Abstract
Adenosine deaminase acting on RNA 1 (ADAR1) is a double-stranded RNA-editing enzyme that converts adenosine (A) to inosine (I), and essential for normal development. In this study, we reported an essential role of ADAR1 in the survival and maintenance of intestinal stem cells and intestinal homoeostasis by suppressing endoplasmic reticulum (ER) stress and interferon (IFN) signaling. ADAR1 was highly expressed in the Lgr5+ cells, and its deletion in adult mice led to a rapid apoptosis and loss of these actively cycling stem cells in the small intestine and colon. ADAR1 deletion resulted in a drastic expansion of progenitors and Paneth cells but a reduction of three other major epithelial lineages. Moreover, loss of ADAR1 induced ER stress and activation of IFN signaling, and altered expression in WNT targets, followed by intestinal inflammation. An ER stress inhibitor partially suppressed crypt apoptosis. Finally, data from cultured intestinal crypts demonstrated that loss of ADAR1 in the epithelial cells is the primary cause of these effects. These results support an essential role of ADAR1 and RNA editing in tissue homeostasis and stem cells.
Keywords: ADAR1, RNA editing, intestinal stem cell, apoptosis, ER stress, interferon signaling
The intestinal epithelium is the most rapidly self-renewing tissue in adult mammals, which renews every 3–5 days in mice.1 Homeostasis of the intestinal epithelium is dependent on intestinal stem cells (ISCs) and a balance among cell proliferation, migration, differentiation, and cell death.1, 2 Several ISC populations with possible overlap have recently been identified in mouse, including the actively cycling columnar cells at the crypt base (CBCs) marked by Lgr5 or CD133, and the slow cycling cells in the +4 area marked by Bmi1, DCLK-1 (also known as DCAMKL-1), or mTERT. Ablation of certain lineage revealed inter-conversion of ISC populations and re-establishment of more diverse ISC populations upon repair.3 Epithelial injury and compromised repair have been linked to a wide range of diseases, including chronic inflammation, autoimmunity, and cancer. Developmental pathways, such as Wnt/β-catenin, BMP, PI3K, and Notch, are important in the maintenance of ISCs and intestinal proliferation and differentiation,4 whereas the role of other pathways remain largely unexplored and can help illuminate pathogenesis of these diseases.
Adenosine-to-inosine (A-to-I) editing of primary transcripts by ADAR enzymes (adenosine deaminase acting on RNA) is the most prevalent RNA-editing mechanism in higher eukaryotes.5, 6 Three mammalian adenosine deaminase acting on RNA (ADAR) genes are found in mammals, which encode two active deaminases (ADAR1 and ADAR2) and one inactive deaminase (ADAR3).5, 6 ADARs act on double-stranded RNA (dsRNA) structures in both messenger and noncoding RNAs. The A–to-I conversion in dsRNA alters the stabilities of the dsRNA helix, and can also lead to codon changes as I is decoded as guanosine (G) during translation that increases transcriptomic diversity.6, 7 ADARs also edit noncoding RNAs to affect microRNA processing,7 as well as sites scattered in Alu repeat elements within human pre-mRNAs.8 ADARs also catalyze hyper-editing of long dsRNAs, whereby up to 50% of adenosines are converted to inosine (I).9
ADAR1 is mutated in a rare autosomal-dominant human-inherited skin pigmentation disease called dyschromatosis symmetrica hereditaria, and over 90 mutations have been identified in patients.10 Most of these mutations are predicted to produce truncated proteins. In mice, ADAR1 deletion causes widespread apoptosis, defective hematopoiesis, and embryonic lethality, suggesting an essential role of ADAR1 for embryo development and cell proliferation and differentiation.11, 12 Using conditional knockout models, ADAR1 was found to be required for the maintenance of hematopoietic stem cells and hematopoietic progenitor cells13, 14 by suppression of interferon (IFN) signaling.13, 15 Loss of ADAR1 in the skin led to massive cell death in the epidermis and loss of hair follicles.16
In this study, we investigated the role of ADAR1 in the intestinal tract using a conditional knockout mouse model. ADAR1 was expressed highly in the Lgr5+ population, and its deletion in adult mice induced a rapid apoptosis and loss of stem cells in the small intestine and colon. ADAR1 deletion led to marked changes in intestinal differentiation, an expansion of transient amplification (TA) cells, and severe intestinal injury and inflammation. Loss of ADAR1 induced endoplasmic reticulum (ER) stress and IFN signaling in intestinal epithelial cells. Data from cultured crypts strongly suggest that ADAR1 suppresses epithelial cell death through a cell-autonomous manner. These data reveal a critical role of ADAR1 in maintaining intestinal homeostasis and ISC survival, and a potential involvement of RNA editing in digestive diseases.
Results
ADAR1 loss leads to crypt apoptosis and intestinal damage in mice
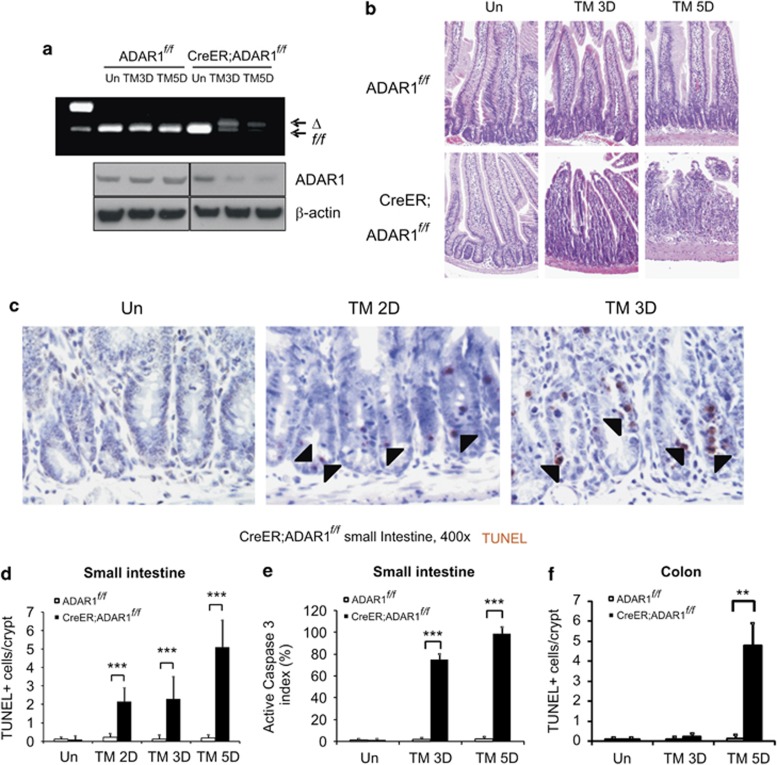
To investigate the role of ADAR1 in the small intestine, 6- to 10-week-old CreER; ADAR1f/f mice were administered with 120 mg/kg tamoxifen on day 0, 1, 3, and 4 to excise floxed ADAR1 alleles (Supplementary Figure 1A). ADAR1f/f mice treated with tamoxifen were used as controls in all experiments. The deletion efficiency of ADAR1 in the small intestine mucosa was over 50 and 70% on day 3 and 5, respectively in CreER; ADAR1f/f mice (Figure 1a). Western blotting confirmed reduction of ADAR1 protein (Figure 1a). ADAR1 deletion can be detected on day 1 (1 × TM), whereas the deletion efficiency on day 2 and 3 (2x TM) were similar as expected (Figure 1a and Supplementary Figure 1B). Based on these observations and a 3–5 day renewal cycle of intestinal epithelium,1, 2 we examined tissue harvested on or before day 5 in our following analyses.
