Abstract
Polycomb group proteins are critical to maintaining gene repression established during Drosophila development. Part of this group forms the PRC2 complex containing Ez that catalyzes methylation of histone H3 lysine 27 (H3K37me2/3), marks repressive to transcription. We report that the mammalian homologs Ezh1 and Ezh2 form similar PRC2 complexes but exhibit contrasting repressive roles. While PRC2-Ezh2 catalyzes H3K27me2/3 and its knockdown affects global H3K27me2/3 levels, PRC2-Ezh1 performs this function weakly. In accordance, Ezh1 knockdown was ineffectual on global H3K27me2/3 levels. Instead, PRC2-Ezh1 directly and robustly represses transcription from chromatinized templates and compacts chromatin in the absence of the methyltransferase cofactor SAM, as evidenced by electron microscopy. Ezh1 targets a subset of Ezh2 genes, yet Ezh1 is more abundant in non-proliferative adult organs while Ezh2 expression is tightly associated with proliferation as evidenced when analyzing aging mouse kidney. These results might reflect sub-functionalization of a PcG protein during evolution.
INTRODUCTION
The Polycomb group (PcG) and Trithorax group (TrxG) of proteins have long been recognized as regulators that maintain the gene expression pattern established during development (Schuettengruber et al., 2007). Moreover, PcG and TrxG proteins perform opposing functions by safeguarding the silenced or active transcriptional states, respectively (Ringrose and Paro, 2007; Schuettengruber et al., 2007).
Three families of complexes containing PcG proteins have been identified in Drosophila to date: Polycomb Repressive Complex 1 and 2 (PRC1 and PRC2) and PhoRC. PRC2 is composed of four core components, the mammalian counterparts of which are: Ezh2, Suz12, RbAp46/48 and Eed. Ezh2 is the catalytic subunit and harbors histone lysine methyltransferase activity within its SET domain that gives rise to di- and tri-methylated versions of lysine residue 27 within histone H3 (H3K27me2/3) (Schuettengruber et al., 2007). The other core components are required for such Ezh2 enzymatic activity. However, whether they play additional roles independent of Ezh2 remains unclear. The core components of Polycomb Repressive Complex 1 (PRC1) include HPC, HPH, Bmi1/Mel18 and Ring1A/B (Levine et al., 2002). PRC1 prevents the ATP dependent remodeling activity of Swi/Snf in vitro (Shao et al., 1999) and is able to condense chromatin in the absence of histone tails (Francis et al., 2004). In addition, PRC1 has a mono-ubiquitylase activity directed towards lysine residue 119 of histone H2A (H2AK119) and this is mediated by the E3 ligase activity of its RING1B component (Wang et al., 2004a). Of note, HPC and its mammalian homologs contain a chromodomain that specifically binds the product of PRC2-catalysis, H3K27me2/3 (Bernstein et al., 2006; Kuzmichev et al., 2002; Wang et al., 2004b). Given this, PRC1 was proposed to act downstream of PRC2 (Wang et al., 2004b). Yet this scenario does not seem to be universal as Xist RNA can recruit PRC1 in the absence of PRC2 and chromatin regions depleted of H3K27me3 can be bound by PRC1 (Schoeftner et al., 2006). Finally, a third polycomb group complex, PhoRC was characterized recently in Drosophila, however its exact function and mammalian counterpart are not yet clear (Klymenko et al., 2006).
PcG proteins bind to Polycomb Response Elements (PRE) that have been identified and characterized in Drosophila. Several DNA binding proteins were shown to be required for PcG recruitment such as GAF, Pipsqueak, Zeste or PHO. Surprisingly, no mammalian counterparts were found for these recruiters and, despite extensive searches, PREs have not been identified to date in mammals (Schuettengruber et al., 2007). Nonetheless, genome-wide analyses identified genes targeted by PRC2 in a variety of cell lines and animal models (Boyer et al., 2006; Bracken et al., 2006; Lee et al., 2006; Schwartz et al., 2006; Squazzo et al., 2006; Tolhuis et al., 2006). As expected, a strong overlap between PRC2, PRC1 and H3K27me2/3 was observed (Boyer et al., 2006; Bracken et al., 2006; Lee et al., 2006; Schwartz et al., 2006). Moreover, PRC target genes were found to extend far beyond the historically recognized HOX loci. Gene ontology of the target genes revealed a strong enrichment for developmental factors as perhaps expected, although glycoprotein and immunoglobulin related genes were also identified depending on the cell model analyzed (Squazzo et al., 2006).
With few exceptions, invertebrates such as Drosophila or sea urchins have only one copy of PcG genes (Whitcomb et al., 2007). However, vertebrates have several paralogs of most PcG genes. Interestingly, among the PRC2 components two genes were not duplicated: Suz12 and Eed. However, different isoforms of Eed do arise from alternative translation start sites (Denisenko and Bomsztyk, 1997) and these might play an important role in creating diversity among the PRC2 complexes (Kuzmichev et al., 2004; Kuzmichev et al., 2005){R.M. and D.R., unpublished results}. Although Drosophila E(z) and its closest mammalian homolog Ezh2 have been well characterized, very little is known about mammalian Ezh1 although it was the first Ez homolog to be cloned (Abel et al., 1996). The RNA levels of Ezh1 and Ezh2 appear to be inversely correlated in that Ezh1 is highly expressed in kidney, brain and skeletal muscle tissues where Ezh2 RNA is barely detectable (Laible et al., 1997). However, two other studies analyzing Ezh1 expression in tissues reported slightly divergent results (Ogawa et al., 1998; van Lohuizen et al., 1998). Ezh1 was also shown to interact with Eed in vitro (Han et al., 2007; Jones et al., 1998; van Lohuizen et al., 1998).
We investigated the cellular role of Ezh1 relative to that of Ezh2. Here, we show that Ezh1 is ubiquitously expressed whereas Ezh2 expression is associated with proliferating tissues. Ezh1 is part of a PRC2 complex quite similar to the one containing Ezh2 and they share an overlapping set of target genes that they appear to co-occupy. Yet surprisingly and in contrast to PRC2-Ezh2, PRC2-Ezh1 exhibits low levels of histone methyltransferase (HKMT) activity. On the other hand, PRC2-Ezh1 efficiently represses transcription and compacts chromatin, in contrast to PRC2-Ezh2. These distinct functional roles for PRC2-Ezh1 and PRC2-Ezh2 in repression might pertain to their differential expression and to sub-functionalization of Ez during evolution.
RESULTS
Ezh1 is a nuclear protein that interacts with Suz12 and Eed
To begin the functional characterization of Ezh1, we generated an antibody directed against its amino terminus (aa1-226) as this region exhibits less conservation with Ezh2 relative to the C-terminal region. The antibody did not exhibit cross-reactivity with Ezh2 as evidenced by western blot analyses of purified proteins (Figure S1A) or nuclear extracts from cell lines over-expressing HA-tagged versions of Ezh1 (Ezh1-HA) or Ezh2 (Ezh2-HA) (Figure S1B). We next gauged the expression levels of Ezh1 in a variety of cell lines: HeLa, Jurkat, HEK293, C2C12, RAG, NIH-3T3, mES and F9 (Figure 1A, and data not shown). Given that Ezh1 and Ezh2 antibodies detect similar amounts of the respective recombinant proteins (data not shown, Supplementary Figure 1), and that four times more nuclear extract was required on average to detect an Ezh1 signal, the levels of Ezh1 were rather low in comparison to those of Ezh2. We focused on RAG, NIH-3T3 and F9 cells as they exhibited a higher level of Ezh1 protein compared to Jurkat cells (Figure 1A). It was previously reported that Ezh1 (Enx-2) localized to the cytoplasm of Jurkat cells where it interacts with ZAP70 (Ogawa et al., 2003). We therefore fractionated F9 cells into cytoplasmic, nuclear, chromatin soluble and insoluble fractions and analyzed each by western blot for Ezh1 and Ezh2 levels. Both were enriched in the chromatin soluble and insoluble fractions and nearly absent from the nuclear fraction (Figure 1A). Immunofluorescence performed in F9 cells confirmed that Ezh1 and Ezh2 overlap with the nuclear marker DAPI but not with cytoplasmic tubulin (Figure S1C).
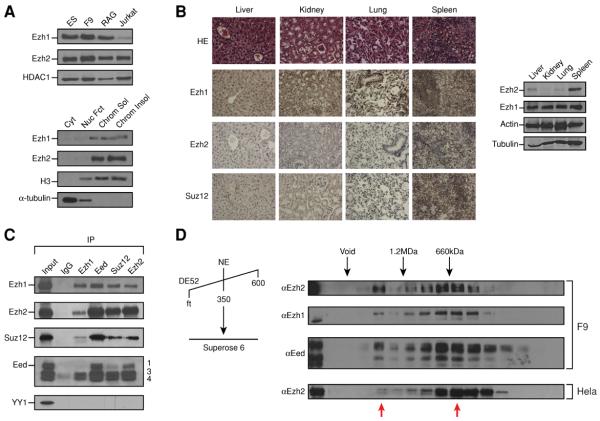
Figure 1. Ezh1 is a nuclear protein expressed in adult mouse tissue and partners with Suz12, Eed and RbAP46/48.
