium, to about 2 years for humans. Despite several efforts the process has not yet been completely automated (Thiele and Palsson 2010). Following basic steps are involved in reconstructing an organism specific metabolic network:
Annotating the genome of the organism to identify the enzymes encoded by its genes,
Assembling the reactions catalyzed by those enzymes,
Verifying the correctness and completeness of the assembled reaction network.
These steps depict an iterative process (DeJongh et al. 2007). Several approaches have been proposed to analyze metabolic networks for newly sequenced genomes. The most common approach makes use of metabolic pathway databases through which it can be ascertained if a particular pathway enzymes are encoded in the genome of the organism (Boyer and Viari 2003).
In the present reconstruction process, following steps were undertaken:
Step1: Retrieval of annotated genome of Pichia pastoris CBS7435
Reconstruction process relies heavily on genome annotation, therefore it becomes extremely important to download or retrieve the most recent version of the genome (Thiele and Palsson 2010). Pichia pastoris CBS7435 is the parent strain of all P. pastoris recombinant protein production hosts. Its genome sequence was annotated automatically to yield 5,007 protein-coding genes, 124 tRNAs and 29 rRNAs and the complete DNA sequence of the first mitochondrial genome of a methylotrophic yeast (Kuberl et al. 2011).
The annotated genome sequence of P. pastoris CBS7435 comprises of 4 chromosomes (GenBank IDs: FR839628.1, FR839629.1, FR839630.1 and FR839631.1) and a mitochondrial DNA sequence (GenBank ID: FR839632.1). The Institute of Molecular Biotechnology of the Graz University of Technology, Austria, sequenced the genome of P. pastoris CBS7435 and the Institute of Genomics and Bioinformatics, Graz University of Technology, registered it in the Genome database of NCBI (www.ncbi.nlm.nih.gov/genome). The accession number of the genome in NCBI is PRJEA62483, and ID is 62483.
Step 2: Enzymes involved in the carbohydrate metabolism and N-glycosylation pathways from the pathway databases
For reconstructing metabolic networks, it is necessary to identify the completely balanced enzyme catalyzed reactions. The metabolic pathway databases like KEGG (Kanehisa et al. 2006) provide comprehensive information about the enzyme catalyzed reactions, chemical information about the enzymes, stoichiometries of the reaction, etc. (Durot et al. 2008).
The enzymes involved in the pathways of the carbohydrate metabolism and the N-glycosylation pathways were retrieved using the information related with metabolic pathways on KEGG PATHWAY database (www.genome.jp/kegg/pathway). The carbohydrate metabolism in KEGG comprises of 15 pathways, namely, glycolysis, gluconeogenesis, TCA cycle, pentose phosphate pathway, fructose and mannose metabolism, starch and sucrose metabolism, amino sugar and nucleotide sugar metabolism, galactose metabolism, inositol phosphate metabolism, glyoxalate and dicarboxylate metabolism, pyruvtae metabolism, butanoate metabolism, propanoate metabolism, ascorbate and aldarate metabolism, C5 branched dibasic acid metabolism and pentose and glucuronate conversions. The N-glycosylation pathways are classified under the category of glycan biosynthesis and metabolism. The names and E.C. numbers of all the enzymes involved in each reference pathway were retrieved from the KEGG ENZYME list via LinkDB search (http://www.genome.jp/dbget-bin/get_linkdb) (Table 1).
Table 1.
List of number of enzymes involved in target pathways
| Pathway | Number of enzymes |
|---|---|
| Glycolysis and gluconeogenesis | 45 |
| Citric acid cycle | 22 |
| Pentose phosphate pathway | 40 |
| Pentose and glucuronate interconversions | 60 |
| Fructose and mannose metabolism | 65 |
| Galactose metabolism | 38 |
| Ascorbate and aldarate metabolism | 45 |
| Starch and sucrose metabolism | 71 |
| Amino sugar and nucleotide sugar metabolism | 103 |
| Pyruvate metabolism | 64 |
| Glyoxylate and dicarboxylate metabolism | 66 |
| Propanoate metabolism | 47 |
| Butanoate metabolism | 50 |
| C5-branched dibasic acid metabolism | 18 |
| Inositol phosphate metabolism | 43 |
| N-glycan biosynthesis | 34 |
| Various types of N-glycan biosynthesis | 25 |
Step 3: Location of enzymes in the genome of Pichia pastoris CBS7435
After all the relevant enzymes were obtained the genome of P. pastoris was browsed to find the chromosomal locations of these enzymes, their coding sequence regions as well as the genes encoding those enzymes. All the chromosomes were thoroughly explored to locate the respective E.C. numbers of the enzymes involved in the target pathways. The locus tag of P. pastoris genome is PP7435 hence every chromosomal location is prefixed with this locus tag. It was a time consuming step since a lot of enzymes were associated with every pathway and all of them had to be searched in the entire genome.
Step 4: Identification of enzyme catalyzed reactions and enzyme compartments
The next step was to identify the enzyme catalyzed reactions alongwith the cellular location of those enzymes. The reactions were retrieved from the ENZYME database of the expert protein analysis system (ExPasy) server of the Swiss Institute of Bioinformatics (SIB). ExPasy provides access to a number of bioinformatics tools and databases centered on proteins and proteomics (Gasteiger et al. 2003).
The ENZYME database of ExPasy was queried using the E.C. numbers of the enzymes to obtain the reactions catalyzed by them. Since the database is cross linked to UniProt (Universal Protein Resource) (Bairoch et al. 2005), sub-cellular localization of enzyme could be ascertained by selecting the appropriate gene name of the corresponding enzyme as present in yeast from the available list on UniProt. This particular step therefore helps in acquisition of organism specific enzyme information, i.e., enzyme name, gene name, alternative gene name, reaction, pathway, description, sub cellular location, sequence, etc.
Step 5: Enlisting the transporters involved in the target pathways
In eukaryotes, intra-cellular transport of metabolites between cellular compartments occurs in case of multi-compartment networks. A compartment in the metabolic network corresponds to a pool of metabolites and their reactions (Schellenberger et al. 2010). Transport reactions are added to the networks to emphasize on the transfer of metabolites from one compartment to another and hence maintain the continuity of the network. The transporters could either be proteins or simple channels (in case of extracellular transport). Various databases and literature evidences impart information about the transporters carrying out the metabolite transport inside the cell.
To retrieve the transporters involved in the target pathways YMDB (Jewison et al. 2012) was accessed (http://www.ymdb.ca). A simple metabolite search is carried out in YMDB to obtain all the metabolite related information, including its transporters. These transporters are further located in the genome of P. pastoris CBS7453 to extract their chromosomal locations and coding sequences. In cases where YMDB could not provide any specific information about the transporters, a thorough literature survey was resorted to for the elucidation of the transport mechanisms.
Step 6: Draft reconstruction
All the relevant information about the pathways and their components, based on genome annotation, pathway databases and other computational tools, was compiled to generate a draft network reconstruction. Draft reconstruction is a preliminary network based on genome annotation and pathway databases like KEGG. All the metabolic reactions of a particular pathway were aligned in a path-by-path fashion to construct a network using KEGG pathway maps as template. Apart from KEGG, certain pathways were also reconstructed using Metacyc (Karp et al. 2002), another pathway database. There existed a few pathways in Metacyc which were absent from KEGG even though they occurred in P. pastoris as well as its nearest phylogenetic neighbor S. cerevisiae , the evidence for which was supported by literature. Such pathways again served as templates to reconstruct the metabolic network. The entire procedure of draft reconstruction was repeated for Metacyc pathways.
