Summary
Hepatitis C virus (HCV) infection is a major cause of chronic liver disease and affects nearly 170 million people worldwide. Whereas the previous standard of care with pegylated interferon and ribavirin had a modest effectiveness, the recent approval of two highly potent protease inhibitors and the ongoing development of dozens of direct acting antivirals (DAAs) constitute a major milestone for HCV therapy. Mathematical modeling of viral kinetics under treatment has played an instrumental role in improving our understanding of virus pathogenesis and in guiding drug development. Here, we review the current state of HCV kinetic modeling, and challenges to the standard biphasic viral decline model that arise when fitting viral kinetic models to data obtained with DAAs.
Introduction
Chronic HCV infection affects nearly 3% of the worldwide population [1]. HCV is divided into 6 genotypes (1–6) with several subtypes within each genotype. The goal of treatment is to achieve a sustained virologic response (SVR), a marker of viral eradication, assessed by the absence of detectable virus six months after cessation of therapy. Treatment outcome with pegylated interferon (peg-IFN) and ribavirin (RBV) is correlated with HCV genotype and SVR is only achieved in approximately 50% of HCV genotype 1 patients, the hardest to treat and most prevalent genotype in western countries [2]. In the last two decades, research on understanding the mechanisms of HCV replication has resulted in the development and the clinical implementation of a large number of direct acting antivirals (DAAs). The recent approval of two protease inhibitors (PI), telaprevir and boceprevir, in combination with peg-IFN/RBV, has raised expectations that a sustained viral response (SVR) can be achieved in more than 70% of treatment naïve HCV genotype 1 patients [3, 4]. Although representing an undisputable milestone in HCV therapy, the success of protease inhibitor-based therapy is tempered by the existence of severe side effects and the emergence of resistance to treatment [5]. New drugs are still needed, and dozens of compounds targeting all stages of viral replication are currently in different phases of clinical development [6].
Mathematical modeling of viral kinetics aims at understanding and quantifying the biological mechanisms that govern the changes in the viral load and related biomarkers, such as alanine aminotransferase (ALT) levels, both before and after therapy [7]. Initiated in the mid-90’s to characterize decline in human immunodeficiency virus (HIV) during treatment with highly active antiretroviral therapy [8–10], it was successfully applied a few years later to understand HCV RNA kinetics during therapy [11]. This approach has provided valuable insights into deciphering the modes of action of peg-IFN/RBV and estimating key parameters of the HCV life cycle, thus offering an efficient tool for the prediction of treatment outcome [12–14]. Here we review recent modeling efforts in the context of DAAs and show how these new models challenge some of our understanding of HCV obtained by analyzing the effects of peg-IFN/RBV on viral load.
The basis of HCV mathematical modeling: the biphasic viral decline during high daily dose IFN therapy
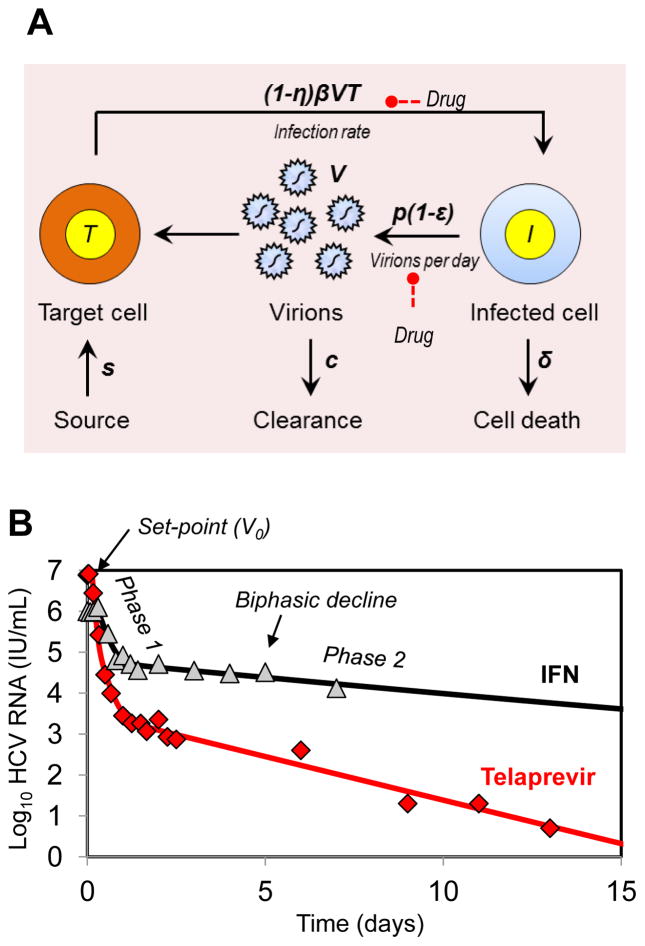
In patients given high daily doses of standard IFN, HCV RNA generally declines in a biphasic manner, where a first phase lasting between 1 and 2 days leads to a 0–2 log fall in virus load, followed by a slower but persistent phase of decline. Similar to what had been done for HIV [9], Neumann et al. [11] developed the following mathematical model to explain the biphasic decline in HCV (Fig. 1A):
| (1) |
where T and I represent the susceptible and the infected cell density, respectively, and V is the virus concentration in the serum. Susceptible cells are generated at a rate s and die at a rate d. Infected cells, which are generated from the interaction of virus with target cells with rate constant β, produce virus at per capita rate p and are eliminated at rate δ per cell. At treatment initiation (i.e., at t=0) the infection is assumed to be in steady state with a constant baseline viral load V0. Treatment affects the viral load after a pharmacologic delay t0. The effectiveness of the drug in decreasing new infections is, whereas the effectiveness in blocking virion production is ε, where the effectiveness is assumed to have a value between 0 (no effect of the drug) and 1 (100% effective). This model is often simplified by assuming that (i) the susceptible cells T(t) after treatment initiation remain constant and equal to their pretreatment value T0 and (ii) the parameter values do not change over time. Under these assumptions the model, Eq. 1, can be solved to yield [11]:
| (2) |
where
Fig. 1.
