Abstract
Using a screening strategy, we identified the tetratricopeptide repeat (TPR) motif protein, Tetratricopeptide repeat domain 5 (TTC5, also known as stress responsive activator of p300 or Strap) as required for the survival of human acute myeloid leukemia (AML) cells. TTC5 is a stress-inducible transcription cofactor known to interact directly with the histone acetyltransferase EP300 to augment the TP53 response. Knockdown (KD) of TTC5 induced apoptosis of both murine and human AML cells, with concomitant loss of clonogenic and leukemia-initiating potential; KD of EP300 elicited a similar phenotype. Consistent with the physical interaction of TTC5 and EP300, the onset of apoptosis following KD of either gene was preceded by reduced expression of BCL2 and increased expression of pro-apoptotic genes. Forced expression of BCL2 blocked apoptosis and partially rescued the clonogenic potential of AML cells following TTC5 KD. KD of both genes also led to the accumulation of MYC, an acetylation target of EP300, and the form of MYC that accumulated exhibited relative hypoacetylation at K148 and K157, residues targeted by EP300. In view of the ability of excess cellular MYC to sensitize cells to apoptosis, our data suggest a model whereby TTC5 and EP300 cooperate to prevent excessive accumulation of MYC in AML cells and their sensitization to cell death. They further reveal a hitherto unappreciated role for TTC5 in leukemic hematopoiesis.
Keywords: TTC5, EP300, MYC, leukemia
Acute myeloid leukemia (AML) is a genetically heterogeneous clonal disorder characterized by the accumulation of poorly differentiated myeloid precursor cells in the bone marrow (BM) and blood.1 Epigenetic dysfunction is central to the molecular pathology of the disease, as evidenced by the recurrent mutations in genes coding for proteins that directly or indirectly regulate the structure and function of chromatin.2 Commonly occurring exemplar mutations in myeloid cancers include those targeting the gene coding for the histone H3K4 methyltransferase MLL, either by chromosomal translocation or partial tandem duplication; those inactivating the enzymatic activities of DNA methyltransferase 3A or the methylcytosine dioxygenase TET2; or those targeting isocitrate dehydrogenase 1 or 2, which result in the production by the mutant enzyme of a neometabolite, 2-hydroxyglutarate, which inhibits the activities of TET-family proteins and Jumonji-domain histone demethylases.2 Further understanding of the structure and function of chromatin in myeloid leukemia cells may lead to the identification of novel therapeutic targets and strategies.
In the course of our studies, we performed a lentiviral short hairpin RNA (shRNA) genetic knockdown screen to identify regulators of chromatin structure and function required for the proliferation or survival of human AML cells (Huang et al, 2013, manuscript submitted). The screen identified tetratricopeptide repeat domain 5 (TTC5; also known as stress responsive activator of p300, Strap) as required for leukemia cell survival. TTC5 is a tetratricopeptide repeat (TPR) motif protein known to have important roles in DNA damage and heat-shock responses. It was originally discovered in complex with an EP300-interacting protein called junction mediating and regulatory protein (JMY), and the histone acetyltransferase (HAT) EP300 (E1A-binding protein p300; also known as p300) itself.3 TTC5 augments the interaction of JMY and EP300, increasing the HAT activity of EP300. This leads to enhanced TP53 acetylation, stability and transcriptional activation during the DNA damage response.3 TTC5 is phosphorylated by ataxia telangiectasia mutated (ATM) at S203 and checkpoint kinase 2 (CHK2) at S221. These phosphorylations promote its stability and nuclear accumulation, also augmenting TP53 activity and promoting apoptosis in etoposide-treated cells.4, 5 In the heat shock response, TTC5 associates with heat shock factor 1 (HSF1) in a chromatin-bound complex that includes EP300. The complex is located at the promoters of heat shock genes and is required to prevent apoptosis.6 A physical interaction of TTC5 with Protein Methyltransferase 5 (PRMT5), which arginine methylates TP53, has been reported,7 as has a regulatory role for TTC5 in steroid hormone signaling where it acts as a cofactor for the glucocorticoid and estrogen receptors.8
While TTC5 is ubiquitously expressed, including in hematopoietic cells (data not shown), to date there is no information as to any functional role for TTC5 in normal or leukemic hematopoiesis. Here, we present evidence demonstrating a critical role for TTC5 in prevention of apoptosis of human AML cells.
Results
TTC5 is required to prevent apoptosis of AML cells
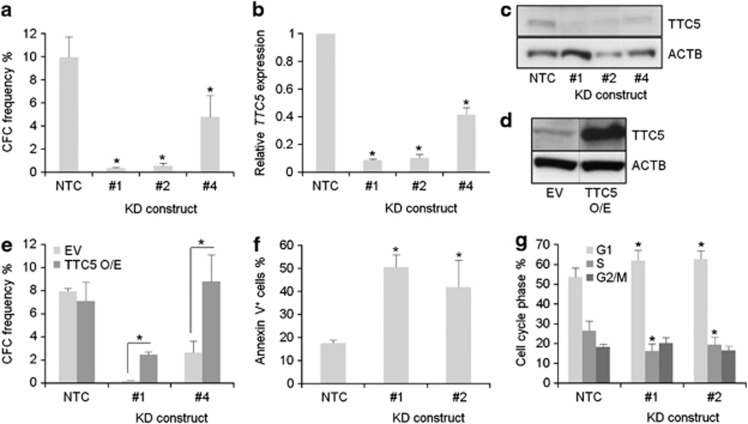
To confirm the preliminary results from our knockdown screen (data not shown), human THP1 AML cells were infected with lentiviral vectors targeting TTC5 for KD. THP1 cells exhibit a t(9;11) translocation, which is the cytogenetic hallmark of MLL-AF9, a fusion oncogene found in 4–5% of patients with AML.1 Successfully transduced cells, as indicated by puromycin drug resistance, were then cultured in semisolid conditions to assess their clonogenic potential. Untransduced THP1 cells treated with puromycin are killed by drug within 48 h (data not shown). While control cells that were infected with a vector expressing a non-targeting hairpin formed plentiful colonies, cells infected with vectors expressing hairpins targeting TTC5 for KD formed significantly fewer colonies (Figure 1a). TTC5 KD was confirmed at both the mRNA and protein levels, and the extent of TTC5 KD correlated with the observed reduction in clonogenic potential (Figures 1b and c). To confirm that the phenotype was an on-target effect of TTC5 KD, we performed repeat experiments using THP1 cells that stably expressed a version of human TTC5 that lacked the 3′-UTR sequences targeted by KD constructs #1 and #4 (Figure 1d). A partial (construct #1) or complete (construct #4) rescue of clonogenic potential was observed (Figure 1e), providing strong additional evidence that the observed phenotype was specific for TTC5. The loss of clonogenic potential following TTC5 KD was in large part due to induction of apoptosis (Figure 1f), although a modest but statistically significant G1 arrest with reduction in the proportion of cells in S phase was also observed (Figure 1g). Thus, TTC5 is required to prevent apoptosis and maintain the proliferative and clonogenic potential of human AML cells.
