Abstract
CCAAT/enhancer-binding protein alpha (C/EBPα) is an important transcription factor involved in granulocytic differentiation. Here, for the first time we demonstrate that E6-associated protein (E6AP), an E3 ubiquitin ligase targets C/EBPα for ubiquitin-mediated proteasome degradation and thereby negatively modulates its functions. Wild-type E6AP promotes ubiquitin dependent proteasome degradation of C/EBPα, while catalytically inactive E6-associated protein having cysteine replaced with alanine at amino-acid position 843 (E6AP-C843A) rather stabilizes it. Further, these two proteins physically associate both in non-myeloid (overexpressed human embryonic kidney epithelium) and myeloid cells. We show that E6AP-mediated degradation of C/EBPα protein expression curtails its transactivation potential on its target genes. Noticeably, E6AP degrades both wild-type 42 kDa CCAAT-enhancer-binding protein alpha (p42C/EBPα) and mutant isoform 30 kDa CCAAT-enhancer-binding protein alpha (p30C/EBPα), this may explain perturbed p42C/EBPα/p30C/EBPα ratio often observed in acute myeloid leukemia (AML). We show that overexpression of catalytically inactive E6AP-C843A in C/EBPα inducible K562- p42C/EBPα-estrogen receptor (ER) cells inhibits β-estradiol (E2)-induced C/EBPα degradation leading to enhanced granulocytic differentiation. This enhanced granulocytic differentiation upon E2-induced activation of C/EBPα in C/EBPα stably transfected cells (β-estradiol inducible K562 cells stably expressing p42C/EBPα-ER (K562-C/EBPα-p42-ER)) was further substantiated by siE6AP-mediated knockdown of E6AP in both K562-C/EBPα-p42-ER and 32dcl3 (32D clone 3, a cell line widely used model for in vitro study of hematopoietic cell proliferation, differentiation, and apoptosis) cells. Taken together, our data suggest that E6AP targeted C/EBPα protein degradation may provide a possible explanation for both loss of expression and/or functional inactivation of C/EBPα often experienced in myeloid leukemia.
Keywords: C/EBPα, E6AP, myeloid leukemia, differentiation, ubiquitination
CCAAT-enhancer-binding protein alpha (C/EBPα) is a key transcription factor required for differentiation of multiple cell types including myeloid cells.1 In hematopoiesis, C/EBPα regulates myeloid differentiation and granulopoiesis.2 C/EBPα mediates its functions by binding to promoters of its target genes and interacting with agonist (co-activators) and antagonist (repressor) proteins in a cell-type-specific manner.3, 4 C/EBPα function is also modulated by post-translational modifications such as phosphorylation, SUMOylation, and ubiquitination.5, 6, 7, 8 The functional activity of wild-type C/EBPα (42 kDa wild-type CCAAT-enhancer-binding protein alpha (p42C/EBPα)) is frequently perturbed in myeloid leukemia resulting in differentiation blockade.2, 4 Notably, loss of function or expression of C/EBPα provides a platform for acute myeloid leukemia (AML) development.9 C/EBPα is mutated in ∼9% of AML cases; reported mutations in C/EBPα comprise point mutations in basic leucine-zipper domain or N-terminus frame-shift mutations leading to formation of a dominant negative C/EBPα isoform (30 kDa mutant CCAAT-enhancer-binding protein alpha (p30C/EBPα)) encoded by same CEBPA from different translation start site.2, 10, 11, 12 p30C/EBPα inhibits myeloid differentiation by exerting dominant negative functions over p42C/EBPα.11 Further, recent studies suggest that p42C/EBPα function is antagonized by protein–protein interactions. AML1/ETO binds C/EBPα, suppresses its transcriptional activity and thereby interferes with C/EBPα promoter autoregulation13 leading to reduced C/EBPα expression; c-Jun promotes proliferation and prevents differentiation by inhibiting C/EBPα DNA binding via interacting with leucine-zipper domain.14 Additionally, AML specific Flt3 mutations downregulate C/EBPα expression and contribute to leukemogenesis.15 Recently, we and others have shown C/EBPα regulation at protein level via ubiquitin-mediated proteasome degradation.7, 16 Phosphorylated JNK stabilizes C/EBPα, while inactive JNK facilitates its ubiquitination.7 Keeshan et al.,17 showed that Trib2 associates with p42C/EBPα and promotes its proteasome-mediated degradation leading to increased p30C/EBPα/p42C/EBPα ratio, commonly seen in AML. However, only two E3 ubiquitin ligases for C/EBPα are known till date; E3 ligase TRIM21 interacts with TRIB2 to downregulate C/EBPα in lung tumors, while Fbwx7 targets it in preadipocytes.18, 19 Here, for the first the time we report that E6-associated protein (E6AP), a homologous to E6AP carboxy terminus domain containing E3 ubiquitin ligase also targets C/EBPα for ubiquitin-mediated proteasome degradation.
E6AP, a 100-kDa cellular protein is a member of functionally related E3-ubiquitin-protein ligases defined by the domain homologous to the carboxy terminus hect domain.20 E3 ligases ubiquitinate and degrade several regulatory proteins including p53, p27, promyelocytic leukemia retinoic acid receptor α and others, which serve as tumor suppressors and cell-cycle inhibitors.21, 22, 23, 24 Recently, using mass spectrometry based proteomics approach we identified E6AP as a target of tamoxifen.25 Moreover, Tamoxifen is reported to enhance C/EBPα expression in HeLa cells leading to apoptosis induction.26 As tamoxifen downregulates E6AP and induces C/EBPα expression in cell-type-specific manner, we hypothesized E6AP might be an E3 ubiquitin ligase for C/EBPα. This assumption was also based on our previous finding that C/EBPα can be destabilized by ubiquitin-mediated proteasome pathway.7
Henceforth, we demonstrate that E6AP promotes C/EBPα ubiquitination leading to its proteasome-mediated degradation and thus functional inactivation. In contrast, E6AP inhibition in K562-C/EBPα-p42-estrogen receptor (ER) cells either by siE6AP or overexpression of catalytically inactive mutant (E6-associated protein having cysteine replaced with alanine at amino-acid position 843 (E6AP-C843A)) promotes myeloid differentiation. Further, in a more physiologically relevant myeloid system U937 and 32Dcl3 cells, we show E6AP inhibition promotes enhanced C/EBPα expression leading to robust differentiation. Thus, our data suggests that E6AP may negatively regulate granulopoiesis by targeting C/EBPα for degradation via ubiquitin proteasome pathway.
Results
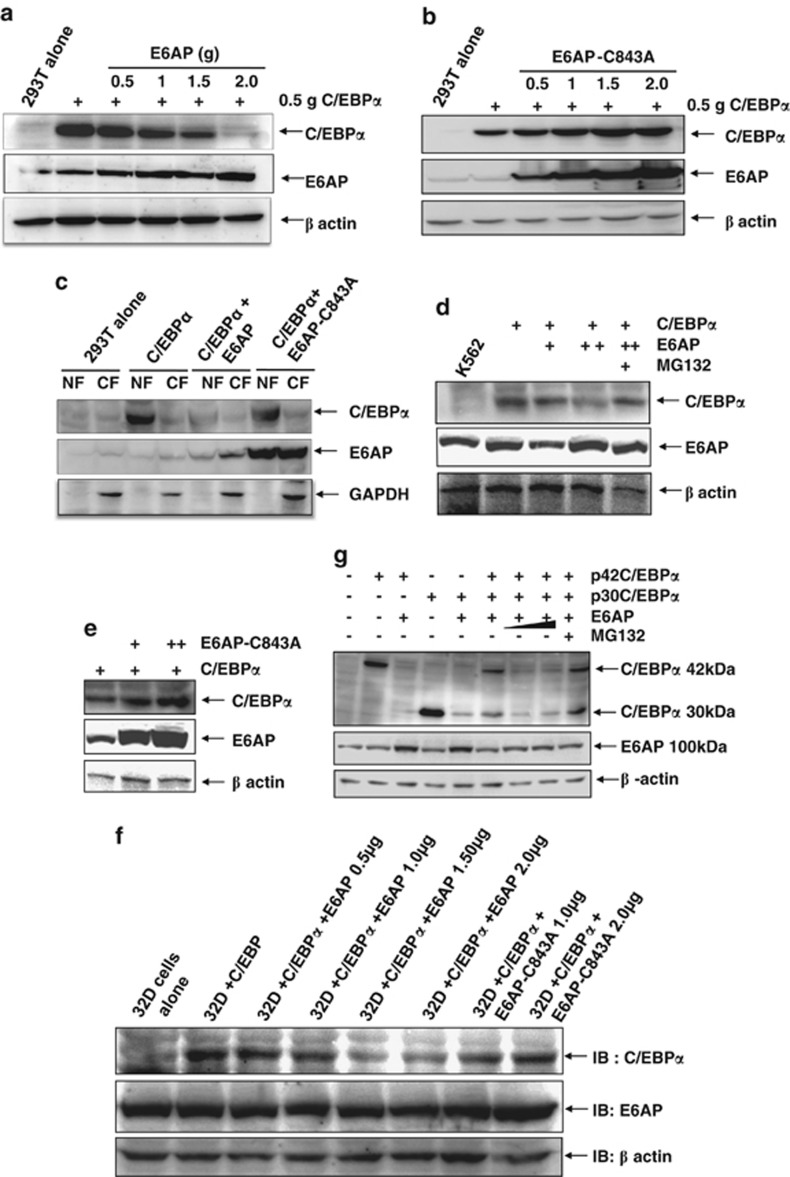
E6AP degrades C/EBPα
We and others have previously shown that C/EBPα can be ubiquitinated and degraded via proteasome-mediated pathway.7, 16 We, therefore, asked if E6AP modulates C/EBPα stability by targeting it for degradation. In order to evaluate the effects of E6AP on C/EBPα protein steady-state levels, we used human embryonic kidney epithelium (HEK293T) cells which have no endogenous C/EBPα expression. These cells were transfected with 0.5 μg C/EBPα together with increasing amounts of E6AP or E6AP-C843A. 24-h post transfection whole-cell extracts (WCEs) were prepared and resolved on 10% SDS-PAGE. Immunoblot with C/EBPα and E6AP antibody showed that E6AP drastically downregulate C/EBPα (Figure 1a) while a marked contrast was seen with E6AP-C843A (Figure 1b). Strikingly, E6AP-C843A rather stabilized C/EBPα possibly due to its dominant negative nature over endogenous E6AP. This data demonstrates that E6AP downregulates C/EBPα protein expression possibly by promoting its degradation. As C/EBPα is a nuclear protein, we asked if E6AP degrades C/EBPα in the nucleus. For this, 293T cells were transfected either with C/EBPα alone or together with E6AP and E6AP-C843A. Twenty-four hour post transfection nuclear extracts were prepared and resolved on 10% SDS-PAGE. Immunoblot with C/EBPα antibody showed E6AP degrades C/EBPα in the nucleus, while E6AP-C843A instead stabilizes it (Figure 1c). To further demonstrate the relevance of our findings in a more physiological setting, we used myeloid cell lines K562 and 32Dcl3, where C/EBPα promotes granulocytic differentiation.27, 28, 29 These cells were transfected with C/EBPα, together with increasing amounts of either E6AP or E6AP-C843A. WCEs were prepared after 24-h transfection and resolved on 10% SDS-PAGE. Immunoblot probed with C/EBPα followed by E6AP antibody confirmed that E6AP inhibits C/EBPα steady-state levels, while E6AP-C843A rather stabilizes (Figures 1d).
