Abstract
Myotubular myopathy (MIM#310400), the X-linked form of Centronuclear myopathy (CNM) is mainly characterized by neonatal hypotonia and inability to maintain unassisted respiration. The MTM1 gene, responsible for this disease, encodes myotubularin – a lipidic phosphatase involved in vesicle trafficking regulation and maturation. Recently, it was shown that myotubularin interacts with desmin, being a major regulator of intermediate filaments. We report the development of a locus-specific database for MTM1 using the Leiden Open Variation database software (http://www.lovd.nl/MTM1), with data collated for 474 mutations identified in 472 patients (by June 2012). Among the entries are a total of 25 new mutations, including a large deletion encompassing introns 2–15. During database implementation it was noticed that no large duplications had been reported. We tested a group of eight uncharacterized CNM patients for this specific type of mutation, by multiple ligation-dependent probe amplification (MLPA) analysis. A large duplication spanning exons 1–5 was identified in a boy with a mild phenotype, with results pointing toward possible somatic mosaicism. Further characterization revealed that this duplication causes an in-frame deletion at the mRNA level (r.343_444del). Results obtained with a next generation sequencing approach suggested that the duplication extends into the neighboring MAMLD1 gene and subsequent cDNA analysis detected the presence of a MTM1/MAMLD1 fusion transcript. A complex rearrangement involving the duplication of exon 10 has since been reported, with detection also enabled by MLPA analysis. It is thus conceivable that large duplications in MTM1 may account for a number of CNM cases that have remained genetically unresolved.
Keywords: locus-specific database, MTM1, novel mutations
Introduction
Congenital myopathies are a heterogeneous group of diseases, generally characterized by muscle weakness, and with onset at birth or during infancy. These myopathies have characteristic histological hallmarks in muscle biopsy allowing the differential classification in distinct entities: centronuclear myopathy (CNM), core myopathy (centralcore and minicore diseases) and nemaline rod myopathy.1, 2 In CNM, the most prominent histopathological features include hypotrophy of type 1 fibers and a high frequency of centrally located nuclei with perinuclear halos lacking myofilaments and occupied by mitochondrial and glycogen aggregates.1 Several genes are reported to be associated with CNM; these include MTM1 in the X-linked form,3, 4 DNM2 and MTMR14 in the autosomal dominant forms,4, 5, 6 BIN1 and RYR1 associated with the autosomal recessive forms.4,7, 8, 9
X-linked myotubular myopathy (XLMTM; MIM 310400) has a prevalence of approximately 1/50 000 males and is characterized by severe hypotonia present at birth and inability to maintain sustained spontaneous respiration.10 Different authors have proposed that patients be classified according to their phenotype, as: (i) severe – characteristic facial features, markedly delayed motor milestones and requiring prolonged ventilatory support (>12 h); (ii) moderate – more rapid acquirement of motor milestones and independent respiration for >12 h per day; (iii) mild – motor milestones slightly delayed and independent spontaneous respiratory function achieved after the neonatal period.11, 12 Carrier females are usually asymptomatic, however there are several records of manifesting heterozygotes due to skewed X chromosome inactivation.13, 14, 15, 16, 17, 18
The MTM1 gene (in Xq28) is composed of 15 exons and has an open reading frame of 1.8 kb encoding the myotubularin protein. Structurally, myotubularins are constituted by four characteristic domains: the protein tyrosine phosphatase (PTP), the predicted GRAM (glucosyltransferases, Rab-like GTPases activators and myotubularins), the RID (Rac-induced recruitment domain) and SID (SET-interaction domain). Functionally, myotubularin is a phosphatase acting specifically on PtdIns3P and PtdIns(3,5)P2, two phosphoinositides (PIs). PIs participate in the regulation of various cellular mechanisms by direct binding to PI-binding domains of effector proteins (that control membrane/vesicular trafficking) and subsequent recruitment/activation of these at specific membrane sites. PtdIns3P and PtdIns(3,5)P2 have a direct role in the endosomal-lysosomal pathway.19 The PTP-catalytic domain of myotubularins is responsible for the phosphoester hydrolysis of the 3-phosphate of PIs. This hydrolysis involves two residues of cysteine and arginine located on a Cys-X5-Arg motif, characteristic for the PTP domain.20 The loss of phosphatase activity or the production of truncated proteins as a result of MTM1 mutations could lead to abnormal dephosphorylation of PtdIns3P/PtdIns(3,5)P2 and subsequent abnormal trafficking of the effector proteins of the endosomal-lysosomal pathway.19 Similar results were observed with mutations located in the GRAM domain of myotubularin. This domain of about 70 amino acids is responsible for PtdIns(3,5)P2 binding.21 Recently mitochondrial homeostasis in muscle fibers and regulation of the desmin cytoskeletal system was attributed to mytobularin. It was experimentally demonstrated that myotubularin interacts with desmin, and that this complex is disrupted by specific mutations in the MTM1 gene.22
It is recognized that, despite the genetic advances in this field and the large number of cases reported,23 a significant number of CNM cases remain genetically unresolved. This may be explained either by the involvement of further gene loci or by the presence of mutations in known genes, that are not detectable by routine techniques. During the development of a locus-specific database (LSDB) for the MTM1 gene we noticed that no large duplications (involving one or more exons) had been reported for this gene. This observation led us to investigate the possibility of their occurrence in molecularly unresolved CNM patients. Accordingly, we report the first multi-exonic duplication in MTM1 (exons 1–5) detected in a male patient with a mild XLMTM phenotype. In line with this finding, a complex rearrangement involving the duplication of exon 10 was recently published.24
Materials and methods
MTM1-LOVD database development
The MTM1 mutation database was implemented using LOVD v2.0 software,25 and is integrated in Leiden Muscular Dystrophy pages (http://www.dmd.nl/). Currently (by 29th June 2012), this large database installation displays a total of 141 genes with 70 512 variants (9265 unique) described in 55 775 individuals. The MTM1 database is subdivided into two main tables, for variant data (n=20) and for patient/clinical items (n=22), the latter being shared among the different LSDBs in the Leiden Muscular Dystrophy pages (example of an entry in Supplementary Figure S1). Data were retrieved from peer-reviewed literature and new variants were directly submitted by different sources (laboratories and clinicians). The curators' tasks included confirmation of information concerning the sequence variants, especially with regard to their descriptions following the recommendations of the Human Genome Variation Society (HGVS),26 using the cDNA reference sequence NM_000252.2. Classification of the clinical phenotype (severe, moderate or mild) was added to the patient's entry when adequate information was reported or submitted.
New MTM1 variants
Variants submitted to MTM1-LOVD that had not been described previously, are presented here. These were reported from different centers (Belgium, Germany and Portugal) and submitted to MTM1-LOVD. Information regarding the patient's phenotype and the mutation origins was also collected. Bioinformatic analysis of sequence variants, in particular missense and splicing mutations, was performed using Polyphen v2 (http://genetics.bwh.harvard.edu/pph2/)27 and the Human Splicing Finder v2.3 (http://www.umd.be/HSF/).28 Phylogenetic conservation analysis of the affected residues and population screening of variants were also performed.