A) Western blot of Ezh1, Ezh2 and HDAC1 in various cell lines as indicated (left). 10 μg of nuclear extract was loaded for Ezh2 and HDAC1 and 40 μg for Ezh1. Equal amounts of F9 cell fractions (cytoplasmic, nuclear fraction, chromatin soluble and insoluble) were loaded. α-tubulin and histone H3 served as cytoplasmic and nuclear markers, respectively (right). B) 6 month-old adult mouse tissue sections stained with H&E or imunohistochemistry with the antibodies indicated (Left). Tissue extracts were prepared concomitantly and proteins were detected by western blot using the same amount of extract in all cases. C) Immunoprecipitation from F9 high salt nuclear extract using antibody against Ezh1, Ezh2, Suz12 and Eed and probed with the same set of antibodies and anti-YY1. IgG served as control. D) High salt nuclear extract from F9 cells was loaded onto a DE52 column, proteins were eluted with BC350 and then run on a Superose 6 sizing column. Every other fraction was loaded. Molecular weights are indicated on top. Complexes containing Ezh proteins are indicated by red arrows.
Previous studies using immunohistochemistry (IHC) indicated that Ezh2 is barely detectable in normal adult tissues (Bachmann et al., 2006), although similar analyses with Ezh1 have not been reported. We analyzed Ezh1 and Ezh2 expression in a set of mouse tissues by IHC (Figure 1B, left panel). Interestingly, a strong signal was detected for Ezh1 in all the tissues analyzed (liver, kidney, lung and spleen) whereas Ezh2 was detected mainly in spleen. This result was not a reflection of antibody performance in this particular assay as similar expression patterns were seen using western blots (Figure 1B, right panel). Of note, the same amount of extract was loaded when probing for Ezh2 and Ezh1 indicating that Ezh1 expression is higher in tissues compared to cell line models.
As expected given their extensive homology, both Ezh1 and Ezh2 pulled-down the PRC2 components Suz12 and Eed in immunoprecipitation experiments performed with high salt extracts from F9 cells (Figure 1C). However surprisingly, Ezh1 and Ezh2 interacted with each other (Figure 1C). This interaction was independent of the presence of nucleic acids (Figure S1D) and was not due to antibody cross-reactivity as we could recapitulate it in vitro using Flag-tagged versions of the proteins and the Flag epitope for IP (Figure S1E). We next sought to determine whether Ezh1 is part of a complex or interacts only transiently with the other PRC2 components. High salt nuclear extracts of F9 cells were fractionated on a DE52 column (Figure 1D). Both Ezh1 and Ezh2 bound to this resin and were eluted with buffer containing 350 mM salt (data not shown). This eluate was then applied to a Superose 6 gel filtration column and fractions were analyzed by western blot. We observed a main peak around 500kDa for Eed and Ezh2 as expected, and for Ezh1 as well. Surprisingly, a second high molecular weight complex associated with the three proteins was also detected. We repeated this experiment with HeLa nuclear extracts and found that the presence of this large complex was less evident suggesting that it might be cell-type specific (Figure 1D). We also observed that Ezh1 interacted with SirT1 and PHF1 (Figure S2A), proteins that we previously reported as being associated with PRC2 complex containing Ezh2 and that affect PRC2 activity (Kuzmichev et al., 2005; Sarma et al., 2008). Hence Ezh1 interacts with the core components of PRC2 as well as with those factors less stably associated with it.
Ezh1 is part of a PRC2-type complex
To further assess Ezh1 interacting partners, we generated a baculovirus expressing Ezh1 with a Flag tag. Flag-Ezh1 interacted directly with Eed and Suz12 and indirectly with RbAp48 through Suz12 (Figure S2B), similar to the case for Ezh2 (Ketel et al., 2005). Sf9 cells were co-infected with baculovirus expressing the four components: Flag-Ezh1, Suz12, Eed and RbAP48. Anti-Flag immunoprecipitation was performed and the eluate analyzed on a Superose 6 sizing column. Western blot and silver staining showed that the four components are part of a complex eluting around 500kDa similar to PRC2-Ezh2 (Figure 2A, and data not shown). The high degree of similarity between the SET domains of Ezh1 and Ezh2 prompted us to test PRC2-Ezh1 for histone methyltransferase (HMT) activity. Using various histone substrates, we found that PRC2-Ezh1 targets H3K27 but preferentially methylates octamers relative to native or recombinant nucleosomal substrates (Figure 2B). Similar to PRC2-Ezh2, PRC2-Ezh1 was also able to target the linker histone H1 in vitro (Figure 2B). We next compared the substrate preferences of PRC2 containing either Ezh1 or Ezh2 using peptides that recapitulate the different methylation states of H3K27. Both PRC2-Ezh1 and PRC2-Ezh2 preferentially utilized H3K27me1 for catalysis (Figure 2C). Given that ES cells depleted of either Suz12 or Eed are also depleted of H3K27me2/3 but not of H3K27me1 (Pasini et al., 2004; Schoeftner et al., 2006), this suggests that PRC2-Ezh complexes initiate methylation on mono-methylated histones and add one or two additional methyl marks. Of note, a methylation signal was also observed around 100kDa, corresponding to methylation of Suz12 and Ezh1 (Figure 2C) (Muller et al., 2002).
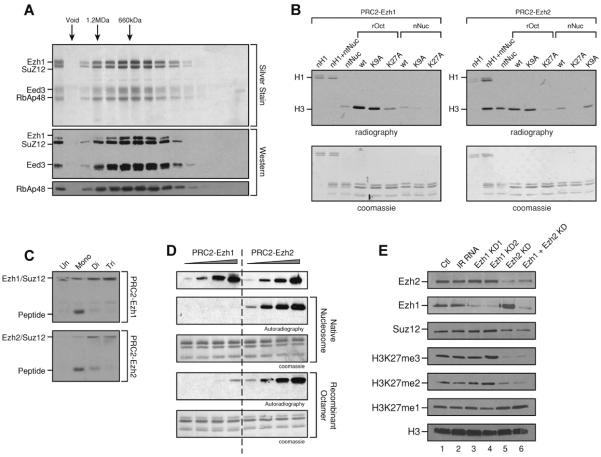
Figure 2. Reconstitution and characterization of PRC2-Ezh1.
A) Immunoprecipitated PRC2-Ezh1 was loaded onto a Superose 6 sizing column and the eluted fractions analyzed by SDS-PAGE followed by western blot and silver staining. B) Histone methyltransferase (HMT) assay using purified PRC2-Ezh2 or PRC2-Ezh1 with various substrates as indicated on top. ntNuc (native nucleosomes), rOct (recombinant octamers) and rNuc (recombinant nucleosomes). C) HMT assay with PRC2-Ezh1 and PRC2-Ezh2 using peptides either unmethylated, mono-, di- or tri-methylated on lysine 27. D) Quantitative HMT assay. Equal amounts of PRC2-Ezh1 and PRC2-Ezh2 were loaded as evidenced by Suz12 levels in the western blot. E) RNAi-mediated knockdown of Ezh1 and of Ezh2 in NIH-3T3 cells. Nuclear extracts were prepared 72 hr after transfection and analyzed by western blot.
In spite of their similar substrate preferences, larger amounts of PRC2-Ezh1 relative to PRC2-Ezh2 were required to obtain comparable HKMT activity even towards recombinant octamers. Using Suz12 as a loading control in western blots, we quantified the activity of both complexes on recombinant octamers and native nucleosomes (Figure 2D). PRC2-Ezh1 activity was considerably lower than that of PRC2-Ezh2. When recombinant octamers were used as substrate, the HKMT activity of PRC2-Ezh1 was roughly 20-fold weaker than that of PRC2-Ezh2 as quantified with liquid scintillation counting (Figure S2C). This marked discrepancy could signify that PRC2-Ezh1 requires additional factor(s) to achieve robust HKMT activity in vitro. Therefore, to appraise the functional role of Ezh1 in catalyzing H3K27 methylation at the global level, Ezh1 was knocked-down in NIH-3T3 cells via RNA interference using two oligonucleotides that efficiently target Ezh1 and the results compared to those obtained with another oligonucleotide that was previously characterized as targeting Ezh2 (Etchegaray et al., 2006). Nuclear extracts were prepared 72 hours after transfection and protein accumulation was analyzed by western blot. Interestingly, Ezh2 knockdown resulted in a marked increase in Ezh1 levels (Figure 2E). This effect occurred at the protein level, as Ezh1 mRNA levels were unaffected (Figure S2D). Analysis of the global levels of histone methylation showed that the contribution of Ezh1 to H3K27me2/3 is minor as there were no significant changes after Ezh1 knockdown (Figure 2E, compare lanes 1 and 2 to lanes 3 and 4, and lane 5 to lane 6). On the other hand, Ezh2 knockdown resulted in decreased levels of H3K27me2/3 and a slight increase in H3K27me1 even though Ezh1 levels were markedly elevated (Figure 2E compare lanes 1 and 5).
PRC2-Ezh1 and -Ezh2 complexes are recruited to the same set of target genes dependent on the presence of the SET domain
Given our findings that Ezh1 resides in a PRC2 complex (Figures 1 and 2) and that it interacts with PRC2 core components (Figure 2), we next tested if Ezh1 can recruit the core PRC2 components using a cell model, derived from 293 T-Rex, in which a Gal4 fusion protein, Gal4-Ezh1 in this case, can be induced for expression and artificially tethered to an integrated luciferase reporter containing Gal4 response elements (Sarma et al., 2008; Vaquero et al., 2004). As expected, upon doxycycline induction of its expression, Gal4-Ezh1 was targeted to the transgene (Figure 3A). This resulted in the recruitment of Suz12, but not of Ezh2, indicating that while a small but detectable portion of Ezh1 and Ezh2 interact (Figure 1C and S1C/D), they are present in distinct complexes in vivo (Figure 3A). Similar results were obtained with cell lines expressing an Ezh1 point mutant within its SET domain that is expected to abolish its activity (designed by alignment with a previously described Ezh2 mutant), or an Ezh1 mutant deleted of its SET domain (Figure 3A).
