Summary
A new study (Zemach et al., 2013) suggests that the chromatin remodeling ATPase, DDM1 is specifically required for cytosine methylation at linker histone H1-associated heterochromatin, facilitating access by three cytosine methyltransferases, including a previously uncharacterized CHH methylase, CMT2.
Modulating gene activity in eukaryotes through cytosine methylation is an ancient molecular adaptation that predates the divergence of fungi, plants and animals. Although some eukaryotes have found ways to do without DNA methylation, those species that do methylate their DNA, including mammals and plants, require cytosine methylation for proper development and for keeping transposons, retroviruses and other potentially mobile repetitive elements transcriptionally silenced and in check (Law and Jacobsen, 2010). Determining how genomic patterns of cytosine methylation are established and maintained is an area of active investigation. In the latest issue of Cell, Zemach et al. (Zemach et al., 2013) provide new insights into why cytosine methylation of transposons and other heterochromatic repeats should specifically require DDM1 (DECREASE IN DNA METHYLATION 1) (Vongs et al., 1993), an Arabidopsis SWI2/SNF2-related nucleosome remodeling ATPase whose mammalian ortholog, LSH1 (LYMPHOID SPECIFIC HELICASE 1, also known as HELLS) plays a similar role in animal cells (Muegge, 2005).
In plants, as in animals, most cytosine methylation occurs at CG motifs and is accomplished primarily by MET1, the Arabidopsis ortholog of mammalian DNMT1 (Law and Jacobsen, 2010). CGs are symmetrical in the sense that a CG motif is also present on the paired DNA strand. A mechanism conserved between plants and mammals maintains symmetrical CG methylation following DNA replication via VIM (plant)/UHRF (mammal) proteins that recognize hemi-methylated CG sites and recruit MET1/DNMT1. Symmetrical CHG (where H is A, T, or C) methylation can be maintained in plants, too, involving CMT3 (CHROMOMETHYLASE 3), a member of a plant-specific family of cytosine methyltransferase that have a chromo domain as well as a BAH domain (Law and Jacobsen, 2010). The chromo and BAH domains both allow CMT3 recruitment to nucleosomes bearing Histone H3 that is dimethylated on lysine 9 (H3K9me2). In turn, the major H3K9 methyltransferase has an SRA domain that binds to methylated CHG, such that CHG DNA methylation and H3K9 histone methylation specify and maintain one another in a self-reinforcing loop. Asymmetric CHH methylation also occurs in plants, and prior to the new paper by Zemach et al. was thought to be almost entirely attributable DRM2, a de novo cytosine methyltransferase that is the Arabidopsis ortholog of mammalian DNMT3a/b (Law and Jacobsen, 2010). DRM2 methylation primarily affects transposons and is guided by a process known as RNA-directed DNA methylation (RdDM) (Zhang and Zhu, 2011). The RdDM pathway begins with multisubunit RNA polymerase IV (Pol IV) (Haag and Pikaard, 2011), which synthesizes precursors or 24 nt siRNAs that associate with ARGONAUTE 4 (AGO4) and guide the complex to sites of transcription by multisubunit RNA polymerase V (Pol V) (Haag and Pikaard, 2011). Through protein-protein interactions that are not fully understood, DRM2 is recruited and cytosines in all sequence contexts are methylated. CG and CHG methylation can then be perpetuated by maintenance methylation. However, asymmetric CHH methylation by DRM2 is not maintained in the absence of siRNA.
In ddm1 mutants, ~70% of all genomic methylation is lost, affecting CG, CHG and CHH methylation primarily within transposons and heterochromatic repeats for reasons that have been unclear (Vongs et al., 1993). Zemach et al. provide several new insights. The first is that ddm1 cytosine methylation losses are suppressed in null mutants for histone H1, suggesting that chromatin remodeling by DDM1 is specifically needed to overcome decreased nucleosome mobility and higher order nucleosome packaging facilitated by H1 (Robinson and Rhodes, 2006). A second insight is that DDM1 enables the maintenance of cytosine methylation by three different DNA methyltransferases: MET1 for CG, CMT3 for CHG, and CMT2 for CHH methylation. A third insight is that DDM1-dependent methylation and DRD1-dependent methylation together account for essentially all transposon cytosine methylation. Whereas DDM1 facilitates CG, CHG and CHH (via CMT2) methylation within long transposons that are enriched for heterochromatic features and localize primarily within pericentromeric regions, DRD1 and the RdDM pathway account for methylation of small transposons and the edges of long transposons, in generally euchromatic regions of the chromosome arms.