Figure 1.
TTC5 prevents apoptosis of THP1 MLL-AF9 AML cells. THP1 cells were infected with lentiviruses expressing shRNAs targeting TTC5 for KD, or a non-targeting control shRNA (NTC), with puromycin drug resistance as the selectable marker. Next day, successfully infected cells were drug-selected in puromycin (3 μg/ml) for 48 h. Bar charts show (a) mean±S.E.M. colony forming cell (CFC) frequencies of drug-resistant cells enumerated after 10 days in semisolid culture (n=3); and (b) mean±S.E.M. TTC5 mRNA levels in KD cells relative to control cells, 72 h after lentiviral infection. Representative western blots show expression of (c) endogenous TTC5 and ACTB in KD and control cells, 7 days after lentiviral infection or (d) endogenous or ectopically expressed TTC5 in THP1 cell lines stably infected with the indicated vectors. Vertical dashed line indicates intervening gel lanes excised for clarity of presentation. Bar charts show (e) mean±S.E.M. CFC frequencies in control or KD cells with or without ectopic expression of human TTC5 (n=3); (f) mean±S.E.M. percentage of apoptotic cells, as determined by annexin V binding, 7 days after lentiviral infection of THP1 cells (n=3) and (g) mean±S.E.M. percentage of control or KD cells in the indicated phase of the cell cycle (n=5). S.E.M.=standard error of the mean; EV=Empty Vector; TTC5 O/E=force-expressed TTC5; * indicates P⩽0.05 for comparison of KD versus control conditions, or for the indicated comparisons, using one-way ANOVA and Fisher's least significant difference post hoc test for (a), (b), (f) and (g) and an unpaired t-test for (e)
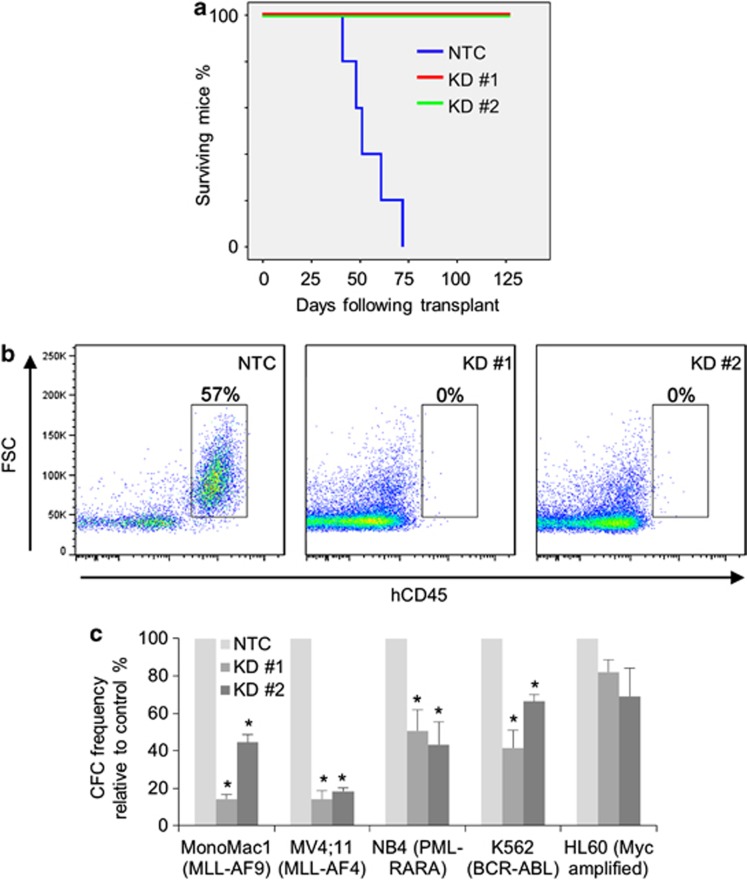
To determine whether TTC5 is required for the leukemia-initiating potential of human AML cells, we transplanted control or KD THP1 AML cells into sublethally irradiated immune-deficient mice. All mice injected with control cells died of short latency AML, whereas mice injected with TTC5 KD cells did not succumb to leukemia (Figures 2a and b). There was no evidence of leukemic engraftment in any of the mice euthanized at the termination of the experiment (Figure 2b and data not shown). To determine whether the observed dependency on TTC5 was specific to the MLL molecular subtype of AML or a more general feature of myeloid leukemia cells, TTC5 KD was initiated in additional human AML cell lines. Significant loss of clonogenic potential of AML cells was observed following TTC5 KD in both MLL mutated lines (MonoMac1 and MV4;11) and lines representative of other AML molecular subtypes (NB4 and K562) (Figure 2c). There was also a reduction in the clonogenic potential of TTC5 KD HL60 cells, although this did not reach statistical significance. These data demonstrate that TTC5 is required for the leukemia-initiating potential of human AML cells and that the requirement for TTC5 is not specific to the MLL mutated molecular subtype.
Figure 2.
