Abstract
We use genotype data from the Marshfield Clinical Research Foundation Personalized Medicine Research Project to investigate genetic similarity and divergence between Europeans and the sampled population of European Americans in Central Wisconsin, USA. To infer recent genetic ancestry of the sampled Wisconsinites, we train support vector machines (SVMs) on the positions of Europeans along top principal components (PCs). Our SVM models partition continent-wide European genetic variance into eight regional classes, which is an improvement over the geographically broader categories of recent ancestry reported by personal genomics companies. After correcting for misclassification error associated with the SVMs (<10%, in all cases), we observe a >14% discrepancy between insular ancestries reported by Wisconsinites and those inferred by SVM. Values of FST as well as Mantel tests for correlation between genetic and European geographic distances indicate minimal divergence between Europe and the local Wisconsin population. However, we find that individuals from the Wisconsin sample show greater dispersion along higher-order PCs than individuals from Europe. Hypothesizing that this pattern is characteristic of nascent divergence, we run computer simulations that mimic the recent peopling of Wisconsin. Simulations corroborate the pattern in higher-order PCs, demonstrate its transient nature, and show that admixture accelerates the rate of divergence between the admixed population and its parental sources relative to drift alone. Together, empirical and simulation results suggest that genetic divergence between European source populations and European Americans in Central Wisconsin is subtle but already under way.
Keywords: population structure, genetic ancestry, admixture, support vector machine, principal component analysis
Introduction
Immigration from around the world to the same region of the United States effectively collapses the isolating distances and geographic barriers responsible for generating current patterns of worldwide population structure. In some cases, the collapsed distances are vast. For example, groups such as African-, Mexican-, and Puerto Rican-Americans are the product of admixture between two or three highly differentiated populations. These and other similarly admixed groups are frequently studied, most often to map variants responsible for disease, detect natural selection, and infer population history.1, 2, 3, 4 Importantly, many local US populations sustain admixture between substantially less divergent sources, often between a variety of European nationalities. The population genetic consequences of this admixture dynamic are seldom examined.
Admixture in the United States can produce highly complicated, locally conditional patterns of population structure (see, eg, Sloan et al5). In these cases, high-resolution ancestry inference may be difficult or impossible, because it requires mapping genetic variation back to multiple source populations of limited differentiation. Indeed, methods that explicitly quantify admixture by identifying the ancestries of chromosomal segments are almost exclusively applied to populations whose ancestral inputs are separate species or historically allopatric populations.6, 7, 8, 9, 10 In general, the resolution of ancestry inference in the United States will be conditional on the level of admixture in the target population (Figure 1a). High rates of admixture will efface detailed genetic connections between a local US population and its ancestral sources. On the other hand, limits to admixture in the United States will preserve genome-wide genetic similarity to a single ancestral population, thereby simplifying the task and increasing the resolution of ancestry inference. At least two factors limit the prevalence of admixture in the United States. First, any positive assortative mating based on sociocultural criteria will constrain admixture. Second, the recency of immigration to the United States implies that an appreciable fraction of Americans will by chance remain largely unadmixed with respect to a non-American source population.
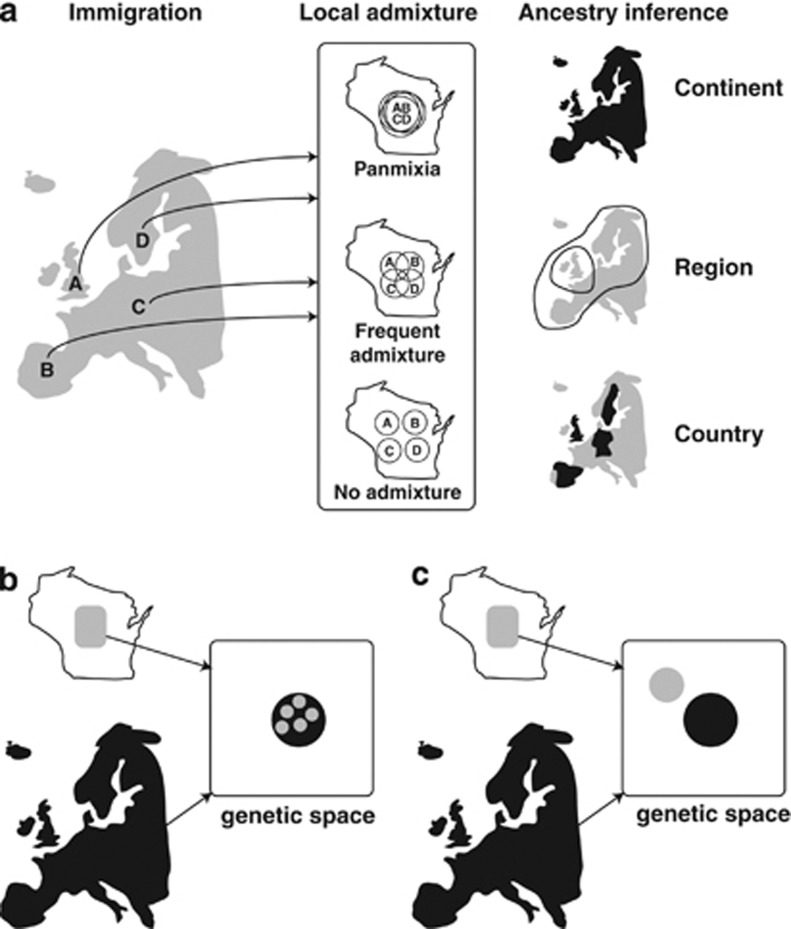
Figure 1.
(a) The resolution of ancestry inference in a local US population is dependent on the level of admixture in the population. For example, in a population of European Americans, the resolution of ancestry inference might range from continent (low resolution), when the local population is panmictic, to country (high resolution), when there is little or no admixture within the local population. (b) Initially, sink (US) and source (European) populations are coincident in genetic space as determined by methods such as PCA. (c) After some period of time, sink and source populations grow divergent because of genetic drift and admixture; the actual pattern of divergence in genetic space will vary depending on, at least, the level of admixture within the sink population.
Here, we focus on a local population of European Americans in Central Wisconsin, the subject of the Marshfield Clinical Research Foundation Personalized Medicine Research Project (PMRP).11 The population sampled by the PMRP is a complex, idiosyncratic mixture of closely related ancestral sources. Historical immigration to Wisconsin as a whole was predominated by Germans. By 1880, Wisconsin possessed the greatest percentage of German-born residents in the United States at 53.8%.12 In all counties sampled by the PMRP, >25% of individuals claim some German ancestry.13 Other common European ancestries in the Central Wisconsin population are Czech, Dutch, English, French, Irish, Norwegian, Polish, and Swedish. The ancestral composition of the Central Wisconsin population is therefore complex but mostly limited to groups that are not highly differentiated at the genetic level. Thus, the PMRP sample is ideal for studying the limits to fine-scale ancestry inference in the United States as well as admixture among closely related groups.