Step 7: Manual curation
Manual curation is the second major stage after draft reconstruction in the metabolic network reconstruction process. Manual curation of the draft network involves its re-evaluation and refinement. The need to curate the draft network arises from the fact that the data present in the databases used so far might contain a certain amount of non-specific information, especially the reactions and their cellular compartments. Hence for the authenticity of the reconstructed network organism-specific metabolites and reactions had to be added (Montagud et al. 2010). The draft network might also contain several dead end compounds, or what we call as missing links, which might be due to a faulty annotation or some missing reaction that links these compounds or metabolites with the rest of the pathway. It’s a sort of gap in the network and thus refinement of the network through gap analysis becomes mandatory (Ates et al. 2011). The term gap in metabolic reconstruction can also be used to describe metabolites that cannot be produced or consumed by any of the reaction or that cannot be imported or exported by an uptake system (Kumar et al. 2007). Gap filling is a tedious process that requires browsing organism-specific databases, literature survey, etc. to obtain highly organism-specific information about its metabolism, like reaction compartment and its directionality (Feist et al. 2009).
The reconstructed networks can be refined by finding the missing links in the network and thereby filling the gaps. Gap filling can be achieved by following approaches:
-
Sequence homology—a faulty genome annotation process might lead to what is known as ‘missing gene problem’. These missing genes are established by evidences indicating the presence of the metabolic pathway in the organism, one or more enzymes of which are missing from the reconstructed draft network, and identifying the genes encoding those enzymes (Osterman and Overbeek 2003). In such a case sequence homology for the missing gene sequence can be performed using sequence alignment tools like BLAST (Altschul et al. 1997).
The nucleotide sequences of enzymes present in the KEGG pathway maps of P. pastoris GS115 were aligned with the nucleotide sequence of all four chromosomes of P. pastoris CBS7435 to find a homologous sequence using nucleotide BLAST (Blastn) (http://blast.ncbi.nlm.nih.gov/). If a match was found the amino acid sequence of the matched gene was retrieved and aligned with the amino acid sequence of the missing enzyme using protein BLAST (Blastp) (http://blast.ncbi.nlm.nih.gov/).
-
Comparative analysis—If organism-specific information has insufficient data then information from phylogenetic neighbors can also be used (Thiele and Palsson 2010). Missing enzymes can also be established through comparative analysis between related species. For instance if a pathway is present in P. pastoris as well as its phylogenetic ancestor S. cerevisiae then it’s most likely that the genes encoding the enzymes of that pathway will be present in both the genomes and the missing enzyme or protein in P. pastoris having highest sequence homology to the orthologous S. cerevisiae protein most probably carries out the missing reactions (Seo and Lewin 2009).
Saccharomyces cerevisiae, being the nearest phylogenetic neighbor of P. pastoris CBS7435, can provide interesting insights into the metabolism of P. pastoris. Hence the enzymes missing in the metabolic network of P. pastoris were picked, if present, from the pathway map of S. cerevisiae in KEGG. If a sequence, homologous to the sequence of S. cerevisiae enzyme was found, the reaction catalyzed by that particular enzyme was introduced into the network. Also saccharomyces genome database (SGD) (Cherry et al. 2012) was used to decipher additional pathway reactions in P. pastoris which were otherwise not a part of KEGG PATHWAY database.
Manual Insertion—Certain important enzymes missing from the reconstructed network without which the pathway cannot progress can be manually inserted based on rigorous literature evidence (Milne et al. 2011).
Step 8: Reconstruction of refined/curated network
A good metabolic network reconstruction is the first step towards understanding genotype-phenotype interactions of an organism (Francke et al. 2005). After the draft network has been curated the final refined metabolic network is reconstructed. Many metabolic networks have been reconstructed using various tools, like Pathway Tools. A recent addition to these tools is MetNetMaker (Forth et al. 2010).
The reactions of the carbohydrate metabolism and N-glycosylation were retrieved from MetNetMaker by simply entering the E.C. numbers of the enzymes catalyzing those reactions. Another alternative to obtain required reactions is by entering the reaction IDs which are same as that of KEGG. The reaction compartment and directionality are also mentioned, gathered through extensive literature survey. In certain cases the reaction to be added, or the compounds/metabolites involved in a reaction were not a part of KEGG LIGAND database, to which MetNetMaker is linked. In such cases reactions and metabolites with an unique reaction and compound ID respectively, were created. The compartments for such reactions and the metabolites involved in them were also mentioned. Such created reactions were added to the selected reaction table that carried information for reaction ID, E.C. number of the enzymes, reaction compartment, maximum and minimum possible flux for each reaction, etc.
Step 9: Visualization of the reconstructed networks
The metabolic networks created are specific for P. pastoris CBS7435. Hence their visualization would provide an easy insight into the metabolism of this particular organism. Visualization also enables users to investigate the pathways step by step and to compare them with proposed pathways. There are many network visualization tools present but we chose Cytoscape for our analysis (Shannon et al. 2003). The tool MetNetMaker used for the reconstruction of the metabolic networks provides a link to Cytoscape that enables the reconstructed networks to be visualized properly. The most appropriate layout of the network from Cytoscape was selected for this purpose.
Results and discussion
During the data collection stage it was found that in few pathways one or two additional enzymes, which were not a part of P. pastoris specific pathways of KEGG, were being encoded in the genome of P. pastoris CBS7435 (Table 2).
Table 2.
Novel enzymes of target pathways in P. pastoris CBS7435 genome
| S. no. | Metabolism | Enzyme | E. C. no. | Chromosomal location | CDS | Reaction |
|---|---|---|---|---|---|---|
| 1. | Fructose and mannose metabolism | d-glucitol: NAD+2-oxido reductase | 1.1.1.14 | PP7435_Chr1-0597 | 1089776–1090822 | d-Sorbitol + NAD+ <=> d-Fructose + NADH + H+ |
| 2. | Butanoate metabolism | Succinate semi aldehyde dehydro genase | 1.2.1.16 | PP7435_Chr1-0018 | 45335–46822 | Succinate semi aldehyde + NAD(P)(+) + H(2)O <=> succinate + NAD(P)H |
| 3. | Starch and sucrose metabolism | Cellulase | 3.2.1.4 | PP7435_Chr4-0326 | 535677–537521 | Cellulose + H2O <=> Cellulose + Cellobiose |
| 4. | Starch and sucrose metabolism | 1,4-alpha-glucan branching enzyme | 2.4.1.18 | PP7435_Chr1-0029, PP7435_Chr1-1064 | 62426..64528, 1919047..1919889 | Amylose <=> Starch |
| 5. | Inositol phosphate metabolism | 1-phosphatidylinositol-3-phosphate 5-kinase | 2.7.1.150 | PP7435_Chr3-0252 | 453935..459913 | ATP + 1-phosphatidyl-1D-myo-inositol 3-phosphate → ADP + 1-phosphatidyl-1D-myo-inositol 3,5-bisphosphate |
The genes encoding these additional enzymes were present in its genome with their complete annotation. Therefore these enzymes were added in the reaction table. This is a slight deviation from the KEGG pathway maps and implies that P. pastoris GS115 (organism in KEGG) and P. pastoris CBS7435 which happens to be the wild type or the parent strain, differ from each other with respect to certain pathway reactions.
