Summary
Cre-loxP technology enables specific examination of the function and development of individual nuclei in the complex brain network. However, for most brain regions, the utilization of this technique has been hindered by the lack of mouse lines with Cre expression restricted to these regions. Here, we identified brain expressions of three transgenic Cre lines previously thought to be pancreas-specific. Cre expression driven by the rat-insulin promoter (Rip-Cre) was found mainly in the arcuate nucleus, and to a lesser degree in other hypothalamic regions. Cre expression driven by the neurogenin 3 promoter (Ngn3-Cre mice) was found in the ventromedial hypothalamus. Cre expression driven by the pancreas-duodenum homeobox 1 promoter (Pdx1-Cre) was found in several hypothalamic nuclei, the dorsal raphe and inferior olivary nuclei. Interestingly, Pdx1-Cre mediated deletion of vesicular GABA transporter led to postnatal growth retardation while Ngn3-Cre mediated deletion had no effects, suggesting a role for Pdx1-Cre neurons, but not pancreas, in the regulation of postnatal growth. These results demonstrate the potential for these Cre lines to study the function and development of brain neurons.
Keywords: Pdx1, Ngn3, Rip-Cre, hypothalamus, pancreas
The Cre-loxP technique has proven to be one of the most efficient approaches to tackle the complexity of physiologic processes (Gaveriaux-Ruff and Kieffer, 2007). In the brain, studies based on this technology have shed new lights on important functions of individual groups of neurons in neural pathways underlying various brain functions (Balthasar, 2006; Morozov et al., 2003). In addition, Cre-loxP technology has provided an unprecedented advantage in developmental lineage studies in peripheral tissues (Gu et al., 2003) and in the brain (Kim and Dymecki, 2009). However, for most brain regions, the usage of Cre-loxP technology has been greatly hindered by the lack of mouse lines with specific Cre expression in these regions. Generation and identification of mouse lines with specific Cre expression in novel groups of neurons would greatly facilitate studies on brain development and function.
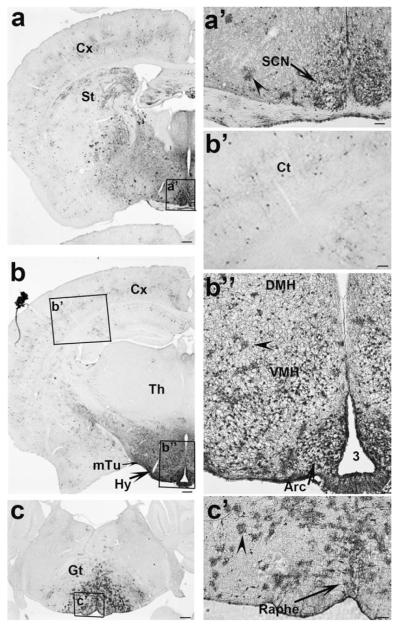
Recent studies have shown that mice with Cre expression driven by the rat insulin promoter (Rip-Cre mice) express Cre in the hypothalamus and suggested a possible role for these neurons in the regulation of energy homeostasis (Choudhury et al., 2005; Covey et al., 2006; Kubota et al., 2004; Lin et al., 2004; Mori et al., 2009). However, the expression pattern of Cre in the hypothalamus of Rip-Cre mice has not been described in detail. To investigate this, we mated Rip-Cre mice with E/ZG mice, in which the expression of green fluorescent protein (GFP) is dependent on Cre activity (Novak et al., 2000). GFP-positive neurons were mainly located in the hypothalamus (Figs. 1a,b). Scattered GFP-positive neurons were also found in the cortex and striatum (Figs. 1a,b,b’). Within the hypothalamus, GFP-positive neurons were located in the suprachiasmatic nucleus (SCN) (Fig. 1a’), the ventral medial hypothalamus (VMH), the dorsal medial hypothalamus (DMH) (Fig. 1b”), the medial tuberal region (mTu) (Fig. 1b), and were particularly concentrated in the arcuate nucleus of the hypothalamus (Arc) (Fig. 1b”). Interestingly, some glia-like structures in the hypothalamus (arrowheads in Fig. 1a’, b”), the gigantocellular region and the raphe area of brain stem were also positive for GFP (Fig. 1c and arrowheads in Fig. 1c’).
FIG. 1.
