Abstract
Runt-related transcription factor 2 (RUNX2) is the best known as an essential protein for osteoblast differentiation. In this study, we have found for the first time that RUNX2 acts as a negative regulator for p53 in response to DNA damage. On DNA damage mediated by adriamycin (ADR) exposure, p53 as well as RUNX2 was induced at protein and mRNA level in human osteosarcoma-derived U2OS cells in association with a significant upregulation of various p53-target genes. Indirect immunostaining and co-immunoprecipitation experiments demonstrated that RUNX2 colocalizes with p53 in cell nucleus and forms a complex with p53 following ADR treatment. Chromatin immunoprecipitation assays revealed that RUNX2/p53 complex is efficiently recruited onto p53-target promoters in response to ADR, suggesting that RUNX2 might be involved in the regulation of transcriptional activation mediated by p53. Indeed, forced expression of RUNX2 resulted in a remarkable downregulation of p53-target genes. Consistent with these observations, knockdown of RUNX2 enhanced ADR-mediated apoptosis and also elevated p53-target gene expression in response to ADR. On the other hand, depletion of RUNX2 in p53-deficient human lung carcinoma-derived H1299 cells had an undetectable effect on p53-target gene expression regardless of ADR treatment, indicating that RUNX2-mediated downregulation of p53-target genes is dependent on p53. Furthermore, RUNX2/p53 complex included histone deacetylase 6 (HDAC6) and HDAC6 was also recruited onto p53-target promoters following ADR exposure. Of note, HDAC6-specific chemical inhibitor tubacin treatment enhanced ADR-mediated upregulation of p53-target gene expression, indicating that deacetylase activity of HDAC6 is required for RUNX2-mediated downregulation of p53-target gene. Taken together, our present findings strongly suggest that RUNX2 inhibits DNA damage-induced transcriptional as well as pro-apoptotic activity of p53 through the functional collaboration with HDAC6 and therefore might be an attractive therapeutic target for cancer treatment.
Keywords: apoptosis, DNA damage, HDAC6, p53, RUNX2
The representative tumor-suppressor p53 is a sequence-specific transcription factor, which regulates the expression of a variety of its direct target genes implicated in the induction of cell cycle arrest and/or apoptosis.1, 2 Extensive mutation searches revealed that over 50% of human primary tumors carry p53 mutations and the majority of them are detectable within the genomic region encoding its DNA-binding domain.3, 4 p53 mutants exhibit a prolonged half-life, lack the sequence-specific transactivation ability and then acquire the oncogenic function.1, 2 In addition, p53 mutants show a dominant-negative behavior toward wild-type p53 and tumors with p53 mutations sometimes display the chemo-resistant phenotypes.5, 6, 7 Indeed, p53-deficient mice developed spontaneous tumors.8 Based on these observations, it is likely that the sequence-specific transactivation activity of p53 is tightly linked to its tumor-suppressive function.9
Under normal conditions, p53 is maintained at a quite low level. MDM2 that acts as an E3 ubiquitin protein ligase for p53, binds to NH2-terminal transactivation domain of p53, catalyzes its ubiquitination at COOH-terminal lysine residues and thereby promoting its ubiquitin-dependent proteasomal degradation.10, 11 As MDM2 is a direct p53-target gene product, MDM2 and p53 create a negative-feedback loop in which p53 transactivates MDM2, which in turn downregulates p53.12 In response to cellular stresses such as DNA damage, p53 is quickly induced to accumulate in cell nucleus.1 DNA damage-mediated sequential post-translational modifications including phosphorylation (Ser-15, Ser-20 and Ser-46) and acetylation (Lys-373/382) contribute to the induction of p53.13, 14 For example, NH2-terminal phosphorylation of p53 promotes to dissociate MDM2 from p53 (ref.15) and COOH-terminal acetylation of p53 attenuates the ubiquitination mediated by MDM2.16 Thus, these modifications repress the ubiquitin-dependent proteasomal degradation of p53 and thereby p53 becomes stable.
On repairable DNA damage, p53 transactivates its cell cycle-related target genes including p21WAF1 and 14-3-3σ to arrest cell cycle progression at G1/S and/or G2/M boundary to save time to repair damaged DNA and then cells with repaired DNA re-enter into the normal cell cycle. When cells receive severe DNA damage, p53 instead promotes the irreversible apoptosis through transactivating its pro-apoptotic target genes such as BAX, NOXA and PUMA and eliminates cells with seriously damaged DNA.1, 13, 14 Therefore, the proper DNA damage response, which monitors and ensures the genomic integrity, has been considered to be a critical barrier to tumorigenesis and p53 stands at the cross-road between cell survival and death following DNA damage.
In addition to DNA damage-mediated post-translational modifications, p53 is also regulated by protein–protein interaction. It has been described that ASPP1/ASPP2 interacts with central DNA-binding domain of p53 and enhances its pro-apoptotic activity.17 Roe et al.18 demonstrated that VHL binds to p53 and stimulates its pro-apoptotic activity through suppressing MDM2-mediated p53 ubiquitination. Lopez-Mateo et al.19 found that CREBZF, which is a member of ATF/CREB family, associates with p53 and enhances its transactivation ability. Yang et al.20 described that polycomb group protein PHF1 binds to COOH-terminal region of p53 and stimulates its pro-apoptotic activity. In contrast, we found that polo-like kinase 1 interacts with central DNA-binding domain of p53 and inhibits its transcriptional as well as pro-apoptotic activity.21 Recently, Yuan et al.22 reported that ATDC/TRIM29 represses p53-dependent transcriptional activation through its sequestration outside of the nucleus. In addition, Li et al.23 demonstrated that SIRT1 binds to p53 and inhibits its pro-apoptotic activity through deacetylation of p53 at Lys-382.
The mammalian RUNX family of nuclear hetero-dimeric transcription factors includes three members, Runt-related transcription factor 1 (RUNX1), RUNX2 and RUNX3, which have pivotal roles in the regulation of normal development and tumorigenesis.24 The RUNX family is highly conserved in their runt homology domain, which is utilized for DNA-binding and hetero-dimerization with the common cofactor CBFβ.25 In addition to runt domain, RUNX family also possesses the other subdomains including a large transactivation domain in COOH-terminal part and an inhibitory domain at COOH-terminal end of the transactivation domain. RUNX1 is essential for the establishment of the hematopoietic stem cells and is a frequent target of chromosomal gene translocations in hematopoietic malignancies.26 Consistent with these observations, RUNX1 stimulates transcription of a variety of myeloid and lymphoid-related genes.27, 28 Recently, we have found that RUNX1 acts as a scaffold protein to elevate DNA damage-mediated acetylation of p53 and thereby enhancing its pro-apoptotic activity.29
RUNX3 acts as a tumor suppressor for human gastric cancer. RUNX3-deficient mice display hyperplasia in gastric mucosa caused by reduced apoptosis and the stimulated growth of the gastric epithelial cells. Over 60% of human primary gastric cancer specimens express significantly lower levels of RUNX3, which might be due to a combination of hemizygous deletion of RUNX3 gene and hypermethylation of its promoter region.30 In addition to human gastric cancer, Jiang et al.31 found that RUNX3 is downregulated in human breast cancer tissues relative to their surrounding normal tissues and this reduction is correlated to the hypermethylation of its promoter region. Nicole Tsang et al.32 described that prolyl isomerase Pin1 is often overexpressed in human breast cancer tissues and its expression level is inversely correlated with RUNX3. According to their results, Pin1 bound to RUNX3 and suppressed its transcriptional activity through the induction of ubiquitin-dependent proteasomal degradation of RUNX3. He et al.33 also demonstrated that RUNX3 is markedly reduced in human clear cell renal cell carcinoma (CCRCC) tissues as compared with their matched adjacent noncancerous tissues and forced expression of RUNX3 represses the tumorigenicity of CCRCC cells. These observations strongly suggest that the functional inactivation of RUNX3 is frequently detectable in a variety of human tumors. Recently, we have found that RUNX3 interacts with p53 in response to DNA damage and enhances its ATM-dependent phosphorylation at Ser-15.34
RUNX2 acts as a master regulator of both osteoblast and terminal chondrocyte differentiation and is essential for bone formation and mineralization in vivo.35, 36 In addition to RUNX2, bone morphogenetic protein 2 is one of the most important cytokines in the regulation of osteoblast differentiation37 and induces RUNX2 through activating Smad1/5/8 signaling.38 Like RUNX1 and RUNX3, RUNX2 is a sequence-specific transcription factor. In a good agreement with these observations, RUNX2 transactivates a number of osteogenic markers such as type 1 collagen, osteopontin and osteocalcin.37, 39 On the other hand, oncogenic nature of RUNX2 has been shown in several human tumors.40, 41, 42, 43, 44, 45 For example, it has been described that gene amplification of RUNX2 is frequently observed in human osteosarcoma and RUNX2 regulates the expression of genes implicated in cell motility and adhesion in human osteosarcoma.45, 46 Browne et al.47 described that RUNX2-mediated induction of anti-apoptotic Bcl-2 contributes to the acquisition of apoptosis-resistant phenotype in human prostate cancers. Although these studies have suggested the oncogenic property of RUNX2, the underlying molecular mechanism(s) is poorly understood. Intriguingly, Blyth et al.48 described that RUNX2 provides a strong anti-apoptotic signal even in the presence of functional p53, indicating that RUNX2 neutralizes p53. Westendorf et al.49 demonstrated that RUNX2 represses p21WAF1 expression. Thus, it is likely that these exists a functional interaction between RUNX2 and p53.