Figure 3. PRC2-Ezh1 and PRC2-Ezh2 share target genes.
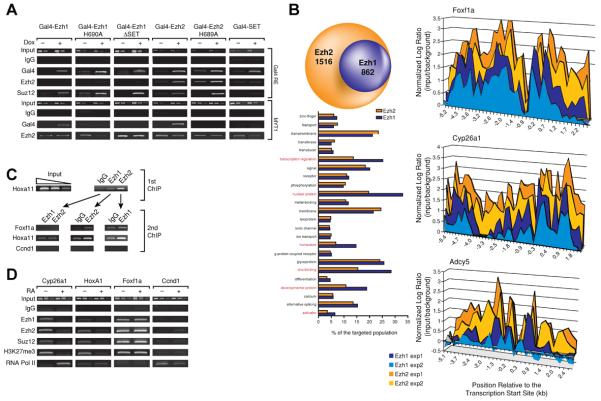
(A) Chromatin immuno-Precipitations (ChIP) were performed in different cell lines expressing Gal4-Ezh1, Gal4-Ezh2 or Gal4-SET under a doxycycline-regulated promoter in 293 T-Rex cells. Ezh1, Ezh2 and Suz12 recruitment at a stably integrated luciferase reporter construct and at endogenous MYT1were analyzed 24 hr after doxycycline addition. B) ChIP-chip experiments were performed in F9 cells using a mouse promoter array covering −5.5 kb to 2.5 kb of the entire mouse genome. Left (top), comparison of Ezh1 and Ezh2 targets. Left (bottom), comparative gene ontology for Ezh1 and Ezh2 target genes. Right, binding patterns of Ezh1 and Ezh2 at three different promoters. Foxf1a and Cyp26a1 are detected as Ezh1 and Ezh2 target genes. Adcy5 is considered an Ezh2 specific target gene. C) ChIP experiments performed in F9 cells. Chromatin was first IP'd with IgG, or Ezh1 or Ezh2 antibodies and then with the antibodies indicated on top. D) F9 cells were treated with Retinoic Acid or vehicle for 16 hr and ChIP was performed at known RAR and Ezh2 target genes (Cyp26a1 and HoxA1), Ezh2 target gene (Foxf1a) or control (CcnD1).
We next analyzed the capacity of Ezh1 or its mutant versions to be recruited to the promoter of the endogenous MYT1 gene, a well characterized Ezh2 target gene (Kirmizis et al., 2003). Gal4-Ezh2 was targeted as expected and similar results were obtained with Gal4-Ezh1, suggesting that both proteins might share a common set of target genes (Figure 3A). Surprisingly, neither the Ezh1 nor the Ezh2 point or deletion mutants in the SET domain were targeted to this gene. To rule out the possibility that these SET domain mutants might be defective in protein folding and consequently cell localization, we fractionated extracts from cells expressing the Gal4-Ezh2, either wild type or point mutant in its SET domain. Both proteins were found in the same subcellular fraction indicating that the mutants lost specific gene recruitment but not their affinity for chromatin (Figure S3A). Of note, the SET domain of Ezh2 by itself was not targeted to endogenous genes (Figure 3A, and see below).
To determine if Ezh1 and Ezh2 share a common set of target genes at the genome-wide level, we performed ChIP-on-chip experiments. Previous reports indicated that 95% of Ezh2 targets are within 1 kb of the transcription start site (Lee et al., 2006). Therefore, we used arrays that span the promoter region (−5.5 to +2.5 kb) of the entire mouse genome with 60-mer probes spaced on average 250 bp apart (Agilent technologies). Experiments were performed in duplicate with independent isolations of chromatin from F9 cells and different batches of antibodies. Only target genes common to both experiments and with a P value less than or equal to 0.01 were considered. With these parameters, 898 target genes were identified for Ezh1 and 2378 for Ezh2 (Table S1). The result is consistent with previous estimates of 8% of the genome being targeted by Ezh2 (Lee et al., 2006). Additionally, we found that each Ezh1 target gene was also bound by Ezh2. Gene ontology analysis of the common genes indicated that Ezh1 targets are specifically enriched for the nuclear and developmental related function of PRC2-Ezh2 (Figure 3B). For instance, almost 30% of Ezh1 target genes are involved in transcription regulation while fewer than 20% of Ezh2 target genes are so classified. The binding profiles of Ezh1 and of Ezh2 at two representative genes (Foxf1a and Cyp26a1) versus the binding profile of Ezh2 at an Ezh2-specific locus (Adcy5) indicate a highly coincident binding pattern for Ezh1 and Ezh2 (Fig. 3B, right panel) and suggest a similar recruitment pathway. While Ezh1 was enriched to some extent on Adcy5, its binding did not pass stringent criteria. When we averaged Ezh2 enrichment at Ezh1 and Ezh2 target genes versus Ezh2-specific genes, the latter was significantly less enriched for Ezh2 (Figure S3B, top). Moreover Ezh1 enrichment plotted as a function of Ezh2 enrichment, revealed a good correlation at genes targeted by these proteins (Figure S3B, bottom). Therefore, we might have underestimated the overlap between Ezh1 and Ezh2, as Ezh1 is less abundant in F9 cells.
To evaluate if Ezh1 and Ezh2 are present simultaneously at their common target genes, we performed consecutive ChIP experiments in which chromatin was immunoprecipitated with Ezh1 antibody and then with Ezh2 antibody or vice versa. Ezh1 and Ezh2 were clearly present simultaneously at a defined promoter (Figure 3C). Of note, it was previously shown that upon retinoic acid induced differentiation of F9 cells, PRC2-Ezh2 was released from RAR target genes preceding transcription (Gillespie and Gudas, 2007; Lee et al., 2007). We confirmed this result by ChIP and importantly, found that the same was true for Ezh1 suggesting that both PRC2-Ezh1 and PRC2-Ezh2 are removed from the promoter to achieve conditions conducive to gene activation (Figure 3D).
Ezh2 but not Ezh1 expression is associated with proliferative tissue and Ezh1 gene targeting is independent of Ezh2
Previous reports indicated that Ezh2 mRNA levels are regulated during development, while those of Suz12 appear to be constant (Gunster et al., 2001; Metsuyanim et al., 2008). This might reflect the role of Suz12 as a component of PRC2-Ezh1. Therefore we compared the levels of Ezh1 and Ezh2 in mouse kidney as a function of development, from newborn to 9 month-old mice in IHC and western blot analyses (Figure 4A and 4B). We observed a dramatic reduction in Ezh2 levels after birth whereas those of Ezh1 were constant. The age dependent increase in the levels of the PRC1 component Bmi-1 suggests that not only PRC2 but also PRC1 is altered during the aging process. Yet, Ezh2 down-regulation in aging organs was not a general phenomenon as its expression was similar in newborn and 9 month-old mouse spleen (Figure S3C). Of note, H3K27me3 global levels were stable despite the absence of Ezh2, and this was also the case in adult tissues from different organs (Figure 1B and data not shown).
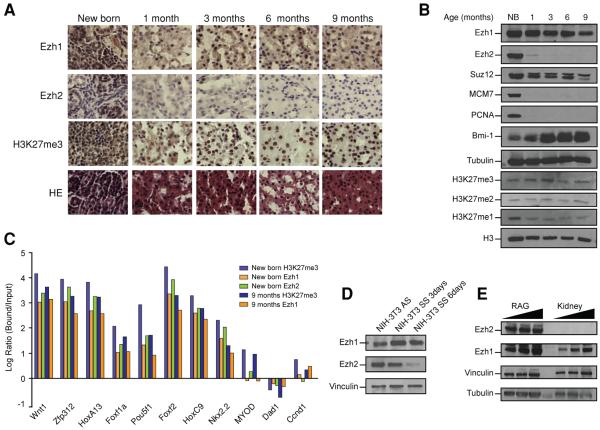
Figure 4. Ezh1/2 regulation in aging mouse kidney.
(A) Tissue sections from mouse females harvested from birth to 9 months and stained with H&E or by immunohistochemistry using the antibodies indicated. B) Tissue extracts were performed concomitantly with the IHC. The same amount of extracts was loaded in all cases. C) ChIP performed from newborn and 9 month-old female tissues. After ChIP, elution and input were amplified by LM-PCR. Equal amounts of DNA were used for qPCR. Log values of the enrichment are the average of two independent experiments. D) Nuclear extracts from NIH-3T3 cells grown either asynchronously or under conditions of serum starvation (media containing 0.5% bovine serum for the indicated period of time) were loaded for western blot analysis using 10 μg for Ezh2 detection and 40 μg for Ezh1 detection. E) Equal amount of whole cell/tissue extracts from mouse renal adenocarcinomal RAG cells or kidney from 6 month-old females were loaded side by side.
We took advantage of this model to study Ezh1 gene targeting in the absence of Ezh2 by performing ChIP on extracts of kidneys isolated from newborn and 9 month-old mice. Due to the limited starting material for newborn kidney, we amplified the ChIP by LM-PCR. In agreement with our results in the previous section, Ezh1 and Ezh2 were present at the same set of target genes and associated with a strong enrichment for H3K27me3, in this case in kidney from newborn mice (Figure 4C). The absence of significant Ezh2 recruitment in kidney from 9 month-old mice was confirmed by conventional ChIP (data not shown). Despite a slight decrease in H3K27me3 enrichment at some targeted genes, Ezh1 recruitment was not appreciably affected by the absence of Ezh2 (Figure 4C).
