Abstract
Recent findings in colon cancer cells indicate that inhibition of the mitochondrial H+-adenosine triphosphate (ATP) synthase by the ATPase inhibitory factor 1 (IF1) promotes aerobic glycolysis and a reactive oxygen species (ROS)-mediated signal that enhances proliferation and cell survival. Herein, we have studied the expression, biological relevance, mechanism of regulation and potential clinical impact of IF1 in some prevalent human carcinomas. We show that IF1 is highly overexpressed in most (>90%) of the colon (n=64), lung (n=30), breast (n=129) and ovarian (n=10) carcinomas studied as assessed by different approaches in independent cohorts of cancer patients. The expression of IF1 in the corresponding normal tissues is negligible. By contrast, the endometrium, stomach and kidney show high expression of IF1 in the normal tissue revealing subtle differences by carcinogenesis. The overexpression of IF1 also promotes the activation of aerobic glycolysis and a concurrent ROS signal in mitochondria of the lung, breast and ovarian cancer cells mimicking the activity of oligomycin. IF1-mediated ROS signaling activates cell-type specific adaptive responses aimed at preventing death in these cell lines. Remarkably, regulation of IF1 expression in the colon, lung, breast and ovarian carcinomas is exerted at post-transcriptional levels. We demonstrate that IF1 is a short-lived protein (t1/2 ∼100 min) strongly implicating translation and/or protein stabilization as main drivers of metabolic reprogramming and cell survival in these human cancers. Analysis of tumor expression of IF1 in cohorts of breast and colon cancer patients revealed its relevance as a predictive marker for clinical outcome, emphasizing the high potential of IF1 as therapeutic target.
Keywords: H+-ATP synthase, ATPase inhibitory factor 1, energy metabolism, mitochondria, cancer prognosis, ROS signaling
Introduction
Downregulation of oxidative phosphorylation and concurrent activation of aerobic glycolysis is a hallmark feature of proliferating cells and of many different human carcinomas.1, 2, 3 An enhanced aerobic glycolysis provides the metabolic intermediates required to sustain proliferation.1, 4 Several genetically-driven mechanisms directly promoting glycolysis, the inhibition of mitochondrial function or both have been proposed in order to explain energy metabolism in cancer cells and tumors (for review see Cuezva et al.1 and Cairns et al.5). Moreover, it has been suggested that the APC/C–Cdh1 complex that controls the levels of PFKFB3, and hence, the rate of glucose consumption might participate in sustaining glycolysis in some types of human carcinomas.6 However, epigenetic mechanisms7, 8 and the tumor microenvironment5, 9 also have relevant roles in cancer development and progression by regulating the bioenergetic phenotype of cancer cells.10
The H+-adenosine triphosphate (ATP) synthase is a master regulator of energy metabolism and cell fate. It is the mitochondrial protein complex of oxidative phosphorylation that catalyzes the synthesis of ATP using as driving force the proton gradient generated by the respiratory chain.11 The H+-ATP synthase is also required for efficient execution of cell death.12, 13 In fact, cells that are unable to perform oxidative phosphorylation have an apoptotic-resistant phenotype.13, 14, 15 Conversely, activation of the bioenergetic function of mitochondria prevents tumor development.16, 17
A compromized bioenergetic activity of mitochondria, as assessed by genomic,18 transcriptomic,10 proteomic1, 19 and functional studies,20 is involved in tumor progression and in chemotherapeutic resistance.21, 22, 23, 24 Specifically, it has been described: (i) the downregulation of the cellular abundance of the mRNAs that encode rate-limiting subunits of the H+-ATP synthase by either promoter hypermethylation of the ATP5B gene23 or by genetic deletion of ATP5A118; (ii) the masking of the translation of β-F1-ATPase mRNA25, 26 through the binding of repressor proteins27 that impede ribosome recruitment and translation and (iii) the overexpression in cancer cells and tumors of the ATPase inhibitory factor 1 (IF1) that inhibits the activity of the mitochondrial H+-ATP synthase.28 Furthermore, recent findings indicate that IF1 has additional functions in colon cancer cells by promoting a reactive oxygen species (ROS)-mediated adaptive cellular response that triggers proliferation and resistance to cell death.29
In this investigation, we have addressed (i) the study of the expression level of IF1 in different prevalent human carcinomas, (ii) the metabolic and signaling events that mediate IF1 overexpression in the lung, breast and ovarian cancer cells, (iii) the mechanisms that mediate IF1 overexpression in tumors and (iv) its relevance as a prognostic marker in breast and colon cancer patients. Overall, the results indicate that IF1 has a very short half-life being highly overexpressed in the colon, lung, breast and ovarian carcinomas by mechanisms that are regulated at post-transcriptional levels. In contrast, normal tissues that overexpress IF1, such as endometrium and kidney, reveal no relevant changes in IF1 triggered by carcinogenesis. The overexpression of IF1 promotes the activation of aerobic glycolysis and concurrently confers a ROS-mediated resistance to staurosporine (STS)-induced cell death to the lung, breast and ovarian cancer cells. Moreover, the tumor expression of IF1 is a predictive marker for clinical outcome in breast and colon cancer patients. As IF1 masters the reprogramming of energy metabolism and signals cell-death resistance in cancer cells, we suggest that IF1 offers a relevant molecule with high potential as a new therapeutic target for treatment of prevalent human carcinomas.
Results
IF1 is overexpressed in most prevalent human carcinomas
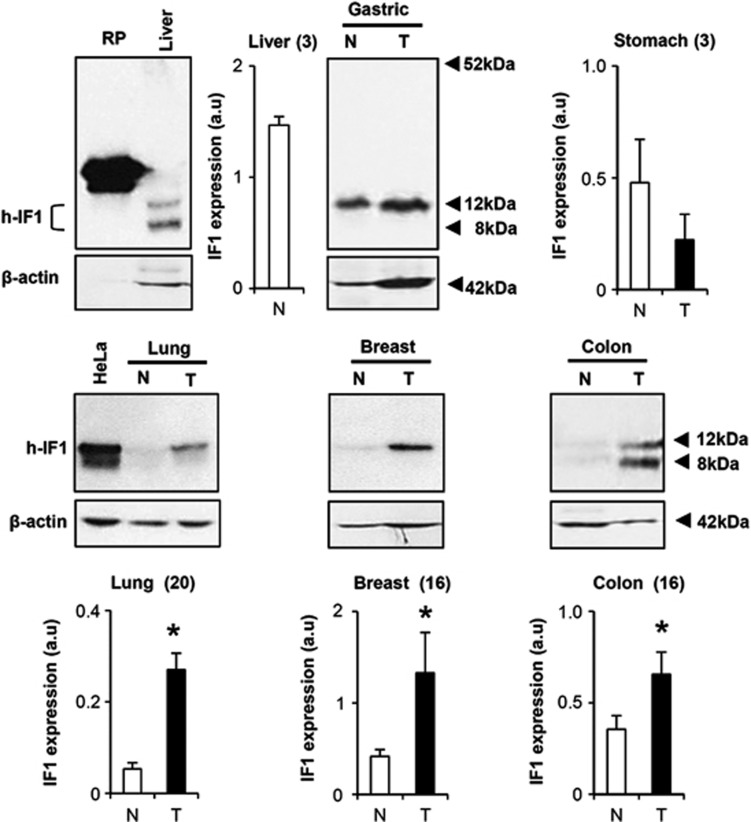
Western blots in Figure 1 illustrate that the monoclonal antibody28 recognizes the recombinant, as well as the two major isoforms (∼12 and ∼8 kDa) of native IF1 in normal human liver and stomach extracts (Figure 1). The expression of IF1 is negligible in the normal breast, colon and lung (Figure 1), but sharply increases in their corresponding carcinomas (Figure 1). Colon carcinomas also express the short ∼8 kDa IF1 isoform (Figure 1). Interestingly, normal stomach shows a high expression of IF1 and revealed no relevant changes by carcinogenesis (Figure 1). It should be noted that heart (not shown) and liver (Figure 1) are the normal human tissues with highest expression of IF1.
Figure 1.
Expression of human IF1 in normal and tumor tissues. Western blots reveal the expression of IF1 (h-IF1) in SDS–polyacrylamide gel electrophoresis fractionated proteins from normal (N) and tumor (T) biopsies of different human tissues. The antibody recognizes the recombinant protein, as well as the two major 12- and 8-kDa protein isoforms of IF1. β-actin expression is shown as loading control. For comparison purposes, the expression level of IF1 in human tissues (histograms) is normalized relative to its expression in HeLa cells (a.u., arbitrary unit) assayed in the same blot. The number of paired normal and tumor biopsies analyzed is indicated in parenthesis.