In this study, we have found that RUNX2 interacts with p53 in response to DNA damage and thereby inhibiting p53 in collaboration with HDAC6.
Results
RUNX2 is induced in response to adriamycin (ADR)-mediated DNA damage
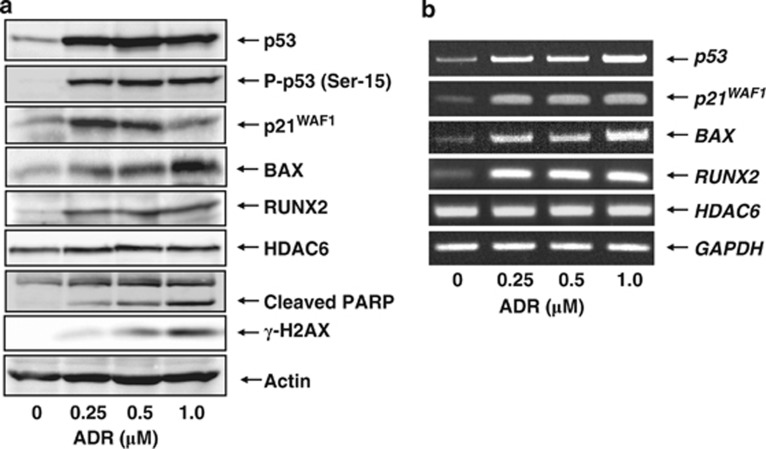
To examine the expression pattern of RUNX2 following DNA damage, p53-proficient human osteosarcoma-derived U2OS cells were exposed to ADR, which leads to DNA double-strand breaks. As described,34 U2OS cells underwent apoptosis in response to ADR as examined by MTT and Trypan blue exclusion assays (Supplementary Figure S1). Under these experimental conditions, cell lysates and total RNA were prepared and subjected to immunoblotting and RT-PCR, respectively. As shown in Figure 1a, ADR treatment significantly stimulated p53 accumulation and phosphorylation at Ser-15 in association with a remarkable upregulation of the numerous p53-target gene products. The expression level of γ-H2AX and cleavage of poly(ADP-ribose)polymerase (PARP) were also examined as a DNA damage and an apoptosis marker, respectively. It was worth noting that, like p53, RUNX2 is also induced in response to ADR. As described,49 histone deacetylase 6 (HDAC6) was involved in the regulation of RUNX2 activity. Under our experimental conditions, HDAC6 remained unchanged regardless of ADR treatment. RT-PCR experiments demonstrated that ADR-mediated induction of RUNX2 is regulated at mRNA level (Figure 1b). In addition, time course experiments revealed that RUNX2 is induced following ADR exposure in a time-dependent manner (Supplementary Figure S2). Similar results were also obtained in p53-proficient human colon carcinoma-derived HCT116 cells (Supplementary Figure S3), indicating that ADR-dependent induction of RUNX2 is not restricted to U2OS cells.
Figure 1.
ADR-mediated induction of RUNX2. (a and b) Expression level of RUNX2 in response to ADR. U2OS cells were exposed to the indicated concentrations of ADR. Twenty-four hours after ADR treatment, cell lysates and total RNA were prepared and processed for immunoblotting (a) and RT-PCR (b), respectively. Actin was used as a loading control and GAPDH served as an internal control
Interaction between p53 and RUNX2 in response to ADR
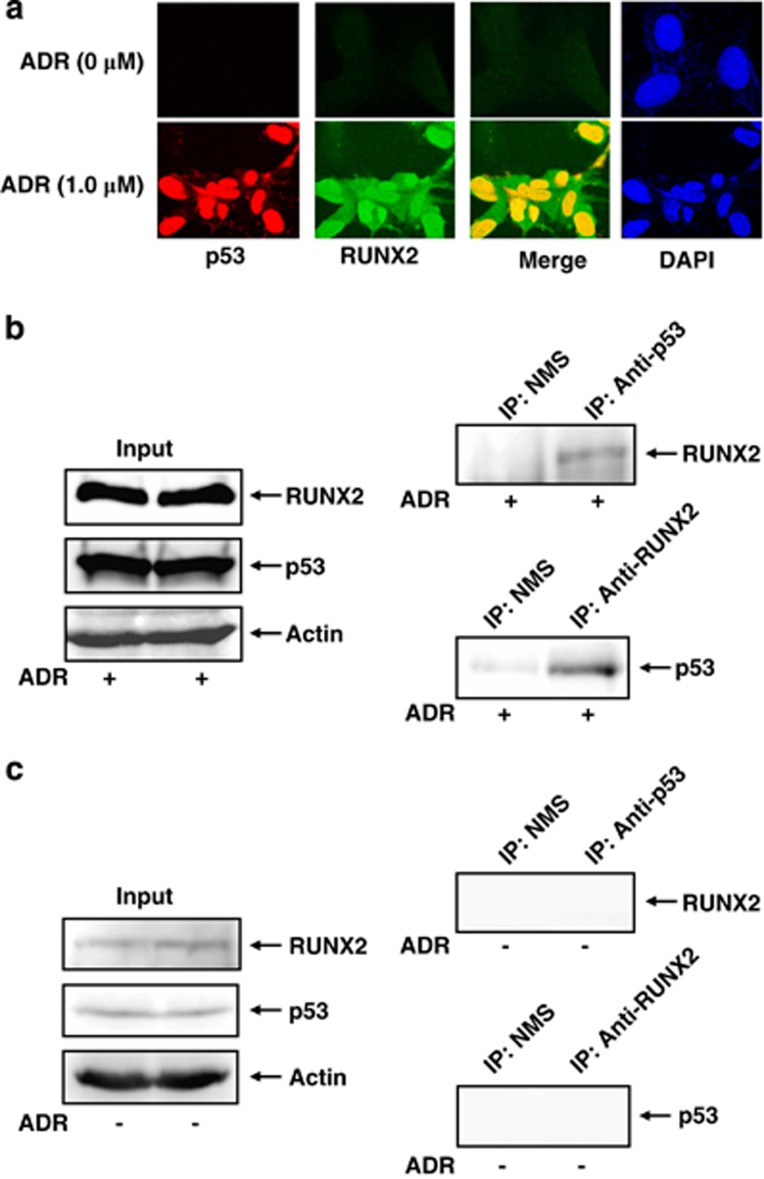
Given that RUNX1 as well as RUNX3 interacts with p53 and modulates its activity,29,34 we sought to investigate whether RUNX2 could also bind to p53. To this end, U2OS cells were treated with ADR or left untreated. Twenty-four hours after ADR treatment, cells were simultaneously incubated with anti-RUNX2 and anti-p53 antibodies. As shown in Figure 2a, RUNX2 and p53 were induced to accumulate in cell nucleus and their nuclear colocalization was detectable in the presence of ADR, indicating that RUNX2 might interact with p53 in cell nucleus following ADR treatment. ADR-mediated nuclear accumulation of RUNX2 was also shown in immunoblotting using nuclear and cytoplasmic fractions (Supplementary Figure S4). To further confirm the presence of their interaction, we performed co-immunoprecipitation experiments. U2OS cells were exposed to ADR. Twenty-four hours after ADR treatment, cell lysates were prepared and immunoprecipitated with normal mouse serum (NMS) or with monoclonal anti-p53 antibody followed by immunoblotting with anti-RUNX2 antibody. As seen in Figure 2b, the anti-p53 immunoprecipitates contained the endogenous RUNX2. Consistent with these results, the reciprocal experiments using NMS or monoclonal anti-RUNX2 antibody demonstrated that the endogenous p53 is co-immunoprecipitated with RUNX2. However, we could not detect RUNX2/p53 complex by co-immunoprecipitation experiments using cell lysates prepared from untreated cells (Figure 2c).
Figure 2.
