Abstract
Choline kinase alpha (ChoKα) is regarded as an attractive cancer target. The enzyme catalyses the formation of phosphocholine (PCho), an important precursor in the generation of phospholipids essential for cell growth. ChoKα has oncogenic properties and is critical for the survival of cancer cells. Overexpression of the ChoKα protein can transform noncancer cells into cells with a cancerous phenotype, and depletion of the ChoKα protein can result in cancer cell death. However, the mechanisms underlying the tumourigenic properties of ChoKα are not fully understood. ChoKα was recently demonstrated to associate with other oncogenic proteins, raising the possibility that a non-catalytic protein scaffolding function drives the tumourigenic properties of ChoKα rather than a catalytic function. In order to differentiate these two roles, we compared the impact on cancer cell survival using two tools specific for ChoKα: (1) small interfering RNA (siRNA) to knockdown the ChoKα protein levels; and (2) compound V-11-0711, a novel potent and selective ChoKα inhibitor (ChoKα IC50 20 nℳ), to impede the catalytic activity. Both treatments targeted the endogenous ChoKα protein in HeLa cells, as demonstrated by a substantial reduction in the PCho levels. siRNA knockdown of the ChoKα protein in HeLa cells resulted in significant cell death through apoptosis. In contrast, compound V-11-0711 caused a reversible growth arrest. This suggests that inhibition of ChoKα catalytic activity alone is not sufficient to kill cancer cells, and leads us to conclude that there is a role for the ChoKα protein in promoting cancer cell survival that is independent of its catalytic activity.
Keywords: choline kinase alpha, apoptosis, scaffolding, inhibitor, siRNA
Introduction
Choline kinase alpha (ChoKα) is a cytosolic enzyme that catalyses the Mg.ATP-dependent phosphorylation of choline to generate phosphocholine (PCho) as the first step in the Kennedy pathway.1 PCho has been reported to be a mitogen required for DNA synthesis induced by growth factors.2 The Kennedy pathway is also the major source of phosphatidylcholine (PtdCho). As well as being a major structural component of mammalian cellular membranes, PtdCho serves as a precursor for the production of lipid second messengers that can activate growth and survival pathways.3 There is compelling literature, suggesting that ChoKα is a good cancer target. This is based on the role of ChoKα (and its downstream products, PCho and PtdCho) in survival signalling,3 and also on the fact that ChoKα depletion causes cancer cell death.4, 5, 6 Furthermore, there is direct evidence that ChoKα is oncogenic. Overexpression of ChoKα is sufficient to transform cells, inducing both anchorage independent growth and promoting tumour proliferation.7 Depletion of the protein using small interfering RNA (siRNA) affects its oncogenic capability and prevents the formation of tumours when cells are implanted in mice.4 Increased ChoKα expression and activity have been detected in human tumour samples,8, 9, 10, 11, 12 and this increased enzyme expression has been associated with high histological tumour grade and poor clinical outcome.9, 13 For these reasons ChoKα has been proposed as a prognostic marker for cancer progression and as a molecular target for cancer therapeutic agents.14
The mechanisms underlying the tumourigenic properties of ChoKα are not understood, and the importance of the catalytic activity has not been fully explored. A common method used to assess the role of ChoKα in cancer has been depletion of the ChoKα protein using siRNA/short hairpin RNA techniques. These studies have resulted in reduced intracellular PCho levels, which has been coupled to a decrease in cancer cell viability.4, 5, 6 Such studies, however, do not allow a determination as to whether the catalytic activity of the enzyme, or a non-catalytic scaffolding role of the protein contributes to the survival of cancer cells. There is accumulating evidence that non-catalytic properties of kinases are essential for cell growth and survival, as recently reviewed by Rauch et al.15 These non-catalytic functions include scaffolding of protein complexes, competition for protein interactions, allosteric effects on other enzymes and subcellular targeting. It is feasible that the ChoKα protein itself, rather than the enzymatic activity, may has a crucial role in survival signalling pathways in cancer. This non-catalytic, ‘scaffolding role' of ChoKα would be consistent with the recent description by Miyake and Parsons16 of a novel complex formation of ChoKα with EGFR, which may contribute to promoting cancer cell survival. These observations make it clear that a better understanding of the role of ChoKα with respect to potential anticancer therapy is needed. In this study, we used two tools to investigate the role of ChoKα in cancer cell survival. First, siRNA knockdown was used to assess the impact of removing the ChoKα protein, thus disrupting both its interaction with other proteins and its catalytic activity. Second, a novel potent and selective small-molecule inhibitor, V-11-0711, was used to inhibit the catalytic activity of the enzyme without affecting the protein levels. Our results show that HeLa cells exhibit different phenotypes depending on the tool used: siRNA caused apoptosis, while V-11-0711 caused no cell death. These results suggest a role for the ChoKα protein in promoting cancer cell survival that is independent of its catalytic activity.