Mathematical modeling of HCV kinetics. (A) Schematic of the standard model for HCV infection described in the text. (B) Representative example of viral decline observed during daily IFN (triangle, in [9]) and the corresponding best-fit prediction using the standard model (black curve). A representative viral decline observed during treatment with telaprevir +peg-IFN 2a for 14 days (diamond) [29] and best fit prediction using the standard model (red curve) of the viral kinetic model are shown. Compared to the decline generated by IFN therapy, telaprevir generates a larger first phase decline and a more rapid second phase decline.
| (3) |
This model provides a simple conceptual framework for the understanding of the biphasic viral decline after treatment initiation (Fig. 1B). Although an effect on other parameters cannot be ruled out, assuming that the major effect of IFN is blocking viral production is necessary to reproduce a biphasic viral decline [11]. As a result of IFN reducing viral production, HCV RNA initially rapidly declines with a rate λ1≈εc, and if treatment is potent, i.e. ε ≈ 1, the viral load declines with a rate close to its natural clearance rate, c. This phase of decline continues until viral load reaches a value V1 that reflects a new lower equilibrium between virus production and clearance (V1=(1−ε)V0). At the end of the first phase there is less virus in serum and hence less infection of new cells. Thus, as infected cells are lost with rate δ they are less efficiently replaced and there is a net loss of infected cells, which causes the second phase decline, with rate λ2, which, if ε is close to 1, will be approximately equal to εδ. Since the loss rate of infected cells is smaller than the viral clearance rate (δ<c), the second phase of viral decline will be slower than the first phase. On the other hand, assuming that treatment blocks only cell infection (η>0, ε=0) results in viral load declining linearly with a rate λ2 which, if η is close to 1, will be approximately equal to ηδ. Thus, for an entry inhibitor, modeling predicts that no rapid first phase decline due to blocking of viral production should be observed.
Parameter value and variability in the population
By fitting this model to HCV RNA data after initiation of IFN treatment, estimates of c, ε and δ could be obtained [11]. Consistently, c has been found on an average to be approximately 6 day−1 (varying between 3.6 and 11.2 day−1), implying a viral half-life in serum (denoted as t1/2) of about 2.7 hours. Contrastingly, ε and δ are very variable in the population, which is largely accounted by HCV genotype, polymorphism in the IL28B, ethnicity, baseline viral load, baseline IP-10 levels and histological factors [15–20].
Modeling changes in drug effectiveness (peg-IFN)
With frequent dosing, such as daily administration of standard IFN, the plasma concentration of drug is nearly constant [21]. Under such conditions the drug effectiveness ε should be nearly constant as assumed in the standard model. However, when drug is administered once a week, as is the case with peg-IFN, the concentration of drug in plasma initially increases and then decreases over time, resulting in weekly fluctuations in HCV RNA levels. These fluctuations in viral load can be captured by relaxing the assumption of constant drug effectiveness and taking into account the pharmacokinetic/pharmacodynamics (PK/PD) of peg-IFN [22, 23]. The average antiviral effectiveness and the kinetic parameters are similar to those found with standard IFN [22, 23]. Some recent reviews discuss the modeling of drug effectiveness in detail [21, 24].
Modeling long-term viral kinetics: relaxing the assumption of constant target cells and the notion of critical drug effectiveness
Even when constant treatment effectiveness is a reasonable assumption, the viral load does not necessarily decline continuously, as predicted by the biphasic model. Viral rebound can occur due to increases in target cells and can be captured in viral kinetic models by relaxing the assumption of constant target cells. In this case, the long-term viral kinetics depend on the ability of treatment to overcome a certain threshold called the critical effectiveness εc [25]. If ε< εc, enough new infections occur that the viral load eventually stops declining. Depending on the parameter values, there can be almost no decline (as seen in non-responders) or a biphasic decline where the second phase is flat (partial responders) or a biphasic decline followed by a rebound where viral load establishes a new set-point value (rebounders). Lastly, a second flat phase followed by a renewed decline of viral load has been reported in some patients [26, 27]. This triphasic pattern can be captured by including the proliferation of infected cells [26]. However, little is known about the rate of hepatocyte proliferation in vivo during chronic HCV infection.
Direct acting antivirals (DAAs)
DAAs have ushered in a new era of HCV. Below we review recent attempts to fit viral kinetics during DAA therapy, and we discuss how the models developed for this purpose challenge the previous models developed for analyzing viral decline during peg-IFN/RBV.
Modeling viral kinetics with protease inhibitors: rapid biphasic viral decline
Like IFN-based therapy, a biphasic viral decline is observed during protease inhibitor (PI) monotherapy (Fig. 1B). PIs have a high antiviral effectiveness (ε), which has been estimated to be higher than during IFN-based therapy (Table 1). Surprisingly, the second phase viral decline has been consistently found to be more rapid with PIs than with IFN (Figure 1B), which in the standard model is attributed to an elevated loss rate of infected cells, δ. Using the standard biphasic model to fit 3 day therapy data from 44 patients [28] treated with various regimens of telaprevir [29–31], Adiwijaya et. al. [28] found the median infected cell loss rate δ=1.2 day−1 for telaprevir monotherapy, i.e., about 10 times higher than during IFN-based treatment [11]. Although Guedj et al. [29], refitting these data using a model that accounts for the increase in drug effectiveness over the successive doses obtained a lower estimate of δ=0.58 day−1, this estimate is still 4 times larger than during IFN-based therapy. Interestingly, other PIs exhibit a more modest second phase viral decline than telaprevir, with mean values on the order of 2–3 times faster than IFN (Table 1).