RUNX2 associates with p53 in response to ADR. (a) Indirect immunostaining experiments. U2OS cells were treated with 1.0 μM of ADR or left untreated. Twenty-four hours after ADR treatment, cells were simultaneously incubated with monoclonal anti-p53 and polyclonal anti-RUNX2 antibodies followed by the incubation with rhodamine-conjugated anti-mouse IgG (red) and fluorescein isothiocyanate (FITC)-conjugated anti-rabbit IgG (green). Cell nuclei were stained with DAPI (blue). Merged images (yellow) indicate the colocalization of RUNX2 with p53 in cell nucleus. (b and c) Co-immunoprecipitation experiments. U2OS cells were exposed to 0.5 μM of ADR (b) or left untreated (c). Twenty-four hours after ADR treatment, cell lysates were prepared and immunoprecipitated with NMS or with monoclonal anti-p53 antibody. The immunoprecipitates were analyzed by immnublotting with monoclonal anti-RUNX2 antibody. The reciprocal experiments and 1/20 of inputs were also shown
p53/RUNX2 complex is efficiently recruited onto p53-target promoters following ADR treatment
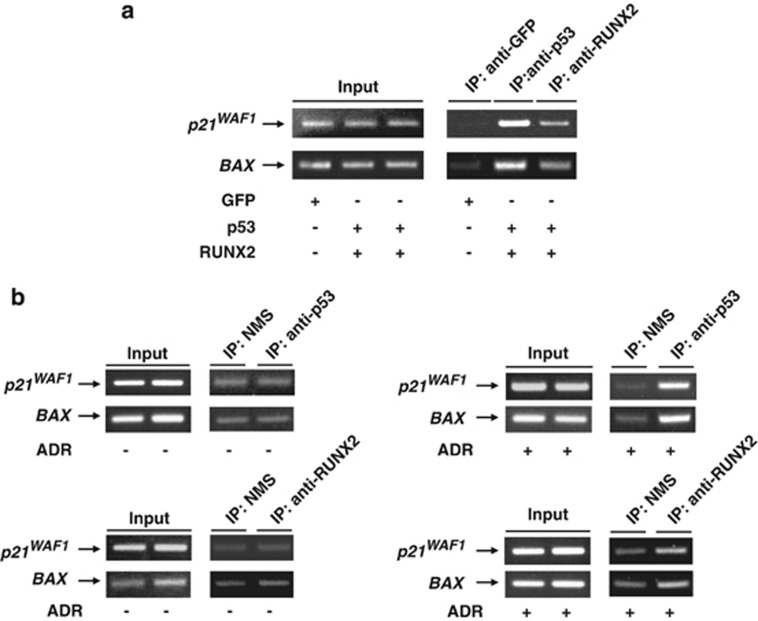
Considering that RUNX2 interacts with p53, we asked whether RUNX2 and p53 could be recruited onto p53-target promoters. For this purpose, we used chromatin immunoprecipitation (ChIP) assay. U2OS cells were transfected with the expression plasmids for green fluorescence protein (GFP) or with p53 plus RUNX2. Forty-eight hours after transfection, cells were fixed in formaldehyde and soluble chromatin was immunoprecipitated with anti-GFP, anti-p53 or with anti-RUNX2 antibody. Genomic DNA fragments were purified from the immunoprecipitates and then subjected to PCR using p21WAF1 or BAX promoter-specific primer sets flanking their p53-responsive elements. As shown in Figure 3a, p53 as well as RUNX2 was detected on p21WAF1 and BAX promoters, whereas GFP was not.
Figure 3.
p53 as well as RUNX2 is recruited onto p53-target promoters. (a) Co-occupancy of p53 and RUNX2 on p53-target promoters. U2OS cells were transfected with the expression plasmids for GFP or with p53 plus RUNX2. Forty-eight hours after transfection, cells were fixed in formaldehyde and incubated with SDS lysis buffer. The extracted chromatin fragments were immunoprecipitated with monoclonal anti-GFP, monoclonal anti-p53 or with monoclonal anti-RUNX2 antibody and the precipitated genomic DNA was analyzed by PCR using primer sets for p21WAF1 and BAX promoter regions containing their p53-responsive elements. PCR amplification was also performed before immunoprecipitation for the input control. (b) ADR-mediated recruitment of p53 and RUNX2 onto p53-target promoters. U2OS cells were treated with 0.5 μM of ADR (right panels) or left untreated (left panels). Twenty-four hours after ADR treatment, cells were fixed in formaldehyde and incubated with SDS lysis buffer. Soluble chromatin was subjected to immunoprecipitation with NMS, monoclonal anti-p53 or with monoclonal anti-RUNX2 antibody. Immunoprecipitated genomic DNA was PCR amplified using primer sets that span p53-responsive elements of p21WAF1 and BAX promoters. Input corresponded to 2.5% of the soluble chromatin that was subjected to immunoprecipitation
Next, we assessed whether the recruitment of p53/RUNX2 complex onto p53-target promoters could be induced in response to ADR. U2OS cells were treated with or without ADR. Twenty-four hours after ADR treatment, cells were fixed and protein-DNA complex was immunoprecipitated with NMS, anti-p53 or with anti-RUNX2 antibody. The immunoprecipitates were then analyzed for p21WAF1 and BAX promoters by PCR. As seen in Figure 3b, binding of RUNX2 and p53 to p21WAF1 and BAX promoters was detectable in the presence of ADR, whereas we could not detect RUNX2 and p53 on these promoters without ADR, suggesting that the efficient recruitment of RUNX2 and p53 onto p53-target promoters is regulated in an ADR-dependent manner.
Our results obtained from the above-mentioned ChIP assays raised a question whether RUNX2 could directly bind to p53-target gene promoters. To address this issue, p53-deficient H1299 cells were transfected with the expression plasmids for p53, RUNX2 or p53 plus RUNX2. Forty-eight hours after transfection, soluble chromatin was prepared, immunoprecipitated with anti-p53 or with anti-RUNX2 antibody and the immunoprecipitates were analyzed by PCR. As shown in Supplementary Figure S5, RUNX2 alone did not bind to p21WAF1 and BAX promoters, indicating that RUNX2 is recruited onto p53-target promoters in collaboration with p53.
RUNX2 represses the transcriptional activity of p53
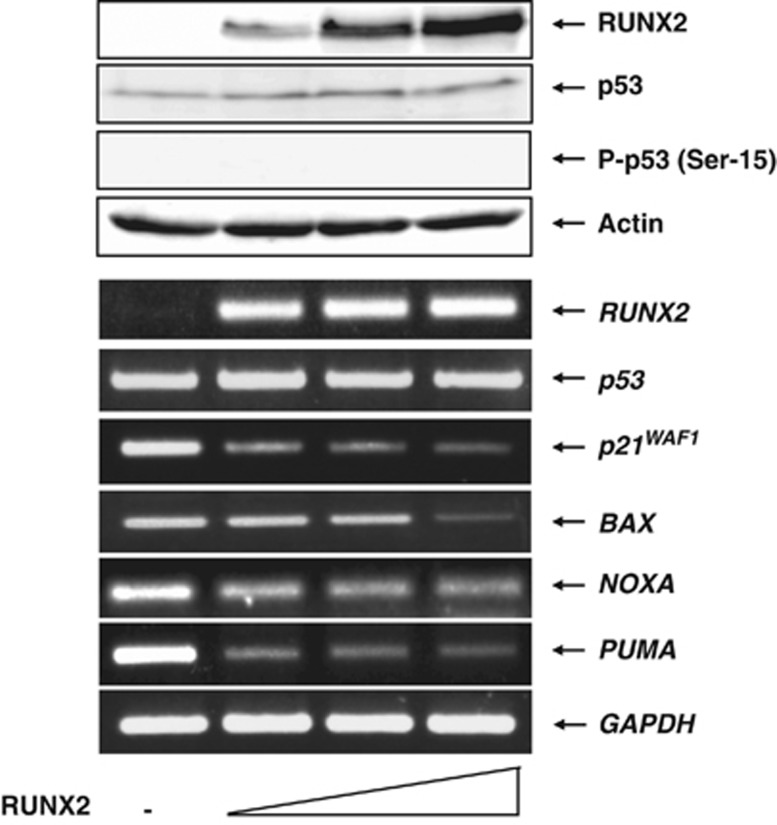
As p53/RUNX2 complex binds to p53-target promoters, we asked whether RUNX2 could affect p53-target gene expression. U2OS cells were transfected with the empty plasmid or with the increasing amounts of the expression plasmid for RUNX2. Forty-eight hours after transfection, cell lysates and total RNA were extracted and analyzed by immunoblotting and RT-PCR, respectively. As shown in upper panels of Figure 4 a dose-dependent expression of RUNX2 was detectable and RUNX2 had an undetectable effect on p53. RT-PCR analysis demonstrated that forced expression of RUNX2 significantly suppresses the expression of p53-target genes (lower panels of Figure 4). Considering that the recruitment of RUNX2 onto p53-target promoters is dependent on p53, these observations strongly suggest that RUNX2 inhibits the transcriptional activity of p53. To gain further evidence showing that RUNX2-mediated downregulation of p53-target gene expression could be indeed p53 dependent, we utilized H1299 cells. H1299 cells were transfected with the empty plasmid or with the expression plasmid for RUNX2. Forty-eight hours after transfection, total RNA was extracted and analyzed by RT-PCR. As shown in Supplementary Figure S6, forced expression of RUNX2 in H1299 cells had a negligible effect on p53-target gene expression. These results firmly supported that RUNX2 inhibits the transcriptional activity of p53.