Studies performed in patient 1
Clinical description
Patient 1 (P1) is a 7-year-old boy born to a non-consanguineous couple. He has a healthy younger sister. Prenatal manifestations included oligohydramnios and signs of premature birth. Delivery was at 35 weeks by cesarean section, with an Apgar score of 8/9, weight 2465 g, height 46 cm and head circumference 33 cm. During the neonatal period the only clinically relevant sign was facial paresis. There were no feeding or respiratory difficulties. During the first month his pediatrician noticed that he was hypotonic and started global stimulation. He started to walk at 21 months. At the age of two he was referred to a pediatric neurology clinic. He was shy and had poor language skills. There was limitation in the abduction of the right eye, facial diparesis with the left side more exacerbated, with a lagophtalmos. Proximal tetraparesis was detected; the patient could not raise his arms completely and was unable to stand up from the floor without bilateral help. He had a waddling gait and could not run. CK levels were normal (36 U/l). Biopsy was performed at age 3. Over the last 3 years muscle weakness progressed and there has been a gradual loss of motor skills.
Multiple ligation-dependent probe amplification (MLPA) analysis
Screening for duplications and deletions in MTM1 was performed by the MLPA technique using the P309-A1 Probe Set (MRC-Holland, Amsterdam, The Netherlands). This contains 16 probes for the MTM1 gene, 7 probes for the MTMR1 gene, 3 probes located on Xq28 (DKC1 and FLNA genes) and 11 reference probes for distinct regions on the X chromosome (Supplementary Table S2). gDNA samples of P1 and four healthy controls (150 ng each) were used in the procedure. Products were separated by capillary electrophoresis on an ABI 3130xl genetic analyzer (Applied Biosystems, Foster City, CA, USA). Data analysis was conducted using GeneMarker software (SoftGenetics LLC, State College, PA, USA). Population normalization method was selected and data were plotted using probe ratio.
Southern blotting and hybridization
gDNA samples from P1, his mother and controls were digested with EcoRI (New England Biolabs, Beverly, MA, USA) and resolved on a 0.8% agarose gel. DNA fragments were vacuum-transferred to a nylon membrane using a saline method. A cDNA probe recognizing MTM1 exons 2–7 was prepared using digoxigenin (DIG) DNA Labeling Kit (Roche Applied Science, Indianapolis, IN, USA) and incubated overnight using the Easy Hyb Buffer (Roche Applied Science). The membrane was prepared with DIG Wash and Block Buffer Set (Roche Applied Science), incubated with Anti-DIG-AP conjugate (Roche Applied Science), and the DIG-labeled probe detected with ready-to-use CDP-Star (Roche Applied Science).
cDNA analysis
Total RNA was extracted from muscle samples of P1 and controls using the PerfectPure RNA Fibrous Tissue Kit (5 PRIME, Hamburg, Germany) and converted to cDNA using the High Capacity RNA-to-cDNA Kit (Applied Biosystems). MTM1 transcripts were subjected to PCR amplification of the region(s) encompassing exons 2–7, using the specific primers (2F: 5′-TCCAGGATGGCTTCTGCATC-3′ and 7R: 5′-CAAGCCCTGCCTCCTGTATTC-3′). For the detection of the MTM1/MAMLD1 fusion transcript total RNA was extracted from blood using the PerfectPure RNA Blood Kit (5 PRIME). After cDNA conversion, PCR amplification was performed using the primer mentioned above for exon 2 of MTM1 and a reverse primer for exon 5 of MAMLD1 (cMAMLD1-5R: 5′-AGTCTGGCCTGAGTGTGAGAGG-3′). PCR products were purified, sequenced using BigDye Terminator Cycle Sequencing Kit v1.1 (Applied Biosystems) and resolved on an ABI 3130xl genetic analyzer (Applied Biosystems).
Next generation sequencing
gDNA was Covaris (S-series) sheared to an average size of 300 to 400 bp. An Illumina sequencing library was prepared using the NEBNext kit (New England Biolabs) without modifications and using Illumina Truseq adapters. No library pre-amplification was performed. Sequencing data was generated on an Illumina Hiseq2000 (Illumina, San Diego, CA, USA) using standard instrument settings for flowcell clustering and paired-end sequencing of 2 × 100 bp reads on a flowcell with v3 reagents. Fastq files were generated using the Illumina Casava v1.8 pipeline. Stampy (v1.0.13) was used to align the sequence data to the human genome (Hg19) using standard parameters. The total number of aligned reads per 10 000 bp bins across the whole-genome was calculated using the Bedtools package.29 Bins overlapping more than 1% with simple-repeat regions were excluded from further processing. For sample comparison of tag coverage in the MTM1 locus, the bin counts were normalized relative to total number of reads aligned to the chromosome X. For visualization purposes, BED tracks were generated for the patient, control and the ratio patient/control, and were uploaded to the UCSC genome browser.30
Studies performed in patient 2
Clinical description
Patient 2 (P2) was the first child of healthy parents, with no consanguinity or family history of note. After delivery he was very hypotonic and required immediate respiratory support with continuous positive airway pressure and naso jujenal feeding. At 2 months of age he underwent tracheal endoscopy and aryepiglottoplasty after which he needed ventilatory support. After extubation he was transferred for intensive care and was diagnosed with congenital nystagmus and pyloric stenosis. At 3 months he was referred for a neurology opinion. He had a frog like posture, absent antigravity movement in the upper limbs, with some in the lower limbs and areflexia. At pyloromyotomy a quadriceps muscle biopsy was taken, which confirmed the diagnosis of CNM. From 5 months respiratory function improved and by 7 months he was off all respiratory support. He began to lift each leg off the cot for 1–2 min and the nystagmus disappeared, with normal eye movements, but he had persisting marked neck weakness. Following an episode of lung collapse he began to show restriction of lateral and vertical eye movements. He was able to lift his arms against gravity and bring his hands together, as well as to hold a rattle, and was able to lift his legs momentarily against gravity. He had a marked scoliosis, a bell shaped chest and there was early hip and knee flexion contractures. Placed prone he was unable to turn his head or push himself up on his arms. At 8 months he developed bronchiolitis and pulmonary collapse, as well as a brief cardiac arrest. Following this he was re-intubated and given ventilatory support. After recovery he was much less active and lost antigravity limb movement and eye movements. At the age of 10 months he became unwell again and died.
Molecular analysis
Initially gDNA was PCR amplified for subsequent sequencing. As no products were obtained for exons 3–14, a large hemizygous MTM1 deletion was suspected. Deletion was confirmed by MLPA analysis, as described for P1. The breakpoints of the large deletion encompassing exons 3–14 were determined by long-range PCR. Amplification of gDNA was performed using the BIO-X-ACT Long DNA Polymerase Kit (Bioline, Taunton, MA, USA) and primers complementary to introns 2 and 14 of MTM1 (2iF: 5′-GAAAGGTTGCTGAAGGACATACTG-3′ and 14iR: 5′-GCCTTGGGTATGAATGCTGG-3′). PCR products were resolved on a 2% agarose gel and purified with QIAquick Gel Extraction Kit (QIAGEN, Valencia, CA, USA), followed by sequencing.