Apart from certain additional enzymes, gaps or missing links leading to discontinuities in a network were also reported (Table 3).
Table 3.
List of missing enzymes of carbohydrate and N-glycosylation pathways in P. pastoris CBS7435
| S. no. | Metabolism | Enzyme | E.C. no. | Reaction | Cellular location | Missing link | Reference |
|---|---|---|---|---|---|---|---|
| 1. | Inositol Phosphate metabolism | Inositol oxygenase | 1.13.99.1 | Myo-inositol + O(2) <=> d-glucuronate + H(2)O | Membrane | Present as a hypothetical protein | KEGG pathway map specific for P. pastoris |
| 2. | Inositol Phosphate metabolism | 1-phosphatidyl inositol-4-phosphate-5-kinase | 2.7.1.68 | ATP + 1-phosphatidyl-1D-myo-inositol 4-phosphate <=> ADP + 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate | Membrane | Present as a hypothetical protein | KEGG pathway map specific for P. pastoris |
| 3. | Inositol Phosphate metabolism | 1-phosphatidyl inositol-4,5- biphosphate phospho diesterase | 3.1.4.11 | 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate + H(2)O <=> 1D-myo-inositol 1,4,5-trisphosphate + diacylglycerol | Membrane | Present as a hypothetical protein | KEGG pathway map specific for P. pastoris |
| 4. | Starch and sucrose metabolism | Trehalose phospahatase | 3.1.3.12 | alpha,alpha’-Trehalose 6-phosphate + H2O <=> alpha,alpha-Trehalose + Ortho phosphate | Cytosol | Present as a hypothetical protein. | KEGG pathway map specific for P. pastoris |
| 5. | Starch and sucrose metabolism | Glucoamylase | 3.2.1.3 | Starch + H2O <=> alpha-d-Glucose + Starch, Dextrin + H2O <=> alpha-d-Glucose + Dextrin | Cytosol | Present as a hypothetical protein | KEGG pathway map specific for P. pastoris |
| 6. | Butanoate metabolism | 4-aminobutyrate amino transferase | 2.6.1.19 | 4-amino butanoate + 2-oxoglutarate <=> succinate semialdehyde + l-glutamate | Cytosol | Present as an orthologous enzyme (S)-3-amino-2-methyl propionate trans aminase (E.C. No 2.6.1.22) | KEGG pathway map specific for P. pastoris |
| 7. | Butanoate metabolism | HMG-CoA synthase | 2.3.3.10 | Acetyl-CoA + H(2)O + acetoacetyl-CoA <=> (S)-3-hydroxy-3-methylglutaryl-CoA + CoA | Cytosol | Present as an orthologous protein called pleiotropic drug resistance protein-1 | KEGG pathway map specific for P. pastoris |
| 8. | Propanoate metabolism | Malonate semialdehyde dehydrogenase (acetylating) | 1.2.1.18 | 3-Oxo propanoate + CoA + NADP + <=> Malonyl-CoA + NADPH + H+ | Mitochondria | Present as an orthologous enzyme methyl malonate semi aldehyde dehydro genase (E.C. No. 1.2.1.27) | KEGG pathway map specific for P. pastoris |
| 9. | N-glycan biosynthesis | Dolichyl phosphatase | 3.6.1.43 | Dolichyl diphosphate + H(2)O <=> dolichyl phosphate + phosphate | Endoplasmic Reticulum (ER) | Present as a putative membrane protein | KEGG pathway map specific for P. pastoris |
| 10. | Pyruvate metabolism | d-lactate dehydrogenase | 1.1.2.4 | (R)-Lactate + 2 Ferri cytochrome c <=> Pyruvate + 2 Ferro cytochrome c + 2 H+ | Mitochondria | Present as a hypothetical protein | KEGG pathway map specific for P. pastoris |
| 11. | Pyruvate metabolism | Hydroxyacyl glutathione hydrolase | 3.1.2.6 | (R)-S-Lactoyl glutathione + H2O <=> Glutathione + (R)-Lactate | Cytosol | Present as a hypothetical protein | KEGG pathway map specific for P. pastoris |
| 12. | Glycogen degradation | Glycogen debranching enzyme | 2.4.1.25 | limit dextrin → limit dextrin with short branches | Cytosol | Present as orthologous enzyme 4α- glucano transferase (E.C. No. 3.2.1.33) | MetaCyc pathway map for S. cerevisiae |
| 13. | Fructose and mannose metabolism | Aldehyde reductase | 1.1.1.21 | Alditol + NAD(P)(+) <=> aldose + NAD(P)H | Cytosol | Present as a hypothetical protein | KEGG pathway map specific for S. cerevisiae |
| 14. | Pentose and glucuronate interconversion | Aldehyde reductase | 1.1.1.21 | Xylitol + NADP + <=> d-Xylose + NADPH + H+ | Cytosol | Present as a hypothetical protein | KEGG pathway map specific for S. cerevisiae |
| 15. | Pentose and glucuronate interconversion | d-xylulose reductase | 1.1.1.9 | Xylitol + NAD + <=> d-Xylulose + NADH + H+ | Cytosol | Present as orthologous enzyme polyol dehydro genase (E.C. No. 1.1.1.14) | KEGG pathway map specific for S. cerevisiae |
| 16. | Gluconeogenesis | Alcohol dehydrogenase (NADP +) | 1.1.1.2 | Ethanol + NADP + <=> Acetaldehyde + NADPH + H+ | Cytosol | Present as a hypothetical protein | KEGG pathway map specific for P. pastoris |
The problem of missing enzymes can arise due to faults in genome annotation. The sequence homology search between the enzyme sequence of P. pastoris GS115 and P. pastoris CBS7435 and in a few cases S. cerevisiae, using BLASTn and BLASTp, yielded 100 % identity between the two nucleotide and protein sequences respectively for almost every missing enzyme. The dot matrix or dot plot showing maximum similarity was also provided in the result. The exact alignments between proteins further authenticated the results of Blastn indicating that the protein products (enzymes) of the found genes (nucleotide sequences) are also highly related or most similar to each other, thus confirming our sequence homology results. This way all missing enzymes were located in the genome of P. pastoris CBS7435. However, none of the enzymes involved in galactose metabolism were encoded by the genome of P. pastoris CBS7435, indicating that galactose is not assimilated by it. There have been previous reports of non-occurrence of galactose metabolism in P. pastoris GS115 in the past where it was proved that it cannot assimilate galactose (Kurtzman 2005). Since P. pastoris CBS7435 happens to be its parent strain it’s quite obvious that the assimilatory pathway of galactose does not occur in it as well. However attempts have been made to genetically engineer P. pastoris strains to use galactose as a carbon source (Davidson et al. 2012).
