Abstract
The gastrointestinal (GI) epithelium is constantly renewing, depending upon the intestinal stem cells (ISC) regulated by a spectrum of transcription factors (TFs), including Myb. We noted previously in mice with a p300 mutation (plt6) within the Myb-interaction-domain phenocopied Myb hypomorphic mutant mice with regard to thrombopoiesis, and here, changes in GI homeostasis. p300 is a transcriptional coactivator for many TFs, most prominently cyclic-AMP response element-binding protein (CREB), and also Myb. Studies have highlighted the importance of CREB in proliferation and radiosensitivity, but not in the GI. This prompted us to directly investigate the p300–Myb–CREB axis in the GI. Here, the role of CREB has been defined by generating GI-specific inducible creb knockout (KO) mice. KO mice show efficient and specific deletion of CREB, with no evident compensation by CREM and ATF1. Despite complete KO, only modest effects on proliferation, radiosensitivity and differentiation in the GI under homeostatic or stress conditions were evident, even though CREB target gene pcna (proliferating cell nuclear antigen) was downregulated. creb and p300 mutant lines show increased goblet cells, whereas a reduction in enteroendocrine cells was apparent only in the p300 line, further resembling the Myb hypomorphs. When propagated in vitro, crebKO ISC were defective in organoid formation, suggesting that the GI stroma compensates for CREB loss in vivo, unlike in MybKO studies. Thus, it appears that p300 regulates GI differentiation primarily through Myb, rather than CREB. Finally, active pCREB is elevated in colorectal cancer (CRC) cells and adenomas, and is required for the expression of drug transporter, MRP2, associated with resistance to Oxaliplatin as well as several chromatin cohesion protein that are relevant to CRC therapy. These data raise the prospect that CREB may have a role in GI malignancy as it does in other cancer types, but unlike Myb, is not critical for GI homeostasis.
Keywords: p300, Myb, CREB, colorectal cancer, ApcMin/+
The epithelium of the small intestine (SI) and colon is the fastest self-renewing tissue in the body. Self-renewal is facilitated by intestinal stem cells (ISCs) near the bottom of the crypts. Gut ISC produce highly proliferating transit-amplifying cells. Maturation of these cells coincides with upward migration and differentiation into either secretory or mucin-secreting goblet cells, hormone peptide-secreting enteroendocrine cells and in the SI a fourth cell type; bactericidal and ISC niche-supporting Paneth cells (PC). When the differentiated cells reach the top of the SI villi or the surface of the colon mucosa, anoikis into the lumen follows.1 Most importantly, the gastrointestinal epithelium (GI) is highly sensitive to radio- and chemotherapy,2 and is subject to radiation-induced malignancy.3
Several pathways involved in the homeostasis of gut have been identified. The best documented is the Wnt pathway definitively shown to be involved in proliferation, the Notch pathway in cell fate and the Ssh/BMP-4 pathway, suggested to contribute to cell survival and -renewal.1, 4, 5 The adenomatous polyposis coli (APC) gene product serves as the key cancer ‘gatekeeper' that prevents the development of colorectal cancer (CRC) and aberrant activation of the Wnt pathway.1 APC mutations occur in ∼80% of CRC.6 Nevertheless, the precise molecular networks involved in the proliferation and differentiation of GI epithelium under normal, stressed and malignant transformation are far from being defined.
We and others have explored the role of the transcription factor (TF) cyclic-AMP responsive element-binding protein (CREB), finding a correspondence of active phosphor-CREB (pCREB) in proliferating cells in different tissues from diverse organisms. Deletion of the highly evolutionarily conserved creb gene alone or in combination with the related family member, crem, has shown the need for these TFs in developing and adult brain and in T-cell recovery in irradiated mice.7, 8, 9 In addition, in vitro and in vivo studies show that CREB also has a role in oncogenesis influencing melanoma, T-cell and myeloid leukemia, hepatocellular carcinoma, clear cell sarcoma, lung adenocarcinoma,10 as well as CRC.11
CREB is a member of the basic region leucine zipper (bZIP) family of TFs. bZIPs bind to the c-AMP responsive element (CRE), a conserved consensus sequence, as homodimers or heterodimers with the closely related family members CREM or ATF1 (Figure 1).12 All members contain a kinase-inducible domain (KIX) where the phosphorylation sites are located, a basic domain essential for DNA binding and a leucine zipper dimerization domain.13
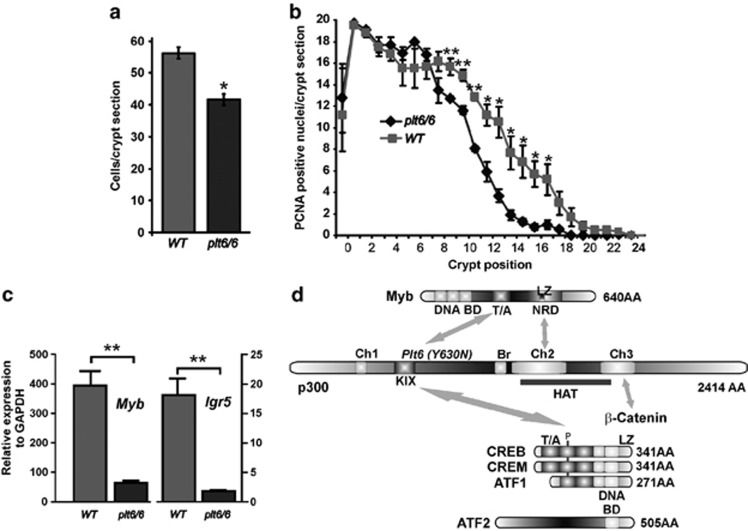
Figure 1.
p300 hypomorphic mutant (plt6/6) mouse intestinal defects phenocopy Myb mutant mice. (a) Like Myb hypomorphs, plt6/6 mutant mice have shorter crypts than WT mice (P=0.03). (b) This reduced crypt length corresponds to reduced proliferation, as determined by PCNA staining at crypts positions from the base (P<0.05–0.01). (c) Expression of ISC gene Myb is significantly reduced in colonic crypts isolated from plt6/6 mutants compared with WT as is Myb target and ISC gene, lgr5, P<0.01. (d) p300 and Myb can be viewed as being part of a larger complex of interacting TFs, including β-catenin, and most particularly, closely related CREB family members (CREB, CREM and ATF1) and more distantly related, ATF2. Linear protein lengths are shown in amino acid (AA) residues, and key interaction and function domains (DNA-binding domain (BD), transactivation domain (T/A), negative regulation domain (NRD) with leucine zipper-like domain (LZ), C=conserved homology domains (Ch), kinase interaction domain (KIX) and key conserved phosphorylation site (P). Means±S.E.M. one-tailed t-test
Multiple external stimuli induce CREB activation via several kinase pathways, including stress pathways.14 These signalling pathways lead to phosphorylation of CREB (pCREB) at a conserved serine residue (Ser133), triggering interaction with co-activators, including CREB-binding protein (CBP) or p300.15, 16 Importantly, p300 and CBP are not only CREB cofactors but also activate additional TFs, including Myb.
An in vivo interaction between Myb and p300 was revealed by the identification of the plt6 mouse strain containing a mutation in the KIX domain.17 This mutant mouse phenocopies three distinct hypomorphic Myb mutant lines with regard to elevated platelet numbers.18 The p300KIX domain interacts with the transactivation domain of Myb. Thus, the interaction between p300 and Myb was shown to be essential for cellular differentiation and lineage hierarchies in hematopoiesis.17, 19 Myb is an important TF for proliferation and differentiation of the intestinal epithelium.20 The transcriptional coregulator p300 might, therefore, integrate the co-activation and transcriptional activities of both pCREB and Myb, orchestrating finely tuned cellular responses in the GI. In this study, an inducible creb knockout (KO) model was used to define the role of CREB in the GI of adult mice in proliferation and differentiation under homeostasis and stress. Extensive pCREB activation was found in ApcMin/+ mouse adenomas. Moreover, in CRC pCREB was detected in most cells associated with elevated CREB-target genes, including CRC-relevant chemotherapy drug Oxaliplatin multidrug transporter, MRP2.