Figure 1.
E6AP inhibits C/EBPα steady state levels. (a) HEK 293T cells were transfected with C/EBPα (0.5 μg) along with increasing amounts of E6AP (0.5–2.0 μg). This was followed by immunoblotting with C/EBPα, E6AP and β-actin antibodies. (b) 293T cells were transfected with C/EBPα (0.5 μg) along with increasing concentrations of E6AP-C843A (0.5–2.0 μg) and were followed by immunoblotting with C/EBPα, E6AP and β-actin antibodies. (c) 293T cells were transfected with C/EBPα (0.5 μg), E6AP (2.0 μg) and E6AP mutant C843A (2.0 μg) as indicated. Post 24 h nuclear extracts were prepared, resolved on 10% SDS-PAGE and probed with C/EBPα, E6AP and GAPDH antibodies. GAPDH was used a control for cytoplasmic protein extract. (d, e) K562 cells were transfected with C/EBPα (0.5 μg), E6AP (1.0, 2.0 μg) and E6AP-C843A (1.0, 2.0 μg). In the indicated condition, cells were treated with 25 μM MG132 3 h prior to cell lysate preparation. The blot was probed with C/EBPα, E6AP and β-actin antibodies. (f) 32Dcl3 cells were transfected with C/EBPα (0.5 μg), E6AP (0.5, 1.0, 1.5 and 2.0 μg) and E6AP-C843A (1.0, 2.0 μg). The blot was probed with C/EBPα, E6AP and β-actin antibodies. (g) 293T cells were transfected with p42C/EBPα (0.5μg), p30C/EBPα (0.5 μg) and E6AP (1.0–2.0 μg) as indicated. Cells were treated with 25 μM MG132 3 h prior to lysate preparation in one of the conditions. Lysates were resolved on 10% SDS-PAGE and probed with C/EBPα, E6AP and β-actin antibodies
As C/EBPα also has a shorter isoform p30C/EBPα (30 kDa), we asked whether p30C/EBPα is also regulated by E6AP or it selectively targets only p42C/EBPα. For this, we co-transfected 293T cells with expression plasmids for p42C/EBPα and p30C/EBPα, together with indicated amounts of E6AP. Twenty-four hours post transfection WCEs were prepared and resolved on 10% SDS-PAGE followed by immunoblotting with C/EBPα and E6AP antibody, which showed E6AP potentially degrades both forms of C/EBPα (Figure 1g). Notably, MG132 treatment inhibited the degradation of both forms, suggesting this degradation to be mediated via proteasome pathway.
To further examine C/EBPα degradation in the presence of E6AP in the nucleus, we performed immuno-fluorescence assay in 293T cells, which showed gradual degradation of C/EBPα in a dose-dependent manner (Supplementary Figure S1). Together, these data suggest that E6AP degrades C/EBPα in the nucleus.
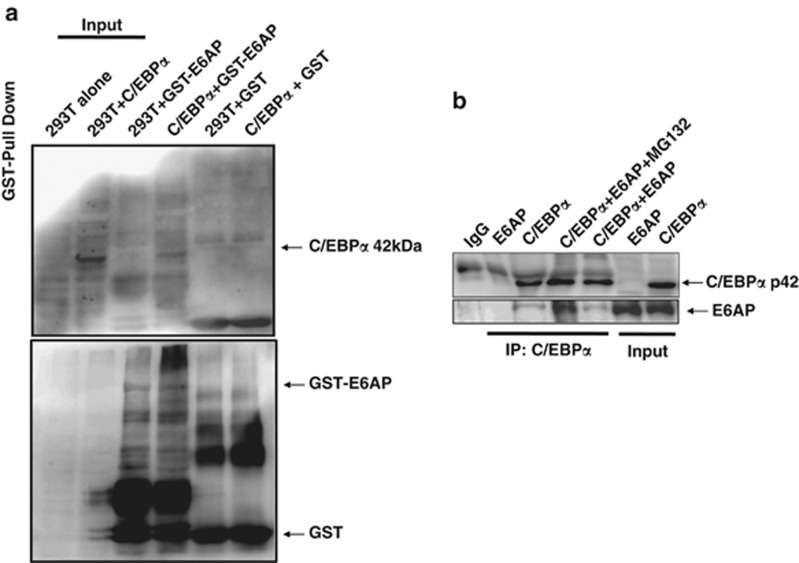
E6AP and C/EBPα physically interact with each other
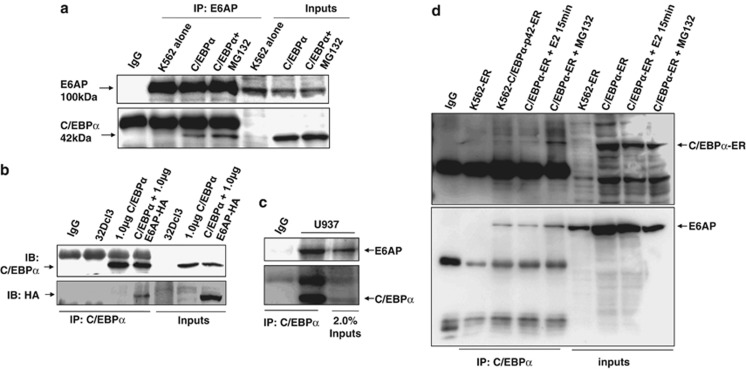
As E6AP targets C/EBPα for degradation, we hypothesized that these two proteins may physically interact; For this, we performed in vitro glutathione sepharose transferase (GST) pull down using bacterially purified GST-E6AP and 293T nuclear extracts transfected with C/EBPα. After pull down, bead bound GST-E6AP with its interacting proteins from the lysates were resolved on 10% SDS-PAGE and probed with C/EBPα antibody. As shown in Figure 2a, C/EBPα does interact with GST-E6AP, while no interaction was observed with GST alone. To further assess in vivo interaction, we performed C/EBPα co-immunoprecipitation from nuclear extracts of 293T cells co-transfected with C/EBPα and E6AP. Co-immunoprecipitates were lysed in SDS buffer and resolved on 10% SDS-PAGE. Immunoblot with C/EBPα followed by E6AP antibody confirmed in vivo interaction between C/EBPα and E6AP. For co-immunoprecipitation studies, we also treated C/EBPα and E6AP transfected cells with proteasome inhibitor. Interestingly, prominent interaction was observed between C/EBPα and E6AP in the presence of MG132, apparently due to inhibition of proteasome pathway (Figure 2b).
Figure 2.
