Abstract
Neurofibromatosis type 1 (NF1) and its related disorders (NF1-Noonan syndrome (NFNS) and Watson syndrome (WS)) are caused by heterozygous mutations in the NF1 gene. Pulmonary stenosis (PS) occurs more commonly in NF1 and its related disorders than in the general population. This study investigated whether PS is associated with specific types of NF1 gene mutations in NF1, NFNS and WS. The frequency of different NF1 mutation types in a cohort of published and unpublished cases with NF1/NFNS/WS and PS was examined. Compared with NF1 in general, NFNS patients had higher rates of PS (9/35=26% vs 25/2322=1.1%, P value<0.001). Stratification according to mutation type showed that the increased PS rate appears to be driven by the NFNS group with non-truncating mutations. Eight of twelve (66.7%) NFNS cases with non-truncating mutations had PS compared with a 1.1% PS frequency in NF1 in general (P<0.001); there was no increase in the frequency of PS in NFNS patients with truncating mutations. Eight out of eleven (73%) individuals with NF1 and PS, were found to have non-truncating mutations, a much higher frequency than the 19% reported in NF1 cohorts (P<0.015). Only three cases of WS have been published with intragenic mutations, two of three had non-truncating mutations. Therefore, PS in NF1 and its related disorders is clearly associated with non-truncating mutations in the NF1 gene providing a new genotype–phenotype correlation. The data indicate a specific role of non-truncating mutations on the NF1 cardiac phenotype.
Keywords: neurofibromatosis type 1, neurofibromatosis–Noonan syndrome, Watson syndrome, non-truncating mutations, genotype–phenotype correlation
Introduction
Neurofibromatosis type 1 (NF1, MIM #162200) is the most common dominantly inherited neurocutaneous disease with a birth incidence of at least 1:2500 worldwide.1 The disease is characterized by multiple café au lait spots (CALS), skin-fold freckling, iris Lisch nodules and neurofibromas. In addition, there are a large number of disease complications, which can affect any body system.2 NF1 is caused by heterozygous mutations in the NF1 gene (NF1, NM_001042492.1), located at chromosome17q11.2. As the great majority of the detected NF1 gene mutations in NF1 patients are truncating mutations, and as about 4–5% of the patients have a whole NF1 gene deletion,3, 4, 5 it is clear that loss-of-function is the causative mechanism of NF1. Neurofibromin acts as a Ras-specific GTPase-activating protein and has a role in the Ras–MAPK (mitogen-activated protein kinase) pathway as a negative regulator of Ras.6, 7
One of the challenges in counseling families with NF1 is the variability of the disease even within families. Genotype–phenotype correlations in NF1 are limited. The first group of patients with a specific genotype–phenotype correlation identified were those with large deletions involving the NF1 gene.8 These patients have deletions of both the NF1 and a variable number of flanking genes. The deletions are usually associated with a more consistently severe phenotype, including facial dysmorphism, marked learning problems and increased neurofibroma burden.8 More recently, a specific mutation has been found to be associated with a much milder NF1 phenotype with a lack of dermal neurofibromas, and increased rate of pulmonary stenosis (PS)—the NF1 exon 17 3-bp in-frame deletion (c.2970_2972delAAT).9 In addition, heterozygous loss-of-function mutations in another gene, SPRED1, have been described to cause NF1-like disease associated with CALS and inguinal/axillary freckles but no neurofibromas or Lisch nodules (Legius syndrome (MIM# #611431)).10, 11 Similarly to neurofibromin, SPRED1 is a negative regulator of RAS–MAPK signaling.12
Before the cloning of the NF1 gene two other related disorders, Watson syndrome (WS) and NF1–Noonan syndrome (NFNS), had been described clinically, both have been shown to be caused by NF1 mutations. Watson13 described autosomal dominant inheritance of PS, multiple CALS spots and intelligence at the lower end of the normal range. A few similar families have since been reported. Follow-up of the original Watson patients confirmed that their phenotype did appear distinct from NF1, as adult patients had few, if any, neurofibromas.14 Molecular analysis has shown WS is caused by mutations in NF1.9, 15, 16 At the time of most of the WS reports it was not recognized that PS occurs at an increased frequency in NF1. Today, sporadic cases with multiple CAL spots and PS are usually just considered to have NF1.