The expression of Rip-Cre in the brain. GFP immunostaining was performed in brain sections of Rip-Cre, Z/EG mice and visualized with a secondary antibody conjugated to avidin-biotin complex and further development in diaminobenzidine solution. Note that GFP immunoreactivity is mainly located in the hypothalamus. a: At Bregma level of −0.34 mm, a subset of SCN neurons, and scattered neurons in the cortex and striatum are positive for GFP. (a’) The enlarged view of the boxed area in a showing GFP-positive neurons in the SCN. Note that the arrowhead points to glia-like immunoreactive structure. (b) At Bregma level of −1.82 mm, a subset of VMH neurons, mTu neurons and DMH neurons, numerous Arc neurons and scattered neurons in the cortex and striatum are positive for GFP. (b’) The enlarged view of the boxed area in the cortex in b showing GFP-positive neurons in the cortex. (b”) The enlarged view of the boxed area in b showing numerous Arc neurons and a subset of VMH neurons are GFP positive. The arrowhead points to glial-like structure. (c) At Bregma level of −6.12 mm, a subset of glia-like structures in the gigantular and raphe areas of the brain stem are also positive for GFP. (c’) The enlarged view of GFP-positive glia-like structures in the boxed area in c. The arrowhead points to glial-like structure. Cx, cortex; St, striatum; SCN, suprachiasmatic nucleus; Th, thamalus; Hy, hypothalamus; VMH, ventromedial hypothalamus; mTu, medial tuberal region; DMH, dorsal medial hypothalamus; Arc, arcuate nucleus; Gt, gigantular area. 3: the third ventricle. Scale bar = 100 μM (in a–c) or 25 μM (in a’, b’, and b”) or 12.5 μM (in c’).
Neurogenin 3 (Ngn3) is a transcriptional factor required for the development of the pancreas and is turned on in progenitors that develop into endocrine cells of the pancreas (Gu et al., 2003; Herrera et al., 2002). Pdx1 is a transcriptional factor required for the development of the pancreas and foregut (Gu et al., 2003). Both Ngn3-Cre and Pdx1-Cre mice were thought to express Cre only in these peripheral regions (Gu et al., 2002, 2003), and were extensively used to manipulate gene expressions in the pancreas (Lee et al., 2007; Lu et al., 2004; Vincent et al., 2009; Wells et al., 2007). However, both Ngn3 (Sommer et al., 1996; Wang et al., 2001) and Pdx1 (Perez-Villamil et al., 1999; Schwartz et al., 2000) have been reported to be expressed in the brain. These results prompted us to speculate the possibility that Ngn3-Cre and Pdx1-Cre also have brain expression. To test this, we examined GFP expression in the brains of Ngn3-Cre and Pdx-Cre mice crossed with Z/EG mice.
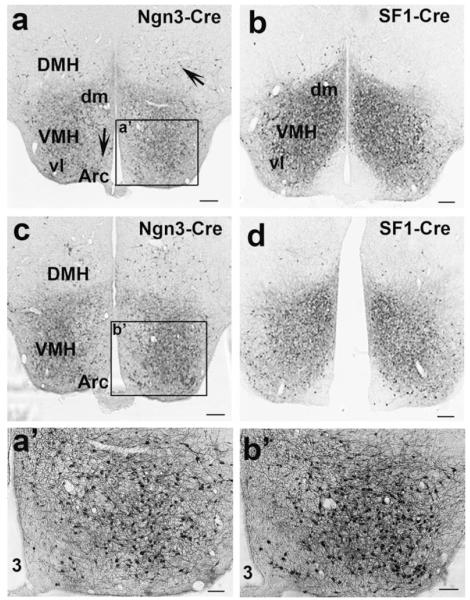
In Ngn3-Cre,Z/EG mice, GFP immunoreactivity was only found in the hypothalamus. Within the hypothalamus, numerous neurons in the VMH and very scattered neurons in the Arc and DMH were positive for GFP (Fig. 2a,c). SF1-Cre is known to specifically express Cre in VMH neurons (Tong et al., 2007). As expected, SF1-Cre expression was limited to the VMH (Fig. 2b,d). However, SF1-Cre positive neurons were more concentrated in the dorsal medial part of the VMH whereas Ngn3-Cre positive neurons were more concentrated in the ventral lateral part (see comparison between Fig. 2a,b; Fig. 2c,d). As shown in the micrograph with higher magnification, within the VMH of Ngn3-Cre mice, numerous neurons were GFP positive (Fig. 2a’,b’). It would be interesting to know whether SF1-Cre and Ngn3-Cre express in 2 distinct groups, or two different groups of VMH neurons with partial overlap. Nonetheless, using a combination of Ngn3-Cre and SF1-Cre will achieve more targeting of VMH neurons.
FIG. 2.
The expression of Ngn3-Cre and SF1-Cre in the hypothalamus. GFP immunostainings were performed in Z/EG mice bred with Ngn3-Cre or SF1-Cre mice. a and b show GFP immunoreactivity at Bregma level of −1.70 mm; c and d show GFP immunoreactivity at Bregma level of −1.94 mm. a and c show GFP immunoreactivity in Ngn3-Cre, Z/EG mice whereas b and d show GFP immunoreactivity in SF1-Cre, Z/EG mice. In Ngn3-Cre, Z/EG mice, some scattered neurons in the DMH and Arc are also positive for GFP (arrows in a). a’ and b’: The enlarged views of the boxed areas in a and c, respectively, showing GFP-positive neurons in the VMH. Note that, within the VMH, Ngn3-Cre expressing neurons are more concentrated in the ventrolateral part of the VMH whereas SF1-Cre expressing neurons are more concentrated in the dorsal medial part of the VMH. VMH, ventral medial hypothalamus; Arc, arcuate nucleus; DMH, dorsal medial hypothalamus; Dm, dorsal medial; vl, ventral lateral; 3, the third ventricle. Scale bar = 100 μM (in a-d) or 25 μM (in a’ and b’).