Figure 1.
ADAR1 loss leads to crypt apoptosis and epithelial damage in the small intestine and colon in mice. ADAR1f/f and CreER; ADAR1f/f mice were subjected to 120 mg/kg tamoxifen (TM) administration(s) as in Supplementary Figure 1. Intestinal tissues were harvested at the indicated days (D). (a) Upper: ADAR1 deletion in the intestinal mucosa detected by genomic PCR in ADAR1f/f and CreER; ADAR1f/f. The upper and lower bands indicated the deletion of ADAR1 (Δ) or intact or floxed (f/f) ADAR1, respectively. Lower: ADAR1 and β-actin protein levels were analyzed by western blotting. (b) Representative H&E staining, magnification × 200. (c) Representative TUNEL IHC in the small intestine. Arrow heads indicate examples of TUNEL+ cells. (d) Quantification of TUNEL staining in the crypts. (e) Qualification of active caspase 3 staining in crypts. (f) Quantification of TUNEL staining in the colonic crypts. **P<0.01, ***P<0.001
There was no discernable difference in the histology of the small intestine between ADAR1f/f and CreER; ADAR1f/f mice without tamoxifen treatment (Figure 1b). However, tamoxifen treatment resulted in shortened villi in CreER; ADAR1f/f mice on day 3, and a severe loss of crypts and epithelial integrity on day 5. Terminal deoxynucleotidyl transferase-mediated deoxyuridinetriphosphate nick end labeling (TUNEL) staining and active caspase 3 immunohistochemical (IHC) indicated apoptosis in the intestinal crypts (Figures 1c–e). Notably, apoptosis was concentrated in the bottom of crypts (Supplementary Figures 1C and D) and could be detected as early as day 2 in the CBC area, whereas little or no apoptosis was detected in the villi (Supplementary Figures 1E and F). ADAR1 deletion resulted in significant crypt loss accompanied by crypt fission in the colon on day 5 (Supplementary Figure 2A), which was not pronounced on day 3. TUNEL staining confirmed induction of cell death in the colonic crypts (Figure 1f, Supplementary Figures 2B and C).
Furthermore, we examined the effect of ADAR1 deletion on the stomach, bone marrow, and spleen. Significant cell death in CreER; ADAR1f/f was detected on day 5, accompanied by apparent hypo-cellularity and damage (Supplementary Figure 3). Little or no structural changes or cell death were detected in all organs examined in ADAR1f/f mice (Supplementary Figure 3). These results demonstrate that ADAR1 has an important role in the homeostasis of GI tract and hematopoietic system, which are highly proliferative and contain a well-defined stem cell compartment.
ADAR1 is required for maintenance of ISCs
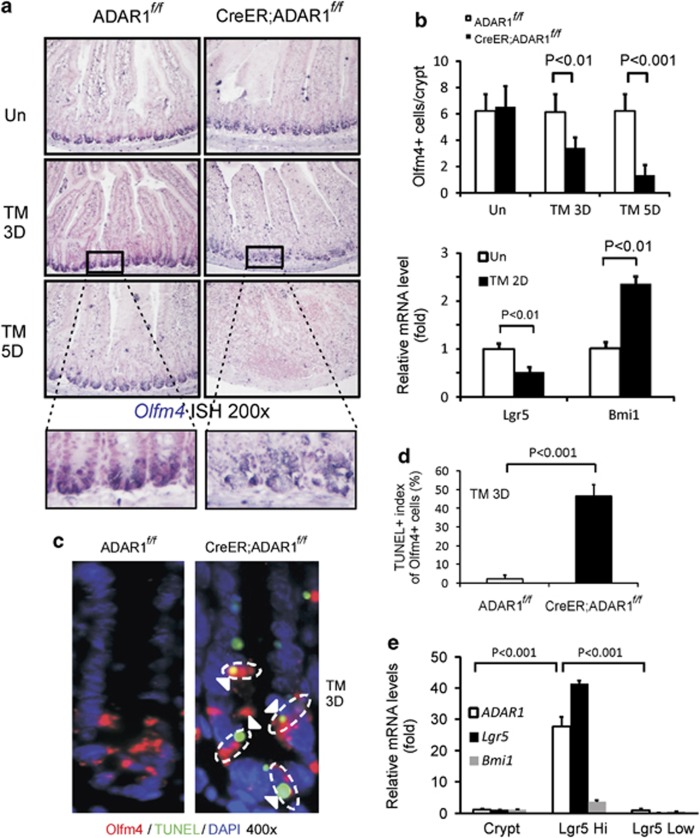
To probe a potential role of ADAR1 in ISC survival, we analyzed the expression of a robust CBC marker Olfm4 by RNA in situ hybridization (ISH).17 Olfm4 expression (blue) was restricted in crypt bottom and CBCs areas in CreER; ADAR1f/f and control mice (Figure 2a). Tamoxifen treatment induced a significant loss of Olfm4 signals, especially in between the Paneth cells in CreER; ADAR1f/f mice on day 3 and by 80% on day 5 (Figures 2a and b). The mRNA level of Lgr5 (Leucine-rich repeat-containing G-protein coupled receptor 5), but not that of the +4 cell-marker Bmi1, was decreased on day 2, before massive epithelial injury was detected (Figure 2b). Importantly, TUNEL staining co-localized with Olfm4 ISH (Figures 2c and Supplementary Figure 4A). RT-PCR analysis showed that ADAR1 was expressed highly in Lgr5 high population compared with Lgr5 low or unfractionated crypts (Figure 2e). Loss of Paneth cells under certain conditions can lead to loss of Lgr5+ or Olfm4+ cells.18 However, cell death was not detected in Paneth cells on day 3 or 5 (Supplementary Figure 4B and data not shown). Loss of Olfm4+ or increase in TUNEL+ cells was not observed in control mice (Figure 2 and Supplementary Figure 4). These data demonstrate that ADAR1 selectively suppresses cell death in actively cycling ISCs.
Figure 2.