TTC5 is required for the leukemia-initiating potential of AML cells. THP1 human AML cells were infected with lentiviruses expressing shRNAs targeting TTC5 for KD, or a non-targeting control (NTC) shRNA, with puromycin drug resistance as the selectable marker. (a) Survival curves of sublethally irradiated immune-deficient mice transplanted with 5000 viable drug resistant control or TTC5 KD THP1 cells (n=5 per cohort). (b) Representative FACS plots show percentage human CD45+ THP1 AML cells in the BM of recipient mice either at death (NTC) or at termination of the experiment on day 125 (TTC5 KD). (c) Bar chart shows mean±S.E.M. colony-forming cell (CFC) frequencies of drug-resistant TTC5 KD cells relative to control cells enumerated after 10 days in semisolid culture for the indicated AML cell lines. * Indicates P⩽0.05 for comparison of KD versus control conditions using one-way ANOVA and Fisher's least significant difference post hoc test
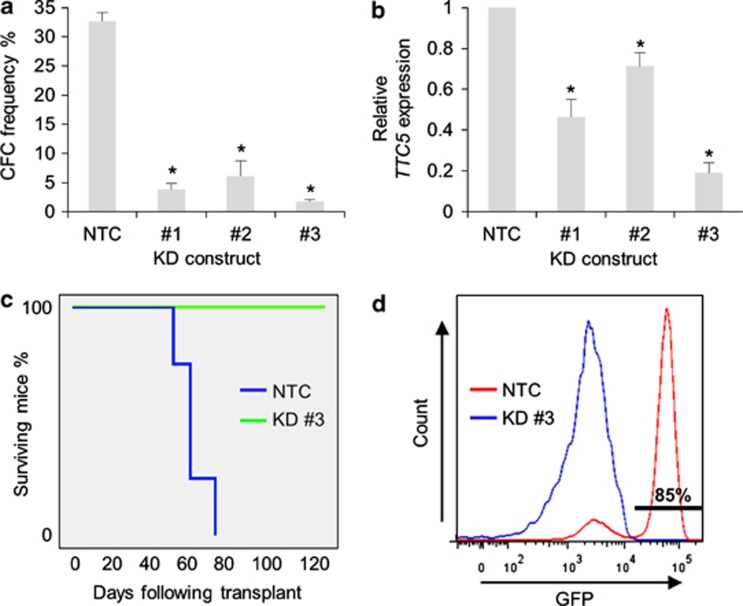
To confirm and expand these observations in a separate species, we performed similar experiments using a mouse model of human MLL-AF9 leukemia, which faithfully recapitulates many of the clinical and pathological features of the human disease.9 Three separate lentivirally expressed shRNAs were used to target murine Ttc5 in murine MLL-AF9 AML cells, this time with GFP as the selectable marker. The extent of transcript KD, as determined by Q-PCR, correlated once more with significant loss of clonogenic potential (Figures 3a and b). To determine whether the leukemia stem cell (LSC) compartment of the leukemia clone was targeted by Ttc5 KD, we performed secondary transplantation of FACS-purified control or Ttc5 KD MLL-AF9 AML cells 48 h following lentiviral infection.10 As for human THP1 cells, all mice receiving control AML cells died of short latency disease, whereas mice transplanted with Ttc5 KD AML cells survived until the termination of the experiment (Figure 3c), and at autopsy exhibited no evidence of leukemic engraftment, as determined by flow cytometry of bone marrow cells. By contrast, engraftment of donor GFP+ AML cells in the BM of leukemic mice was readily confirmed (Figure 3d). These data confirm that, as for human AML cells, Ttc5 is essential for murine MLL-AF9 leukemia-initiating cells.
Figure 3.
Ttc5 is required to maintain the leukemia-initiating potential of murine MLL-AF9 AML cells. MLL-AF9 cells were infected with lentiviruses expressing shRNAs targeting Ttc5 for KD or a non-targeting control (NTC) shRNA, with GFP as the selectable marker. After 48 h, cells were FACS-purified for GFP expression. Bar charts show (a) mean±S.E.M. CFC frequencies of control and Ttc5 KD cells after 5 days in semisolid culture (n=3) and (b) mean±S.E.M. Ttc5 mRNA levels in KD cells relative to control cells 48 h following lentiviral infection (n=3). (c) Survival curves of sublethally irradiated syngeneic mice secondarily transplanted with 1000 control or Ttc5 KD AML cells (n=4 mice per cohort). (d) Representative FACS plots show the presence of GFP+ donor AML cells in the BM of a leukemic mouse transplanted with control cells (Day 60) and absence of AML cells in a mouse transplanted with Ttc5 KD cells at termination of the experiment (Day 120). * Indicates P⩽0.05 for comparison of KD versus control conditions using one-way ANOVA and Fisher's least significant difference post hoc test
TTC5 sustains expression of BCL2 in AML cells
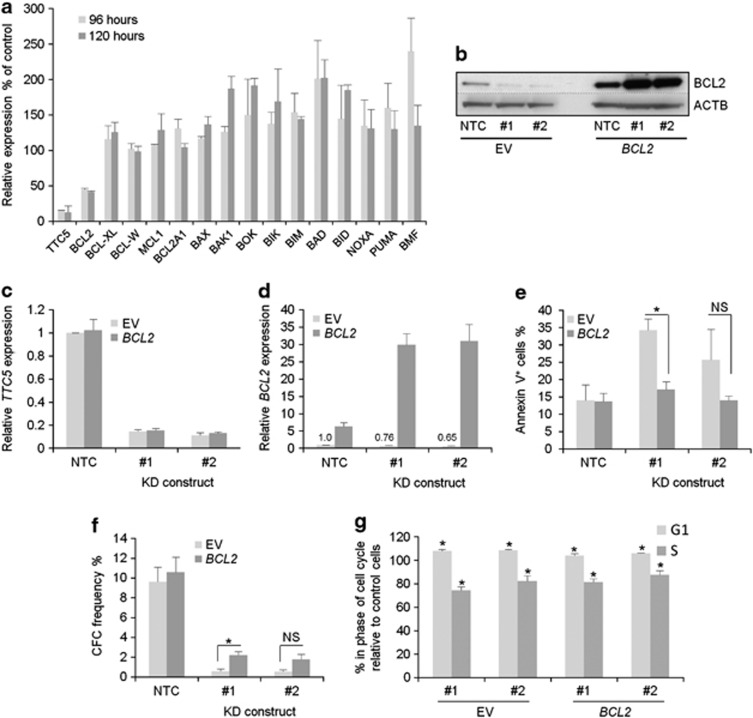
To address the mechanism by which TTC5 KD cells undergo apoptosis, we determined transcript levels of a panel of genes coding for proteins that regulate cell death 96 and 120 h following lentiviral infection of THP1 AML cells. Relative to that of ACTB, expression of the anti-apoptotic gene BCL2 decreased by >50% and expression of pro-apoptotic genes such as BAK1, BOK, BIK, BAD, BID, PUMA (also known as BBC3) and BMF increased by >50% in TTC5 KD cells by comparison with control cells at one or the other time point (Figure 4a). The change in expression of other genes was less marked. Reduced BCL2 expression was confirmed in repeat experiments with two separate TTC5 KD constructs at both protein (Figure 4b) and transcript levels (Figures 4c and d). To determine whether reduced expression of BCL2 contributes to apoptosis of AML cells following TTC5 KD, we force-expressed BCL2 in THP1 cells and repeated KD. Interestingly, we observed an increase in both BCL2 transcripts and protein in BCL2-overexpressing THP1 cells following TTC5 KD (Figures 4b and d), suggesting that TTC5 may directly or indirectly contribute to repression of the integrated retroviral long terminal repeat promoter. Forced expression of BCL2 (Figures 4b–d) blocked apoptosis and partially rescued the loss of clonogenic potential consequent upon TTC5 KD (Figures 4e and f). However, forced expression of BCL2 did not rescue the modest but significant G1 arrest and reduction of the proportion of cells in S phase, likely explaining why only a partial rescue was observed (Figure 4g). Thus, TTC5 selectively and positively regulates expression of BCL2 in human AML cells, either directly or indirectly, and downregulation of BCL2 following TTC5 KD contributes to apoptosis and loss of clonogenic potential.