Results
MTM1-LOVD
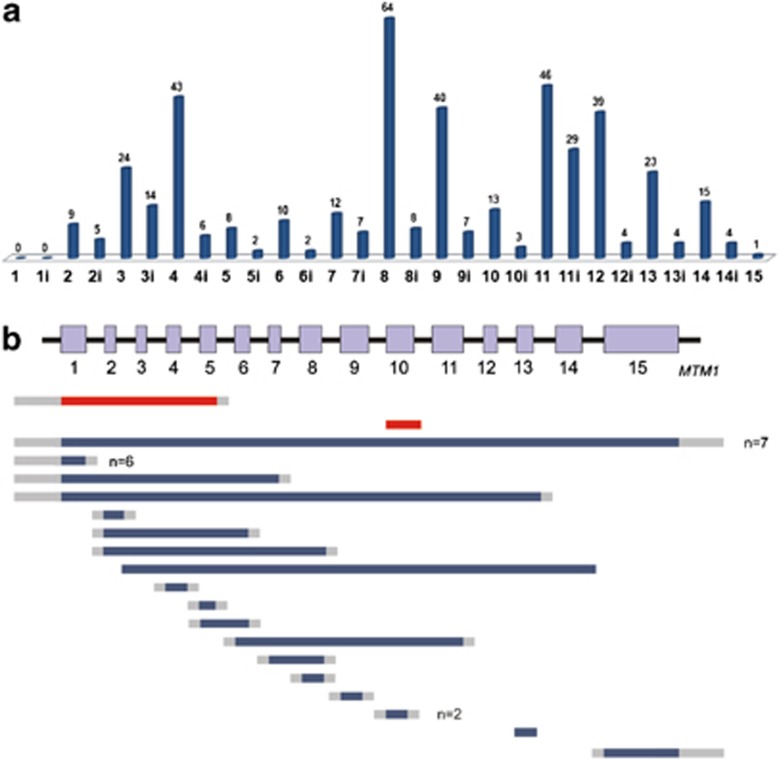
A total of 496 variants are currently (June 2012) listed in the MTM1 LSDB. Besides non-pathogenic variants (n=18) or sequence changes with unknown significance (n=4), this database includes 474 MTM1 mutations identified in 472 XLMTM patients. The mutation profile of the MTM1 gene, according to LOVD-MTM1 data, is summarized in Table 1 and Figure 1. The most represented group of mutations is single-nucleotide substitutions (68.8% of total mutations), which can be subdivided as: missense (n=145), nonsense (n=91), those affecting splicing (n=85) or translation (n=5). Deletions were identified in 102 patients. These include ‘small' deletions reported in 72 patients (15.2% of total mutations) the majority of which (76.4%) predictably induce premature termination codons (PTC). Large deletions encompassing one or more MTM1 exons account for 6.3% of mutations reported. The eighteen different large deletions reported to date (identified in 30 cases) are detailed in Figure 1b. Small duplications were identified in 6.5% of total mutations, all but one case predictably creating PTC. Mutations that consist in a deletion associated with sequence insertion (delins) were identified in seven cases (1.5%), whereas simple insertions were reported in six patients (1.3%). Only two cases (0.4%) involving large (exonic) duplications were reported to date. One patient was reported as having a complex rearrangement that involves the duplication of exon 10.24 Patient P1 presented in detail in this work is the only recorded case with a multi-exonic duplication of MTM1. A total of 55 cases were not reported in the literature and were submitted directly to the MTM1-LOVD.
Table 1. Overview of mutations in MTM1-LOVD.
| DNA | RNAa | Proteinb | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| del/dup/ins/delins | ||||||||||||
| Mutation type | r.(0?) | del/dup/ins/subs | r.(spl?) | r.(?) | p.0? | Missense | Nonsense | IF | OF | MP | p.(?) | Total |
| Substitutions | — | 26 | 62 | 238 | 5 | 145 | 91 | 35 | 8 | 3 | 39 | 326 (152) |
| Deletions | — | 2 | 7 | 63 | — | — | 3 | 10 | 52 | — | 7 | 72 (50) |
| Duplications | — | — | 1 | 30 | — | — | 7 | — | 23 | — | 1 | 31 (23) |
| Insertions | — | 1 | — | 5 | — | — | — | 1 | 5 | — | — | 6 (6) |
| Deletion/insertions | — | — | 2 | 5 | — | — | 1 | — | 4 | — | 2 | 7 (7) |
| Large deletionsc | 15 | 2 | — | 13 | 18 | — | — | 10 | 1 | — | 1 | 30 (18) |
| Large duplicationsc | — | 2 | — | — | — | — | — | 2 | — | — | — | 2 (2) |
| 474 (258) | ||||||||||||
Abbreviations: r.(0?), predictably no expression at RNA level; del, deletion; dup, duplication; ins, insertion; subs, substitution; r.(spl?), predicted to affect splicing; r.(?), RNA effect unknown; p.0?, predictably no protein production; delins, deletion and insertion; IF, in-frame; OF, out-of-frame; MP, multiple polypeptides; p.(?), effect at protein level unknown.
Data extracted from MTM1-LOVD (29/06/2012). Numbers between brackets indicate the total count of unique (different) mutations.
Experimentally tested at RNA level (del, dup, ins, subs) or predicted (remaining columns).
Predicted protein changes inferred from DNA or RNA sequence.
Involving one or more exons.
Figure 1.
Mutations registered in MTM1-LOVD as distributed along the MTM1 gene. (a) Total number of small mutations is shown for each exon and intron of MTM1. (b) Large deletions and duplications identified in MTM1. Solid bars represent regions involved (blue – deletions; red – duplications); gray bars represent undelineated breakpoints. In cases with more than one independent report the total number (n) of patients is indicated.
New MTM1 variants
The 25 novel point mutations identified in CNM male patients are described in Table 2. Among these, 11 are single-nucleotide substitutions located within exonic sequences: c.2T>A, c.32C>A, c.323G>A, c.469G>T, c.595C>A, c.637C>T, c.659G>C, c.1241T>C, c.1247A>G, c.1318C>T and c.1600T>C. With the exception of c.2T>A that predictably affects the initiation codon of myotubularin (p.0?), the majority of these substitutions are predicted to be missense mutations (p.Gly108Asp, p.Pro199Thr, p.Leu213Phe, p.Arg220Thr, p.Phe414Ser, p.His416Arg and p.Trp534Arg). The remaining three are nonsense mutations (p.Ser11*, p.Glu157* and p.Gln440*). A condensed view of all data corroborating the pathogenicity of missense changes is presented in Supplementary Information (Supplementary Table S3). Briefly, missense variants were considered pathogenic when affecting phylogenetically conserved residues and/or were not detected in ethnically matched control chromosomes. Three single-nucleotide substitutions are located in intronic donor splice consensus sequences and predictably affect splicing: c.231+1G>T, c.342+5G>A and c.867+1G>A. In one of these changes (c.342+5G>A) cDNA studies demonstrated that it promotes exon 4 skipping (r.232_342del) leading to an in-frame deletion at the protein level (p.Ser79_Asp115del). In addition, four deletions (c.1088_1089del, c.1328_1331del, c.1509_1510del and c.1519_1522del), two duplications (c.509_528dup and c.596dup) and two deletion/insertion mutations (c.765_767delinsGG and c.1319_1321delinsTA) were also detected. All of these variants are predicted, at the RNA and protein level, to be frame-shift mutations that generate PTC. An insertion of 376 bp in exon 13 (c.1388_1389ins376, GenBank JQ403527) was detected in a patient with a severe phenotype. Using the Repbase database,31 this insertion was identified as an AluYa5 sequence. Further studies revealed that this alteration affects exon 13 splicing (Supplementary Figure S4), predictably resulting in an in-frame deletion of 38 amino acids (p.Phe452_Gln489del) located in the SID region of myotubularin. The remaining two novel mutations are the large deletion and duplication presented below in more detail.