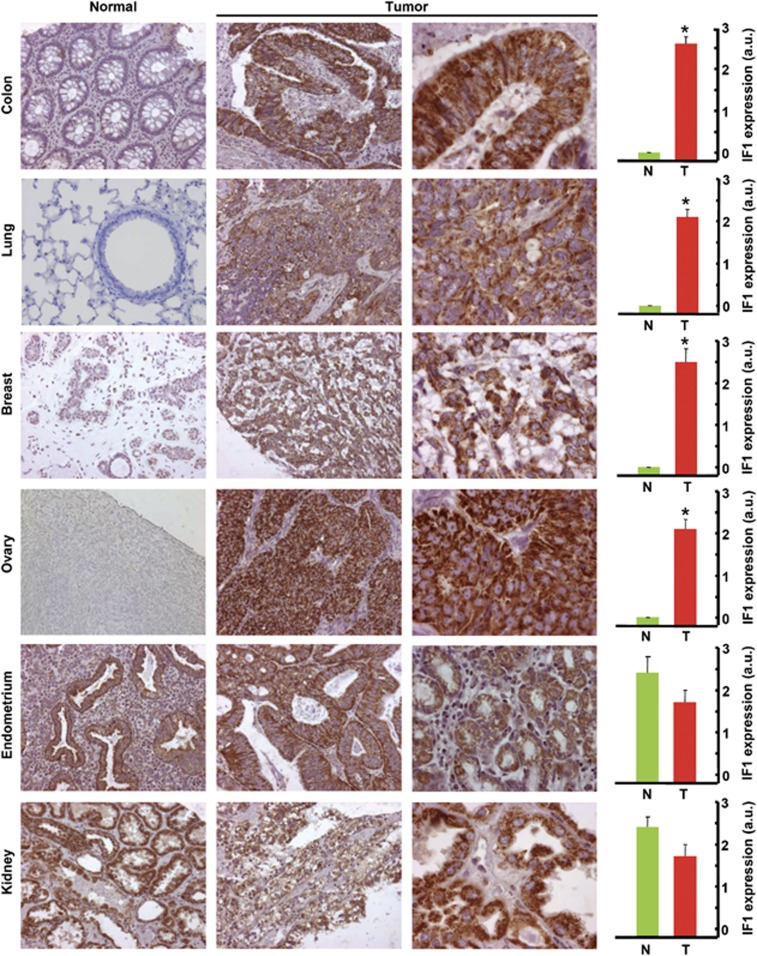
The expression of IF1 was also assessed by immunohistochemistry in a different cohort of normal human tissues and carcinomas (Figure 2). Consistent with western blot data (Figure 1), immunohistochemistry of cancer tissue microarrays confirmed that the expression of IF1 is negligible in the normal colon, lung, breast and ovary (Figure 2). In contrast, carcinomas in these tissues showed a highly significant increase in the granular cytoplasmic immunostaining of IF1 (Figure 2). Contrary to these findings, the expression of IF1 in normal epithelial cells of the endometrium and kidney was very high (Figure 2) and its content did not show significant changes by carcinogenesis (Figure 2).
Figure 2.
IF1 is upregulated in some prevalent human carcinomas. Representative immunohistochemistries of IF1 expression in normal and tumor tissue of the colon, lung, breast, ovary, endometrium and kidney. Magnification × 20, × 40 and × 63. Histograms to the right of the pictures show the quantification of IF1 expression in normal (N, green, n=5) and tumor (T, red, n=10) specimens expressed as arbitrary units (a.u.). The results shown are the mean±s.e.m. *P<0.05 when compared with normal by Student's t-test. Note that whereas normal epithelial cells from the colon, lung, breast and ovary show low or negligible expression of IF1, endometrial and kidney cells have very-high expression of IF1.
Post-transcriptional regulation of IF1 expression in cancer
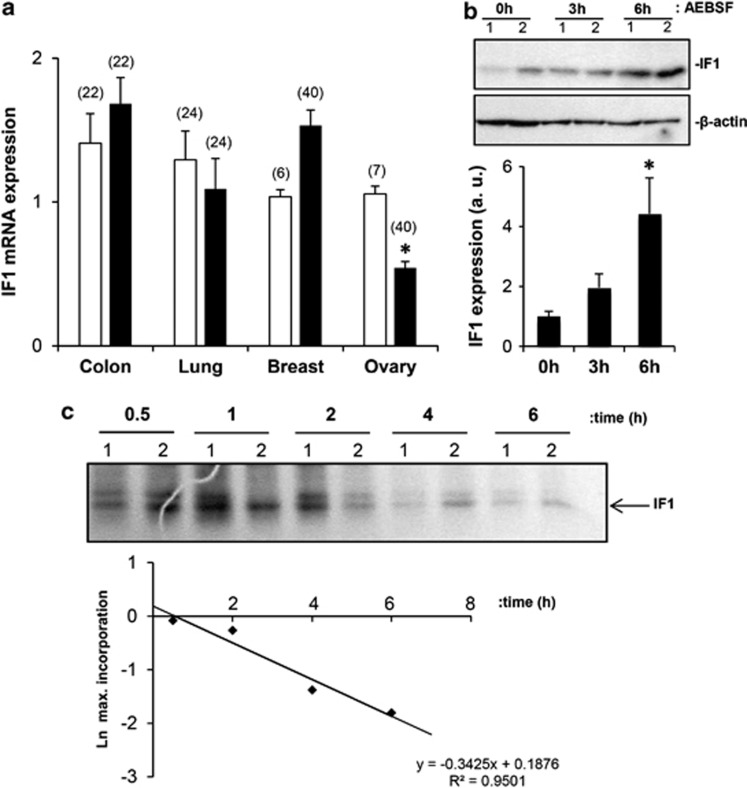
Details on the analysis of the promoter region of the human IF1 gene (ATPIF1) and its possible relationship with the expression of hypoxia-inducible factor-1α30 are provided in Supplementary Figure S1 and Supplementary Figure S2, respectively. In any case, the cellular availability of IF1 mRNA was not significantly different in the colon, lung and breast carcinomas when compared with that in the corresponding normal tissues (Figure 3a). Moreover, IF1 mRNA was significantly reduced in ovarian carcinomas (Figure 3a).
Figure 3.
IF1 expression is regulated at post-transcriptional levels. (a) The colon (HCRT103), lung (HLRT104), breast (BCRT101) and ovarian (HORT102) TissueScan Tissue quantitative PCR Arrays were used to determine the expression of IF1 mRNA in different normal (open bars) and tumor (closed bars) tissue specimens. The number of studied patients is indicated in brackets. The results shown are the mean±s.e.m. *P<0.001 when compared with normal by Student's t-test. (b) HCT116 cells were treated with 400 μℳ of the serine-proteases inhibitor 4-(2-aminoethyl) benzenosulfonyl fluoride hydrochloride for the indicated time and the expression of IF1 and β-actin (loading control) analyzed by western blot. Lanes 1 and 2, show different experiments of the same condition. Bars are the mean±s.e.m. of four experiments. *P<0.05 when compared with 0 h by Student's t-test. (c) After metabolic labeling with 35S-methionine IF1 was immunoprecipitated from HCT116 cells at the indicated time. Lanes 1 and 2, show different experiments of the same condition of the chase. The fluorogram reveals the migration of both the precursor and mature IF1 (arrow) 35S-labeled immunoprecipitated proteins. The plot shows the first order rate kinetics of the decay of IF1. The t1/2 for IF1 is in the 105–120 min range.
The immediate early response gene (IER3) has been shown to target IF1 for degradation by a mitochondrial protease.31 Studies aimed at characterizing the relevance of protein stability in the expression of IF1 in colon cancer cells revealed a very rapid accumulation of the protein in response to the serine-protease inhibitor 4-(2-aminoethyl) benzenosulfonyl fluoride hydrochloride (Figure 3b), suggesting the participation of either the ATP-dependent Lon protease or ClpXP32 in the degradation of IF1. The short half-life of IF1 was further demonstrated by pulse-chase experiments (Figure 3c) that indicated a t1/2 for the protein of∼100 min. Overall, these findings indicate the relevance of translational and post-translational regulatory mechanisms for the expression of IF1 in cancer.