C/EBPα interacts with E6AP. (a) GST pulldown using GST-E6AP and C/EBPα transfected 293T protein lysates was performed. Washed beads were harvested with SDS loading buffer and resolved on 10% SDS-PAGE, followed by immunoblotting with C/EBPα and GST antibody. (b) Co-immunoprecipitation with C/EBPα antibody was performed using 293T lysates co-transfected with C/EBPα and E6AP as indicated. In one of the C/EBPα and E6AP-transfected conditions, cells were treated with 25 μM MG132 3 h prior to lysate preparation. The blot was first probed with E6AP antibody and later the same blot was stripped and probed with C/EBPα antibody
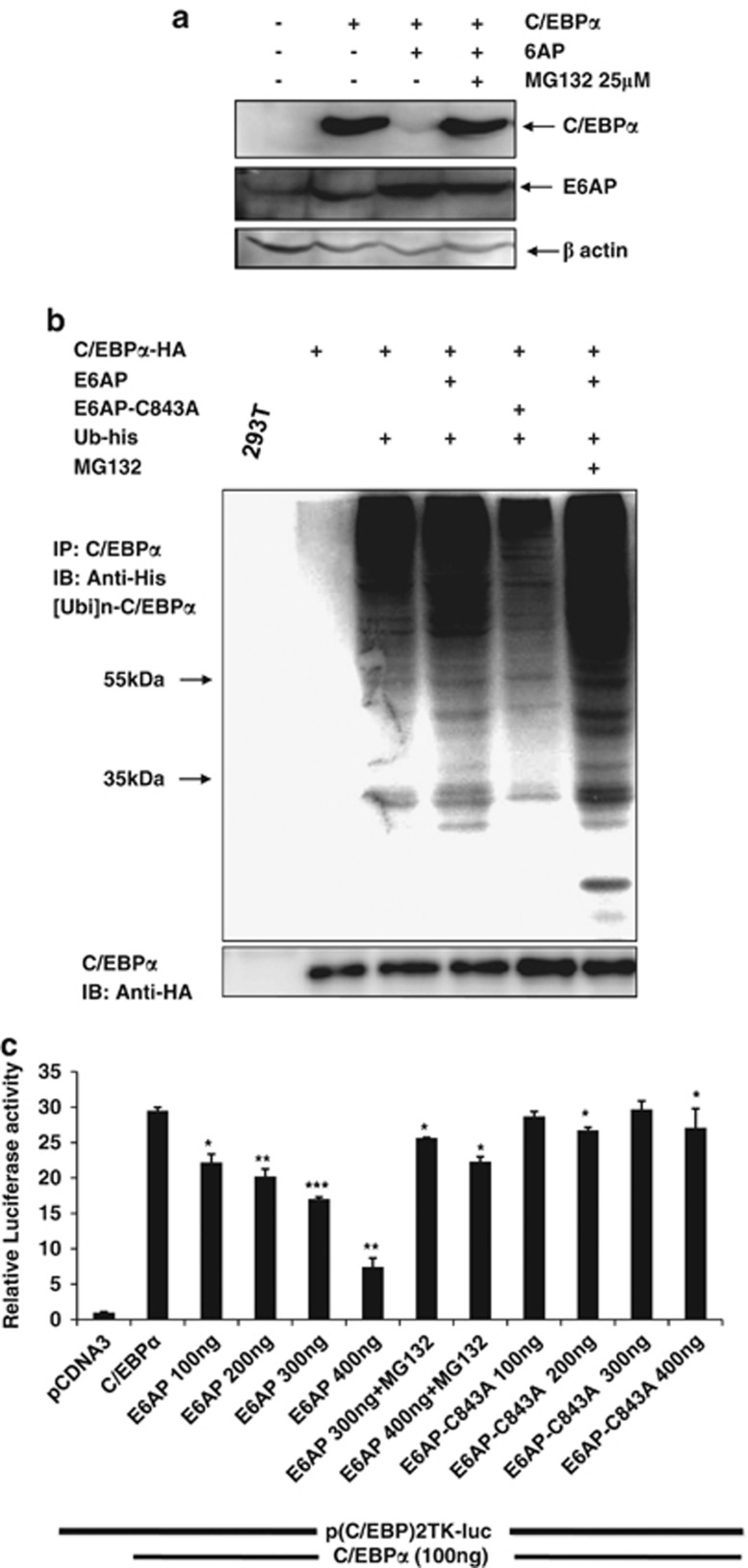
E6AP degrades C/EBPα via ubiquitin proteasome pathway and negatively affects its transactivation capacity
We showed E6AP destabilizes C/EBPα and moreover, these two proteins also physically interact with each other. We, therefore, asked whether it involves ubiquitin-mediated proteasome degradation. For this, 293T cells were transiently transfected either with C/EBPα or together with E6AP expression plasmids as indicated in Figure 3a. Twenty-four hours post transfection cells were treated with MG132 in C/EBPα and E6AP co-transfected condition followed by lysate preparation. Immunoblot with C/EBPα and E6AP antibody nicely showed that C/EBPα degradation is dramatically restored upon MG132 treatment (Figure 3a). This strongly suggests that E6AP promotes proteasomal degradation of C/EBPα. However, to further support the notion that E6AP-mediated C/EBPα degradation involves ubiquitin proteasome pathway, we performed in vivo ubiquitination assay by transfecting 293T cells with C/EBPα-HA, Ubiquitin-His, E6AP and E6AP-C843A. Twenty-four hours post transfection, cells were harvested; C/EBPα was co-immunoprecipitated and probed with His antibody. Strong C/EBPα ubiquitination ladder was observed in C/EBPα, E6AP and His-Ubiquitin co-transfected cells, while mild ubiquitination was seen in cells co-transfected with C/EBPα, His-Ubiquitin and E6AP-C843A, suggesting E6AP indeed promotes ubiquitination of C/EBPα. Interestingly, more intense ubiquitinated ladder pattern was observed in cells treated with MG132, which apparently stabilized the ubiquitinated C/EBPα by inhibiting proteasome machinery (Figure 3b). This data affirms that ligase activity of E6AP promotes C/EBPα ubiquitination leading to its degradation via proteasome pathway.
Figure 3.
E6AP targets C/EBPα for ubiquitin proteasome pathway and negatively affects its transactivation capacity. (a) 293T cells were transfected with C/EBPα (0.5 μg) and E6AP (2.0 μg); cells were treated with 25 μM MG132 3 h prior to cell harvesting. Lysates were resolved on 10% SDS-PAGE and probed with C/EBPα, E6AP and β-actin antibodies after stripping and reprobing the same blot. (b) 293T cells were transfected with expression plasmids for C/EBPα-HA (1.0 μg), E6AP (2.0 μg), E6AP-C843A (2.0 μg) and ubiquitin-His (1.0 μg) as indicated. Twenty four hours post transfection, cells were treated with 25 μM MG132 3 h prior to harvesting in the indicated lanes. C/EBPα was co-immunoprecipitated and probed with His antibody. The same membrane was stripped and probed with HA antibody (lower panel). (c) 293T cells were transiently transfected with pTK-C/EBP-luc reporter and expression plasmids for C/EBPα, E6AP and E6AP-C843A. Twenty four hours post transfection, luciferase activity was measured. 25 μM MG132 treatment was given 3 h prior to cell harvesting for luciferase activity measurement. Data are representative of three independent experiments. Results are given as standard error of mean (±S.E.M.); *P<0.05; **P<0.001, ***P<0.0001
As E6AP targets C/EBPα for degradation, we further asked if it affects C/EBPα transactivation potential. For this, we performed luciferase reporter assay on a minimal pTK promoter containing two C/EBP-binding sites. Indicated amounts of reporter vector and expression plasmids for C/EBPα, E6AP and E6AP-C843A were transfected in 293T cells. Twenty-four hours post transfection luciferase activity was measured, which showed co-transfection of E6AP with C/EBPα significantly inhibited C/EBPα transactivation potential in a dose-dependent manner (Figure 3c). Further, MG132 treatment efficiently restored C/EBPα transactivation potential even in the presence of E6AP. Additionally, co-transfection of E6AP-C843A with C/EBPα did not inhibit C/EBPα transactivation capacity. This clearly indicates that catalytically active E6AP negatively modulates C/EBPα protein stability and its biological functions.
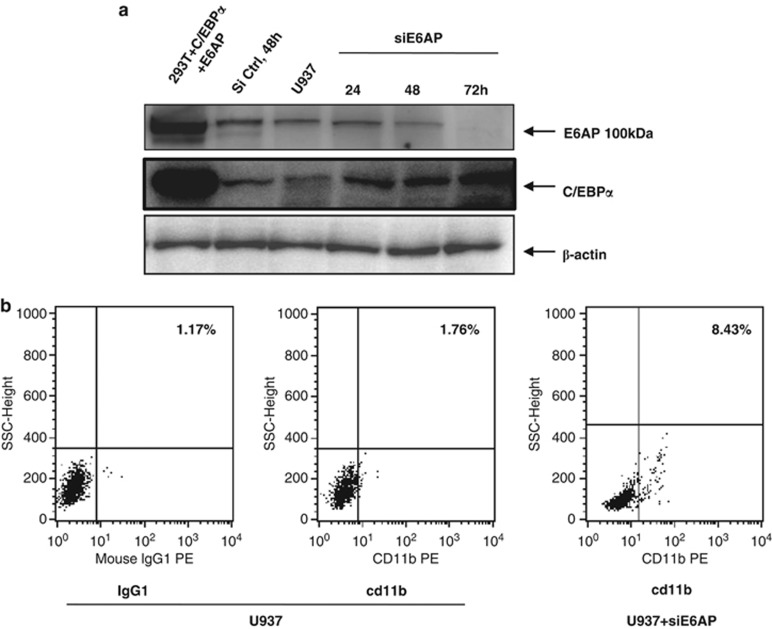
E6AP knockdown enhances C/EBPα expression and promotes differentiation in U937 cells
C/EBPα is a key regulator of granulocyte development.30, 31 Moreover, differentiation arrest in myeloid leukemia subtypes is attributed to functional inactivation of C/EBPα.32 As we show that E6AP targets C/EBPα for proteasomal degradation, we sought to assess the protein expression levels of E6AP in some of the representative myeloid leukemia cell lines (U937, HL60, K562 and Kasumi-1) having diminished expression levels of C/EBPα. WCEs of indicated cell lines probed with E6AP antibody clearly showed ample expression of E6AP in these cells with K562 showing greatest expression (Supplementary Figure S4). To establish functional correlation between E6AP and C/EBPα expression in myeloid leukemia cells, we sought to assess C/EBPα protein levels in myeloid leukemia cell U937 after inhibiting E6AP by small interfering RNAs (siRNA). Notably, there is ample expression of E6AP in U937 cells (Supplementary Figure S4) and as a matter of fact it has reduced C/EBPα expression. We transiently transfected U937 cells with control siRNA and siE6AP (Figure 4a) (siE6AP efficacy was validated in MCF7 and K562 cells; Supplementary Figure S5). After indicated time points WCEs were prepared, resolved on 10% SDS-PAGE and immunoblotted with E6AP, C/EBPα and β-actin antibodies. Expectedly, it showed persistent decrease in E6AP protein levels (in siE6AP transfected lanes) with concomitant increase in C/EBPα expression (Figure 4a). In fact, FACS flow cytometer analysis of U937cells transiently transfected with siE6AP for 72 h also showed enhanced expression of myeloid differentiation marker cd11b (Figure 4b), suggesting E6AP inhibition in myeloid leukemia cells may overcome differentiation blockade,33 a common phenomenon observed in several leukemia subtypes.