Table 2. Novel MTM1 mutations submitted to the MTM1-LOVD.
| Patient ID | DNA mutation | Gene location | Type of mutation | cDNA effect | Protein effect | Origin of mutation | Phenotype | Geographic origin |
|---|---|---|---|---|---|---|---|---|
| 18914 | c.=/(?_-76)_342 +?dup | Exons 1–5 | Large duplication | r.[=, 343_444del] | p.Asp115_Leu148del | De novo, somatic | Mild | Portugal |
| 10959 | c.63+834_1645-2104del | Intron 2–15 | Large deletion | r.(?) | p.(Thr22_Gln548del) | Inherited (MC) | Severe | United Kingdom |
| 24703 | c.2T>A | Exon 2 | Affects initiation codon | r.(?) | p.0? | Inherited (de novo, in mother) | Severe (at birth) | Germany |
| 21000 | c.32C>A | Exon 2 | Nonsense | r.(?) | p.(Ser11*) | de novo | Severe | Germany |
| 24706 | c.231+1G>T | Intron 4 | Donor splice site disruptiona | r.(spl?) | p.(?) | Inherited (MGC) | Severe (died at 10 months) | Germany |
| 20980 | c.323G>A | Exon 5 | Missense | r.(?) | p.(Gly108Asp)b | de novo | Mild (24 years old, still walking) | Germany |
| 18861 | c.342+5G>A | Intron 5 | Donor splice site disruption | r.232_342del | p.Ser79_Asp115del | Inherited (MC) | Severe | Germany |
| 20960 | c.469G>T | Exon 7 | Nonsense | r.(?) | p.(Glu157*) | Inherited (MC) | Severe | United Kingdom |
| 20961 | c.509_528dup | Exon 7 | Out-of-frame duplication | r.(?) | p.(Gly177Trpfs*16) | Unknown | Severe (long-term survivor) | Germany |
| 20933 | c.595C>A | Exon 8 | Missense | r.(?) | p.(Pro199Thr)b | Inherited (MC) | Unknown (neonatal death) | Portugal |
| 24711 | c.596dup | Exon 8 | Out-of-frame duplication | r.(?) | p.(Ala200Cysfs*12) | Inherited (MC) | Severe | Germany |
| 24702 | c.637C>T | Exon 8 | Missense | r.(?) | p.(Leu213Phe)b | Inherited (MC) | Mild (diagnosed at 24 years of age) | Germany |
| 24707 | c.659G>C | Exon 8 | Missense | r.(?) | p.(Arg220Thr)b | Unknown | Severe (at birth) | Germany/Albania |
| 20951 | c.765_767delinsGG | Exon 9 | Out-of-frame deletion | r.(?) | p.(Asp256Valfs*28) | Inherited (MC) | Severe | Turkey |
| 11124 | c.867+1G>A | Intron 9 | Donor splice site disruptiona | r.(spl?) | p.(?) | Inherited (MC) | Unknown | Portugal |
| 16734 | c.1088_1089del | Exon 11 | Out-of-frame deletion | r.(?) | p.(Lys363Serfs*14) | Inherited (MC) | Severe | Germany |
| 11170 | c.1241T>C | Exon 11 | Missense | r.(?) | p.(Phe414Ser)b | Unknown | Unknown | Portugal |
| 12852 | c.1247A>G | Exon 11 | Missense | r.(?) | p.(His416Arg)b | Unknown | Severe | Germany |
| 20944 | c.1318C>T | Exon 12 | Nonsense | r.(?) | p.(Gln440*) | Inherited (MC) | Severe | Germany |
| 20955 | c.1319_1321delinsTA | Exon 12 | Out-of-frame deletion/insertion | r.(?) | p.(Gln440Leufs*24) | Inherited (MC) | Severe | Germany |
| 24712 | c.1328_1331del | Exon 12 | Out-of-frame deletion | r.(?) | p.(Asp443Valfs*20) | Unknown | Severe | Germany |
| 14208 | c.1388_1389ins376, GenBank JQ403527 | Exon 13 | In-frame exon skipping | r.1354_1467del | p.Phe452_Gln489del | Inherited (de novo, in mother) | Severe | Turkey |
| 20938 | c.1509_1510del | Exon 14 | Out-of-frame deletion | r.(?) | p.(Asn503Lysfs*2) | Unknown | Severe | United Kingdom |
| 20952 | c.1519_1522del | Exon 14 | Out-of-frame deletion | r.(?) | p.(Glu507Asnfs*28) | Inherited (MC) | Severe | Germany |
| 24705 | c.1600T>C | Exon 14 | Missense | r.(?) | p.(Trp534Arg)b | Inherited (MGC) | Severe (at birth) | Germany |
Abbreviations: ID, patient identification in database; MC, mother carrier; MGC, maternal grandmother carrier.
Variants are described according to the reference sequence NM_000252.2, using HGVS nomenclature guidelines.
Predicted to affect splicing by bioinformatic analysis.
Pathogenicity assessment of missense variants in Supplementary Information (S3).
Multi-exonic duplication in patient 1
Patients included in this work were initially studied by routine MTM1 gene analysis. From our molecularly unresolved CNM patient cohort, six males and two females were selected for quantitative studies by MLPA analysis of MTM1, based on clinical and histological criteria that were compatible with CNM. Of these eight candidates, a single male patient (P1) was found to be positive, with a multi-exonic duplication in MTM1. A second male patient was subsequently found to carry a DNM2 mutation and the remainder are as yet uncharacterized.
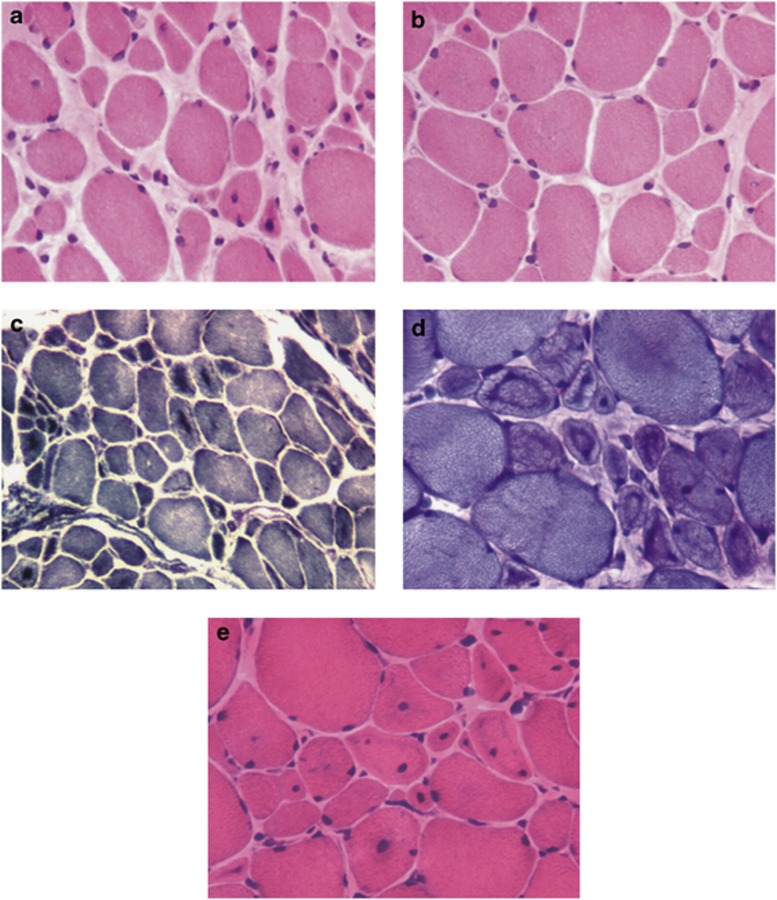
Muscle biopsy of P1 showed fiber size variation with atrophy, numerous central nuclei and endomysium fibrosis (Figure 2a). In some areas muscle was better preserved, with scanty central nuclei and atrophic fibers (Figure 2b). There was central dark staining with NADH-TR, SDH and PAS, reflecting aggregation of mitochondria and glycogen (Figures 2c and d). ‘Necklace'-fibers were easily identified with routine histological stains (Figure 2e), PAS (Figure 2d) and histoenzimological stains. ATPase histoenzymology showed the presence of only type 1 fibers.