ADAR1 is required for the maintenance and survival of intestinal stem cells. ADAR f/f or CreER; ADARf/f mice were subjected to tamoxifen (TM) administration(s) as in Supplementary Figure 1. Intestinal tissues were harvested at the indicated days (D). (a) Olfm4 ISH staining (blue), magnification × 200. Lower panels, enlarged views of the selected areas. (b) Upper, quantification of Olfm4 ISH in crypts. Lower, expression of Lgr5 and Bmi1 transcripts in isolated crypts was analyzed by real-time RT-PCR. Values are means±S.D. n=3 mice in each group. (c) Representative pictures of staining of Olfm4 ISH, TUNEL, and DAPI (nuclei) in crypts on day 3, magnification × 400. Arrow heads indicate examples of Olfm4+/TUNEL+ cells. (d) Quantification of TUNEL staining in the Olfm4+ cells in crypts. (e) Expression of ADAR1, Lgr5, and Bmi1 transcripts in intestinal crypts, Lgr5 high, or Lgr5 low cells was analyzed by real-time RT-PCR. Values are means±S.D.
ADAR1 loss impairs intestinal differentiation
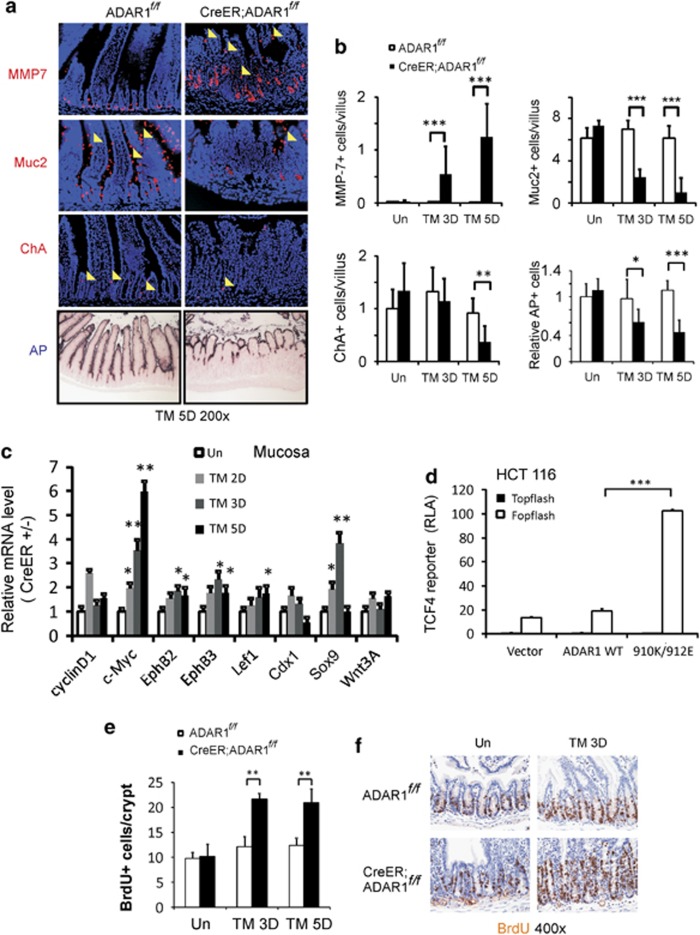
Differentiaton can be used to measure stem cell quality. We examined four major differentiated intestinal epithelial lineages using established markers. Tamoxifen treatment induced a sharp decrease in the numbers of enterocytes, goblet cells, and enteroendocrine cells in CreER; ADAR1f/f mice on days 3 and 5 (Figures 3a, and Supplementary Figure 5). In contrast, a drastic expansion and mislocalization of Paneth cells was observed (Figures 3a and b, and Supplementary Figure 6). These changes were absent in ADAR1f/f mice, suggesting ADAR1 deletion impairs differentiation of progenitor or TA cells.
Figure 3.
ADAR1 loss impairs intestinal differentiation. ADAR f/f or CreER; ADARf/f mice were subjected to tamoxifen (TM) administration(s) as in Supplementary Figure 1. Intestinal tissues were harvested at the indicated days (D). (a) Representative pictures of Paneth cells, Goblet cells, enteroendocrine cells, and enterocytes examined by differentiation markers MMP7, Muc2, chromogranin A (ChA), and alkaline phosphotase (AP), respectively, magnification × 200. Arrow heads indicate examples of staining positive cells. (b) Quantification of MMP7, Muc2, ChA, AP (relative height) staining in the villus. *P<0.05, **P<0.01, ***P<0.001. (c) Expression of Wnt-regulated genes in the mucosa was analyzed by real-time RT-PCR. *P<0.05, **P<0.01. Values are means±S.D. n=3 mice in each group. The values were normalized to the ratio of CreER; ADARf/f/ADAR f/f (ER+versus ER- as 1) on day 0 (un). (d) TCF4 reporter assays following expression of murine WT ADAR1 and editing mutant 910K/912E at 48 h in HCT 116 colon cancer cells. The TCF-specific relative luciferase activity (RLA) was calculated following normalization to the transfection control and that of the reporter containing mutant TCF-binding sites (Fopflash) set as 1. (e) Quantification of BrdU staining. **P<0.01. (f) Representative pictures of BrdU incorporation (brown), magnification × 400
Given a key role of Wnt/β-catenin pathway in intestinal homeostasis, proliferation, and differentiation,19 we examined the effects of ADAR1 deletion on the expression of several Wnt targets using quantitative RT-PCR. These include cyclinD1, c-Myc, EphB2, EphB3, Lef1, Cdx1, Sox9, and Wnt 3A. Among them, c-Myc, Sox9, EphB2, EphB3, and Lef1, were significantly induced in the mucosa of CreER; ADAR1f/f mice by day 3, compared with ADAR1f/f mice. The increased expression of Sox9 and c-Myc was most notable (Figure 3c). Immunostaining revealed a drastic expansion of Sox9+ cells (Figure 3d and Supplementary Figure 6D) and bromodeoxyuridine+ (BrdU+) cells (Figures 3e and f). In addition, the expression of editing defective ADAR1 mutant 910K/912E significantly activated the TCF4 reporter in HCT 116 colon cancer cells. However, the expression of WT ADAR1 did not suppress elevated basal activity of TCF reporter because of APC mutation in these cells20 (Figure 3d and Supplementary Figure 6E). These data suggest that ADAR1 deletion leads to an expansion of progenitors and skewed Paneth cell fate, associated with abnormal Wnt signaling.