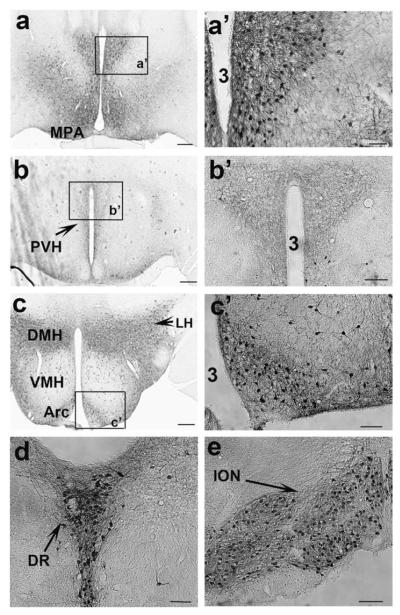
In Pdx1-Cre, Z/EG mice, numerous Cre-expressing neurons were found in the medial preoptic area (MPA) (Fig. 3a,a’), in the Arc, the DMH, the lateral hypothalamus (LH) (Fig. 3c,c’), the raphe nucleus (Fig. 3d) and the inferior olivary nucleus (ION) (Fig. 3e). Notably, none of Pdx1-Cre positive neurons were found in the paraventricular hypothalamus (PVH) or SCN neurons (Fig. 3b), and only a negligible number of neurons were positive in the VMH (Fig. 3c). However, numerous GFP-positive fibers were observed in the PVH (Fig. 3b,b’), suggesting a direct projection from Pdx1-Cre neurons to this region.
FIG. 3.
Expression of Pdx1-Cre in the brain. GFP immunostaining was performed in Pdx1-Cre, Z/EG mice. (a–e) GFP-positive structures in brain sections from rostral to caudal regions. GFP positive neurons were found in the anterior part of the MPA (a); the Arc, DMH, and LH, but not in VMH (c); in the raphe nucleus (d) and in the ION (e). GFP positive fibers were found in the PVH (b). a’–c’: The enlarged views of boxed areas in a, b, and c, respectively, showing GFP-positive neurons (a’ and c’) and fibers (b’). Note that Pdx1-Cre expressing neurons are not found in the PVH or the SCN (c). MPA, medial preoptic area; PVH, paraventricular hypothalamus; SCN, suprachiasmatic nucleus; Arc, arcuate nucleus; VMH, ventromedial hypothalamus; DMH, dorsal medial hypothalamus; LH, lateral hypothalamus; DR, dorsal raphe; ION, inferior olivary nucleus; 3, the third ventricle. Scale bar = 100 μM (in a–c) or 25 μM (in a’–c’).
The ION is the sole source of climbing fibers, which synapse on the dendrites of Purkinjie neurons in the cerebellar cortex (Sugihara, 2006). The abundant expression of Pdx1-Cre provides a system to visualize the projection from the ION to the cerebellar cortex. Indeed, in Pdx1-Cre,Z/EG mice, GFP-positive fibers constituting the inferior cerebellar peduncle (ICP) were observed emanating from the ION (Fig. 4a). These fibers then extended dorsally toward the cerebellum (Fig. 4b) and diffused into cerebellar structures (Fig. 4c). Notably, numerous GFP-positive fibers were observed running transversely in the cerebellar cortex (Fig. 4d), indicative of climbing fibers. Thus, Pdx1-Cre mice provide a useful tool to study the function and development of climbing fibers.
FIG. 4.
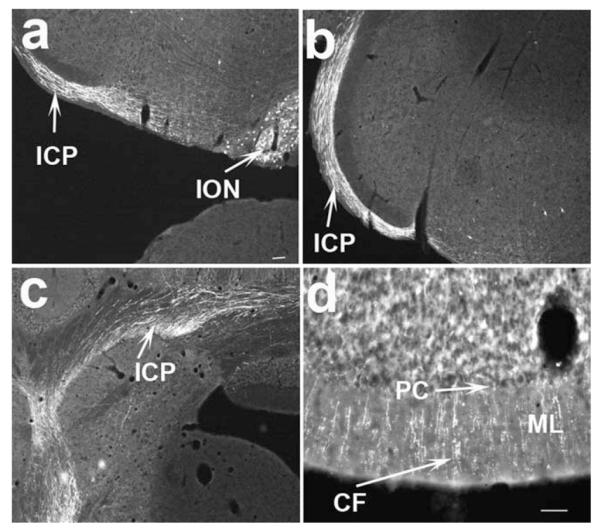
Projections from the ION to the cerebellum. GFP immunostaining was performed in PDX1-Cre, Z/EG mice and developed with flurorescein-conjugated secondary antibody. The immunoreactive structures were then converted into a white pseudocolor when the pictures were taken using a fluorescent microscope. GFP immunoreactive neurons and fibers emanating from ION (a), GFP immunoreactive fibers extended toward the cerebellum (b) and diffused into the cerebellum (c). Numerous GFP positive fibers were found running transversely through the cerebellar cortex reminiscent of climbing fibers (d). ICP, inferior cerebellar peduncle; ION, inferior olivary nucleus; CF, climbing fibers; ML, molecular layer; PC, purkinjie cell layer. Scale bar = 100 μM.