Figure 2.
Histological features of patient 1. Histological analysis of the left deltoid muscle biopsy performed using staining with hematoxylin and eosin (a, b and e), histochemical reactions for NADH-TR (c), and PAS (d). Abnormal fiber size variability is seen on all stainings. The key feature of the pathology is the central nuclei (more evident on H&E image a and e). The dark central staining areas accompanying the central nuclei are another striking feature (c and d). The ‘necklace fibers', recently described as a histopathological marker of MTM1, can be best appreciated on PAS staining (d) and faintly on H&E (e).
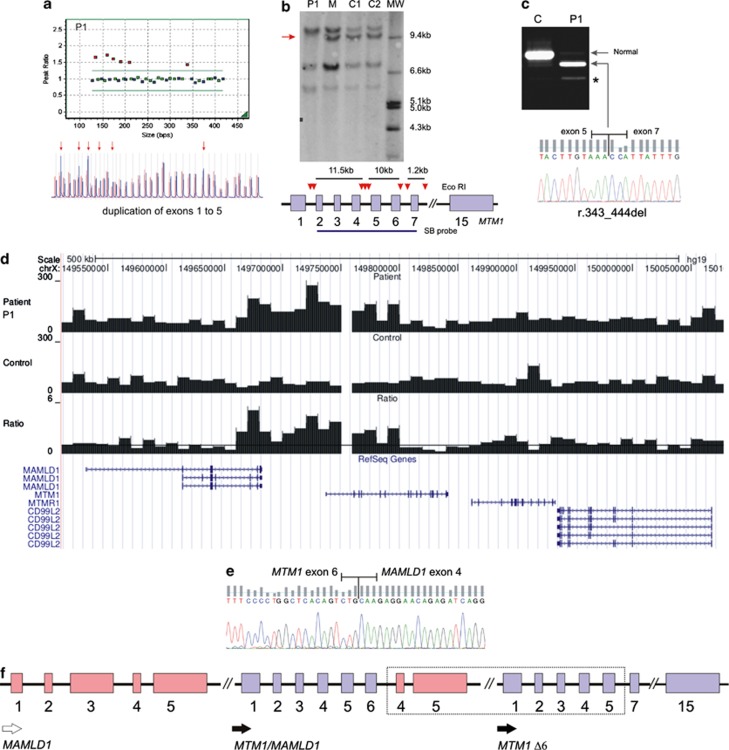
Following the detection of a large duplication spanning exons 1–5 of the MTM1 gene (Figure 3a), the 3′-breakpoint of the duplication was narrowed down by Southern blot analysis (Figure 3b), which revealed the absence of a ∼10 kb band corresponding to exons 5 and 6 of the gene. cDNA studies were carried out on reminiscent muscle tissue, in order to further characterize the rearrangement and evaluate its impact at the mRNA level. This analysis revealed a residual amount of normal transcript and a predominant mutant isoform lacking exon 6 (r.343_444del) (Figure 3c). It is predictable that this abnormal transcript will originate an in-frame deleted myotubularin lacking 34 amino acids of the GRAM domain.
Figure 3.
Large genomic duplication detected in patient 1. (a) MLPA analysis in patient 1 (P1) shows duplication of exons 1–5 of the MTM1 gene. Red arrows indicate the duplicated probes. (b) Southern blot technique using a cDNA probe spanning exons 2–7 (SB probe); red arrow-heads represent EcoRI restriction sites; red arrow highlight the absence of a band in the patient, corresponding to exons 5 and 6. (c) RT-PCR analysis of mRNA obtained from muscle, revealing a mutated isoform lacking exon 6; (*) – smaller faint band corresponding to an alternative splicing product (Δ exon 5 and 6, ENSEMBL transcript ID: ENST00000424519). (d) Low coverage NGS analysis of a 500 kb interval within the X chromosome. Histograms correspond to the total number of reads aligned per 10 kb interval for the patient (top), an unrelated normal control sample (middle) and ratio between the patient and control (bottom) where the line corresponds to ratio value of 1. (e) Partial sequence of the fusion transcript, showing MTM1 exon 6 adjacent to MAMLD1 exon 4. (f) Schematic representation of duplicated region (dashed rectangle); resulting transcripts are indicated by black arrows – observed (filled) and predicted (unfilled). P1, patient; C/C1/C2, controls; M, patient's mother; MW, molecular weight marker.
In order to gain more insight into the structural rearrangement at the MTM1 locus, we performed a whole-genome low coverage analysis by next generation sequencing. After alignment, the total number of reads aligned per 10 kb interval was calculated for the patient and an unrelated normal control sample. The ratio of aligned reads in the 10 kb bins between patient and control is indicative for amplification (ratio >1) or deletion (ratio <1) events. Multiple 10 kb bins in the 5'-region of the MTM1 locus showed amplification (Figure 3d). Interestingly, the amplification appeared to extend into the 3'-region of the neighboring MAMLD1 gene. An in depth analysis of the orientation of the aligned read pairs, as well as several long-range PCR experiments, failed to provide further information regarding the breakpoints of the structural rearrangement. Considering the NGS results, we postulated that the duplication could originate a MTM1/MAMLD1 fusion transcript. In order to test this possibility, additional expression experiments were performed using RNA obtained from whole blood. Results revealed that a MTM1/MAMLD1 fusion transcript was indeed generated (Figure 3e). Based on these results, a schematic representation of the duplication is presented (Figure 3f).
Large deletion in patient 2
During routine MTM1 gDNA sequencing in P2, no symmetrical PCR amplification was obtained for the majority of exons (3–14). This led us to suspect the presence of a large intragenic deletion, which was subsequently confirmed by MLPA (Supplementary Figure S5a). This mutation was further characterized by long-range PCR, in order to predict its impact at the protein level and to facilitate carrier screening and prenatal diagnosis. Sequencing of the junction fragment obtained by long-range PCR revealed breakpoints located in introns 2 and 14 (∼75 kb sized deletion) and enabled us to determine the full description of the mutation as c.63+834_1645-2104del (Supplementary Figure S5b). At the protein level, if efficient translation occurred, this would correspond to the loss of ∼87% of primary protein sequence including all of the functionally relevant myotubularin domains.
Discussion
MTM1-LOVD
Providing evidence for pathogenicity of variants is costly and time-consuming, so diagnostic laboratories rely on previous reports of variants detected in patients. For MTM1 however, such data has been dispersed in the literature, in various formats. These difficulties prompted us to develop an LSDB for the MTM1 gene. We also engaged in this task because the scientific community has expressed the importance and the need for establishing a dedicated XLMTM database (mutational and clinical).23 The implementation and curation of this LSDB followed guidelines reported elsewhere,32, 33 and is registered with the HGVS (LSDB list, http://www.hgvs.org/dblist/glsdb.html).
The main objective for this freely accessible database is the description of MTM1 sequence variants with phenotypic impact, collected from different sources. These data ultimately will allow further insights with respect to the mutational spectrum of the MTM1 gene and epidemiology, abetting the molecular diagnosis and research in this field. Currently >11% of the entries included in this database have not been previously described in the literature. This means that this database contributes to the visibility of new MTM1 mutations within the public domain that would otherwise have a limited possibility to be released as single or individual case reports in the literature.