ADAR1 deletion induces ER stress and activation of IFN signaling in the intestinal epithelium
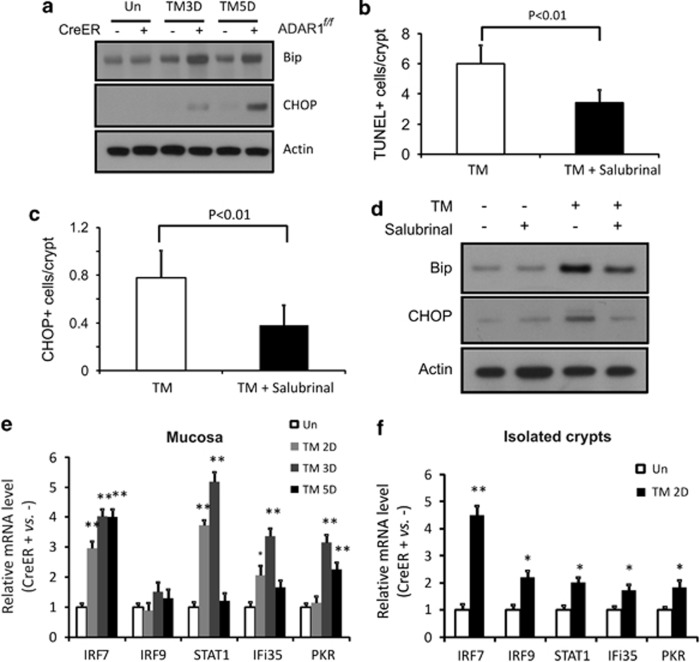
ER stress can induce death in the intestinal epithelial cells,21 which is associated with decreased number of goblet cells, and expansion and mislocalization of Paneth cells.22, 23, 24, 25 To test a potential role of ER stress in cell death following ADAR1 deletion, we measured several markers of ER stress such as glucose-regulating peptide 78 (Bip) and C/EBP homologous protein (CHOP).21 Tamoxifen administration induced Bip and CHOP in the mucosa of CreER; ADAR1f/f mice, compared with control mice (Figure 4a). Furthermore, administration of an ER stress inhibitor Salubrinal blunted cell death (Figure 4b) and the expression of Bip and CHOP (Figures 4c and d).
Figure 4.
ADAR1 deletion rapidly induces endoplasmic reticulum (ER) stress and activation of interferon signaling in intestinal epithelium. ADAR f/f or CreER; ADARf/f mice were subjected to 120 mg/kg tamoxifen (TM) administration(s) or the ER stress inhibitor Salubrinal at 1 mg/kg daily, alone, or in combination. Intestinal tissues were harvested at the indicated days (D). (a) Bip, CHOP, and β-actin levels in the small intestinal mucosa were analyzed by western blotting. (b) Quantification of TUNEL staining in the crypts, with or without Salubrinal. (c) Quantification of CHOP staining in the crypts, with or without Salubrinal. (d) Bip, CHOP, and β-actin levels in the small intestinal mucosa at day 3, with or without Salubrinal, were analyzed by western blotting. (e) Expression of the indicated interferon-responsive genes in mucosa was analyzed by real-time RT-PCR. **P<0.001, *P<0.01, and compared with the control (day 0, Un). (f) Expression of the indicated interferon-responsive genes in isolated crypts was analyzed by real-time RT-PCR. **P<0.001, *P<0.01, and compared with the control (day 0, Un)
ADAR1 suppresses IFN signaling in hematopoietic stem cells,13 whereas IFNs can induce apoptosis through the ER stress pathway.26, 27, 28 We therefore examined a panel of IFN-regulated genes by quantitative RT-PCR in intestinal mucosa and isolated crypts, following ADAR1 deletion. These genes include the signal transducers and activators of transcription 1 (STAT), the IFN regulatory factors IRF7 and IRF9, the RNA-activated protein kinase PKR, and the IFN-induced proteins Ifi35.13 IRF-7, STAT1, and PKR, but not IRF9 and Ifi35, were significantly induced in the crypts of CreER; ADAR1f/f mice as early as day 2 or day 1, compared with control mice (Figures 4e and Supplementary Figure 7), which preceded apoptosis (Figure 1). These data indicate that ADAR1 deletion lead to ER stress and activation of IFN signaling to induce intestinal epithelial cell death.
ADAR1 deletion induces intestinal inflammation
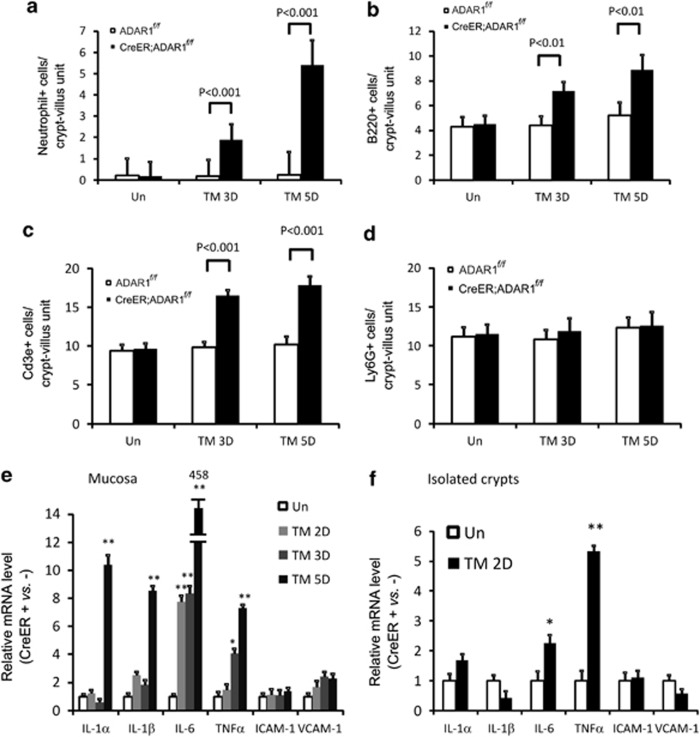
ADAR1 deletion induced progressive intestinal epithelial damage and inflammation in the lamina propria and submucosa (Figure 1b). To rule out immune cell infiltration as a primary cause of the IFN signaling or ER stress, we examined major immune cell types in the intestine by immunostaining, including neutrophils, T (Cd3e+), B (B220+), and myeloid (Ly6G+) cells. Increased infiltration of neutrophils, T, and B cells, but not myeloid cells, was present in the lamina propria of CreER; ADAR1f/f mice on day 3 and 5 after tamoxifen treatment (Figure 5 and Supplementary Figure 8). Consistent with intestinal inflammation, the expression of inflammatory cytokines IL-1α, IL-1β, IL-6, and TNFα was significantly elevated in the small intestinal mucosa on day 5 (Figures 5e and f). Interestingly, induction of IL-6 and TNFα was detected as early as day 2 in both mucosa and isolated crypts, before immune cell infiltration (Figures 5e and 5f, and data not shown). The expression of leukocyte adhesion molecules ICAM-1 and VCAM-1 did not change (Figures 5e and f). Little or no change in intestinal inflammation was present in ADAR1f/f mice (Figure 5 and Supplementary Figure 8). These results demonstrated that immune cell infiltration occurs primarily after ISC apoptosis following ADAR1 deletion and likely exacerbates intestinal injury.
Figure 5.