Hypothalamic Cre expression of these lines offers an opportunity to study the function of Cre-expressing neurons in the hypothalamus using Cre-loxP technology. Indeed, Rip-Cre mice have been used in a number of studies to examine the function of hypothalamic neurons in energy homeostasis (Choudhury et al., 2005; Covey et al., 2006; Lin et al., 2004; Mori et al., 2009). However, due to concurrent deletion in pancreatic islets, data interpretation of these studies was confounded by possible roles from pancreatic islets. Given the common expression in pancreatic islets and differential expression in the brain of Rip-Cre and Ngn3-Cre mice, these Cre lines may be used in a binary fashion to identify gene functions in brain neurons. Deletion mediated by Ngn3-Cre can be used to eliminate potential confounding effects from the pancreas in the above mentioned studies. On the other hand, Pdx1-Cre mice have been used to examine the function of various genes in the pancreas related to glucose homeostasis (Lee et al., 2007; Lu et al., 2004; Morioka et al., 2007). As brain regions with Pdx1-Cre expression are also involved in glucose homeostasis (Coppari et al., 2005; Kokorovic et al., 2008; Lam et al., 2005; Obici et al., 2002; Pocai et al., 2005), it might be possible that deletion mediated by Pdx1-Cre in brain neurons contributes to the glucose phenotype described in these studies. Therefore, further investigations are required to distinguish the function of the pancreas from the hypothalamus.
The hypothalamus consists of diverse groups of neurons with distinct functions. How the diversity evolves is less understood. Rip-Cre, Ngn3-Cre, and Pdx1-Cre have been extensively used for cell lineage studies in pancreas development. The expression of these Cre lines in the hypothalamus will be useful to study developmental lineages of these hypothalamic Cre-expressing neurons. In this regard, Ngn3-Cre targets a different group of neurons in the VMH from SF1-Cre neurons (with possible overlap), and it would be interesting to determine cell lineages of Ngn3-Cre and SF1-Cre neurons.
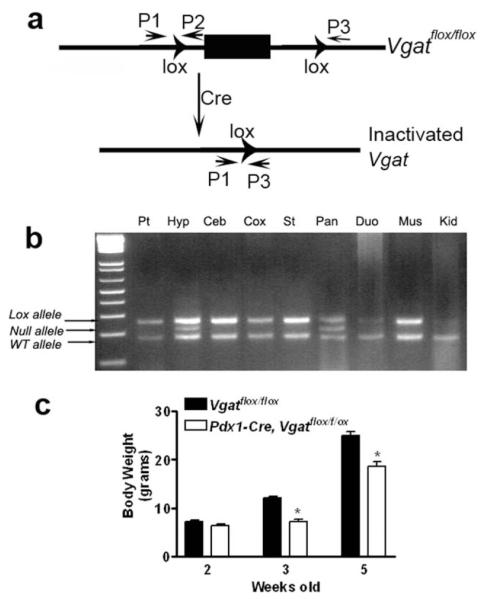
To demonstrate the usage of these Cre lines in a binary fashion, we crossed both Pdx1-Cre and Ngn3-Cre with mice bearing the conditional allele of vesicular GABA transporter (Vgatflox/flox). Vgatflox/floxmice were generated previously to achieve neuron-specific ablation of release of GABA, the major inhibitory neurotransmitter in the brain (Tong et al., 2008). To confirm Cre-mediated deletion of Vgat in the hypothalamus, we performed genomic PCR for the presence or absence of lox P flanked genomic sequence in Pdx1-Cre, Vgatflox/+ mice (Fig. 5a). A band that corresponds to the null allele of Vgat (i.e., deletion of the loxP flanked genomic sequence) was observed in both hypothalamus and pancreas (Fig. 5b). A similar genomic deletion was also observed in Ngn3-Cre, Vgatflox/+ mice (data not shown). These results suggest concurrent deletion of Vgat in the hypothalamus and pancreas mediated by both Cre lines. Interestingly, while these mice had normal body weight at two weeks old, Pdx1-Cre, Vgatflox/flox mice showed greatly reduced body weight at three and five weeks old (Fig. 5c), indicative of a specific postnatal development defect. Consistently, these mice exhibited a shorter statue (data not shown). Ngn3-Cre, Vgatflox/flox mice, in contrast, showed completely normal growth, compared with controls (Fig. 5d). Taken together, these results demonstrate that GABA release from Pdx1-Cre expressing neurons, but not pancreatic islets, is required for normal postnatal development.
FIG. 5.