Table 1.
Average viral dynamic parameter estimates obtained using the standard mathematical model (Eqs. 1) for different treatments.
| Treatment | N | ε | c (day−1) | δ (day−1) | References |
|---|---|---|---|---|---|
| Telaprevir (VX-950) monotherapy* | 36 | 0.999 | 13.4 | 0.58 | [29] |
| Ciluprevir (BILN 2061) monotherapy* (2×500 mg/day) | 8 | 0.999 | 8 | 0.36 | [59] |
| TMC-435* (200 mg/day) | 6 | 0.998 | 6.25 | 2.8 | [60] |
| Mericitabine (RG7128) monotherapy (2×1500 mg/day) | 8 | 0.98 | 6 (fixed) | 0.03 | [50] |
| Daclatsavir (BMS-79002) monotherapy (10 or 100 mg/day) | 5 | 0.997 | 23.1 | 1.06 | [49] |
| Silibinin monotherapy (20 mg/kg/day dose) | 17 | 0.89 | 6 (fixed) | 0.77 | [54] |
| IFN-α2b monotherapy | 14 | 0.91 | 5.95 | 0.14 | [11] |
| IFN-α2b+RBV | 31 | 0.92 | 8.0 | 0.14 | [12] |
Protease inhibitors
What does the second phase decline during protease inhibitor therapy represent?
According to the standard model, the elevated second phase viral decline during PI therapy is due to a more rapid elimination of infected cells. However, no elevation of transaminases, such as ALT, a marker of cell necrosis, has been reported during PI therapy. Therefore the assumption that the enhanced second phase reflects an elevation in the cell death rate is unlikely. Moreover, the observation of a linear correlation between the infected cell elimination rate δ and the log transformed treatment effectiveness log10(1−ε) during telaprevir [29–31] and peg-IFN treatment [16, 32] supports the assumption that a high antiviral effectiveness not only affects the magnitude of the first phase decline but also results in an acceleration of the second phase viral decline. This feature can be explained using a model that considers both extracellular and intracellular levels of infection. Guedj and Neumann [33] suggested the existence of a critical intracellular drug effectiveness εIC, which characterized the ability for vRNA to be maintained inside an infected cell. If treatment is highly potent such that ε > εIC, vRNA is predicted to be progressively eliminated from infected cells. As a result, the second slope of viral decline is rapid and reflects the loss rate of vRNA due to the combined effect of loss of infected cells and continuously decreasing vRNA levels within the remaining infected cells and concomitant continuing decrease in viral production. Although this remains speculative, some experiments using the replicon system support the suggestion that vRNA not only initially declines by the factor (1−ε), but also continues to decline under protease inhibitor or IFN treatment [34, 35]. Hence the higher chance for attaining SVR observed in patients with a rapid viral response could in part be due to the progressive elimination of intracellular replication complexes resulting from potent antiviral treatment.
The duration of treatment needed to eradicate drug-sensitive virus
The rapid second phase decline observed with protease inhibitors suggests that the duration of therapy needed to clear drug-sensitive virus might be considerably shortened with protease inhibitors as compared to IFN based treatments [31]. Conceptually, viral eradication can be considered as achieved when the predicted total HCV RNA is less than 1 copy in the entire extracellular fluid volume (which in a 70 kg human is about 15 L), corresponding to a viral concentration of 6.7 × 10−5 HCV RNA/mL [12]. Thus for an individual with a baseline viral load above 106 HCV RNA/mL viral eradication requires over a 10 log decline from baseline. A more conservative assumption is to consider that SVR is achieved when the predicted number of infected cells is lower than 1 [13], which, typically adds another 2–4 logs of decline [29].
Since PIs exhibit a 2–4 fold more rapid second phase viral decline than IFN (Table 1), the treatment duration with a PI should be able to be roughly reduced 2–4 fold from that used with IFN. Using parameters obtained in 44 treatment naïve patients during the first 3 days of telaprevir treatment, Guedj et al. [29] estimated the empirical distribution of the time needed to achieve SVR. The model predicted that in fully compliant patients, the last virus particle could be eradicated within 7 to 8 weeks for 95% and 99% patients, respectively [29], whereas another 3 weeks would be necessary to clear the last infected cell. The latter estimate seems closer to the clinical observation where a higher rate of virologic failure after week 12 (10% vs 5%) with drug-sensitive virus was observed in patients that had received telaprevir for 8 instead of 12 weeks [36]. However, with the emergence of highly resistant virus whose eradication essentially relies on peg-IFN/RBV, treatment duration might be significantly extended [4].