Analysis of the expression of IER3 in the lung, breast and colon carcinomas revealed that it is overexpressed in 16 out of the 18 carcinomas analyzed (Supplementary Figure S3A). Moreover, partial silencing of the protein in colon cancer cells revealed no relevant effect on the expression of IF1 (Supplementary Figure S3B), what suggests that degradation of IF1 is a complex process.
IF1 regulates energy metabolism in the lung, breast and ovarian cancer cells
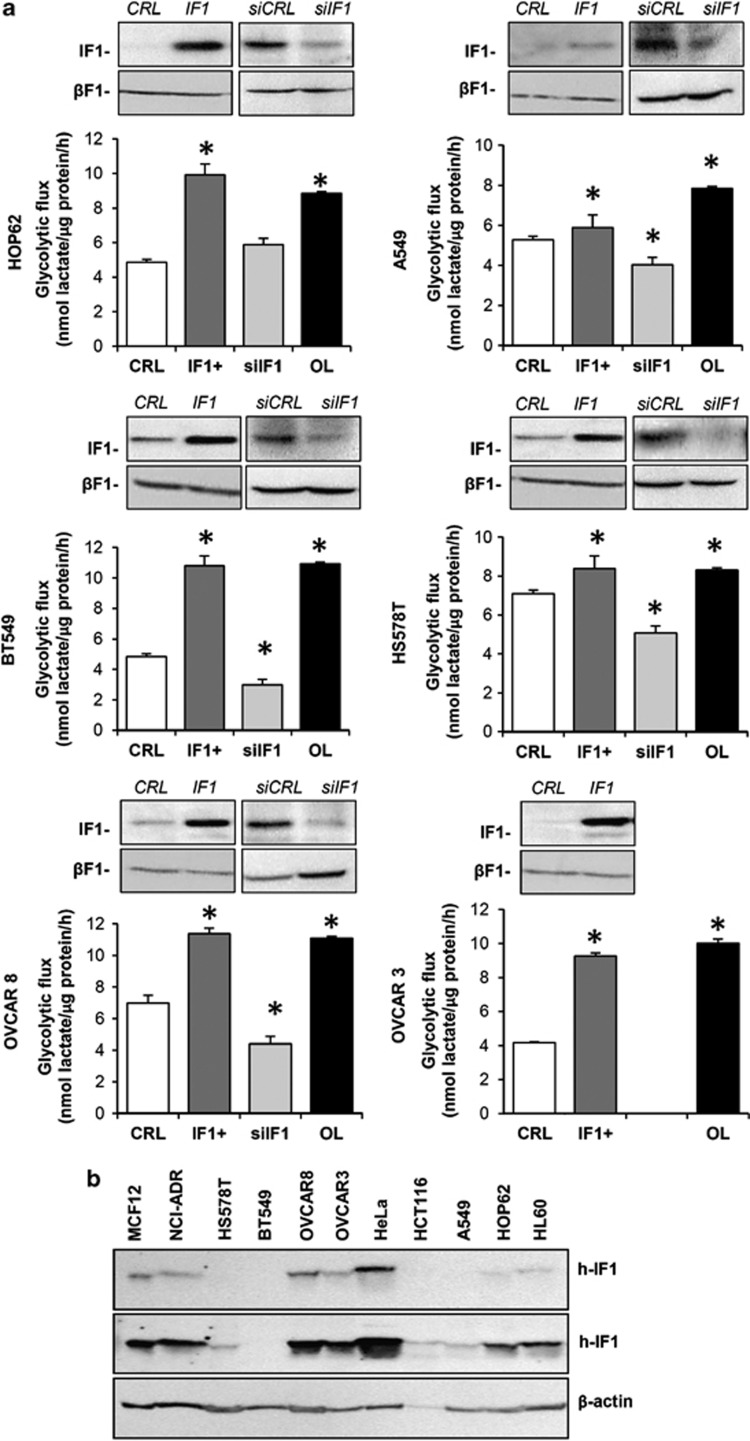
Consistent with previous reports in colon cancer cells,29 the overexpression of IF1 in the lung, breast and ovarian cancer cells promoted a significant increase in the rates of aerobic glycolysis (Figure 4a). The increase in glycolysis was similar to that exerted by incubation of the cells with oligomycin, a pharmacological inhibitor of the H+-ATP synthase (Figure 4a). Conversely, small interfering RNA-mediated silencing of IF1 promoted a significant reduction in the rates of glycolysis in most cancer cells except in HOP62 (Figure 4a). The expression of IF1 varies largely between cancer cells (Figure 4b), whereas the basal rates of aerobic glycolysis were found to be quite similar (Figure 4a), indicating that glycolysis is regulated by other factors in addition to the expression level of IF1.33
Figure 4.
IF1 regulates the activity of aerobic glycolysis in the lung (HOP62, A549), breast (BT549, HS578T) and ovarian (OVCAR8, OVCAR3) cancer cells. (a) Cells transfected with CDL-GFP-β-3′UTR were co-transfected with control (CRL and siCRL, open bars), IF1 plasmid (IF1+, dark gray bars) or siIF1 small interfering RNA (siIF1, light gray bars) to regulate the expression of IF1 for the determination of the rates of aerobic glycolysis. The effect of 6 μℳ oligomycin (OL, closed bars) is shown. Representative blots of IF1 and β-F1-ATPase (β-F1) expression. Bars are the mean±s.e.m. of six different samples. *P<0.05 when compared with CRL by Student's t-test. (b) IF1 (h-IF1) expression in 20 μg of protein from different human cell lines. Two different exposures of the IF1 film are presented. β-actin expression is shown as loading control.
IF1 overexpression generates a mitochondrial ROS signal in the lung, breast and ovarian cancer cells
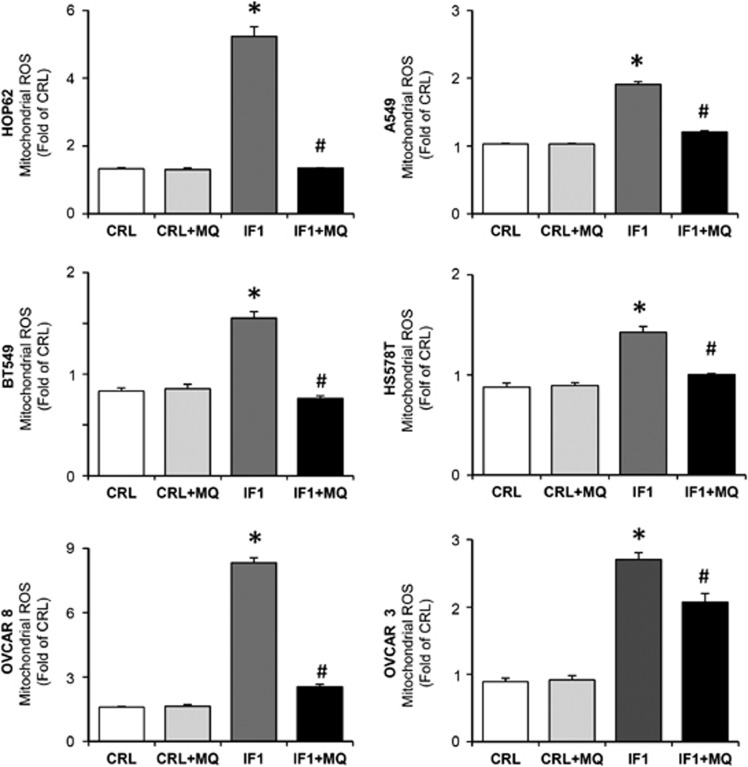
The overexpression of IF1 in the lung, breast and ovarian cancer cells promoted a significant increase in the production of superoxide radical (Figure 5). The mitochondrial scavenger MitoQ (MQ)34 was able to quench the production of superoxide in all cell lines studied (Figure 5), supporting the role of IF1 as a general ROS-mediating signaling molecule in mitochondria.29 We should mention that the IF1-mediated ROS signal generated in mitochondria is of mild intensity because neither the cellular hydrogen peroxide levels nor the GSH/GSSG ratio have been found to be altered by IF1 overexpression.29
Figure 5.
IF1 regulates mitochondrial ROS production in the lung (HOP62, A549), breast (BT549, HS578T) and ovarian (OVCAR8, OVCAR3) cancer cells. Cells transfected with CDL-GFP-β-3′UTR were co-transfected with control (CRL, open and light gray bars) or IF1 plasmid (IF1, dark gray and closed bars) in the absence or presence (+MQ) of 5 nℳ of the mitochondrial ROS scavenger MitoQ (MQ). The superoxide radical was determined by fluorescence-activated cell sorting analysis using MitoSox. The data shown are mean±s.e.m. of 12 different samples. *P<0.05 and #P<0.05 when compared with CRL or IF1 by Student's t-test, respectively.