Figure 4.
siE6AP-mediated inhibition of E6AP enhances C/EBPα expression in the myeloid leukemia cell line U937. (a) U937 cells were transfected with 50 nM siE6AP and cell lysate was prepared after the indicated time points. Lysate was resolved on 10% SDS-PAGE followed by immunoblotting with E6AP, C/EBPα and β-actin antibodies. (b) U937 cells were transfected with siE6AP for 72 h. Cells were washed and labeled with cd11b and its respective IgG-PE-conjugated antibodies for FACS analysis
E6AP and C/EBPα-ER physically interact in K562 cells stably expressing C/EBPα-ER
As we showed C/EBPα and E6AP physically interact both in vitro (Figure 2a) and in vivo (Co-Immunoprecipitation (Co-IP), Figure 2b), we further validated their interaction in physiologically relevant myeloid leukemia cells. We transiently transfected K562 and 32Dcl3 cells with C/EBPα and E6AP either alone or together as indicated. Twenty-four hours post transfection, lysates were prepared and C/EBPα was immunoprecipitated using C/EBPα antibody. Co-immununoprecipitates were resolved on 8% SDS-PAGE, probed with C/EBPα followed by E6AP antibody after stripping the same blot, which again confirmed in vivo interaction in K562 and 32Dcl3 cells (Figures 5a and b; Supplementary Figure S6 shows uncroped Figure 5a). To further endorse their physical association, we assessed in vivo interaction between endogenous C/EBPα and E6AP from U937 cells. C/EBPα was co-immunoprecipitated using C/EBPα antibody from U937 WCEs and resolved on 8% SDS-PAGE (Figure 5c). Immunoblotting with C/EBPα and E6AP antibodies clearly showed in vivo interaction between these two proteins (Figure 5d). As we used β-estradiol inducible K562 cells stably expressing p42C/EBPα-ER (K562-C/EBPα-p42-ER) (detailed in Supplementary fig.S3), as a model cell line to further address biological relevance of their physical interaction, we, therefore, also confirmed interaction between these two proteins in these stable cells. We performed co-immunoprecipitation using C/EBPα antibody from WCEs of K562-ER, K562-p42C/EBPα-ER, and β-estradiol (E2)-induced K562-C/EBPα-p42-ER (induced for short period to avoid fast degradation of C/EBPα-ER) as indicated (Figure 5d). Immunoblot with E6AP followed with C/EBPα antibody after stripping the same blot nicely showed physical interaction between C/EBPα and E6AP. As expected prominent interaction was observed in MG132-treated conditions, obviously due to stabilization of C/EBPα (Figures 5a and d). Taken together, these results suggest that E6AP physically interacts with C/EBPα in myeloid cells and may promote its ubiquitin-mediated degradation.
Figure 5.
C/EBPα and E6AP physically interact in vivo in myeloid cells. (a) K562 cells were transiently transfected with C/EBPα and 24 h post transfection in one of the conditions cells were treated with 25 μM MG132 prior to cell lysate preparation. Co-immunoprecipitation using E6AP antibody was performed and the blot was probed with C/EBPα followed by E6AP antibody after stripping the same blot. (b) Co-immunoprecipitation using C/EBPα antibody was performed in lysates prepared from C/EBPα and E6AP co-transfected 32Dcl3 murine cells as indicated. Co-immunoprecipitates were resolved on 8% SDS-PAGE and probed with E6AP antibody followed by C/EBPα antibody after stripping the same blot. (c) Endogenous C/EBPα was co-immunoprecipitated from 2.5 mg whole cell lysates of U937 cells using C/EBPα antibody, resolved on 8% SDS-PAGE and probed with E6AP antibody followed by C/EBPα antibody after stripping the same blot. (d) K562-p42 C/EBPα-ER (C/EBPα-ER) stable cells were induced with 5 μM E2 for the indicated time points. Co-immunoprecipitation was performed with C/EBPα antibody. In one of the conditions 25 μM MG132 treatment was given for 3 h prior to cell lysate preparation. K562 cells transfected with empty vector were used as a control. The blot was probed with E6AP antibody. The same membrane was stripped and probed with C/EBPα antibody. Results are representative of minimum three independent experiments
E6AP inhibition in β-estradiol (E2) inducible K562 cells stably expressing C/EBPα-ER enhances granulocytic differentiation
As E6AP expression was higher in K562 cells where there are non-detectable levels of C/EBPα,34 we generated β-estradiol inducible K562 cells stably expressing C/EBPα-ER (Supplementary Figure S3) to demonstrate E6AP actions over C/EBPα functions. These stable cells (K562-p42C/EBPα-ER) express p42C/EBPα fused with ER ligand binding domain while control cells express only ER domain (K562-ER). These K562-C/EBPα-p42-ER cells are useful model system to study the dynamics of C/EBPα dependent granulocytic differentiation because they respond to β-estradiol treatment by nuclear translocation. Upon nuclear translocation, C/EBPα binds and activates its target genes such as GCSFR3 and CEBPE.35, 36 In stably transfected cells, this fusion protein stays in the cytoplasm bound with heat shock proteins (HSPs). Stimulation with E2 activates it by binding to ER domain of this fusion protein and this ligand binding relieves the HSPs bound to C/EBPα-ER, which then migrates to the nucleus and activates its target genes.
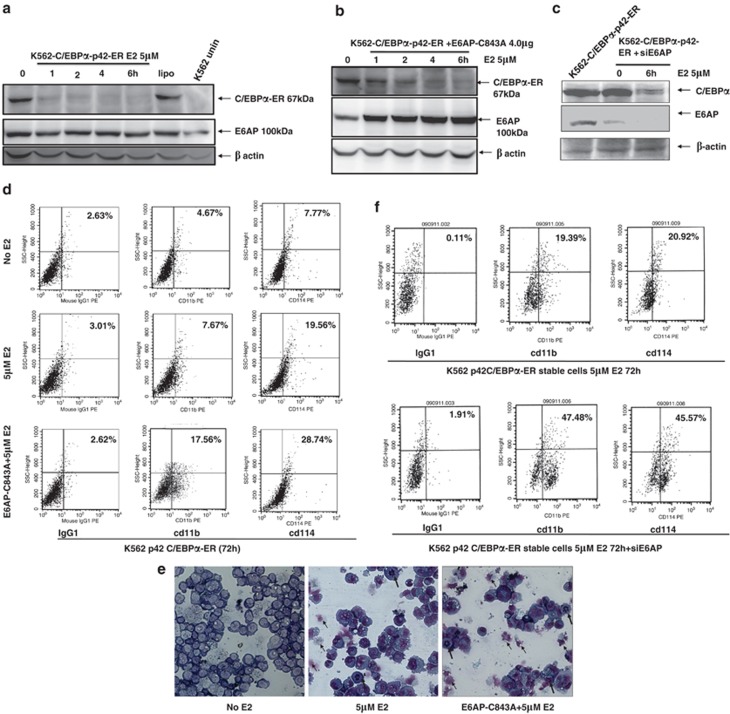
Notably, E2 binding to this fusion protein also promotes its degradation, and therefore, upon nuclear translocation C/EBPα-ER protein is rapidly eliminated within 24 h (Figure 6a). To verify that E6AP is involved in this rapid degradation of ligand bound C/EBPα-ER, we overexpressed E6AP-C843A in these cells. Twenty-four hours post transfection cells were stimulated with E2 and harvested after indicated time points. As shown in Figure 6b the rate of C/EBPα degradation is substantially inhibited in the presence of E6AP-C843A and C/EBPα protein is stabilized for longer duration. In fact, siE6AP transfection in these cells also caused significant reduction in rate of C/EBPα degradation (Figure 6c). This clearly indicates that E6AP promotes C/EBPα degradation, which can be modestly restored either by overexpression of E6AP-C843A or inhibition of E6AP by siE6AP; however, as E6AP inhibition is unable to completely restore C/EBPα expression; it is very likely that other E3 ligases may also regulate C/EBPα protein stability.
Figure 6.