Results and discussion
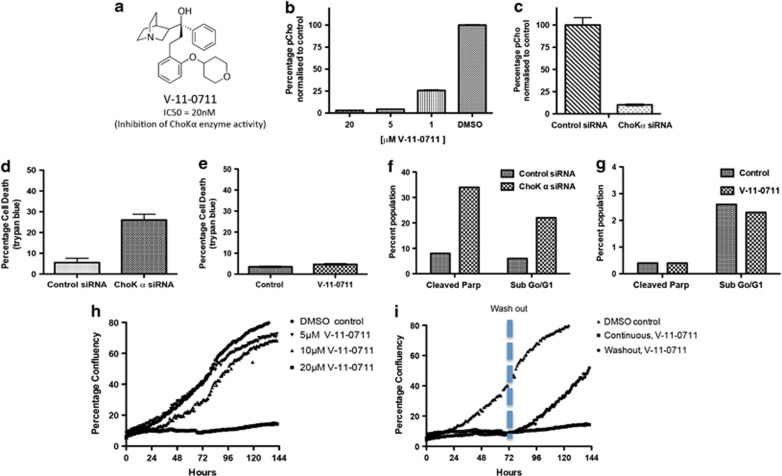
The aim of our study was to investigate the role of the ChoKα protein, compared with its catalytic activity in carcinogenesis. A small-molecule inhibitor of enzyme activity was required in addition to using ChoKα siRNA. We therefore developed compound V-11-0711 as a novel potent and selective inhibitor of ChoKα. Compound V-11-0711 (Figure 1a) is the product of a structure-directed lead optimisation programme. V-11-0711 inhibited recombinant human ChoKα with an IC50 of 20 nℳ, and showed 11 fold less activity against ChoKβ (IC50 220 nℳ). V-11-0711 exhibited excellent selectivity against a panel of 50 kinases (Merck Millipore KinaseProfiler service, Dundee, UK), with very little inhibition at 2 μℳ (see Supplementary Table S1). We then assessed the ability of V-11-0711 to inhibit the production of PCho in intact cells. V-11-0711 reduced the level of PCho in HeLa cells in a concentration-dependent manner, with an IC50 of <1 μℳ (Figure 1b). Depletion of ChoKα by siRNA in HeLa cells (which resulted in a 68% depletion of ChoKα protein) also led to a substantial reduction in the levels of PCho (Figure 1c). This leads us to believe that V-11-0711 is a good tool with which to probe the role of ChoKα catalytic activity in cancer cell survival. There have been very few potent inhibitors of ChoKα reported in the literature. Published ChoKα inhibitors, such as MN58b and CK-37, have an intriguing disconnect between their weak effects on ChoKα enzyme activity, and their more potent cellular and in vivo activities.12, 17, 18, 19 As the full selectivity profiles of MN58b and CK-37 are not known, the interpretation of their effects on cells is confounded.
Figure 1.