Ezh2 expression appears to be associated with actively dividing cells (Bracken et al., 2003). In accordance with this, we observed that Ezh2 expression displays the same pattern of regulation as a matter of age as PCNA and MCM7, two proteins associated with DNA replication. Hence, Ezh2, PCNA, and MCM7 were barely detectable in kidney isolated from mice one-month and older (Figure 4B). In order to expand this observation, we compared the levels of Ezh1 and Ezh2 in NIH-3T3 cells before and after being made quiescent by serum starvation and also in normal versus cancerous kidney cells. Upon serum starvation, Ezh2 protein levels were reduced in NIH-3T3 cells whereas those of Ezh1 were not significantly affected (Figure 4D). Ezh2, usually below the detection limit in extracts from normal kidney tissue, was very highly expressed in the proliferative RAG kidney cancer cell line (Figure 4E). Therefore, based on three related parameters: age, quiescence versus proliferation, and non-dividing versus tumorigenic cells, our results underscore that Ezh2, but not Ezh1, expression is tightly associated with cell proliferation.
PRC2-Ezh1 represses transcription in vivo and in vitro
Gal4-Ezh1 can mediate transcriptional repression of the luciferase transgene and surprisingly so too can the Gal4 version of Ezh1 mutant in its SET domain (Figure 5A). Thus both Ezh1-mediated recruitment of Suz12 (Figure 3A) and gene repression occur independently of its having an intact SET domain. Consistent with this, in both cases a Gal4 fusion protein containing only the Ezh1 SET domain was ineffectual (Figures 3A and 5A). This suggests that the histone lysine methyltransferase activity of Ezh1 is not required for transcriptional repression. We previously reported that Gal4-Ezh2 recruitment results in robust increases in H3K27me2 whereas PRC1 binding required H3K27me3 as observed when Ezh2 was recruited through PHF1 (Sarma et al., 2008). Yet in the case of Gal4-Ezh1 recruitment, there were no significant changes in H3K27me2/3 levels (Figure 5B) consistent with the Ezh1-SET domain being dispensable and in accordance with the lack of enrichment of the PRC1 component Bmi-1 (Figure 5B).
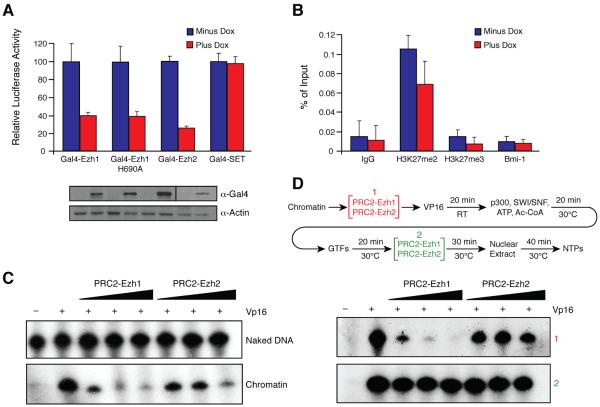
Figure 5. PRC2-Ezh1 represses transcription independently of histone methylation.
(A) Top, luciferase activity in cell lines stably transfected with a luciferase reporter and expressing the indicated Gal4 fusion proteins in a doxycycline dependent manner. Bottom, expression control for the Gal4 fusion proteins. Luciferase values are the mean ± SD (standard deviation) of three independent biological replicates. B) ChIPs were performed with or without Gal4-Ezh1 expression. Histone methylation enrichment and Bmi-1 recruitment at the transgene were analyzed by qPCR. qPCR values are the mean ± SD (standard deviation) of three independent biological replicates. C) In vitro transcription assay performed on naked DNA or chromatinized templates using Gal4-VP16 as activator. Similar amounts of PRC2-Ezh1 and PRC2-Ezh2 were titrated in the assay. A representative result of three independent experiments is shown. D) “Bypass” in vitro transcription assay was performed as indicated in the scheme at the top. PRC2-Ezh1/2 was added as indicated between brackets. A representative result of three independent experiments is shown.
To reconcile the observed Ezh1-mediated repression with the lack of concordant production of the repressive H3K27me2/3 marks, we first examined Ezh1-mediated repression using in vitro transcription assays. Increasing amounts of PRC2-Ezh1 and PRC2-Ezh2 (see Figure 2D for complex titration) had no effect on transcription from naked DNA templates (Figure 5C). In contrast, PRC2-Ezh1 significantly repressed transcription, and more efficiently than PRC2-Ezh2, using chromatinized templates. Similar results were obtained when the assay was performed in the presence of Sadenosyl methionine (SAM) (data not shown). The weak repressive activity of PRC2-Ezh2 might reflect the requirement for additional factors, such as PHF1, to achieve appreciable H3K27me3 activity and/or the absence of PRC1 in this assay. To further analyze the mechanism of PRC2-Ezh1 mediated repression we used the protocol shown in Figure 5D, whereby PRC2-Ezh1 or PRC2-Ezh2 complex was added prior to or after the formation of a transcription initiation complex. The results demonstrated that PRC2-Ezh1 effectively repressed transcription if added prior to the formation of the transcription pre-initiation complex (PIC). However, repression was thwarted if PRC2-Ezh1 was added after PIC formation (Figure 5D). In contrast, PRC2-Ezh2 had little effect regardless of the time of its addition, similar to its ineffectualness with naked DNA as shown in Figure 5C.
PRC2-Ezh1 compacts chromatin even in the absence of SAM yet histone tails are required
Since PRC2-Ezh1-mediated repression does not involve the covalent modification of histone H3K27 as is requisite in the case of PRC2-Ezh2, yet both repress transcription only in the context of chromatin, we investigated if PRC2-Ezh1 does so by directly altering chromatin structure.
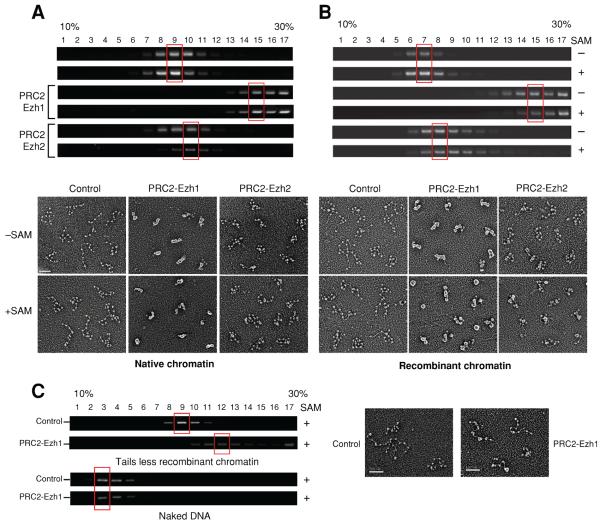
We reconstituted chromatin using a DNA fragment containing 12 nucleosome positioning sites (Dorigo et al., 2004) and hyperacetylated core histones purified from HeLa cell nuclei. We then probed for changes in chromatin structure as a function of the presence of PRC2-Ezh1 and SAM using sucrose density gradient centrifugation and electron microscopy (EM) (Sims et al., 2007). While DNA alone remained close to the top (Figure 6C, fraction 3,), chromatin migrated towards the center of the sucrose gradient (Figure 6A, fraction 9) and this was unaffected by pre-incubation with SAM. Interestingly, when the chromatin was incubated with PRC2-Ezh1, a dramatic shift toward the densest fractions was observed. This shift occurred to nearly the same extent in the absence of SAM suggesting that PRC2-Ezh1 is able to compact chromatin independently of histone methylation. We repeated the experiment with PRC2-Ezh2 using the same amount of complex. We observed a slight shift in the absence of SAM (peak at fractions 9 and 10) and one fraction shift in its presence (fraction 10). Of note, increasing amounts of PRC2-Ezh2 in the absence of SAM did not affect the sedimentation of the chromatin (Figure S4A).
Figure 6. PRC2-Ezh1 compacts chromatin and binds to tail-less chromatin but not to DNA.
Chromatin reconstituted with (A) hyperacetylated native octamers or (B) bacterially expressed and purified recombinant octamers was incubated with PRC2-Ezh1 or PRC2-Ezh2 in the presence or absence of SAM as indicated. The complexes were loaded on a 10–30% sucrose gradient and peak fractions (highlighted in red) were analyzed by EM. Pictures of representative molecules are shown. Compaction events were close to 100% in the presence of PRC2-Ezh1. C) As in (B), but using recombinant tail-less chromatin or naked DNA with PRC2-Ezh1.
We then analyzed the material in the main peaks (shown within red rectangles) by EM. Chromatin alone displayed a “beads on a string” structure typical of an open configuration (Figure 6A, bottom panel). In the presence of PRC2-Ezh1, we observed a dramatic change with the chromatin being now completely compacted regardless of the presence of SAM; we did not observe any large aggregates. Addition of PRC2-Ezh2 to the chromatin resulted in a slight compaction. The possibility existed that the chromatin compaction observed after PRC2-Ezh1 addition resulted from its bridging histone marks, a property previously reported for L3-MBTL1 (Trojer et al., 2007). To test this possibility, we reconstituted chromatin with histones that were expressed and purified from bacteria (recombinant histones) and therefore lacking in all post-translational modifications that exist in HeLa-derived histones. Again incubation of chromatin with PRC2-Ezh1 resulted in a robust compaction (Figure 6B). PRC2-Ezh2 was also able to compact chromatin to some extent. This result does not necessarily mean that histone marks do not play a role in chromatin compaction mediated by PRC2-Ezh1, but they are not required for this process in vitro. Furthermore, careful analysis of the chromatin compacted by PRC2-Ezh1 revealed that the degree of compaction is of a more heterogeneous nature with recombinant chromatin than with the native counterpart (Figure 6B, bottom panel).