Function of Pdx1-Cre neurons in growth regulation. (a) Vgatflox/flox allele and the cre-mediated inactivation; (b) genomic PCR for the presence for loxP and the deletion of loxP flanked genomic sequence in Vgatflox/flox allele using primers indicated in a in various types of tissues indicated. P1 and P2 are the primers for detecting the presence of the loxP site whereas P1 and P3 for detecting the deletion of the loxP flanking sequence. Pt, pituitary; Hyp, hypothalamus; Ceb, cerebellum; Cox, cortex; St, striatum; Pan, pancreas; Duo, duodenum; Mus, muscle; Kid, kidney. c; body weight of Pdx1-Cre, Vgatflox/flox mice and their controls at two, three, and five weeks old, *<0.05.
It is unknown whether brain expressions of Cre in these transgenic lines reflect the endogenous activities of these promoters. Notably, VMH-specific expression of Ngn3-Cre is consistent with previously reported expression of Ngn3 in the ventral medial part of the hypothalamus (Sommer et al., 1996). Pdx1 was also reported to be expressed in the hypothalamus (Perez-Villamil et al., 1999; Schwartz et al., 2000). To determine the onset of Pdx1-Cre expression in the brain during development, we crossed Pdx1-Cre with a Cre-dependent expression of red fluorescent protein (RFP) reporter mouse line Ai9 (Madisen et al., 2010) and looked for RFP expression in embryonic days 7 (E7) and 10 (E10). The RFP expression in Ai9 reporter line can be directly visualized under the fluorescent microscope (Madisen et al., 2010). At E7, no RFP signal was observed (data not shown) while at E10, a specific subset of neurons with strong RFP signal was observed in brain region (Fig. 6a,b), suggesting Pdx1-Cre was turned on between E7 and E10. The expression of insulin in the brain seems to be controversial although studies have suggested insulin expression in the brain (Gerozissis, 2003; Grunblatt et al., 2007; Hrytsenko et al., 2007; Madadi et al., 2008). To examine whether the insulin promoter is intrinsically active in the brain and whether it is active in the adult brain, we used another Cre transgenic line driven by the same insulin promoter as in the Rip-Cre transgene: Rip-CreER. In this line, Cre is sequestered in the cytoplasm due to its fusion with an inactive estrogen receptor (ER). Upon Tamoxifen (Tam) administration, activation of ER will translocate Cre from the cytoplasm to the nucleus. Therefore, cleavage of genomic sequence can be induced by Tam administration at any desired time point (Danielian et al., 1998). Rip-CreER, Ai9 mice were treated with either Tam or vehicle at eight weeks old. With vehicle administration, these mice showed only a few neurons positive for RFP in the hypothalamus (Fig. 6c), suggestive of low level of leaky expression. However, with Tam administration, these mice showed substantial number of RFP positive neurons in the hypothalamus (Fig. 6d), reminiscent of Rip-Cre expression. These results suggest that Rip-CreER is expressed in the adult hypothalamus, and therefore can be used to study the function of adult brain neurons. Taken together, the finding that Rip-Cre (Rip-CreER), Ngn3-Cre and Pdx1-Cre all express Cre in the hypothalamus suggests that hypothalamic activities of these promoters might be intrinsic. According to the developmental paradigm of the pancreas, insulin-expressing β cells are a subset of Ngn3-expressing endocrine cells, and the latter are a subset of the endocrine part of Pdx1-expressing pancreas (Gu et al., 2003). However, in the hypothalamus, Rip-Cre expresses in the Arc, SCN, VHM, DMH and mTu; Ngn3-Cre only expresses in the VMH whereas Pdx1-Cre expresses in the Arc, DMH, LH, and MPA, but not in the VMH or SCN. Therefore, it seems that the transcriptional profiles of these promoters in the hypothalamus are distinct from those in the pancreas.
FIG. 6.
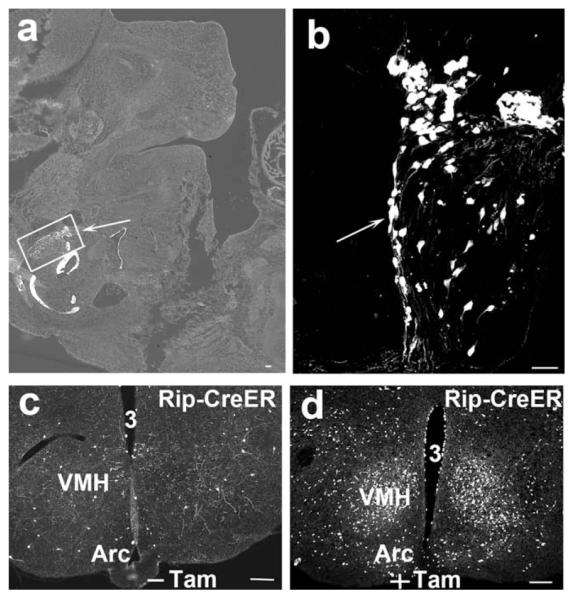
Developmental expression of Pdx1-Cre and adult expression of Rip-CreER. Both Pdx1-Cre and Rip-CreER mice were bred with Ai9 mice. In Pdx1-Cre, Ai9 mice, RFP was looked for in the sections of embryos E10. A specific group of neurons in the brain region of the embryo express intense RFP (a, arrow). (b) The amplified picture of the boxed area in panel a, demonstrating RFP-positive neurons (arrow). Rip-CreER, Ai9 mice were administrated with either vehicle or Tam at eight weeks old. A few neurons were RFP positive with vehicle treatment (c) while abundant neurons were found to express RFP with Tam treatment (d). VMH, ventromedial hypothalamus; Arc, arcuate nucleus; 3, the third ventricle; −Tam, without Tam treatment; +Tam, with Tam treatment. Scale bar = 100 μM.