Figure 4.
Forced expression of RUNX2 reduces the expression levels of various p53-target genes. U2OS cells were transfected with the empty plasmid (2.0 μg) or with the increasing amounts of the expression plasmid encoding RUNX2 (0.5, 1.0 or 2.0 μg). Total amounts of the expression plasmid were kept constant (2.0 μg) with pcDNA3. Forty-eight hours after transfection, cell lysates and total RNA were isolated and subjected to immunoblotting with the indicated antibodies (upper panels) and RT-PCR with the indicated primer sets (lower panels), respectively. Immunoblotting with anti-actin antibody was used as a loading control. Expression levels of an internal control GAPDH were also monitored by RT-PCR
Knockdown of RUNX2 enhances ADR-mediated apoptosis
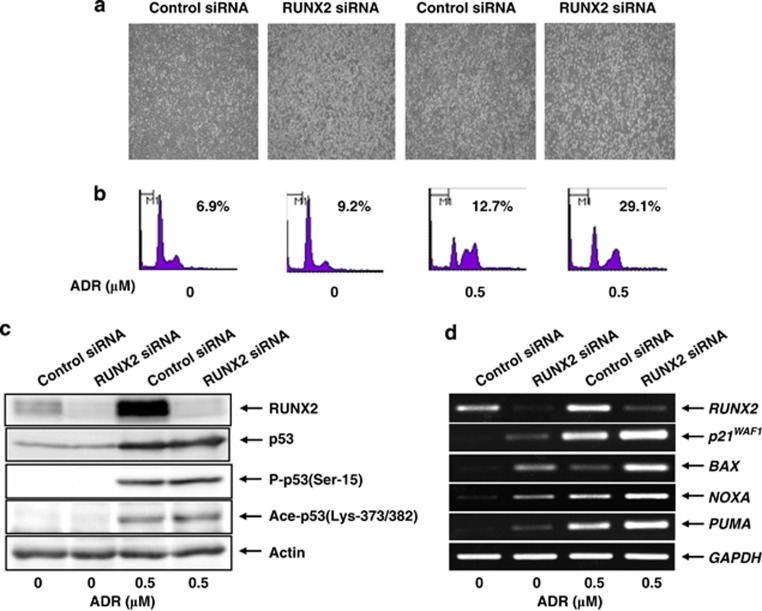
To further confirm the notion that RUNX2 has an inhibitory role on p53, we performed small interference RNA (siRNA)-mediated knockdown of RUNX2. U2OS cells were transfected with control siRNA or with siRNA against RUNX2. Twenty-four hours after transfection, cells were treated with ADR or left untreated. As shown in Figure 5a, number of attached cells was apparently decreased in RUNX2 knockdown cells exposed to ADR as compared with that in ADR-treated control cells. In support of these results, fluorescence-activated cell sorter (FACS) analysis demonstrated that knockdown of RUNX2 results in a remarkable increase in number of cells with sub-G1 DNA content following ADR exposure (Figure 5b), indicating that RUNX2 attenuates p53-dependent apoptosis in response to ADR.
Figure 5.
Knockdown of RUNX2 enhances ADR-mediated apoptosis. U2OS cells were transfected with control siRNA or with siRNA against RUNX2. Twenty-four hours after transfection, cells were treated with 0.5 μM of ADR or left untreated. Twelve hours after ADR exposure, images were taken (a) and floating and attached cells were collected, fixed in 70% ethanol, stained with propidium iodide (PI) and subjected to FACS analysis (b). Cells with sub-G1 DNA content were considered dead. Under the same experimental conditions, cell lysates and total RNA were prepared and analyzed by immunoblotting (c) and RT-PCR (d), respectively. Actin and GAPDH were a loading control for immunoblotting and an internal control for RT-PCR, respectively
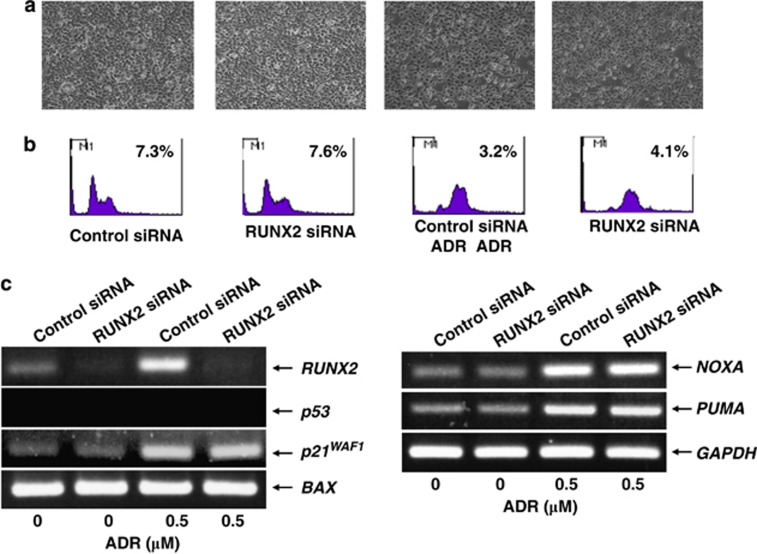
Under these experimental conditions, cell lysates and total RNA were isolated and subjected to immunoblotting and RT-PCR, respectively. As shown in Figure 5c, ADR treatment led to a significant accumulation of p53 in association with the obvious induction of p53 phosphorylation and acetylation at Ser-15 and at Lys-373/382, respectively. Although knockdown of RUNX2 had no substantial effect on ADR-mediated modifications of p53, RT-PCR analysis revealed that ADR-mediated induction of p53-target genes is further enhanced in RUNX2 knockdown cells (Figure 5d). In contrast, knockdown of RUNX2 did not promote apoptosis in H1299 cells following ADR exposure (Figures 6a and b) and also did not elevate ADR-mediated induction of p53-target gene expression (Figure 6c). Taken together, these observations indicate that RUNX2 prohibits p53-dependent apoptosis in response to ADR through the downregulation of p53-target genes.
Figure 6.