Results
p300 plt6/plt6 intestines phenocopy Myb hypomorphs
As part of a comprehensive ENU mutagenesis screen focused on thrombopoiesis18 or hematopoiesis,19 Myb emerged as a central factor in these processes.21 These observations were consistent with observations made in MybKO mice where definitive hematopoiesis was ablated and embryos died at ed15.22 The utility of the Myb hypomorphs came to the fore as they allowed the examination of other tissues that also expressed robust levels of Myb, notably the colon and SI in viable adults. Three distinct Myb hypomorphs showed defects in the GI, establishing an essential role for Myb in the GI,20 and later it was directly required for GI stem cell function.23 Two hypomorphs had mutations that impinged upon interaction between Myb and p300/CBP. When the p300plt6 mutation emerged from the ENU screen with a similar phenotype to the Myb hypomorphs,17 we posed the question whether p300 was also important for homeostasis in the GI.
The expression of p300 and CBP in the GI has been reported, and there are varying views about the relative importance of one over the other in development and homeostasis.24, 25 Therefore, we first examined the expression of both p300 and CBP proteins in the GI. (Supplementary Figure 1) Both factors are expressed in the majority of cells in the epithelial layer including post-mitotic cells, highlighting the importance of these transcriptional co-activators in both immature and differentiated cells.
Examination of the intestines of p300plt6/plt6 mice showed that the colonic crypts have 30% fewer cells per cross-section than littermate controls (Figure 1a), and there was a premature decline in PCNA+ve (proliferating cells) (Figure 1b). A similar proliferative defect was noted in the SI (data not shown). Of greater interest, however, was that these defects phenocopy those reported by us in Myb hypomorphs.20 Myb RNA expression in purified crypts was significantly reduced in the p300 hypomorph as was its target gene lgr523 (Figure 1c). As p300 interacts with many other TFs, we were mindful that this p300 mutation might be affecting other KIX-dependent interactions, most notably with the CREB family of TFs. These interactions are depicted in Figure 1d, raising the question of whether CREB, which is expressed in gut epithelium, might be similarly important in the GI for proliferation and crypt homeostasis.
CREB recombination is specifically and efficiently induced in the GI tract
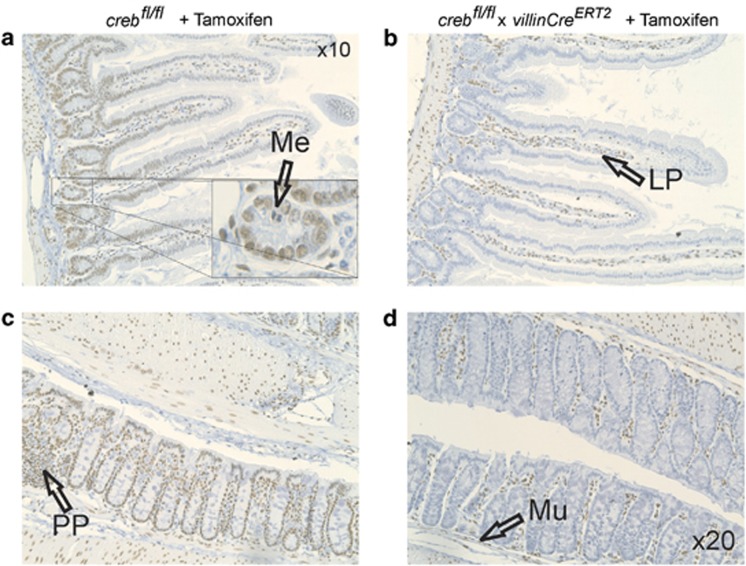
In view of studies demonstrating that CREB contributes to proliferation and differentiation of different cell types in multiple non-GI tissue compartments, we deleted CREB in the intestine using a Tamoxifen-inducible CRE-mediated intestine-specific crebKO mice line. In these mice, exon 10 was flanked by loxP sites, and earlier studies indicated that the deletion of exon 10 results in loss of all CREB polypeptides.9 To assess whether CREB deletion occurred in the gut epithelium after 4 weeks of Tamoxifen treatment, immunohistochemistry (IHC) was employed using antibodies against CREB. Control littermates (crebfl/fl+Tamoxifen) showed CREB protein throughout the colonic crypts and the SI extending into the villi (Figures 2a and c). Enterocytes were the predominant CREB+ve cell type, but cells in metaphase were CREB−ve. Importantly, CREB was evident in the lamina propria, lymphoid-rich Peyer's patches and basal stromal regions, particularly where endothelial cells would be expected to reside. The muscularis also contained a few CREB+ve cells. When epithelial cells were examined after Tamoxifen treatment, all crypt and villi exhibited loss of CREB expression (Figures 2b and d). These data indicate that the creb deletion (from this point called crebKO) was epithelial-specific, pervasive and uniform, but resulted in only modest effects on GI architecture.
Figure 2.
Complete intestinal epithelial deletion of CREB. crebfl/fl mice (a and c) and crebfl/fl x villinCreERT2 mice (b and d) were proved chow with Tamoxifen ad libitum for 4 weeks before cull, and preparation of colon and SI sections. These were subjected to IHC with an antibody specific to CREB to show complete loss of antigen in epithelial cells associated with crypts, but not lamina propria (LP), Peyer's patches (PP) or muscle (Mu). (Insert panel) Metaphase (Me) figures within the epithelial compartments were CREB−ve (arrow)
Effect of CREB loss on crypt proliferation and cell fate
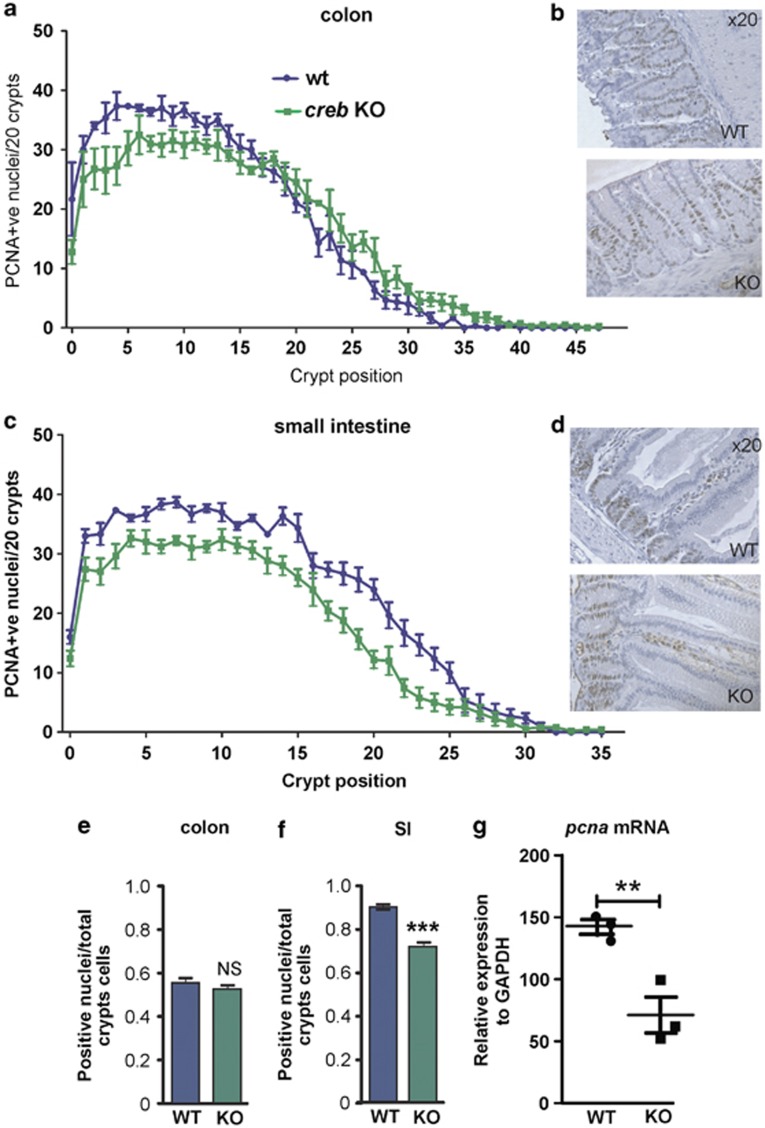
As crypts have a high turnover and pcna has been reported to be a CREB target gene,26 we examined the proliferative status in WT and crebKO intestines by IHC (Figures 3a–d). Overall, there was a reduction in PCNA+ve cells at the crypt base in SI and colon (Figures 3a and c), but this was only significant in the SI when considering the total number of PCNA+ve nuclei (Figures 3e and f) and was confirmed by evaluation of mRNA by qRT-PCR (Figure 3g). To further investigate the crebKO colon and SI architecture, we explored the relative numbers of Chromogranin A+ve cells, as CREB regulates the ChrA gene27, 28 and neuroendocrine cell function.29, 30, 31, 32 However, the number of these cells appeared elevated in the SI and significantly increased in the colon in crebKO mice. Goblet cells and mucin production, as judged by period acid shift (PAS) histochemistry, was more intense in the crebKO colons (Supplementary Figure 2).