METHODS
Mice
Rip-Cre transgenic mice (B6.Cg-Tg(Ins2-cre)25Mgn/J), Ngn3-Cre transgenic mice (B6.FVB(Cg)-Tg(Neurog3-cre)-C1Able/J), Z/EG transgenic mice (Tg(CAG-Bgeo/GFP)21Lbe/J), Ai9 reporter mice (B6.Cg-Gt(ROSA)26-Sortm9(CAG-tdTomato)Hze/J) and Rip-CreER (Tg(Ins2-cre/Esr1)1Dam/J) were purchased from the Jackson Laboratory. Pdx1-Cre mice were obtained from Dr. Doug Melton of Harvard University. To permit the identification of Cre-expressing neurons in the brain, these Cre lines were mated either with a Cre-dependent expression of GFP reporter line E/ZG mice or with a Cre-dependent expression RFP reporter line Ai9 mice. The Cre genotyping primers were: 5′GCG GTC TGG CAG TAA AAA CTA TC 3′ and 5′ GTG AAA CAG CAT TGC TGT CAC TT 3′. Z/EG genotyping primers were 5′ AAG TTC ATC TGC ACC ACC G 3′ and 5′ TCC TTG AAG AAG ATG GTG CG 3′. Pdx1-Cre or Ngn3-Cre mice were first crossed with Vgatflox/flox mice. The resulting mice with both Cre and Vgatflox/+ positive (Cre,Vgatflox/+) were further crossed with Vgatflox/flox mice to generate Cre,Vgatflox/flox mice. Study subjects were littermates of breeding pairs of Cre,Vgatflox/flox mice and Vgatflox/flox mice. All animal experiments were approved by the Institutional Animal Care and Use Committee of Beth Israel Deaconess Medical Center and the Animal Welfare Committee of the University of Texas Health Science Center at Houston.
Immunostaining and Histology
Adult Mouse (eight weeks old) brains were obtained after transcardiac perfusion with 10% neutral buffered formalin and sectioned into five series of 25 μM thickness (Sliding Microtome, Leica). For Z/EG reporter mice crossed with various Cre lines, one of the series was immunostained for GFP with an anti-GFP serum (Invitrogen, Carlsbad, CA) and visualized with a secondary antibody conjugated to avidin-biotin complex and further development in diaminobenzidine solution (brown) (Jackson ImmunoResearch Lab, West Grove, PA), or with a secondary antibody conjugated to alexa 488 (green flurorescein) (Jackson ImmunoResearch Lab, West Grove, PA). For Ai9 reporter mice crossed with various Cre lines, red fluorescent protein (RFP) expression was directly visualized under the fluorescent microscope. Results were visualized and recorded by fluorescence or bright-field microscopy (Zeiss Axioskop Microscope).
Embryo Collection and Tamoxifen Injection
For embryo collection, adult males and females (eight weeks old) were paired for one night. The morning of visual appearance of vaginal plug was designated as embryonic day 0.5 (E0.5). E7 and E10 embryos were obtained and genotyped for the presence of both Cre and reporter transgenes. The bigenic embryos were sectioned using cryostat microtome and examined for reporter expression. For Tamoxifen (Tam, Sigma) injection, eight weeks old Rip-CreER,Ai9 mice were administrated intraperitoneally with either Tam (dissolved in corn oil) at the concentration of 20 mg/kg or vehicle (corn oil) once a day for continuous three days. After one week of last injection, the mice were perfused, the brain was obtained for sectioning and RFP expression was examined.
ACKNOWLEDGMENTS
The authors thank Dr. Bradford B Lowell for his training and support.