Mathematical modeling of emergence of drug resistance to telaprevir treatment
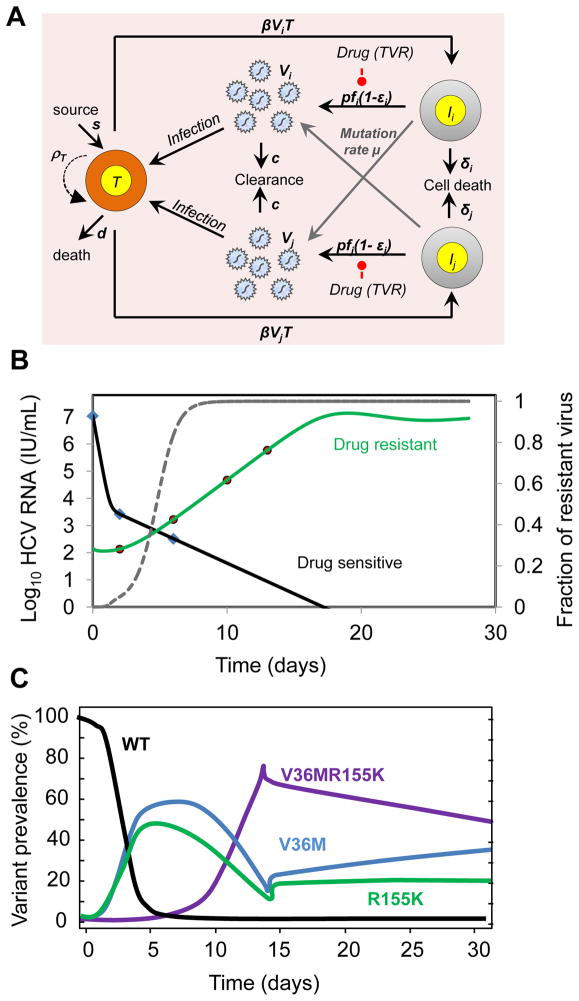
For the protease inhibitors telaprevir and boceprevir emergence of drug resistant variants and viral breakthrough hinders their applicability as monotherapy agents [5]. The existence of resistant variants is primarily due to the high replication rate of HCV and the error rate (μ) of the NS5B RNA dependent RNA polymerase (RdRp) estimated to be approximately 10−5 to 10−4 per nucleotide per replication cycle [37, 38]. Using a conservative base substitution rate of μ=10−5, the probability of 0, 1 and 2 mutations per replication cycle is estimated to be 91%, 8.7% and 0.42% respectively [39]. Given, that an infected patient typically produces 1012 virions/day [11], approximately 8.7×1010 and 4.2×109 mutants with single and double-nucleotide changes, respectively, i.e., all possible single and double mutants, are expected to be produced every day. In absence of treatment, viral variants with lower replication fitness than wild-type are not competitive and therefore may not grow to detectable levels. However, in the presence of selective pressure, the preexistence of these variants, even at low levels may explain why patients treated with telaprevir monotherapy may exhibit a large proportion of resistant virus as early as day 2 of treatment [40].
A more quantitative understanding of the kinetics of growth and decline of resistant virus during and after therapy can be obtained by expanding the standard model (Eq.1) to include viral competition. Following what had been done for HIV, Rong et al. showed that a model with two populations of virus (i.e., drug-sensitive and drug-resistant) could fit well the viral kinetics observed in the first two weeks following the initiation of telaprevir as monotherapy or in combination with peg-IFN [39]. When the kinetics of several known variants is measured, for instance by clonal sequencing, a more comprehensive picture of viral competition can be obtained by using a model that directly includes the various viral variants (Fig. 2A). Using such a model, Adiwijaya et al. [30] provided an estimate of the in vivo fitness of the principal single and double mutants observed during two weeks of telaprevir monotherapy. In a subsequent study [31], the authors applied their model to various phase 2/3 studies where telaprevir was given in combination with peg-IFN/RBV for longer times. Their model could predict the observed SVR rates for various regimens of telaprevir and peg-IFN/RBV, suggesting that modeling might be a relevant approach to design treatment strategies.
Fig. 2.
Mathematical models for the emergence of drug resistance during treatment with telaprevir. (A) Schematic representation of the multi-variant viral dynamic model of Adiwijaya et al. [30]. The model of Rong et. al. [39] is a special case in which only two strains are considered. In the multi-strain model by Adiwijaya et al. [30], target cells T, are infected by viral variant Vi, with a characterized amino-acid substitution with a certain sensitivity to telaprevir, to form a variant infected cell, Ii, which produces variants at rate pfi, a fraction of which mutate at a rate μ to generate variant j. Viral production is suppressed by factor 1−εi in the presence of telaprevir. In the Adiwijaya et al. model proliferation of target cells is not considered (ρT=0). The two-strain model of Rong et. al. [39] is a specific case of the multi-variant model, where i=1 and 2 correspond to drug-sensitive (Vs) and drug-resistant virus (VR) viral populations, and the drug-sensitive (IS) and drug-resistant (IR) infected cell population respectively. Also, the Rong et al. allows proliferation of target cells at maximum rate ρT. (B) The Rong et. al. [39] model predicts a long-term viral decline and viral rebound when target cells are allowed to expand by proliferation (primary y-axis). Even though both wild-type and resistant virus levels fall, the more profound and longer fall of the wild-type uncovers the pre-existing resistant variants and allows them to reach 5–20% of the viral population by day 2. The dashed grey line is the fraction of resistant virus VR/(VR+VS) is shown on the secondary y-axis. (C) Model prediction of evolution of wild-type (WT) and variant strains with single point mutations (V36M, R155K) and double point mutations (V36MR155K) (re-drawn from Adiwijaya et al. [30]).
An important assumption of these models [30, 39] is that the growth of resistant virus is supported by the rapid elimination of infected cells and their rapid replacement by new susceptible cells. Although both of these models provide a good fit of the data, the origin of replication space needed to support the growth of virus remains to be discovered. If the rapid decline of sensitive virus represents the intracellular elimination of virus rather than the elimination of infected cells per se, part of the replication space could come from infection with resistant virus of cells that were previously infected with wild-type virus [33].