Figure 4.
TTC5 prevents apoptosis of THP1 cells through regulation of BCL2 expression. THP1 cells were infected with lentiviruses expressing shRNAs targeting TTC5 for KD, or a non-targeting control (NTC) shRNA, with puromycin drug resistance as the selectable marker. Bar chart shows (a) mean±S.E.M. transcript expression levels of the indicated genes in KD cells relative to control cells at the indicated time points following lentiviral infection (n=2). (b) Representative western blots show expression of the indicated proteins, 7 days after lentiviral infection. Bar charts show (c) mean±S.E.M. TTC5 or (d) BCL2 transcript levels in control (EV, empty vector) or BCL2 overexpressing TTC5 KD THP1 cells relative to control cells, 72 h after lentiviral infection (n=3). Bar charts show (e) mean±S.E.M. percentage of apoptotic cells, as determined by annexin V binding, 7 days after lentiviral infection (n=3) and (f) mean±S.E.M. colony-forming cell (CFC) frequencies of drug-resistant cells enumerated following 10 days in semisolid culture (n=3). * Indicates P⩽0.05 (Mann–Whitney U-test) for comparison of KD versus control conditions, or for the indicated comparisons. NS=not significant. (g) Bar chart shows mean±S.E.M. percentage of TTC5 KD cells in G1 or S phase of cell cycle relative to that observed in control cells (for BCL2 over expressing and control THP1 lines) 96 h after initiation of KD (n=3). * Indicates P⩽0.05 for comparison of KD versus control conditions using one-way ANOVA and Fisher's least significant difference post hoc test
EP300 sustains the expression of BCL2 in AML cells
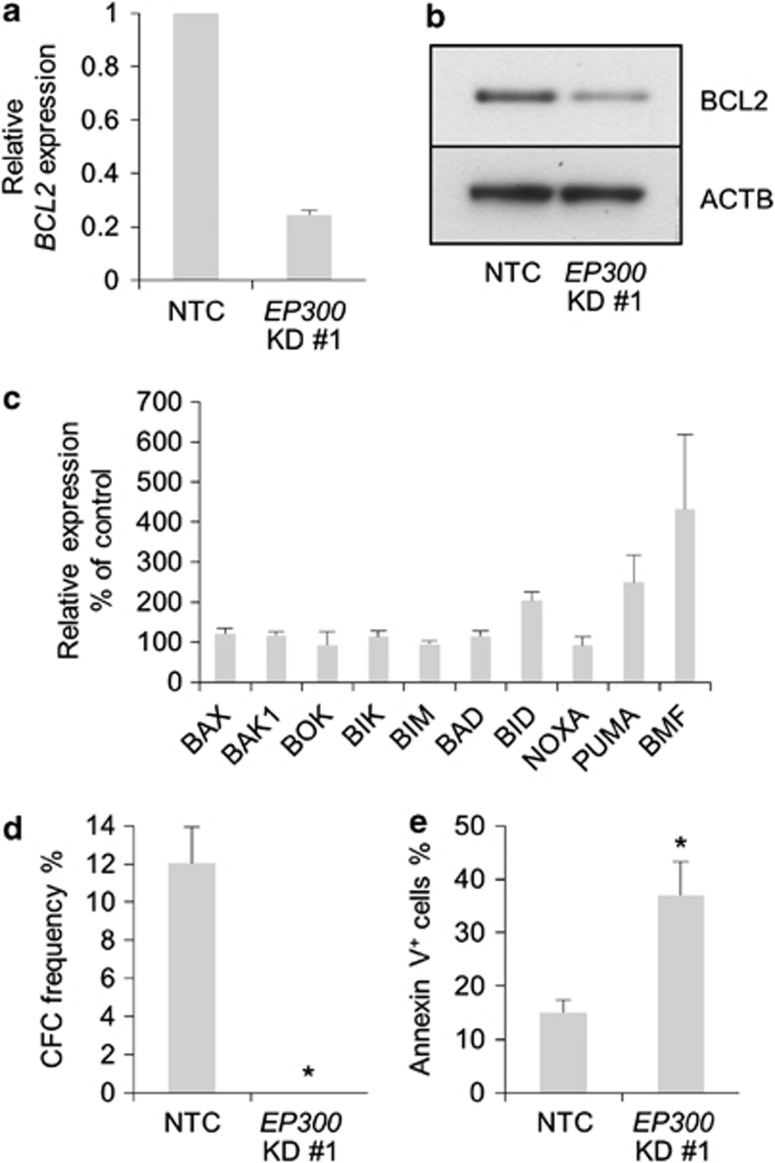
TTC5 was originally identified in complex with EP300 and an EP300-interacting protein JMY,3 and was also found to augment EP300 histone acetyltransferase activity in U2OS osteosarcoma cells.6 We therefore hypothesized that TTC5 might similarly regulate EP300 activity in human THP1 AML cells to enhance cell survival. Consistent with this, EP300 KD to 51±8% (mean±S.E.M.; n=3) of control levels induced a similar phenotype to TTC5 KD: EP300 KD cells exhibited reduced BCL2 expression (Figures 5a and b), a >50% increase in expression of the pro-apoptotic genes BID, PUMA and BMF (which were also induced by TTC5 KD) (Figure 5c), and a failure to form colonies in semisolid culture (Figure 5d) due to induction of apoptosis (Figure 5e). These data suggest that TTC5 and EP300 cooperate, directly or indirectly, to maintain expression of BCL2 in AML cells and repress pro-apoptotic genes such as BID, PUMA and BMF, thus preventing leukemia cell apoptosis.
Figure 5.