Contract grant sponsor: Boston Obesity Nutrition Research Center, Contract grant number: 5P30 DK46200-15; Contract grant sponsor: AHA award; Contract grant number: 10SDG3280017; Contract grant sponsor: Young Investigator Award (NAASO)
LITERATURE CITED
- Balthasar N. Genetic dissection of neuronal pathways controlling energy homeostasis. Obesity (Silver Spring) 2006;14(Suppl 5):222S–227S. doi: 10.1038/oby.2006.313. [DOI] [PubMed] [Google Scholar]
- Choudhury AI, Heffron H, Smith MA, Al-Qassab H, Xu AW, Selman C, Simmgen M, Clements M, Claret M, Maccoll G, Bedford DC, Hisadome K, Diakonov I, Moosajee V, Bell JD, Speakman JR, Batterham RL, Barsh GS, Ashford ML, Withers DJ. The role of insulin receptor substrate 2 in hypothalamic and beta cell function. J Clin Invest. 2005;115:940–950. doi: 10.1172/JCI24445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppari R, Ichinose M, Lee CE, Pullen AE, Kenny CD, McGovern RA, Tang V, Liu SM, Ludwig T, Chua SC, Jr, Lowell BB, Elmquist JK. The hypothalamic arcuate nucleus: A key site for mediating leptin’s effects on glucose homeostasis and locomotor activity. Cell Metab. 2005;1:63–72. doi: 10.1016/j.cmet.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Covey SD, Wideman RD, McDonald C, Unniappan S, Huynh F, Asadi A, Speck M, Webber T, Chua SC, Kieffer TJ. The pancreatic beta cell is a key site for mediating the effects of leptin on glucose homeostasis. Cell Metab. 2006;4:291–302. doi: 10.1016/j.cmet.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- Gaveriaux-Ruff C, Kieffer BL. Conditional gene targeting in the mouse nervous system: Insights into brain function and diseases. Pharmacol Ther. 2007;113:619–634. doi: 10.1016/j.pharmthera.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Gerozissis K. Brain insulin: Regulation, mechanisms of action and functions. Cell Mol Neurobiol. 2003;23:1–25. doi: 10.1023/A:1022598900246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunblatt E, Salkovic-Petrisic M, Osmanovic J, Riederer P, Hoyer S. Brain insulin system dysfunction in streptozotocin intracere-broventricularly treated rats generates hyperphosphorylated tau protein. J Neurochem. 2007;101:757–770. doi: 10.1111/j.1471-4159.2006.04368.x. [DOI] [PubMed] [Google Scholar]
- Gu G, Brown JR, Melton DA. Direct lineage tracing reveals the ontogeny of pancreatic cell fates during mouse embryogenesis. Mech Dev. 2003;120:35–43. doi: 10.1016/s0925-4773(02)00330-1. [DOI] [PubMed] [Google Scholar]
- Gu G, Dubauskaite J, Melton DA. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129:2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- Herrera PL, Nepote V, Delacour A. Pancreatic cell lineage analyses in mice. Endocrine. 2002;19:267–278. doi: 10.1385/ENDO:19:3:267. [DOI] [PubMed] [Google Scholar]
- Hrytsenko O, Wright JR, Jr, Morrison CM, Pohajdak B. Insulin expression in the brain and pituitary cells of tilapia (Oreochromis niloticus) Brain Res. 2007;1135:31–40. doi: 10.1016/j.brainres.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Kim JC, Dymecki SM. Genetic fate-mapping approaches: New means to explore the embryonic origins of the cochlear nucleus. Methods Mol Biol. 2009;493:65–85. doi: 10.1007/978-1-59745-523-7_5. [DOI] [PubMed] [Google Scholar]
- Kokorovic A, Cheung GW, Rossetti L, Lam TK. Hypothalamic sensing of circulating lactate regulates glucose production. J Cell Mol Med. 2008;13:4403–4408. doi: 10.1111/j.1582-4934.2008.00596.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota N, Terauchi Y, Tobe K, Yano W, Suzuki R, Ueki K, Takamoto I, Satoh H, Maki T, Kubota T, Moroi M, Okada-Iwabu M, Ezaki O, Nagai R, Ueta Y, Kadowaki T, Noda T. Insulin receptor substrate 2 plays a crucial role in beta cells and the hypothalamus. J Clin Invest. 2004;114:917–927. doi: 10.1172/JCI21484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam TK, Gutierrez-Juarez R, Pocai A, Rossetti L. Regulation of blood glucose by hypothalamic pyruvate metabolism. Science. 2005;309:943–947. doi: 10.1126/science.1112085. [DOI] [PubMed] [Google Scholar]
- Lee JY, Gavrilova O, Davani B, Na R, Robinson GW, Hennighausen L. The transcription factors Stat5a/b are not required for islet development but modulate pancreatic beta-cell physiology upon aging. Biochim Biophys Acta. 2007;1773:1455–1461. doi: 10.1016/j.bbamcr.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Taguchi A, Park S, Kushner JA, Li F, Li Y, White MF. Dysregulation of insulin receptor substrate 2 in beta cells and brain causes obesity and diabetes. J Clin Invest. 2004;114:908–916. doi: 10.1172/JCI22217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Herrera PL, Guo Y, Sun D, Tang Z, LeRoith D, Liu JL. Pancreatic-specific inactivation of IGF-I gene causes enlarged pancreatic islets and significant resistance to diabetes. Diabetes. 2004;53:3131–3141. doi: 10.2337/diabetes.53.12.3131. [DOI] [PubMed] [Google Scholar]