(a) Structure and enzyme activity of V-11-0711. Recombinant ChoKα or ChoKβ proteins were produced in Escherichia coli strain BL21(DE3), and purified using glutathione affinity purification followed by size exclusion via Superdex-200 26/60 (GE Healthcare, Buckinghamshire, UK). Enzyme activity was assayed in buffer (100 mℳ Tris-HCl pH 7.5, 100 mℳ KCl and 10 mℳ MgCl2) using a ultraviolet spectroscopic assay.22 Inhibitor IC50 was determined using 400 μℳ ATP and 200 μℳ choline. (b,c). PCho levels in HeLa cells were depleted to the same extent following treatment with compound V-11-0711 or ChoKα siRNA. HeLa cells (ATCC) were grown in Dulbecco's modified eagle's medium medium supplemented with 10% foetal bovine serum, 1% penicillin–streptomycin solution, 1% nonessential amino acids and 1% L-glutamine (Sigma, Poole, UK). Compound was dissolved in dimethylsulphoxide (DMSO, Sigma) before addition to cultures, with a final DMSO concentration not exceeding 0.25%. HeLa cells were treated with different concentrations of V-11-0711 for 24 h and cell pellet prepared. Cell pellets underwent small organic molecule extraction, and PCho content was determined by liquid chromatography-tandem mass spectrometry. In brief, cell pellets were flash frozen immediately after harvesting, and were treated by adding Buffer A, containing internal standards (Buffer A: 80% ACN/12.7% H2O/6.8% EtOH/0.3% 1 ℳ NH4AOc/0.2% AA and valine d8 at 25 μℳ). Samples were vortexed, incubated at −20 °C for 20 min, and centrifuged at 16 000 rcf for 20 min at 4 °C. Supernatants were diluted in Buffer A, and analysed using a 4000QTRAP(AB/SCIEX, Foster City, CA, USA) spectrometer and an 1100 HPLC system (Agilent, Santa Clara, CA, USA). Protein contents (2D Quant Kit, Amersham Biosciences, Buckinghamshire, UK) were used to normalise measured data for small organic molecules. Results are expressed as a percentage of the control; (c) HeLa cells were transfected with ChoKα or non-targeting (control) siRNA, and PCho content was determined as in (b). siRNA studies were performed as follows: siRNA for ChoKα and non-targeting siRNA (Dharmacon, Lafayette, CO, USA) were resuspended at 20 μℳ in molecular biology grade water (Sigma). HeLa cells were seeded into 6-well plates at a density of 0.45 × 105 per well. Transfection was carried out 24 h later using oligofectamine (Invitrogen, Paisley, UK) according to the manufacturers instructions, using 80 nℳ siRNA on cells. Cells were used in studies either 96 or 120 h after transfection. ChoKα knockdown was assessed by western blot after 96 h, and typically achieved >60% reduction in ChoKα protein. (d) ChoKα depletion by siRNA resulted in significant cancer cell death in contrast to incubation with V-11-0711. HeLa cells were transfected with ChoKα or non-targeting (control) siRNA, then analysed for cell death using trypan blue exclusion. Briefly, cells were harvested by trypsinisation, washes were pooled and the resulting pellet resuspended in a fixed volume of phosphate-buffered saline (PBS). Trypan blue was added at a ratio of 1:1, and the cells counted using a haemocytometer. Results are expressed as a percentage of total cell number; (e) HeLa cells were incubated with 10 μℳ V-11-0711 for 72 h, and analysed as described for (d); (f) The extent of apoptosis in HeLa cells was measured after cells were transfected with ChoKα or non-targeting siRNA. Cells were stained with propidium iodide (PI) and a fluorescently-labelled antibody targeting the 89 kDa cleaved fraction of PARP, and analysed using flow cytometry. Cell pellets were washed in PBS and fixed over night using ice-cold methanol (90%). Cells were assessed for DNA content using 20 μg/ml PI (Sigma), and for PARP cleavage using anticleaved PARP (Asp214) (clone F21-852, BD Pharmingen, Oxford, UK). Briefly, cells were incubated in blocking buffer (PBS, 0.2% tween-20, 5% goat serum) for 30 min, then incubated for 40 min with primary antibody in blocking buffer, washed with PBS-T followed by incubation with secondary antibody (Alexafluor 488, Invitrogen) in PBS containing 200 μg/ml DNase free RNase (Sigma) and 20 μg/ml PI (Sigma). Analysis was carried out using a FACs Canto (BD, Oxford, UK) flow cytometer. (g) HeLa cells were incubated with 10 μℳ V-11-0711 (or just DMSO in the control) for 72 h, and stained as described for (f). (h) Inhibition of ChoKα with V-11-0711 results in cytostasis. HeLa cells were incubated with different concentrations of V-11-0711, and growth was assessed over 144 h using the IncuCyte cell imager. HeLa cells were seeded onto 24-well plates and incubated at 37 °C, 5% CO2 and 95% humidity for 24 h in growth medium. Compound was added at a final concentration of 20, 10 or 5 μℳ. Percentage confluence was calculated from images recorded every hour using the IncuCyte Live-Cell imaging System (Essen Instruments, Welwyn Garden City, UK). For extended time-course studies, wells were washed three times after 72 h and then replaced with fresh medium with or without compound. (i) HeLa cells were incubated with 20 μℳ V-11-0711 as in (h). At 72 h cells were washed and either fresh medium plus compound were added (‘continuous'), or fresh medium was added with no additional compound (‘washout'). Data are expressed as percentage confluency.