Figure 3.
Modestly reduced proliferation in CREB−ve intestinal crypts despite uniform epithelial KO. (a and b) PCNA IHC was performed and determined at each crypt position from the base, showing a modest reduction in proliferation in the transit-amplifying region of the colonic crypt (d and e) and throughout the SI crypt. (e) Overall, the total PCNA staining in the colon was not significantly different (f), whereas a statistically reduced number of PCNA+ve nuclei were found in the SI. (g) Relative levels of mRNA for CREB target gene pcna was found to be statistically reduced following KO. Means±S.E.M., two-tailed t-test. *P=0.01; ***P=0.001
Transcriptionally Active pCREB is not compensated for by other family members
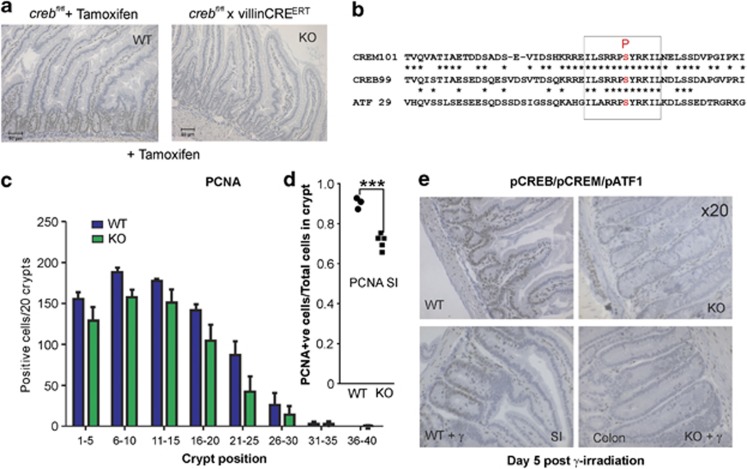
In its active form, CREB is phosphorylated on a key serine residue (ser133)14 (Figure 1d), and pCREB correlates best with proliferation.33 Accordingly, GI sections were investigated for the presence of pCREB, where we found a similar distribution of PCNA+ve and pCREB within nuclei. No pCREB staining epithelial cells were evident in the crebKO sections (Figure 4a), whereas this active form of CREB, unlike the CREB protein in general, is restricted to the crypts in the SI, consistent with the location of the proliferating transit-amplifying cell and stem cell compartment.34 Importantly, due to close structural similarity, the pCREB antibody also recognizes the kinase-inducible domains shared by family members, CREM and ATF135 (Figure 4b), which allows the conclusion to be drawn that the active form of these other members are not co-expressed in the GI epithelium, and therefore, unable to compensate for CREB loss. To extend this analysis further, we also employed an antibody against a more distantly related TF, ATF2 (Figure 1d), finding it to be expressed in a similar manner to pCREB, but in the crebKO this antigen is expressed to a weaker extent compared with WT crypts (Supplementary Figure 3).
Figure 4.
No apparent compensation for CREB loss by family members, CREM and ATF1, including following irradiation. (a) WT and KO sections were subjected to IHC with phospho-CREB antibodies to show that under steady state KO, SI epithelial cell showed no signal. (b)This antibody also detects closely related phospho-CREM and phospho-ATF1. (c and d) When mice were exposed to whole-body irradiation and intestines processed, 5 days later it was apparent that the small but significant difference in proliferation observed in unirradiated mice in the SI was not exacerbated with radiation treatment. (e) It was notable that the extent of phospho-CREB/CREM/ATF1 was more restricted to the base of crypts in WT mice (SI) and that all three CREB members were not present in the KO crypts (colon) with or without radiation, indicating that these were not induced by radiation damage at this time point
Defects in proliferation are sometimes revealed or exacerbated by stressing the GI with radiation, as observed with Myb mutant mice,21 although Myb expression itself was unaffected in the unirradiated CREBKO GI (Supplementary Figure 4). Indeed, CREB has been implicated indirectly influencing radiosensitivity, and its target gene pcna is intimately involved in DNA repair.36 Mice were thus whole-body irradiated with 13 Gy and allowed to recover for 5 days, at which time point, the mice were killed and GI sections prepared. PCNA IHC showed that in a similar manner to the unirradiated control, there was a significant, but apparently no greater reduction in proliferation in the crebKO mice SI (Figure 4c). Even under these stress conditions, pCREB/pCREM/pATF1 were not induced in irradiated GI (Figure 4e), suggesting that PCNA expression under these conditions is in part under the influence of pCREB but that CREB loss does not lead to GI hypersensitivity to radiation, challenging previous views based upon using dominant negative forms of CREB.36
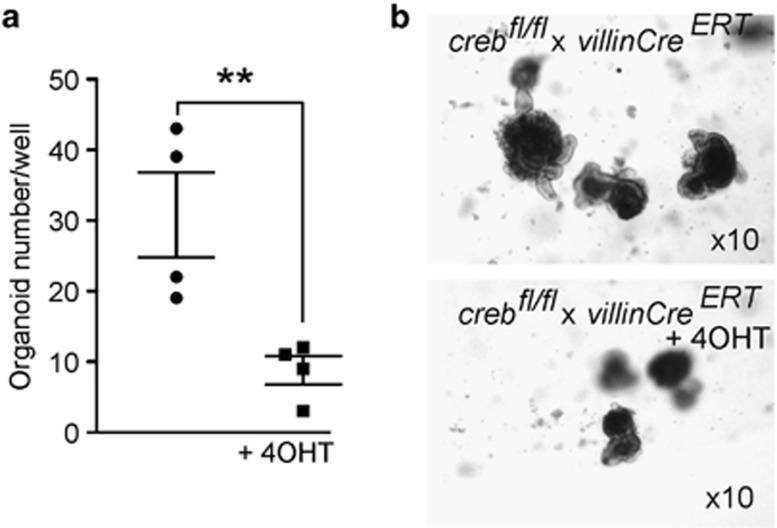
In vitro deletion of CREB impedes SI organoid formation
Having found only modest defects in proliferation of crebKO crypts under homeostasis or following challenge with radiation, we decided to quantify the ability of crypt stem/progenitor cells to initiate and form organoids ex vivo.37 Crypt nests were isolated from creb/villinCreERT2 mice that had not been exposed to Tamoxifen and were plated in phenol-free Matrigel with one group receiving 4-hydroxy-tamoxifen (4OHT) throughout the 7-day-culture period. Previous experience had shown that 4OHT had no demonstrable effect on organoid formation for crypt nests isolated from WT mice (n=15; data not shown). MTT growth assay (Figure 5) showed that crebKO significantly impedes organoid growth. These effects were similar to those observed in Myb hypomorph (MybPlt4/Plt4) organoid formation,23 suggesting that CREB loss does affect stem/progenitor cell proliferation when evaluated ex vivo.
Figure 5.
In vitro culture of SI organoids reveals a defect in colony-forming ability in KO mice. (a) Organoid cultures were initiated from crebfl/fl x villinCreERT2 mice and these were exposed to 4OHT to show reduced colony-forming efficiency in the KO cultures; Means±S.E.M. two-tailed t-test; **P<0.01. (b) Representative images are shown, (mag= × 10)
When candidate cell cycle genes reported to be CREB targets were examined by qRT-PCR of total RNA isolated from parallel organoid cultures, no evidence of changes in CyclinD1, CyclinA1, Erg138 or Bcl-239 expression were observed (data not shown).