The other phenotype is NFNS, where patients have overlapping features of both NF1 and Noonan syndrome. Although originally reported as a distinct condition, subsequent clinical17 and molecular analysis has shown that the majority of NFNS patients have only NF1 gene mutations, with a significantly higher prevalence of non-truncating mutations, particularly in-frame deletions, than in typical NF1.18, 19 Two cases have been reported with both NF1 and PTPN11 mutations. In one, the patient inherited the PTPN11 gene mutation from a parent and had a de novo NF1 mutation,20 and in the other a de novo PTPN11 gene mutation with inherited NF1.21 Although molecular diagnostics have added to our understanding of these conditions, there remains considerable overlap: the same mutations have been seen in NF1 with no features of NFNS.18 Some of the cases with the c.2970_2972delAAT exon 17 mutation were clinically diagnosed as NFNS or WS.9
Individuals of all these three phenotypes: NF1, WS and NFNS, fulfill the NIH criteria for NF1 and have mutations in the NF1 gene. Therefore, they all may be considered as NF1 gene-related disorders. The only reason persisting with the distinction is whether they have a distinct natural history. To date, there is only a suggestion for this for WS, where the adult patients have had few, if any, neurofibromas.14
The other important factor in our understanding of the pathogenesis of NF1-related phenotypes is the fact that the genes for NF1, Legius syndrome and NS are all in the Ras–MAPK pathway along with other syndromes with overlapping phenotypes. The group of disorders is now referred to as the Rasopathies.22 The overlapping phenotypes of Rasopathies include PS, short stature, pectus abnormalities and learning problems.
Cardiac defects are uncommon in NF1, affecting only 54/2322 of the patients.23 PS is the most common cardiac defect in NF1 and represented almost 50% of the cardiac malformations in the largest series of NF1 cases with cardiac problems (25/54).23 At this frequency, PS is six times more prevalent in NF1 patients compared with the general population. No clear genotype–phenotype correlation for the cardiac phenotype in NF1 and/or NFNS has been suggested, although a trend of association was found between heart defects, NFNS and in-frame/single amino-acid substitution.18
Given the previous observations of an increased prevalence of non-truncating mutations in NFNS,18 of an excess of PS in patients with NF1,23 particularly in patients with the non-truncating 3 bp exon 17 deletion,9 we hypothesized that non-truncating mutations in NF1 may be responsible for particular disease features, especially those which overlap with other Ras–MAPK disorders. Here, we report our analysis of mutation type in patients with PS and NF1, WS or NFNS.
Materials and methods
We performed a search of electronic bibliographic data using Pubmed (http://www.ncbi.nlm.nih.gov/pubmed/); the search was restricted to English language publications involving humans and combining the terms: NF1, Neurofibromatosis, Watson Syndrome, NF1-Noonan syndrome, (NF1+noonan syndrome), (neurofibromatosis+Noonan syndrome), pulmonary stenosis, cardiac defect and Heart. Cases of NF1, WS, NFNS, which had both a description of PS and intragenic mutations in the NF1 gene, were included in this study. In familial cases, we included families in which at least one individual with the disease had PS.
We then looked at the mutations that had been found in a series of unpublished cases of individuals with clinical diagnosis of NF1 (according to the NIH criteria), PS and intragenic NF1 gene mutations. These came from three sources: cases that had had diagnostic mutation testing in Tel Aviv and Manchester, and from the research mutation database in Cardiff. Ethical committee approval has been obtained for the research cases. Mutation anlysis was done either by DNA-based methods or by using the combined RNA/DNA approach developed by Messiaen et al.3
Statistical analysis
Categorical variables were analyzed and compared by Fisher's exact test. Results are expressed as P values and likelihood ratios (LR). P<0.01 was considered significant.