EP300 KD phenocopies TTC5 KD. THP1 cells were infected with lentiviruses expressing shRNAs targeting EP300 for KD, or a non-targeting control shRNA (NTC), with puromycin drug resistance as the selectable marker. (a) Bar chart shows mean±S.E.M. BCL2 mRNA levels in EP300 KD cells relative to control cells, 72 h after lentiviral infection (n=3). (b) Representative western blot shows expression of the indicated proteins, 72 h after lentiviral infection (n=3). (c) Bar chart shows mean±S.E.M. transcript levels of the indicated genes in EP300 KD cells relative to control cells, 72 h after lentiviral infection (n=3). Bar charts show (d) mean±S.E.M. colony-forming cell (CFC) frequencies of drug-resistant cells enumerated following 10 days in semisolid culture (n=3) and (e) mean±S.E.M. percentage of apoptotic cells, as determined by annexin V binding, 7 days after lentiviral infection (n=3). * indicates P⩽0.05 (Mann–Whitney U-test) for comparison of KD versus control conditions
TTC5 and EP300 regulate turnover of MYC in AML cells
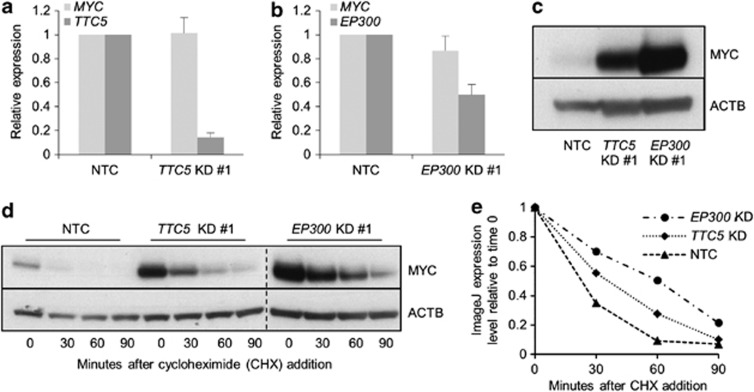
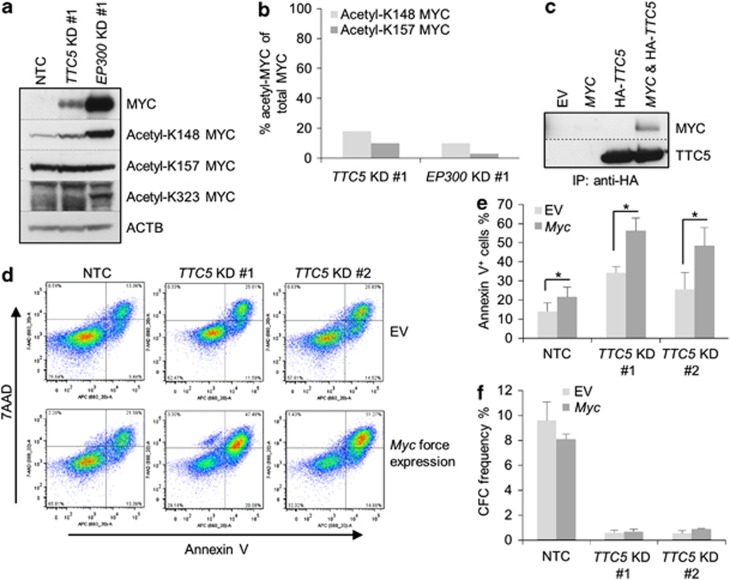
The pattern of change in expression of BCL2 family genes following TTC5 KD (Figure 4a), with reduced expression of BCL2 and increased expression of pro-apoptotic genes, was reminiscent of the pattern we have previously observed in AML cells in which Myc was force-expressed (Huang et al, 2013, manuscript submitted). Furthermore, EP300 regulates the stability of MYC through both acetylation-dependent and acetylation-independent mechanisms.11 These data prompted us to investigate whether TTC5 or EP300 KD altered the expression of MYC. Neither TTC5 KD nor EP300 KD in THP1 AML cells led to a significant change in the expression of MYC transcripts (Figures 6a and b). By contrast, both TTC5 KD and EP300 KD led to a robust accumulation of MYC protein (Figure 6c), which was explained by an increase in its cellular half-life from <30 min (control cells) to >60 min (EP300 KD) (Figures 6d and e). Given that EP300 directly acetylates MYC at K148, K157 and K323,11 we next investigated whether acetylation of MYC at these residues was perturbed in TTC5 or EP300 KD cells. In contrast to the robust accumulation of total MYC protein, levels of acetyl-K157 MYC did not change following either TTC5 or EP300 KD, and levels of acetyl-K148 MYC increased only modestly (Figure 7a); this was confirmed by semiquantitative assessment of western blot bands using ImageJ analysis (Figure 7b). By contrast, acetylation of MYC at K323 was not readily detected in control or TTC5 KD THP1 cells, although it was detected in EP300 KD cells. Thus, following TTC5 or EP300 KD, the form of MYC that accumulated in human AML cells is relatively hypoacetylated at K148 and K157. These data are consistent with previously described roles for EP300 in MYC acetylation and turnover11 and TTC5 in regulation of the HAT activity of EP300.3, 6
Figure 6.
TTC5 and EP300 promote MYC turnover. THP1 cells were infected with lentiviruses expressing shRNAs targeting TTC5 or EP300 for KD, or a non-targeting control shRNA (NTC), with puromycin drug resistance as the selectable marker. Bar charts show (a) mean±S.E.M. mRNA levels of MYC and TTC5 in TTC5 KD cells and (b) mean±S.E.M. mRNA levels of MYC and EP300 in EP300 KD cells relative to control cells, 72 h after lentiviral infection (n=3). Representative western blots show (c) expression of MYC after EP300 or TTC5 KD (n=3) and (d) MYC half-life, as determined by incubation of control or KD cells with 10 μg/ml cycloheximide for the indicated times, 72 h after lentiviral infection (n=2). Vertical dashed line demarcates separate gels, although exposure time was identical. (e) MYC half-life was semiquantified by densitometric analysis using ImageJ software
Figure 7.
Myc accumulation and apoptosis after TTC5 or EP300 KD. THP1 cells were infected with lentiviruses expressing shRNAs targeting TTC5 or EP300 for KD, or a non-targeting control shRNA (NTC), with puromycin drug resistance as the selectable marker. (a) Representative western blots show expression of MYC, acetylated MYC and ACTB after EP300 or TTC5 KD, 72 h after lentiviral infection. (b) Bar chart shows semiquantitative analysis of acetyl-K148 MYC or acetyl-K157 MYC signal as a percentage of total MYC signal, as determined by ImageJ analysis of the bands shown in (a). (c) 293FT cells were transiently transfected with the indicated constructs. After 48 h, cells were harvested and lysates were immunoprecipitated with an HA-tag antibody. The immunoprecipitated complex was analyzed by western blotting using MYC or TTC5 antibodies. IP=immunoprecipitation. (d–f) Control and Myc-overexpressing THP1 cells were infected with lentiviruses expressing shRNAs targeting TTC5 for KD or a NTC shRNA, with puromycin drug resistance as the selectable marker. (d) Representative FACS plots show annexin V and 7-aminoactinomycin (7-AAD) binding to control and KD cells, 7 days after lentiviral infection. Bar charts show (e) mean±S.E.M. percentage of apoptotic cells, as determined by annexin V binding, 7 days after lentiviral infection (n=3); and (f) mean±S.E.M. colony-forming cell (CFC) frequencies of drug-resistant cells enumerated following 10 days in semisolid culture (n=3). EV=empty vector. * indicates P⩽0.05 (paired t-test) for comparison of KD versus control conditions
To determine whether TTC5 and MYC interact, co-immunoprecipitation experiments were performed in 293FT cells: an HA-tagged version of TTC5 readily pulled down co-expressed MYC in (Figure 7c). Finally, to determine whether MYC accumulation might contribute to induction of apoptosis, we performed TTC5 KD in THP1 cells where murine Myc was force-expressed. Expression of murine Myc transcripts was detected in Myc-expressing human THP1 cells, but was absent in control cells (data not shown). Consistent with a role for MYC in sensitizing cells to apoptosis,12 THP1 cells force-expressing Myc exhibited significantly greater levels of apoptosis by comparison with control cells, both when infected with non-targeting control vectors (albeit modestly) and when infected with vectors targeting TTC5 for KD (Figures 7d and e). The substantially greater proportion of TTC5 KD versus control cells undergoing apoptosis in the context of Myc forced expression furthermore suggested a quantitative link between the amount of cellular MYC and the rapidity of onset of apoptosis. There was no significant difference in the clonogenic potential of control or Myc force-expressing TTC5 KD cells (Figure 7f). Together, these data indicate a role for TTC5 in the regulation of MYC protein turnover in leukemia cells and suggest that MYC accumulation might contribute to the induction of apoptosis through altering the balance of expression of anti- versus pro-apoptotic proteins.