E6AP inhibition in β-estradiol (E2) inducible K562 cells stably expressing C/EBPα-ER enhances granulocytic differentiation. (a) K562-p42 C/EBPα-ER stable cells were induced with 5 μM E2 for the indicated time points. Lysates were resolved on 10% SDS-PAGE and probed with C/EBPα antibody. The same blot was stripped and reprobed with E6AP and β-actin antibody. (b) K562-p42 C/EBPα-ER stable cell line was transfected with E6AP-C843A and, post 24 h of transfection, was induced with 5 μM E2 for the indicated time points. Lysates were resolved on 10% SDS-PAGE and probed with C/EBPα, E6AP and β-actin antibodies. (c) K562-p42-C/EBPα-ER cells were transiently transfected with siE6AP. Post 24 h of transfection cells were induced with β-estradiol (5 μM) for the indicated time points (0 and 6 h). Cell lysates were prepared followed by immunoblotting with E6AP, C/EBPα and β-actin antibodies. (d) K562 p42-C/EBPα-ER stable clones were transfected with E6AP-C843A and were induced with 5 μM estradiol. After 72 h of induction cells were washed and labelled with cd11b and cd114-PE-conjugated antibodies for the FACS analysis. (e) Giemsa staining showing granulocyte-like morphology upon E2 induction: May-Grünwald and Giemsa staining was performed after 72 h in the K562-C/EBPα-p42-ER stable cells transfected with E6AP-C843A and induced with 5 μM E2 (bold arrows=granulocytes/neutrophils; simple arrows=apoptotic cells). (f) K562-p42-C/EBPα-ER stable clones were transfected with 50 nM siE6AP and were induced with 5 μM E2. After 72 h of induction cells were washed and labeled with cd11b and cd114-PE-conjugated antibodies for the FACS analysis. Results are representative of minimum three independent experiments
As transfection of E6AP-C843A in K562-C/EBPα-p42-ER cells inhibited the rate of C/EBPα-ER degradation, we assumed that overexpression of E6AP-C843A would enhance granulocytic differentiation upon E2 induction. For this, K562-C/EBPα-p42-ER cells were transfected with E6AP-C843A and 24-h post transfection, cells were induced with E2 for further 72 h and then subjected to FACS analysis for cd11b and cd114 expression, which shows percentage of cells undergoing differentiation substantially increased in E6AP-C843A-transfected cells treated with E2 (Figure 6d). Further, cellular morphology of these cells was also assessed by cytospin and Wright's-Giemsa staining post 72 h E2 treatment, which showed some cells exhibited cytoplasmic blebbing (an early sign of apoptosis; middle panel), while some had multi-lobed nuclei and some displayed more eccentric and indented nuclei, a feature of differentiation (dark arrow).28 However, in cells transfected with E6AP-C843A followed by induction with E2 (Right panel) post 24 h transfection, large number of small cells in late apoptosis with multi-segmented nuclei (reminiscent of mature granulocytes) were observed suggesting E6AP-C843A potently drives E2-induced K562-C/EBPα-p42-ER to granulocytic differentiation (Figure 6e). As E6AP inhibition by siE6AP efficiently restored C/EBPα expression in K562-C/EBPα-p42-ER cells, we sought to assess the differentiation potential of K562-C/EBPα-p42-ER cells upon E6AP knockdown. For this, K562-C/EBPα-p42-ER cells were transiently transfected with siE6AP; 24 h post transfection cells were stimulated with E2 and were assayed for cd11b and cd114 under FACS flow cytometer after 72 h of E2 induction. This clearly demonstrated that E6AP knockdown in these cells potentially alleviates myeloid differentiation (Figure 6f). Together, these data indicate that E6AP inhibition in myeloid cells may stabilize C/EBPα expression leading to enhanced myeloid differentiation.
E6AP knockdown or ectopically expressed E6AP-C843A triggers differentiation in 32Dcl3 cells
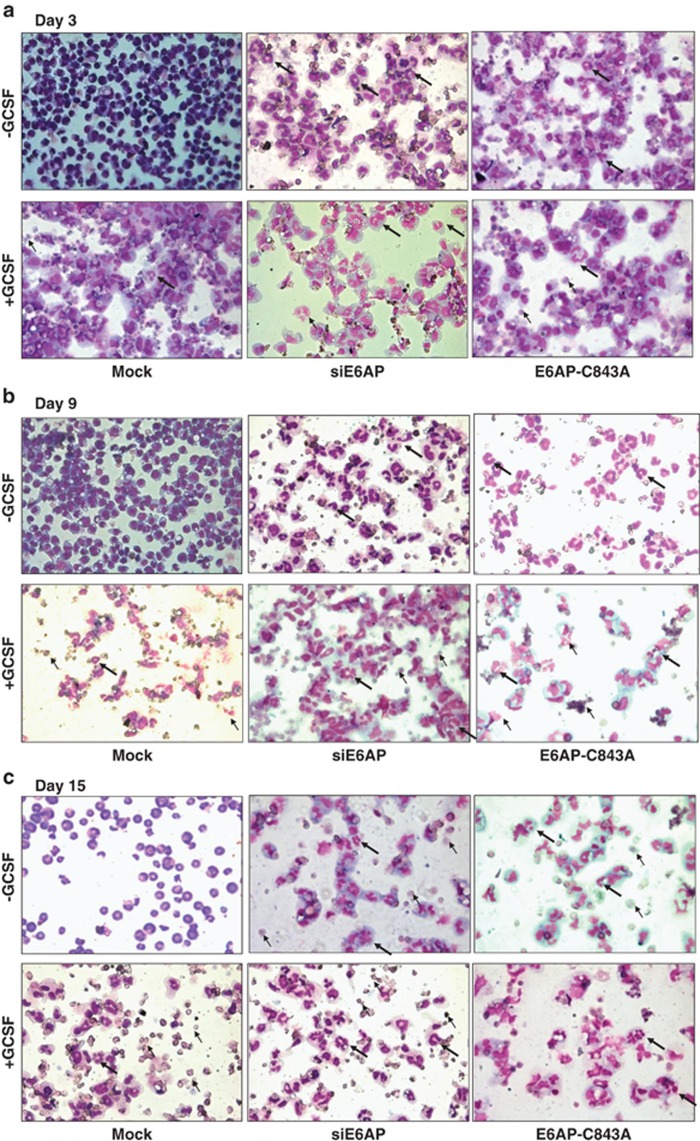
E6AP inhibition either by transient transfection of siE6AP or E6AP-C843A leading to induction of differentiation in K562-C/EBPα-p42-ER cells was also validated in yet another myeloid system 32Dcl3 cells. Notably, IL-3 replacement with G-CSF stimulates proliferation of 32Dcl3 cells for 4–5 days followed by growth arrest and apparent neutrophil-like morphology by 12 days. Moreover, G-CSF alone can drive 32Dcl3 cells to differentiation.37, 38 To assess differentiation via E6AP inhibition, 32Dcl3 cells were either mock transfected or with siE6AP and E6AP-C843A separately; grown in IL-3 free medium containing G-CSF. Post 3, 9 and 15 days of culture, Giemsa-stained cells were visualized for morphological changes. Interestingly, on day 3, few granule-like nuclear morphology was observed in cells transfected with siE6AP and E6AP-C843A even in G-CSF-untreated cells (Figure 7a). Moreover, by day 9, cells attained myelocyte morphology with oval-shaped nucleus with obvious differentiation-like morphology even in G-CSF-untreated mock transfected cells in addition to siE6AP and E6AP-C843A-transfected cells (Figure 7b). At day 15, cells appeared more like metamyelocytes with elongated, dense horseshoe or ring-shaped nucleus, while few demonstrated polymorphonuclear neutrophil-like morphology with segmented nucleus (Figure 7c).39 Together, these results indicate that E6AP inhibition may stabilize C/EBPα leading to enhanced granulocytic differentiation in 32Dcl3 cells.
Figure 7.
E6AP inhibition leads to granulocytic differentiation in 32Dcl3 cells. 32Dcl3 cells were transfected with siE6AP and E6AP-C843A and were induced with G-CSF for the indicated time points. After (a) 3 days, (b) 9 days and (c) 15 days of induction cells were washed, cytospun and stained with May-Grünwald and Giemsa staining (bold arrows=granulocytes/neutrophils; simple arrows=apoptotic cells)
Discussion
Most cancers are caused by activating mutations in proto-oncogenes and/or inactivating mutations in tumor suppressor genes rendering corresponding proteins functionally inactive. Additionally, perturbed stability of these regulatory proteins is a major cause for functional impairment. The target proteins are usually degraded tightly by proteasome machinery, which involves ubiquitin attachment to the target proteins through a series of enzymatic reactions where E3 ligases are key players.17, 24, 40 E6AP is one such E3 ubiquitin ligase implicated in the degradation of tumor suppressor protein p53 in conjunction with HPV viral E6 protein in some cancers.23 p53 degradation affects apoptosis and cell-cycle as it is chiefly involved in the regulation of these pathways. Like p53, there are several other cellular proteins reported to be regulated by E6AP.41, 42, 43, 44, 45, 46
C/EBPα is a key regulator of cellular processes, such as proliferation arrest, adipocyte differentiation, and granulopoiesis in particular.34, 47, 48, 49 We and others have previously shown that C/EBPα can be degraded via ubiquitin proteasome pathway; however, except Fbwx7, the E3 ligases involved in its degradation has largely remained elusive.19 Here, for the first time we report that E6AP can also target C/EBPα for proteasomal degradation. We explored the role of E6AP in the ubiquitination of C/EBPα, their physical interaction and consequent relevance in myeloid differentiation. Here, we provide several lines of evidence that indicate E6AP is an E3 ubiquitin ligase for C/EBPα. First, E6AP promotes proteasomal degradation of C/EBPα (Figures 1a and Figures 3a and b). Further, we show E6AP also degrades p30C/EBPα (Figure 1f). Second, a catalytically inactive E6AP-C843A inhibits C/EBPα degradation (Figure 1b c and d). Third, we show that C/EBPα and E6AP physically interact in myeloid cells (Figures 2 and 5). Fourth, degradation of C/EBPα is via ubiquitin proteasome pathway (Figures 3a and b). Thus, these data implicate a direct role of E6AP in the proteasomal degradation of C/EBPα.
As a consequence of C/EBPα protein degradation mediated by E6AP, C/EBPα transactivation potential is compromised (Figure 3c), which showed E6AP may inhibit functional activity of C/EBPα. E6AP-mediated negative effects on the functional activity of C/EBPα were further addressed in K562-C/EBPα-p42-ER cells stably expressing C/EBPα-ER fusion protein. As shown in Supplementary Figure S3, there is expression of p42C/EBPα-ER in the K562 stable clones, which induces granulocytic differentiation upon E2 induction.