Prediction of the disease-related effect of the missense mutations
Prediction of the disease-related effect of the missense mutations was performed using the SNP &Go server (http://snps-and-go.biocomp.unibo.it/snps-and-go/index.html, Bologna Biocomputing Grou, university of Bolognia).24
Prediction of splicing effect
We have utilized Site Finder-like, MaxEntScan, NNSPLICE, GeneSplicer and Human Splicing Finder tools to assess splicing effect for missense mutations for which RNA analysis was not available.
Results
For the analysis we have used the frequency of different cardiac defects reported from the NF1 international database (Lin et al 23). They reported 2550 patients, 228 of whom did not satisfy NIH NF1 diagnostic criteria. Of the 2322 patients that did satisfy criteria, 25 had PS. The frequency of different mutation types in NF1 from the large series is reported by Messiaen and Wimmer.4 In their series of 1770 unrelated patients 27% had splicing mutations, 26% frameshift, 21% nonsense,18% missense or 1-multi AA del/dup, 5% large deletions, 2% had an intragenic copy number change and 1% had unique complex mutations. In our literature review we identified 35 cases of NFNS with mutation data, five patients with WS and only one case of NF1 with PS. We report 11 new cases of NF1 and PS with their mutation data.
Reported patients with NFNS and PS
Review of published clinical data identified 35 cases of NFNS, 9 of which had PS. The clinical and mutation data is summarized in Tables 1, 2 and Figure 1. The reported molecular analyses of this group was based on DNA analyses except for a single case in which combined DNA/RNA approach was utilized.19 The disease association predictive tool (SNP &Go24) suggested a disease causative prediction with maximal reliability score in all of the mutations associated with NFNS and PS. In addition, using Splice Site Finder-like, MaxEntScan, NNSPLICE, GeneSplicer and Human Splicing Finder tools no effect on splicing was predicted for these mutations.
Table 1. Summary of cardiac status and mutation type in published cases of individuals with NFNS (n=35 individuals).
| Yes | No | Overall | |
|---|---|---|---|
| Mutation identified | 30 | 5 | 35 |
| Intragenic mutation | 29 | 1 (Large deletion) | 30 |
| Known cardiac status of individuals with intragenic mutation | 28 | 1 | 29 |
| Individuals with known cardiac status and truncating mutations | 16 | 12 (Non-truncating mutations) | 28 |
| PS among individuals with truncating mutations | 1 | 15 | 16 |
| PS among individuals with non-truncating mutations | 8 | 4 | 12 |
Table 2. NF1 mutations among published cases with NFNS and pulmonary stenosis.
| Number | Reference | Original ID | Mutation detected | Type of mutation |
|---|---|---|---|---|
| 1 | De Luca et al18 | NFNS4a | c.1862delC | Truncating |
| 2 | De Luca et al18 | NFNS6a | c.2970delAAT | In-frame |
| 3 | De Luca et al18 | NFNS8 | 4267A>G p.K1423E | Missense |
| 4 | De Luca et al18 | NFNS12 | c.4312delGAA | In-frame |
| 5 | Hüffmeier et al 25 | 1 | c.3587T>G p.L1196R | Missense |
| 6 | Baralle et al 26 | 5 | c.4312delGAA | In-frame |
| 7 | Bertola et al 20 | NA | c.2531A>G p.L844R | Missense |
| 8 | Nyström et al 19 | NA | c.4168C>T p.L1390F | Missense |
| 9 | Stevenson et al 27 | NA | 2970delAAT | In-frame |
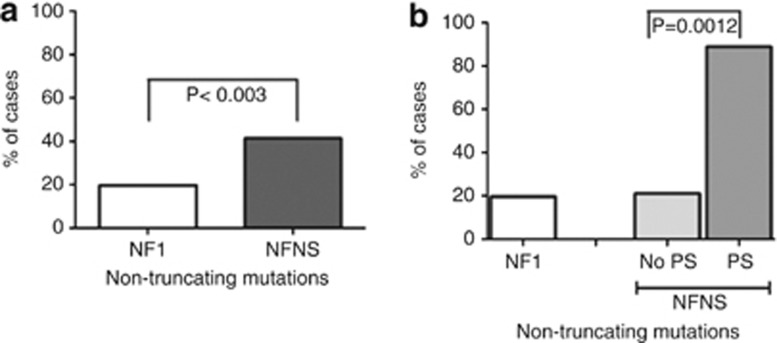
Figure 1.