Having shown that PRC2-Ezh1 can compact recombinant chromatin, we next asked whether “tail-less” chromatin or naked DNA would suffice. Although PRC2-Ezh1 retained the ability to bind to tail-less chromatin as shown by the shift in sucrose gradients (Figure 6C, top of left panel), the complex could not compact this chromatin (Figure 6C, right panel). Finally, unlike PRC1 (Francis et al., 2004), PRC2-Ezh1 was unable to bind to naked DNA (Figure 6C, left panel).
Full compaction requires all components of PRC2-Ezh1 that brings 3 to 4 nucleosomes together
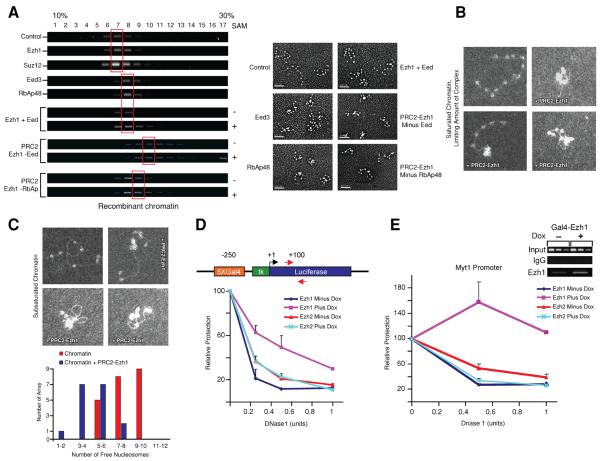
The difference in PRC2-Ezh1 and -Ezh2 with respect to chromatin compaction was noteworthy in that both complexes share three of their four components and the distinguishing components, Ezh1 and Ezh2, are well conserved in amino acid sequence (65% identity overall and 94% for the SET domain). To understand the basis of this, we analyzed the ability of individual PRC2 components and of partial PRC2-Ezh1 complexes (Figures S4C and S4D, respectively) to bind and compact chromatin. Figure 7A shows that whereas the independent addition of Suz12 or Ezh1 did not impact chromatin significantly, the independent addition of Eed or of Rbap48 resulted in a one-fraction shift on the sucrose gradient. This shift was most likely due to protein binding as compaction of the chromatin was not detectable by EM (Figure 7A, right panel). This result was expected as previous studies have shown that Eed interacts with the histone H3 tail (Tie et al., 2007), and that RbAp46/48 interacts with the histone H4 tail (Verreault et al., 1998). We next examined if all components of PRC2-Ezh1 were necessary for compaction by comparing Ezh1/Eed, and PRC2-Ezh1 either without Eed or without Rbap48 (Figure S4D and 7A). Indeed, all components were required for compaction as none of these partial complexes could recapitulate the effect obtained after addition of PRC2-Ezh1. Interestingly in the absence of Eed, the PRC2-Ezh1 complex was able to compact the chromatin to a minor extent. This observation highlights the distinctive contribution of each PRC2 component to compaction and to HKMT activity. For example, whereas omitting Eed from PRC2-Ezh2 resulted in a lack of HKMT activity (data not shown), in its absence PRC2-Ezh1 retained some ability to compact chromatin. In contrast, omitting RbAp46/48 impeded the compaction mediated through PRC2-Ezh1 but only partially reduced PRC2-Ezh2 HKMT activity (Cao and Zhang, 2004).
Figure 7. All components of PRC2-Ezh1 are required for full compaction with one PRC2-Ezh1 complex bringing together 3–4 nucleosomes.
A) Individual PRC2-Ezh1 components or partially reconstituted PRC2-Ezh1 complexes were purified and analyzed for their ability to shift recombinant chromatin on sucrose gradients (left). Peak fractions (highlighted in red) were analyzed by EM (right). B) Native hyperacetylated chromatin was incubated with a limiting amount of PRC2-Ezh1 (roughly 35% of the arrays displayed compaction) and analyzed by EM (dark field method). A control and three representative compacted arrays are shown. C) Top, native hyperacetylated chromatin was reconstituted with a limiting amount of histones (an average of 8 nucleosomes per array) and analyzed by EM (dark field method). A control and three representative compacted arrays are shown. Bottom, quantification of the number of free nucleosomes per array. D and E) Chromatin accessibility of an integrated Gal4-Luciferase reporter (D) and of an endogenous PRC2 target gene (E). Top D representation of the transgene with arrows indicating the primers used. Top E, ChIP as described in Figure 3A, and the MYT1 gene was analyzed by PCR. Isolated nuclei were treated with increasing amounts of DNAse 1 and protection was quantified by qPCR. Results are given as percentages of control after normalization to an undigested region of the control SYN1 gene and are the mean ± SD (standard deviation) of three independent biological replicates.
To better understand the mechanism involved in PRC2-Ezh1-mediated compaction, we performed additional EM experiments using limiting amounts of complex or limiting amounts of histones (Figures 7B and 7C, respectively) and the dark field method (Nikitina et al., 2007) to improve image resolution. Limiting amounts of complex allowed us to observe a wider range of chromatin compaction. PRC2-Ezh1 preferentially brings contiguous nucleosomes together. Furthermore in the presence of chromatin with sub-saturating amounts of histones, PRC2-Ezh1 gathers nucleosomes together as evidenced by the formation of loops of naked DNA. In order to estimate how many nucleosomes PRC2-Ezh1 is able to bind, we determined the average number of nucleosomes per array and compared this value to the number of free nucleosomes remaining after addition of limiting amounts of PRC2-Ezh1 (Figure 7D). An average of 4 to 5 nucleosomes were detected within an array reconstituted with an average of 8 nucleosomes, therefore PRC2-Ezh1 could bring together 3 to 4 nucleosomes at a time. As a control, we compared PRC2-Ezh1 complex alone to the complex bound to chromatin (Figure S4B). This result indicated that in most of the compaction events we analyzed, only one complex was bound.
Finally, to determine if this compaction is relevant in vivo, we probed for changes in chromatin accessibility as a function of the presence of PRC2-Ezh1 or PRC2-Ezh2 (Figures 3A and 5A). Nuclei were isolated from the stable cell lines expressing inducible GAL4 versions of the Ezh subunits and containing the integrated Gal4-luciferase reporter (see Figure 7D, top diagram). The accessibility of the latter to DNase1 digestion was quantified by qPCR and normalized to that of the SYN1 gene that we found to be inaccessible to DNase1 digestion under the conditions tested. PRC2-Ezh1 recruitment led to a clear reduction in chromatin digestion, while there was no appreciable change upon PRC2-Ezh2 recruitment (Figure 7D). We also performed this analysis at the known PRC2 target gene MYT1 (Figures 3A and 7E), and obtained similar results (Figure 7E). While overexpression of GAL4-Ezh1 resulted in a clearly detectable increase in its occupancy at the MYT1 gene, there was no significant change in the case of GAL4-Ezh2. This is likely a consequence of the markedly elevated levels of Ezh2 relative to Ezh1 present endogenously in dividing 293 cells. That PRC2-Ezh1 recruitment gave rise to chromatin that was more refractory to DNase1 digestion is in accordance with its observed affect on chromatin structure in the EM studies above. We conclude that PRC2-Ezh1 functions in transcriptional repression by compacting nucleosomal arrays in vitro and in vivo.
DISCUSSION
We have characterized a new PRC2 complex containing Ezh1, Suz12, Eed and RbAP46/48 (PRC-Ezh1). This complex can be reconstituted with all components of the previously described PRC2 complex containing Ezh2 (Cao et al., 2002; Kuzmichev et al., 2002), and displays a similar molecular weight. However, unlike PRC2-Ezh2, this complex exhibits low HKMT activity and accordingly, and in contrast with Ezh2, knockdown of Ezh1 does not result in a global change in H3K27me2/3 levels. Yet PRC2-Ezh1 does repress transcription in vivo and from a chromatinized template in vitro. Remarkably, PRC2-Ezh1 elicits such repression through its ability to compact chromatin as shown in vitro by EM and in vivo by the decreased nuclease accessibility of a Gal4-luciferase reporter to which it is targeted.
Interestingly, PRC2-Ezh1-mediated chromatin compaction is quite distinct from that mediated by PRC1 or L3MBT-L1. L3MBT-L1 performs chromatin compaction by bridging either identical histone marks or a combination of two different marks (Trojer et al., 2007). PRC1 compacts chromatin through interaction with nucleosomes regardless of the presence of histone tails (Francis et al., 2004). PRC2-Ezh1 takes the middle ground, by binding to nucleosomes in the absence of tails but requiring their presence to achieve compaction. However whereas one protein, or even a fragment thereof, is involved in L3MBT-L1 or PRC1 mediated compaction, all four subunits are required in the case of PRC2-Ezh1. Three of them can potentially interact directly with the histone tails (Eed, RbAp46/48 and Ezh1), and this could explain the ability of PRC2-Ezh1 to bring an average of 3 to 4 nucleosomes together. The structure of Nurf55 (RbAP48) was solved (Song et al., 2008). The authors described a histone H4 binding pocket whose function is important when RbAp48 is in complex with HAT1. However, they also showed that point mutations in this pocket impede the integration of RbAp48 into the PRC2-Ezh2 complex suggesting that depending on the complex, the same domain has distinct roles. The structure of Eed was also solved recently (Han et al., 2007), and appears to be quite similar to that of Nurf55 with both proteins forming a seven-bladed β–propeller structure. While the domain of interaction between Ezh2 and Eed was described, the N-terminal region of Eed that interacts with histone H3 has not yet been solved (Tie et al., 2007). An important question is how two highly similar complexes, PRC2-Ezh1 and PRC-Ezh2, can manage to perform such different functions. Indeed, three of the four components are identical and the fourth one is composed of two well-conserved homologs. A finding likely to be pertinent in this regard is that a single point mutation outside of the SET domain of Ezh2 or on other subunits that do not affect complex integrity did disrupt enzymatic activity (Ketel et al., 2005). Also to be considered is that one protein domain might have different functions depending on the complex it composes, as in the case of the binding pocket of RbAp48. Recently, it was reported that Ezh2 and PRC1 are required for genomic compaction at the imprinted Kcnq1 locus (Terranova et al., 2008). Although we show here that PRC2-Ezh1 mediates local compaction, it remains to be determined whether it can also contribute to this kind of long-range effect. We speculate that Ezh1 which is present in non-dividing differentiated cells, may not mediate this function as such cells may have compacted the chromatin into a more stable structure.