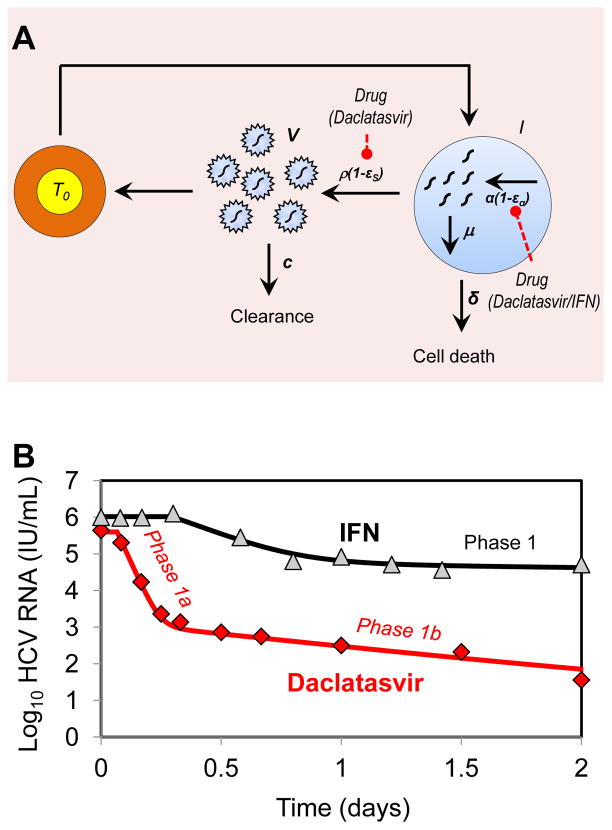
Viral kinetics seen with the NS5A inhibitor daclatasvir (BMS-790052)
Although in vitro studies suggested an essential role of NS5A for both viral replication [41–43] and assembly/release of infectious particles [44–47], the lack of a known enzymatic function has long limited the search for compounds targeting HCV NS5A. An innovative screening approach identified daclatasvir (BMS-790052) as a potent NS5A inhibitor [48]. In a single ascending dose study daclatasvir showed high antiviral effectiveness that decreased HCV RNA levels by ~3 orders of magnitude within 8–12 hours after administration [48]. Although in most patients viral load rapidly rebounded due to drug elimination, 5 patients (1 treated with 10 mg and 4 treated with 100 mg) had a sustained viral decline until day 3 post-dosing. In these patients, viral decline was biphasic, which allowed using the standard model to fit the data. Interestingly the mean antiviral effectiveness in blocking viral production/secretion, ε, was found equal to 0.997. Further, the rapid viral decline was attributed to high mean rates of elimination of free virus, c, and of infected cells, δ, estimated to 23.3 day−1 and 1.06 day−1, respectively (Table 1) [49].
In order to understand the high estimate of c and δ and how this might be related to daclatasvir’s mode of action, Guedj et al. [49] extended the standard model to incorporate essential features of intracellular viral replication that may be targeted by daclatasvir. In this model (Fig. 3A), levels of intracellular viral RNA (vRNA, denoted R) depend on the time a cell has been infected (denoted a) and are governed by the equation:
| (4) |
where α, μ and ρ are the (intracellular) rates of vRNA production, degradation and assembly/secretion as virions into the circulation, respectively. Unlike the standard model, the effects of blocking viral production (i.e., reducing α) and reducing assembly/secretion of virus (i.e., reducing ρ) can be distinguished.
Fig. 3.
Mathematical model of HCV kinetics in response to daclatasvir treatment. (A) Schematic representation of the multiscale model used by Guedj et al. [49] for fitting the early HCV viral load decline observed during treatment with daclatasvir (100 mg/day). Here, α represents the rate of viral RNA production, μ represents the rate of degradation of intracellular viral RNA and ρ is rate of export of vRNA as virions. The model assumes viral RNA production is suppressed by factor 1−eα and viral RNA packaging/secretion is suppressed by factor 1−εS in the presence of daclatasvir. (B) Representative example of viral decline observed after one dose (100 mg) of daclatasvir (diamonds) and the corresponding best fit prediction using the multiscale model (red curve) (100 mg/day) [49]. After treatment with a single dose of daclatasvir the usual 1–2 day first phase decline seen with IFN (triangles, black curve, [11]) is replaced by an early biphasic viral decline with phases 1a and 1b (diamonds, red curve).
This model provides a more precise understanding of the determinants of early viral decline (Fig. 3B). What is commonly called the “first phase” of viral decline is governed by two different processes: i) blocking of virion assembly/secretion induces an almost immediate decline of HCV RNA in the circulation with a rate c that reflects clearance of virus particles (phase 1a); ii) blocking of vRNA production induces a slower viral decline reflecting the fact that infected cells containing less vRNA will produce fewer virus particles (phase 1b). According to the model, to observe both phases a drug needs to block both assembly/secretion and vRNA production with high efficacy. If this is not the case, only one of these phases will be observed.
Within the context of this new model, the rapid early biphasic decline seen over the first two days of therapy suggests that daclatasvir has a dual mode of action, i.e., that it efficiently blocks viral assembly/secretion and vRNA production, with in vivo effectivenesses estimated to be 99.8% and 99.3%, respectively. If a drug does not effectively block virion assembly/secretion then the continuing release of virus after drug is administered will counterbalance virion clearance and lead to an underestimate of the true virion clearance rate. Therefore, the virion clearance rate estimated with daclatasvir, which corresponds to a virion half-life of 45 minutes is likely to represent an improved estimate of the HCV half-life [49]. With IFN a single early phase of viral decline is seen, which is significantly slower than that seen with daclatasvir. Guedj et al. [49] attribute this to a low effectiveness of IFN (about 50%) in blocking virion assembly/secretion.