PS among individuals with NFNS according to mutation type. (a) An increased rate of non-truncating mutations is seen in individuals with NFNS compared with the general NF1 population (12/29 (41%) of patients with NFNS compared with 19.4% (326/1681) in a large NF1 cohort.4 (b) Stratifying the NFNS patients by their mutation type showed that among individuals with NFNS and PS 88.9% (8/9) had non-truncating mutations, whereas only 21% (4/19)4 of individuals with NFNS without PS had non-truncating mutations.
PS is about 20 times more prevalent in the NFNS group compared with the NF1 group (9/35=26%) vs 1.1% (25/2322); P<0.001, LR=40).
Thirty of the published NFNS group had mutations identified, 9 of whom had PS, 29 of them had an intragenic mutation, whereas 1 had a whole gene deletion. In 28 of them, information regarding the cardiac status was available. The difference in PS prevalence between the NFNS group and the NF1 group, was shown to be driven by the NFNS patients with non-truncating mutation: 66.7% (8/12) of NFNS with non-truncating mutation patients had PS vs 1.1% (25/2322)23 in NF1 (P<0.001, LR=55); only 6.2% (1/16) NFNS patients with a truncating mutation had PS, not significantly different from NF1 patients: 1.1% (P=0.164) (Table 1).
Analyzing the patient groups by mutation type showed that overall 12/29 (41.4%) patients with NFNS and intragenic mutation had a non-truncating intragenic mutation compared with 326/1681 (19.4%,4) individuals with NF1, (P<0.003, LR=7.2; Figure 1a). Eight out of nine (88.9%) patients with NFNS and PS had a non-truncating mutation compared with only 4/19 (21%) NFNS patients without PS. The former group, therefore, had a very significant excess (88.9% vs 19.4% (P<0.0001, LR=20.2), whereas the non-PS NFNS cases had a similar frequency of non-truncating mutations to the large NF1 cohort (21% vs 19.4%, P=0.7; Figure 1b).
Reported patients with WS
WS includes, by definition, NF1 phenotype and PS. Given that, we have decided next to test the frequency of non-truncating mutations among patients with WS. However, as of now, only five patients/families with WS and NF1 molecular analysis have been reported (Table 3). Moreover, only three of these families had characterized intragenic mutations, two with in-frame mutations, and one with nonsense mutation. Two families had deletions within the NF1 gene, with no available information regarding the effect of these changes on the reading frame. Thus, from the published cases there is a potential increase in non-truncating mutations in WS too (2/3) as seen in NFNS with PS; however, given the low number of WS cases, a statistical analysis has not been performed, and these results could still be an accidental finding.
Table 3. NF1 mutations in published cases with WS.
Reported cases with NF1 and PS cases
Having established a genotype–phenotype correlation between the mutation type and the existence of PS among NFNS patients and a similar trend among WS patients, we then looked for such a correlation among individuals with NF1 and PS. As noted, there is a lack of published data regarding the molecular analysis of patients with NF1 and PS. The largest study concerning cardiac defects in individuals with NF1 provides molecular data only in one case out of 25, of those who were concurrently afflicted by PS.23 That individual had an entire gene deletion, which may cause by itself heart defects due to contiguous gene deletion.