Knockdown of RUNX2 has a negligible effect on H1299 cells in response to ADR. (a and b) RUNX2 has an undetectable effect on H1299 cells following ADR exposure. H1299 cells were transfected with control siRNA or with siRNA targeting RUNX2. Twenty-four hours after transfection, cells were treated with 0.5 μM of ADR or left untreated. Twenty-four hours after ADR treatment, images were taken (a). Floating and attached cells were then collected and subjected to FACS analysis (b). (c) RUNX2 has a negligible effect on p53-target gene expression in response to ADR. H1299 cells were treated as in (a). Twenty-four hours after ADR treatment, total RNA was prepared and processed for RT-PCR. GAPDH was used as an internal control
Functional interaction among RUNX2, p53 and HDAC6
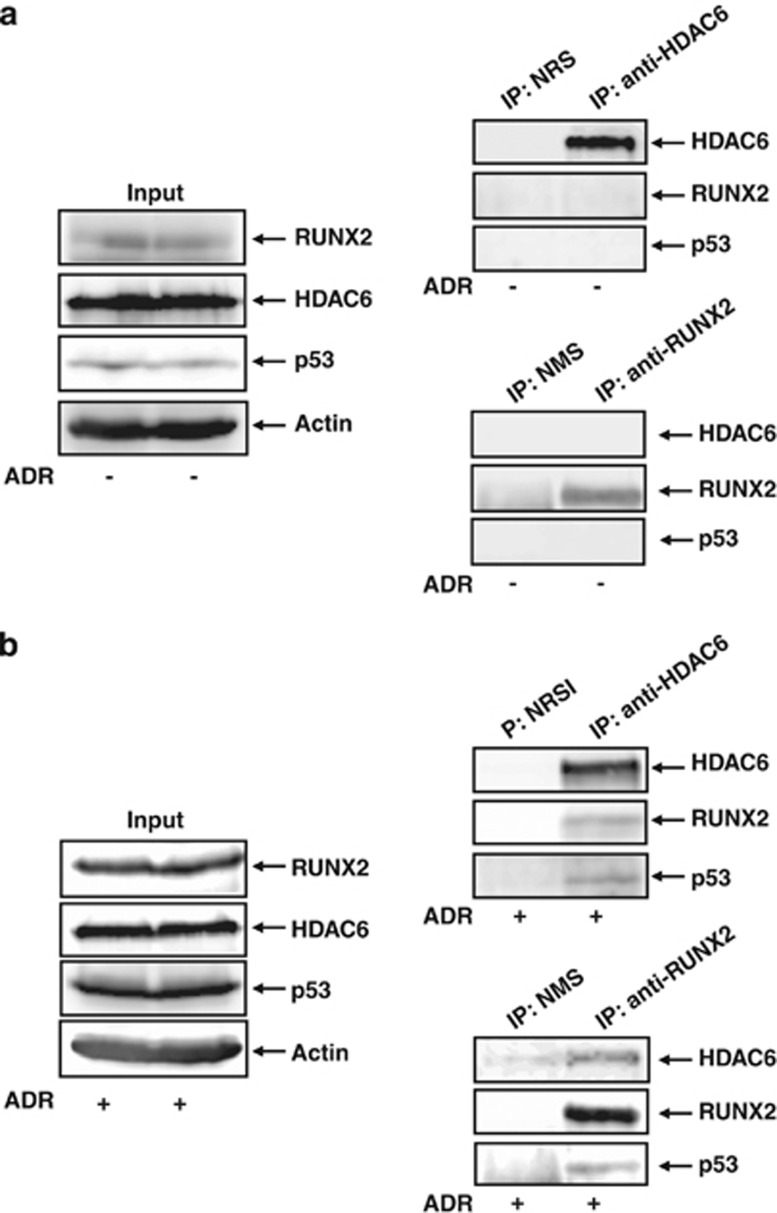
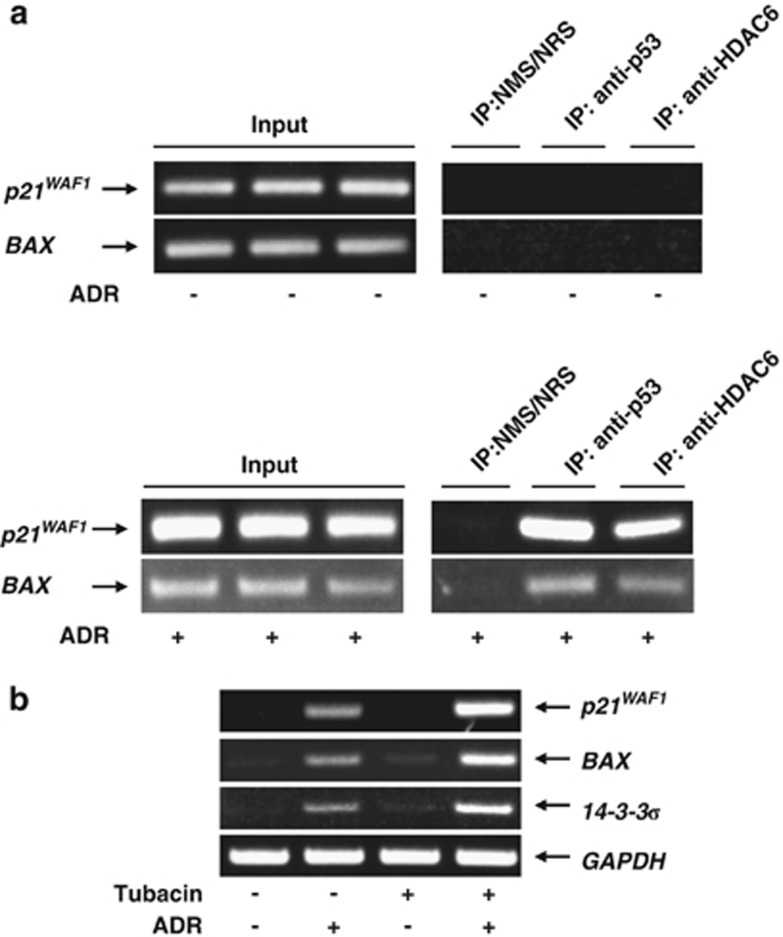
As described,49 HDAC6 bound to RUNX2 and was required for the maximal repression of p21WAF1 promoter caused by RUNX2. These findings led us to investigate whether HDAC6 could be included in p53/RUNX2 complex. The initial co-immunoprecipitation experiments to detect RUNX2/p53/HDAC6 complex using cell lysates prepared from untreated U2OS cells were failed (Figure 7a). We then prepared cell lysates from ADR-treated U2OS cells and performed co-immunoprecipitation experiments using normal rabbit serum (NRS) or polyclonal anti-HDAC6 antibody followed by immunoblotting with the indicated antibodies. As shown in Figure 7b, the anti-HDAC6 immunoprecipitates contained the endogenous RUNX2 and p53. The reciprocal experiments using NMS or monoclonal anti-RUNX2 antibody demonstrated that the endogenous HDAC6 and p53 are co-immunoprecipitated with RUNX2. Notably, ChIP assays using ADR-treated U2OS cells revealed that, like RUNX2, HDAC6 is detectable on p21WAF1 and BAX promoter but not in untreated cells (Figure 8a), indicating that HDAC6 is involved in RUNX2-mediated inhibition of p53 transcriptional response. To address this issue, U2OS cells were treated with or without HDAC6-specific chemical inhibitor tubacin50 followed by ADR exposure. As shown in Figure 8b, tubacin treatment resulted in a remarkable enhancement of ADR-induced induction of p53-target genes. Collectively, these findings suggested that HDAC6 has a critical role in the regulation of RUNX2-mediated inhibition of p53.
Figure 7.
Interaction among p53, RUNX2 and HDAC6. Co-immunoprecipitation. U2OS cells were left untreated (a) or treated with 0.5 μM of ADR (b). Twenty-four hours after ADR treatment, cell lysates were prepared and immunoprecipitated with NRS or with polyclonal anti-HDAC6 antibody followed by immunoblotting with the indicated antibodies. Reciprocal immunoprecipitation experiments using NMS or monoclonal anti-RUNX2 and 1/20 of input were also shown
Figure 8.
HDAC6 contributes to RUNX2-mediated repression of p53. (a) HDAC6 binds to p53-target gene promoters in the presence of ADR. U2OS cells were exposed to 0.5 μM of ADR (lower panels) or left untreated (upper panels). Twenty-four hours after ADR treatment, cells were fixed in formaldehyde and incubated with SDS lysis buffer. Cross-linked chromatin was immunoprecipitated with the indicated antibodies and collected with protein A-agarose beads. After reversal of the cross-link and digestion with proteinase K, genomic DNA was purified by ethanol precipitation followed by PCR amplification using primer sets that span p53-responsive elements of p21WAF1 and BAX promoters. (b) Tubacin enhances ADR-mediated upregulation of p53-target gene expression. U2OS cells were exposed to HDAC6 inhibitor tubacin (at a final concentration of 5 μM) or left untreated. Twenty-four hours after tubacin treatment, cells were exposed to 0.5 μM of ADR or left untreated. Twenty-four hours after ADR exposure, total RNA was prepared and subjected to RT-PCR. GAPDH was used as an internal control
Discussion
Here, we have found that RUNX2 interacts with p53 and represses p53-dependent apoptosis in response to ADR. During ADR-mediated apoptosis, RUNX2 was efficiently recruited onto p53-target promoters in a p53-dependent manner and prohibited p53 transcriptional response in collaboration with HDAC6. Thus, RUNX2 might be an attractive molecular target for cancer therapy.
Accumulating evidence demonstrated that an aberrant regulation of RUNX2 expression is frequently detectable in a variety of human tumors including osseous (osteosarcoma) and nonosseous tumors (prostate, thyroid, breast, colon and pancreatic cancers),40, 41, 42, 43, 44, 45 suggesting that RUNX2 has an oncogenic function in tumor etiology. In accordance with this notion, Sase et al.44 described that RUNX2 expression level is strongly associated with a poor clinical outcome of colon carcinoma patients and Sadikovic et al.51 found that RUNX2 overexpression is closely correlated with poor response to chemotherapy in human osteosarcoma. However, the precise molecular mechanism(s) how RUNX2 could contribute to tumorigenesis remained elusive. Given that RUNX2 is a sequence-specific transcription factor, van der Deen et al.45 sought to identify physiological RUNX2-target genes to gain insight into understanding the molecular pathway(s), which is perturbed in human osteosarcoma. Based on their results, RUNX2 induced various genes related to cell adhesion and motility such as focal adhesion kinase PTK2/FAK and TLN1, indicating that RUNX2 is involved in the regulation of cellular movement. Consistent with these results, it has been shown that RUNX2 expression is increased in highly metastatic human breast and prostate tumors.52 Pratap et al.53 described that RUNX2 regulates metalloproteinase and VEGF expression, which contribute to the metastatic properties of malignant tumors. These observations might provide a clue to understand the molecular basis of oncogenic property of RUNX2.