Database content analysis
Although large genomic deletions contributed to narrowing down the XLMTM candidate region and ultimately the identification of the MTM1 gene,34 large intragenic deletions appear to be a relatively rare cause of myotubular myopathy (6.3% of all cases) compared with point mutations. In fact, these deletions are generally identified in single case reports, with the exception of exon 1 and full MTM1 gene deletions. The majority are associated with a severe disease outcome, even in deletions that are predictably in-frame. However, it should be noted that besides the large deletion reported in this work (P2) only a few mutations, namely the deletion of exon 9 and the deletion of exon 13, have been studied in enough detail to allow a more accurate prediction of their impact at the protein level.10, 35
The largest proportion (93.3%) of pathogenic sequence variants described to date in MTM1 is that comprising small mutations. Since the last published MTM1 mutation update,23 there has been a disproportional increase of missense and splicing mutations reported in MTM1. This might be attributed to the recent identification of additional XLMTM cases with a milder phenotype, often associated with these types of sequence variants.36, 37, 38, 39 Analysis of the distribution of point mutations reveals a higher incidence in exons 8, 11, 9, 4 and 12 (in decreasing order of frequency), and this should be taken into consideration when conducting MTM1 gene mutation screening. Altogether, mutations in these exons account for almost half of all reported XLMTM cases.
A significant number of MTM1 point mutations coincide with the hypermutable CpG dinucleotides that are amenable to methylation-mediated deamination. This mechanism could explain the recurrence, hence higher frequency, of some variants (Table 3). In fact six mutations (c.109C>T, c.141_144delAGAA, c.205C>T, c.1261-10A>G, c.1261C>T and c.1262G>A) were each detected in nine or more patients, accounting for ∼24% of total cases in the MTM1-LOVD database. The most frequent variant in the MTM1-LOVD is the splicing mutation c.1261-10A>G that activates a cryptic splice site, promoting the inclusion of 9 bp in the open reading frame. This cryptic splice site is preferentially used in MTM1 transcripts detected in skeletal muscle.40, 41 The change is predicted to include three amino acids in the core of the PTP (active) site. It was identified in several patients from different ethnic origins (US, Japan and Europe) and associated with a severe disease outcome in male patients. The cause of this recurrent mutational event is still unknown.
Table 3. Frequent mutations in MTM1-LOVD.
| Number of DB entries (n) | Gene region | Mutation (DNA) | Mutation (protein) | Sequence context | Possible cause |
|---|---|---|---|---|---|
| 12 | Exon 3 | c.109C>T | p.Arg37* | TCCTCGACT | CpG |
| 13 | Exon 4 | c.141_144delAGAA | p.Glu48Leufs*24 | CAAAGAAGT | Slippage |
| 10 | Exon 4 | c.205C>T | p.Arg69Cys | TTATCGTCT | CpG |
| 9 | Exon 8 | c.614C>T | p.Pro205Leu | GTTCCGTAT | CpG |
| 13 | Exon 9 | c.721C>T | p.Arg241Cys | TGTGCGTTG | CpG |
| 28 | Intron 11 | c.1261-10A>G | p.Ser420_Arg421insPheIleGln | ATCAATTTA | Unknown |
| 9 | Exon 12 | c.1261C>T | p.Arg421* | TCAGCGAAT | CpG |
| 18 | Exon 12 | c.1262G>A | p.Arg421 Gln | CAGCGAATA | CpG |
Abbreviations: DB, database; CpG, dinucleotide mutational hotspot.
Description of mutations with nine or more independent entries. Nucleotides affected by the mutation are underlined. Variants are described according to the reference sequence NM_000252.2, using HGVS nomenclature guidelines.
In a previous study,12 the analysis of a large number of patients revealed a statistically significant association between non-truncating mutations and the mild phenotype, as opposed to the intermediate/severe phenotype associated with mutations that give rise to PTC. We performed a similar analysis of all data available in MTM1-LOVD with the aim of obtaining further correlation between genotype and phenotype. In line with previous reports, it was evident that truncating mutations are predominantly associated with a severe phenotype. A total of 188 male patients were reported to have mutations that originate PTC. Clinical classification was available in 146 of these (77.7%). The majority (n=139, 95.2%) were reported as severe. Only four patients were reported as mild or mild/moderate, and the PTC inducing mutations were c.836delC, c.1558C>T (n=2) and c.1792delC. Besides these, there are three patients with a moderate phenotype, one associated with a splicing mutation (c.137-7T>G) and the other two with nonsense mutations which had also been found in patients with a severe disease outcome. Overall, the registries show a clear bias towards the severest end of the disease, which may reflect either a specific biological pattern related to defects in myotubularin or the patient selection criteria used to conduct MTM1 gene analysis.
As the majority of non-truncating mutations are of the missense type (∼80%) we centered our analysis on these variants and their localization on the protein (Table 4). Nonsense mutations were also included for comparative purposes. Most missense mutations (96%) are located in myotubularin regions with known function (representing 71% of the total protein sequence), whereas only 59% of nonsense mutations are located in these domains. This suggests that missense changes are not randomly scattered in the protein. Moreover, 85% of mutations associated with a severe phenotype are located in the PTP (catalytic) domain and the RID/desmin-binding region. In terms of its relative proportion, mild/moderate missense mutations are more frequent in the GRAM domain and in the remaining regions of myotubularin. However, it should be noted that there is some degree of phenotypic variability; four missense mutations were classified as ‘inconsistent' in terms of the reported clinical severity. The presence of more than one sequence variant in a patient can hamper attempts at genotype/phenotype correlations. In fact two independent publications reported male patients with double MTM1 mutations. In the first case a patient classified as having a severe phenotype (died at 6 weeks of age) two missense changes were identified (p.Asp431Asn and p.Asp433Asn).42 The second patient was reported with severe neonatal hypotonia requiring assisted ventilation due to respiratory failure. Mutational analysis revealed the presence of two MTM1 changes that create PTC: c.109C>T (p.Arg37*) and c.386_387insA (p.Ser129Argfs*7).43
Table 4. Analysis of MTM1-LOVD missense mutations.
| Missense mutations | Nonsense mutations | ||||||
|---|---|---|---|---|---|---|---|
| Phenotype reported a | |||||||
| Protein domain | Residues involved (% of total protein) | n (% of total) | Severe | Mild/Moderate | Inconsistent | Unknown | n (% of total) |
| GRAM | aa 34–149 (19.1%) | 9 (12.3%) | 2 (22.2%) | 5 (55.6%) | 0 | 2 (22.2%) | 5 (15.6%) |
| RIDb | aa 162–265 (17.1%) | 28 (38.4%) | 15 (53.6%) | 7 (25.0%) | 3 (10.7%) | 3 (10.7%) | 6 (18.8%) |
| PTP | aa 274–434 (26.5%) | 28 (38.4%) | 18 (64.3%) | 5 (17.9%) | 1 (3.5%) | 4 (14.3%) | 3 (9.4%) |
| SID | aa 435–486 (8.5%) | 5 (6.8%) | 3 (60.0%) | 1 (20.0%) | 0 | 1 (20.0%) | 5 (15.6%) |
| Other | (28.8%) | 3 (4.1%) | 1 (33.3%) | 2 (66.7%) | 0 | 0 | 13 (40.6%) |
| Total | 73 | 39 | 21 | 4 | 9 | 32 | |
Abbreviations: aa, amino acid position within myotubularin protein (reference sequence NP_000243.1); GRAM, glucosyltransferase, Rab-like GTPase activators and myotubularins; PTP, protein tyrosine phosphatase; RID, Rac-induced recruitment domain; SID, SET-interacting domain.
Only phenotypes reported in male patients were considered for this analysis; the mild and moderate phenotypes were combined as several reports do not distinguish the two clinical classifications.