Unpublished cases
In Table 4 we summarize the clinical and mutational data for 11 unpublished cases with a clinical diagnosis of NF1, according to the NIH criteria, and PS. Some of these individuals were clinically diagnosed as having NFNS or WS and this is summarized in Table 4. Reflecting our current clinical practise of treating all of these individuals as having NF1 we analyzed the mutational data as a group. Four patients had missense mutations; two cases shared the same missense mutation c.3827G>A. Disease association predictive tool (SNP &Go24) in all of these mutations revealed a disease-causative prediction with maximal reliability score. RNA analyses for both the c.3827G>A mutation32 and the c.3572C>G mutation showed no effect on splicing for these two mutations. The molecular analysis of one case was based on solely DNA analysis (c.4277A>G). No effect on splicing was predicted for this mutation using the following in silico tools; Splice Site Finder-like, MaxEntScan, NNSPLICE, GeneSplicer and Human Splicing Finder. Two had in-frame 3 bp deletions, one being a further case of c.2970delAAT.9 One patient had a deletion of exon 14, which should not affect the reading frame. Three patients had truncating mutations: one of them had a nonsense mutation, another had out-of-frame deletion, and another mutation was a splice site mutation expecting to result in out-of-frame mutation.4
Table 4. NF1 mutations in unreported cases of NF1 with PS.
| Number | Phenotype | Family history | Mutation detected | Type of mutation |
|---|---|---|---|---|
| 1 | NF1 | Positive | c.3586C>T p.L1196F | Missense |
| 2 | NFNS | Positive | c.2970delAAT | In-frame |
| 3 | NFNS | Negative | Exon 14 deletion | Deletion (in-frame) |
| 4 | NF1 | Positive | c.539_540dupTA | Truncating |
| 5 | NFNS | Positive | 7702C>T p.Q2568X | Nonsense |
| 6 | NFNS | Negative | c.7324del CTT | In-frame |
| 7 | WS | Negative | c.4277A>G p.Q1426R | Missense |
| 8 | WS | Positive | c.3827G>A p.R1276Q | Missense – no splicing effect32 |
| 9 | WS | Negative | c.1466A>G p.Y489C | Splicing, out-of-frame4 |
| 10 | WS- family C in Watson's original report12, 13 | Positive | c.3827 C>G p.R1276Q | Missense – no splicing effect32 |
| 11 | NF1 | Positive | c.3572C>G p.T1191R | Missense – no splicing effect |
Abbreviations: NF1, neurofibromatosis type 1; NFNS, NF1–Noonan syndrome; WS, Watson syndrome.
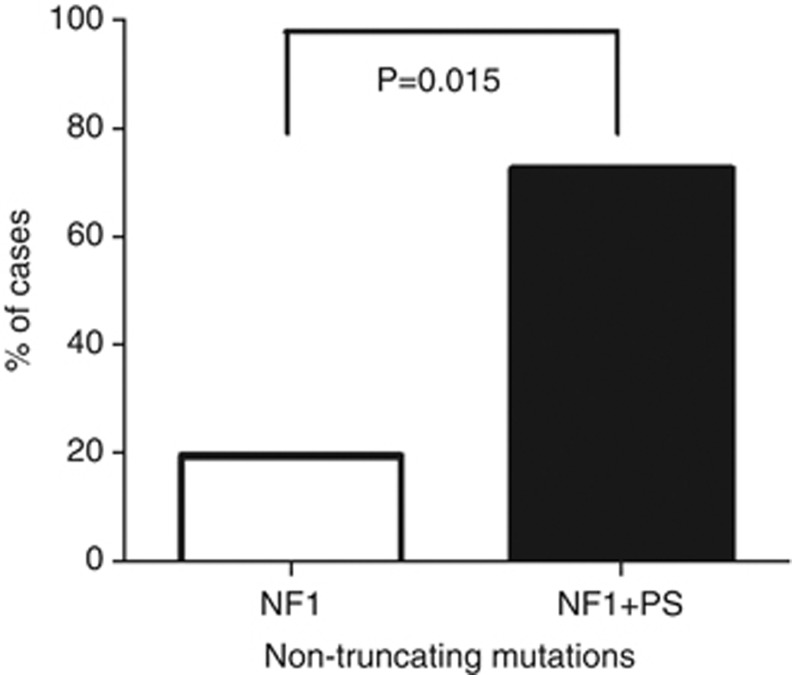
Overall, 8/11 (73%) of individuals with NF1 and PS had non-truncating mutations compared with 19.4%4 of the general population of individuals with NF1 (P=0.015) (Figure 2).
Figure 2.