Several aspects of this new model still need to be explored. First, the Guedj et al. model [49], assumes the rate of viral production is proportional to the amount of intracellular vRNA, however, other relationships may hold and need to be explored. Second, the model was developed to study the first 2 days of viral decline following one dose of daclatasvir. To fit longer term viral kinetics the model may need to be modified, for example, by relaxing the assumption that vRNA production occurs at a constant rate during therapy. As a consequence of the low genetic barrier of daclatasvir, information about long-term viral declines will need to be attained from analysis of data obtained from combination therapy studies. This will also be true for the study of other DAAs with a low barrier to resistance. Third, the new estimate of HCV’s half-life of 45 minutes seems incontrovertible, since measurement of the rate of viral decline during the first hours after administration of drug yields this half-life. However, whether this is a consequence of daclatasvir blocking virion assembly/secretion (revealing a previously underestimated rate of virion clearance), or whether daclatasvir has other modes of action that lead to an off-target enhancement of the viral clearance rate needs to be examined.
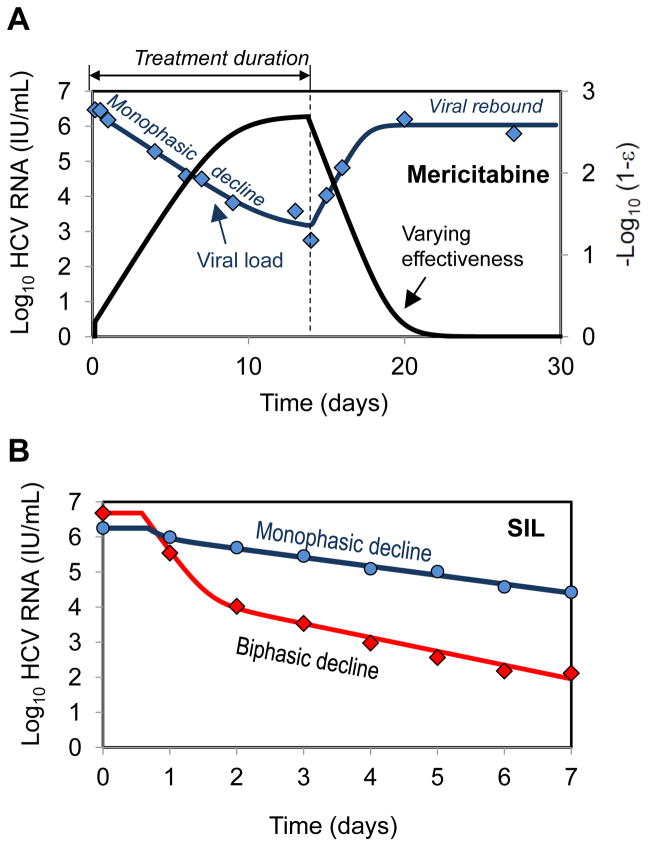
Modeling HCV kinetics with the nucleoside polymerase inhibitor mericitabine (RG7128)
As discussed previously, the rapid and efficient blocking of viral production attributed to IFN and PIs causes a rapid decline of HCV RNA during the first 1–2 days of treatment. However, when the nucleoside polymerase inhibitor mericitabine (RG7128) was given to 32 patients as monotherapy for 14 days, a much slower viral decline was observed (Fig. 4A) [50]. Further about 40% of the patients in this study did not show the typical biphasic pattern, but rather had a monophasic viral decline, regardless of dosing group (Fig. 4A). Because mericitabine, as all nucleoside polymerase inhibitors, needs to be triphosphorylated in order to become an active drug, one can ask whether the observed slow viral decline is related to the rate at which active drug is generated intracellularly.
Fig. 4.
Mathematical models of HCV kinetics in response to DAA treatment. (A) A monophasic decline of measured HCV RNA (diamond) and best-fit prediction of the viral kinetic model (blue curve, primary y-axis) [50] during a 14 day treatment with mericitabine monotherapy (1500 mg-BID). Follow up showed viral rebound as drug was eliminated from the body. In the model of Guedj et al. [50] the effectiveness of mericitabine (1500 mg-BID) changes with time during treatment (black curve, secondary y-axis). It is the slow buildup of effectiveness that is responsible for the monophasic decay. (B) A biphasic decline of measured HCV RNA and best-fit prediction of the viral kinetic model during a 7 day treatment with silibinin monotherapy (20 mg/kg/day) [54]. Patients demonstrated both biphasic (diamonds, red curve) and monophasic (circles, blue curve) viral declines.
Guedj et al. [50] showed that a slow viral decline can be reproduced in the standard viral dynamic model by allowing the drug effectiveness, ε, to gradually increase over time until some final effectiveness is reached, and that such a model agrees well with the viral kinetic data obtained during mericitabine monotherapy [50]. Using this model the final effectiveness reached by mericitabine with BID dosing was found to be high (mean 750–mg and 1500-mg: 98.0% and 99.8%, P=0.018).
The observation that the pyrimidine nucleotide PSI-7977 induces a more rapid first phase of viral decline [51] than mericitabine, even though the active species of PSI-7977, a uridine triphosphate, is the same uridine triphosphate produced by mericitabine suggests that the first phosphorylation may be the step limiting the rapid buildup of mericitabine’s antiviral effectiveness. Interestingly, ribavirin, which also needs to be triphosphorylated, when given as monotherapy, also induces a monophasic viral decline [52]. Thus, the assumption that ribavirin needs time to become fully effective might also explain why, in combination with IFN, ribavirin was found to enhance the second but not the first phase of viral decline [32].
Mode of action of silibinin using viral kinetics modeling
Because of the high cost and toxicity of the currently approved anti-HCV drugs, finding a low cost natural product that has anti-HCV activity would be highly desirable. Silymarin, an extract made from the seeds of the milk thistle plant, has been used since the time of the ancient Egyptians for the treatment of liver aliments. More recently silibinin (SIL), the major active component of silymarin has undergone clinical trials to assess its activity against HCV. In a pivotal study led by P. Ferenci, SIL monotherapy demonstrated a strong antiviral effectiveness against HCV genotype 1 or 4 in 25 patients that were administered 10, 15 or 20 mg/kg over 4 hr infusions each day for seven days [53].