Recent proliferation of direct-to-consumer personal genomics companies in the United States suggests high demand for knowledge of personal genetic ancestry.14, 15 Although the working definition of genetic ancestry used by companies is frequently difficult to discern,16, 17 consumers seem most interested in obtaining knowledge of recent ancestry at higher resolution than European or Southeast Asian.18, 19 Continuous population structure among ancestral populations and admixture in the United States make it difficult to meet this demand. Personal genomics companies therefore limit services to inference of ancient ancestry based on analysis of non-recombining DNA and coarse inference of recent ancestry using SNP chip data.17 For example, the company 23andMe currently classifies the recent ancestry of chromosomal segments as African, Asian, and European, though expansion to numerous other broad geographic regions is anticipated.19
Using genotype data from PMRP participants, we aimed to determine if modifying current methodology might improve resolution of European ancestry estimates in European Americans. In particular, we use support vector machines (SVMs) trained on the results of principal component analysis (PCA) to classify individuals. We find the PCA+SVM approach attractive because it provides an objective means to classify the continuous variation of PC plots, produces reusable models that can be applied to newly sampled individuals, and potentially provides the framework for more difficult classification tasks, such as diagnosing specific admixed ancestries. In this study, application of the PCA+SVM approach to the PMRP sample allows us to quantify discrepancies between self-reports of insular ancestry and model-based predictions as well as increase the resolution of insular ancestry inference to eight ancestral classes within Europe. We note, however, that the current implementation of this approach is limited to inference of insular ancestry. Furthermore, although we define ancestral regions (classes) in a systematic manner, the performance of the PCA+SVM approach is dependent on the quality of class definitions.
For some time following European colonization of the focal Central Wisconsin population, immigrants and their descendants will overlap with European populations in genetic/PC space (Figure 1b). However, assuming low gene flow from Europe to Central Wisconsin following the initial wave of European immigration, the Central Wisconsin population should diverge from Europe via genetic drift (Figure 1c). To investigate the details of incipient divergence, we used simulations that broadly capture the recent peopling of Central Wisconsin and future Wisconsin/Europe divergence. We were specifically interested in identifying any signs of nascent divergence between the contemporary PMRP sample and Europe. To this end, our simulations focused on the elevated rate of admixture between individuals of various European ancestries in Wisconsin relative to Europe. We reasoned that if a pattern of nascent divergence was detectable in the PMRP sample, it was likely to originate from the relatively high frequency of admixture in the United States, which distinguishes mating in Wisconsin from that in Europe. Interestingly, both empirical and simulation results suggest increased variance along higher-order PCs as an indicator of nascent divergence. Our results suggest that the studied Central Wisconsin population is already actively diverging from ancestral populations in Europe.
Materials and methods
Subjects and genotype data
The PMRP is a population-based DNA biobank organized and operated by the Marshfield Clinic Research Foundation, Marshfield, WI, USA.11 The biobank includes >20 000 participants, sampled from 19 communities in Central Wisconsin, USA. Self-reports of ancestry by PMRP participants was limited to combinations of ‘Other Ancestry' and nine Northern European ancestries: Czech, Dutch, English, French, German, Irish, Norwegian, Polish, and Swedish (see Supplementary Text for more detailed information). German was the most commonly reported ancestry, with 77% of genotyped participants reporting either insular or admixed German ancestry. Nearly 4000 participants have been genotyped at >500 000 SNPs, including 3903 European Americans (PMRPunabridged), of which 2009 reported insular ancestry (PMRPinsular). Because close relatives and/or uneven sample sizes generate skewed PCA results,20 we created another data set by removing close relatives based on pedigree data and 847 of 1047 individuals reporting insular Germany ancestry (PMRPabridged). Finally, PMRPinsular-abridged included the 546 Wisconsinites from PRMPabridged who reported insular ancestry. The Population Reference Sample (POPRES) includes several thousand participants from around the world genotyped at 500 000 SNPs.21 We only used individuals whose country of origin was the same as the reported ancestry of their maternal and paternal grandparents. English and Swiss nationalities are overrepresented in POPRES. Therefore, we randomly eliminated 702 (of 902) individuals sampled from England and 1300 (of 1500) individuals samples from Switzerland. This resulted in a sample of 1247 Europeans (POPRESEurope), including 474 individuals from the nine countries specifically reported as ancestral by participants of the PMRP (Supplementary Table S1).
Genome-wide genotyping of PMRP individuals was performed at the Center for Inherited Disease Research (CIDR; John Hopkins University, Baltimore, MD, USA) using the Illumina 660W-Quad Platform (Illumina, San Diego, CA, USA). Genotyping calls were made at CIDR using BeadStudio version 3.3.7 (Illumina). POPRES individuals were genotyped on the Affymetrix Gene Chip 500K Array (Santa Clara, CA, USA). Hardy–Weinberg equilibrium (HWE) tests were performed separately for PMRPabridged and POPRESEurope (exact test implemented in PLINK22; HW disequilibrium when P-value <0.05). Only SNPs in HWE in both samples were used. The set of SNPs that met this criterion were pruned for LD, after which 75 301 SNPs common to both platforms remained (LD pruning performed in PLINK22 using parameter values: 50 SNP window, 5 SNP shift, and variance inflation factor=2). We removed eight SNPs with excessive values of FST >0.1 (POPRESEurope vs PMRPabridged). Another set of 106 FST outliers (FST>0.9) were identified as G/C polymorphisms for which the reported allele was switched between platforms. In these cases, we reversed PMRP genotypes to match POPRES calls (see Supplementary Figure S1 for final FST distribution). All analyses used the remaining 75 293 SNPs; average spacing between SNPs was 38 kb. Frequency spectra of the PMRPabridged and POPRESEurope data sets were nearly identical (Supplementary Figure S1). The fraction of missing genotypes was substantially higher in POPRES than PMRP (on average, 2.47 vs 0.02% missing/individual, respectively). We assessed the effect of this disparity by generating an artificial PMRP data set in which 1861 (2.47%) random genotypes were coded as missing data in each individual. Analyses using the artificial and original data sets produced qualitatively similar results. The position of missing SNPs was not correlated across individuals of either sample.
Combined and projection PCA
We refer to PCA on a PMRPabridged+POPRESEurope data set as combined PCA. We ran PCA in SMARTPCA (EIGENSOFT v3.0),23 with five iterations of outlier removal using the first 10 PCs to identify outliers. Further iterations of outlier removal did not alter results substantially. To further assess the possibility that nonrandom missing data might mislead inference of population structure, we also performed PCA runs in which only patterns of missing data were used to compute PCs. We did not find evidence of informative missing data. We refer to projection of a PMRP data set onto PCs computed from the POPRESEurope sample as projection PCA. PC scores of projected individuals are often biased toward zero relative to individuals used to compute PCs. This has the effect of shrinking the plot of projected data. We used the algorithm of Lee et al24 to calculate the ‘shrinkage factor' for each PC of interest. We multiplied projected PC scores by the reciprocal of the shrinkage factor to correct for projection bias.