The frequency of non-truncating mutations in 11 unreported cases NF1 and (+) PS. 73% (8/11) of individuals with NF1 and PS were found to have intragenic non-truncating mutations compared with 19% in the general NF1 population.4
Discussion
Genotype–phenotype correlations have a vital role in clinical genetics. Such correlations, when detected, may affect patient care and surveillance. Moreover, genotype–phenotype correlations may increase our understanding regarding the normal gene function, the pathophysiology of a disease, and assist to prevent and to develop novel therapeutic approaches.
NF1 is characterized, however, by poor genotype–phenotype correlations. Accordingly, it may serve as one of the best examples of a disorder with striking clinical variability, not only between unrelated individuals with similar mutations, but even among affected individuals within the same family. Here, we describe a genotype–phenotype correlation between non-truncating disease causing mutations in the NF1 gene and PS. Such a correlation has been observed before in a cohort of a particular in-frame NF1 gene mutation (c.2970-2972 delAAT)9, 26 and we report a further case with this mutation.
NF1 is caused, in the great majority of the cases, by truncating mutations or whole gene deletions, expected to cause decay or absence of the RNA generated from the mutated allele. The fact that the total amount of protein is translated from a single allele creates a dosage effect. In addition, in such situations the cells are vulnerable for the effect of somatic mutation occurring at the other allele. It is intriguing, therefore, that only non-truncating mutations predicted to be associated with the presence of abnormally functioning protein are associated with PS. This implies that the PS is caused by an additional effect of the mutated protein rather than the common loss-of-function effect.
PS is a classic feature of other Rasopatheis such as Noonan syndrome and multiple lentigines syndrome (Leopard syndrome). As NF1 gene has a role in the Ras–MAPK pathway as well, we believe that PS in NF1, similar to the PS phenotype seen in Noonan syndrome and other Ras–MAPK disorders is related to the role of NF1 in the Ras–MAPK pathway.
In conclusion, it might be possible that the truncating and non-truncating mutations in the NF1 gene may have a similar role in some aspects of the disease, but different roles in other aspects. The fact that PS is associated with non-truncating mutations that, unlike the truncating mutations, do not result in loss of the NF1 protein suggests that the abnormal protein has a special cardiac effect. That effect may be similar to the cardiac effect of Noonan syndrome and other rasopathies and may represent a specific role of these mutations in the Ras–MAPK pathway.
Ras–MAPK inhibitors have been recently suggested to have a therapeutic role for NF1-related learning problems and in Rasopathies in general.22 If indeed, different NF1 mutations affect the activity of the Ras–MAPK pathway differently, than the effect of such drugs may be variable on the patients, based on their specific mutation type. It may be that other features of NF1 and its related disorders may be associated with specific effects of the NF1 mutation type, particularly those that are shared with other Rasopathies such as Noonan-like facies, pectus abnormalities, webbed neck and short stature.
Our findings may not only enable a better understanding of the disease processes and the NF1 gene physiological effect, but may have some practical aspects related to improvement patients' care and surveillance as well. Careful cardiac examination is already part of routine NF1 care in early childhood. If our findings are confirmed in larger studies, perhaps echocardiograms should be performed on children with NF1 and non-truncating mutations.
Finally, the findings of increasing numbers of possible genotype–phenotype correlations in small series of NF1 patients19, 29, 30, 31 suggest the need for renewed interest in this area with further large prospective studies of well-characterized patients.
The authors declare no conflict of interest.