We showed that both PRC2-Ezh1 and PRC2-Ezh2 repress transcription in vivo. However, in vitro, PRC2-Ezh1 is a significantly more potent repressor. This suggests that PRC2-Ezh2-mediated repression might involve other factors. For instance, we demonstrated previously that PHF1 can stimulate specifically the H3K27me3 activity of PRC2-Ezh2, so that PHF1 (or one of its homologs) might be required for optimal PRC2-Ezh2-mediated transcriptional repression by elevating H3K27me3 levels and thereby PRC1 recruitment. In the case of Ezh1, our studies in vitro indicated that the formation of a transcription competent initiation complex was sufficient to impede repression suggesting that transcription-mediated alteration/repositioning of nucleosomal arrays thwarted their targeting or their compaction by PRC2-Ezh1.
To support the complementary role of PRC2-Ezh1 and PRC2-Ezh2 in gene repression, we performed gene expression profiling after knockdown of Ezh1, Ezh2 or both (data not shown). However, the results were not interpretable due to their cross-regulation. For instance, Ezh2 knockdown resulted in Ezh1 up-regulation, but at the same time the Suz12 protein was reduced. A clear picture of the involvement of each of these complexes in gene regulation likely requires Ezh1 knockdown in kidney from adult mice or Ezh2 knockdown in an Ezh1−/− background.
We demonstrated that PRC2-Ezh2 and PRC2-Ezh1 share an overlapping set of target genes. In mammals very little is known about how PRC2 is recruited. Recent work has implicated RNA in this process (Rinn et al., 2007), however this might not be a general mechanism as RNAse treatment of U2OS cells did not affect the global localization of Ezh2 or Suz12 as judged by immunofluorescence (Aoto et al., 2008). Here we showed that the SET domain of Ezh proteins is required for their recruitment to target genes. The introduction of a single point mutation in the SET domain resulted in the loss of the complex from target gene promoters. Whether this is a consequence of impaired targeting or reduced stability of the complex at the gene promoters remains to be determined. However, this mutation does not affect complex formation (data not shown). We speculate that either the SET domain is essential for interaction with putative DNA binding factors or is important for recruitment by “sensing” the chromatin environment for nucleosomes to methylate or compact. Although not the subject of study in this report, it is also possible that the Ezh1 SET domain might target a non-chromatin protein(s) as suggested in the case for Ezh2 (Su et al., 2005). Nevertheless, PRC2 gene targeting is probably a multifactor mechanism as evidenced by cells that are depleted of Eed exhibiting a severe impediment in PRC2 recruitment even though Ezh1 and Ezh2 are still expressed (data not shown). In this case however, it is unclear whether Ezh1 recruitment is impaired because of the absence of H3K27me3 or because of the direct involvement of Eed in this process.
Another important difference between Ezh1 and Ezh2 that we report in this study is their expression profiles. We detected Ezh1 in all tissues analyzed and a moderate difference was observed between normal tissue and a cancer cell line from the same origin. However, Ezh2 was barely detectable in non-proliferative tissue, being expressed during development as shown in the case of kidney. Interestingly, whereas deletion of Ezh2 in pro-B cells resulted in a dramatic decrease in H3K27 methylation (Su et al., 2003), we did not observe a clear correlation between Ezh2 expression and H3K27me3 in kidney from aging mice or in adult tissues (data not shown). Considering these data together, it is tempting to speculate that Ezh1 and Ezh2 are recruited to a specific set of genes during development by specific RNA or transcription factors in a cell-type dependent manner. In this scenario, PRC2-Ezh2 would then flag its targeted genes with the H3K27me2/3 mark. When the cells stop dividing, Ezh2 protein levels would be down-regulated. The chromatin would then be kept in gene repression mode by the concerted action of PRC2-Ezh1 and PRC1. The presence of the H3K27me2/3 mark in non-proliferative tissue could be a consequence of its inherent stability, its protection from demethylases through chromatin compaction mediated by PRC1 and PRC2-Ezh1, and possibly the low, but ubiquitous presence of H3K27me2/3 activity associated with PRC2-Ezh1.
Ezh1 and Ezh2 are two paralogs and whereas Drosophila has only one gene, duplication of the ancestor Ez gene occurred relatively early in evolution since two paralogs are also found in Zebrafish (Whitcomb et al., 2007). Sequence alignment suggests that Ezh2 is the closest homolog to Ez (data not shown). Various evolutionary models have been proposed and the main question regarding gene duplication is whether or not this leads to a new function (neo-functionalization) or fosters a pre-established function (sub-functionalization) (Prince and Pickett, 2002). Several histone methyltransferases have undergone gene duplication during evolution, for example Suv4-20h1/h2 and Suv39h1/h2, however in these cases both orthologs display a similar HMT activity (O'Carroll et al., 2000; Schotta et al., 2004). Consequently there is a high degree of redundancy as illustrated by the absence of phenotype associated with a knockout of either Suv39h1 or Suv39h2. This is in contrast to the severe phenotype associated with the double mutant (Peters et al., 2001). In striking opposition to these cases, knockout of Ezh2 is embryonic lethal (O'Carroll et al., 2001). In conjunction with this, PRC2-Ezh2 and PRC2-Ezh1 have distinct functions in vitro and in vivo. To address the evolutionary basis of this, a paramount issue is whether Drosophila PRC2-Ez that exhibits HMKT activity similar to that of PRC2-Ezh2 can also exhibit chromatin compaction similar to that of PRC2-Ezh1. We hypothesize that duplication of Ez corresponds to a sub-functionalization phenomenon with one paralog being dedicated to the transmission of the repressive mark during replication (Ezh2) and the other directly involved in repression (Ezh1). That Ezh1 and potentially its homologs, can directly repress transcription within the context of PRC2, might pertain to both the means by which PRC2 can target genes in the absence of PRC1 (Boyer, 2006) and function in organisms lacking PRC1 (C. Elegans or Oikopleura Dioica, Schuettengruber, 2007). In the case of Ezh1 and Ezh2, sharing the task might facilitate a tighter regulation of Ezh2. This could be an asset as Ezh2 mis-regulation might be of consequence to inheritable gene deregulation, as observed in cancer.
EXPERIMENTAL PROCEDURES
Cell Culture and Stable Cell Lines
F9, NIH-3T3, RAG and Jurkat cells were purchased from ATCC and grown in DMEM supplemented with 10% FCS. 293F and Hela cells were routinely maintained in the lab. 293T-rex were purchased from Invitrogen and grown according to the manufacturer's protocol. Ezh1, Ezh2 or mutated versions s were subcloned into pCDNA4-T0 with the addition of an HA tag (C-ter) and Gal4DBD (N-ter) and stably transfected in 293 T-Rex. Individual clones were selected for each stable cell line. The 5XGal4RE-tk-Luc-neo construct was stably integrated (G418 at 250 μG/ml), and the selected clone was subsequently transfected with pCDNA4-TO containing Gal4-Ezh1 or -Ezh2, HA-wt or -mutant and selected with Zeocin (50 μG/ml final).
Nuclear extracts and Immunoprecipitation
For high salt nuclear extract preparation, cells were incubated in buffer A (10 mM Hepes pH 7.9, 5 mM MgCl2, 0.25 M Sucrose, 0.1% NP40, DTT and PMSF) for 10 min on ice, centrifuged at 8000g, resuspended in buffer B (25 mM Hepes pH 7.9, 20% glycerol, 1.5 mM MgCl 1.5, 0.1 mM EDTA 0.1, 700 mM NaCl, DTT and PMSF), extensively sonicated, centrifuged at 14000 rpm and then dialyzed against BC350 containing 0.1% NP40. Immunoprecipitations were incubated and washed in the same buffer. Elution was performed with 0.2 M glycine at pH 2.6 for endogenous IP or with 0.2 mg/ml HA/Flag peptide.
Cell Fractionation
Cell fractionation was done essentially as described (Wysocka et al., 2001) except that after microccocal nuclease digestion, the pellet was resuspended in buffer B, sonicated, spun down and loaded as chromatin insoluble.
Antibodies
Antibody specific to Ezh2, Suz12 and H3K27me2/3 were previously described (Kuzmichev et al., 2004). Antibody specific to Ezh1 (Enx-2) was raised against aa 1-226 expressed in bacteria using the pET102 plasmid (Invitrogen) and affinity purified using GST-Ezh1 1-226 (pGEX 6P1). Anti-Eed was raised against aa 95-174 of mEed expressed using the pET102 plasmid and affinity purified using a GST-Eed 95-174 (pGEX 6P1). Antibodies against total H3, H3K27me3 (for IHC and ChiP, ab6002) and MCM7 were purchased from Abcam. Anti-H3K27me3 for western blots was a generous gift from Dr Thomas Jenuwein. Antibodies specific for H3K27me1, Gal4-DBD for ChIP and HDAC1 were purchased from Upstate (Millipore). Those for PCNA, Bmi-1, Gal4-DBD (for western blot) and YY1 were purchased from Santa Cruz. Anti-HA, -FLAG, -Actin, -α-tubulin and vinculin were purchased from Sigma. Anti-His was purchased from Qiagen.