Using SVMs to predict ancestry of Wisconsinites claiming insular ancestry
SVM is a form of supervised learning applied to classification problems.25 Each training datum is of the form (xi, yi). The feature vector xi characterizes individual i in n dimensions, whereas yi is a discrete variable that indicates to which of k classes individual i belongs. The training data set is used to identify support vectors – a subset of the xi that help define boundaries between classes. The support vectors and other parameter estimates form a model (the SVM), which can be used to predict the class of an individual not used in training the SVM.
We used POPRESEurope as the training data set. Each xi consisted of PC1 and PC2 scores for individual i (combined or projection PCA), whereas each corresponding yi identified the European region from which the individual was sampled. Inclusion of higher-order PCs in xi resulted in very modest increases in training accuracy. Therefore, to simplify visualization we only used the first two PCs. The test data set was either PMRPinsular (projection PCA) or PMRPinsular-abridged (combined PCA). We used the R26 e1071 package27 to train SVMs and perform SVM-based ancestry prediction. For each European class, we first eliminated outliers using a one-class SVM. In one-class SVM, the parameter η is the proportion of individuals to be removed as outliers. Because the choice of η is subjective, we performed the full SVM analysis using η=0.1, 0.2, and 0.4 to assess the effects of different values of η. Parameter γ was set to the reciprocal of sample size, a standard value. We then trained a multiclass SVM using only nonoutlier Europeans (C-classification, radial basis function kernel). We used the tune.svm function to identify optimal values of hyperparameters γ and c. Ten-fold cross-validation was used to estimate class-specific and overall misclassification error for each model.
We assessed resolution of ancestry inference by training SVMs using various partitions of POPRESEurope as class definitions. The highest tested resolution treated each European country as a class. Lower-resolution classes grouped European countries into more inclusive classes. We used the set of class definitions that included the greatest number of classes, yet yielded an overall misclassification rate of <10% for both projection and combined PCA trainers. This optimal class set contained eight regional classes: British Isles=Ireland+Great Britain+Scotland; Scandinavia=Denmark+Norway+Sweden; Northeast=Czech Republic+Hungary+Poland+Slovakia; West=Belgium +France+Switzerland; Central=Austria+Germany+Netherlands; Iberia=Spain+Portugal; Mediterranean=Italy+Greece; Southeast=Albania+Bosnia and Herzegovina +Bulgaria+Croatia+Macedonia +Romania+Serbia +Slovenia.
We used function predict.svm to predict the ancestry of PMRP participants who reported insular ancestry. It is not possible to model admixed classes in an SVM based on PC data alone because admixed individuals lie intermediate to source populations along top PCs.23 Discrepancies between reported ancestry and estimated ancestry were quantified for each ancestry i as: % discrepancy=(100/n)[di–(di × MCEi)], where di is the number of individuals (out of n sampled individuals) who reported ancestry i but were predicted (by SVM) to have a different ancestry and MCEi is the misclassification error specific to ancestral class i.
FST and correlations between genetic and geographic distances
To calculate FST and genetic–geographic correlations, we treated the countries of Europe as populations. Europeans identified as outliers by one-class SVM were excluded from calculations. Moreover, we only included Wisconsinites whose self-reports of insular ancestry were confirmed by SVM. Only groups represented by ≥10 individuals were included. We calculated FST for each SNP28 and report the arithmetic mean of FST at all SNPs. Correlations between genetic and geographic distances were assessed using Mantel tests (P-values based on 100 000 permutations). We measured genetic distance as (1) Euclidean distances between group means on the PC1/PC2 biplot (projection PCA of PMRPinsular+POPRESEurope), or (2) standardized FST=FST/1−FST.29 Geographic distance was calculated as the natural log of great-circle distance between the geographical centers of the relevant countries in Europe.
Simulating Wisconsin/Europe data sets
We simulated SNP chip data from a source continent analogous to Europe using coalescent simulations embedded in MARKSIM.30 A total of 75 293 unlinked SNPs were simulated to match the number of SNPs in the empirical data set. The source continent comprised five populations exchanging frequent migrants (see Supplementary Text). We used a sample of 100 diploids from each population to estimate allele frequencies of each source population. We assumed migration-drift equilibrium on the source continent – that is, allele frequencies remained the same for each source population throughout the simulation. The first generation of the sink population (analogous to the local Central Wisconsin population) consisted of 1000 diploids of insular ancestry (40% population A, 20% populations B and C, 15% population D, and 5% population E). Each genotype of an individual immigrant was simulated via binomial sampling based on estimated allele frequencies of the relevant source population. Subsequent generations in the sink population consisted of two steps. First, new immigrants arrived (Supplementary Table S2). Second, we simulated reproduction by randomly choosing mating pairs with replacement from the population and a maternal and paternal allele at random. Total population size only grew in response to the influx of new immigrants. We also ran comparative simulations in which reproduction was not allowed between individuals whose lineages traced back to different source populations (ie, no admixture). We emphasize that the interpretation of these simulations is mostly qualitative. Our primary aim was to identify patterns of genetic differentiation (as captured by PCA) that arise in recently founded populations subject to frequent admixture between closely related sources.
Results
PCA
Combined PCA
We retained the first 10 PCs, all of which were statistically significant (Tracy–Widom statistic,23 P≪0.0001). However, each of the top 10 PCs accounted for <0.25% of the variance (Supplementary Figure S2), which agrees with a previous analysis of population structure in the European POPRES sample.31 Plots of PCs 1–3 showed that: (1) PCs 1 and 2 roughly correspond to N-S and E-W geographic axes, respectively (Figure 2a); (2) the PMRP sample is mostly Northern European (Figure 2a); (3) Wisconsinites of self-reported insular origin generally cluster around appropriate European means, although outliers are evident (Supplementary Figure S3); (4) Wisconsinites and Northern Europeans occupy the same extent of PC space along these axes of variation (Figure 2a and Supplementary Figure S4). Higher-order PCs did not correspond to geography in a straightforward manner. However, dispersion of Wisconsinites along PCs 5–10 was noticeably greater than that of northern Europeans (Figure 2b and Supplementary Figures S5 and S6). This was true even after accounting for differences in missing data between the two data sets (Supplementary Figure S7). None of the outlying Wisconsinites on PCs 5–10 (Figure 2b) were outliers on PCs 1 or 2, and most reported admixed ancestry. Combined PCA of Wisconsinites and individuals from around the world also revealed PMRP outliers along higher-order PCs (Supplementary Figure S8).