The newly defined role for CMT2 in CHH methylation helps explain several previous observations. For instance, CHH methylation found to persist in drm1 drm2 cmt3 triple mutant implicated an undefined cytosine methyltransferase (Cokus et al., 2008), which can now be safely presumed to be CMT2. Likewise, in a recent study of pol iv or pol v mutants, the total number of methylated genomic CHH sites was similar to wild-type, with half of all sites unchanged and the remainder occurring at ectopic locations, primarily within pericentromeric regions (Wierzbicki et al., 2012). CMT2 seems likely to account for the methylated CHH sites that were maintained, whereas the ectopic methylation sites could be the work of DRM2 no longer guided by Pol IV or Pol V-dependent RNAs. Genetic tests of these hypotheses should be straightforward.
An important implication of the study by Zemach et al. is that DDM1 and CMT2-dependent CHH methylation can be maintained, in the absence of small RNAs, via cross-talk with histone methylation. CMT2, like CMT3, has a chromo and a BAH domain, both of which can bind H3K9me2. In a mutant (ibm) defective for the removal of H3K9 methylation, which accumulates within gene bodies as a consequence of transcription, H3K9me2 recruits CMT3, resulting in high levels of CHG methylation not observed in wild-type plants (Miura et al., 2009). Zemach et al. demonstrate that CHH methylation also increases in ibm mutants suggesting that CMT2 might be recruited by histone modifications in the same way as CMT3.
Another noteworthy point reinforced by study by Zemach et al. is that maintenance methylation probably does not take place on naked DNA immediately following passage of the DNA replication fork, as is often depicted in review articles. The presence of binding domains for methylated H3K9 in mammalian UHRF proteins and plant CMT2 and CMT3, and cytosine methyltransferases’ need for help from nucleosome remodelers such as DDM1 or LSH1 is compelling evidence that cytosine methylation occurs in a nucleosomal context involving both core and linker histones.
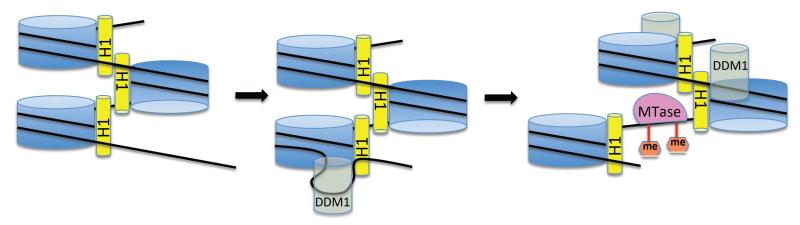
A model for nucleosome sliding facilitated by DNA translocase activity of DDM1. Linker Histone H1 (yellow) is thought to decrease nucleosome mobility by interacting with DNA at the entry and exit points of each nucleosome core particle (blue) and promoting tight packing of nucleosomes. Genetic evidence of Zemach et al. suggests that DDM1 is needed to counteract the effects of H1 for cytosine methylation. The model depicts one possibility to explain the genetic results, consistent with other chromatin remodeling ATPases, whereby DDM1overcomes the DNA binding energy of H1 to loop out DNA from the surface of nucleosome. Translocation of the looped out DNA around the nucleosome transiently generates a longer linker on the other side of the nucleosome, where DNA methyltransferases (MTAse) might gain access and methylate the DNA (orange hexagons). Concerted action of multiple DDM1 translocases could effectively slide DNA through the nucleosomes like rope through a series of pulleys.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References cited
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag JR, Pikaard CS. Nature Rev. Mol. Cell Biology. 2011;12:483–492. doi: 10.1038/nrm3152. [DOI] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura A, Nakamura M, Inagaki S, Kobayashi A, Saze H, Kakutani T. EMBO J. 2009;28:1078–1086. doi: 10.1038/emboj.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muegge K. Biochem Cell Biol. 2005;83:548–554. doi: 10.1139/o05-119. [DOI] [PubMed] [Google Scholar]
- Robinson PJ, Rhodes D. Curr Opin Struct Biol. 2006;16:336–343. doi: 10.1016/j.sbi.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Vongs A, Kakutani T, Martienssen RA, Richards EJ. Science. 1993;260:1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- Wierzbicki AT, Cocklin R, Mayampurath A, Lister R, Rowley MJ, Gregory BD, Ecker JR, Tang H, Pikaard CS. Genes Dev. 2012;26:1825–1836. doi: 10.1101/gad.197772.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, Kim MY, Hsieh P-H, Coleman-Derr D, Williams L. Eshed, Thao K, Harmer SL, Zilberman D, D. Cell. 2013 doi: 10.1016/j.cell.2013.02.033. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhu JK. Curr Opin Plant Biol. 2011;14:142–147. doi: 10.1016/j.pbi.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]