CREB is elevated in intestinal adenomas and adenocarcinoma cells
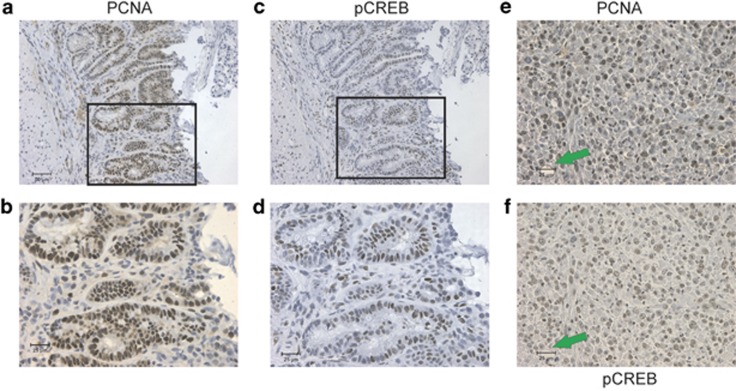
As CREB loss leads to reduced proliferation in the GI and CREB has been shown to activate PCNA expression36 and impart oncogenic properties in some tissues,35, 40 we investigated CREB expression in tumor tissue. Intestinal adenomas from the Apcmin/+ mouse were examined by IHC (Figures 6a and d) to show concordant PCNA and pCREB in the aberrant epithelial cells that align through consecutive sections. Furthermore, this relationship is also evident in mouse MC38 CRC cells (Figures 6e and f).
Figure 6.
Proliferation and phospho-CREB in intestinal adenomas and colon adenocarcinoma are concordant. (a and b) Apcmin/+ mice develop intestinal adenomas, which contain a large fraction of cells that are highly proliferative as shown by PCNA staining. (c and d) The pattern of PCNA staining mirrors that for phosphor-CREB. (e and f) MC38 colon adenocarcinoma cells form large tumors in syngeneic mice (C57Bl/6), which show high levels of PCNA+ve and pCREB+ve nuclei (green arrows indicate a focus to align nuclei in each micrograph)
Gene expression analysis using RNAseq
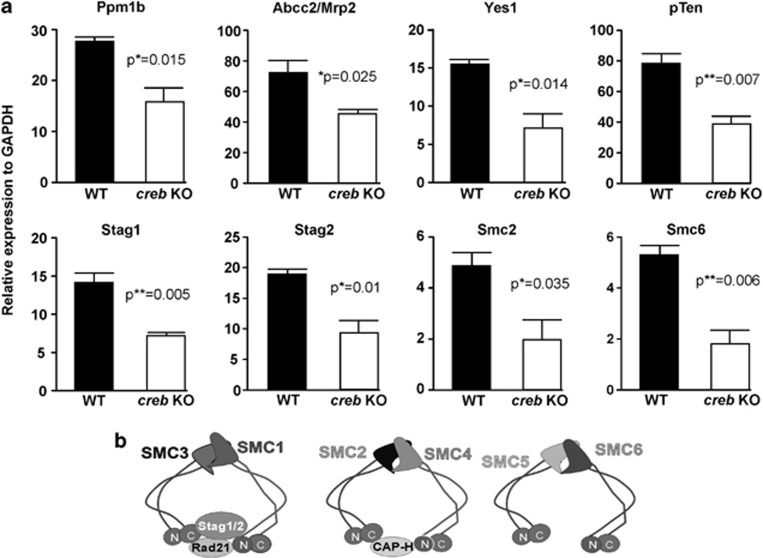
As the anticipated gene expression effects that might be expected in crebKO tissue were not observed, we isolated SI crypts for gene expression studies (these were depleted of the villi) from three creb/villinCreERT2 and three crebfl/flcontrol mice where each cohort had been subjected to 4-week Tamoxifen treatment. This was important for both groups, as Tamoxifen influences transcription.41, 42 This strategy was also considered to most closely parallel that used to characterize the GI morphology, to examine the GI cells most likely to express pCREB and to capture unanticipated gene expression changes. This unbiased approach confirmed our previous data regarding no changes in the anticipated cell cycle genes (Supplementary Data File). However, several other genes showed significant changes that were confirmed by qRT-PCR, some of which had previously been suggested to be CREB target genes. Most notable was the lower expression of pcna, and of course creb itself. Among the genes showing the differential expression were phosphatases, Ppm1b and PTEN, tyrosine kinase Yes1 and cytotoxic chemotherapy drug transporter ABCC2/MRP2. We also noted that several chromatin cohesin genes stood out as being differentially reduced in the crebKO SI. These genes were thus evaluated by qRT-PCR as shown in Figure 7.
Figure 7.
Messenger RNA expression changes in selected genes identified by RNAseq studies validated by qRT-PCR. (a) Significant changes in mRNA levels for phosphatases, Pmm1b, pTen, multidrug resistance gene Abcc/Mrp2 and tyrosine kinase gene, Yes1. Four genes that encode proteins engaged in chromosome cohesion, Stag1, Stag 2, Smc2 and Smc6, are similarly lower in the crebKO small intestine crypts. Cartoons depicting the structural roles and general theme of interactions of the cohesion molecules are shown (b). Models of the structural convergence of several putative CREB target gene products, Stag1, Stag 2, Smc2 and Smc6, all of which are involved in cohesion functions, are shown
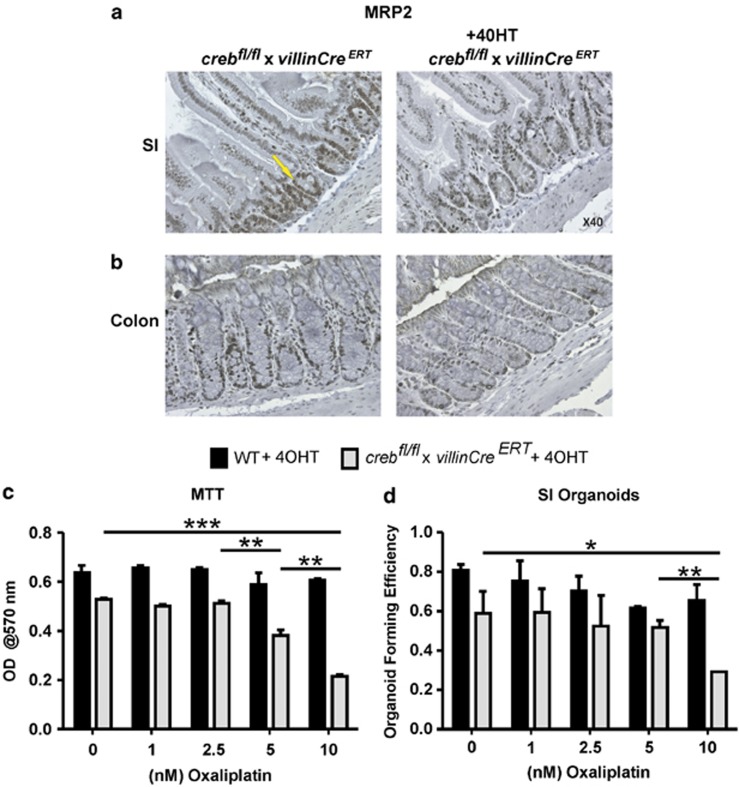
To extend the analysis of the two genes most directly implicated in carcinogenesis that were expressed differentially in the crebKO SI, we employed antibodies to tumor suppressor protein, PTEN and drug transporter MRP2. Supplementary Figure 5 indicates that PTEN is expressed at a lower level in the crebKO, and most notably, in the cytoplasm and apical membranes in WT villi. MRP2 expression, however, was restricted to crypts in both the SI and the lower half of colonic crypts in WT mice and this signal was markedly diminished in KO mice (Figures 8a and b). Although MRP2 is reported to be apically expressed,43, 44 this was not overtly evident in WT sections. MRP2, unlike other MRPs, is elevated in CRC at the mRNA level45 and when adenomas and MC38 sections were examined for MRP2 protein, abundant expression was observed (Supplementary Figure 6). MRP2 is of substantive interest as frontline CRC cytotoxic drugs, Oxaliplatin, 5-Fluorouracil and Camptothecin are substrates or are influenced by MRP2 overexpression.44, 45, 46
Figure 8.
Multidrug transporter MRP2 is underexpressed in creb KO intestine. (a) Small intestine, and (b) colon sections were stained with αMRP2 antibodies (yellow arrow) to reveal reduced expression in crebfl/fl/villinCreERT2crypts compared with WT, following 4-weeks exposure to Tamoxifen. (c and d) Organoid cultures were established in the presence of 4OHT for 3 days, and subsequently subjected to treatment with Oxaliplatin after cultures were allowed to proceed for another 7 days. These results show that as expected, the growth (MTT) and organoid-forming ability was impeded by crebKO alone, but these measures were significantly reduced compared with WT in the presence of Oxaliplatin. Means±S.E.M., two-tailed t-test. *P=0.01; ***P=0.001
To explore this relationship more directly, we established organoid cultures from WT and crebf/f/villinCreERT2mice, allowing the culture to establish in the presence of 4OHT for 3 days prior to the addition of Oxaliplatin. As expected, the crebKO organoids grew less efficiently than the WT organoids, however, beyond this baseline difference, colony numbers and growth assessed by the MTT assay indicated that loss of CREB further sensitizes primary intestinal cells to the cytotoxic effects of the CRC chemotherapeutic drug (Figure 8).