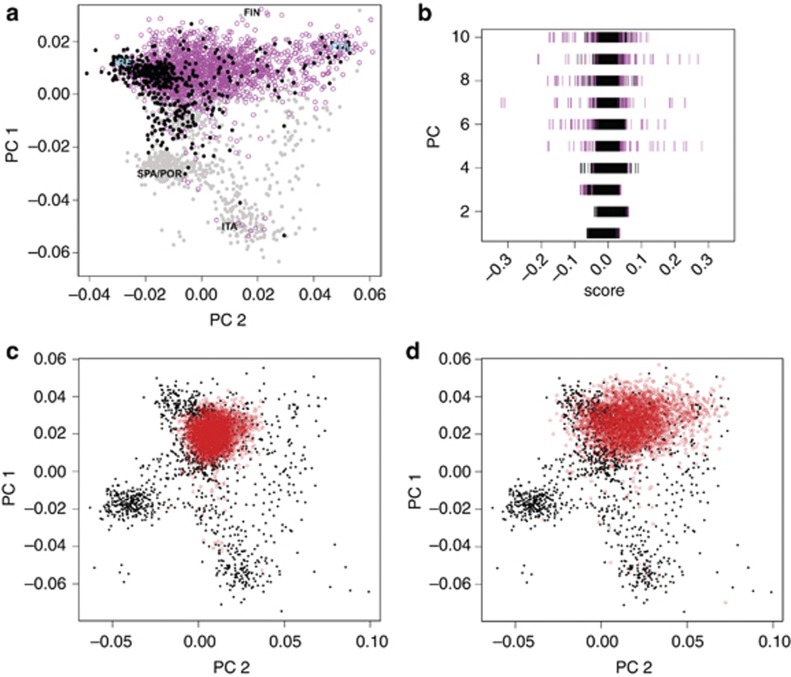
Figure 2.
Empirical PCA results. (a) The PC1/PC2 biplot from combined PCA corroborates the Northern European ancestry of most European-American participants of the PMRP. Shown are Europeans from POPRES (filled circles, black for individuals from one of the nine countries reported in the PMRP sample, gray circles for others), and Wisconsinites (open, magenta circles). Abbreviations mark the mean position of POPRESEurope ancestries (IRE: Ireland; FIN: Finland; POL: Poland; SPA/POR: Spain+Portugal; ITA: Italy). (b) Scores for PCs 1–4 from combined PCA are equally variable in Europeans (black bars) and Wisconsinites (magenta bars); however, Wisconsinites show visibly greater variation along PCs 5–10. (c) PMRPabridged individuals (open, red circles) projected onto PCs 1 and 2 computed from POPRESEurope data only. (d) The same as (c) after PMRP PC scores were corrected for projection bias following Lee et al.32
Projection PCA
Bias associated with projection PCA is visibly evident in Figure 2c, which shows little overlap between projected Wisconsinites and Europeans sampled from Ireland, the Czech Republic, and Poland despite hundreds of Wisconsinites reporting insular ancestry from these countries. Multiplying projected PC scores by the reciprocals of their shrinkage factors corrected for projection bias. Boundaries of the corrected PMRP sample matched the boundaries of the European sample in the PC1/PC2 biplot (Figure 2d). A more direct demonstration of the efficacy of projection bias correction using POPRES individuals is shown in Supplementary Figure S9, and results from projection PCA can help resolve conflicts in reported ancestry between close relatives (Supplementary Figure S10). Unlike combined PCA, we did not observe greater dispersion of projected Wisconsinites along higher-order PCs.
Testing claims of insular ancestry in Wisconsinites
Following outlier removal by one-class SVM (η=0.2), 976 European individuals from the projection PCA results remained. We used these individuals to tune and train an eight-class SVM (SVMprojection). Estimated misclassification error for the overall model was 0.079, and 163 of 976 individuals were used as support vectors (Figure 3c and Supplementary Table S3). We used the SVMprojection model to predict the ancestry of all 2009 Wisconsinites claiming insular ancestry (Table 1). After correcting for class-specific misclassification errors, we observed a discrepancy between reported and predicted ancestry in 22.7% of individuals claiming insular ancestry. Discrepant reports were particularly common in individuals claiming Czech, Dutch, and French ancestry (>30% in all cases). According to the SVMprojection model, 23% of the 1558 Wisconsinites claiming insular German ancestry were predicted to be mistaken (Figure 3d). After outlier removal by one-class SVM on combined PCA results (η=0.2), 979 European individuals remained. We used these individuals to train a separate eight-class SVM (SVMcombined). Estimated misclassification error for the overall model was 0.094, and 179 of 979 individuals were used as support vectors (Figure 3a; Supplementary Table S3). Because Wisconsinites from the PMRP sample were used to compute PCs, we used PMRPinsular-abridged in combined PCA. Thus, the ancestries of only 546 Wisconsinites were tested using SVMcombined (Table 1). After correcting for class-specific misclassification errors, we observed a discrepancy between reported and predicted ancestry in 14.4% of individuals using the SVMcombined model. Discrepant reports were particularly common for individuals claiming Dutch or French ancestry: 43 and 57%, respectively. Only 9% of the 165 included Wisconsinites claiming insular German ancestry provided discrepant reports (Figure 3b). Here, we focus on results where η=0.2 in one-class SVMs. However, the proportion of discrepant reports was highly similar across all tested values of η (SVMcombined and SVMprojection; Supplementary Tables S3 and S4). The only exceptions involved the Scandinavian regional class in the SVMcombined analysis, likely because of difficulty estimating misclassification error by cross-validation using a small training data set (only 14 Scandinavian individuals remained after one-class outlier removal when η=0.4).
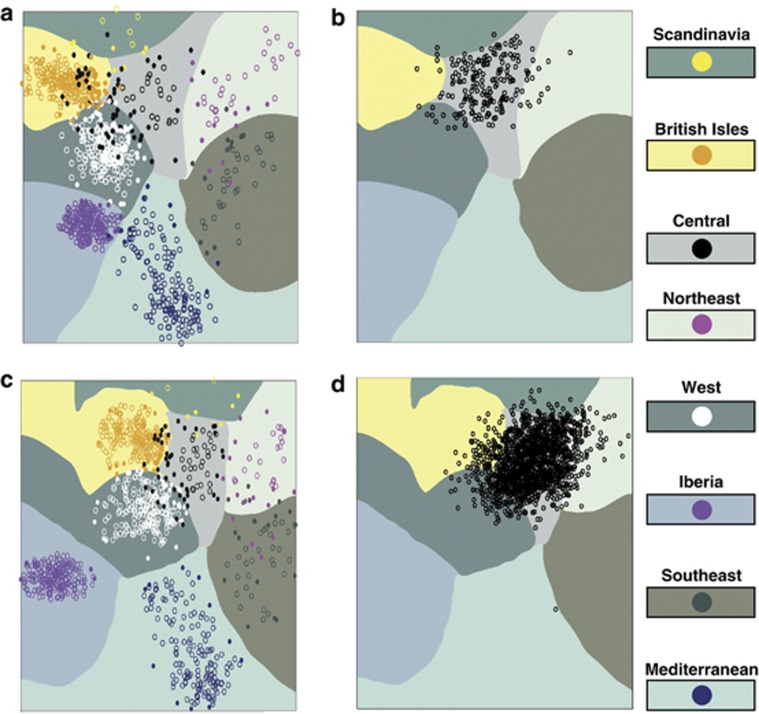
Figure 3.