Literature studies have shown that reduced PCho levels can impact cancer cell viability.4, 5, 6 We observed that inhibition of PCho production by siRNA depletion of ChoKα resulted in increased HeLa cell death (Figure 1d), in agreement with published data.4, 6 We therefore postulated that the inhibition of PCho production by V-11-0711 would also cause cancer cell death. In contrast to siRNA treatment, V-11-0711 did not cause the death of HeLa cells (Figure 1e), and V-11-0711 was equally ineffective on three additional cancer cell lines (see Supplementary Tables S2 and S3). In addition, V-11-0711-treated cells and ChoKα-siRNA treated cells exhibited different levels of apoptotic markers. Treatment of HeLa cells with ChoKα siRNA resulted in a large proportion of cells undergoing apoptosis (Figure 1f): 22% of cells were in the sub G0/1 phase, and 34% of cells were positive for cleaved poly(ADP-ribose)polymerase (PARP). Similar results were obtained using two different ChoKα siRNA oligonucleotides, strongly suggesting that the results obtained were not due to an off-target effect. High levels of cleaved PARP, such as that observed here, is indicative of cell death via an apoptotic pathway. The extent of apoptosis in HeLa cells, measured by PARP cleavage, increased over time, as did the number of dead cells, reaching 50% at 144 h post ChoKα siRNA transfection. The majority of the cleaved PARP-positive cells appeared in the G1 phase. In contrast, HeLa cells treated with V-11-0711 displayed low levels of apoptotic markers (Figure 1g): 2.3% of cells were in the sub G0/1 phase, and 0.4% of cells were positive for cleaved PARP. Although V-11-0711 did not kill HeLa cells, it did inhibit their growth. A detailed analysis of the cell cycle shows that there is a trend towards G1 accumulation, consistent with a slow down in cell growth. HeLa cells treated with different concentrations of V-11-0711 did not divide at the same rate as control cells, and the population remained static at high concentrations of compound throughout the 144 h course of the experiment (Figure 1h). On washout of V-11-0711 after 72 h, the cells resumed growth at a similar rate to control cells (Figure 1i). Therefore, the inhibition of ChoKα in HeLa cells with V-11-0711 led to a reversible growth arrest, which is in contrast to the apoptosis observed following siRNA depletion of the ChoKα protein. These contrasting phenotypes occurred with similar levels of PCho reduction.
Although V-11-0711 appears to be selective for ChoKα, we cannot rule out the possibility that an unknown cross-reactivity of V-11-0711 is somehow protecting the cells from apoptosis. An alternative explanation for the HeLa cell death using ChoKα siRNA could be attributed to the combined stress of the siRNA transfection procedure with concomitant ChoKα enzyme inhibition. However, this is unlikely, as in control experiments, in which HeLa cells were incubated with V-11-0711 in the presence of transfection reagent alone, we did not observe cell death. The results for V-11-0711 are in contrast to published data for MN58b, where this compound induced cancer cell death.20, 21 Possible reasons for this difference are discussed above. It is also possible that MN58b could disrupt interactions with other proteins, resulting in cancer cell death.