In sharp contrast with the findings in colon cancer cells,29 the IF1-mediated ROS signal was unable to stimulate cellular proliferation as assessed by the incorporation of 5-ethynyl-2′-deoxyuridine (EdU) into cellular DNA in the lung, breast and ovarian cancer cells (Supplementary Figure S4).
Mitochondrial ROS protect the lung, breast and ovarian carcinomas from STS-induced cell death
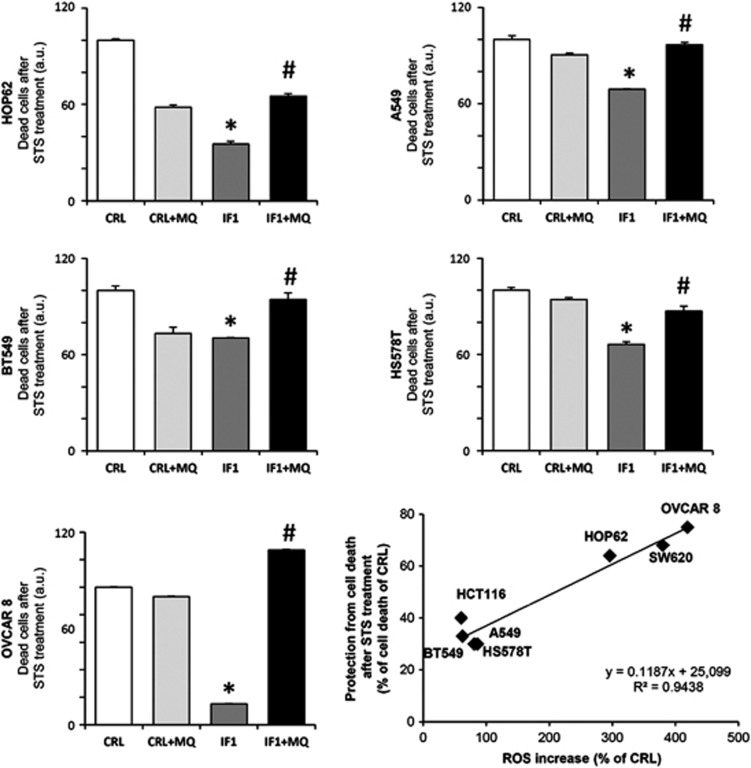
The IF1-mediated ROS signal was able to protect the lung, breast and ovarian cancer cells from STS-induced cell death (Figure 6). Quenching the ROS signal with MQ prevented protection against STS-induced cell death (Figure 6). Interestingly, we observed that the intensity of the ROS-mediated response to IF1 overexpression (Figure 5) correlated with the degree of protection against STS-induced cell death (plot in Figure 6), suggesting a relevant role for mitochondrial ROS signaling in promoting survival pathways in all cancer cells.
Figure 6.
IF1 regulates the cell-death response in the lung (HOP62, A549), breast (BT549, HS578T) and ovarian (OVCAR8) cancer cells. Cells transfected with CDL-GFP-β-3′UTR were co-transfected with control (CRL, open and light gray bars) or IF1 plasmid (IF1, dark gray and closed bars) in the absence or presence (+MQ) of 5 nℳ of the mitochondrial ROS scavenger MitoQ (MQ). Twenty-four hours after transfection, the cells were treated with 1 μℳ STS and 24 h later the cells were stained with propidium iodide and the green population of cells was analyzed by flow cytometry to evaluate the percentage of sub-G0 cells. The data shown are mean±s.e.m. of six–nine different samples. *P<0.05 and #P<0.05 when compared with CRL or IF1 by Student's t-test, respectively. The plot shows the linear correlation that exists between the IF1-mediated ROS-response and the observed cell death.
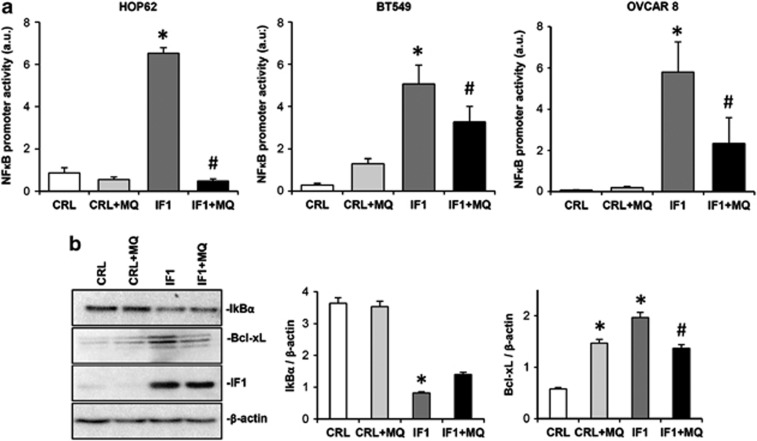
For instance, and in support of this idea, transcriptional activity of the nuclear factor kappa-light-chain-enhancer of activated B cells (NFκB) promoter in the lung, breast and ovarian carcinomas was greatly enhanced by IF1 overexpression (Figure 7a) and partially quenched by the mitochondrial ROS scavenger MQ (Figure 7a). The ROS-mediated activation of the NFκB pathway in HOP62 lung cancer cells is supported by a reduction in the expression of the NFκB repressor IκBα (nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha) and the concurrent increase in the expression of the antiapoptotic Bcl-xL (Figure 7b). These changes were partially reversed by the mitochondrial ROS scavenger MQ (Figure 7b). However, the same studies in breast (BT549, HS578T) and ovarian (OVCAR8) cancer cells did not provided similar findings (data not shown) emphasizing the complexity of the survival pathways that are activated in cancer cells in response to mitochondrial ROS signaling.
Figure 7.
IF1 triggers transcriptional activation of the NFκB promoter in the lung (HOP62), breast (BT549) and ovarian (OVCAR8) cancer cells. Cells transfected with CDL-GFP-β-3′UTR were co-transfected with control (CRL, open and light gray bars) or IF1 (IF1, dark gray or closed bars) plasmid in the absence or presence (+MQ) of 5 nℳ of the mitochondrial ROS scavenger MitoQ (MQ). (a) A luciferase reporter plasmid of the NKκB promoter was co-transfected and the luciferase activity was determined in cellular extracts after 24-h transfection. The data shown are mean±s.e.m. of 10 samples. *P<0.05 and #P<0.05 when compared with CRL or IF1 by Student's t-test, respectively. (b) Cellular proteins of HOP62 were fractionated on SDS–polyacrylamide gel electrophoresis and processed for western blotting with the indicated primary antibodies. Representative blots are shown. The results are mean±s.e.m. of three experiments. *P<0.05 and #P<0.05 when compared with CRL or IF1 by Student's t-test, respectively.
Clinical relevance of IF1 in breast cancer
A next question was to assess the potential relevance of IF1 in the clinics. We determine the expression level of IF1 in a cohort of tumors of breast cancer patients operated from invasive carcinomas for which the bioenergetic signature or bioenergetic cellular index (β-F1-ATPase/Hsp60/GAPDH ratio) and follow-up information is available.35, 36 The results in Supplementary Table S1 summarize the clinicopathological characteristics of the cohort of patients studied and the expression level of IF1 in the carcinomas according to the clinical information. It should be noted that the expression of IF1 in normal breast biopsies was negligible (Figures 1 and 2). Although the tumor expression of IF1 did not show significant differences between patients with clinical-pathological markers relevant for tumor progression, such as nodal involvement, tumor size and histological grade (Supplementary Table S1), it was significantly diminished in the poor prognosis groups of lobular and hormone receptor negative carcinomas when compared with ductal and hormone receptor positive carcinomas, respectively (Supplementary Table S1).
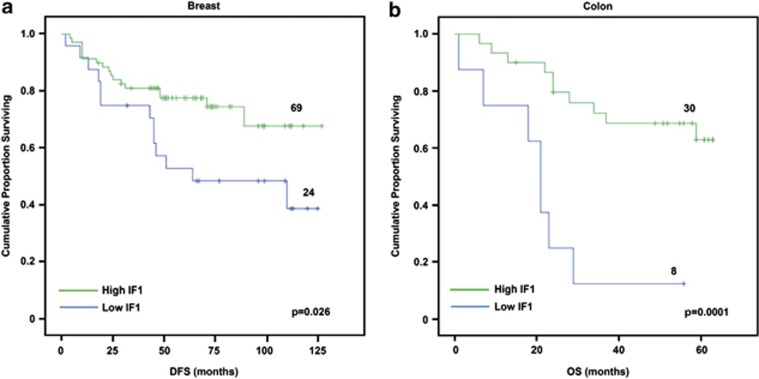
From the molecular point of view, the expression of IF1 inversely correlated with the bioenergetic cellular index35 of the tumors (R=−0.437; P=0.0001), suggesting that the increased expression of IF1 parallels the program of repression of the bioenergetic activity of mitochondria in cancer cells. Surprisingly, Kaplan–Meier survival analysis revealed that a low tumor expression of IF1 predicted a higher rate for disease-recurrence in breast cancer patients (Figure 8a).