To understand the MOA of SIL, Guedj et. al. [54] modeled HCV RNA kinetics measured in the Ferenci et al. study using the standard model of viral kinetics. Viral load data could be well fitted (Fig. 4B) by assuming that SIL blocks viral production only with a dose-dependent effectiveness ε=0.69 for 10+15 mg/kg/day and ε=0.91 for 20 mg/kg/day [28].
A rapid second phase between day 2 and 7 was observed in all patients (mean 0.29 logIU/mL/day), which is comparable to estimate made from the early decline seen with telaprevir (0.25 logIU/mL/day) [29]. This demonstrates that a drug can generate a rapid second phase decline even if it has a modest first phase decline. Thus, the relationship that was established [28, 29] between the amplitude of the first phase decline and the slope of the second phase decline in patients treated with telaprevir may be drug specific.
Further, 40% of patients exhibited a monophasic viral decline with a rate of viral decline over the first 2 days that led to a < 1 log decline and which was roughly similar to the rate of decline after day 2 seen in all patients (Fig. 4B). This observation contradicts the hypothesis that SIL only blocks viral production, as the absence of a profound first phase of viral decline should in this case result in a large number of new cell infections that should hamper the second phase of viral decline. Indeed assuming that SIL inhibits both viral production and viral infection yielded a statistically improved fit to the patient data. This suggested that SIL potentially acts via both these modes of action, though blocking viral production was the more pronounced effect, which is consistent with in vitro experimental data [55–58]. However, the fact that blocking viral infection has a much less pronounced effect than blocking viral production did not make it possible to estimate this effect precisely. Moreover, although SIL concentrations in serum did not increase over time, the hypothesis that the monophasic viral decline results from a gradual increase in drug effectiveness intracellularly, as predicted for mericitabine, could not be ruled out.
Conclusions
Mathematical modeling of HCV kinetics during IFN-based therapy has provided significant insights. In the new era of DAA based therapy, mathematical models continue to hold the potential to provide biologically relevant explanations of HCV kinetics under therapy. In the future, the goal for HCV treatment is to eliminate both IFN and RBV and generate SVR rates approaching 100% as well as shorten treatment duration. The most likely treatment will entail the use of DAA combination therapy. A major emphasis of such treatment regimens would be to reduce drug resistance and toxicity, as well as having a pharmacokinetic profile that would allow once a day dosing of an all oral combination. To achieve this goal would require optimizing drug combinations. Mathematical models may offer a convenient method for rationally designing therapy based on the properties of single agents. As far as we are aware this has not yet been achieved, but studies as the ones reported here on single DAAs are the first step in understanding the properties of drug combinations.
Although the conceptual framework of the biphasic model brought valuable insights into the origin of viral declines observed during treatment with high daily doses of standard IFN [11] and in combination with RBV [12], extensions of this model are needed to understand the patterns of viral decline during DAA therapy. Here we have shown that under telaprevir monotherapy and in combination with peg-IFN/RBV second phase slopes are up to four times faster than seen with IFN-based therapies. Further, the second phase slope increases with increasing drug effectiveness, as measured by the magnitude of the first phase log decline. Patients treated with the NS5A inhibitor daclatsvir (BMS-790052) show a first phase decline that is 4-fold faster than that seen with IFN-based therapies, suggesting that the HCV half-life in serum may be 45 minutes rather than the 2–3 hrs previously estimated. Also, the analysis of viral declines seen with daclatasvir suggest that the standard first phase viral decline seen over the first two days of therapy may have two parts, a phase 1a decay due to virion clearance and a phase 1b decay due to loss of intracellular viral RNA. Viral kinetics observed during a 14 day study with the nucleoside polymerase inhibitor mericitabine and in a 7 day study with the natural product silibinin are frequently monophasic rather than biphasic. Data on other DAAs is needed to provide a more complete picture of viral kinetics with the different agents that might be combined into novel therapies.
In the future, we expect to see more sophisticated and detailed models being developed that take into consideration both intracellular HCV dynamics (such as key biological steps involved during viral entry, replication, and viral assembly and release) as well as the kinetics of serum HCV RNA change. A better understanding of replication space is needed in order to explain the rapid appearance of drug resistant variants as observed during telaprevir monotherapy. Models of the future will hopefully consider the effects of cell-to-cell spread of HCV and hence include consideration of how HCV spreads spatially in the liver. Such models will be needed to accurately assess the effects of entry inhibitors, which could be another component of a DAA cocktail. Host factors are another potential target of HCV therapy. For example, the cyclophilin inhibitor alisporivir is in phase III trials. Nonetheless, no models have yet appeared in the literature. Understanding the viral kinetic aspects of host targeted therapies is another frontier that modelers hopefully will address.
Acknowledgments
This work was performed under the auspices of the U.S. Department of Energy under the contract DE-AC52-06NA25396, and supported by NIH grants RR006555, AI028433, P20-RR018754, and AI065256 (ASP). We would also like to acknowledge support of the U.S. Department of Energy through the LDRD Program for providing partial funding for AC.