ADAR1 deletion leads to intestinal inflammation. ADAR f/f or CreER; ADARf/f mice were subjected to tamoxifen (TM) administration(s) as in Supplementary Figure 1. Intestinal tissues were harvested at the indicated days (D). Quantification of staining of (a) neutrophil antigen, (b) B220 (B-cell), (c) Cd3e (T-cell), and (d) myeloid lineage Ly-6G (Gr-1). (e) Expression of the indicated inflammatory cytokines in the small intestinal mucosa was analyzed by real-time RT-PCR. (f) Expression of the indicated inflammatory cytokines in isolated crypts was analyzed by real-time RT-PCR. Values are means±S.D. n=3 mice in each group. The values were normalized to the ratio of CreER; ADARf/f/ADAR f/f at day 0 (ER+ versus ER− as 1, Un). (e and f) **P<0.001, *P<0.01, and compared with the control (day 0)
Loss of ADAR1 induces apoptosis in isolated intestinal crypt cells
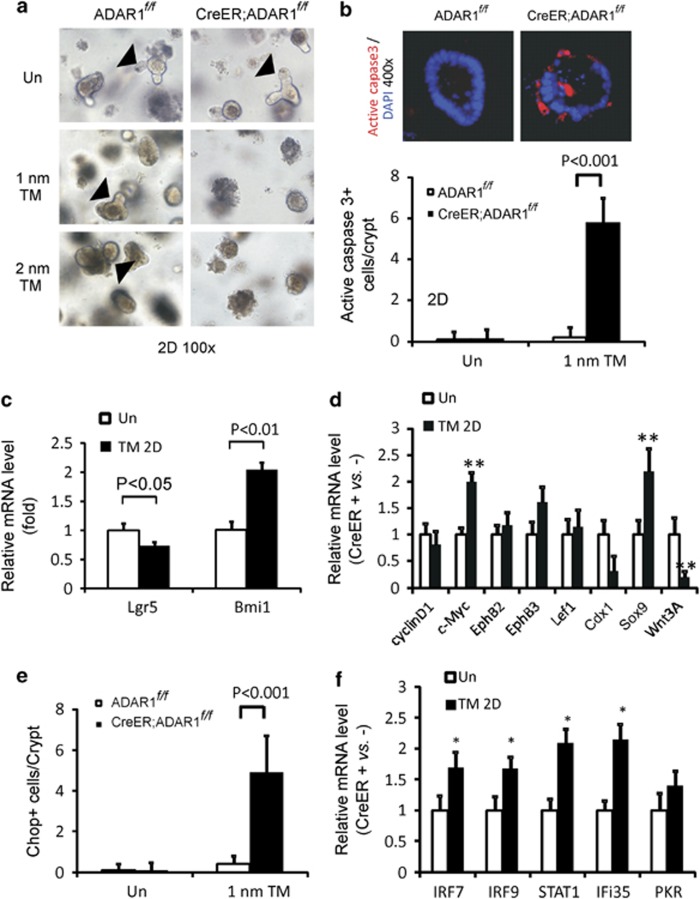
To directly investigate a role of ADAR1 in the intestinal epithelial cells, we cultured intestinal crypts isolated from mice, and induced ADAR1 deletion in vitro. Tamoxifen treatment (1 or 2 nM) induced massive apoptosis in cultured crypts from CreER; ADAR1f/f mice within 24 h, but not in those from the control mice (Figure 6a and Supplementary Figure 9A; Figure 6b and Supplementary Figure 9B). Apoptosis was correlated with decreased expression of Lgr5, but not Bmi1 (Figure 6c). Consistent with our in vivo data, ablation of ADAR1 in cultured crypts also led to a dramatic increase in c-Myc and Sox9 (Figure 6d), CHOP (Figure 6e and Supplementary Figure 9C), and induction of IFN-regulated genes (Figure 6f). A significant reduction in Wnt 3A was also observed following ADAR1 deletion (Figure 6d), and could potentially contribute to ISC loss or the lack of crypt growth.18 Moreover, changes in Wnt targets including c-Myc, Sox9, and Wnt 3A following ADAR1 deletion differed significantly in the intestinal crypts, MEFs, and hepatocytes (Supplementary Figure 10). These data strongly suggest that ADAR1 suppresses ER stress and IFN signaling to maintain crypt survival and intestinal lineage commitment.
Figure 6.
Deletion of ADAR1 in vitro leads to rapid crypt apoptosis. Small intestinal crypts were isolated from ADAR f/f or CreER; ADARf/f mice and plated into Matrigel for 24 h (day 0) as described in methods. Cultured crypts were subjected to DMSO (Un), or 1 nM or 2 nM of tamoxifen (TM) and analyzed 48 h (2 days, 2D) later. (a) Representative pictures of cultured intestinal crypts, magnification × 100. Arrow heads indicate crypt budding or growth, which is lacking in CreER; ADARf/f culture with TM. (b) Quantification of apoptosis in cultured crypts by active caspase 3 staining (red). Values are means±S.D. n=3 different wells from two different mice in each group. A representative picture of active caspase 3/DAPI staining, magnification × 400. (c) The expression of Lgr5 and Bmi1 transcripts in the cultured crypt was analyzed by real-time RT-PCR. Values are means±S.D. n=3 different wells from three different mice in each group. (d) Expression of the indicated Wnt-regulated genes was analyzed by real-time RT-PCR. **P<0.01 compared with the control. Values are means±S.D. n=3 mice in each group. (e) Quantification of CHOP staining in the cultured crypts. Values are means±S.D. n=3 mice in each group. (f) Expression of the indicated interferon-regulated genes were analyzed by real-time RT-PCR. *P<0.01 compared with the control (DMSO). The values of mRNA levels were normalized to the ratio of CreER; ADARf/f/ADAR f/f at day 0 (ER+versus ER− as 1, Un)
Discussion
Intestinal epithelial injury, particularly that related to the stem cells29 and ER stress,21 is implicated in common digestive diseases, including inflammatory bowl diseases and inflammation-associated cancer.30 Developmental pathways such as Wnt/β-catenin, BMP, and Notch are important in the maintenance of ISCs and intestinal homeostasis. In contrast, the p53/PUMA/p21 axis has a key role in the survival and regeneration of ISCs following radiation injury, but is largely dispensable for ISC survival and homeostasis.2, 31, 32 In this study, we discovered an essential role of the RNA-editing enzyme ADAR1 in maintaining intestinal homeostasis and stem cells using whole mouse and isolate crypts. To our knowledge, this is the first report on the requirement of RNA-editing enzymes in intestinal homeostasis and supports that active cycling CBCs are responsible for epithelial renewal under homeostasis.1 Furthermore, ADAR1 appears to have an important role in the maintenance of active stem cells and early progenitors in hematopoietic system13, 14 and skin,16 whereas its role in more quiescent stem cells remains to be explored.