C/EBPα is required for granulocytic differentiation, and restoration of its proper function can enable leukemic stem and progenitor cells in AML and chronic myeloid leukemia myeloid blast crisis to overcome differentiation blockade and mature into functional effector cells.27, 50 It acts as a master switch between uncommitted proliferating progenitors and differentiated cells.51 Thus, the downregulation and functional inactivation of C/EBPα is involved in tumorigenesis. Growing evidences reveal that alterations of the key myeloid transcription factor C/EBPα is involved in the pathogenesis of AML.52, 53, 54 Importantly, different mechanisms lead to decreased C/EBPα function in different AML subtypes.3, 4 Therefore, enhancing/stabilizing C/EBPα protein expression or function in myeloid leukemia cells can be beneficial from therapeutic perspective. As restoration of C/EBPα in K562 cells promotes granulocytic differentiation,27 we assumed stabilization of C/EBPα in these stable cells would have similar effects. For this, we overexpressed E6AP-C843A in K562-C/EBPα-p42-ER stable cells and 24 h post transfection induced them with β-estradiol. Similarly, we knocked down E6AP in these cells using siE6AP and 24 h post transfection induced them with β-estradiol. Post 72 h E2 induction, FACS analysis for cd11b and cd114 expression confirmed that E6AP-C843A overexpression and/or siE6AP-mediated knockdown of E6AP in K562-C/EBPα-p42-ER stable cells substantially increases the percentage of cells undergoing differentiation in the presence of E2 (Figure 6). Besides, E6AP knockdown in U937 cells also led to enhanced C/EBPα protein levels and subsequent myeloid differentiation (Figure 4). Moreover, biological effects of E6AP inhibition validated in a yet another myeloid differentiation model 32Dcl3 cells, also resulted in increased granulocytic differentiation (Figure 7), which further consolidated our data that E6AP targets C/EBPα for degradation and E6AP knock down or its functional inhibition may stabilize C/EBPα, leading to enhanced differentiation.
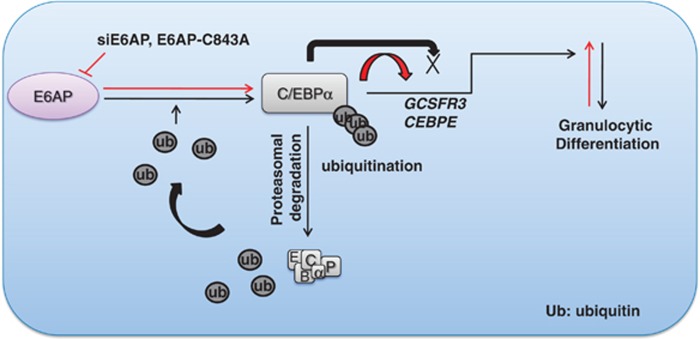
Henceforth, we propose a hypothetical model (Figure 8), which suggests that E6AP targets C/EBPα for degradation via ubiquitin proteasome pathway. As loss of C/EBPα expression is associated with leukemogenesis, our results predict a correlation between elevated levels of E6AP and loss of C/EBPα expression, as well as function in leukemic cells. Furthermore, inhibition of E6AP either via siE6AP or dominant negative E6AP-C843A stabilizes C/EBPα leading to enhanced granulopoiesis. Thus, targeting E6AP can have therapeutic implications in myeloid leukemia and other cancers where C/EBPα is a crucial cellular factor.
Figure 8.
E6AP negatively regulates granulocytic differentiation by targeting C/EBPα for degradation. The figure depicts a hypothetical model suggesting degradation of C/EBPα by E6AP, thereby attenuating its transactivation potential and hence granulocytic differentiation
Materials and Methods
Cell culture and expression plasmids
HEK293T and K562 cells obtained from ATCC were cultured in DMEM and phenol red free RPMI-1640, respectively, supplemented with 10% fetal bovine serum (FBS) and antibiotics. K562-C/EBPα-p42-ER stable clones were maintained in phenol red free RPMI-1640 supplemented with 2.0 μg/ml puromycin, 10% charcoal stripped FBS and antibiotics. IL-3 dependent murine myeloid 32Dcl3 cells were obtained from ATCC. 32Dcl3 cells were maintained in RPMI-1640 medium supplemented with 10% FBS, 1% PenStrep (Gibco, Grand Island, NY, USA) and 10 ng/ml murine IL-3 (Prospec, East Brunswick, NJ, USA) at 37 °C and 5% CO2. Notably, hnRNPs inhibit CEBPA mRNA translation in K562 cells and hence no detectable levels of C/EBPα are seen in these cells. Interestingly, ectopic expression of C/EBPα in these cells drives them to granulopoietic differentiation.27 In contrast, 32Dcl3 cells are murine myeloid precursor cells that differentiate to mature granulocytes in response to G-CSF. Interestingly, ectopically expressed C/EBPα promotes and accelerates G-CSF driven differentiation of 32Dcl3 cells.
Plasmids and siRNA
Expression plasmids for pCDNA3.1-C/EBPα–HA,48 pCDNA3.1-E6AP,21 pCAG-HA-E6AP, pCAG-HA-E6AP-C843A,46 pGEX4T-GST-E6AP55 were kind gifts from G. J. Darlington, Nihar Jana, Ikuo Shoji and Zafar Nawaz respectively; while pCDNA3-p42C/EBPα, Ubiquitin-his, p(C/EBP)2TK-luc and pCDNA3-p30C/EBPα are previously described.6, 7 The siE6AP and scrambled siRNA, as well as siRNA transfection reagent Dhermafect were purchased from Dharmacon RNA Technologies (Lafayette, CO, USA). E6AP-C843A is a catalytically inactive form of E6AP where active site cysteine residue is substituted with alanine (C843A). This cysteine residue present in the catalytic domain transfers ubiquitin directly to the substrate via ubiquitin-enzyme cascade leading to their degradation.
Generation of stable cell line
pBabe-Puro-C/EBPα p42-ER and pBabe-Puro empty vector constructs were used for generating stable clones as previously described.28, 56 For this, K562 cells were transfected with pBabe-Puro-p42C/EBPα-ER and empty vector, selection of cells was performed in RPMI-1640 supplemented with 10% FBS, 1 × antibiotic solution and 2.0 μg/ml puromycin. Cells resistant to puromycin (2.0 μg/ml) were further cultured in puromycin supplemented medium for next 2 weeks. In total, six clones were selected by the serial dilution of cells. These six clones were cultured for another 2 weeks in RPMI-1640 supplemented with puromycin (2.0 μg/ml) and subsequently confirmed for C/EBPα-ER expression by immunoblotting (Supplementary Figure S2).
Western blotting
Cells were harvested after indicated time points using RIPA buffer (1% NP40, 0.5% sodium deoxycholate, 0.1% SDS, 0.15 M NaCl, 5 mM EDTA and 50 mM Tris pH8.0) and equal amount of proteins were separated on 10% SDS-PAGE as previously described.7, 57 Subsequently, proteins were transferred and immunoblotted using primary antibodies against C/EBPα, His, GST, β-actin, GAPDH (Santacruz Biotechnology, SantaCruz, CA, USA) and E6AP (Sigma-Aldrich, St. Louis, MO, USA).
Immunofluorescence microscopy
HEK293T cells were plated in chamber slide one day before transfection. Next day cells were transfected with C/EBPα and E6AP plasmids. Twenty-four hours after transfection cells were washed with phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde for 10 min, permeabilized with 0.5% Triton X-100 in PBS for 5 min, washed with PBS, and then blocked with 1% BSA in PBS for 1 h. The cells were then incubated with primary antibodies C/EBPα and E6AP (1 : 200) overnight at 4 °C. Next day, cells were washed thrice with PBS, incubated with Alexa Flour 594 and 488 secondary antibodies (1 : 250 dilutions) for 1 h; Again washed thrice with PBS followed by 4′, 6-diamidino-2-phenylindole staining (Sigma-Aldrich). Cells were then mounted with vectashield (Vector Laboratories, Burlingame, CA, USA) and were visualized using a confocal microscope (Leica, Wetzlar, Germany).
Luciferase reporter assay
1 × 105 HEK 293T cells/well were plated 1 day before transfection. Next day cells were transfected with pTK-C/EBP-luc promoter, C/EBPα, E6AP and E6AP mutant (E6AP-C843A). Twenty-four hours post transfection cell extracts were assayed for luciferase activity, using luciferase assay reagent (Promega, Madison, WI, USA). Data are presented as means of triplicate values obtained from representative experiments.
Co-Immunoprecipitation assay
For Co-IP cell lysates were prepared in RIPA buffer (1% (v/v) NP40, 0.5% (w/v) sodium deoxycholate, 0.1% (w/v) SDS, 0.15 M NaCl, 5 mM EDTA, 1 mM DTT and protease inhibitors). Protein lysates after preclearing with IgG were incubated with C/EBPα or E6AP antibody and Protein Agarose G or A beads (Millipore Company, Bedford, MA, USA) for 3 h at 4 °C in IP buffer (1% TBS, 0.5% NP40, Protease inhibitors). After incubation beads were washed with IP buffer three times and bound protein were eluted in Laemmli buffer. Samples were separated on 10% SDS-PAGE and were subsequently immunoblotted with C/EBPα and E6AP antibody.
GST-Pull down
For GST-Pull down assay GST-E6AP protein was overexpressed in E.coli and was subsequently purified using immobilized glutathione sepharose beads (Amersham Bioscience/GE Healthcare, Pittsburg, PA, USA) in NETN buffer (20 mM Tris pH 8.0,100 mM NaCl,1 mM EDTA,0.5% NP40). Cell lysate of C/EBPα overexpressed in HEK293T cells were prepared in RIPA buffer. For pull down experiments, GST purified proteins were incubated with WCEs in NETN buffer for 3 h at 4 °C on a rotating shaker. After pull down, protein bound GST sepharose beads were washed three times with NETN buffer. SDS loading dye was added to the beads and was resolved on 10% SDS-polyacrylamide gel, immunoblotted with GST and C/EBPα antibody to confirm the interaction.