Presence of genes like OCH1 (Ha et al. 2011) and alg3 (Bobrowicz et al. 2004) in P. pastoris CBS7435 which play a vital role in the humanization process of N-linked glycosylation suggests that the N-glycosylation pathway of P. pastoris CBS7435 can also be engineered to produce human-like glycoproteins. This would mean that production of recombinant therapeutics, safe for human consumption, is possible in this particular strain also.
This study also reveals the possibility of existence of cellulose and xylose degradation pathways in P. pastoris CBS7435. Till now there has not been any report of existence of cellulose degradation in either S. cerevisiae or P. pastoris GS115 (the most studied strain of P. pastoris). This is because they both lack the very first enzyme of the pathway, i.e., endoglucanase (E.C. 3.2.1.4) which breaks down cellulose into cellobiose and 1,4-β-d-glucan. S. cerevisiae also lacks the second enzyme converting the products of first reaction into β-d-glucose. This enzyme is β-glucosidase (E.C. 3.2.1.21). However both these enzymes are encoded in the genome of P. pastoris CBS7435. This implies that there might be a possibility of occurrence of cellulose degradation pathway in it. No yeast so far has been reported to exhibit an efficient cellulose digesting activity, except Trichosporon species. Other yeasts like S. cerevisiae have been genetically engineered for this purpose because they lack the natural capacity to do so (Van Rensburg et al. 1998; Zhang et al. 2012). Similarly, the enzymes required for assimilation of xylose, a five-carbon sugar, were found to exist in the genome of P. pastoris CBS7435. So far none of the strains of this species have been reported to degrade xylose which is a major component of lignocellulosic biomass hydrolysates. It only occurs in Pichia stipitis amongst all methylotrophic yeasts. Out of the four enzymes involved in its degradation, two were already annotated in the genome while the remaining two had to be located using sequence homology. If this pathway is validated experimentally researchers would be able to do away with the cumbersome task of expressing the genes of these enzymes from different organisms in P. pastoris to bring about xylose degradation in this species. Subsequently its own genes can be over-expressed to degrade lignocelluloses.