Discussion
TTC5 was originally described as an EP300-interacting transcription cofactor that accumulates following etoposide treatment of U2OS osteosarcoma cells, and which augments the TP53 response.3 Subsequent work has identified more general roles for TTC5 in the regulation of the cellular response to stress, for example, following heat shock.6 Our studies using cells from two different species now reveal roles for TTC5 in the prevention of apoptosis of leukemia cells and maintenance of their clonogenic capacity, through regulation of MYC and BCL2 expression. Syngeneic and xenogeneic transplantation experiments confirm an in vivo requirement for TTC5 in leukemia initiation, as well as its in vitro requirement. Importantly, our observations are not explained by an effect on TP53 because THP1 cells exhibit a frame-shift deletion of TP53 from R17413 and, as expected, we were unable to detect TP53 expression in these cells (data not shown). While the dependency of AML cells on TTC5 was discovered in the context of the MLL mutated molecular subtype of AML, the in vitro clonogenic activity of AML cell lines representative of other molecular subtypes was likewise dependent on TTC5, suggesting a wide role for the protein in leukemic hematopoiesis.
Knockdown of either TTC5 or EP300 in AML cells similarly alters the balance of expression of pro- versus anti-apoptotic genes, consistent with the physical and functional interaction of their gene products. The decrease in expression of the anti-apoptotic gene BCL2 and increase in expression of pro-apoptotic genes such as BID, PUMA and BMF likely alters the cellular apoptotic rheostat in favor of apoptosis. Such changes might be explained by localized, specific and direct effects of a TTC5/EP300 complex at gene promoters to repress or activate expression respectively. Alternatively, changes in gene expression might be consequent upon altered expression of a protein target of the histone acetyltransferase activity of the TTC5/EP300 complex.
In support of the latter interpretation, we have previously found that forced expression of Myc in AML cells induces expression of pro-apoptotic genes (Huang et al, 2013, manuscript submitted). In contrast, forced expression of Myc in THP1 cells reduces expression of BCL2 relative to ACTB to 44±12% of control levels (mean±S.E.M., n=3). The similarity in the change in expression of pro- versus anti-apoptotic genes observed following Myc-overexpression or TTC5/EP300 KD prompted our investigation of MYC as an intermediary of the observed gene expression changes. MYC associates directly with and is acetylated by EP300 on multiple residues, including K148, K157 and K323.11 Acetylation of MYC by EP300 promotes its turnover.11 In keeping with these observations, we observed that EP300 KD led to robust accumulation of a form of MYC that was relatively hypoacetylated at both K148 and K157; TTC5 KD gave similar results. Together, these data support a role for TTC5 in promoting the turnover of MYC though its regulation of the acetyltransferase activity of EP300. Given that TPR-motif proteins function as scaffolding proteins, it is possible that TTC5 could stabilize the interaction between EP300 and MYC.14
While physiologic levels of MYC are required for normal cell cycle progression, accumulation of MYC above a certain threshold is associated with induction of apoptosis,12 through a transcriptional mechanism that results in leak of cytochrome c from the mitochondrion into the cytoplasm.15 We found a modest but significant increase in apoptosis in THP1 cells where Myc was force-expressed. However, following TTC5 or EP300 KD, the extent of apoptosis was much greater, consistent with the observed significant and substantial accumulation of endogenous MYC. Nevertheless, our observations of a role for TTC5 in the regulation of MYC turnover do not rule out other important roles for TTC5 in leukemic hematopoiesis. For example, because TTC5 is an EP300-interacting protein, and given the varied and critical roles of EP300 in hematopoiesis, it is possible that TTC5 regulates other hematopoietic transcriptional complexes.16, 17
Materials and Methods
Cell lines, culture and clonogenic assays
THP1 and MonoMac-1 cells were from DMSZ (Braunschweig, Germany). MV(4;11), NB4, HL60 and K562 cells were gifts from Dr. Vaskar Saha and were cultured as recommended (www.dmsz.de). Clonogenic assays of cell lines were performed in methylcellulose medium (H4320, Stem Cell Technologies, Vancouver, BC, Canada) with no supplemental growth factors. Liquid culture of murine AML cells was in RPMI 1640 containing 20% fetal calf serum and 5% X63 supernatant.18 Clonogenic assays of murine AML cells were performed in methylcellulose medium (M3231, Stem Cell Technologies) containing 20 ng/ml SCF, 10 ng/ml IL6, 10 ng/ml GM-CSF and 10 ng/ml IL3 (Peprotech, London, UK).
Reagents, plasmids and lentiviral production
Puromycin, cycloheximide and lentiviral vectors were purchased from Sigma (Poole, UK). Vectors were as follows: non-targeting control pLKO.1 (SHC002); TTC5 KD pLKO.1 (TRCN0000156809 (#1), TRCN0000156289 (#2) and TRCN0000152678 (#4)); Ttc5 KD pLKO.1 (TRCN0000176549 (#1), TRCN0000178101 (#2) and TRCN0000181231 (#3)) and EP300 KD pLKO.1 (TRCN0000039886). For the murine Ttc5 KD constructs, the PGK-puromycin cassette was exchanged for an SFFV-eGFP cassette (amplified from pHR′SIN.cPPT-SEW, a gift from Dr. Adrian Thrasher), to generate eGFP versions. To generate a lentiviral vector for expression of TTC5, TTC5 cDNA was PCR amplified using pcDNA3-HA-TTC5 as a template (a gift from Marija Krstic-Demonocas), incorporating Cla1 and Xba1 restriction sites, using the following primers:
5′ ATAATATCGATATGATGGCTGATGAAGAGGAAGAAGTC,
3′ATAATTCTAGATCATTCACACTGTGGTCGCGATG.