Recently, we have found that RUNX1 and RUNX3 enhance DNA damage-mediated pro-apoptotic activity of p53 through induction of p300-dependent acetylation and ATM-dependent phosphorylation of p53, respectively.29, 34 These observations prompted us to investigate whether RUNX2 could also be involved in p53-dependent DNA damage response. Here, we showed that forced expression of RUNX2 inhibits p53-dependent transcriptional activation of its target genes and knockdown of RUNX2 enhances ADR-mediated pro-apoptotic activity of p53 in association with a further stimulation of p53-target gene expression following ADR exposure, suggesting that, in contrast to RUNX1 and RUNX3, RUNX2 has an inhibitory role in the regulation of p53. As pro-apoptotic activity of p53 is tightly linked to its sequence-specific transcriptional activity, RUNX2 might inhibit p53 transcriptional machinery. Indeed, our ChIP assays revealed that RUNX2/p53 complex is efficiently recruited onto p53-target promoters in response to ADR. Lutterbach et al.54 found that p21WAF1 promoter contains three putative RUNX-binding sites and the proximal element is required for the downregulation of p21WAF1 in NIH3T3 cells, indicating that RUNX2 directly binds to this element and thereby reducing p21WAF1 expression. Under our experimental conditions, however, RUNX2 had an undetectable effect on p53-target genes including p21WAF1 in p53-deficient H1299 cells regardless of ADR treatment, which might rule out a possibility that RUNX2 alone is involved in the repression of p21WAF1 gene expression. In addition, ADR-mediated recruitment of RUNX2 onto p21WAF1 promoter was dependent on p53. Considering that NIH3T3 cells carry wild-type p53, it is likely that RUNX2-mediated inhibitory effect on p21WAF1 gene expression is dependent on p53.
We also demonstrated that HDAC6 participates in RUNX2-mediated downregulation of p53-target genes in response to ADR. As described,49, 55 HDAC6 and HDAC3 interacted with RUNX2. It has been shown that HDACs are the enzymatic components of multi-protein complexes, which are recruited onto specific DNA elements together with transcription factors and then remove acetyl residues from nucleosomal histones as well as other substrates leading to chromatin condensation and gene repression.56, 57 Schroeder et al.54 reported that HDAC3/RUNX2 complex associates with RUNX2-target promoter and HDAC3 represses RUNX2-mediated induction of its direct target genes. Westendorf et al.49 demonstrated that RUNX2 binds to HDAC6 and inhibits the transcription of p21WAF1 through the deacetylase activity of HDAC6. According to their results, RUNX2 efficiently recruited HDAC6 into cell nucleus and suppressed the transcription of p21WAF1. Intriguingly, Lee et al.58 found that HDAC6 is responsible for efficient oncogenic transformation and tumor formation. In accordance with these observations, it has been shown that inhibition of HDAC6 enhances the chemo-sensitivity of transformed cells.51 Similarly, Wang et al.59 reported that depletion of HDAC6 increases cispaltin-induced apoptosis. Based on our present results, HDAC6 was included in RUNX2/p53 complex and bound to p53-target promoters in response to ADR. Moreover, HDAC6 inhibitor tubacin treatment further enhanced ADR-mediated induction of p53-target genes, raising a possibility that there is a functional collaboration among RUNX2, p53 and HDAC6 and deacetylase activity of HDAC6 is required for RUNX2-mediated repression of p53 transcriptional activity. As RUNX2 knockdown had little effect on ADR-dependent acetylation of p53 at Lys-373/392, it is plausible that HDAC6 does not affect the acetylation status of p53 following ADR exposure. However, we could not exclude a possibility that the other acetylation site(s) of p53 could be regulated by HDAC6. To adequately address this issue, further studies should be required.
Collectively, our present findings provided strong evidence that RUNX2 prohibits p53-mediated apoptosis in response to DNA damage through the collaboration with HDAC6 and might imply that RUNX2 is a potential therapeutic target for cancer treatment.
Materials and Methods
Cell culture and transfection
Human osteosarcoma-derived U2OS, human colon carcinoma-derived HCT116 and human lung carcinoma-derived H1299 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA) and penicillin–streptomycin at 37 °C in 5% CO2. For transfection, cells were transiently transfected with the indicated combinations of the expression plasmid by using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen).
MTT cell survival assay
Cells were seeded in triplicate at a density of 5 × 103 cells per 96-well plates. The next day, cells were treated with the indicated concentrations of ADR or left untreated. Twenty-four after ADR treatment, 10 μl of a modified 3-(4,5-dimethylthiazol-2-yl) 2,5-diphenyl-tetrazolium bromide solution (Dojindo, Kumamoto, Japan) were added to the culture and reaction mixtures were incubated at 37 °C for 2 h. The absorbance readings for each well were carried out at 570 nm using the microplate reader (Model 450, Bio-Rad Laboratories, Hercules, CA, USA).
Trypan blue exclusion assay
Cells were exposed to the indicated concentrations of ADR. Twenty-four hours after ADR treatment, the floating dead cells in the medium and the cells that remained attached to the plates were collected by trypsinization and counted using a hemacytometer in the presence of 0.4% Trypan blue reagent (Sigma, St. Louis, MO, USA).
FACS analysis
For sub-G1 analysis, cells were exposed to the indicated concentrations of ADR. Twenty-four hours after ADR exposure, floating and attached cells were collected, washed in ice-cold phosphate-buffered saline (PBS) and fixed in 70% ethanol at −20 °C. Cells were then treated with RNase A (at a final concentration of 200 μg/ml) and 25 μg/ml of propidium iodide at room temperature for 1 h in the dark. Stained nuclei were analyzed by a FACScan machine (Becton Dickinson, Mountain View, CA, USA).
Reverse transcription (RT)-PCR
Total RNA was prepared from cells by using RNeasy kit according to the manufacturer's instructions (QIAGEN, Valencia, CA, USA), and checked on an agarose gel for intact rRNA bands. RT was carried out with random primers and a SuperScript II reverse transcriptase according to the manufacturer's protocol (Invitrogen). The resultant cDNA was diluted with nuclease-free sterile water and subjected to PCR analysis. The number of cycles required for amplification in the linear range was determined for each primer set and template. The PCR products were electrophoresed on 1.5% agarose gels and their amounts were evaluated by staining with ethidium bromide. Glyderaldehyde-3-phosphate dehydrogenase was used as an internal control. Primer sequences used in the present study are available on request.
Immunoblotting
Cells were rinsed twice in ice-cold PBS and lysed in a lysis buffer containing 25 mM Tris-HCl pH 7.5, 137 mM NaCl, 2.7 mM KCl, 1% Triton X-100 and protease inhibitor cocktail (Sigma). Whole-cell lysates were clarified by centrifugation at 12 000 r.p.m. at 4 °C for 20 min. Protein concentrations were determined by Bradford's reagent (Bio-Rad Laboratories). Equal amounts of whole-cell lysates (100 μg of protein) were resolved on a 10% standard 10% polyacrylamide gel, electro-transferred onto an immobilon membrane (Millipore, Billerica, MA, USA). The membrane was blocked with 5% dry milk in Tris-buffered saline (TBS) plus 0.1% Tween 20 (TBS-T) at 4 °C overnight. After blocking, the membrane was incubated with monoclonal anti-p53 (DO-1, Santa Cruz Biotechnologies, Santa Cruz, CA, USA), monoclonal anti-RUNX2 (8G5, Medical and Biological Laboratories, Nagoya, Japan), polyclonal anti-phospho-p53 at Ser-15 (Cell Signaling Technologies, Beverly, MA, USA), polyclonal anti-acetyl-p53 at Lys-373/382 (Millipore), polyclonal anti-p21WAF1 (H-164, Santa Cruz Biotechnologies), polyclonal anti-BAX (Cell Signaling Technologies), polyclonal anti-PARP (Cell Signaling Technologies), polyclonal anti-HDAC6 (Cell Signaling Technologies) or with polyclonal anti-actin antibody (20-33, Sigma) at room temperature for 1 h. After washing with TBS-T, the membrane was then incubated with HRP-conjugated anti-mouse IgG or with HRP-conjugated anti-rabbit IgG (Invitrogen) at room temperature for 1 h. The membrane was extensively washed with TBS-T and the immunoblotted proteins were then visualized by enhanced chemiluminescence (ECL, Amersham Biosciences, Piscataway, NJ, USA).