Assimilation of xylose and cellulose by P. pastoris might also pave way for its use in biofuel industry. Biofuel is the need of the hour because it is cost effective, obtained from renewable resources, is environment friendly, etc. It is obtained as a by-product of degradation of various agricultural wastes by microorganisms and therefore involves their natural metabolic capacity to assimilate cellulosic and lignocellulosic wastes of plants and crops. Bioethanol produced by yeasts via glucose fermentation is a common example of biofuel. Other important substrates used for this purpose are cellulose and xylose. Their fermentation is essential for production of biofuels. However most of the yeast species are unable to utilize them. Although P. pastoris is largely considered as non-fermentative, formation of ethanol as a by-product on being fed with excess glycerol, has been reported by Inan and Meagher (2001). Presence of genes encoding the enzymes of cellulose and xylose degradation pathways in P. pastoris indicates the possible assimilation of these substrates for biofuel production. If this hypothesis is validated through experiments it might be major breakthrough in the field of biofuel production using yeast. It can then further be genetically engineered for this purpose. It would further lend P. pastoris CBS7435 an additional advantage of being an efficient producer of biofuels apart from its established role as an expression system and heterologous protein production host.
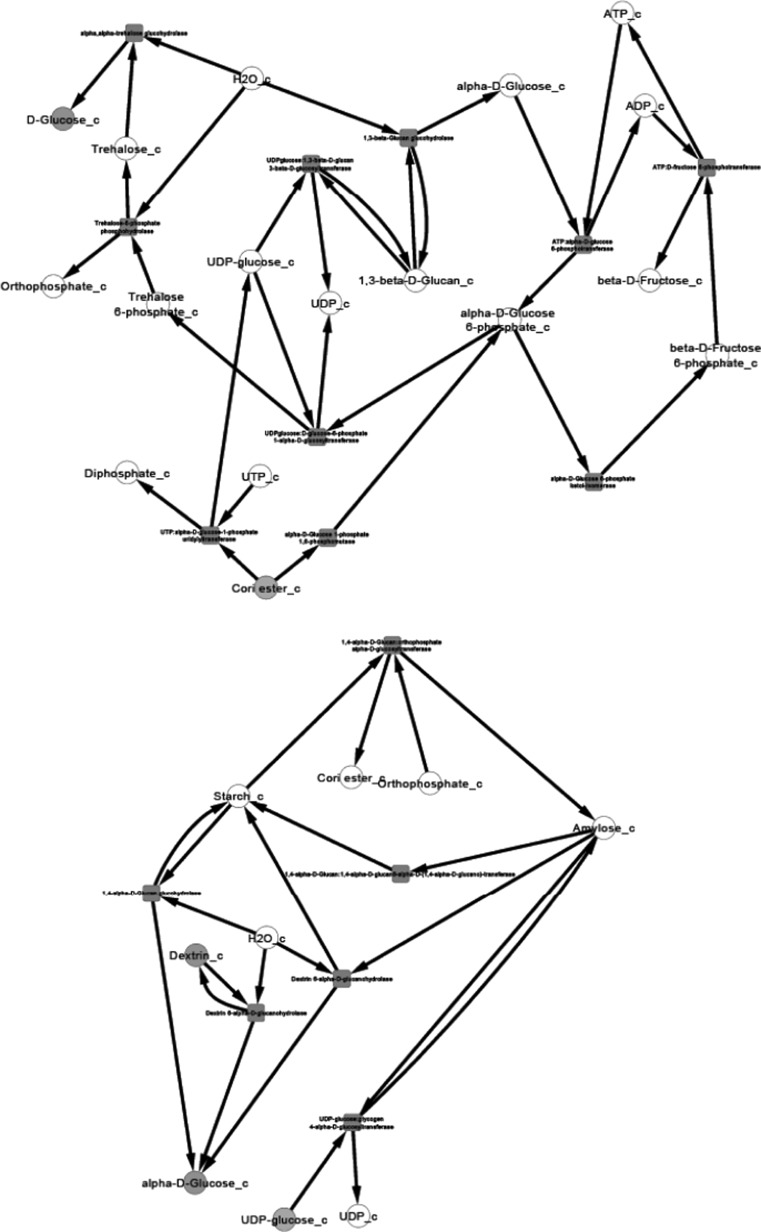
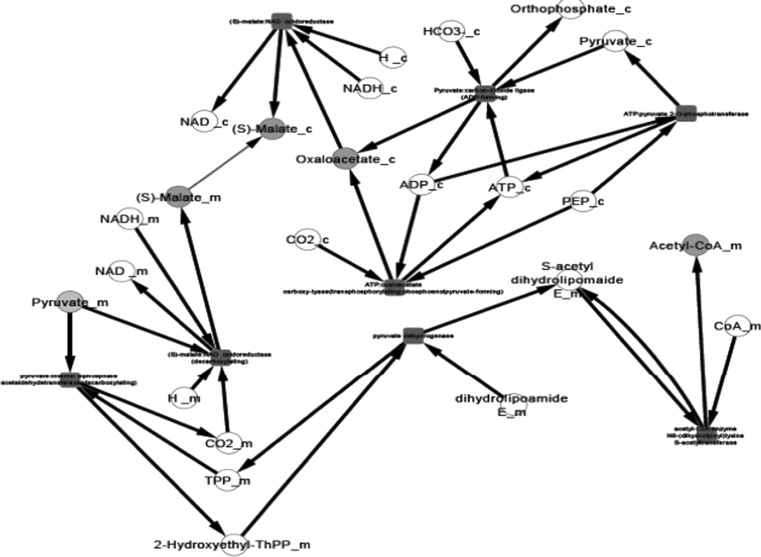
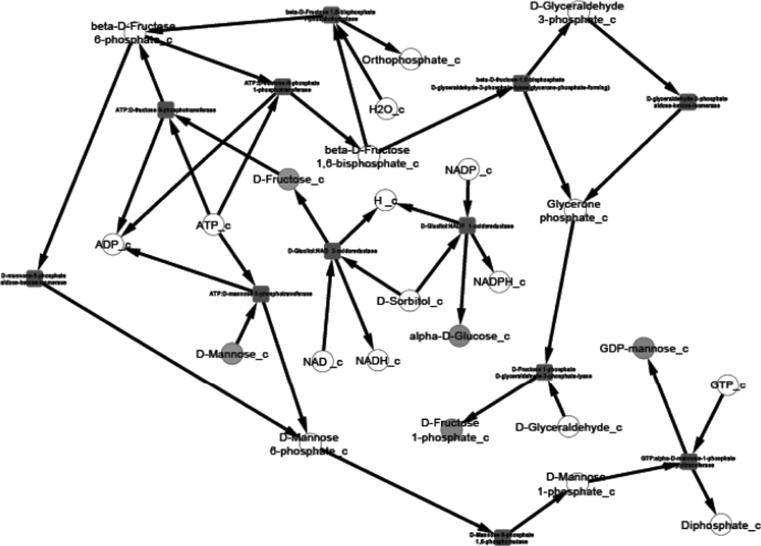
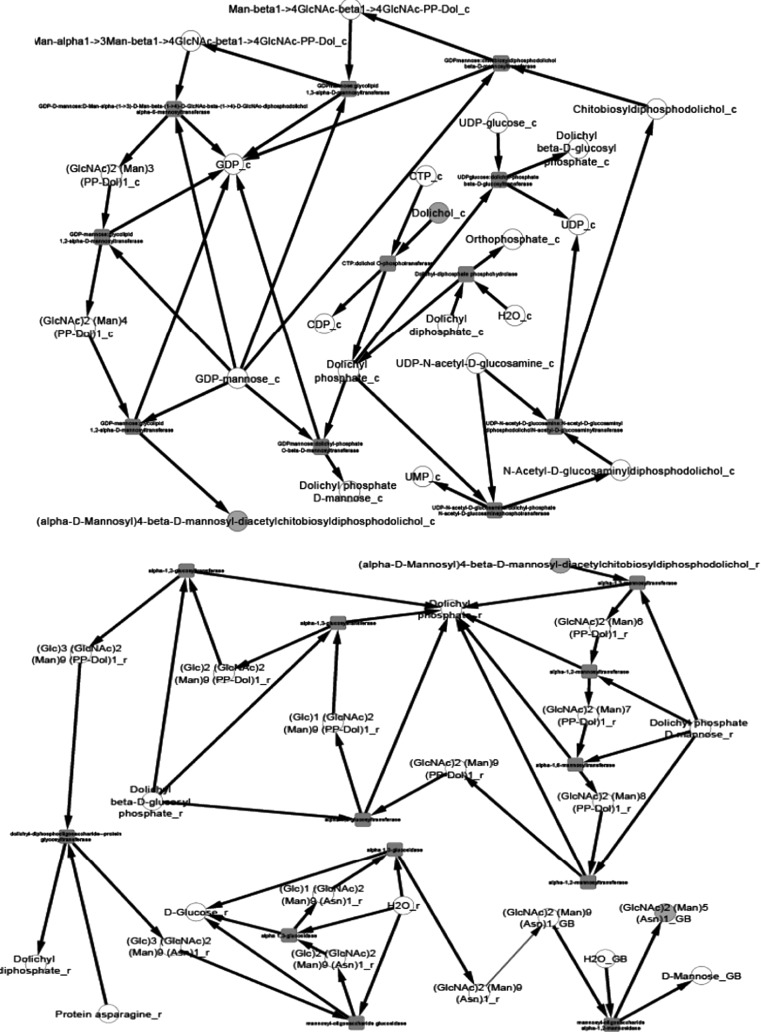
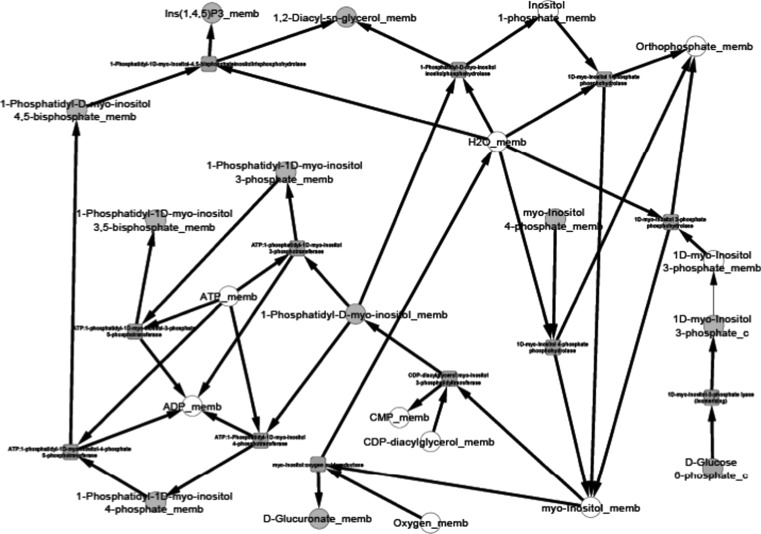
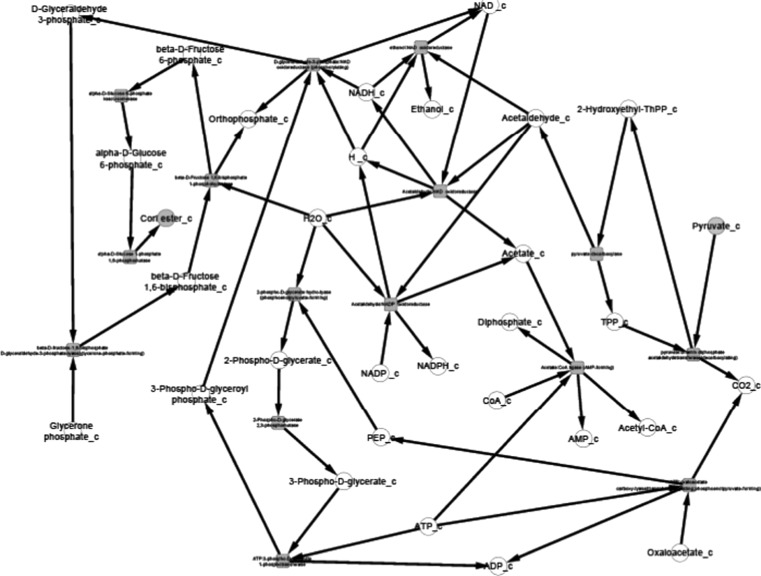
After the compilation of all the relevant data and their organization into pathway networks the latter were finally visualized. The reconstructed networks are highly organism-specific. The layout of the networks in Cytoscape given here is the grid view. The reactions are aligned in a pathway manner and represented with black edges. The blue color nodes represent the starting metabolites/compounds in a pathway while orange nodes represent the end products. The transport reactions are depicted with a purple edge. The enzymes are highlighted with the red blocks. The direction of edges from the starting metabolite to the end product enable tracing of the pathway in the organism. For further simplifying the view of the networks some of the pathways were divided based on compartments in case of multi compartment pathways. This was especially done for the N-glycosylation pathways and various types of N-glycosylation pathways, where the portion of cycle occurring in Endoplasmic Reticulum was separated from that occurring in Golgi body for the ease of visualization.