ADAR1 and cell death
Both IFN signaling13 and ER stress are likely involved in the cell death triggered by ADAR1 deletion, and linked to a variety of human conditions including intestinal epithelial injury and inflammation.21, 33 In mice, targeted deletion of the ER stress response transcription factor X-box-binding protein 1, ER stress protein inositol-requiring enzyme 1β, or missense mutations of Muc2 or deletion of Anterior Gradient 2 (AGR2) cause ER stress and death of intestinal epithelial cells in mice.22, 23, 24, 25 Editing-defective tRNA synthetase causes protein misfolding and ER stress in neurons.34 The sensitivity of intestinal epithelium to ER stress might be explained by the rapid proliferation of TA cells or progentiors, the presence of several secretory lineages and polarized enterocytes, demanding a highly funtional secretory pathway. The mechanism by which ADAR1 suppresses IFN signaling is not known, and both RNA editing7, 10, 15 or non-editing35 functions might be involved. Recently, hyper-edited dsRNAs (IU-dsRNAs) was shown to suppress the induction of IFN-stimulated genes and apoptosis induced by poly(IC),9 which can cross talk with ER stress pathway.26, 27, 28 It will be interesting to determine whether IFN signaling engages ER stress to promote ISC death or whether these two pathways act independently upon the loss of ADAR1.
ADAR1 and differentiation
The Wnt/β-catenin pathway is the master regulator of intestinal homeostasis.36 Loss of ADAR1 led to upregulation of Wnt targets c-Myc and Sox9 in the intestinal epithelium and crypts. It is possible that c-Myc is involved in both the proliferation and cell death36 following ADAR1 deletion. Sox9 is required for the differentiation of Paneth cells.37 Expansion of Sox9+ cells might help explain the large increase of Paneth cells and TA cells, whereas goblet, enteroendocrine, and enterocyte lineages were diminished. However, the increase in TA cells might also reflect a compensatory proliferation of progenitors or quiescent stem populations trigged by CBC depletion. The minor but distinct skin pigmentation phenotypes in the dyschromatosis symmetrica hereditaria individuals attributable to the loss of melanocytes10 perhaps suggest another lineage-specific effect of ADAR1.
ADAR1 in intestinal injury and inflammation
Apoptosis of ISCs and impaired differentiation can certainly contribute to intestinal inflammation following ADAR1 deletion. Persistent IFN signaling, production of inflammatory cytokines, ER stress and possibly a feed-forward loop among them are likely to exacerbate intestinal inflammation. Our data suggest that the intestinal epithelial or crypt cells, rather than the immune cells, are the primary source of these signals. However, we could not exclude the possibility that ADAR1 deletion in other cell types contributes to or exacerbates ISC loss. Given the dynamic nature of microbial interactions with host immune and epithelial cells, further exploration of ADAR1 conditional and intestinal inflammation models should shed more light on the roles of the unfolded protein response/ER stress and IFN signaling in inflammatory bowel disease.21, 33
The role of A-to-I editing in the etiology or progression of human diseases is emerging,7, 10 and so is that of epithelial and stem cell apoptosis38 and ER stress in IBD.21 Our studies demonstrated an essential role of ADAR1 in ISC survival and maintenance, and intestinal homeostasis, likely requiring RNA editing much like other systems.5, 39 Future work is needed to understand the precise role and mechanisms of ADAR1 in the GI epithelium, which might lead to new targets for disease intervention or treatment.
Materials and Methods
Mice and treatment
The procedures for all animal experiments were approved by the Institutional Animal Care and Use Committee at the University of Pittsburgh. CreER; ADAR1flox/flox (CreER; ADAR1f/f ) and ADAR1f/f littermates were generated from CreER; ADAR1f/f and ADAR1f/f crosses, and genotyping was performed as described.16 CreER (CAG-CreER is expressed in various developing and adult tissues, Jackson laboratory, Bar Harbor, ME, USA, stock 004453); ADAR1f/f16 mice were generated via two backcrosses with C57BL/6J (B6-F2). The Lgr5-EGFP (Lgr5-EGFP-IRES-creERT2) mice have been described previously.40 Mice were housed in micro-isolator cages in a room illuminated from 0700 to 1900 hours (12:12-h light–dark cycle) and allowed access to water and chow ad libitum.
For tamoxifen treatment, tamoxifen (cat# T5648, Sigma, St. Louis, MO, USA; 120 mg/kg) was injected intraperitoneally (i.p.) into 6- to 8-week-old mice on day 1, 2, 4, and 5. Mice were killed at various times to collect the small intestine, colon, liver, stomach, bone marrow, and spleen for histology analysis. Mice receiving the eIF2α inhibitor, Salubrinal (cat#2347, R&D, Minneapolis, MN, USA),41, 42 were injected with 1 mg/kg i.p. once daily starting 1 day before tamoxifen treatment and continued until 3 days after tamoxifen treatment. All mice were injected i.p. with 100 mg/kg of BrdU (cat# 858811, Sigma) 2 h before killing to label cells in S-phase.
Intestinal crypt culture and isolation of ISCs
The crypt was isolated and cultured in Matrigel with supplementation of growth factors in 24-well plates for 1 day as previously described,43, 44 and subjected to tamoxifen treatment for 1 or 2 days. The intestinal crypts and Lgr5+ cells were isolated from 7- to 8-week-old Lgr5-EGFP-IRES-creERT2 mice as previously described.40, 43 Analyses on the growth of cultured crypt, immunostaing, and RT-PCR were performed. Detailed methods are found in Supplementary Materials.
Western blotting
Total protein was prepared from freshly isolated small intestine as described.38, 45, 46, 47 Extracts were analyzed by NuPage gel (Invitrogen, Carlsbad, CA, USA) electrophoresis.48 Primary antibodies included ADAR1,12 Bip (cat#3177, Cell signaling, Danvers, MA, USA), CHOP (cat# 2895, Cell signaling), and β-actin (cat#5441, Sigma).
Total RNA extraction and real-time reverse transcriptase PCR
Total RNA was prepared for tissues, isolated crypts, cultured crypts, or isolated ISCs, and cDNAs were then generated for real-time PCR analysis.46
Histological analysis, TUNEL, and BrdU staining
Tissue was fixed in 10% formalin for 24 h and stored in 70% ETOH before processing. Sections (5 μm) from paraffin-embedded tissue were subjected to hematoxylin and eosin staining for histological analysis and various staining. Protocols for TUNEL and BrdU staining have been described.31, 46 The apoptotic or BrdU index was scored in 100 crypts or villus/mouse and reported as mean±S.D. Three or more mice were used in each group.
Alkaline phosphatase staining
Paraffin-embedded tissue was sectioned (5-μm thick), rehydrated, and incubated with NBT/BCIP solution (B1911, Sigma) for 30 min. When staining was completed, slides were washed in water and mounted in Clear Mount (cat# 17985-16, Electron Microscopy Sciences, Hatfield, PA, USA). The height of the staining in the crypt-villus units were measured and plotted relative to that in the untreated control mice.