Discussion
A growing collective of TFs are involved in achieving intestinal crypt homeostasis and previously we have specifically shown a role of Myb,20 exploiting several Myb hypomorphic mutant mice,18, 19 sustained survival of which allowed the analysis of adult tissues not possible with embryonically lethal MybKO mice22 and that complemented the inducible intestinal-specific MybKO studies.20 Our discovery that the p300plt6/plt6 mutant in part phenocopied the Myb hypomorphs17 and indeed led to a substantial reduction in Myb expression highlights the utility of these mutants. That the p300 mutation resides within the KIX domain raised a parallel question about the role of the archetypical p300/CBP-binding partner CREB in intestinal biology. Indeed, as transcription is mostly an orchestrated process of many TFs, we explored the interplay of p300 with Myb and CREB in regulating intestinal biology.
Protein (histone) acetyl transferases p300 and CBP are transcriptional co-activators and central to transcriptional activation of many genes through direct interaction with partner TFs that directly bind to enhancer elements within target gene promoter regions.47 Several interaction faces within p300/CBP are implied, with the KIX box being the best characterized.48 Of key relevance here is the kinase-inducible domain (KID) in CREB and the transactivation domain in Myb, which share structural similarities49 implying that Myb and CREB may even directly compete for KIX domains in p300/CBP. The intriguing observation that even though CBP expression is coincident with p300, it does not compensate for the defects associated with the p300 hypomorphic mutation, is also evident in the hematopoietic system.50
Evidence that p300 might be important in the intestines has been reported,25 with mRNA analysis in the rat indicating that p300 is expressed in excess of CBP.24 Here, we have shown by IHC that p300 is expressed to a similar extent to its close relative protein, CBP, within the crypt base and in the transit-amplifying regions. There is some debate about the relative contributions of each of these acetyl transferases, but the most convincing case is made for p300.25 Thus, the defects in the p300plt6/plt6 mutant leading to reduced proliferation, distorted differentiation and indeed shorter crypts are completely consistent with a central role for p300, and not for CBP.
When we examined the role of CREB in the intestines by tissue-specific deletion, it was anticipated that crypts would be either lost or replaced by crypts in which CREB was not deleted. This view was informed by our previous studies in high-proliferation tissues in mice and zebrafish,8 and other work that collectively shows that CREB is important in the maintenance of proliferation and protection from apoptosis. The villinCre-driven deletion of CREB in both the colon and SI was essentially absolute and restricted to the epithelial cells, sparing cells in the lamina propria and muscularis. Nevertheless, near-normal looking crypts were evident, and directed investigation into proliferation and specific cell lineages revealed relatively subtle effects. For instance, by evaluating the PCNA+ve nuclei at each crypt cell position it became evident that proliferation could be maintained, but to a reduced extent. Such an effect was seen in the hypomorphic p300plt6/plt6 and Myb mutant mice,20 although these were not full loss of function mutants, whereas the crebKO was null. Similarly, an increase in mucin production and the presence of more ChrA+ve cells suggested that perhaps goblet and enteroendocrine cells may be abnormally increased, however, only goblet cells were increased and enteroendocrine cells were reduced in the hypomorphs.20 Together these data indicate that Myb has a more dominant role in crypt proliferation compared with CREB, and that p300 is required for Myb to prosecute this role, whereas CREB has mostly a distinct effect on the differentiation of secretory lineages.
The question remains as to why CREB loss has such a modest effect on this highly proliferative tissue? We were concerned that CREB loss might be compensated for by family members, CREM and ATF1, as reported in brain.9 However, these other family members were not expressed in the GI under homeostasis or when stressed, excluding compensation. Nevertheless, using RNAseq and by validation using qRT-PCR and IHC, a range of gene expression changes suggested that CREB regulates an important group of genes most relevant to carcinogenesis, for example, PTEN, MDR2 and, most interestingly, related proteins involved in chromatin/chromosome cohesion (for example, both STAG1 and 2 (SCC3; SA1 and 2) and SMC2, as well as SMC6). The established role of CREB in regulating these cohesion proteins in the context of DNA repair certainly warrants further investigation.
With regard to PTEN, it is fascinating that it is expressed at reduced levels in the crebKO crypts, as it is reported that the CREB protein is a target for PTEN phosphatase action,51 which opens up the prospect that a regulatory feedback loop may involve pCREB and PTEN.
Much has been made of the role of CREB in radiation sensitivity.36 Accordingly, we were surprised to find that deletion of CREB in vivo in a highly radiation-sensitive tissue like GI was innocuous. One clear difference between these contrasting observations might be that the radiosensitivity studies were done in cell lines with dominant negative forms of CREB36 and this may reflect that other mechanisms might be at play.
Reducing p300 function might be predicted to shift TF interactions to those with CBP as is argued for β-catenin,52 and this in turn would favor an expansion of non-differentiated (and perhaps) stem and progenitor cells. However, the opposite appears to be the case for the p300plt6/plt6 hypomorph, where reduced proliferation and enhanced differentiation occur. Overall, the data presented here suggest that much of the effects exerted by p300 in the GI are mediated through Myb and are also apparent in the hematopoietic system,17, 19, 53, 54 and, by contrast, CBP seems less important in both of these highly proliferative compartments.
The modest roles of CREB and, by implication, CREM and ATF1 in the GI was a genuine surprise, but there was evidence that pCREB/CREM/ATF1 is elevated in early-stage and advanced adenocarcinoma in the mouse. Commensurate with this association is the increased expression of a key cytotoxic drug transporter, MRP2, which may have a role in the response of CRC to frontline drugs, Oxaliplatin, 5-Flurouracil and Camptothecin. Based upon other studies where increased MRP2 may invoke resistance to cytotoxic drugs,46 it is reasonable to speculate that the elevated pCREB evident in mouse adenomas and CRC may also occur in human CRC and have a role in compromising the therapeutic impact of these drugs. Overall, the data presented here indicate that Myb is central to the action of p300 in the GI and that if CREB is working through p300, it is more engaged in potentially important gene regulation that impinges upon CRC but not GI homeostasis or radiation response.
Materials and Methods
Mice
creb/villinCreERT2 mice were generated by crossing crebf/f mice, as described,9 to villinCreERT2mice55 on a C57BL/6J background. Mice were housed in a specific pathogen-free (SPF) facility and fed a standard animal diet in Peter MacCallum Cancer Centre. For induction of creb recombination, 5-week old creb/villinCreERT2 mice were fed chow ad libitum supplemented with 0.1 g Tamoxifen citrate salt (Sigma, Australia, Castle Hill, NSW)/120 g food for 4 weeks. Littermates continued to be fed with a standard chow. The p300plt6/plt6 mutant mice were generated as described17 and maintained at the Walter and Eliza Hall Institute. ApcMin/+ mice56 were maintained on a C57BL/6J background. All animal experiments were approved by the Animal Ethics Committee of the Peter MacCallum Cancer Centre. The carcinogen-induced colon carcinoma cell line MC38 is described elsewhere, and was injected subcutaneously in C57Bl/6 mice to generate tumors.57
Antibodies
Primary antibodies used in this study were: αCREB (1 : 500) (Cell Signaling, USA 9197), αpCREB (1 : 250) (Cell Signaling, Danvers, MA, USA, 9198), αChromogranin A (ChrA), (1 : 100) (Santa Cruz Biotechnology, Santa Cruz, CA, USA H-300), αPCNA (1 : 100) (BD Biosciences, Bedford, MA, USA), αNeurogenin 3 (NGN3), (1 : 700) (Santa Cruz Biotechnology, M-80), αATF2 (1:200) (Cell signaling), αCREM (1 : 100) (Abcam, Cambridge, MA, USA), αCBP, (1 : 100) (Santa Cruz Biotechnology, C-1), αMyb1.1 (Upstate Biotech, Lake Placid NY, USA), αp300 (1 : 800) (Santa Cruz Biotechnology, C-20), αMRP2 (1 : 500) (Santa Cruz Biotechnology, no. SC-5770), αpTen (1 : 300) (Cell Signaling, CS-9559). Secondary antibodies used were: anti-rabbit Impress reagent kit peroxidase (Vector labs, Burlingame, CA, USA), Envision system labeled polymer-HRP anti-mouse (Dako, Campbellfield, VIC, Australia), donkey-anti-goat IgG-HRP (Santa Cruz Biotechnology).