Pyruvate metabolism, another complex pathway, was divided into two based on the choice of starting metabolite. All reactions beginning with or forming pyruvate were clubbed into one network and all reactions beginning with acetyl CoA were a part of the second network. For starch and sucrose metabolism, the reactions for formation of glycogen and d-glucose formed one network, and the reactions involving breakdown of glycogen formed another network. The citric acid cycle was also divided into two parts for an easy visualization of the networks.
The reconstructed pathways in the present study differ from those already present in KEGG and Metacyc. The pathway maps presented here, unlike KEGG or Metacyc, highlight the reaction compartments, transport reactions in case of multi compartment metabolic reactions, and some additional reactions that are not incorporated in pathway maps existing in other metabolic pathway databases. Also the pathways of P. pastoris present in KEGG are not strain specific, i.e., they only encompass the metabolic reactions occurring in P. pastoris GS115. Therefore pathways presented in this work differ from KEGG in being specific for P. pastoris CBS7435.
We present the pictorial depiction of some of the pathways of P. pastoris CBS7435 below, which are distinct from other strains of this species or for which we performed gap-filling (Figs. 1, 2, 3, 4, 5, 6, 7, 8, 9 and 10). This carbohydrate and N-glycosylation metabolism visualization will form an integral part of systems biology studies for P. pastoris CBS7435.
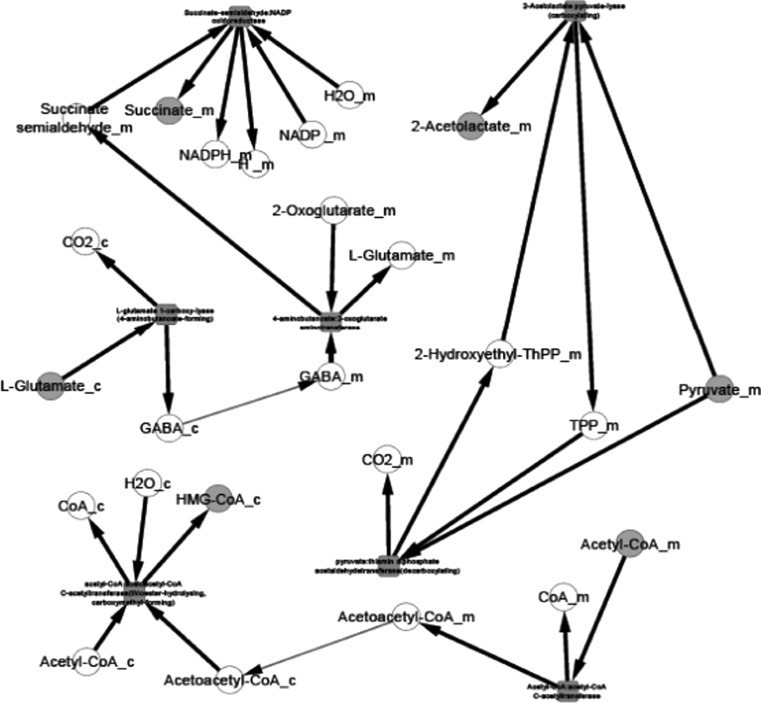
Fig. 1.
Butanoate metabolism
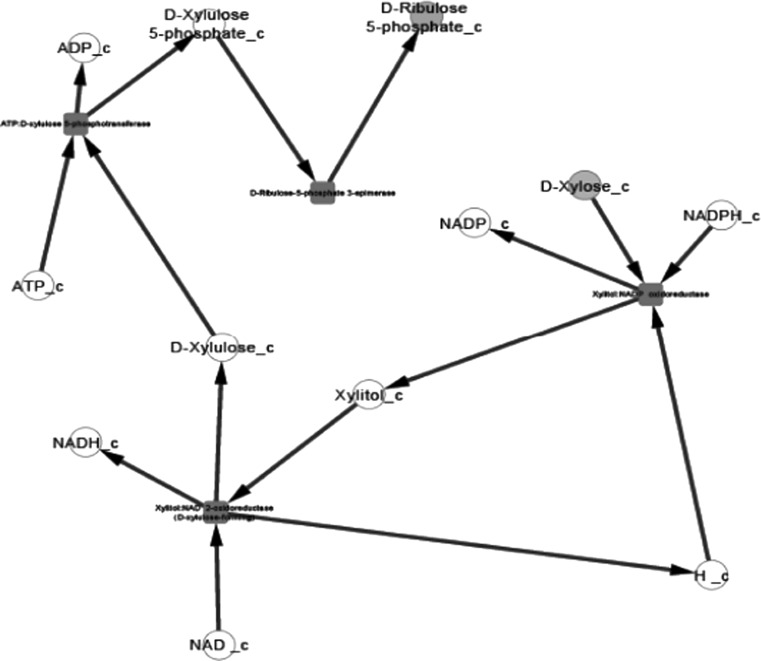
Fig. 2.
Xylose metabolism
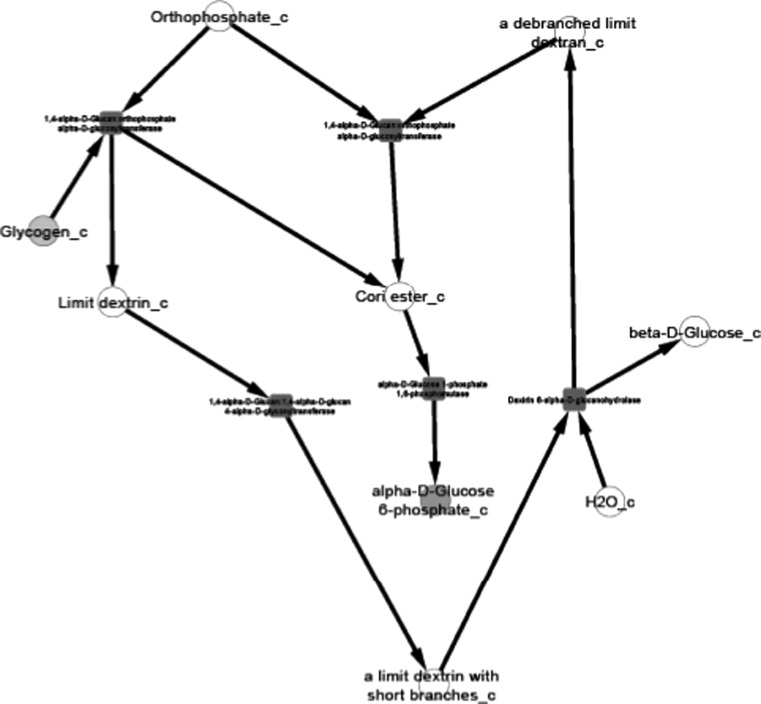
Fig. 3.