Multiclass SVM models trained on PC1 and PC2 scores from combined and projection PCA. (a) SVM trained on combined PCA results. The underlying contour map roughly outlines the decision boundaries for the eight regional classes. Data points are colored according to the class of the training datum. Filled circles are support vectors. (b) Positions of PMRPinsular-abridged individuals claiming German insular ancestry superimposed on the SVM based on combined PCA. (c) SVM trained on projection PCA results. (d) Positions of PMRPunabridged individuals claiming German insular ancestry superimposed on the SVM based on projection PCA.
Table 1. Ancestry prediction based on multiclass SVM of eight ancestral European classes (see Materials and Methods for countries included in each class).
| Irish | English | French | Swedish | Norwegian | Dutch | German | Czech | Polish | |
|---|---|---|---|---|---|---|---|---|---|
| Iberia | 0/0 | 0/0 | 0/1 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 |
| British Isles | 39/29 | 61/62 | 8/6 | 3/1 | 24/7 | 8/11 | 99/5 | 1/0 | 0/0 |
| West | 1/0 | 3/1 | 12/8 | 0/0 | 1/1 | 2/1 | 177/11 | 0/0 | 0/0 |
| Scandinavia | 0/0 | 1/2 | 0/0 | 13/18 | 28/50 | 2/4 | 45/8 | 0/0 | 2/0 |
| Central | 4/1 | 31/13 | 3/3 | 9/2 | 27/2 | 12/5 | 1051/165 | 14/8 | 8/5 |
| Mediterranean | 0/0 | 0/0 | 1/0 | 0/0 | 0/0 | 0/0 | 1/0 | 0/0 | 0/0 |
| Northeast | 0/0 | 4/3 | 0/2 | 1/0 | 0/0 | 1/1 | 185/9 | 22/21 | 105/80 |
| Southeast | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 1/0 | 0/0 |
| n | 44/30 | 100/81 | 23/19 | 26/21 | 80/59 | 25/22 | 1558/198 | 38/29 | 115/85 |
| di | 5/1 | 39/19 | 11/12 | 13/3 | 52/10 | 13/17 | 507/33 | 16/8 | 9/5 |
| Est. MCE | 0.02/0.02 | 0.02/0.02 | 0.01/0.04 | 0.78/0.35 | 0.78/0.35 | 0.29/0.45 | 0.29/0.45 | 0.22/0.15 | 0.22/0.15 |
| di(corr) | 4.9/1.0 | 38.4/18.6 | 10.9/11.5 | 2.9/2.0 | 11.5/6.5 | 9.2/9.4 | 358.5/18.2 | 12.4/6.8 | 7.0/4.3 |
| est. % discrepant | 11/3 | 38/23 | 48/57 | 11/9 | 14/11 | 37/43 | 23/9 | 33/23 | 6/5 |
Columns are self-reported insular ancestries of Central Wisconsinites and rows are predicted European ancestry. di is the raw number of discrepancies between reported and predicted ancestry, Estimated MCE is class-specific misclassification error estimated in SVM training, and di(corr) is the corrected (by MCE) number of discrepancies. Results of projection analysis are shown before slash and results of combined analysis after slash. Bold numbers indicate cells where reported ancestry and ancestral class match. Results correspond to analyses where η=0.2 in preparatory one-class SVM.
Genetic differentiation and genetic–geographic correlation
Comparable values of pairwise FST were strikingly similar in Wisconsin and Europe (Table 2). For example, FST between Poland and Germany was 0.00127 whereas FST between insular Wisconsinites of Polish and German ancestry was 0.00125. In the three cases where sample size allowed direct comparison, differentiation was nearly nonexistent. FST equaled 0.00016, 0.00039, and 0 for England–English, Germany–German, and Poland–Polish comparisons, respectively (Table 2c). The first two values were, however, significantly different than zero (permutation test; P<0.0001 in both cases).
Table 2. (a) Intra-Europe, (b) intra-Wisconsin, and (c) intercontinental F ST ( × 1000) values.
| (a) | Ireland | England | France | Germany | Poland | |
|---|---|---|---|---|---|---|
| Ireland | — | 0.48 | 1.32 | 1.67 | 2.65 | |
| England | — | 0.63 | 0.62 | 1.74 | ||
| France | — | 0.91 | 2.29 | |||
| Germany | — | 1.27 | ||||
| Poland | — | |||||
| (b) | English | Norwegian | Swedish | German | Czech | Polish |
| English | — | 0.66 | 0.17 | 0.24 | 0.97 | 2.11 |
| Norwegian | — | 0 | 0.82 | 1.29 | 2.36 | |
| Swedish | — | 0.28 | 0.73 | 1.58 | ||
| German | — | 0.22 | 1.25 | |||
| Czech | — | 0.24 | ||||
| Polish | — | |||||
| (c) | Ireland | England | France | Germany | Poland | |
| English | 0.62 | 0.16 | 0.53 | 0.62 | 1.74 | |
| Norwegian | 1.53 | 0.96 | 1.93 | 1.17 | 1.97 | |
| Swedish | 1.56 | 0.71 | 1.44 | 0.81 | 1.14 | |
| German | 1.49 | 0.56 | 0.82 | 0.39 | 0.81 | |
| Czech | 2.16 | 1.28 | 1.05 | 0.77 | 0 | |
| Polish | 3.28 | 2.46 | 2.82 | 1.88 | 0 |
Only groups represented by ≥10 individuals were considered. Bold values indicate a direct comparison between a European nationality and Wisconsinites self-reporting insular ancestry from that nation.
Using a PC-based measure of genetic distance, we found a significant correlation between genetic distances in Europe and Wisconsin (Table 3; P<0.0001). The correlation between PC-based genetic distances in Wisconsin and European geographic distances was significant (Mantel's r=0.57; P=0.0054) and nearly identical to the correlation between European genetic and geographic distances (Mantel's r=0.59; P=0.0029). Regressing PC-based genetic distance on European geographic distance, we found an identical relationship in Wisconsin and Europe (Supplementary Figure S11; slopes not statistically different, F-test, P=0.136). Interestingly, all but two of the comparisons falling above the trend lines in Supplementary Figure S11 include the Czech Republic or Poland, which suggests a discontinuity in gene flow between the Northeast ancestral region we defined and other regions of Northern Europe. Using standardized FST as the measure of genetic distance, correlation between European genetic and geographic distances was nearly double the correlation using Wisconsin genetic distances (Mantel's r=0.88 and 0.48, respectively; Table 3). Both correlations were significant, although P-values were notably higher than for the same comparisons using PC-based genetic distance. Note that because of insufficient sample size, many ancestral groups were not included in the comparisons using standardized FST.
Table 3. Mantel tests for correlation between matrices of genetic and geographic distances.
| Distance matrices compared | Genetic distance metric | Mantel's r | P-value |
|---|---|---|---|
| EURgen vs EURgeo | PC biplot | 0.59 (0.438, 0.853) | 0.0029 |
| WIgen vs EURgeo | PC biplot | 0.57 (0.455, 0.780) | 0.0054 |
| EURgen vs WIgen | PC biplot | 0.89 (0.845, 0.936) | 0.00008 |
| EURgen vs EURgeo | FST/(1−FST) | 0.88 (0.782, 0.938) | 0.0166 |
| WIgen vs EURgeo | FST/(1−FST) | 0.48 (0.354, 0.778) | 0.049 |
Abbreviations: EURgen: European genetic distance; Wigen: Wisconsin genetic distance; EURgeo: European geographic distances (natural log of distance between geographic centers).