Baculovirus
Ezh2, Ezh1 were subcloned into pAcHLTc baculovirus expression plasmid, and Eed (95-535) and Suz12 were subcloned in pFast-Bac. Sf9 cells were grown in Grace media (Gibco) supplemented with 10% heat inactivated FCS and antimycotic/antibiotic (Gibco). Cells were harvested 72 hr after infection, resuspended in BC350 containing 0.1% NP40 and 0.2 mM PMSF, sonicated, centrifuged and used for IP. Superose 6 sizing columns were run in the same buffer after addition of 1mM final DTT.
Chromatin Immunoprecipitation and ChIP on chip
Experiments were done essentially as previously described (Blais et al., 2005), except that the hybridization step was modified according to the manufacturer's instructions (Agilent). ChIP on chip experiments were done two times with two different preparations of chromatin and two batches of affinity purified antibodies. Slides were read using either the agilent scanner or the axon genepix (4000). Data were processed using feature extraction and ChIP analytics with the whitehead 2.0 error model. Only probes with a P value <0.01 in both experiments were considered as bound. Re-Chip was performed as previously described (Metivier et al., 2003). The list of targeted sequences is provided in Supplementary Data (Table 1), and additional information is available on request (complete, raw data).
To prepare chromatin from tissues, newborn or 9 month-old females were sacrificed, kidneys were harvested, kept in PBS plus protease inhibitors and finely chopped with razor blades. Formaldehyde was added to 1% final concentration and samples were kept for 15 min at RT before addition of glycine at 0.125 M final. Cross-linked cells were washed with PBS and transferred to a type A dounce homogenizer. After 8 strokes, samples were centrifuged and pellets were processed for ChIP on chip. For qPCR, 2.5 ng of DNA was used as template for the specific IP or for the input.
Sucrose Gradient and Electron Microscopy
Chromatin was reconstituted by salt dialysis of histones and DNA through a linear gradient (2.24M NaCl to 0.4M NaCl) for 20 hrs, followed by a step dialysis against TE. For each reconstitution, histone and DNA were titrated, analyzed on sucrose gradients (10–30%) and by electron microscopy (EM). Chromatin and purified proteins or complexes were incubated together prior to sucrose gradients in HEB buffer (25 mM Hepes, 40 mM KCl, 0.2 mM EDTA, 1 mM DTT) in the presence or absence of cold SAM at 100 uM final for 1 hr at RT. The amount of complex or protein was titrated and at the highest concentration, a molar ratio of 1 to 2 (protein to nucleosome), was used. Sucrose gradients were prepared using a Gradient maker (Biocomp) according to the manufacturer's instruction and centrifuged for 16 hr at 22000 rpm in a Beckmann 60Ti rotor. Fractionation was done manually (250 uL fraction) and samples were loaded with SDS on a 0.8% agarose gel, stained with Ethidium Bromide and destained in water.
For EM, samples were fixed with 0.6% glutaraldehyde for 30 minute on ice, and dialyzed against TE after addition of 10 mM final Tris pH 8.0. For rotary shadowing, protein-nucleosome complexes were mixed with a buffer containing spermidine to a final concentration of 2 mM, adsorbed to glow-charged carbon-coated grids, washed stepwise with increasing amounts of water/ethanol and rotary shadow cast with tungsten (Griffith and Christiansen, 1978). Samples were examined using a CM 12 transmission electron microscope. Darkfield EM of nucleosomal arrays was done as described (Nikitina et al., 2007).
HMT Assay
HMT assays were performed in HMT buffer (50mM Tris pH 8.8, 2.5 mM MgCl2, and 2.5 mM DTT) as previously described (Nishioka et al., 2002). Peptides used cover amino acids 20 to 30 of histone H3.
Immunohistochemistry
Immunohistochemistry was performed as previously described (Taneja et al., 2005). Briefly, 5 μM tissue sections were dewaxed and rehydrated, peroxidase was quenched with 3% H2O2, and antigen retrieval was done by boiling the slides in Antigen Unmasking Solution (VectorLabs) in the microwave. Once cooled, the slides were blocked in Normal Goat Serum (VectorLabs) and incubated with the primary antibody overnight at 4° C in a humid chamber. Secondary antibodies (VectorLabs) were incubated for 1 hr, followed by ABC treatment and staining using the Vector NovaRed substrate Kit (VectorLabs). Antibody dilution and developing time were kept constant for all tissues.
Tissue Etxtract
Organs were dissected, rinsed with PBS, and lysis buffer was added (50 mM Tris pH 7.4, 450 mM NaCl, 0.2 mM EDTA, 5 mM NaF, aprotinin, leupeptin, sodium orthovanadate, PMSF and DTT). Tissues were homogenized using a tissue tearor, incubated on ice for 15 min, centrifuged and lysates were carefully recovered from the top fat layer and bottom cell pellet.
Chromatin Assembly and in vitro Transcription Assays
Chromatin was assembled and purified with RSF/NAP-1 as described previously (Pavri et al., 2006). The transcription template (pG5MLP 5S array) has been described (An and Roeder, 2004). In vitro transcription assays were performed as described previously (Orphanides et al., 1998). For transcription, 50 ng of naked DNA or purified chromatin reconstituted with native core histones was incubated with various amounts of PRC2-Ezh1 or PRC2-Ezh2 complexes, the transcription reactions were assembled and incubated at 30°C for 45 min followed by the addition of 20 ng of Gal4-VP16 and 100 μg of Hela cell nuclear extract. For the bypass transcription assays, the reaction scheme is shown in Figure 5D. The RNA product was extracted by phenol/chloroform, precipitated with ethanol and analyzed on 6% acrylamide denaturing gels.
DNAse 1 protection assay
The assay was performed essentially as described previously (O'Donnell et al., 2008) except that proteinase K digestion was performed for 1 hour at 55°C. The results from DNAse 1 digestion were normalized to the −520 to −370 region of the SYN1 gene that was inaccessible to DNAse1 digestion in the cell lines used.
qPCR
qPCR was done essentially as previously described (Sarma et al., 2008). For primer sequence see Table S2.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Lynne Vales for comments on the manuscript. We would like to thank Dr. Thomas Jenuwein for providing Ezh1 cDNA and H3K27me3 antibody and Dr. Thimoty J Richmond for the P177-12 DNA. We are very grateful to Deborah Hernandez for excellent technical assistance. Also, John Mallen St. Clair, Rachel Ruoff and Susan Logan provided with valuable technical support for the immunohistochemistry and tissue extract preparation, and Chris Van Oevelen helped us with the ChIP on chip data formatting. This work was supported by grants from DOD PC050535 (R.M.), NIH GM64844 (D.R.), the New Jersey Cancer Institute (D.R.) and the HHMI (D.R.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abel KJ, Brody LC, Valdes JM, Erdos MR, McKinley DR, Castilla LH, Merajver SD, Couch FJ, Friedman LS, Ostermeyer EA, et al. Characterization of EZH1, a human homolog of Drosophila Enhancer of zeste near BRCA1. Genomics. 1996;37:161–171. doi: 10.1006/geno.1996.0537. [DOI] [PubMed] [Google Scholar]
- An W, Roeder RG. Reconstitution and transcriptional analysis of chromatin in vitro. Methods in enzymology. 2004;377:460–474. doi: 10.1016/S0076-6879(03)77030-X. [DOI] [PubMed] [Google Scholar]
- Aoto T, Saitoh N, Sakamoto Y, Watanabe S, Nakao M. Polycomb group protein-associated chromatin is reproduced in post-mitotic G1 phase and required for S-phase progression. The Journal of biological chemistry. 2008;283(27):18905–18915. doi: 10.1074/jbc.M709322200. [DOI] [PubMed] [Google Scholar]
- Bachmann IM, Halvorsen OJ, Collett K, Stefansson IM, Straume O, Haukaas SA, Salvesen HB, Otte AP, Akslen LA. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J Clin Oncol. 2006;24:268–273. doi: 10.1200/JCO.2005.01.5180. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Molecular and cellular biology. 2006;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais A, Tsikitis M, Acosta-Alvear D, Sharan R, Kluger Y, Dynlacht BD. An initial blueprint for myogenic differentiation. Genes & development. 2005;19:553–569. doi: 10.1101/gad.1281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes & development. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. The EMBO journal. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science (New York, N.Y. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Molecular cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- Denisenko ON, Bomsztyk K. The product of the murine homolog of the Drosophila extra sex combs gene displays transcriptional repressor activity. Molecular and cellular biology. 1997;17:4707–4717. doi: 10.1128/mcb.17.8.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorigo B, Schalch T, Kulangara A, Duda S, Schroeder RR, Richmond TJ. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science (New York, N.Y. 2004;306:1571–1573. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- Etchegaray JP, Yang X, DeBruyne JP, Peters AH, Weaver DR, Jenuwein T, Reppert SM. The polycomb group protein EZH2 is required for mammalian circadian clock function. The Journal of biological chemistry. 2006;281:21209–21215. doi: 10.1074/jbc.M603722200. [DOI] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science (New York, N.Y. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- Gillespie RF, Gudas LJ. Retinoid regulated association of transcriptional co-regulators and the polycomb group protein SUZ12 with the retinoic acid response elements of Hoxa1, RARbeta(2), and Cyp26A1 in F9 embryonal carcinoma cells. Journal of molecular biology. 2007;372:298–316. doi: 10.1016/j.jmb.2007.06.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith JD, Christiansen G. Electron microscope visualization of chromatin and other DNA-protein complexes. Annual review of biophysics and bioengineering. 1978;7:19–35. doi: 10.1146/annurev.bb.07.060178.000315. [DOI] [PubMed] [Google Scholar]