Glycogen degradation II pathway
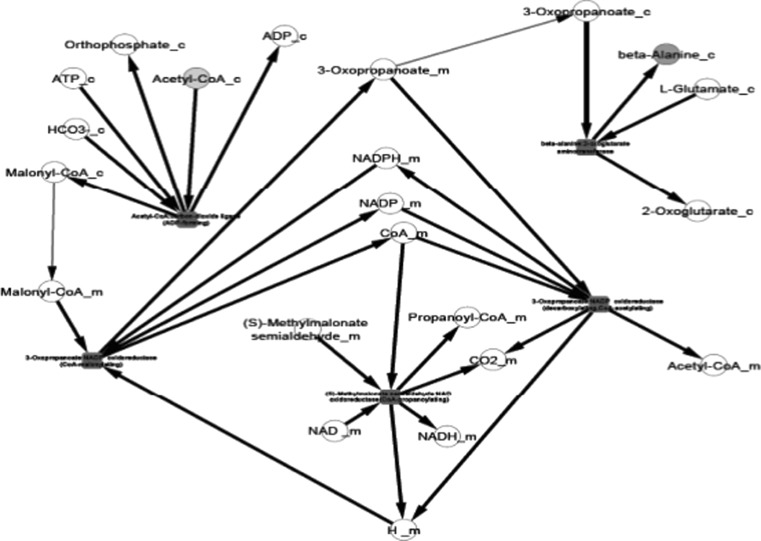
Fig. 4.
Propanoate metabolism
Fig. 5.
Starch and sucrose metabolism (divided into two)
Fig. 6.
Pyruvate metabolism
Fig. 7.
Fructose and mannose metabolism
Fig. 8.
N-glycan biosynthesis (divided into two for ER and cytosol)
Fig. 9.
Inositol phosphate metabolism
Fig. 10.
Gluconeogenesis
Conclusion
Since an organism’s metabolism is a key factor in understanding its physiology, we have reconstructed the metabolic networks of pathways of carbohydrate metabolism and N-glycosylation pathways of the methylotrophic yeast P. pastoris CBS7435. The growing importance of the organisms of this species as potent expression systems as well as hosts for recombinant protein production compelled us to explore certain primary metabolic pathways of the parent strain of this species, i.e., P. pastoris CBS7435. The present study focuses on the significance of the carbohydrate metabolism and N-glycosylation, which lead to high biomass yields and post-translational modification of the recombinant proteins, respectively, the two major advantages of P. pastoris over other expression systems. The reconstructed metabolic networks of these already important pathways offer further scope of their improvement.
The networks presented in this study are organism-specific, and hence provide complete insight into its metabolic events and requirements. Because of their comprehensive nature these networks can easily be modulated or engineered at genetic levels to produce better results in terms of better product yields and better overall performance. The carbohydrate metabolism helps in understanding the nutritional requirements of the organism, and can therefore help in designing of the growth medium. Through the reconstructed networks for carbohydrate metabolism we are able to assess and analyze the sugars or carbohydrates that can be assimilated by P. pastoris CBS7435 for growth, and then compare and choose the best carbon source out of them for achieving higher biomass yields. Carbohydrates present in the culture medium also influence the performance of the promoter systems employed for recombinant protein expression. They also provide an insight into the possible use of P. pastoris CBS7435 for biofuel production. The network can also be modulated or metabolically engineered at a crucial step to produce favorable growth of P. pastoris. Similarly, N-glycosylation pathways of P. pastoris CBS7435 can be re-engineered to produce human like glycosylated proteins, as accomplished in the past using other strains for the production of therapeutic proteins. This can be made possible by metabolically engineering the pathway once we understand the steps and the chemical moieties (enzymes, metabolites) involved in it, and metabolic network reconstruction of N-glycosylation pathway for P. pastoris CBS7435 is the first step towards it. Thus our work provides the researchers with the metabolic networks specific for P. pastoris CBS7435, which can be used by them for future endeavors like metabolic engineering, simulations, flux balance analysis, biofuel production, etc.
Acknowledgments
The authors are thankful to Department of Biotechnology, IET, Lucknow & TERI University, New Delhi for providing the facility and technical support during the preparation of manuscript.
Footnotes
Akriti Srivastava and Pallavi Somvanshi share first authorship.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ates O, Oner ET, Arga KY. Genome-scale reconstruction of metabolic network for a halophilic extremophile, Chromohalobacter salexigens DSM 3043. BMC Syst Biol. 2011;5:12. doi: 10.1186/1752-0509-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A, Apweiler R, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Natale DA, O’Donovan C, Redaschi N, Yeh LL. The universal protein resource (UniProt) Nucleic Acids Res. 2005;33:154–159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balamurugan G, Reddy VR, Suryanarayan VVS. Pichia pastoris: a notable heterologous expression system for the production of foreign proteins—vaccines. IJBT. 2007;6:175–186. [Google Scholar]
- Bobrowicz P, Davidson RC, Li H, Potgeiter TI, Nett JH, Hamilton SR, Stadheim TA, Meile RG, Bobrowicz B, Mitchell T, Rausch S, Renfer E, Wildt S. Engineering of an artificial glycosylation pathwayblocked in core oligosaccharide assembly in yeast Pichia pastoris: production of complex humanized glycoproteins with terminal galactose. Glycobiology. 2004;14(9):757–766. doi: 10.1093/glycob/cwh104. [DOI] [PubMed] [Google Scholar]
- Boyer F, Viari A. Ab initio reconstruction of metabolic pathways. Bioinformatics. 2003;19:26–34. doi: 10.1093/bioinformatics/btg1055. [DOI] [PubMed] [Google Scholar]
- Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, Fisk DG, Hirschman JE, Hitz BC, Karra K, Krieger CJ, Miyasato SR, Nash RS, Park J, Skrzypek MS, Simison M, Weng S, Wong ED. Saccharomyces genome database: the genomics resource of budding yeast. Nucleic Acids Res. 2012;40:700–705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung BKS, Selavarasu S, Camattari A, Ryu J, Lee H, Ahn J, Lee H, Lee DY. Genome scale metabolic reconstruction and in silico analysis of methylotrophic yeast Pichia pastoris for strain improvement. Microb Cell Fact. 2010;9:50. doi: 10.1186/1475-2859-9-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson RC, Bobrowicz P, Zha D. (2012) Metabolic engineering of a galactose assimilation pathway in the glycoengineered yeast Pichia pastoris. U.S. Patent 20120003695, January 5, 2012