Genetic distances are either the Euclidean distance between country means on a projection PC1/PC2 biplot (PC biplot) or standardized FST.
Simulating source–sink dynamics between Europe and Wisconsin
We simulated five source populations connected by high levels of gene flow (Supplementary Figure S12). After establishment of the sink population, source and sink individuals showed roughly equal dispersion along all PCs (Figure 4a and c, generation 0). After five generations of immigration and ubiquitous admixture, however, FST(sink–source) increased to 0.0005 and dispersion along PC6 was noticeably greater in the sink population (Figure 4c); the latter result is qualitatively similar to empirical results summarized in Figure 2b. Although admixture among individuals of European descent in Wisconsin is a recent phenomenon, we traced simulated source–sink populations for 75 generations to obtain data relevant to a future Central Wisconsin population. We were particularly interested in the longevity of the higher-order PC pattern and if sink and source populations would eventually diverge along top PCs. After 75 generations of immigration and admixture, source and sink populations did form discrete clusters along PC1 (Figure 4b) and FST(sink–source) had risen to 0.004. Greater dispersion of PC6 scores within the sink population was still evident at 50 generations after colonization, but not after 75 generations (Figure 4c). This result is qualitatively similar to analytical results that show that variance of admixture proportions in an admixed population decreases rapidly after contributions from source populations diminish.32
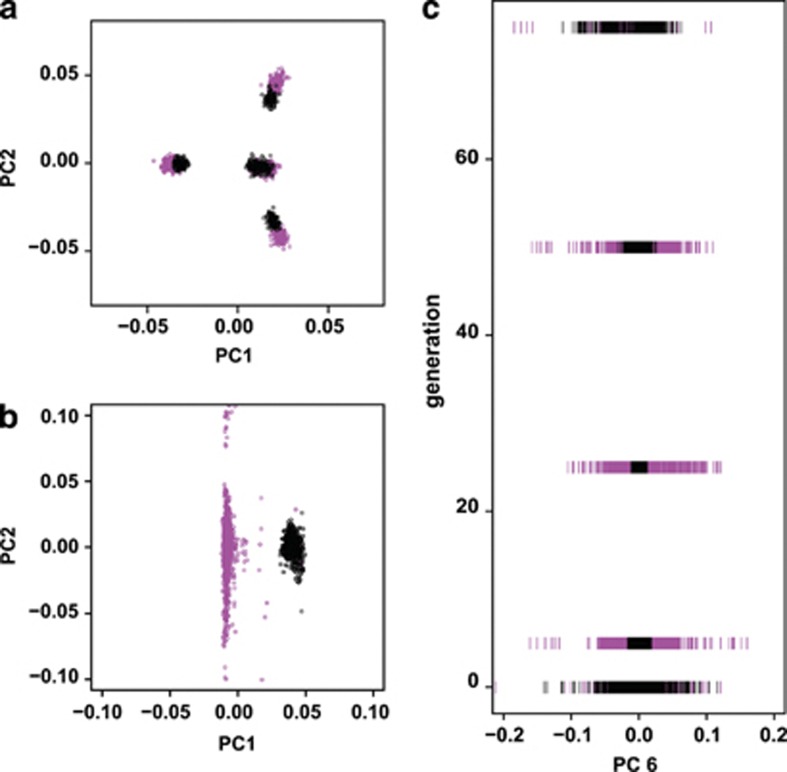
Figure 4.
Simulation PCA results (admixture plus drift). (a) PC1/PC2 biplot of source (black) and sink (magenta) populations immediately after the founding of the sink population. At this point, sink and source populations are largely coincident with differences likely due to founder effects. (b) PC1/PC2 biplot of source and sink populations 75 generations after colonization. Source and sink populations are divergent, forming discrete clusters of individuals. (c) PC6 through time. PC6 scores from sink and source individuals are equally variable upon founding of the sink population. At 5, 25, and 50 generations after colonization, however, the sink population shows much greater variation, reminiscent of patterns in Figure 2b. By 75 generations after colonization, sink and source populations once again show equal variation along PC6.
A qualitatively different pattern of divergence between sink and source populations emerged in the absence of admixture. Individuals descended from distinct source populations formed mostly discontinuous clusters on the PC1/PC2 biplot (Supplementary Figure S13). By comparison, under the admixture plus drift scenario, the sink population diverged from source populations as a single, discrete cluster (Figure 4b and Supplementary Figure S13). Under the drift-only scenario, sink individuals still exhibited greater dispersion along higher-order PCs than source individuals (Supplementary Figure S13). However, average pairwise distance between source and sink individuals at 40 generations after colonization was nearly twice as great under the admixture+drift scenario than under the drift-only scenario (0.05 and 0.027, respectively).
Discussion
PCA, SVMs, and inference of American ancestry
Projection PCA seems well-suited to the task of inferring the ancestry of Americans. Superimposition of Americans on genetic maps computed from worldwide genetic variation provides a graphical and highly intuitive method for diagnosing and visualizing genetic ancestry. Indeed, companies such as 23andMe33 use results from projection PCA to illustrate an individual's relationship to worldwide variation. Attractively, because reference PCs are computed separately, even close relatives may be included in the projected sample. Thus, ancestry of families or larger groups of closely related individuals may be assayed (Supplementary Figure S10). However, we emphasize that projected PC scores must be corrected for projection bias.24 It is not obvious that previous examples of projection PCA corrected for projection bias.34, 35 We also note that although combined PCA suffers from the need to remove closely related individuals, broadly similar results are obtained (Table 1 and Supplementary Tables S3 and S4).
An important advantage to analyzing human genetic variation via PCA is that the method implicitly acknowledges the continuous nature of most human population structure. Plots such as those found in Figure 2 graphically demonstrate the continuity and make it easy to intuit genetic distances between individuals. Still, consumers prefer labeled ancestries to those defined as a relative position on a map of genetic distances.17 To this end, we can discretize continuous PC (genetic) space and assign individuals to the resulting ancestral classes. Several methods have been used to do this. Using top PC scores as input, discriminant analysis assigns individuals to clusters a posteriori.36 The SVM approach used here requires a priori definitions of classes. However, these were rooted in the political geography of Europe, which is preferable in the context of ancestry inference. For example, a European American is more likely to have interest in whether or not he/she is a Scandinavian than if he/she belongs to the third objective cluster from the right. The k-nearest neighbor (k-NN) algorithm37 identifies the k nearest neighbors of an individual (on the PC map) and infers his/her ancestry as the average geographic sampling location of the k neighbors. Attractively, k-NN also does not require a priori class definitions. However, the accuracy of k-NN depends on the accuracy of two underlying transformations. The first transformation is from PC coordinates to geographic sampling location, which is not problematic when genetic–geographic correlation is high, as it is here. However, k-NN also depends on accurate transformation from sampling location to fine-scale geographic ancestry. This is often not available. For example, the sampling locations of POPRES individuals are mostly limited to country of origin.21 In contrast, SVM classification only requires that members of the training set belong to the relatively broad geographic classes they represent. Finally, Heath et al38 present a method of ancestry inference in which the probability of an unknown individual belonging to a particular ancestry is weighted by his or her position along all PCs showing separation between the mean positions of input countries. Although not reported here, output from SVM prediction includes similar probabilities, which could be used to refine ancestry estimates. Furthermore, SVMs trained on the results of projection PCA are reusable models. If satisfied that the reference (training) data set is representative of the ancestral classes, the model itself can be distributed rather than requiring each research group to reanalyze the raw reference data.