- Gunster MJ, Raaphorst FM, Hamer KM, den Blaauwen JL, Fieret E, Meijer CJ, Otte AP. Differential expression of human Polycomb group proteins in various tissues and cell types. Journal of cellular biochemistry. 2001;(Suppl 36):129–143. doi: 10.1002/jcb.1093. [DOI] [PubMed] [Google Scholar]
- Han Z, Xing X, Hu M, Zhang Y, Liu P, Chai J. Structural basis of EZH2 recognition by EED. Structure. 2007;15:1306–1315. doi: 10.1016/j.str.2007.08.007. [DOI] [PubMed] [Google Scholar]
- Jones CA, Ng J, Peterson AJ, Morgan K, Simon J, Jones RS. The Drosophila esc and E(z) proteins are direct partners in polycomb group-mediated repression. Molecular and cellular biology. 1998;18:2825–2834. doi: 10.1128/mcb.18.5.2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketel CS, Andersen EF, Vargas ML, Suh J, Strome S, Simon JA. Subunit contributions to histone methyltransferase activities of fly and worm polycomb group complexes. Molecular and cellular biology. 2005;25:6857–6868. doi: 10.1128/MCB.25.16.6857-6868.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirmizis A, Bartley SM, Farnham PJ. Identification of the polycomb group protein SU(Z)12 as a potential molecular target for human cancer therapy. Molecular cancer therapeutics. 2003;2:113–121. [PubMed] [Google Scholar]
- Klymenko T, Papp B, Fischle W, Kocher T, Schelder M, Fritsch C, Wild B, Wilm M, Muller J. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes & development. 2006;20:1110–1122. doi: 10.1101/gad.377406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Jenuwein T, Tempst P, Reinberg D. Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Molecular cell. 2004;14:183–193. doi: 10.1016/s1097-2765(04)00185-6. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Margueron R, Vaquero A, Preissner TS, Scher M, Kirmizis A, Ouyang X, Brockdorff N, Abate-Shen C, Farnham P, Reinberg D. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:1859–1864. doi: 10.1073/pnas.0409875102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes & development. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laible G, Wolf A, Dorn R, Reuter G, Nislow C, Lebersorger A, Popkin D, Pillus L, Jenuwein T. Mammalian homologues of the Polycomb-group gene Enhancer of zeste mediate gene silencing in Drosophila heterochromatin and at S. cerevisiae telomeres. The EMBO journal. 1997;16:3219–3232. doi: 10.1093/emboj/16.11.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ER, Murdoch FE, Fritsch MK. High histone acetylation and decreased polycomb repressive complex 2 member levels regulate gene specific transcriptional changes during early embryonic stem cell differentiation induced by retinoic acid. Stem cells (Dayton, Ohio) 2007;25:2191–2199. doi: 10.1634/stemcells.2007-0203. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine SS, Weiss A, Erdjument-Bromage H, Shao Z, Tempst P, Kingston RE. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Molecular and cellular biology. 2002;22:6070–6078. doi: 10.1128/MCB.22.17.6070-6078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- Metsuyanim S, Pode-Shakked N, Schmidt-Ott KM, Keshet G, Rechavi G, Blumental D, Dekel B. Accumulation of Malignant Renal Stem Cells is Associated with Epigenetic Changes in Normal Renal Progenitor Genes. Stem cells (Dayton, Ohio) 2008 doi: 10.1634/stemcells.2007-0322. [DOI] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O'Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- Nikitina T, Shi X, Ghosh RP, Horowitz-Scherer RA, Hansen JC, Woodcock CL. Multiple modes of interaction between the methylated DNA binding protein MeCP2 and chromatin. Molecular and cellular biology. 2007;27:864–877. doi: 10.1128/MCB.01593-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka K, Rice JC, Sarma K, Erdjument-Bromage H, Werner J, Wang Y, Chuikov S, Valenzuela P, Tempst P, Steward R, et al. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Molecular cell. 2002;9:1201–1213. doi: 10.1016/s1097-2765(02)00548-8. [DOI] [PubMed] [Google Scholar]
- O'Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Molecular and cellular biology. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Carroll D, Scherthan H, Peters AH, Opravil S, Haynes AR, Laible G, Rea S, Schmid M, Lebersorger A, Jerratsch M, et al. Isolation and characterization of Suv39h2, a second histone H3 methyltransferase gene that displays testis-specific expression. Molecular and cellular biology. 2000;20:9423–9433. doi: 10.1128/mcb.20.24.9423-9433.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell A, Yang SH, Sharrocks AD. MAP kinase-mediated c-fos regulation relies on a histone acetylation relay switch. Molecular cell. 2008;29:780–785. doi: 10.1016/j.molcel.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Hiraoka Y, Aiso S. The Polycomb-group protein ENX-2 interacts with ZAP-70. Immunology letters. 2003;86:57–61. doi: 10.1016/s0165-2478(02)00293-6. [DOI] [PubMed] [Google Scholar]
- Ogawa M, Hiraoka Y, Taniguchi K, Aiso S. Cloning and expression of a human/mouse Polycomb group gene, ENX-2/Enx-2. Biochimica et biophysica acta. 1998;1395:151–158. doi: 10.1016/s0167-4781(97)00156-5. [DOI] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Molecular and cellular biology. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. The EMBO journal. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- Prince VE, Pickett FB. Splitting pairs: the diverging fates of duplicated genes. Nat Rev Genet. 2002;3:827–837. doi: 10.1038/nrg928. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development (Cambridge, England) 2007;134:223–232. doi: 10.1242/dev.02723. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma K, Margueron R, Ivanov A, Pirrotta V, Reinberg D. Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo. Molecular and cellular biology. 2008;28:2718–2731. doi: 10.1128/MCB.02017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, Wutz A. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. The EMBO journal. 2006;25:3110–3122. doi: 10.1038/sj.emboj.7601187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes & development. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nature genetics. 2006;38:700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu CT, Bender W, Kingston RE. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Molecular cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JJ, Garlick JD, Kingston RE. Structural basis of histone H4 recognition by p55. Genes & development. 2008;22:1313–1318. doi: 10.1101/gad.1653308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squazzo SL, O'Geen H, Komashko VM, Krig SR, Jin VX, Jang SW, Margueron R, Reinberg D, Green R, Farnham PJ. Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome research. 2006;16:890–900. doi: 10.1101/gr.5306606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su IH, Basavaraj A, Krutchinsky AN, Hobert O, Ullrich A, Chait BT, Tarakhovsky A. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nature immunology. 2003;4:124–131. doi: 10.1038/ni876. [DOI] [PubMed] [Google Scholar]
- Taneja SS, Ha S, Swenson NK, Huang HY, Lee P, Melamed J, Shapiro E, Garabedian MJ, Logan SK. Cell-specific regulation of androgen receptor phosphorylation in vivo. The Journal of biological chemistry. 2005;280:40916–40924. doi: 10.1074/jbc.M508442200. [DOI] [PubMed] [Google Scholar]
- Terranova R, Yokobayashi S, Stadler MB, Otte AP, van Lohuizen M, Orkin SH, Peters AH. Polycomb Group Proteins Ezh2 and Rnf2 Direct Genomic Contraction and Imprinted Repression in Early Mouse Embryos. Developmental cell. 2008 doi: 10.1016/j.devcel.2008.08.015. [DOI] [PubMed] [Google Scholar]
- Tie F, Stratton CA, Kurzhals RL, Harte PJ. The N terminus of Drosophila ESC binds directly to histone H3 and is required for E(Z)-dependent trimethylation of H3 lysine 27. Molecular and cellular biology. 2007;27:2014–2026. doi: 10.1128/MCB.01822-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, de Wit E, Muijrers I, Teunissen H, Talhout W, van Steensel B, van Lohuizen M. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nature genetics. 2006;38:694–699. doi: 10.1038/ng1792. [DOI] [PubMed] [Google Scholar]
- Trojer P, Li G, Sims RJ, 3rd, Vaquero A, Kalakonda N, Boccuni P, Lee D, Erdjument-Bromage H, Tempst P, Nimer SD, et al. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell. 2007;129:915–928. doi: 10.1016/j.cell.2007.03.048. [DOI] [PubMed] [Google Scholar]
- van Lohuizen M, Tijms M, Voncken JW, Schumacher A, Magnuson T, Wientjens E. Interaction of mouse polycomb-group (Pc-G) proteins Enx1 and Enx2 with Eed: indication for separate Pc-G complexes. Molecular and cellular biology. 1998;18:3572–3579. doi: 10.1128/mcb.18.6.3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Molecular cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr Biol. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004a;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS. Hierarchical recruitment of polycomb group silencing complexes. Molecular cell. 2004b;14:637–646. doi: 10.1016/j.molcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Whitcomb SJ, Basu A, Allis CD, Bernstein E. Polycomb Group proteins: an evolutionary perspective. Trends Genet. 2007;23:494–502. doi: 10.1016/j.tig.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Wysocka J, Reilly PT, Herr W. Loss of HCF-1-chromatin association precedes temperature-induced growth arrest of tsBN67 cells. Molecular and cellular biology. 2001;21:3820–3829. doi: 10.1128/MCB.21.11.3820-3829.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.