This domain partially overlaps desmin-binding region.
Novel multi-exonic duplication
We describe two patients with large rearrangements in the MTM1 gene, one of which (P1) carries the first multi-exonic duplication described to date. The clinical presentation of this patient is milder than the classical form of XLMTM – although he had delayed motor development skills and a progressive tetraparesis, at age 7 he remains ambulant and there is no report of respiratory impairment. Analysis of the patient's muscle biopsy revealed variation of fiber size and typical central nuclei, which lead to the diagnosis of CNM. Additionally, ‘necklace' fibers were identified. These structures, characterized by a basophilic ring deposit following the contour of the cell in which the nuclei are aligned, were initially described as a particular feature of older CNM cases.1 It is now evident that ‘necklace' fibers can also be found in early onset cases with a milder phenotype. Using a variety of experimental methods, we have shown that the duplication encompassing exons 1–5 in MTM1 has an unexpected effect at the mRNA level, resulting in an in-frame deletion of the sequence corresponding to exon 6 (r.343_444del). At the gDNA level, using a genome wide low coverage analysis of the patient's genome we were able to confirm the amplification within the MTM1 gene and obtained evidence that it extended into the MAMLD1 gene, known to be associated with hypospadias.44 However, unlike intragenic duplications, it is foreseeable that because this duplication affects only the 3′-end of the MAMLD1 gene, a normal copy would be left intact, hence the duplication should have little or no phenotypic consequence, thereby explaining why no genital abnormalities were found in the patient.
MLPA analysis had shown that the average ratio of duplicated MTM1 probes (1.6) was significantly lower than expected for a male (∼2.0). This, together with the presence of a residual amount of normal transcript and the finding that the patient's mother was not a carrier, provides strong evidence for somatic mosaicism in the patient – the first such case described to date – which might explain the patient's relatively mild phenotype in relation to what could be predicted from the genotype.
Standard routine methods for XLMTM molecular diagnosis, such as genomic analysis of MTM1 by Sanger sequencing, do not allow the identification of duplications and may also fail to detect mosaics. The only other description of a duplication involving a complex MTM1 rearrangement concerns a male infant with characteristic clinical and histopathologic findings of XLMTM.24 The patient had severe neonatal hypotonia and respiratory insufficiency. Other XLMTM compatible findings included the absence of deep tendon reflexes, cryptorchidism and elongated fingers and toes. There was a progressive respiratory deterioration culminating in death at 1 month of age, due to respiratory failure. It is conceivable that there may be a considerable number of CNM cases with MTM1 duplications, and it is therefore advisable to perform MLPA analysis in all CNM cases, after excluding point mutations.
Future perspectives
It would be important to gather more MTM1 mutational data that are presently dispersed among other non-public databases (such as UMD-MTM1 and the Cardiff database for XLMTM),45 as well as in private registries of diagnostic laboratories around the world. According to the meeting report of the 6th ENMC workshop on centronuclear (myotubular) myopathies, efforts are currently being made to develop a patient registry for CNM with the support of TREAT-NMD and the Myotubular Trust.45 Until now the harmonized clinical items to be included in this registry are not yet in the public domain. After full release of LOVD software v3.0 the clinical items to be included will follow these international recommendations for the MTM1 registry, ultimately allowing the data integration, which at this point in time is a recognized difficulty.
Acknowledgments
We thank Dr António Guimarães for the histopathology studies in Patient 1 and Roel Brekelmans (MRC-Holland) for the MLPA kit design.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Romero NB. Centronuclear myopathies: a widening concept. Neuromuscul Disord. 2010;20:223–228. doi: 10.1016/j.nmd.2010.01.014. [DOI] [PubMed] [Google Scholar]
- Nance JR, Dowling JJ, Gibbs EM, Bönnemann CG. Congenital myopathies: an update. Curr Neurol Neurosci Rep. 2012;12:165–174. doi: 10.1007/s11910-012-0255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte J, Hu LJ, Kretz C, et al. A gene mutated in X-linked myotubular myopathy defines a new putative tyrosine phosphatase family conserved in yeast. Nat Genet. 1996;13:175–182. doi: 10.1038/ng0696-175. [DOI] [PubMed] [Google Scholar]
- Biancalana V, Beggs AH, Das S, et al. Clinical utility gene card for: Centronuclear and myotubular myopathies Eur J Hum Genet 2012. doi: 10.1038/ejhg.2012.91 [DOI] [PMC free article] [PubMed]
- Bitoun M, Maugenre S, Jeannet PY, et al. Mutations in dynamin 2 cause dominant centronuclear myopathy. Nat Genet. 2005;37:1207–1209. doi: 10.1038/ng1657. [DOI] [PubMed] [Google Scholar]
- Tosch V, Rohde HM, Tronchère H, et al. A novel PtdIns3P and PtdIns(3,5)P2 phosphatase with an inactivating variant in centronuclear myopathy. Hum Mol Genet. 2006;15:3098–3106. doi: 10.1093/hmg/ddl250. [DOI] [PubMed] [Google Scholar]
- Nicot AS, Toussaint A, Tosch V, et al. Mutations in amphiphysin 2 (BIN1) disrupt interaction with dynamin 2 and cause autosomal recessive centronuclear myopathy. Nat Genet. 2007;39:1134–1139. doi: 10.1038/ng2086. [DOI] [PubMed] [Google Scholar]
- Wilmshurst JM, Lillis S, Zhou H, et al. RYR1 mutations are a common cause of congenital myopathies with central nuclei. Ann Neurol. 2010;68:717–726. doi: 10.1002/ana.22119. [DOI] [PubMed] [Google Scholar]
- Bevilacqua JA, Monnier N, Bitoun M, et al. Recessive RYR1 mutations cause unusual congenital myopathy with prominent nuclear internalization and large areas of myofibrillar disorganization. Neuropathol Appl Neurobiol. 2011;37:271–284. doi: 10.1111/j.1365-2990.2010.01149.x. [DOI] [PubMed] [Google Scholar]
- Jungbluth H, Wallgren-Pettersson C, Laporte J. Centronuclear (myotubular) myopathy. Orphanet J Rare Dis. 2008;3:26. doi: 10.1186/1750-1172-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman GE, Finegold M, Zhao W, de Gouyon B, Metzenberg A. Medical complications in long-term survivors with X-linked myotubular myopathy. J Pediatr. 1999;134:206–214. doi: 10.1016/s0022-3476(99)70417-8. [DOI] [PubMed] [Google Scholar]
- McEntagart M, Parsons G, Buj-Bello A, et al. Genotype-phenotype correlations in X-linked myotubular myopathy. Neuromuscul Disord. 2002;12:939–946. doi: 10.1016/s0960-8966(02)00153-0. [DOI] [PubMed] [Google Scholar]