Figure 8.
Kaplan–Meier survival analyses reveal the clinical relevance of IF1. (a) Protein samples from breast tumor biopsies were analyzed by western blot for the expression level of IF1. Kaplan–Meier disease-free survival analysis for 93 breast cancer patients stratified by the tumor expression level of IF1. The plot shows a significant association of low IF1 expression with a poor outcome for the patients. The log-rank test P-value is shown. (b) Protein samples from normal and tumor biopsies from a cohort of patients with colorectal cancer were analyzed by reverse phase protein microarrays to quantify IF1 (see Supplementary Figure S4). Kaplan–Meier overall survival analysis for 38 colon cancer patients stratified by the tumor quantity of IF1. The plot shows a significant association of low IF1 expression with a poor outcome for the patients. The log-rank test P-value is shown.
Clinical relevance of IF1 in colon cancer
Next, we determine the quantity of IF1 in normal and tumor biopsies in a cohort of colon cancer patients in which the markers of energy metabolism have been previously quantified.37 The amount of IF1 was determined using reverse phase protein microarrays (Supplementary Figure S5).37 The results in Supplementary Table S2 summarize the clinicopathological characteristics of the cohort of patients studied and the quantity of IF1 in the carcinomas according to the clinical information. Colon carcinomas showed a significant twofold increase in the amount of IF1 when compared to normal colon biopsies (Supplementary Table S2). The tumor content of IF1 did not show significant differences between patients with clinical-pathological markers relevant for tumor progression (Supplementary Table S2).
In colon cancer, the expression of IF1 also inversely correlated with the bioenergetic cellular index37 of the tumors (R=−0.526; P=0.001), once again suggesting that the increased expression of IF1 in colon cancer cells parallels the program of metabolic reprogramming experienced by these carcinomas. Kaplan–Meier survival analysis also revealed that a low tumor expression of IF1 predicted a worst overall prognosis for colon cancer patients (Figure 8b).
Discussion
We show that IF1 is highly overexpressed in all human carcinomas of the colon, lung, breast and ovary showing negligible expression in their corresponding normal tissues. IF1 promotes the activation of aerobic glycolysis and generates a concurrent ROS signal in mitochondria that activates cell-type specific adaptive responses aimed at preventing death in the lung, breast and ovarian cancer cells. Mechanistically, IF1 mimics the effects of oligomycin, an inhibitor of the H+-ATP synthase.13, 29 Remarkably, we show that the regulation of the expression of IF1 in carcinomas of the colon, lung, breast and ovary is exerted at post-transcriptional levels. In fact, we demonstrate that IF1 is a short-lived protein that is degraded by mitochondrial serine-proteases. Overall, we provide the first demonstration supporting that regulation of the synthesis and/or degradation of a mitochondrial protein involved in the control of oxidative phosphorylation has a master role in metabolic rewiring and in signaling cell-death resistance in prevalent human carcinomas. Moreover, we show that IF1 expression has relevance as a predictive marker for clinical outcome in breast and colon cancer patients.
The expression level of IF1 varies greatly within normal human tissues. Consistent with the role of IF1 as inhibitor of the H+-ATP synthase,28, 29 a high expression of IF1 in normal tissues such as heart, liver and kidney would imply the partial mass-action-mediated inhibition of oxidative phosphorylation and thus a limitation in cellular ATP availability. This situation is obviously not possible because these tissues have a very-high metabolic demand. Thus, our findings suggest that in addition to the well characterized pH regulated binding of IF1 to the H+-ATP synthase,38 a mechanism should exist in tissues with high metabolic demand to promote the mass-action-mediated inhibition of IF1 on the H+-ATP synthase activity.28 In this regard, it has been described that IF1 also binds other membrane proteins of mitochondria in a pH and ΔΨm independent manner39, 40 hampering its activity as an inhibitor of the ATPase.40 Alternatively, potential tissue-specific post-translational modifications of IF1 could explain the differential activity exerted by IF1 in different human tissues.
At variance with other mitochondrial proteins41 we show that IF1 is a short-lived protein with a turnover in the range of minutes. This finding is of utmost importance to understand the bioenergetic activity of mitochondria. In fact, a rapid turnover of the protein would allow cells to quickly adapt the output of ATP by oxidative phosphorylation to changing physiological cues. One can speculate that mutations in cancer genes or other epigenetic events of the tumor microenvironment switch-on the mechanisms that promote a high expression of IF1 in the tumor. We suggest that an increase in translation of IF1 mRNA and/or in the stability of the protein should occur in human carcinomas to provoke the overwhelming expression of IF1 observed in these tumors. The control of IF1 translation is presently unknown and it might involve regulatory proteins and microRNAs as we have shown for β-F1-ATPase mRNA.27, 42, 43
IER3 gene has been shown to suppress ROS production and to render IF1 prone to proteolytic digestion.31 Paradoxically, and in contrast with findings in ovarian44 and pancreatic45 cancer our results indicate that IER3 is overexpressed in the lung, breast and colon cancer. Moreover, we found no correlation between IER3 and IF1 expression suggesting that the control of the degradation of IF1 is more complex than originally anticipated and that the participation of IER3 might depend on the cell-type analyzed. An alternative explanation for the accumulation of IF1 in carcinomas could be the partial inactivation of the mitochondrial serine-protease involved in its turnover. In this regard, the Lon protease has been reported to diminish its activity during ageing.46 Moreover, cancer cells exhibit high basal levels of oxidative stress5 and the Lon protease is particularly vulnerable to inactivation by ROS.47 The implication of ClpXP in human pathology is scant despite its relevance in the degradation of proteins involved in metabolic reprogramming.48 As previously discussed, potential oncogene- and/or metabolic-driven post-translational modifications of IF1 could also explain its accumulation in carcinomas if such changes hamper the mitochondrial pathway of IF1-degradation.
Despite the large structural and molecular differences of mitochondria in mammalian cells,49 but consistent with previous findings in colon cancer,29 we show that the IF1-mediated inhibition of the H+-ATP synthase switches on aerobic glycolysis and generates a mitochondrial ROS signal in all cancer cells studied. Mitochondrial ROS signaling represents a pathway of retrograde communication to the nucleus of the cell that influences adaptive cellular responses.50 The results herein indicate that whereas mitochondrial ROS signaling triggers protection against cell death in all cellular types studied, it is unable to stimulate proliferation in the lung, breast and ovarian carcinomas, what is at variance with colon cancer cells.29 These findings indicate the existence of common and cell-type specific programs of nuclear response to mitochondrial ROS signaling. The effect of ROS on the cellular response depends on the level51 and site52 at which they are being produced. Moreover, ROS interact with diverse signaling pathways being transcription factor NFκB, a crucial regulator of adaptive responses related with survival.53, 54 In colon cancer cells, IF1 mediated the activation of the canonical NFκB pathway of survival.29 The results herein support that although IF1 is able to trigger the ROS-mediated transcriptional activation of the NFκB promoter as a common response to the lung, breast and ovarian cancer cells, only lung carcinomas seem to activate the same NFκB-mediated survival pathway. These results emphasize the critical function that NFκB has in signaling lung tumor development, in agreement with previous findings in a mouse model of lung carcinogenesis,55 and highlight the need of specific studies aimed at unveiling the IF1-mediated pathways of survival that are activated in breast and ovarian cancer cells.