IHC, immunofluorescent, and RNA ISH
Slides were deparaffinized, rehydrated, and treated with 3% hydrogen peroxide. Antigen retrieval was performed by boiling the sections for 10 min in 0.1 M citrate buffer antigen retrieval solution (pH 6.0). Non-specific antibody binding was blocked using 20% goat serum for 30 min. For double staining, TUNEL staining was performed after RNA ISH for Olfm417, 38 or MMP7 IF staining.49 Cells with positive staining were scored in at least 100 crypts or villa and reported as mean±S.D. Three or more mice were used in each group. Detailed information is found in Supplementary Materials.
Transfection and reporter assays
Transfection of HCT116 human colorectal cancer cells with lipofectamine 2000 (Invitrogen) was performed using TCF4 luciferase reporter plasmids, modified Topflash and Fopflash,50 transfection control pCMVβ (Promega, Madison, WI, USA),51 and WT ADAR1, editing defective mutant (H910K/A912E) ADAR1, or empty vector. All reporter experiments were performed in triplicate and repeated on at least three independent occasions. Details of transfection and ADAR1 expression constructs are found in Supplementary Materials.
Statistical analysis
Statistical analysis was carried out using GraphPad Prism V software (San Diego, CA, USA). Data are presented as mean±S.D. Statistical significance was calculated with a Student's t-test. P<0.05 was considered to be significant. The means±1 S.D. are shown in the figures where applicable.
Acknowledgments
We thank Kazuko Nishikura (The Wistar Institute) for ADAR1flox/flox mice, Hans Clevers and Nick Barker (Hubrecht Institute, the Netherlands) for Lgr5-EGFP mice, Olfm4 plasmid, Linheng Li and Fengchao Wang (Stowers Institute for Medical Research) and Scott T Magness (University of North Carolina at Chapel Hill) for advice on crypt culture, Hongtao Liu and Andrea Sebastini for breeding mice and genotyping, members in the Yu lab for helpful discussion and critical reading. This work is supported in part by NIH grants UO1-DK085570, CA129829, American Cancer Society grant RGS-10-124-01-CCE and FAMRI (J Yu), and NIH grants CA106348, CA121105, and American Cancer Society grant RSG-07-156-01-CNE (L Zhang) and R21AI078094 (Q Wang). The Yu laboratory is a member of the Intestinal Stem Cell Consortium, supported by NIDDK and NIAID (UO1). This project used the UPCI shared glassware, animal, and cell and tissue imaging facilities that were supported in part by award P30CA047904.
Author contributions
JY designed and supervised all experiments in this study. WQ, XW, MB, KH, and RS designed and performed experiments. QW and LZ provided key reagents and experimental designs. WQ, XW, MB, KH, RS, QW, and JY analyzed data. WQ, XW, MB, and JY wrote the paper.
Glossary
- ADAR1
adenosine deaminase acting on RNA 1
- TUNEL
terminal deoxynucleotidyl transferase-mediated deoxyuridinetriphosphate nick end labeling
- ER
endoplasmic reticulum
- CHOP
C/EBP homologous protein
- PBS
phosphate-buffered saline
- WT
wild type
- KO
knockout
- IFN
interferon
- ISC
intestinal stem cell
- CBC
columnar cells at the crypt base
- dsRNA
double-stranded RNA
- TA
transient amplification
- BrdU
bromodeoxyuridine
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by Y Shi
Supplementary Material
References
- Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327:542–545. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potten CS, Booth C, Pritchard DM. The intestinal epithelial stem cell: the mucosal governor. Int J Exp Pathol. 1997;78:219–243. doi: 10.1046/j.1365-2613.1997.280362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda N, Jain R, LeBoeuf MR, Wang Q, Lu MM, Epstein JA. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334:1420–1424. doi: 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoville DH, Sato T, He XC, Li L. Current view: intestinal stem cells and signaling. Gastroenterology. 2008;134:849–864. doi: 10.1053/j.gastro.2008.01.079. [DOI] [PubMed] [Google Scholar]
- Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizasa H, Nishikura K. A new function for the RNA-editing enzyme ADAR1. Nat Immunol. 2009;10:16–18. doi: 10.1038/ni0109-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo A, Galardi S. A-to-I RNA editing and cancer: from pathology to basic science. RNA Biol. 2008;5:135–139. doi: 10.4161/rna.5.3.6739. [DOI] [PubMed] [Google Scholar]
- Li JB, Levanon EY, Yoon JK, Aach J, Xie B, Leproust E, et al. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–1213. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- Vitali P, Scadden AD. Double-stranded RNAs containing multiple IU pairs are sufficient to suppress interferon induction and apoptosis. Nat Struct Mol Biol. 2010;17:1043–1050. doi: 10.1038/nsmb.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas S, Kawahara Y, Tamburro KM, Nishikura K. A-to-I RNA editing and human disease. RNA Biol. 2006;3:1–9. doi: 10.4161/rna.3.1.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Miyakoda M, Yang W, Khillan J, Stachura DL, Weiss MJ, et al. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J Biol Chem. 2004;279:4952–4961. doi: 10.1074/jbc.M310162200. [DOI] [PubMed] [Google Scholar]
- Wang Q, Khillan J, Gadue P, Nishikura K. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science. 2000;290:1765–1768. doi: 10.1126/science.290.5497.1765. [DOI] [PubMed] [Google Scholar]
- Hartner JC, Walkley CR, Lu J, Orkin SH. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat Immunol. 2009;10:109–115. doi: 10.1038/ni.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XuFeng R, Boyer MJ, Shen H, Li Y, Yu H, Gao Y, et al. ADAR1 is required for hematopoietic progenitor cell survival via RNA editing. Proc Natl Acad Sci USA. 2009;106:17763–17768. doi: 10.1073/pnas.0903324106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller CK, Li Z, George CX, Samuel CE. Protein kinase PKR and RNA adenosine deaminase ADAR1: new roles for old players as modulators of the interferon response. Curr Opin Immunol. 2011;23:573–582. doi: 10.1016/j.coi.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R, Wang Y, Zhou P, Steinman RA, Wang Q. An essential role of RNA editing enzyme ADAR1 in mouse skin. J Dermatol Sci. 2011;64:70–72. doi: 10.1016/j.jdermsci.2011.06.013. [DOI] [PubMed] [Google Scholar]
- van der Flier LG, Haegebarth A, Stange DE, van de Wetering M, Clevers H. OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology. 2009;137:15–17. doi: 10.1053/j.gastro.2009.05.035. [DOI] [PubMed] [Google Scholar]
- Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fevr T, Robine S, Louvard D, Huelsken J. Wnt/beta-catenin is essential for intestinal homeostasis and maintenance of intestinal stem cells. Mol Cell Biol. 2007;27:7551–7559. doi: 10.1128/MCB.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, et al. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- McGuckin MA, Eri RD, Das I, Lourie R, Florin TH. ER stress and the unfolded protein response in intestinal inflammation. Am J Physiol Gastrointest Liver Physiol. 2010;298:G820–G832. doi: 10.1152/ajpgi.00063.2010. [DOI] [PubMed] [Google Scholar]
- Bertolotti A, Wang X, Novoa I, Jungreis R, Schlessinger K, Cho JH, et al. Increased sensitivity to dextran sodium sulfate colitis in IRE1beta-deficient mice. J Clin Invest. 2001;107:585–593. doi: 10.1172/JCI11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood CK, Cook MC, Eri R, Price GR, Tauro SB, Taupin D, et al. Aberrant mucin assembly in mice causes endoplasmic reticulum stress and spontaneous inflammation resembling ulcerative colitis. PLoS Med. 2008;5:e54. doi: 10.1371/journal.pmed.0050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, Tilg H, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F, Edwards R, Dizon D, Afrasiabi K, Mastroianni JR, Geyfman M, et al. Disruption of Paneth and goblet cell homeostasis and increased endoplasmic reticulum stress in Agr2−/− mice. Dev Biol. 2010;338:270–279. doi: 10.1016/j.ydbio.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Suzuki O, Haruyama T, Akaike T. Interferon-gamma induces reactive oxygen species and endoplasmic reticulum stress at the hepatic apoptosis. J Cell Biochem. 2003;89:244–253. doi: 10.1002/jcb.10501. [DOI] [PubMed] [Google Scholar]
- Lin W, Harding HP, Ron D, Popko B. Endoplasmic reticulum stress modulates the response of myelinating oligodendrocytes to the immune cytokine interferon-gamma. J Cell Biol. 2005;169:603–612. doi: 10.1083/jcb.200502086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki K, Kawamura T, Watanabe Y. Control of ER stress by a chemical chaperone counteracts apoptotic signals in IFN-gamma-treated murine hepatocytes. Apoptosis. 2009;14:309–319. doi: 10.1007/s10495-009-0318-x. [DOI] [PubMed] [Google Scholar]
- Casali A, Batlle E. Intestinal stem cells in mammals and Drosophila. Cell Stem Cell. 2009;4:124–127. doi: 10.1016/j.stem.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Quante M, Wang TC. Inflammation and stem cells in gastrointestinal carcinogenesis. Physiology (Bethesda) 2008;23:350–359. doi: 10.1152/physiol.00031.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibowitz BJ, Qiu W, Liu H, Cheng T, Zhang L, Yu J. Uncoupling p53 functions in radiation-induced intestinal damage via PUMA and p21. Mol Cancer Res. 2011;9:616–625. doi: 10.1158/1541-7786.MCR-11-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potten CS. Radiation, the ideal cytotoxic agent for studying the cell biology of tissues such as the small intestine. Radiat Res. 2004;161:123–136. doi: 10.1667/rr3104. [DOI] [PubMed] [Google Scholar]
- Kaser A, Blumberg RS. Endoplasmic reticulum stress and intestinal inflammation. Mucosal Immunol. 2010;3:11–16. doi: 10.1038/mi.2009.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Beebe K, Nangle LA, Jang J, Longo-Guess CM, Cook SA, et al. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443:50–55. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- Nie Y, Ding L, Kao PN, Braun R, Yang JH. ADAR1 interacts with NF90 through double-stranded RNA and regulates NF90-mediated gene expression independently of RNA editing. Mol Cell Biol. 2005;25:6956–6963. doi: 10.1128/MCB.25.16.6956-6963.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Mori-Akiyama Y, van den Born M, van Es JH, Hamilton SR, Adams HP, Zhang J, et al. SOX9 is required for the differentiation of paneth cells in the intestinal epithelium. Gastroenterology. 2007;133:539–546. doi: 10.1053/j.gastro.2007.05.020. [DOI] [PubMed] [Google Scholar]
- Qiu W, Wu B, Wang X, Buchanan ME, Regueiro MD, Hartman DJ, et al. PUMA-mediated intestinal epithelial apoptosis contributes to ulcerative colitis in humans and mice. J Clin Invest. 2011;121:1722–1732. doi: 10.1172/JCI42917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, Kasher PR, Forte GM, Mannion NM, Greenwood SM, Szynkiewicz M, et al. Mutations in ADAR1 cause Aicardi-Goutieres syndrome associated with a type I interferon signature. Nat Genet. 2012;44:1243–1248. doi: 10.1038/ng.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Saxena S, Cabuy E, Caroni P. A role for motoneuron subtype-selective ER stress in disease manifestations of FALS mice. Nat Neurosci. 2009;12:627–636. doi: 10.1038/nn.2297. [DOI] [PubMed] [Google Scholar]
- Boyce M, Bryant KF, Jousse C, Long K, Harding HP, Scheuner D, et al. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science. 2005;307:935–939. doi: 10.1126/science.1101902. [DOI] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Stelzner M, Helmrath M, Dunn JC, Henning SJ, Houchen CW, Kuo C, et al. A nomenclature for intestinal in vitro cultures. Am J Physiol Gastrointest Liver Physiol. 2012;302:G1359–G1363. doi: 10.1152/ajpgi.00493.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu W, Carson-Walter EB, Kuan SF, Zhang L, Yu J. PUMA suppresses intestinal tumorigenesis in mice. Cancer Res. 2009;69:4999–5006. doi: 10.1158/0008-5472.CAN-09-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu W, Carson-Walter EB, Liu H, Epperly M, Greenberger JS, Zambetti GP, et al. PUMA regulates intestinal progenitor cell radiosensitivity and gastrointestinal syndrome. Cell Stem Cell. 2008;2:576–583. doi: 10.1016/j.stem.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu W, Leibowitz B, Zhang L, Yu J. Growth factors protect intestinal stem cells from radiation-induced apoptosis by suppressing PUMA through the PI3K/AKT/p53 axis. Oncogene. 2009;29:1622–1632. doi: 10.1038/onc.2009.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu W, Leibowitz B, Zhang L, Yu J. Growth factors protect intestinal stem cells from radiation-induced apoptosis by suppressing PUMA through the PI3K/AKT/p53 axis. Oncogene. 2010;29:1622–1632. doi: 10.1038/onc.2009.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu W, Wang X, Leibowitz B, Liu H, Barker N, Okada H, et al. Chemoprevention by nonsteroidal anti-inflammatory drugs eliminates oncogenic intestinal stem cells via SMAC-dependent apoptosis. Proc Natl Acad Sci USA. 2010;107:20027–20032. doi: 10.1073/pnas.1010430107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa LT, He TC, Yu J, Sparks AB, Morin PJ, Polyak K, et al. CDX2 is mutated in a colorectal cancer with normal APC/beta-catenin signaling. Oncogene. 1999;18:5010–5014. doi: 10.1038/sj.onc.1202872. [DOI] [PubMed] [Google Scholar]
- Yu J, Zhang L, Hwang PM, Kinzler KW, Vogelstein B. PUMA induces the rapid apoptosis of colorectal cancer cells. Mol Cell. 2001;7:673–682. doi: 10.1016/s1097-2765(01)00213-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.