Irradiation
creb/villinCreERT2 mice treated with Tamoxifen for 2 weeks were exposed to a sublethal dose of γ-irradiation of 13 Gy. As irradiation compromises the immune system, 2.5 ml of 0.25% (w/v) Neomycin Sulfate and 0.13% (w/v) Polymyxin B Sulfate in dH2O were added to the drinking water of the mice in the days after irradiation. On day 5, the mice were culled, and the SI and colons were isolated and processed for histology.
Histology and Immunohistochemistry (IHC)
SI and colons (creb/villinCreERT2; p300plt6/plt6), and carcinomas from C57Bl/6J mice injected with the MC38 cell line were isolated and fixed in Methacarn (60% methanol, 30% chloroform, 10% acetic acid), dehydrated in a graded series of ethanol from 70 to 100% and then immersed in xylene. The tissues were embedded in paraffin, and 4-μm sections were cut with the paraffin microtome and transferred onto 3-aminopropyl-triethoxy silane-coated glass slides.
For IHC, sections of SI and colon were dewaxed and dehydrated by immersing in xylene and a graded series of ethanol. Slides were boiled either in citrate buffer (10 mℳ tri-sodium citrate, pH 6.0) (CREB, pCREB, PCNA, ChrA), EDTA buffer (1 mℳ EDTA disodium salt dehydrate, pH 8.0) (NGN3, CREM) or TRIS buffer (10 mℳ TRIS, pH 9.0) (p300, CBP) for 3 min at 125 °C and 10 s at 90 °C in the Dako pressure cooker. Endogenous peroxidases were then blocked with 3% (v/v) H2O2 for 10 min. Slides were washed in Tris-buffered saline-Tween-20 (TBS-T) (5 M Tris, 0.15 M NaCl, pH 7.6, with 0.1% (v/v) Tween-20), and for p300, blocked in 5% (w/v) bovine serum albumin in TBS-T for 30 min. Then antibodies were added in TBS-T and incubated for 1 h at RT (PCNA, ChrA, CBP, p300) or O/N at 4 °C (CREB, pCREB, NGN3, CREM). Sections were washed with TBS-T and incubated with the corresponding secondary antibody for 30 min at RT. After washing in TBS-T, sections were exposed to 3,3′-diaminobenzidine tetrachloride (DAB+ substrate chromogen system) (Dako) for 1–10 min, resulting in a positive brown staining visible under the light microscope. For mucin staining, sections were exposed to periodic acid for 10 min followed by incubation with Schiff's reagent (PAS staining) (Australian Biostain, Traralgon, VIC, Australia). Slides were counterstained with hematoxylin for several seconds. After washing in water, slides were dehydrated in a graded series of ethanol, immersed in xylene and mounted with DPX.
Organoid culture
SI of mice (creblox/lox;villinCREERT2 and control littermates; between 3 and 5 weeks old) were isolated, opened longitudinally and cleaned in phosphate-buffered saline (PBS) supplemented with 0.1 mg/ml Nystatin (Sigma-Aldrich, St. Louis, MO, USA) and 0.1 mg/ml Gentamicin (Invitrogen, Carlsbad, CA, USA) (PBS-GN). The isolated intestines were incubated two times 15 min in PBS-GN on a shaker at 4 °C, followed by incubation in PBS containing 2 mℳ EDTA for 30 min at 4 °C. Then the tissues were shaken vigorously in 20 ml cold PBS, of which the supernatant was discarded. The tissues were transferred to 10 ml of cold PBS and shaken vigorously again to obtain fraction 2. This step was repeated to obtain fraction 3. Fractions 2 and 3 were combined and spun down at 200 × g for 3 min after which the supernatant, containing single cells, was removed. The pellet was resuspended in PBS and filtered through a 40-μm filter. The required amount was spun down at 400 × g for 6 min. Five hundred or 1000 viable crypts were seeded in 50 μl Matrigel Basement Membrane Matrix (BD Bioscience) per well of a 24-wells plate. The 24-wells plate was then placed at 37 °C for 30 min to allow polymerization. Control crypts were cultured in crypt culture medium consisting of Dulbecco's Modified Eagle Medium nutrient mixture F-12 (DMEM/F12; Sigma) supplemented with 10% (v/v) fetal calf serum (FCS), 100 U/ml penicillin and 100 U/ml streptomycin, 1 ng/ml recombinant mouse basic-FGF (eBioscience, San Diego, CA, USA), 2 ng/ml murine EGF (Peprotech, Rocky Hill, NJ, USA), 1 ng/ml recombinant mouse R-spondin-1 (R&D System Minneapolis, MN, USA), 0.1 ng/ml murine Noggin (Peprotech), B-27 Supplement Minus AO (Invitrogen). Half of the crypts of each mouse group were cultured in crypt culture medium supplemented with 0.1 μg/ml 4OHT. Medium was changed every 2–3 days, and after 7 days, the cells were isolated by incubation with 50 μl of Accumax (Millipore, Temecula, CA, USA) for 30 min at 37 °C.
Oxaliplatin (Ebewe Pharmaceutical Company, Touristic Area, Egypt) was dissolved in sterile dH2O and was added on day 3 following 4OHT (10−7 M) (Sigma). Organoids were processed as described previously.23
RNA isolation, DNAse treatment and complementary DNA synthesis
For isolation of RNA from cells of cultured organoids RNeasy mini kit (QIAGEN, Hilden, Germany) was used according to the manufacturer's instructions. RNA was DNAse treated by adding 10 μl 10 × DNAse1 Buffer (Promega, Madison, WI, USA), 3 μl RQ1 RNase-Free DNase1 (1 U/μl; Promega), 0.5 μl RNAsin (40 U/μl; Promega), 5 μl DTT (0.1 M; Invitrogen) and 59 μl H2O, prior to incubation at 37 °C for 30 min. Then, 100 μl H2O was added followed by 200 μl RNA-buffered phenol-chloroform (pH 5). The samples were spun down at 13 000 r.p.m. for 5 min, and an equal volume of 4% (v/v) isoamyl ethanol in chloroform was added to the top phase. After another spin at 13 000 r.p.m. for 5 min, the top phase was precipitated with three equal volumes of ethanol/sodium acetate (3% (v/v) 3 M NaAC; pH 5.6; in ethanol) O/N at −80 °C. Two microliter of glycogen (20 mg/ml) was added for visualization of the pellet. After incubation a spin at 13 000 r.p.m. for 15 min followed and the resulting pellet was washed in 70% (v/v) ethanol. Then the samples were spun down at 7800 r.p.m. for 8 min, the pellet was air dried for 5–10 min and dissolved in H2O. RNA concentrations were quantified with the nanodrop.
Complementary DNA (cDNA) was synthesized by incubating 2 μg RNA with 0.5 μl random primers (500 μg/ml) (Promega) and 2 μl 5 mℳ dNTP for 5 min at 65 °C. After incubation on ice for 1 min, a reaction mixture of 4 μl 5 × First strand buffer (Invitrogen), 2 μl 0.1 M DTT, 1 μl RNAsin (40 U/μl) and 1 μl Superscript III Reverse Transcriptase (200 U/μl; Invitrogen) was added and the samples were incubated for 10 min at RT, after which an incubation of 60 min at 50 °C followed. For inactivation of the reverse transcriptase reaction, the mixtures were incubated for 10 min at 70 °C.