References
- Evans DG, Howard E, Giblin C, et al. Birth incidence and prevalence of tumor-prone syndromes: estimates from a UK family genetic register service. Am J Med Genet A. 2010;152A:327–332. doi: 10.1002/ajmg.a.33139. [DOI] [PubMed] [Google Scholar]
- Ferner RE.Neurofibromatosis 1in Ferner RE, Huson SM, Evans DGR (eds): Neurofibromatoses in Clinical Practice London: Springer; 20111–46. [Google Scholar]
- Messiaen LM, Callens T, Mortier G, et al. Exhaustive mutation analysis of the NF1 gene allows identification of 95% of mutations and reveals a high frequency of unusual splicing defects. Hum Mutat. 2000;15:541–555. doi: 10.1002/1098-1004(200006)15:6<541::AID-HUMU6>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Messiaen LM, Wimmer K.NF1 mutation spectrumin Kaufmann D, (ed.): Neurofibromatoses Basel: Karger; 200863–77. [Google Scholar]
- Kluwe L, Siebert R, Gesk S, et al. Screening 500 unselected neurofibromatosis 1 patients for deletions of the NF1 gene. Hum Mutat. 2004;23:111–116. doi: 10.1002/humu.10299. [DOI] [PubMed] [Google Scholar]
- Le LQ, Parada LF. Tumour microenvironement and neurofibromatosis type 1: connecting the GAPs. Oncogene. 2007;26:4609–4616. doi: 10.1038/sj.onc.1210261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouhilahti EM, Peltonen S, Heam AM, Peltonen J. Pathoaetiology of neurofibromatosis type one. Am J Pathol. 2011;178:1392–1399. doi: 10.1016/j.ajpath.2010.12.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mautner VF, Kluwe L, Friedrich RE, et al. Clinical characterisation of 29 neurofibromatosis type one patients with molecularly aseratined 1.4 Mb type one NF1deletions. J Med Genet. 2010;47:623–630. doi: 10.1136/jmg.2009.075937. [DOI] [PubMed] [Google Scholar]
- Upadhyaya M, Huson SM, Davies M, et al. An absence of cutaneous neurofibromas associated with a 3-bp in-frame deletion in exon 17 of the NF1 gene (c.2970-2972 delAAT): evidence of a clinically significant NF1 genotype-phenotype correlation. Am J Hum Genet. 2007;80:140–151. doi: 10.1086/510781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brems H, Chmara M, Sahbatou M, et al. Germline loss-of-function mutations in SPRED1 cause a neurofibromatosis 1-like phenotype. Nat Genet. 2007;39:1120–1126. doi: 10.1038/ng2113. [DOI] [PubMed] [Google Scholar]
- Denayer E, Chmara M, Brems H, et al. Legius syndrome in fourteen families. Hum Mutat. 2011;32:E1985–E1998. doi: 10.1002/humu.21404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phoenix TN, Temple S. Spred1, a negative regulator of Ras-MAPK-ERK, is enriched in CNS germinal zones, dampens NSC proliferation, and maintains ventricular zone structure. Genes Dev. 2010;24:45–56. doi: 10.1101/gad.1839510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson GH. Pulmonary stenosis, café -au-lait spots, and dull intelligence. Arch Dis Child. 1967;42:303–307. doi: 10.1136/adc.42.223.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allanson JE, Upadhyaya M, Watson GH, et al. Watson syndrome: is it a sub-type of neurofibromatosis. J Med Genet. 1991;28:752–756. doi: 10.1136/jmg.28.11.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassabehji M, Strachan T, Sharland M, et al. Tandem duplication within a neurofibromatosis type 1 (NF1) gene exon in a family with features of Watson syndrome and Noonan syndrome. Am J Hum Genet. 1993;53:90–95. [PMC free article] [PubMed] [Google Scholar]
- Upadhyaya M, Shen M, Cherryson A, et al. Analysis of mutations at the neurofibromatosis 1 (NF1) locus. Hum Mol Genet. 1992;1:735–740. doi: 10.1093/hmg/1.9.735. [DOI] [PubMed] [Google Scholar]
- Colley A, Donnai D, Evans DG. Neurofibromatosis/Noonan phenotype: a variable feature of type 1 neurofibromatosis. Clin Genet. 1996;49:59–64. doi: 10.1111/j.1399-0004.1996.tb04328.x. [DOI] [PubMed] [Google Scholar]