A subjective step of importance in the PCA+SVM approach practiced here is the choice of η in the outlier removal stage. Encouragingly, we found that estimated discrepancy between self-reported and predicted ancestry was highly similar across three wide-ranging values of η (Supplementary Table S4). This suggests that η is not likely to affect results negatively as long as class-specific misclassification rates estimated during outlier removal are controlled for in the predictive stage.
In PC space, admixed individuals are often coincident with individuals who do not share their ancestry (Supplementary Figure S10). We sidestepped this confounding effect of admixture by interrogating claims of insular ancestry only. In the future, however, PCA+SVM may provide a means for diagnosing admixed ancestries among closely related source populations. SVM feature vectors can incorporate numerous data types, continuous and discrete. Future reference data sets that include phenotypic variables such as those currently gathered by personal genomics companies39 as well as documented instances of specific admixed ancestries (perhaps based on careful consideration of subjects40 and pedigree analysis) may provide sufficient information to diagnose specific admixture classes. Output from other genetic analyses of the genotype data might also be added to the feature vector. For example, STRUCTURE results on their own are not well-suited to the identification of clusters within a continuous gradient of genetic variation41 (though see Engelhardt and Stephens42). However, estimated admixture proportions considered jointly with PC scores may help distinguish admixed from insular ancestry.
In the United States, multiple generations of admixture have taken place and the physical connection to ancestral populations is lost. Therefore, we might expect self-knowledge of ancestry to be reduced in a population of European Americans. Indeed, in one study, only 49% of 546 sibling pairs from the Upper Midwest could agree on the specific ancestries of both their parents.43 Here, SVMs trained on projection and combined PCA results both found >14% discrepancy between reported and inferred insular ancestry in the PMRP sample. A majority of discrepant reports are likely to be cases in which an individual is unaware of his or her actual genetic ancestry. Despite correction for misclassification error, however, some fraction of discrepancies likely still result from model error. For example, it is unlikely that the high percentage of discrepant reports among individuals reporting Dutch ancestry (Table 1) reflects a systematic bias toward mistakenly reporting Dutch ancestry. This result more likely stems from the small number of Dutch individuals included in the Central class training set (n=16) as well as the geographical position of the Netherlands near the intersection of the British Isles, Central, Scandinavian, and Western classes. Also, we stress that ancestry in a sociological context is not addressed by genetic ancestry prediction.
Similarity and divergence between Central Wisconsin and Europe
A number of studies have documented the genetic similarity between European Americans and Europeans.34, 44, 45 We extend these observations by showing that a local European-American population is already subtly distinct from its European sources. We found that European Americans exhibit visibly greater dispersion along higher-order PCs than Europeans (Figure 2b and Supplementary Figures S5 and S6) despite marginal values of FST (Table 2) and continued coincidence between the groups along top PCs (Figure 2a and Supplementary Figure S4). Simulations of a scenario that resembles the peopling and evolution of the Central Wisconsin population produced qualitatively similar patterns, thereby corroborating the hypothesis of nascent divergence (Figure 4c). The pattern of increased variance we identify here is distinct from the spatial artifacts of PCA identified by Novembre and Stephens.46 However, the pattern we describe comprises a subset of individuals with substantially greater variance. Similar patterns may be generated by poor-quality genotype data.32 Yet, recovery of the same pattern from (1) simulated data that lack genotyping error and (2) empirical data that correct for difference in missing data (Supplementary Figure S9) both suggest that data quality is not causative. Moreover, identification of the pattern in combined PCA rules out projection bias as a possible explanation. It is not clear why the pattern of increased variance in the Wisconsin/sink population is only found in results from combined PCA. However, consider that, similarly, close relatives only affect PCA results when included in a combined PCA and not when projected. Regardless, increased dispersion of recent immigrant populations along higher-order PCs supports a simple test for the presence of nascent divergence not yet detectable in plots of top PCs (see Supplementary Figure S14).
Simulations also suggest that frequent admixture increases the rate of divergence between source and sink populations. At 40 generations after colonization, the pair-group average distance between source and sink individuals on the PC1/PC2 biplot is 0.05 in the case of frequent admixture and only 0.027 otherwise. Despite the small genetic distances separating Northern European populations, our simulation results therefore suggest that admixture between these closely related groups has an appreciable impact on the divergence of the admixed population. Taken together, PCA results from empirical and simulated data suggest that active divergence of a unique American population from its ancestral sources is already under way.