The amplified fragment was sequence-verified and cloned in to the Cla1 and Xba1 sites of pLentiGS downstream of an EF1α promoter (Huang et al, 2013, manuscript submitted). pMSCV-BCL2-IRES-GFP (#8972) and pMSCV-Myc-IRES-GFP (#18770) were purchased from Addgene (Cambridge, MA, USA). Lentiviral and retroviral supernatants were prepared and leukemic human and murine cells infected with viral particles as described.19
Western blotting and immunoprecipitation
For western blotting, cells were lysed in High Salt Lysis Buffer (45 mM HEPES (pH 7.5), 400 mM NaCl, 1 mM EDTA, 10% Glycerol, 0.5% NP40, 6.25 mM NaF, 20 mM β-glycerophosphate, 1 mM DTT, 20 mM sodium butyrate and 1 × Protease Inhibitor cocktail (Roche, Burgess Hill, UK)) and equal amounts of protein were loaded and separated by SDS-PAGE. For co-immunoprecipitation, cells were lysed in NP40 Lysis Buffer (50 mM Tris/HCl (pH 8.0), 50 mM KCl, 10 mM EDTA, 1% NP40, 6.25 mM NaF, 20 mM β-glycerophosphate, 1 mM DTT and 1 × Protease Inhibitor cocktail (Roche)). Lysate was incubated overnight with the appropriate antibody and immune complexes were pulled down with Protein G–Sepharose (Sigma). Samples were washed four times with IP wash buffer (50 mM Tris/HCl (pH 7.5), 150 mM NaCl, 5 mM EDTA and 0.1% Tween 20), eluted and separated by SDS-PAGE. Horseradish peroxidase-linked secondary antibodies (GE Healthcare, Little Chalfont, UK) and ECL (enhanced chemiluminescence; GE Healthcare) or Supersignal (Pierce, Rockford, IL, USA) were used to detect immune complexes. Protein expression levels were quantified using ImageJ software (NIH, USA). Antibodies used for western blotting or immunoprecipitations were as follows: anti-TTC5 (PAB12888, Abnova, Cambridge, UK), anti-ACTB (C4, Millipore, Watford, UK), anti-BCL2 (#124, Dako, Ely, UK), anti-MYC (Y69, Abcam, Cambridge, UK), anti-acetyl MYC K148 (ABE25, Millipore), anti-acetyl MYC K157 (ABE27, Millipore), anti-acetyl MYC K323 (ABE26, Millipore) and anti-HA (12CA5, Abcam).
RNA isolation and analysis
RNA was extracted using QIAshredder spin columns and an RNeasy Plus Micro kit (Qiagen, Manchester, UK). For quantitative PCR, first strand synthesis was performed using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Paisley, UK). Quantitative PCR assays were performed in 384-well MicroAmp optical reaction plates using Taqman Fast Universal PCR Mastermix (Applied Biosystems) and Universal Probe Library System (Roche) designed primers and probes, which are listed in Table 1.
Table 1. Primer and probe sets used for Q-PCR.
| Gene | Q-PCR primer sequences | Roche probe library number |
|---|---|---|
| TTC5 F | ACCGAATTCAGCACAAAGGA | 47 |
| TTC5 R | CTAGCAGGAGGGGCGTCT | — |
| ACTB F | ATTGGCAATGAGCGGTTC | 11 |
| ACTB R | GGATGCCACAGGACTCCAT | — |
| EP300 F | GATCTGTGTCCTTCACCATGAG | 40 |
| EP300 R | AAACAGCCATCACAGACGAA | — |
| MYC F | GCTGCTTAGACGCTGGATTT | 66 |
| MYC R | TAACGTTGAGGGGCATCG | — |
| BCL2 F | AGTACCTGAACCGGCACCT | 75 |
| BCL2 R | GCCGTACAGTTCCACAAAGG | — |
| BCL2L1 (BCL-XL) F | AGCCTTGGATCCAGGAGAA | 66 |
| BCL2L1 (BCL-XL) R | AGCGGTTGAAGCGTTCCT | — |
| MCL1 F | TGGATGGTGGCCTACCTG | 28 |
| MCL1 R | CGTCCCCGTATAGAGCTGT | — |
| BCL2A1 (A1) F | CAGGAGAATGGATAAGGCAAA | 75 |
| BCL2A1 (A1) R | CCAGCCAGATTTAGGTTCAAA | — |
| BCL2L2 (BCL-W) F | TGGATGGTGGCCTACCTG | 28 |
| BCL2L2 (BCL-W) R | CGTCCCCGTATAGAGCTGTG | — |
| BAX F | AGCAAACTGGTGCTCAAGG | 69 |
| BAX R | TCTTGGATCCAGCCCAAC | — |
| BAK F | TGGTCACCTTACCTCTGCAAC | 43 |
| BAK R | ATGTCGTCCCCGATGATG | — |
| BIK F | CCCTATGGAGGACTTCGATTC | 28 |
| BIK R | GGCTCACGTCCATCTCGT | — |
| BCL2L11 (BIM) F | CATCGCGGTATTCGGTTC | 70 |
| BCL2L11 (BIM) R | GCTTTGCCATTTGGTCTTTTT | — |
| BAD F | CGAGTTTGTGGACTCCTTTAAGA | 78 |
| BAD R | CACCAGGACTGGAAGACTCG | — |
| BID F | GTGCTGGGGTCATGATGG | 19 |
| BID R | CGACTCACTCCTGGTTCACA | — |
| BBC3 (PUMA) F | GACCTCAACGCACAGTACGA | 51 |
| BBC3 (PUMA) R | CACCTAATTGGGCTCCATCTC | — |
| PMAIP1 (NOXA) F | GGAGATGCCTGGGAAGAAG | 67 |
| PMAIP1 (NOXA) R | CCTGAGTTGAGTAGCACACTCG | — |
| BMF F | GAGACTCTCTCCTGGAGTCACC | 25 |
| BMF R | CTGGTTGGAACACATCATCCT | — |
| Ttc5 F | TGCAGCAGATGGAGGAAGTA | 98 |
| Ttc5 R | GAGTCACATTCAGTGCCTTCC | — |
| Actb F | TGACAGGATGCAGAAGGAGA | 106 |
| Actb R | CGCTCAGGAGGAGCAATG | — |
| Myc F | CCTAGTGCTGCATGAGGAGA | 77 |
| Myc R | TCCACAGACACCACATCAATTT | — |
Flow cytometry, apoptosis and cell cycle analysis
FACS analyses were performed using a LSR Model II flow cytometer (BD Biosciences, Oxford, UK). Cell sorting experiments were performed using either an Influx or a FACSAria II flow cytometer (both from BD Biosciences). Anti-CD45-PE-Cy7 (HI30) was from eBioscience (Hatfield, UK). Apoptosis was assessed using a BD Pharmingen APC Annexin V Kit (Oxford, UK), according to the manufacturer's instructions. Propidium iodide cell cycle analyses were performed as described.20
Mice and murine experiments
Experiments were approved by the Paterson Institute's Animal Ethics Committee and performed under a project license issued by the United Kingdom Home Office, in keeping with the Home Office Animal Scientific Procedures Act, 1986. C57BL/6 (CD45.2+) mice were purchased from Harlan (Shardlow, UK). NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were purchased from Jackson Laboratories (Bar Harbor, ME, USA) and bred in-house. The cohort of mice with syngeneic MLL-AF9 AML, initiated using a retroviral transduction and transplantation protocol, was generated as described.10, 19
Acknowledgments
We thank Morgan Blaylock, Jeff Barry, Mike Hughes, Gail Bruder and Angela Cooke for technical support; and Marija Krstic-Demonacos and Costas Demonacos for plasmids and helpful discussions. This work was supported by Cancer Research UK grant number C5759/A12328.