Redox regulation has an essential role in malignancies29, 56, 57 but the mechanisms of its actions and its impact in tumor prognosis remain unclear. Contrary to what would be expected for an oncogenic protein,58 we found that breast and colon cancer patients with high tumor expression of IF1 have a better prognosis. There are other examples in the literature illustrating similar paradoxes. For instance, miR-200s that modulate the oxidative stress response increase tumor growth in mouse models.59 However, a high expression of miR-200s is linked to a favorable prognosis,60 whereas downregulation of miR-200s is associated with relapse in patients with ovarian cancer.61 Similarly, isocitrate dehydrogenase mutations are paradoxically associated with better survival in glioma patients.62 As a low expression of IF1 in carcinomas predicts a shorter time for relapse or death of the patient, we suggest that cells with low expression of IF1 are more likely to metastasize. In this regard, it is possible that cells with high expression of IF1, which have a low bioenergetic signature become more vulnerable to metabolic or other forms of stress during detachment and/or become more easily recognized by the immune system.2 Indeed, cells with a low bioenergetic signature are addicted to glucose and are more sensitive to glucose deprivation and the inhibition of glycolysis.24, 63
Overall, we document that the short-lived inhibitor of the mitochondrial H+-ATP synthase is overexpressed in the colon, lung, breast and ovarian cancer mastering the reprogramming of energy metabolism and signaling a cell-death resistance phenotype. Moreover, we support its potential as a marker of clinical outcome in breast and colon cancer patients. We stress that specific studies and animals models are needed to unveil the molecular and cellular biology of IF1 in the different cell types of mammals.
Materials and methods
Patient specimens and protein extraction
Frozen tissue obtained from surgical specimens of untreated cancer patients with primary adenocarcinomas of the breast, colon, stomach, kidney and lung were obtained from the Banco de Tejidos y Tumores, IDIBAPS (Instituto de Investigaciones Biomédicas Pi y Suñer), Hospital Clinic, Barcelona, Spain.64 A collection of frozen tissue sections obtained from surgical specimens of (i) ninety-three patients who had an operation for invasive breast carcinoma at the Hospital Universitario La Paz between 1991 and 200035, 36 and (ii) of untreated cancer patients with primary colorectal adenocarcinomas enrolled in the incident Spanish CRC Epicolon Study and prospectively followed during 5 years37, 65 obtained from the Banco de Tejidos y Tumores, Hospital Meixoeiro, Vigo, Spain were also used (see Supplementary Information for Bioethic details).
Cell cultures, treatments, transfections and small interfering RNA silencing
The Human cervical (HeLa), breast (HS578T, NCI-ADR-RES, BT549, MCF12), lung (A549, HOP62), colon (KM12, HCT116) and ovarian (OVCAR8, OVCAR 3) cells were grown following the supplierś instructions. When needed, cells were left untreated or treated with 200 μℳ CoCl2 (Sigma-Aldrich, St Louise, MO, USA) for 6 h. For inhibition of mitochondrial serine-proteases HCT116 cells were treated with 400 μℳ of 4-(2-aminoethyl) benzenosulfonyl fluoride hydrochloride for the indicated time. Transfection and silencing experiments were performed as recently described.29
Protein fractionation and western blots
Cell lysis and protein fractionation were carried out as described.27 The primary antibodies used were: anti-β-actin (Sigma-Aldrich, 1:20000), anti-β-F1-ATPase (Acebo et al.66 1:20000), anti-IkBα and anti-Bcl-xL (Cell Signaling Technology, Inc., Danvers, MA, USA, 1:1000), anti-hypoxia-inducible factor-1α (Santa Cruz Biotechnology Inc., Dallas, TX, USA, 1:150) and anti-IF1 (Sanchez-Cenizo et al.28 1:200).
Immunohistochemistry
Cancer Survey Tissue Microarrays (OriGene (Rockville, MD, USA)) containing 5 μm sections of formalin-fixed normal and tumor specimens of the breast, colon, lung, kidney, ovarian and endometrial tissues were immunostained using the monoclonal anti-IF1 (1:200) antibody as previously described.19 Sections were counterstained with hematoxylin.
Quantification of mRNA expression
Human β-F1-ATPase and IF1 mRNA levels in normal and tumor tissues were carried out by quantitative PCR using the breast (BCRT101), ovarian (HORT102), colon (HCRT103) and lung (HLRT104) TissueScan Tissue quantitative PCR Arrays from OriGene Technologies, Inc. For detailed pathological information of these patients see Supplementary Table S3. Real-time quantitative PCR was performed as described.27 The following forward (F) and reverse (R) primers were used to amplify human β-F1-ATPase and IF1 cDNAs: F: 5′- CAGCAGATTTTGGCAGGTG-3′, R: 5′-CTTCAATGGGTCCCACCATA-3′ F: 5′-GGGCCTTCGGAAAGAGAG-3′ and R: 5′-TTCAAAGCTGCCAGTTGTTC-3′, respectively.
Metabolic labeling and immunoprecipitation
Metabolic labeling was initiated by addition to the culture medium 0.65 mCi of [35S]-methionine/ml. Duration of the pulse was 1 h. At the indicated time-points IF1 was immunoprecipitated from cellular lysates using G-sepharose precoated with 12 μg of anti-IF1 IgGs.67
Quantification of IF1 in colorectal adenocarcinoma biopsies using reverse phase protein microarrays
IF1 was expressed and purified as detailed.28 Samples from colorectal adenocarcinoma patients were diluted in PBS to a final protein concentration of 1 μg/μl before printing onto nitrocellulose-coated glass slides (FAST Slides, Scheleicher & Schuell BioScience, Inc.) using a BioOdyssey Calligrapher MiniArrayer printer (Bio-Rad Laboratories, Inc., Keene, NH, USA) as recently described.37 Arrays were incubated with anti-IF1 (1 μg/ml) followed by incubation with a donkey anti-mouse conjugated with alexa-488 (Life Technologies, Grand Island, NY, USA). Microarrays were scanned using a Typhoon 9410 scanner (GE Healthcare Bio-Sciences Corp., Piscataway, NJ, USA). The mean fluorescent intensity of the spots was quantified using Image J software (N.I.H., USA) and converted into pg of protein/ng of total protein using the fluorescent intensity units obtained in the respective standard curve of recombinant protein.37
Bioinformatic Search of the ATPIF1 gene
UCSC Genome Browser (http://genome.ucsc.edu/) was used. Chip-sep data was obtained from ENCODE database.
Determination of reactive oxygen species
Where indicated, ∼2 × 105 cells were incubated overnight with 10–20 nℳ of MQ. The intracellular production of superoxide radical was monitored by flow cytometry using 5 μℳ MitoSOX (Invitrogen) incubated 15 min at 37 °C.29 Cells were analyzed in a FACScan. For each analysis 10 000 events were recorded.
Other methods
Details for the determination of aerobic glycolysis, proliferation, cell death and NFκB promoter activity have been recently provided.29 Where indicated 5 nℳ of MQ was added to the incubation.
Statistical analysis
Distribution of molecular markers and other categorical variables were compared by χ2 and Student's t-test. The statistical significance of linear regressions was assessed by Pearson's correlation t-test. To determine the association between the expression level of IF1 with disease-free survival and overall survival the cutoff point used to define high- and low-risk groups was the mean value of protein expression in normal breast or colon samples. Survival curves were derived from Kaplan–Meier estimates and compared by log-rank test. Statistical test were two-side at the 5% level of significance. All computations were carried out using SPSS, version 17.0 (IBM Corporation, Armonk, NY, USA).
Acknowledgments
We are indebted to Professor Manuel González-Barón and other members of the Oncopaz group for support and encouragement. We thank Mrs. Margarita Chamorro, Cristina Núñez de Arenas and Irma Joyce Diaz de Almeida for expert technical assistance. IMR, JGB, FS, IMW and LSC and were supported by predoctoral fellowships from JAE-CSIC (IMR), FPI-MEC (JGB, LSC), FPI-UAM (FS), FPU-MEC (IMW), Spain. LF is supported by a Juan de la Cierva Grant (JCI2009-03918) and Fundación Asociación Española Contra el Cáncer. This work was supported by grants from the Ministerio de Educación y Ciencia (BFU2010-18903), the Centro de Investigación Biomédica en Red de Enfermedades Raras (CIBERER), ISCIII, Madrid and Comunidad de Madrid (S2011/BMD-2402), Spain. The CBMSO receives an institutional grant from the Fundación Ramón Areces.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogenesis website (http://www.nature.com/oncsis).