Acknowledgments
We thank James Weber, Alkes Price, John Hawks, and two anonymous reviewers for helpful comments; and Luke Rasmussen for help with PMRP data. RJH dedicates this work to the memory of his grandmother Dawne Marie Haasl, a loving Irishwoman who lives on in the PMRP data matrix. This research was supported by NIH Grant 1U01HG004608-01 to CAM and NIH Grant HG004498 to BAP. The collections and methods for the Population Reference Sample (POPRES) are described by Nelson et al (2008). The datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000145.v2.p2 through dbGaP accession number phs000145.v2.p2.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Smith MW, Patterson N, Lautenberger JA, et al. A high-density admixture map for disease gene discovery in African Americans. Am J Hum Genet. 2004;74:1001–1013. doi: 10.1086/420856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind JM, Hutcheson-Dilks HB, Williams SM, et al. Elevated male European and female African contributions to the genomes of African American individuals. Hum Genet. 2007;120:713–722. doi: 10.1007/s00439-006-0261-7. [DOI] [PubMed] [Google Scholar]
- Basu A, Tang H, Zhu X, et al. Genome-wide distribution of ancestry in Mexican Americans. Hum Genet. 2008;124:207–214. doi: 10.1007/s00439-008-0541-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Via M, Gignoux CR, Roth LA, et al. History shaped the geographic distribution of genomic admixture on the island of Puerto Rico. PLoS One. 2001;6:e16513. doi: 10.1371/journal.pone.0016513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan CD, Andrew AD, Duell EJ, et al. Genetic population structure analysis in New Hampshire reveals Eastern European ancestry. PLoS One. 2009;4:e6928. doi: 10.1371/journal.pone.0006928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoggart CJ, Shriver MD, Kittles RA, et al. Design and analysis of admixture mapping studies. Am J Hum Genet. 2004;74:965–978. doi: 10.1086/420855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Hattangadi N, Lane B, et al. Methods for high-density admixture mapping of disease genes. Am J Hum Genet. 2004;74:979–1000. doi: 10.1086/420871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankararaman S, Sridhar S, Kimmel G, et al. Estimating local ancestry in admixed populations. Am J Hum Genet. 2008;82:290–303. doi: 10.1016/j.ajhg.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Tandon A, Patterson N, et al. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet. 2009;5:e1000519. doi: 10.1371/journal.pgen.1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S. Population genetics models of local ancestry. Genetics. 2012;191:607–619. doi: 10.1534/genetics.112.139808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty CA, Wilke RA, Giampietro PF, et al. Marshfield Clinic Personalized Medicine Research Project (PMRP): design, methods, and recruitment for a large population-based biobank. Pers Med. 2005;2:49–79. doi: 10.1517/17410541.2.1.49. [DOI] [PubMed] [Google Scholar]
- Everest KA. How Wisconsin Came by Its Large German Element Wisconsin Historical Collections; Madison, WI. 1892;vol 12:299–334. [Google Scholar]
- Voss PR, Vernoff DL, Long DD. Wisconsin's People: A Portrait of Wisconsin's Population on the Threshold of the 21st Century. Wisconsin Blue Book: Madison, WI; 2003-2004. pp. 99–173. [Google Scholar]
- Shriver MD, Kittles RA. Genetic ancestry and the search for personalized genetic histories. Nat Rev Genet. 2004;5:611–618. doi: 10.1038/nrg1405. [DOI] [PubMed] [Google Scholar]
- Via M, Ziv E, Burchard EG. Recent advances of genetic ancestry testing in biomedical research and direct to consumer testing. Clin Genet. 2009;76:225–235. doi: 10.1111/j.1399-0004.2009.01263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss KM, Long JC. Non-Darwinian estimation: my ancestors, my genes' ancestors. Genome Res. 2009;19:703–710. doi: 10.1101/gr.076539.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royal CD, Novembre J, Fullerton SM, et al. Inferring genetic ancestry: opportunities, challenges, and implications. Am J Hum Genet. 2010;86:661–673. doi: 10.1016/j.ajhg.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SSJ, Bolnick DA, Duster T, et al. The illusive gold standard in genetic ancestry testing. Science. 2009;325:38–39. doi: 10.1126/science.1173038. [DOI] [PubMed] [Google Scholar]
- Callaway E. Ancestry testing goes for pinpoint accuracy. Nature. 2012;486:7. doi: 10.1038/486017a. [DOI] [PubMed] [Google Scholar]
- McVean G. A genealogical interpretation of principal components analysis. PLoS Genet. 2009;5:e1000686. doi: 10.1371/journal.pgen.1000686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MR, Bryc K, King KS, et al. The Population Reference Sample, POPRES: a resource for population, disease, and pharmacological genetics research. Am J Hum Genet. 2008;83:347–358. doi: 10.1016/j.ajhg.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neall B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Zou F, Wright FA. Convergence and prediction of principal component scores in high-dimensional settings. Ann Statist. 2010;38:3605–3629. doi: 10.1214/10-AOS821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes C, Vapnik V. Support vector networks. Mach Learn. 1995;20:273–297. [Google Scholar]
- R Development Core Team: R: A language and environment for statistical computing 2011R Foundation for Statistical Computing: Vienna, Austria. URL http://www.R-project.org/ . [Google Scholar]
- Dimitriadou E, Hornik K, Leisch F, et al. e1071: Misc Functions of the Department of Statistics (e1071), TU Wien R Package Version 201115–26.http://cran.r-project.org/web/packages/e1071/index.html . [Google Scholar]
- Weir BS, Cockeram CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Rousset FR. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics. 1997;145:1219–1228. doi: 10.1093/genetics/145.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasl RJ, Payseur BA. Multi-locus inference of population structure: a comparison between single nucleotide polymorphisms and microsatellites. Heredity. 2011;106:158–171. doi: 10.1038/hdy.2010.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novembre J, Johnson T, Bryc K, et al. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdu P, Rosenberg NA. A general mechanistic model for admixture histories of hybrid populations. Genetics. 2011;189:1413–1426. doi: 10.1534/genetics.111.132787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23andMe website http://www.23andme.com/ancestry (Accessed 20 July 2012).
- Lao O, Lu TT, Nothnagel M, et al. Correlation between genetic and geographic structure in Europe. Curr Biol. 2008;18:1241–1248. doi: 10.1016/j.cub.2008.07.049. [DOI] [PubMed] [Google Scholar]
- Price AL, Helgason A, Palsson S, et al. The impact of divergence time on the nature of population structure: an example from Iceland. PLoS Genet. 2009;5:e1000505. doi: 10.1371/journal.pgen.1000505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T, Devillard S, Balloux F. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 2010;11:94. doi: 10.1186/1471-2156-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drineas P, Lewis J, Paschou P. Inferring geographic coordinates of origin for Europeans using small panels of ancestry informative markers. PLoS One. 2010;5:e11892. doi: 10.1371/journal.pone.0011892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath SC, Gut IG, Brennan P, et al. Investigation of the fine structure of European populations with applications to disease association studies. Eur J Hum Genet. 2008;16:1413–1429. doi: 10.1038/ejhg.2008.210. [DOI] [PubMed] [Google Scholar]
- Eriksson N, Macpherson JM, Tung JY, et al. Web-based, participant-driven studies yield novel genetic associations for common traits. PLoS Genet. 2010;6:e1000993. doi: 10.1371/journal.pgen.1000993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winney B, Boumertit A, Day T, et al. People of the British Isles: preliminary analysis of genotypes an surnames in a UK-control population. Eur J Hum Genet. 2012;20:203–210. doi: 10.1038/ejhg.2011.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Wen X, Falush D.2010Documentation for structure software: version 2.3Accessed at http://pritch.bsd.uchicago.edu/structure.html .
- Engelhardt BE, Stephens M. Analysis of population structure: a unifying framework and novel methods based on sparse factor analysis. PLoS Genet. 2010;6:e1001117. doi: 10.1371/journal.pgen.1001117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett MS, Strain KJ, Lesnick TG, et al. Reliability of self-reported ancestry among siblings: implications for genetic association studies. Am J Epidemiol. 2006;163:486–492. doi: 10.1093/aje/kwj057. [DOI] [PubMed] [Google Scholar]
- Price AL, Butler J, Patterson N, et al. Discerning the ancestry of European Americans in genetic association studies. PLoS Genet. 2008;4:e236. doi: 10.1371/journal.pgen.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Kosoy R, Nassir R, et al. European population genetic substructure: further definition of ancestry informative markers for distinguishing among diverse European ethnic groups. Mol Med. 2009;15:371–383. doi: 10.2119/molmed.2009.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novembre J, Stephens M. Interpreting principal component analyses of spatial population genetic variation. Nat Genet. 2008;40:646–649. doi: 10.1038/ng.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.