- DeJongh M, Formsma K, Boillot P, Gould J, Rycenga M, Best A. Towards the automated generation of genome-scale metabolic networks in the SEED. BMC Bioinformatics. 2007;8:139. doi: 10.1186/1471-2105-8-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durot M, Bourguignon PY, Schachter V. Genome-scale models of bacterial metabolism: reconstruction and applications. FEMS Microbiol Rev. 2008;33:164–190. doi: 10.1111/j.1574-6976.2008.00146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feist AM, Scholten JCM, Palsson BO, Brockman FJ, Ideker T. Modeling methanogenesis with a genome-scale metabolic reconstruction of Methanosarcina barkeri. Mol Syst Biol. 2006 doi: 10.1038/msb4100046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feist AM, Herrgard MJ, Thiele I, Reed JL, Palsson BO. Reconstruction of biochemical networks in microbial organisms. Nat Rev Microbiol. 2009;7(2):129–143. doi: 10.1038/nrmicro1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folger O, Jerby L, Frezza C, Gottleib E, Ruppin E, Shlomi T. Predicting selective drug targets in cancer through metabolic networks. Mol Syst Biol. 2011;7:501. doi: 10.1038/msb.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster J, Famili I, Fu P, Palsson BO, Nielsen J. Genome scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res. 2003;13:244–253. doi: 10.1101/gr.234503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forth T, McConkey GA, Westhead DR. MetNetMaker: a free and open source tool for the creation of novel metabolic networks in SBML format. Bioinformatics. 2010;26(18):2352–2353. doi: 10.1093/bioinformatics/btq425. [DOI] [PubMed] [Google Scholar]
- Francke C, Seizen RJ, Teusink B. Reconstructing the metabolic network of a bacterium from its genome. Trends Microbiol. 2005;13:550–558. doi: 10.1016/j.tim.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha S, Wang Y, Rustandi RR. Biochemical and biophysical characterization of humanized IgG1 produced in Pichia pastoris. MAbs. 2011;3(5):453–460. doi: 10.4161/mabs.3.5.16891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F, Li X, Lu J, Mao PH, Jin X, Rao B, Zheng P, Zhou YL, Liu SY, Ke T, Ma XD, Ma LX. A visual method for direct selection of high-producing Pichia pastoris clones. BMC Biotechnol. 2011;11:23. doi: 10.1186/1472-6750-11-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inan M, Meagher MM (2001) Non-repressing carbon sources for Alcohol Oxidase (AOX1) promoter of Pichia pastoris. J Biosci Bioeng 92(6):585–589 [DOI] [PubMed]
- Jewison T, Knox C, Neveu V, Djoumbou Y, Guo AC, Lee J, Liu P, Mandal R, Krishnamurthy R, Sinelnikov I, Wilson M, Wishart DS. YMDB: the yeast metabolome database. Nucleic Acids Res. 2012;40:815–820. doi: 10.1093/nar/gkr916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:354–357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp PD, Riley M, Paley SM, Pellegrini-toole A. The metacyc database. Nucleic Acids Res. 2002;30:59–61. doi: 10.1093/nar/30.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko P, Vitkup D, Church GM. Filling gaps in a metabolic network using expression information. Bioinformatics. 2004;20:178–185. doi: 10.1093/bioinformatics/bth930. [DOI] [PubMed] [Google Scholar]
- Kuberl A, Schneider J, Thallinger GG, Anderl I, Wibberg D, Hajek T, Jaenicke S, Brinkrolf K, Goesmann A, Szczepanowski R, Puhler A, Schwab H, Gleider A, Pichler H. High-quality genome sequence of Pichia pastoris CBS7435. J Biotechnol. 2011;154(4):312–320. doi: 10.1016/j.jbiotec.2011.04.014. [DOI] [PubMed] [Google Scholar]
- Kumar VS, Dasika MS, Maranas CD. Optimization based automated curation of metabolic reconstructions. BMC Bioinformatics. 2007;8:212. doi: 10.1186/1471-2105-8-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzman CP. Description of Komagataella phaffii sp. nov. and the transfer of Pichia pseudopastoris to the methylotrophic yeast genus Komagataella. Int J Syst Evol Microbiol. 2005;55:973–976. doi: 10.1099/ijs.0.63491-0. [DOI] [PubMed] [Google Scholar]
- Mazurie A, Bonchev D, Schwikowski B, Buck GA. Evolution of metabolic network organization. BMC Syst Biol. 2010;4:59. doi: 10.1186/1752-0509-4-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne CB, Eddy JA, Raju R, Ardekani S, Kim PJ, Senger RS, Jin YS, Blaschek HP, Price ND. Metabolic network reconstruction and genome-scale model of butanol-producing strain Clostridium beijerinckii NCIMB 8052. BMC Syst Biol. 2011;5:130. doi: 10.1186/1752-0509-5-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagud A, Navarro E, de Cordoba PF, Urchueguia JF, Patil KR. Reconstruction and analysis of genome-scale metabolic model of a photosynthetic bacterium. BMC Syst Biol. 2010;4:156. doi: 10.1186/1752-0509-4-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notebaart RA, van Enckevort FHJ, Francke C, Seizen RJ, Teusink B. Accelerating the reconstruction of genome-scale metabolic networks. BMC Bioinformatics. 2006;7:296. doi: 10.1186/1471-2105-7-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhardt MA, Palsson BO, Papin JA. Applications of genome-scale metabolic reconstructions. Mol Syst Biol. 2009;5:320. doi: 10.1038/msb.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterman A, Overbeek R. Missing genes in metabolic pathways: a comparative genomics approach. Curr Opin Chem Biol. 2003;7:238–251. doi: 10.1016/s1367-5931(03)00027-9. [DOI] [PubMed] [Google Scholar]
- Schellenberger J, Park JO, Conrad TM, Palsson BO. BiGG: a biochemical genetics and genomics knowledgebase of large scale metabolic reconstructions. BMC Bioinformatics. 2010;11:213. doi: 10.1186/1471-2105-11-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutter KD, Lin YC, Tiels P, Hecke AV, Glinka S, Lehmann JW, Rouze P, de Peer YV, Callewaert N. Genome sequence of recombinant protein production host Pichia pastoris. Nat Biotechnol. 2009;7:561–566. doi: 10.1038/nbt.1544. [DOI] [PubMed] [Google Scholar]
- Seo S, Lewin HA. Reconstruction of metabolic pathways for cattle genome. BMC Syst Biol. 2009;3:33. doi: 10.1186/1752-0509-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele I, Palsson BO. A protocol for generating a high quality genome-scale metabolic reconstruction. Nat Protoc. 2010;5(1):93–121. doi: 10.1038/nprot.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rensburg P, Van Zyl WH, Pretorius IS. Engineering yeast for efficient cellulose degradation. Yeast. 1998;14(1):67–76. doi: 10.1002/(SICI)1097-0061(19980115)14:1<67::AID-YEA200>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Weidner M, Taupp M, Hallam SJ (2010). Expression of recombinant proteins in methylotrophic yeast Pichia pastoris. J Vis Exp. 36 [DOI] [PMC free article] [PubMed]
- Yadava A, Ockenhouse CF. Effect of codon optimization on expression levels of a functionally folded malaria vaccine candidate in prokaryotic and eukaryotic expression systems. Infect Immun. 2003;71(9):4961–4969. doi: 10.1128/IAI.71.9.4961-4969.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Liu C, Wang G, Ma Y, Zhang K, Zou S, Zhang M. Comparison of expression in Saccharomyces cerevisiae of endoglucanase II from Trichoderma reesei and Endoglucanase I from Aspergillus aculeatus. BioResource. 2012;7(3):4031–4045. [Google Scholar]