- Madadi G, Dalvi PS, Belsham DD. Regulation of brain insulin mRNA by glucose and glucagon-like peptide 1. Biochem Biophys Res Commun. 2008;376:694–699. doi: 10.1016/j.bbrc.2008.09.054. [DOI] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H, Inoki K, Munzberg H, Opland D, Faouzi M, Villanueva EC, Ikenoue T, Kwiatkowski D, MacDougald OA, Myers MG, Jr, Guan KL. Critical role for hypothalamic mTOR activity in energy balance. Cell Metab. 2009;9:362–374. doi: 10.1016/j.cmet.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morioka T, Asilmaz E, Hu J, Dishinger JF, Kurpad AJ, Elias CF, Li H, Elmquist JK, Kennedy RT, Kulkarni RN. Disruption of leptin receptor expression in the pancreas directly affects beta cell growth and function in mice. J Clin Invest. 2007;117:2860–2868. doi: 10.1172/JCI30910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozov A, Kellendonk C, Simpson E, Tronche F. Using conditional mutagenesis to study the brain. Biol Psychiatry. 2003;54:1125–1133. doi: 10.1016/s0006-3223(03)00467-0. [DOI] [PubMed] [Google Scholar]
- Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- Obici S, Zhang BB, Karkanias G, Rossetti L. Hypothalamic insulin signaling is required for inhibition of glucose production. Nat Med. 2002;8:1376–1382. doi: 10.1038/nm1202-798. [DOI] [PubMed] [Google Scholar]
- Perez-Villamil B, Schwartz PT, Vallejo M. The pancreatic homeodomain transcription factor IDX1/IPF1 is expressed in neural cells during brain development. Endocrinology. 1999;140:3857–3860. doi: 10.1210/endo.140.8.7048. [DOI] [PubMed] [Google Scholar]
- Pocai A, Lam TK, Gutierrez-Juarez R, Obici S, Schwartz GJ, Bryan J, Aguilar-Bryan L, Rossetti L. Hypothalamic K(ATP) channels control hepatic glucose production. Nature. 2005;434:1026–1031. doi: 10.1038/nature03439. [DOI] [PubMed] [Google Scholar]
- Schwartz PT, Perez-Villamil B, Rivera A, Moratalla R, Vallejo M. Pancreatic homeodomain transcription factor IDX1/IPF1 expressed in developing brain regulates somatostatin gene transcription in embryonic neural cells. J Biol Chem. 2000;275:19106–19114. doi: 10.1074/jbc.M000655200. [DOI] [PubMed] [Google Scholar]
- Sommer L, Ma Q, Anderson DJ. neurogenins, a novel family of atonal-related bHLH transcription factors, are putative mammalian neuronal determination genes that reveal progenitor cell heterogeneity in the developing CNS and PNS. Mol Cell Neurosci. 1996;8:221–241. doi: 10.1006/mcne.1996.0060. [DOI] [PubMed] [Google Scholar]
- Sugihara I. Organization and remodeling of the olivocerebellar climbing fiber projection. Cerebellum. 2006;5:15–22. doi: 10.1080/14734220500527385. [DOI] [PubMed] [Google Scholar]
- Tong Q, Ye C, McCrimmon RJ, Dhillon H, Choi B, Kramer MD, Yu J, Yang Z, Christiansen LM, Lee CE, Choi CS, Zigman JM, Shulman GI, Sherwin RS, Elmquist JK, Lowell BB. Synaptic glutamate release by ventromedial hypothalamic neurons is part of the neurocircuitry that prevents hypoglycemia. Cell Metab. 2007;5:383–393. doi: 10.1016/j.cmet.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Q, Ye CP, Jones JE, Elmquist JK, Lowell BB. Synaptic release of GABA by AgRP neurons is required for normal regulation of energy balance. Nat Neurosci. 2008;11:998–1000. doi: 10.1038/nn.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent DF, Yan KP, Treilleux I, Gay F, Arfi V, Kaniewsky B, Marie JC, Lepinasse F, Martel S, Goddard-Leon S, Iovanna JL, Dubus P, Garcia S, Puisieux A, Rimokh R, Bardeesy N, Scoazec JY, Losson R, Bartholin L. Inactivation of TIF1gamma cooperates with Kras to induce cystic tumors of the pancreas. PLoS Genet. 2009;5:e1000575. doi: 10.1371/journal.pgen.1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chu LT, He J, Emelyanov A, Korzh V, Gong Z. A novel zebrafish bHLH gene, neurogenin3, is expressed in the hypothalamus. Gene. 2001;275:47–55. doi: 10.1016/s0378-1119(01)00648-5. [DOI] [PubMed] [Google Scholar]
- Wells JM, Esni F, Boivin GP, Aronow BJ, Stuart W, Combs C, Sklenka A, Leach SD, Lowy AM. Wnt/beta-catenin signaling is required for development of the exocrine pancreas. BMC Dev Biol. 2007;7:4. doi: 10.1186/1471-213X-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]