The difference in cellular phenotype observed using either siRNA-induced knockdown of ChoKα protein or inhibition of the catalytic activity using a small-molecule inhibitor leads one to conclude that inhibition of ChoKα enzyme activity alone is insufficient to cause cancer cell death. The potential for ChoKα-mediated effects on cancer cells that are independent of the catalytic activity was recently demonstrated by Miyake and Parsons.16 They showed that the overexpression of ChoKα is sufficient to induce cell proliferation, but importantly also showed that cell proliferation could still be induced on overexpression of a catalytically inactive ChoKα D306A mutant. This suggests that ChoKα has a small, but positive effect on cancer cell growth that is independent of its catalytic activity. A non-catalytic, ‘scaffolding role' for ChoKα would also be consistent with additional observations reported by Miyake and Parsons.16 They describe a novel complex formation between ChoKα and EGFR, which contributes to the regulation of cell proliferation and tumourigenesis. It is interesting to speculate that the association of the ChoKα protein with EGFR might have a crucial role in the oncogenic properties of ChoKα.
In summary, we conclude that inhibition of ChoKα enzyme activity alone is insufficient to cause cancer cell death, but instead leads to reversible cytostasis. We speculate that the oncogenic properties of ChoKα and the promotion of cell survival are driven by a non-catalytic protein scaffolding function of ChoKα, rather than its catalytic activity. This challenges the notion that selective small-molecule inhibitors of ChoKα will kill cancer cells, and suggest instead that an approach aimed at destabilising the protein may offer a more powerful antitumour strategy.
Acknowledgments
We thank Rebecca Bordas for the preparation of the ChoK proteins, Mark Fleming and Ye Gu for their help with the phosphocholine detection, and Simon Everitt for the synthesis of compound V-11-0711.
All authors are employees of Vertex Pharmaceuticals and hold shares in the company.
Footnotes
Supplementary Information accompanies the paper on the Oncogenesis website (http://www.nature.com/oncsis).
Supplementary Material
References
- Kennedy EP. Metabolism of lipids. Ann Rev Biochem. 1957;26:119–148. doi: 10.1146/annurev.bi.26.070157.001003. [DOI] [PubMed] [Google Scholar]
- Cuadrado A, Carnero A, Dolfi F, Kimenez B, Lacal JC. Phosphorylcholine: a novel second messenger essential for mitogenic activity of growth factors. Oncogene. 1993;8:2959–2968. [PubMed] [Google Scholar]
- Gibellini F, Smith TK. The Kennedy pathway - De novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB Life. 2010;63:414–428. doi: 10.1002/iub.337. [DOI] [PubMed] [Google Scholar]
- Yalcin A, Clem B, Makoni S, Clem A, Nelson K, Thornburg J, et al. Selective inhibition of choline kinase simultaneously attenuates MAPK and PI3K/AKT signalling. Oncogene. 2009;29:139–149. doi: 10.1038/onc.2009.317. [DOI] [PubMed] [Google Scholar]
- Mori N, Glunde C, Takagi T, Raman V, Bhujwalla Z. Choline kinase down regulation increases the effect of 5-Fluorouracil in breast cancer cell. Cancer Res. 2007;67:11284–11290. doi: 10.1158/0008-5472.CAN-07-2728. [DOI] [PubMed] [Google Scholar]
- Banez-Coronel M, Ramirez de Molina A, Rodriguez-Gonzalez A, Sarmentero J, Ramos MA, Garcia-Cabezas MA, et al. Choline kinase alpha depletion selectively kills tumoral cells. Curr Cancer Drug Targets. 2008;8:709–719. doi: 10.2174/156800908786733432. [DOI] [PubMed] [Google Scholar]
- Ramirez de Molina A, Gallego-Ortega D, Sarmentero J, Banez-Coronel M, Martin-Cantalejo Y, Lacal JC. Choline kinase is a novel oncogene that potentiates RhoA-induced carcinogenesis. Cancer Res. 2005;65:5647–5653. doi: 10.1158/0008-5472.CAN-04-4416. [DOI] [PubMed] [Google Scholar]