Immunostaining
Cells were treated with ADR or left untreated. Twenty-four hours after ADR treatment, cells were rinsed in ice-cold PBS, fixed with 3.7% formaldehyde for 30 min at room temperature, permeabilized with 0.1% Triton X-100 in PBS for 5 min at room temperature, and then blocked with 3% bovine serum albumin in PBS for 1 h at room temperature. After blocking, cells were washed in ice-cold PBS and simultaneously incubated with mouse monoclonal anti-RUNX2 and rabbit monoclonal anti-p53 antibodies (7F5, Cell Signaling Technologies) for 1 h at room temperature. After washing with ice-cold PBS, cells were incubated with fluorescein isothiocyanate-conjugated anti-mouse IgG and rhodamine-conjugated anti-rabbit IgG (Invitrogen) at room temperature for 1 h. Cell nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI; Vector Laboratories, Peterborough, UK). Fluorescent images were captured using a confocal microscope (Leica, Milton Keynes, UK).
Immunoprecipitation
Whole-cell lysates (1 mg of protein) prepared as described above were precleared by adding 30 μl of protein G-Sepharose beads for 1 h at 4 °C. The supernatant was then incubated with NMS or with monoclonal anti-p53 antibody at 4 °C overnight. Subsequently, 30 μl of protein G-Sepharose beads were added to the reaction mixture and incubated for 2 h at 4 °C. The protein G-Sepharose beads were then washed three times with a lysis buffer, boiled in 30 μl of SDS-sample buffer, and the supernatant was analyzed by immunoblotting with monoclonal anti-RUNX2 antibody.
ChIP assay
ChIP experiments were performed essentially as described.29 In brief, U2OS cells were cross-linked with formaldehyde (at a final concentration of 1%) for 15 min at 37 °C, washed in ice-cold PBS twice and incubated with SDS lysis buffer for 10 min at 4 °C. The lysates were sonicated to obtain DNA fragments of 500 bp as determined by agarose gel electrophoresis with ethidium bromide staining. Protein-DNA complexes were subjected to centrifugation at 13 000 r.p.m. for 20 min, and the supernatants were incubated with protein A-agarose beads for 1 h at 4 °C. The supernatants were recovered by a brief centrifugation and then incubated with NMS, monoclonal anti-p53 or with monoclonal anti-RUNX2 antibody overnight at 4 °C. After the incubation, protein A-agarose beads were added to the reaction mixtures and incubated for another 2 h at 4 °C. The immunoprecipitates were sequentially washed with the appropriate buffers and the immune complexes were eluted from the beads with elution buffer containing 1% SDS and 0.1 M NaHCO3. The protein-DNA complexes were then treated with proteinase K at 50 °C for 1 h followed by reverse cross-linking at 65 °C for 4 h. DNA was extracted with phenol–chloroform, precipitated with ethanol, dissolved in 25 μl of Tris-EDTA buffer, and analyzed by standard PCR.
Acknowledgments
This work was supported in part by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology, Japan, a Grant-in-Aid from the Ministry of Health, Labor and Welfare for Third Term Comprehensive Control Research for Cancer, Japan and a Hamaguchi Foundation.
Glossary
- ADR
adriamycin
- ChIP
chromatin immunoprecipitation
- DAPI
4,6-diamidino-2-phenylindole
- FACS
fluorescence-activated cell sorter
- GAPDH
glyderaldehyde-3-phosphate dehydrogenase
- GFP
green fluorescence protein
- HDAC
histone deacetylase
- NMS
normal mouse serum
- NRS
normal rabbit serum
- PARP
poly(ADP-ribose)polymerase
- PBS
phosphate-buffered saline
- RT
reverse transcription
- RUNX2
Runt-related transcription factor 2
- siRNA
small interference RNA
- TBS
Tris-buffered saline
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by RA Knight
Supplementary Material
References
- Vousden KH, Lu X. Live or let die: the cell's response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- Oren M. Decision making by p53: life, death and cancer. Cell Death Differ. 2003;10:431–442. doi: 10.1038/sj.cdd.4401183. [DOI] [PubMed] [Google Scholar]
- Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- Harris CC. p53: at the crossroads of molecular carcinogenesis and risk assessment. Science. 1993;262:1980–1981. doi: 10.1126/science.8266092. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW. p53 function and dysfunction. Cell. 1992;70:523–526. doi: 10.1016/0092-8674(92)90421-8. [DOI] [PubMed] [Google Scholar]
- Velculescu VE, el-Deiry WS. Biological and clinical importance of the p53 tumor suppressor gene. Clin Chem. 1996;42:858–868. [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr, Butel JS, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- Pietenpol JA, Tokino T, Thiagalingam S, el-Deiry WS, Kinzler KW, Vogelstein B. Sequence-specific transcriptional activation is essential for growth suppression by p53. Proc Natl Acad Sci USA. 1994;91:1998–2002. doi: 10.1073/pnas.91.6.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- Prives C, Hall PA. The p53 pathway. J Pathol. 1999;187:112–126. doi: 10.1002/(SICI)1096-9896(199901)187:1<112::AID-PATH250>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Sionov RV, Haupt Y. The cellular response to p53: the decision between life and death. Oncogene. 1999;18:6145–6157. doi: 10.1038/sj.onc.1203130. [DOI] [PubMed] [Google Scholar]
- Piette J, Neel H, Marechal V. Mdm2: keeping p53 under control. Oncogene. 1997;15:1001–1010. doi: 10.1038/sj.onc.1201432. [DOI] [PubMed] [Google Scholar]
- Yuan ZM, Huang Y, Ishiko T, Nakada S, Utsugisawa T, Shioya H, et al. Role for p300 in stabilization of p53 in the response to DNA damage. J Biol Chem. 1999;274:1883–1886. doi: 10.1074/jbc.274.4.1883. [DOI] [PubMed] [Google Scholar]
- Samuels-Lev Y, O'Connor DJ, Bergamaschi D, Trigiante G, Hsieh JK, Zhong S, et al. ASPP proteins specifically stimulate the apoptotic function of p53. Mol Cell. 2001;8:781–794. doi: 10.1016/s1097-2765(01)00367-7. [DOI] [PubMed] [Google Scholar]
- Roe JS, Kim H, Lee SM, Kim ST, Cho EJ, Youn HD. p53 stabilization and transactivation by a von Hippel-Lindau protein. Mol Cell. 2006;5:395–405. doi: 10.1016/j.molcel.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Lopez-Mateo I, Villaronga MA, Llanos S, Belandia B. The transcription factor CREBZF is a novel positive regulator of p53. Cell Cycle. 2012;11:3887–3895. doi: 10.4161/cc.22133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wang C, Zhang P, Gao K, Wang D, Yu H, et al. Polycomb group protein PHF1 regulates p53-dependent cell growth arrest and apoptosis. J Biol Chem. 2012;288:529–539. doi: 10.1074/jbc.M111.338996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando K, Ozaki T, Yamamoto H, Furuya K, Hosoda M, Hayashi S, et al. Polo-like kinase 1 (Plk1) inhibits p53 function by physical interaction and phosphorylation. J Biol Chem. 2004;279:25549–25561. doi: 10.1074/jbc.M314182200. [DOI] [PubMed] [Google Scholar]
- Yuan Z, Villagra A, Peng L, Coppola D, Glozak M, Sotomayor EM, et al. The ATDC (TRIM29) protein binds p53 and antagonizes p53-mediated functions. Mol Cell Biol. 2010;30:3004–3015. doi: 10.1128/MCB.01023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Wang L, Li L, Wang Z, Ho Y, McDonald T, et al. Activation of p53 by SIRT1 inhibition enhances elimination of CML leukemia stem cells in combination with imatinib. Cancer Cell. 2012;21:266–281. doi: 10.1016/j.ccr.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blyth K, Cameron ER, Neil JC. The RUNX genes: gain or loss of function in cancer. Nat Rev Cancer. 2005;5:376–387. doi: 10.1038/nrc1607. [DOI] [PubMed] [Google Scholar]
- Anglin I, Passaniti A. Runx protein signaling in human cancers. Cancer Treat Res. 2004;119:189–215. doi: 10.1007/1-4020-7847-1_10. [DOI] [PubMed] [Google Scholar]
- Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1 the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- Takahashi A, Satake M, Yamaguchi-Iwai Y, Bae SC, Lu J, Maruyama M, et al. Positive and negative regulation of granulocyte-macrophage colony-stimulating factor promoter activity by AML1-related transcription factor, PEBP2. Blood. 1995;86:607–616. [PubMed] [Google Scholar]
- Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae SC, Komori T, et al. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;111:621–633. doi: 10.1016/s0092-8674(02)01111-x. [DOI] [PubMed] [Google Scholar]
- Wu D, Ozaki T, Yoshihara Y, Kubo N, Nakagawara A. Runt-related transcription factor 1 (RUNX1) stimulates tumor suppressor p53 in response to DNA damage through complex formation and acetylation. J Biol Chem. 2012;288:1353–1364. doi: 10.1074/jbc.M112.402594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QL, Ito K, Sakakura C, Fukamachi H, Ki Inoue, Chi XZ, et al. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell. 2002;109:113–124. doi: 10.1016/s0092-8674(02)00690-6. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Tong D, Lou G, Zhang Y, Geng J. Expression of RUNX3 gene, methylation status and clinicopathological significance in breast cancer and breast cancer cell lines. Pathobiology. 2008;75:244–251. doi: 10.1159/000132385. [DOI] [PubMed] [Google Scholar]
- Nicole Tsang YH, Wu XW, Lim JS, Wee Ong C, Salto-Tellez M, Ito K, et al. Prolyl isomerase Pin1 downregulates tumor suppressor RUNX3 in breast cancer. Oncogene. 2012;32:1488–1496. doi: 10.1038/onc.2012.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Zhao X, Wang H, Zhang P, Guo C, Huang C, et al. RUNX3 mediates suppression of tumor growth and metastasis of human CCRCC by regulating cyclin related proteins and TIMP-1. PLoS One. 2012;7:e32961. doi: 10.1371/journal.pone.0032961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada C, Ozaki T, Ando K, Suenaga Y, Inoue K, Ito Y, et al. RUNX3 modulates DNA damage-mediated phosphorylation of tumor suppressor p53 at Ser-15 and acts as a co-activator for p53. J Biol Chem. 2010;285:16693–16703. doi: 10.1074/jbc.M109.055525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- Welch RD, Jones AL, Bucholz RW, Reinert CM, Tjia JS, Pierce WA, et al. Effect of recombinant human bone morphogenetic protein-2 on fracture healing in a goat tibial fracture model. J Bone Miner Res. 1998;13:1483–1490. doi: 10.1359/jbmr.1998.13.9.1483. [DOI] [PubMed] [Google Scholar]
- Lee MH, Kim YJ, Yoon WJ, Kim JI, Kim BG, Hwang YS, et al. Dlx5 specifically regulates Runx2 type II expression by binding to homeodomain-response elements in the Runx2 distal promoter. J Biol Chem. 2005;280:35579–35587. doi: 10.1074/jbc.M502267200. [DOI] [PubMed] [Google Scholar]
- Karsenty G. The genetic transformation of bone biology. Genes Dev. 1999;13:3037–3051. doi: 10.1101/gad.13.23.3037. [DOI] [PubMed] [Google Scholar]
- Brubaker KD, Vessella RL, Brown LG, Corey E. Prostate cancer expression of runt-domain transcription factor Runx2, a key regulator of osteoblast differentiation and function. Prostate. 2003;56:13–22. doi: 10.1002/pros.10233. [DOI] [PubMed] [Google Scholar]
- Kayed H, Jiang X, Keleg S, Jesnowski R, Giese T, Berger MR, et al. Regulation and functional role of the Runt-related transcription factor-2 in pancreatic cancer. Br J Cancer. 2007;97:1106–1115. doi: 10.1038/sj.bjc.6603984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T, Ohta K, Kobayashi T. Expression and function of Cbf a-1/Runx2 in thyroid papillary carcinoma cells. J Clin Endocrinol Metab. 2008;93:2409–2412. doi: 10.1210/jc.2007-2805. [DOI] [PubMed] [Google Scholar]
- Onodera Y, Miki Y, Suzuki T, Takagi K, Akahira J, Sakyu T, et al. Runx2 in human breast carcinoma: its potential roles in cancer progression. Cancer Sci. 2010;101:2670–2675. doi: 10.1111/j.1349-7006.2010.01742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sase T, Suzuki T, Miura K, Shiiba K, Sato I, Nakamura Y, et al. Runt-related transcription factor 2 in human colon carcinoma: a potent prognostic factor associated with estrogen receptor. Int J Cancer. 2012;131:2284–2293. doi: 10.1002/ijc.27525. [DOI] [PubMed] [Google Scholar]
- van der Deen M, Akech J, Lapointe D, Gupta S, Young DW, Montecino MA, et al. Genomic promoter occupancy of runt-related transcription factor RUNX2 in osteosarcoma cells identifies genes involved in cell adhesion and motility. J Biol Chem. 2012;287:4503–4517. doi: 10.1074/jbc.M111.287771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau CC, Harris CP, Lu XY, Perlaky L, Gogineni S, Chintagumpala M, et al. Frequent amplification and rearrangement of chromosomal bands 6p1 2-p2 1 and 17p11.2 in osteosarcoma. Genes Chromosomes Cancer. 2004;39:11–21. doi: 10.1002/gcc.10291. [DOI] [PubMed] [Google Scholar]
- Browne G, Nesbitt H, Ming L, Stein GS, Lian JB, McKeown SR, et al. Bicalutamide-induced hypoxia potentiates RUNX2-mediated Bcl-2 expression resulting in apoptosis resistance. Br J Cancer. 2012;107:1714–1721. doi: 10.1038/bjc.2012.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blyth K, Vaillant F, Hanlon L, Mackay N, Bell M, Jenkins A, et al. Runx2 and MYC collaborate in lymphoma development by suppressing apoptotic and growth arrest pathways in vivo. Cancer Res. 2006;66:2195–2201. doi: 10.1158/0008-5472.CAN-05-3558. [DOI] [PubMed] [Google Scholar]
- Westendorf JJ, Zaidi SK, Cascino JE, Kahler R, van Wijnen AJ, Lian JB, et al. Runx2 (Cbfa1, AML-3) interacts with histone deacetylase 6 and represses the p21(CIP1/WAF1) promoter. Mol Cell Biol. 2002;22:7982–7992. doi: 10.1128/MCB.22.22.7982-7992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namdar M, Perez G, Ngo L, Marks PA. Selective inhibition of histone deacetylase 6 (HDAC6) induces DNA damage and sensitizes transformed cells to anticancer agents. Proc Natl Acad Sci USA. 2010;107:20003–20008. doi: 10.1073/pnas.1013754107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadikovic B, Thorner P, Chilton-Macneill S, Martin JW, Cervigne NK, Squire J, et al. Expression analysis of genes associated with human osteosarcoma tumors shows correlation of RUNX2 overexpression with poor response to chemotherapy. BMC Cancer. 2010;10:202–211. doi: 10.1186/1471-2407-10-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akech J, Wixted JJ, Bedard K, van der Deen M, Hussain S, Guise TA, et al. Runx2 association with progression of prostate cancer in patients: mechanisms mediating bone osteolysis and osteoblastic metastatic lesions. Oncogene. 2010;29:811–821. doi: 10.1038/onc.2009.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratap J, Javed A, Languino LR, van Wijnen AJ, Stein JL, Stein GS, et al. The Runx2 osteogenic transcription factor regulates matrix metalloproteinase 9 in bone metastatic cancer cells and controls cell invasion. Mol Cell Biol. 2005;25:8581–8591. doi: 10.1128/MCB.25.19.8581-8591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutterbach B, Westendorf JJ, Linggi B, Isaac S, Seto E, Hiebert SW. A mechanism of repression by acute myeloid leukemia-1, the target of multiple chromosomal translocations in acute leukemia. J Biol Chem. 2000;275:651–656. doi: 10.1074/jbc.275.1.651. [DOI] [PubMed] [Google Scholar]
- Schroeder TM, Kahler RA, Li X, Westendorf JJ. Histone deacetylase 3 interacts with runx2 to repress the osteocalcin promoter and regulate osteoblast differentiation. J Biol Chem. 2004;279:41998–42007. doi: 10.1074/jbc.M403702200. [DOI] [PubMed] [Google Scholar]
- Lee DY, Hayes JJ, Pruss D, Wolffe AP. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72:73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- Hu E, Chen Z, Fredrickson T, Zhu Y, Kirkpatrick R, Zhang GF, et al. Cloning and characterization of a novel human class I histone deacetylase that functions as a transcription repressor. J Biol Chem. 2000;275:15254–15264. doi: 10.1074/jbc.M908988199. [DOI] [PubMed] [Google Scholar]
- Lee YS, Lim KH, Guo X, Kawaguchi Y, Gao Y, Barrientos T, et al. The cytoplasmic deacetylase HDAC6 is required for efficient oncogenic tumorigenesis. Cancer Res. 2008;68:7561–7569. doi: 10.1158/0008-5472.CAN-08-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Xiang S, Williams KA, Dong H, Bai W, Nicosia SV, et al. Depletion of HDAC6 enhances cisplatin-induced DNA damage and apoptosis in non-small cell lung cancer cells. PLoS One. 2012;7:e44265. doi: 10.1371/journal.pone.0044265. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.