In vivo ubiquitination assay
HEK293T cells were transfected with C/EBPα, E6AP and ubiquitin constructs. Post 24 h of transfection, cells were harvested and RIPA lysates were prepared. Subsequently, co-immunoprecipitation was performed with 2 μg of C/EBPα antibody using protein G Agarose beads (Millipore). After preclearing, protein lysates were incubated with antibody and beads for 3 h. The co-immunoprecipitated proteins were then separated by 10% SDS-PAGE and probed with His antibody.
Giemsa staining
K562-C/EBPα-p42-ER and 32Dcl3 cells were cytospun on slides; air-dried and were stained with May-Grünwald and Giemsa solution. For this, cells were stained with May-Grünwald solution for 5 min followed by washing with 1 × PBS for 2 min. Meanwhile, Giemsa solution was diluted 1 : 20 ratio in PBS and slides were further stained in this diluted solution for 15–20 min. Cells were then washed in running tap water to remove the excess stain, air-dried and subjected to microscopic examinations under light microscope and were photographed.
Acknowledgments
Funding from Center for Research on Anabolic Skeletal Targets in Health and Illness (ASTHI), CSIR (BSC0201), Department of Biotechnology (GAP0068) to AKT and ICMR Senior Research Fellowship to PP is acknowledged. Institutional communication number for this article is 8425.
Glossary
- C/EBPα
CCAAT/enhancer-binding protein alpha
- p42C/EBPα
42 kDa wild-type CCAAT-enhancer-binding protein alpha
- p30C/EBPα
30 kDa mutant CCAAT-enhancer-binding protein alpha
- E6AP
E6-associated protein
- E6AP-C843A
E6-associated protein having cysteine replaced with alanine at amino-acid position 843
- GST
glutathione sepharose transferase
- siRNA
small interfering RNAs
- HEK293T
human embryonic kidney epithelium
- WCEs
whole-cell extracts
- E2
β-estradiol
- HSPs
heat shock proteins
- AML
acute myeloid leukemia
- PBS
phosphate-buffered saline
- K562-C/EBPα-p42-ER
β-estradiol inducible K562 cells stably expressing p42C/EBPα-ER
- 32Dcl3
32D clone 3, a cell line widely used model for in vitro study of hematopoietic cell proliferation, differentiation, and apoptosis
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by G Ciliberto
Supplementary Material
References
- Birkenmeier EH, Gwynn B, Howard S, Jerry J, Gordon JI, Landschulz WH, et al. Tissue-specific expression, developmental regulation, and genetic mapping of the gene encoding CCAAT/enhancer binding protein. Genes Dev. 1989;3:1146–1156. doi: 10.1101/gad.3.8.1146. [DOI] [PubMed] [Google Scholar]
- Nerlov C. C/EBPαlpha mutations in acute myeloid leukaemias. Nat Rev Cancer. 2004;4:394–400. doi: 10.1038/nrc1363. [DOI] [PubMed] [Google Scholar]
- Koschmieder S, Halmos B, Levantini E, Tenen DG. Dysregulation of the C/EBPαlpha differentiation pathway in human cancer. J Clin Oncol. 2009;27:619–628. doi: 10.1200/JCO.2008.17.9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi AK, Pal P, Behre G, Singh SM. Multiple ways of C/EBPαlpha inhibition in myeloid leukaemia. Eur J Cancer. 2008;44:1516–1523. doi: 10.1016/j.ejca.2008.04.022. [DOI] [PubMed] [Google Scholar]
- Ross SE, Radomska HS, Wu B, Zhang P, Winnay JN, Bajnok L, et al. Phosphorylation of C/EBPαlpha inhibits granulopoiesis. Mol Cell Biol. 2004;24:675–686. doi: 10.1128/MCB.24.2.675-686.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geletu M, Balkhi MY, Peer Zada AA, Christopeit M, Pulikkan JA, Trivedi AK, et al. Target proteins of C/EBPαlphap30 in AML: C/EBPαlphap30 enhances sumoylation of C/EBPαlphap42 via up-regulation of Ubc9. Blood. 2007;110:3301–3309. doi: 10.1182/blood-2007-01-071035. [DOI] [PubMed] [Google Scholar]
- Trivedi AK, Bararia D, Christopeit M, Peerzada AA, Singh SM, Kieser A, et al. Proteomic identification of C/EBP-DBD multiprotein complex: JNK1 activates stem cell regulator C/EBPαlpha by inhibiting its ubiquitination. Oncogene. 2007;26:1789–1801. doi: 10.1038/sj.onc.1209964. [DOI] [PubMed] [Google Scholar]
- Behre G, Singh SM, Liu H, Bortolin LT, Christopeit M, Radomska HS, et al. Ras signaling enhances the activity of C/EBP alpha to induce granulocytic differentiation by phosphorylation of serine 248. J Biol Chem. 2002;277:26293–26299. doi: 10.1074/jbc.M202301200. [DOI] [PubMed] [Google Scholar]
- Rosenbauer F, Tenen DG. Transcription factors in myeloid development: balancing differentiation with transformation. Nat Rev Immunol. 2007;7:105–117. doi: 10.1038/nri2024. [DOI] [PubMed] [Google Scholar]
- Gombart AF, Hofmann WK, Kawano S, Takeuchi S, Krug U, Kwok SH, et al. Mutations in the gene encoding the transcription factor CCAAT/enhancer binding protein alpha in myelodysplastic syndromes and acute myeloid leukemias. Blood. 2002;99:1332–1340. doi: 10.1182/blood.v99.4.1332. [DOI] [PubMed] [Google Scholar]
- Pabst T, Mueller BU, Zhang P, Radomska HS, Narravula S, Schnittger S, et al. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-alpha (C/EBPαlpha), in acute myeloid leukemia. Nat Genet. 2001;27:263–270. doi: 10.1038/85820. [DOI] [PubMed] [Google Scholar]
- Kato N, Kitaura J, Doki N, Komeno Y, Watanabe-Okochi N, Togami K, et al. Two types of C/EBPαlpha mutations play distinct but collaborative roles in leukemogenesis: lessons from clinical data and BMT models. Blood. 2011;117:221–233. doi: 10.1182/blood-2010-02-270181. [DOI] [PubMed] [Google Scholar]
- Westendorf JJ, Yamamoto CM, Lenny N, Downing JR, Selsted ME, Hiebert SW. The t(8;21) fusion product, AML-1-ETO, associates with C/EBP-alpha, inhibits C/EBP-alpha-dependent transcription, and blocks granulocytic differentiation. Mol Cell Biol. 1998;18:322–333. doi: 10.1128/mcb.18.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangatia J, Vangala RK, Singh SM, Peer Zada AA, Elsasser A, Kohlmann A, et al. Elevated c-Jun expression in acute myeloid leukemias inhibits C/EBPαlpha DNA binding via leucine zipper domain interaction. Oncogene. 2003;22:4760–4764. doi: 10.1038/sj.onc.1206664. [DOI] [PubMed] [Google Scholar]
- Radomska HS, Basseres DS, Zheng R, Zhang P, Dayaram T, Yamamoto Y, et al. Block of C/EBP alpha function by phosphorylation in acute myeloid leukemia with FLT3 activating mutations. J Exp Med. 2006;203:371–381. doi: 10.1084/jem.20052242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian L, Benson MD, Iniguez-Lluhi JA. A synergy control motif within the attenuator domain of CCAAT/enhancer-binding protein alpha inhibits transcriptional synergy through its PIASy-enhanced modification by SUMO-1 or SUMO-3. J Biol Chem. 2003;278:9134–9141. doi: 10.1074/jbc.M210440200. [DOI] [PubMed] [Google Scholar]
- Keeshan K, He Y, Wouters BJ, Shestova O, Xu L, Sai H, et al. Tribbles homolog 2 inactivates C/EBPαlpha and causes acute myelogenous leukemia. Cancer Cell. 2006;10:401–411. doi: 10.1016/j.ccr.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandinetti KB, Stevens TA, Ha S, Salamone RJ, Walker JR, Zhang J, et al. Overexpression of TRIB2 in human lung cancers contributes to tumorigenesis through downregulation of C/EBPαlpha. Oncogene. 2011;30:3328–3335. doi: 10.1038/onc.2011.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengoechea-Alonso MT, Ericsson J. The ubiquitin ligase Fbxw7 controls adipocyte differentiation by targeting C/EBPαlpha for degradation. Proc Natl Acad Sci USA. 2010;107:11817–11822. doi: 10.1073/pnas.0913367107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse JM, Scheffner M, Beaudenon S, Howley PM. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci USA. 1995;92:5249. doi: 10.1073/pnas.92.11.5249-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A, Godavarthi SK, Jana NR. UBE3A/E6-AP regulates cell proliferation by promoting proteasomal degradation of p27. Neurobiol Dis. 2009;36:26–34. doi: 10.1016/j.nbd.2009.06.010. [DOI] [PubMed] [Google Scholar]
- Talis AL, Huibregtse JM, Howley PM. The role of E6AP in the regulation of p53 protein levels in human papillomavirus (HPV)-positive and HPV-negative cells. J Biol Chem. 1998;273:6439–6445. doi: 10.1074/jbc.273.11.6439. [DOI] [PubMed] [Google Scholar]
- Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- Louria-Hayon I, Alsheich-Bartok O, Levav-Cohen Y, Silberman I, Berger M, Grossman T, et al. E6AP promotes the degradation of the PML tumor suppressor. Cell Death Differ. 2009;16:1156–1166. doi: 10.1038/cdd.2009.31. [DOI] [PubMed] [Google Scholar]
- Lochab S, Pal P, Kanaujiya JK, Tripathi SB, Kapoor I, Bhatt ML, et al. Proteomic identification of E6AP as a molecular target of tamoxifen in MCF7 cells. Proteomics. 2012;12:1363–1377. doi: 10.1002/pmic.201100572. [DOI] [PubMed] [Google Scholar]