RNA sequencing
Large RNA was extracted from SI crypts using the miRNeasy mini kit and RNeasy MinElute Cleanup kit (QIAGEN). RNA concentration and quality was determined to be suitable using NanoDrop spectrophometer (Thermo Scientific, Wilmington, DE, USA) and integrity verified using the RNA 6000 kit (Agilent Technologies, Santa Clara, CA, USA). Library preparations were performed using the TruSeq RNA Sample Preparation protocol (Illumina, San Diego, CA, USA) and correct size established using the DNA 1000 kit (Agilent technologies). Libraries were quantified with qPCR, normalized and pooled to 2 nM before sequencing with single ended 50 bp reads using standard protocols on the HiSeq2000 (Illumina, San Diego, CA, USA). Computational resources were provided by Galaxy Project at Penn State.58, 59, 60
qRT-PCR
A mixture of 0.8 μl of cDNA, synthesized as described before, was combined with 10 μl of SyBr Green PCR Master Mix (Applied Biosystems, Carlsbad, CA, USA), 2 μl of 2 mℳ forward and reverse oligonucleotides (Geneworks, Adelaide, SA, Australia) and 7.2 μl of H2O, and amplified in the StepOnePlus Real-Time PCR system (Applied Biosystems). Amplification conditions used were: 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min, and a final cycle for the melt curve at 95 °C for 15 min, 60 °C for 1 min and 95 °C for 15 s. Expression levels of all genes were compared with GAPDH expression in order to determine relative mRNA levels. Primer sequences used for quantitative RT-PCR are tabulated in Supplementary Table 1
Acknowledgments
This work was supported by Fellowships (WSA, RGR) and Program Grants (1016647; WSA, 487922; JM and RGR) from the National Health and Medical Research Council of Australia. We thank the Molecular Genomics Core for their expert assistance with the RNAseq studies.
Glossary
- AB
alcian blue
- CREB
cyclic-AMP response element-binding protein
- PAS
periodic acid Schiff
- PCNA
proliferating cell nuclear antigen
- SI
small intestine
- ISC
intestinal stem cell
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by G Melino
Supplementary Material
References
- Gregorieff A, Clevers H. Wnt signaling in the intestinal epithelium: from endoderm to cancer. Genes Dev. 2005;19:877–890. doi: 10.1101/gad.1295405. [DOI] [PubMed] [Google Scholar]
- Novak JM, Collins JT, Donowitz M, Farman J, Sheahan DG, Spiro HM. Effects of radiation on the human gastrointestinal tract. J Clin Gastroenterol. 1979;1:9–39. doi: 10.1097/00004836-197903000-00003. [DOI] [PubMed] [Google Scholar]
- Chang ML, Hou JK. Cancer risk related to gastrointestinal diagnostic radiation exposure. Curr Gastroenterol Rep. 2011;13:449–457. doi: 10.1007/s11894-011-0214-8. [DOI] [PubMed] [Google Scholar]
- Ishizuya-Oka A, Hasebe T. Sonic hedgehog and bone morphogenetic protein-4 signaling pathway involved in epithelial cell renewal along the radial axis of the intestine. Digestion. 2008;77 (Suppl 1:42–47. doi: 10.1159/000111487. [DOI] [PubMed] [Google Scholar]
- Lee CS, Perreault N, Brestelli JE, Kaestner KH. Neurogenin 3 is essential for the proper specification of gastric enteroendocrine cells and the maintenance of gastric epithelial cell identity. Genes Dev. 2002;16:1488–1497. doi: 10.1101/gad.985002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- Baumann S, Kyewski B, Bleckmann SC, Greiner E, Rudolph D, Schmid W, et al. CREB function is required for normal thymic cellularity and post-irradiation recovery. Eur J Immunol. 2004;34:1961–1971. doi: 10.1002/eji.200324826. [DOI] [PubMed] [Google Scholar]
- Dworkin S, Heath JK, deJong-Curtain TA, Hogan BM, Lieschke GJ, Malaterre J, et al. CREB activity modulates neural cell proliferation, midbrain-hindbrain organization and patterning in zebrafish. Dev Biol. 2007;307:127–141. doi: 10.1016/j.ydbio.2007.04.026. [DOI] [PubMed] [Google Scholar]
- Mantamadiotis T, Lemberger T, Bleckmann SC, Kern H, Kretz O, Martin Villalba A, et al. Disruption of CREB function in brain leads to neurodegeneration. Nat Genet. 2002;31:47–54. doi: 10.1038/ng882. [DOI] [PubMed] [Google Scholar]
- Siu YT, Jin DY. CREB--a real culprit in oncogenesis. FEBS J. 2007;274:3224–3232. doi: 10.1111/j.1742-4658.2007.05884.x. [DOI] [PubMed] [Google Scholar]
- Nishihara H, Hwang M, Kizaka-Kondoh S, Eckmann L, Insel PA. Cyclic AMP promotes cAMP-responsive element-binding protein-dependent induction of cellular inhibitor of apoptosis protein-2 and suppresses apoptosis of colon cancer cells through ERK1/2 and p38 MAPK. J Biol Chem. 2004;279:26176–26183. doi: 10.1074/jbc.M313346200. [DOI] [PubMed] [Google Scholar]
- Lalli E, Sassone-Corsi P. Signal transduction and gene regulation: the nuclear response to cAMP. J Biol Chem. 1994;269:17359–17362. [PubMed] [Google Scholar]
- Gonzalez GA, Yamamoto KK, Fischer WH, Karr D, Menzel P, Biggs W, 3rd, et al. A cluster of phosphorylation sites on the cyclic AMP-regulated nuclear factor CREB predicted by its sequence. Nature. 1989;337:749–752. doi: 10.1038/337749a0. [DOI] [PubMed] [Google Scholar]
- Shaywitz AJ, Greenberg ME. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem. 1999;68:821–861. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- Lundblad JR, Kwok RP, Laurance ME, Harter ML, Goodman RH. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature. 1995;374:85–88. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- Kauppi M, Murphy JM, de Graaf CA, Hyland CD, Greig KT, Metcalf D, et al. Point mutation in the gene encoding p300 suppresses thrombocytopenia in Mpl−/− mice. Blood. 2008;112:3148–3153. doi: 10.1182/blood-2007-10-119677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpinelli MR, Hilton DJ, Metcalf D, Antonchuk JL, Hyland CD, Mifsud SL, et al. Suppressor screen in Mpl−/− mice: c-Myb mutation causes supraphysiological production of platelets in the absence of thrombopoietin signaling. Proc Natl Acad Sci USA. 2004;101:6553–6558. doi: 10.1073/pnas.0401496101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg ML, Sutton SE, Pletcher MT, Wiltshire T, Tarantino LM, Hogenesch JB, et al. c-Myb and p300 regulate hematopoietic stem cell proliferation and differentiation. Dev Cell. 2005;8:153–166. doi: 10.1016/j.devcel.2004.12.015. [DOI] [PubMed] [Google Scholar]
- Malaterre J, Carpinelli M, Ernst M, Alexander W, Cooke M, Sutton S, et al. c-Myb is required for progenitor cell homeostasis in colonic crypts. Proc Natl Acad Sci USA. 2007;104:3829–3834. doi: 10.1073/pnas.0610055104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay RG, Gonda TJ. MYB function in normal and cancer cells. Nat Rev Cancer. 2008;8:523–534. doi: 10.1038/nrc2439. [DOI] [PubMed] [Google Scholar]
- Mucenski ML, McLain K, Kier AB, Swerdlow SH, Schreiner CM, Miller TA, et al. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell. 1991;65:677–689. doi: 10.1016/0092-8674(91)90099-k. [DOI] [PubMed] [Google Scholar]
- Cheasley D, Pereira L, Lightowler S, Vincan E, Malaterre J, Ramsay RG. Myb controls intestinal stem cell genes and self-renewal. Stem Cells. 2011;29:2042–2050. doi: 10.1002/stem.761. [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Suruga K, Sakaguchi N, Takase S, Goda T. Major intestinal coactivator p300 strongly activates peroxisome proliferator-activated receptor in intestinal cell line, Caco-2. Gene. 2002;291:271–277. doi: 10.1016/s0378-1119(02)00625-x. [DOI] [PubMed] [Google Scholar]