Glossary
- 7-AAD
7-aminoactinomycin
- AML
acute myeloid leukemia
- ATM
ataxia telangiectasia mutated
- BM
bone marrow
- CFC
colony forming cell
- CHEK2
checkpoint kinase 2
- CHX
cycloheximide
- EP300
E1A-binding protein p300
- EV
empty vector
- FACS
fluorescence-activated cell sorter
- HSF1
heat-shock factor 1
- IP
immunoprecipitation
- JMY
junction mediating and regulatory protein
- KD
knockdown
- LSC
leukemia stem cell
- NS
not significant
- NTC
non-targeting control
- PTMs
post-translational modifications
- Q-PCR
quantitative reverse transcriptase PCR
- SEM
standard error of the mean
- shRNA
short hairpin RNA
- TPR
tetratricopeptide repeat
- TTC5
tetratricopeptide repeat domain 5
- TP53
tumor protein 53
- MDM2
murine double minute 2
The authors declare no conflict of interest.
Footnotes
Edited by P Salomoni
References
- Estey E, Döhner H. Acute myeloid leukaemia. Lancet. 2006;368:1894–1907. doi: 10.1016/S0140-6736(06)69780-8. [DOI] [PubMed] [Google Scholar]
- Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer. 2012;12:599–612. doi: 10.1038/nrc3343. [DOI] [PubMed] [Google Scholar]
- Demonacos C, Krstic-Demonacos M, La Thangue NB. A TPR motif cofactor contributes to p300 activity in the p53 response. Mol Cell. 2001;8:71–84. doi: 10.1016/s1097-2765(01)00277-5. [DOI] [PubMed] [Google Scholar]
- Adams CJ, Graham AL, Jansson M, Coutts AS, Edelmann M, Smith L, et al. ATM and Chk2 kinase target the p53 cofactor Strap. EMBO Rep. 2008;9:1222–1229. doi: 10.1038/embor.2008.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demonacos C, Krstic-Demonacos M, Smith L, Xu D, O'Connor DP, Jansson M, et al. A new effector pathway links ATM kinase with the DNA damage response. Nat Cell Biol. 2004;6:968–976. doi: 10.1038/ncb1170. [DOI] [PubMed] [Google Scholar]
- Xu D, Zalmas LP, La Thangue NB. A transcription cofactor required for the heat-shock response. EMBO Rep. 2008;9:662–669. doi: 10.1038/embor.2008.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson M, Durant ST, Cho EC, Sheahan S, Edelmann M, Kessler B, et al. Arginine methylation regulates the p53 response. Nat Cell Biol. 2008;10:1431–1439. doi: 10.1038/ncb1802. [DOI] [PubMed] [Google Scholar]
- Davies L, Paraskevopoulou E, Sadeq M, Symeou C, Pantelidou C, Demonacos C, et al. Regulation of glucocorticoid receptor activity by a stress responsive transcriptional cofactor. Mol Endocrinol. 2011;25:58–71. doi: 10.1210/me.2010-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somervaille TCP, Matheny CJ, Spencer GJ, Iwasaki M, Rinn JL, Witten DM, et al. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4:129–140. doi: 10.1016/j.stem.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somervaille TCP, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 2006;10:257–268. doi: 10.1016/j.ccr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Faiola F, Liu X, Lo S, Pan S, Zhang K, Lymar E, et al. Dual regulation of c-Myc by p300 via acetylation-dependent control of Myc protein turnover and coactivation of Myc-induced transcription. Mol Cell Biol. 2005;25:10220–10234. doi: 10.1128/MCB.25.23.10220-10234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan GI, Wyllie AH, Gilbert CS, Littlewood TD, Land H, Brooks M, et al. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–307. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan R, Ratajczak T. Versatile TPR domains accommodate different modes of target protein recognition and function. Cell Stress Chaperones. 2011;16:353–367. doi: 10.1007/s12192-010-0248-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juin P, Hueber AO, Littlewood T, Evan G. c-Myc-induced sensitization to apoptosis is mediated through cytochrome c release. Genes Dev. 1999;13:1367–1381. doi: 10.1101/gad.13.11.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel GA. CREB-binding protein and p300: molecular integrators of hematopoietic transcription. Blood. 2000;95:745–755. [PubMed] [Google Scholar]
- Rebel VI, Kung AL, Tanner EA, Yang H, Bronson RT, Livingston DM. Distinct roles for CREB-binding protein and p300 in hematopoietic stem cell self-renewal. Proc Natl Acad Sci USA. 2002;99:14789–14794. doi: 10.1073/pnas.232568499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasuyama H, Melchers F. Establishment of mouse cell lines which constitutively secrete large quantities of interleukin 2, 3, 4 or 5, using modified cDNA expression vectors. Eur J Immunol. 1988;18:97–104. doi: 10.1002/eji.1830180115. [DOI] [PubMed] [Google Scholar]
- Harris WJ, Huang X, Lynch JT, Spencer GJ, Hitchin JR, Li Y, et al. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell. 2012;21:473–487. doi: 10.1016/j.ccr.2012.03.014. [DOI] [PubMed] [Google Scholar]
- Somervaille TCP, Linch DC, Khwaja A. Different levels of p38 MAP kinase activity mediate distinct biological effects in primary human erythroid progenitors. Br J Haematol. 2003;120:876–886. doi: 10.1046/j.1365-2141.2003.04204.x. [DOI] [PubMed] [Google Scholar]