- Nakagami K, Uchida T, Ohwada S, Koibuchi Y, Suda Y, Sekine T, et al. Increased choline kinase activity and elevated phosphocholine levels in human colon cancer. Jpn J Cancer Res. 1999;90:419–424. doi: 10.1111/j.1349-7006.1999.tb00764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez de Molina A, Rodriguez-Gonzalez A, Gutierrez R, Martinez-Pineiro L, Sanchez JJ, Bonilla F, et al. Overexpression of choline kinase is a frequent feature in human tumor derived cell lines and in lung, prostate, and colorectal human cancers. Biochem Biophys Res Comms. 2002;296:580–583. doi: 10.1016/s0006-291x(02)00920-8. [DOI] [PubMed] [Google Scholar]
- Iorio E, Ricci A, Bagnoli M, Pisanu ME, Castellano G, Di Vito M, et al. Activation of phosphocholine cycle enzymes in human epithelial ovarian cancer cells. Cancer Res. 2005;65:9369–9376. doi: 10.1158/0008-5472.CAN-09-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabellieri C, Beloueche-babari M, Jamin Y, Payne GS, Leach MO, Eykyn TR. Modulation of choline kinase activity in human cancer cells observed by dynamic 31P NMR. NMR Biomed. 2009;22:456–461. doi: 10.1002/nbm.1361. [DOI] [PubMed] [Google Scholar]
- Hernando E, Sarmentero-Estrada J, Koppie T, Belda-Iniesta C, Ramirez de Molina V, Cejas P, et al. A critical role for choline kinase-α in the aggressiveness of bladder carcinomas. Oncogene. 2009;28:2425–2435. doi: 10.1038/onc.2009.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez de Molina A, Sarmentero-Estrada J, Belda-Iniesta C, Taron M, Ramirez de Molina V, Cejas P, et al. Expression of choline kinase alpha to predict outcome in patients with early stage non- small-cell lung cancer: a retrospective study. Lancet Oncol. 2007;8:889–897. doi: 10.1016/S1470-2045(07)70279-6. [DOI] [PubMed] [Google Scholar]
- Glunde K, Jacobs MA, Bhujwalla Z. Choline metabolism in cancer: Implications for diagnosis and therapy. Expert Rev Mol Diagn. 2006;6:821–829. doi: 10.1586/14737159.6.6.821. [DOI] [PubMed] [Google Scholar]
- Rauch J, Volinsky N, Romano D, Kolch W. The secret life of kinases: functions beyond catalysis. Cell Commun Signal. 2011;9:23. doi: 10.1186/1478-811X-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake T, Parsons SJ. Functional interactions between choline kinase-α, epidermal growth factor receptor and c-Src in breast cancer cell proliferation. Oncogene. 2012;31:1431–1441. doi: 10.1038/onc.2011.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Alcoceba R, Saniger L, Campos J, Nunez MC, Khaless F, Gallo MA, et al. Choline kinase inhibitors as a novel approach for anti-proliferative drug design. Oncogene. 1997;15:2289–2301. doi: 10.1038/sj.onc.1201414. [DOI] [PubMed] [Google Scholar]
- Ramirez de Molina A, Banez-Coronel M, Gutierrez R, Rodriguez-Gonzalez A, Olmeda D, Medias D, et al. Choline kinase activation is a critical requirement for the proliferation of primary human mammary epithelial cells and breast tumor progression. Cancer Research. 2004;64:6732–6739. doi: 10.1158/0008-5472.CAN-04-0489. [DOI] [PubMed] [Google Scholar]
- Clem BF, Clem AL, Yalcin A, Goswami U, Arumugam S, Telang S, et al. A novel small molecule antagonist of choline kinase-α that simultaneously suppresses MAPK and PI3K/AKT signalling. Oncogene. 2011;30:3370–3380. doi: 10.1038/onc.2011.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Gonzalez A, Ramirez de Molina A, Fernandez F, Lacal JC. Choline kinase inhibition induces the increase in ceramides resulting in a specific and highly selective cytotoxic antitumoral strategy as a potential mechanism of action. Oncogene. 2004;23:8247–8259. doi: 10.1038/sj.onc.1208045. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Gonzalez A, Ramirez de Molina A, Banez-Coronel M, Megias D, Lacal JC. Inhibition of choline kinase renders a highly selective cytotoxic effect in tumour cells through a mitochondrial independent mechanism. Int J Oncology. 2005;26:999–1008. [PubMed] [Google Scholar]
- Goldman PR, Northrop DB. Purification and spectrophotometric assay of neomycin phosphotransferase II1. Biochem Biophys Res Commun. 1976;69:230–236. doi: 10.1016/s0006-291x(76)80297-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.