- Tanner SM, Orstavik KH, Kristiansen M, et al. Skewed X-inactivation in a manifesting carrier of X-linked myotubular myopathy and in her non-manifesting carrier mother. Hum Genet. 1999;104:249–253. doi: 10.1007/s004390050943. [DOI] [PubMed] [Google Scholar]
- Hammans SR, Robinson DO, Moutou C, et al. A clinical and genetic study of a manifesting heterozygote with X-linked myotubular myopathy. Neuromuscul Disord. 2000;10:133–137. doi: 10.1016/s0960-8966(99)00073-5. [DOI] [PubMed] [Google Scholar]
- Sutton IJ, Winer JB, Norman AN, Liechti-Gallati S, MacDonald F. Limb girdle and facial weakness in female carriers of X-linked myotubular myopathy mutations. Neurology. 2001;57:900–902. doi: 10.1212/wnl.57.5.900. [DOI] [PubMed] [Google Scholar]
- Jungbluth H, Sewry CA, Buj-Bello A, et al. Early and severe presentation of X-linked myotubular myopathy in a girl with skewed X-inactivation. Neuromuscul Disord. 2003;13:55–59. doi: 10.1016/s0960-8966(02)00194-3. [DOI] [PubMed] [Google Scholar]
- Schara U, Kress W, Tücke J, Mortier W. X-linked myotubular myopathy in a female infant caused by a new MTM1 gene mutation. Neurology. 2003;60:1363–1365. doi: 10.1212/01.wnl.0000058763.90924.fa. [DOI] [PubMed] [Google Scholar]
- Pénisson-Besnier I, Biancalana V, Reynier P, Cossée M, Dubas F. Diagnosis of myotubular myopathy in the oldest known manifesting female carrier: a clinical and genetic study. Neuromuscul Disord. 2007;17:180–185. doi: 10.1016/j.nmd.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Robinson FL, Dixon JE. Myotubularin phosphatases: policing 3-phosphoinositides. Trends Cell Biol. 2006;16:403–412. doi: 10.1016/j.tcb.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Laporte J, Blondeau F, Buj-Bello A, Mandel JL. The myotubularin family: from genetic disease to phosphoinositide metabolism. Trends Genet. 2001;17:221–228. doi: 10.1016/s0168-9525(01)02245-4. [DOI] [PubMed] [Google Scholar]
- Tsujita K, Itoh T, Ijuin T, et al. Myotubularin regulates the function of the late endosome through the gram domain-phosphatidylinositol 3,5-bisphosphate interaction. J Biol Chem. 2004;279:13817–13824. doi: 10.1074/jbc.M312294200. [DOI] [PubMed] [Google Scholar]
- Hnia K, Tronchère H, Tomczak KK, et al. Myotubularin controls desmin intermediate filament architecture and mitochondrial dynamics in human and mouse skeletal muscle. J Clin Invest. 2011;121:70–85. doi: 10.1172/JCI44021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte J, Biancalana V, Tanner SM, et al. Mtm1 Mutations in X-linked myotubular myopathy. Hum Mutat. 2000;15:393–409. doi: 10.1002/(SICI)1098-1004(200005)15:5<393::AID-HUMU1>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Trump N, Cullup T, Muntoni F, Verheij J, Jungbluth H. X-linked myotubular myopathy due to a complex rearrangement involving exon 10 of the myotubularin (MTM1) gene. Neuromuscul Disord. 2012;22:384–388. doi: 10.1016/j.nmd.2011.11.004. [DOI] [PubMed] [Google Scholar]
- Fokkema IF, Taschner PE, Schaafsma GC, Celli J, Laros JF, den Dunnen JT. LOVD v.2.0: the next generation in gene variant databases. Hum Mutat. 2011;32:557–563. doi: 10.1002/humu.21438. [DOI] [PubMed] [Google Scholar]
- den Dunnen JT, Antonarakis SE. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat. 2000;15:7–12. doi: 10.1002/(SICI)1098-1004(200001)15:1<7::AID-HUMU4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmet FO, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, Béroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Celli J, Dalgleish R, Vihinen M, Taschner PE, den Dunnen JT. Curating gene variant databases (LSDBs): Toward a universal standard. Hum Mutat. 2011;33:291–297. doi: 10.1002/humu.21626. [DOI] [PubMed] [Google Scholar]
- Vihinen M, den Dunnen JT, Dalgleish R, Cotton RG. Guidelines for establishing locus specific databases. Hum Mutat. 2011;33:298–305. doi: 10.1002/humu.21646. [DOI] [PubMed] [Google Scholar]
- Dahl N, Hu LJ, Chery M, et al. Myotubular myopathy in a girl with a deletion at Xq27-q28 and unbalanced X inactivation assigns the MTM1 gene to a 600-kb region. Am J Hum Genet. 1995;56:1108–1115. [PMC free article] [PubMed] [Google Scholar]
- Tsai TC, Horinouchi H, Noguchi S, et al. Characterization of MTM1 mutations in 31 Japanese families with myotubular myopathy, including a patient carrying 240 kb deletion in Xq28 without male hypogenitalism. Neuromuscul Disord. 2005;15:245–252. doi: 10.1016/j.nmd.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Biancalana V, Caron O, Gallati S, et al. Characterisation of mutations in 77 patients with X-linked myotubular myopathy, including a family with a very mild phenotype. Hum Genet. 2003;112:135–142. doi: 10.1007/s00439-002-0869-1. [DOI] [PubMed] [Google Scholar]
- Yu S, Manson J, White S, et al. X-linked myotubular myopathy in a family with three adult survivors. Clin Genet. 2003;64:148–152. doi: 10.1034/j.1399-0004.2003.00118.x. [DOI] [PubMed] [Google Scholar]
- Hoffjan S, Thiels C, Vorgerd M, Neuen-Jacob E, Epplen JT, Kress W. Extreme phenotypic variability in a German family with X-linked myotubular myopathy associated with E404K mutation in MTM1. Neuromuscul Disord. 2006;16:749–753. doi: 10.1016/j.nmd.2006.07.020. [DOI] [PubMed] [Google Scholar]
- Bevilacqua JA, Bitoun M, Biancalana V, et al. ‘Necklace' fibers, a new histological marker of late-onset MTM1-related centronuclear myopathy. Acta Neuropathol. 2009;117:283–291. doi: 10.1007/s00401-008-0472-1. [DOI] [PubMed] [Google Scholar]
- de Gouyon BM, Zhao W, Laporte J, Mandel JL, Metzenberg A, Herman GE. Characterization of mutations in the myotubularin gene in twenty six patients with X-linked myotubular myopathy. Hum Mol Genet. 1997;6:1499–1504. doi: 10.1093/hmg/6.9.1499. [DOI] [PubMed] [Google Scholar]
- Nishino I, Minami N, Kobayashi O, et al. MTM1 gene mutations in Japanese patients with the severe infantile form of myotubular myopathy. Neuromuscul Disord. 1998;8:453–458. doi: 10.1016/s0960-8966(98)00075-3. [DOI] [PubMed] [Google Scholar]
- Laporte J, Guiraud-Chaumeil C, Vincent MC, et al. Mutations in the MTM1 gene implicated in X-linked myotubular myopathy. ENMC International Consortium on Myotubular Myopathy. European Neuro-Muscular Center. Hum Mol Genet. 1997;6:1505–1511. doi: 10.1093/hmg/6.9.1505. [DOI] [PubMed] [Google Scholar]
- Tachi N, Kozuka N, Chiba S, Miyaji M, Watanabe I. A double mutation in a patient with X-linked myotubular myopathy. Pediatr Neurol. 2001;24:297–299. doi: 10.1016/s0887-8994(01)00244-2. [DOI] [PubMed] [Google Scholar]
- Ogata T, Wada Y, Fukami M. MAMLD1 (CXorf6): a new gene for hypospadias. Sex Dev. 2008;2:244–250. doi: 10.1159/000152040. [DOI] [PubMed] [Google Scholar]
- Jungbluth H, Wallgren-Pettersson C, Laporte JF:, Centronuclear (Myotubular) Myopathy Consortium 164th ENMC International workshop: 6th workshop on centronuclear (myotubular) myopathies, 16-18th January 2009, Naarden, The Netherlands. Neuromuscul Disord. 2009;19:721–729. doi: 10.1016/j.nmd.2009.06.373. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.