Supplementary Material
References
- Cuezva JM, Ortega AD, Willers I, Sanchez-Cenizo L, Aldea M, Sanchez-Arago M. The tumor suppressor function of mitochondria: translation into the clinics. Biochim Biophys Acta. 2009;1792:1145–1158. doi: 10.1016/j.bbadis.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer's Achilles' heel. Cancer Cell. 2008;13:472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- Almeida A, Bolanos JP, Moncada S. E3 ubiquitin ligase APC/C-Cdh1 accounts for the Warburg effect by linking glycolysis to cell proliferation. Proc Natl Acad Sci USA. 2010;107:738–741. doi: 10.1073/pnas.0913668107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17:330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissell MJ, Hines WC. Why don't we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat Med. 2011;17:320–329. doi: 10.1038/nm.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Arago M, Chamorro M, Cuezva JM. Selection of cancer cells with repressed mitochondria triggers colon cancer progression. Carcinogenesis. 2010;31:567–576. doi: 10.1093/carcin/bgq012. [DOI] [PubMed] [Google Scholar]
- Boyer PD. The ATP synthase. A splendid molecular machine. Annu Rev Biochem. 1997;66:717–749. doi: 10.1146/annurev.biochem.66.1.717. [DOI] [PubMed] [Google Scholar]
- Matsuyama S, Xu Q, Velours J, Reed JC. The mitochondrial F0F1-ATPase proton pump is required for function of the proapoptotic protein Bax in yeast and mammalian cells. Mol Cell. 1998;1:327–336. doi: 10.1016/s1097-2765(00)80033-7. [DOI] [PubMed] [Google Scholar]
- Santamaria G, Martinez-Diez M, Fabregat I, Cuezva JM. Efficient execution of cell death in non-glycolytic cells requires the generation of ROS controlled by the activity of mitochondrial H+-ATP synthase. Carcinogenesis. 2006;27:925–935. doi: 10.1093/carcin/bgi315. [DOI] [PubMed] [Google Scholar]
- Dey R, Moraes CT. Lack of oxidative phosphorylation and low mitochondrial membrane potential decrease susceptibility to apoptosis and do not modulate the protective effect of Bcl-x(L) in osteosarcoma cells. J Biol Chem. 2000;275:7087–7094. doi: 10.1074/jbc.275.10.7087. [DOI] [PubMed] [Google Scholar]
- Tomiyama A, Serizawa S, Tachibana K, Sakurada K, Samejima H, Kuchino Y, et al. Critical role for mitochondrial oxidative phosphorylation in the activation of tumor suppressors Bax and Bak. J Natl Cancer Inst. 2006;98:1462–1473. doi: 10.1093/jnci/djj395. [DOI] [PubMed] [Google Scholar]
- Schulz TJ, Thierbach R, Voigt A, Drewes G, Mietzner B, Steinberg P, et al. Induction of oxidative metabolism by mitochondrial frataxin inhibits cancer growth: Otto Warburg revisited. J Biol Chem. 2006;281:977–981. doi: 10.1074/jbc.M511064200. [DOI] [PubMed] [Google Scholar]
- D'Errico I, Salvatore L, Murzilli S, Lo Sasso G, Latorre D, Martelli N, et al. Peroxisome proliferator-activated receptor-gamma coactivator 1-alpha (PGC1alpha) is a metabolic regulator of intestinal epithelial cell fate. Proc Natl Acad Sci USA. 2011;108:6603–6608. doi: 10.1073/pnas.1016354108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffer M, Bacolod MD, Zuk O, Giardina SF, Pincas H, Barany F, et al. Association of survival and disease progression with chromosomal instability: a genomic exploration of colorectal cancer. Proc Natl Acad Sci USA. 2009;106:7131–7136. doi: 10.1073/pnas.0902232106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuezva JM, Krajewska M, de Heredia ML, Krajewski S, Santamaria G, Kim H, et al. The bioenergetic signature of cancer: a marker of tumor progression. Cancer Res. 2002;62:6674–6681. [PubMed] [Google Scholar]
- Lopez-Rios F, Sanchez-Arago M, Garcia-Garcia E, Ortega AD, Berrendero JR, Pozo-Rodriguez F, et al. Loss of the mitochondrial bioenergetic capacity underlies the glucose avidity of carcinomas. Cancer Res. 2007;67:9013–9017. doi: 10.1158/0008-5472.CAN-07-1678. [DOI] [PubMed] [Google Scholar]
- Shin YK, Yoo BC, Chang HJ, Jeon E, Hong SH, Jung MS, et al. Down-regulation of mitochondrial F1F0-ATP synthase in human colon cancer cells with induced 5-fluorouracil resistance. Cancer Res. 2005;65:3162–3170. doi: 10.1158/0008-5472.CAN-04-3300. [DOI] [PubMed] [Google Scholar]
- Hernlund E, Hjerpe E, Avall-Lundqvist E, Shoshan M. Ovarian carcinoma cells with low levels of beta-F1-ATPase are sensitive to combined platinum and 2-deoxy-D-glucose treatment. Mol Cancer Ther. 2009;8:1916–1923. doi: 10.1158/1535-7163.MCT-09-0179. [DOI] [PubMed] [Google Scholar]
- Li RJ, Zhang GS, Chen YH, Zhu JF, Lu QJ, Gong FJ, et al. Down-regulation of mitochondrial ATPase by hypermethylation mechanism in chronic myeloid leukemia is associated with multidrug resistance. Ann Oncol. 2010;7:1506–1514. doi: 10.1093/annonc/mdp569. [DOI] [PubMed] [Google Scholar]
- Sanchez-Arago M, Cuezva JM. The bioenergetic signature of isogenic colon cancer cells predicts the cell death response to treatment with 3-bromopyruvate, iodoacetate or 5-fluorouracil. J Transl Med. 2011;9:19. doi: 10.1186/1479-5876-9-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Heredia ML, Izquierdo JM, Cuezva JM. A conserved mechanism for controlling the translation of beta-F1-ATPase mRNA between the fetal liver and cancer cells. J Biol Chem. 2000;275:7430–7437. doi: 10.1074/jbc.275.10.7430. [DOI] [PubMed] [Google Scholar]
- Willers IM, Isidoro A, Ortega AD, Fernandez PL, Cuezva JM. Selective inhibition of beta-F1-ATPase mRNA translation in human tumours. Biochem J. 2010;426:319–326. doi: 10.1042/BJ20091570. [DOI] [PubMed] [Google Scholar]
- Ortega AD, Willers IM, Sala S, Cuezva JM. Human G3BP1 interacts with beta-F1-ATPase mRNA and inhibits its translation. J Cell Sci. 2010;123:2685–2696. doi: 10.1242/jcs.065920. [DOI] [PubMed] [Google Scholar]
- Sanchez-Cenizo L, Formentini L, Aldea M, Ortega AD, Garcia-Huerta P, Sanchez-Arago M, et al. Up-regulation of the ATPase inhibitory factor 1 (IF1) of the mitochondrial H+-ATP synthase in human tumors mediates the metabolic shift of cancer cells to a Warburg phenotype. J Biol Chem. 2010;285:25308–25313. doi: 10.1074/jbc.M110.146480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formentini L, Sánchez-Aragó M, Sánchez-Cenizo L, Cuezva JM. The mitochondrial ATPase Inhibitory Factor 1 (IF1) triggers a ROS-mediated retrograde pro-survival and proliferative response. Mol Cell. 2012;45:731–742. doi: 10.1016/j.molcel.2012.01.008. [DOI] [PubMed] [Google Scholar]
- Huang LJ, Chuang IC, Dong HP, Yang RC. Hypoxia-inducible factor 1alpha regulates the expression of the mitochondrial ATPase inhibitor protein (IF1) in rat liver. Shock. 2011;36:90–96. doi: 10.1097/SHK.0b013e318219ff2a. [DOI] [PubMed] [Google Scholar]
- Shen L, Zhi L, Hu W, Wu MX. IEX-1 targets mitochondrial F1Fo-ATPase inhibitor for degradation. Cell Death Differ. 2009;16:603–612. doi: 10.1038/cdd.2008.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppen M, Langer T. Protein degradation within mitochondria: versatile activities of AAA proteases and other peptidases. Crit Rev Biochem Mol Biol. 2007;42:221–242. doi: 10.1080/10409230701380452. [DOI] [PubMed] [Google Scholar]
- Formentini L, Martinez-Reyes I, Cuezva JM. The mitochondrial bioenergetic capacity of carcinomas. IUBMB Life. 2010;62:554–560. doi: 10.1002/iub.352. [DOI] [PubMed] [Google Scholar]
- Kelso GF, Porteous CM, Coulter CV, Hughes G, Porteous WK, Ledgerwood EC, et al. Selective targeting of a redox-active ubiquinone to mitochondria within cells: antioxidant and antiapoptotic properties. J Biol Chem. 2001;276:4588–4596. doi: 10.1074/jbc.M009093200. [DOI] [PubMed] [Google Scholar]