- Shikama N, Lutz W, Kretzschmar R, Sauter N, Roth JF, Marino S, et al. Essential function of p300 acetyltransferase activity in heart, lung and small intestine formation. EMBO J. 2003;22:5175–5185. doi: 10.1093/emboj/cdg502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Mathews MB. Transcriptional coactivator cAMP response element binding protein mediates induction of the human proliferating cell nuclear antigen promoter by the adenovirus E1A oncoprotein. Proc Natl Acad Sci USA. 1997;94:4481–4486. doi: 10.1073/pnas.94.9.4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer T, Juttner S, Plath T, Mergler S, Seufferlein T, Wang TC, et al. Gastrin transactivates the chromogranin A gene through MEK-1/ERK- and PKC-dependent phosphorylation of Sp1 and CREB. Cell Signal. 2008;20:60–72. doi: 10.1016/j.cellsig.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Hocker M, Raychowdhury R, Plath T, Wu H, O'Connor DT, Wiedenmann B, et al. Sp1 and CREB mediate gastrin-dependent regulation of chromogranin A promoter activity in gastric carcinoma cells. J Biol Chem. 1998;273:34000–34007. doi: 10.1074/jbc.273.51.34000. [DOI] [PubMed] [Google Scholar]
- Miller RS, Wolfe A, He L, Radovick S, Wondisford FE. CREB binding protein (CBP) activation is required for luteinizing hormone beta expression and normal fertility in mice. Mol Cell Biol. 2012;32:2349–2358. doi: 10.1128/MCB.00394-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahapatra NR, Mahata M, Ghosh S, Gayen JR, O'Connor DT, Mahata SK. Molecular basis of neuroendocrine cell type-specific expression of the chromogranin B gene: crucial role of the transcription factors CREB, AP-2, Egr-1 and Sp1. J Neurochem. 2006;99:119–133. doi: 10.1111/j.1471-4159.2006.04128.x. [DOI] [PubMed] [Google Scholar]
- Tentler JJ, Hadcock JR, Gutierrez-Hartmann A. Somatostatin acts by inhibiting the cyclic 3',5'-adenosine monophosphate (cAMP)/protein kinase A pathway, cAMP response element-binding protein (CREB) phosphorylation, and CREB transcription potency. Mol Endocrinol. 1997;11:859–866. doi: 10.1210/mend.11.7.9943. [DOI] [PubMed] [Google Scholar]
- Struthers RS, Vale WW, Arias C, Sawchenko PE, Montminy MR. Somatotroph hypoplasia and dwarfism in transgenic mice expressing a non-phosphorylatable CREB mutant. Nature. 1991;350:622–624. doi: 10.1038/350622a0. [DOI] [PubMed] [Google Scholar]
- Dworkin S, Mantamadiotis T. Targeting CREB signalling in neurogenesis. Expert Opin Ther Targets. 2010;14:869–879. doi: 10.1517/14728222.2010.501332. [DOI] [PubMed] [Google Scholar]
- Potten CS, Loeffler M. Stem cells: attributes, cycles, spirals, pitfalls and uncertainties. Lessons for and from the crypt. Development. 1990;110:1001–1020. doi: 10.1242/dev.110.4.1001. [DOI] [PubMed] [Google Scholar]
- Rosenberg D, Groussin L, Jullian E, Perlemoine K, Bertagna X, Bertherat J. Role of the PKA-regulated transcription factor CREB in development and tumorigenesis of endocrine tissues. Ann N Y Acad Sci. 2002;968:65–74. doi: 10.1111/j.1749-6632.2002.tb04327.x. [DOI] [PubMed] [Google Scholar]
- Amorino GP, Mikkelsen RB, Valerie K, Schmidt-Ullrich RK. Dominant-negative cAMP-responsive element-binding protein inhibits proliferating cell nuclear antigen and DNA repair, leading to increased cellular radiosensitivity. J Biol Chem. 2003;278:29394–29399. doi: 10.1074/jbc.M304012200. [DOI] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Raychowdhury R, Schafer G, Fleming J, Rosewicz S, Wiedenmann B, Wang TC, et al. Interaction of early growth response protein 1 (Egr-1), specificity protein 1 (Sp1), and cyclic adenosine 3′5′-monophosphate response element binding protein (CREB) at a proximal response element is critical for gastrin-dependent activation of the chromogranin A promoter. Mol Endocrinol. 2002;16:2802–2818. doi: 10.1210/me.2001-0292. [DOI] [PubMed] [Google Scholar]
- Wilson BE, Mochon E, Boxer LM. Induction of bcl-2 expression by phosphorylated CREB proteins during B-cell activation and rescue from apoptosis. Mol Cell Biol. 1996;16:5546–5556. doi: 10.1128/mcb.16.10.5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantamadiotis T, Papalexis N, Dworkin S. CREB signalling in neural stem/progenitor cells: recent developments and the implications for brain tumour biology. Bioessays. 2012;34:293–300. doi: 10.1002/bies.201100133. [DOI] [PubMed] [Google Scholar]
- Katzenellenbogen BS, Montano MM, Ediger TR, Sun J, Ekena K, Lazennec G, et al. Estrogen receptors: selective ligands, partners, and distinctive pharmacology Recent Prog Horm Res 200055163–193.discussion 194-195. [PubMed] [Google Scholar]
- Frasor J, Chang EC, Komm B, Lin CY, Vega VB, Liu ET, et al. Gene expression preferentially regulated by tamoxifen in breast cancer cells and correlations with clinical outcome. Cancer Res. 2006;66:7334–7340. doi: 10.1158/0008-5472.CAN-05-4269. [DOI] [PubMed] [Google Scholar]
- Keppler D, Konig J. Hepatic canalicular membrane 5: expression and localization of the conjugate export pump encoded by the MRP2 (cMRP/cMOAT) gene in liver. FASEB J. 1997;11:509–516. doi: 10.1096/fasebj.11.7.9212074. [DOI] [PubMed] [Google Scholar]
- Kruijtzer CM, Beijnen JH, Schellens JH. Improvement of oral drug treatment by temporary inhibition of drug transporters and/or cytochrome P450 in the gastrointestinal tract and liver: an overview. Oncologist. 2002;7:516–530. doi: 10.1634/theoncologist.7-6-516. [DOI] [PubMed] [Google Scholar]
- Hinoshita E, Uchiumi T, Taguchi K, Kinukawa N, Tsuneyoshi M, Maehara Y, et al. Increased expression of an ATP-binding cassette superfamily transporter, multidrug resistance protein 2, in human colorectal carcinomas. Clin Cancer Res. 2000;6:2401–2407. [PubMed] [Google Scholar]
- Theile D, Grebhardt S, Haefeli WE, Weiss J. Involvement of drug transporters in the synergistic action of FOLFOX combination chemotherapy. Biochem Pharmacol. 2009;78:1366–1373. doi: 10.1016/j.bcp.2009.07.006. [DOI] [PubMed] [Google Scholar]
- Chan HM, La Thangue NB. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J Cell Sci. 2001;114 (Pt 13:2363–2373. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan I, Perez-Alvarado GC, Parker D, Dyson HJ, Montminy MR, Wright PE. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- Zor T, De Guzman RN, Dyson HJ, Wright PE. Solution structure of the KIX domain of CBP bound to the transactivation domain of c-Myb. J Mol Biol. 2004;337:521–534. doi: 10.1016/j.jmb.2004.01.038. [DOI] [PubMed] [Google Scholar]
- Kasper LH, Boussouar F, Ney PA, Jackson CW, Rehg J, van Deursen JM, et al. A transcription-factor-binding surface of coactivator p300 is required for haematopoiesis. Nature. 2002;419:738–743. doi: 10.1038/nature01062. [DOI] [PubMed] [Google Scholar]
- Gu T, Zhang Z, Wang J, Guo J, Shen WH, Yin Y. CREB is a novel nuclear target of PTEN phosphatase. Cancer Res. 2011;71:2821–2825. doi: 10.1158/0008-5472.CAN-10-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo JL, Kahn M. The Wnt signaling pathway in cellular proliferation and differentiation: A tale of two coactivators. Adv Drug Deliv Rev. 2010;62:1149–1155. doi: 10.1016/j.addr.2010.09.012. [DOI] [PubMed] [Google Scholar]
- Blobel GA. CREB-binding protein and p300: molecular integrators of hematopoietic transcription. Blood. 2000;95:745–755. [PubMed] [Google Scholar]
- Hilton DJ, Kile BT, Alexander WS. Mutational inhibition of c-Myb or p300 ameliorates treatment-induced thrombocytopenia. Blood. 2009;113:5599–5604. doi: 10.1182/blood-2008-12-195255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el Marjou F, Janssen KP, Chang BH, Li M, Hindie V, Chan L, et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39:186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- Moser AR, Pitot HC, Dove WF. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 1990;247:322–324. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- Corbett TH, Griswold DP, Jr., Roberts BJ, Peckham JC, Schabel FM., Jr Tumor induction relationships in development of transplantable cancers of the colon in mice for chemotherapy assays, with a note on carcinogen structure. Cancer Res. 1975;35:2434–2439. [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, et al. Galaxy: a web-based genome analysis tool for experimentalists Curr Protoc Mol Biol 2010Chapter 19(Unit 19.10)1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.