- De Luca A, Bottillo I, Sarkozy A, et al. NF1 gene mutations represent the major molecular event underlying neurofibromatosis-Noonan syndrome. Am J Hum Genet. 2005;77:1092–1101. doi: 10.1086/498454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyström AM, Ekvall S, Allanson J, et al. Noonan syndrome and neurofibromatosis type I in a family with a novel mutation in NF1. Clin Genet. 2009;76:524–534. doi: 10.1111/j.1399-0004.2009.01233.x. [DOI] [PubMed] [Google Scholar]
- Bertola DR, Pereira AC, Passetti F, et al. Neurofibromatosis-Noonan syndrome: molecular evidence of the concurrence of both disorders in a patient. Am J Med Genet A. 2005;136:242–245. doi: 10.1002/ajmg.a.30813. [DOI] [PubMed] [Google Scholar]
- Thiel C, Wilken M, Zenker M, et al. Independent NF1 and PTPN11 mutations in a family with neurofibromatosis-noonan syndrome. Am J Med Genet A. 2011;149A:1263–1267. doi: 10.1002/ajmg.a.32837. [DOI] [PubMed] [Google Scholar]
- Rauen KA, Banerjee A, Bishop WR, et al. Costello and cardio-facio-cutaneous syndromes: moving toward clinical trials in RASopathies. Am J Med Genet C Semin Med Genet. 2011;157:136–146. doi: 10.1002/ajmg.c.30294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin AE, Birch PH, Korf BR, et al. Cardiovascular malformations and other cardiovascular abnormalities in neurofibromatosis 1. Am J Med Genet. 2000;95:108–117. doi: 10.1002/1096-8628(20001113)95:2<108::aid-ajmg4>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Calabrese R, Capriotti E, Fariselli P, Martelli PL, Casadio R. Functional annotations improve the predictive score of human disease-related mutations in proteins. Hum Mutat. 2009;30:1237–1244. doi: 10.1002/humu.21047. [DOI] [PubMed] [Google Scholar]
- Hüffmeier U, Zenker M, Hoyer J, Fahsold R, Rauch A. A variable combination of features of Noonan syndrome and neurofibromatosis type I are caused by mutations in the NF1 gene. Am J Med Genet A. 2006;140:2749–2756. doi: 10.1002/ajmg.a.31547. [DOI] [PubMed] [Google Scholar]
- Baralle D, Mattocks C, Kalidas K, et al. Different mutations in the NF1 gene are associated with neurofibromatosis-Noonan syndrome (NFNS) Am J Med Genet A. 2003;119A:1–8. doi: 10.1002/ajmg.a.20023. [DOI] [PubMed] [Google Scholar]
- Stevenson DA, Viskochil DH, Rope AF, Carey JC. Clinical and molecular aspects of an informative family with neurofibromatosis type 1 and Noonan phenotype. Clin Genet. 2006;69:246–253. doi: 10.1111/j.1399-0004.2006.00576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle B, Baser ME, Huson SM, Cooper DN, Upadhyaya M. Evaluation of genotype-phenotype correlations in neurofibromatosis type 1. J Med Genet. 2003;40:e109. doi: 10.1136/jmg.40.10.e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif S, Upadhyaya M, Ferner R, et al. A molecular analysis of individuals with neurofibromatosis type 1 (NF1) and optic pathway gliomas (OPGs), and an assessment of genotype-phenotype correlations. J Med Genet. 2011;48:256–260. doi: 10.1136/jmg.2010.081760. [DOI] [PubMed] [Google Scholar]
- Messiaen L, Callens T, Williams JB, et al. Genotype-phenotype correlations in spinal NF. (Abstract 985) Presented at the annual meeting of The American Society of Human Genetics, 25 October 2007, San Diego, California. Available from: http://www.ashg.org/genetics/ashg07s/index.shtml .
- Upadhyaya M, Spurlock G, Kluwe L, et al. The spectrum of somatic and germline NF1 mutations in NF1 patients with spinal neurofibromas. Neurogenetics. 2009;10:251–263. doi: 10.1007/s10048-009-0178-0. [DOI] [PubMed] [Google Scholar]
- Thomas L, Richards M, Mort M, Dunlop E, Cooper DN, Upadhyaya M.Assessment of the potential pathogenicity of missense mutations identified in the GTPase-activating protein (GAP)-related domain of the neurofibromatosis type-1 (NF1) gene Hum Mut2012; e-pub ahead of print 16 July 2012; doi: 10.1002/humu.22162 [DOI] [PubMed]