- Cheng J, Yu DV, Zhou JH, Shapiro DJ. Tamoxifen induction of CCAAT enhancer-binding protein alpha is required for tamoxifen-induced apoptosis. J Biol Chem. 2007;282:30535–30543. doi: 10.1074/jbc.M704829200. [DOI] [PubMed] [Google Scholar]
- Tavor S, Park DJ, Gery S, Vuong PT, Gombart AF, Koeffler HP. Restoration of C/EBPαlpha expression in a BCR-ABL+ cell line induces terminal granulocytic differentiation. J Biol Chem. 2003;278:52651–52659. doi: 10.1074/jbc.M307077200. [DOI] [PubMed] [Google Scholar]
- Wang X, Scott E, Sawyers CL, Friedman AD. C/EBPαlpha bypasses granulocyte colony-stimulating factor signals to rapidly induce PU.1 gene expression, stimulate granulocytic differentiation, and limit proliferation in 32D cl3 myeloblasts. Blood. 1999;94:560–571. [PubMed] [Google Scholar]
- Keeshan K, Santilli G, Corradini F, Perrotti D, Calabretta B. Transcription activation function of C/EBPαlpha is required for induction of granulocytic differentiation. Blood. 2003;102:1267–1275. doi: 10.1182/blood-2003-02-0477. [DOI] [PubMed] [Google Scholar]
- Friedman AD, Keefer JR, Kummalue T, Liu H, Wang QF, Cleaves R. Regulation of granulocyte and monocyte differentiation by CCAAT/enhancer binding protein alpha. Blood Cells Mol Dis. 2003;31:338–341. doi: 10.1016/s1079-9796(03)00135-9. [DOI] [PubMed] [Google Scholar]
- Rosmarin AG, Yang Z, Resendes KK. Transcriptional regulation in myelopoiesis: Hematopoietic fate choice, myeloid differentiation, and leukemogenesis. Exp Hematol. 2005;33:131–143. doi: 10.1016/j.exphem.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Leroy H, Roumier C, Huyghe P, Biggio V, Fenaux P, Preudhomme C. CEBPA point mutations in hematological malignancies. Leukemia. 2005;19:329–334. doi: 10.1038/sj.leu.2403614. [DOI] [PubMed] [Google Scholar]
- Tenen DG. Disruption of differentiation in human cancer: AML shows the way. Nat Rev Cancer. 2003;3:89–101. doi: 10.1038/nrc989. [DOI] [PubMed] [Google Scholar]
- Radomska HS, Huettner CS, Zhang P, Cheng T, Scadden DT, Tenen DG. CCAAT/enhancer binding protein alpha is a regulatory switch sufficient for induction of granulocytic development from bipotential myeloid progenitors. Mol Cell Biol. 1998;18:4301–4314. doi: 10.1128/mcb.18.7.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DE, Zhang P, Wang ND, Hetherington CJ, Darlington GJ, Tenen DG. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc Natl Acad Sci USA. 1997;94:569–574. doi: 10.1073/pnas.94.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekstrom-Himes JA. The role of C/EBP(epsilon) in the terminal stages of granulocyte differentiation. Stem Cells. 2001;19:125–133. doi: 10.1634/stemcells.19-2-125. [DOI] [PubMed] [Google Scholar]
- Chakraborty A, Tweardy DJ. Granulocyte colony-stimulating factor activates a 72-kDa isoform of STAT3 in human neutrophils. J Leukoc Biol. 1998;64:675–680. doi: 10.1002/jlb.64.5.675. [DOI] [PubMed] [Google Scholar]
- Valtieri M, Santoli D, Caracciolo D, Kreider BL, Altmann SW, Tweardy DJ, et al. Establishment and characterization of an undifferentiated human T leukemia cell line which requires granulocyte-macrophage colony stimulatory factor for growth. J Immunol. 1987;138:4042–4050. [PubMed] [Google Scholar]
- Guchhait P, Tosi MF, Smith CW, Chakaraborty A. The murine myeloid cell line 32Dcl3 as a model system for studying neutrophil functions. J Immunol Methods. 2003;283:195–204. doi: 10.1016/j.jim.2003.09.013. [DOI] [PubMed] [Google Scholar]
- Bernassola F, Karin M, Ciechanover A, Melino G. The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell. 2008;14:10–21. doi: 10.1016/j.ccr.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Kuhne C, Banks L. E3-ubiquitin ligase/E6-AP links multicopy maintenance protein 7 to the ubiquitination pathway by a novel motif, the L2G box. J Biol Chem. 1998;273:34302–34309. doi: 10.1074/jbc.273.51.34302. [DOI] [PubMed] [Google Scholar]
- Kumar S, Talis AL, Howley PM. Identification of HHR23A as a substrate for E6-associated protein-mediated ubiquitination. J Biol Chem. 1999;274:18785–18792. doi: 10.1074/jbc.274.26.18785. [DOI] [PubMed] [Google Scholar]
- Oda H, Kumar S, Howley PM. Regulation of the Src family tyrosine kinase Blk through E6AP-mediated ubiquitination. Proc Natl Acad Sci USA. 1999;96:9557–9562. doi: 10.1073/pnas.96.17.9557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M, Banks L. Inhibition of Bak-induced apoptosis by HPV-18 E6. Oncogene. 1998;17:2943–2954. doi: 10.1038/sj.onc.1202223. [DOI] [PubMed] [Google Scholar]
- Mani A, Oh AS, Bowden ET, Lahusen T, Lorick KL, Weissman AM, et al. E6AP mediates regulated proteasomal degradation of the nuclear receptor coactivator amplified in breast cancer 1 in immortalized cells. Cancer Res. 2006;66:8680–8686. doi: 10.1158/0008-5472.CAN-06-0557. [DOI] [PubMed] [Google Scholar]
- Shimoji T, Murakami K, Sugiyama Y, Matsuda M, Inubushi S, Nasu J, et al. Identification of annexin A1 as a novel substrate for E6AP-mediated ubiquitylation. J Cell Biochem. 2009;106:1123–1135. doi: 10.1002/jcb.22096. [DOI] [PubMed] [Google Scholar]
- Lane MD, Tang QQ, Jiang MS. Role of the CCAAT enhancer binding proteins (C/EBPs) in adipocyte differentiation. Biochem Biophys Res Commun. 1999;266:677–683. doi: 10.1006/bbrc.1999.1885. [DOI] [PubMed] [Google Scholar]
- Harris TE, Albrecht JH, Nakanishi M, Darlington GJ. CCAAT/enhancer-binding protein-alpha cooperates with p21 to inhibit cyclin-dependent kinase-2 activity and induces growth arrest independent of DNA binding. J Biol Chem. 2001;276:29200–29209. doi: 10.1074/jbc.M011587200. [DOI] [PubMed] [Google Scholar]
- Umek RM, Friedman AD, McKnight SL. CCAAT-enhancer binding protein: a component of a differentiation switch. Science. 1991;251:288–292. doi: 10.1126/science.1987644. [DOI] [PubMed] [Google Scholar]
- Yeamans C, Wang D, Paz-Priel I, Torbett BE, Tenen DG, Friedman AD. C/EBPαlpha binds and activates the PU.1 distal enhancer to induce monocyte lineage commitment. Blood. 2007;110:3136–3142. doi: 10.1182/blood-2007-03-080291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porse BT, Bryder D, Theilgaard-Monch K, Hasemann MS, Anderson K, Damgaard I, et al. Loss of C/EBP alpha cell cycle control increases myeloid progenitor proliferation and transforms the neutrophil granulocyte lineage. J Exp Med. 2005;202:85–96. doi: 10.1084/jem.20050067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taskesen E, Bullinger L, Corbacioglu A, Sanders MA, Erpelinck CA, Wouters BJ, et al. Prognostic impact, concurrent genetic mutations, and gene expression features of AML with CEBPA mutations in a cohort of 1182 cytogenetically normal AML patients: further evidence for CEBPA double mutant AML as a distinctive disease entity. Blood. 2011;117:2469–2475. doi: 10.1182/blood-2010-09-307280. [DOI] [PubMed] [Google Scholar]
- Hollink IH, van den Heuvel-Eibrink MM, Arentsen-Peters ST, Zimmermann M, Peeters JK, Valk PJ, et al. Characterization of CEBPA mutations and promoter hypermethylation in pediatric acute myeloid leukemia. Haematologica. 2011;96:384–392. doi: 10.3324/haematol.2010.031336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulikkan JA, Peramangalam PS, Dengler V, Ho PA, Preudhomme C, Meshinchi S, et al. C/EBPαlpha regulated microRNA-34a targets E2F3 during granulopoiesis and is down-regulated in AML with CEBPA mutations. Blood. 2010;116:5638–5649. doi: 10.1182/blood-2010-04-281600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz Z, Lonard DM, Smith CL, Lev-Lehman E, Tsai SY, Tsai MJ, et al. The Angelman syndrome-associated protein, E6-AP, is a coactivator for the nuclear hormone receptor superfamily. Mol Cell Biol. 1999;19:1182–1189. doi: 10.1128/mcb.19.2.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SM, Trivedi AK, Behre G. C/EBPαlpha S248A mutation reduces granulocytic differentiation in human leukemic K562 cells. Biochem Biophys Res Commun. 2008;369:692–694. doi: 10.1016/j.bbrc.2008.02.090. [DOI] [PubMed] [Google Scholar]
- Pal P, Kanaujiya JK, Lochab S, Tripathi SB, Bhatt ML, Singh PK, et al. 2-D gel electrophoresis-based proteomic analysis reveals that ormeloxifen induces G0-G1 growth arrest and ERK-mediated apoptosis in chronic myeloid leukemia cells K562. Proteomics. 2011;11:1517–1529. doi: 10.1002/pmic.201000720. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.