- Isidoro A, Casado E, Redondo A, Acebo P, Espinosa E, Alonso AM, et al. Breast carcinomas fulfill the Warburg hypothesis and provide metabolic markers of cancer prognosis. Carcinogenesis. 2005;26:2095–2104. doi: 10.1093/carcin/bgi188. [DOI] [PubMed] [Google Scholar]
- Ortega AD, Sala S, Espinosa E, Gonzalez-Baron M, Cuezva JM. HuR and the bioenergetic signature of breast cancer: a low tumor expression of the RNA-binding protein predicts a higher risk of disease recurrence. Carcinogenesis. 2008;29:2053–2061. doi: 10.1093/carcin/bgn185. [DOI] [PubMed] [Google Scholar]
- Aldea M, Clofent J, Nunez de Arenas C, Chamorro M, Velasco M, Berrendero JR, et al. Reverse phase protein microarrays quantify and validate the bioenergetic signature as biomarker in colorectal cancer. Cancer Lett. 2011;311:210–218. doi: 10.1016/j.canlet.2011.07.022. [DOI] [PubMed] [Google Scholar]
- Cabezon E, Montgomery MG, Leslie AG, Walker JE. The structure of bovine F1-ATPase in complex with its regulatory protein IF1. Nat Struct Biol. 2003;10:744–750. doi: 10.1038/nsb966. [DOI] [PubMed] [Google Scholar]
- Ichikawa N, Nakabayashi K, Hashimoto T. A yeast mitochondrial ATPase inhibitor interacts with three proteins that are easy to dissociate from the mitochondrial inner membrane. J Biochem. 2002;132:649–654. doi: 10.1093/oxfordjournals.jbchem.a003269. [DOI] [PubMed] [Google Scholar]
- Lopez-Mediavilla C, Vigny H, Godinot C. Docking the mitochondrial inhibitor protein IF1 to a membrane receptor different from the F1-ATPase beta subunit. Eur J Biochem. 1993;215:487–496. doi: 10.1111/j.1432-1033.1993.tb18058.x. [DOI] [PubMed] [Google Scholar]
- Grisolía S, Hernandez-Yago J, Knecht E. Regulation of mitochondrial protein concentration: a plausible model which may permit assessing protein turnover. Curr Top Cell Regul. 1985;27:387–396. doi: 10.1016/b978-0-12-152827-0.50040-2. [DOI] [PubMed] [Google Scholar]
- Izquierdo JM, Cuezva JM. Control of the translational efficiency of beta-F1-ATPase mRNA depends on the regulation of a protein that binds the 3' untranslated region of the mRNA. Mol Cell Biol. 1997;17:5255–5268. doi: 10.1128/mcb.17.9.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willers IM, Martínez-Reyes I, Martínez-Diez M, Cuezva J. miR-127-5p targets the 3'UTR of human β-F1-ATPase mRNA and inhibits its translation. Biochim Biophys Acta-Bioenergetics. 2012;1817:838–848. doi: 10.1016/j.bbabio.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Han L, Geng L, Liu X, Shi H, He W, Wu MX. Clinical significance of IEX-1 expression in ovarian carcinoma. Ultrastruct Pathol. 2011;35:260–266. doi: 10.3109/01913123.2011.608916. [DOI] [PubMed] [Google Scholar]
- Sasada T, Azuma K, Hirai T, Hashida H, Kanai M, Yanagawa T, et al. Prognostic significance of the immediate early response gene X-1 (IEX-1) expression in pancreatic cancer. Ann Surg Oncol. 2008;15:609–617. doi: 10.1245/s10434-007-9669-0. [DOI] [PubMed] [Google Scholar]
- Bakala H, Delaval E, Hamelin M, Bismuth J, Borot-Laloi C, Corman B, et al. Changes in rat liver mitochondria with aging. Lon protease-like reactivity and N(epsilon)-carboxymethyllysine accumulation in the matrix. Eur J Biochem. 2003;270:2295–2302. doi: 10.1046/j.1432-1033.2003.03598.x. [DOI] [PubMed] [Google Scholar]
- Stanyer L, Jorgensen W, Hori O, Clark JB, Heales SJ. Inactivation of brain mitochondrial Lon protease by peroxynitrite precedes electron transport chain dysfunction. Neurochem Int. 2008;53:95–101. doi: 10.1016/j.neuint.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Flynn JM, Neher SB, Kim YI, Sauer RT, Baker TA. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol Cell. 2003;11:671–683. doi: 10.1016/s1097-2765(03)00060-1. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Bunkenborg J, Olsen JV, Hjerrild M, Wisniewski JR, Stahl E, et al. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115:629–640. doi: 10.1016/s0092-8674(03)00926-7. [DOI] [PubMed] [Google Scholar]
- Butow RA, Avadhani NG. Mitochondrial signaling: the retrograde response. Mol Cell. 2004;14:1–15. doi: 10.1016/s1097-2765(04)00179-0. [DOI] [PubMed] [Google Scholar]
- Hamanaka RB, Chandel NS. Mitochondrial reactive oxygen species regulate cellular signaling and dictate biological outcomes. Trends Biochem Sci. 2010;35:505–513. doi: 10.1016/j.tibs.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T. Signal transduction by reactive oxygen species. J Cell Biol. 2011;194:7–15. doi: 10.1083/jcb.201102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- Morgan MJ, Liu ZG. Crosstalk of reactive oxygen species and NF-kappaB signaling. Cell Res. 2011;21:103–115. doi: 10.1038/cr.2010.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E, Dooley AL, Feldser DM, Shen L, Turk E, Ouyang C, et al. Requirement for NF-kappaB signalling in a mouse model of lung adenocarcinoma. Nature. 2009;462:104–107. doi: 10.1038/nature08462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, Imanishi H, et al. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320:661–664. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- Weinberg F, Hamanaka R, Wheaton WW, Weinberg S, Joseph J, Lopez M, et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci USA. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Arago M, Formentini L, Garcia-Bermudez J, Cuezva JM. IF1 reprograms energy metabolism and signals the oncogenic phenotype in cancer. Cell Cycle. 2012;11:2963–2964. doi: 10.4161/cc.21387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateescu B, Batista L, Cardon M, Gruosso T, de Feraudy Y, Mariani O, et al. miR-141 and miR-200a act on ovarian tumorigenesis by controlling oxidative stress response. Nat Med. 2011;17:1627–1635. doi: 10.1038/nm.2512. [DOI] [PubMed] [Google Scholar]
- Hu X, Macdonald DM, Huettner PC, Feng Z, El Naqa IM, Schwarz JK, et al. A miR-200 microRNA cluster as prognostic marker in advanced ovarian cancer. Gynecol Oncol. 2009;114:457–464. doi: 10.1016/j.ygyno.2009.05.022. [DOI] [PubMed] [Google Scholar]
- Marchini S, Cavalieri D, Fruscio R, Calura E, Garavaglia D, Nerini I, et al. Association between miR-200c and the survival of patients with stage I epithelial ovarian cancer: a retrospective study of two independent tumour tissue collections. Lancet Oncology. 2011;12:273–285. doi: 10.1016/S1470-2045(11)70012-2. [DOI] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelicano H, Martin DS, Xu RH, Huang P. Glycolysis inhibition for anticancer treatment. Oncogene. 2006;25:4633–4646. doi: 10.1038/sj.onc.1209597. [DOI] [PubMed] [Google Scholar]
- Isidoro A, Martinez M, Fernandez PL, Ortega AD, Santamaria G, Chamorro M, et al. Alteration of the bioenergetic phenotype of mitochondria is a hallmark of breast, gastric, lung and oesophageal cancer. Biochem J. 2004;378:17–20. doi: 10.1042/BJ20031541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jover R, Zapater P, Castells A, Llor X, Andreu M, Cubiella J, et al. The efficacy of adjuvant chemotherapy with 5-fluorouracil in colorectal cancer depends on the mismatch repair status. Eur J Cancer. 2009;45:365–373. doi: 10.1016/j.ejca.2008.07.016. [DOI] [PubMed] [Google Scholar]
- Acebo P, Giner D, Calvo P, Blanco-Rivero A, Ortega AD, Fernandez PL, et al. Cancer abolishes the tissue type-specific differences in the phenotype of energetic metabolism. Transl Oncol. 2009;2:138–145. doi: 10.1593/tlo.09106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Reyes I, Sanchez-Arago M, Cuezva JM. AMPK and GCN2-ATF4 signal the repression of mitochondria in colon cancer cells. Biochem J. 2012;444:249–259. doi: 10.1042/BJ20111829. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.