References
- 1.Dahari H, Layden-Almer JE, Kallwitz E, Ribeiro RM, Cotler SJ, Layden TJ, Perelson AS. A mathematical model of hepatitis C virus dynamics in patients with high baseline viral loads or advanced liver disease. Gastroenterology. 2009;136:1402–1409. doi: 10.1053/j.gastro.2008.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Awad T, Thorlund K, Hauser G, Stimac D, Mabrouk M, Gluud C. Peginterferon alpha-2a is associated with higher sustained virological response than peginterferon alfa-2b in chronic hepatitis C: Systematic review of randomized trials. Hepatology. 2010;51:1176–1184. doi: 10.1002/hep.23504. [DOI] [PubMed] [Google Scholar]
- 3.Poordad F, McCone J, Jr, Bacon BR, Bruno S, Manns MP, Sulkowski MS, Jacobson IM, Reddy KR, Goodman ZD, Boparai N, DiNubile MJ, Sniukiene V, Brass CA, Albrecht JK, Bronowicki JP. Boceprevir for untreated chronic HCV genotype 1 infection. N Eng J Med. 2011;364:1195–1206. doi: 10.1056/NEJMoa1010494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwong AD, Kauffman RS, Hurter P, Mueller P. Discovery and development of telaprevir: an NS3-4A protease inhibitor for treating genotype 1 chronic hepatitis C virus. Nat Biotech. 2011;29:993–1003. doi: 10.1038/nbt.2020. [DOI] [PubMed] [Google Scholar]
- 5.Halfon P, Locarnini S. Hepatitis C virus resistance to protease inhibitors. J Hepatol. 2011 doi: 10.1016/j.jhep.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 6.Soriano V, Vispo E, Poveda E, Labarga P, Martin-Carbonero L, Fernandez-Montero JV, Barreiro P. Directly acting antivirals against hepatitis C virus. J Antimicrob Chemother. 2011;66:1673. doi: 10.1093/jac/dkr215. [DOI] [PubMed] [Google Scholar]
- 7.Ribeiro RM, Layden-Almer J, Powers KA, Layden TJ, Perelson AS. Dynamics of alanine aminotransferase during hepatitis C virus treatment. Hepatology. 2003;38:509–517. doi: 10.1053/jhep.2003.50344. [DOI] [PubMed] [Google Scholar]
- 8.Perelson AS, Essunger P, Cao YZ, Vesanen M, Hurley A, Saksela K, Markowitz M, Ho DD. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature. 1997;387:188–191. doi: 10.1038/387188a0. [DOI] [PubMed] [Google Scholar]
- 9.Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD. HIV-1 dynamics in vivo: Virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 10.Perelson AS. Modelling viral and immune system dynamics. Nat Rev Immunol. 2002;2:28–36. doi: 10.1038/nri700. [DOI] [PubMed] [Google Scholar]
- 11.Neumann AU, Lam NP, Dahari H, Gretch DR, Wiley TE, Layden TJ, Perelson AS. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science. 1998;282:103–107. doi: 10.1126/science.282.5386.103. [DOI] [PubMed] [Google Scholar]
- 12.Dixit NM, Layden-Almer JE, Layden TJ, Perelson AS. Modelling how ribavirin improves interferon response rates in hepatitis C virus infection. Nature. 2004;432:922–924. doi: 10.1038/nature03153. [DOI] [PubMed] [Google Scholar]
- 13.Snoeck E, Chanu P, Lavielle M, Jacqmin P, Jonsson EN, Jorga K, Goggin T, Grippo J, Jumbe NL, Frey N. A comprehensive hepatitis C viral kinetic model explaining cure. Clin Pharmacol Ther. 2010;87:706–713. doi: 10.1038/clpt.2010.35. [DOI] [PubMed] [Google Scholar]
- 14.Mihm U, Herrmann E, Sarrazin C, Zeuzem S. Review article: predicting response in hepatitis C virus therapy. Alimentary Pharmacol Ther. 2006;23:1043–1054. doi: 10.1111/j.1365-2036.2006.02863.x. [DOI] [PubMed] [Google Scholar]
- 15.Dahari H, Guedj J, Perelson A, Layden T. Hepatitis C viral kinetics in the era of direct acting antiviral agents and interleukin-28B. Curr Hepat Rep. 2011;10:214–227. doi: 10.1007/s11901-011-0101-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guedj H, Guedj J, Negro F, Lagging M, Westin J, Bochud PY, Bibert S, Neumann AU. The impact of fibrosis and steatosis on early viral kinetics in HCV genotype 1–infected patients treated with Peg-IFN-alfa-2a and ribavirin. J Viral Hepatitis. doi: 10.1111/j.1365-2893.2011.01569.x. in press. [DOI] [PubMed] [Google Scholar]
- 17.Layden-Almer JE, Ribeiro RM, Wiley T, Perelson AS, Layden TJ. Viral dynamics and response differences in HCV infected African American and white patients treated with IFN and ribavirin. Hepatology. 2003;37:1343–1350. doi: 10.1053/jhep.2003.50217. [DOI] [PubMed] [Google Scholar]
- 18.Neumann AU, Lam NP, Dahari H, Davidian M, Wiley TE, Mika BP, Perelson AS, Layden TJ. Differences in viral dynamics between genotypes 1 and 2 of hepatitis C virus. J Infect Dis. 2000;182:28–35. doi: 10.1086/315661. [DOI] [PubMed] [Google Scholar]
- 19.Bochud PY, Bibert S, Negro F, Haagmans B, Soulier A, Ferrari C, Missale G, Zeuzem S, Pawlotsky JM, Schalm S. IL28B polymorphisms predict reduction of HCV RNA from the first day of therapy in chronic hepatitis C. J Hepatol. 2011 doi: 10.1016/j.jhep.2011.01.050. [DOI] [PubMed] [Google Scholar]
- 20.Lagging M, Askarieh G, Negro F, Bibert S, Söderholm J, Westin J, Lindh M, Romero A, Missale G, Ferrari C. Response prediction in chronic hepatitis C by assessment of IP-10 and IL28B-related single nucleotide polymorphisms. PLoS ONE. 2011;6:e17232. doi: 10.1371/journal.pone.0017232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shudo E, Ribeiro RM, Perelson AS. Modeling HCV kinetics under therapy using PK and PD information. Expert Opin Drug Metab Toxicol. 2009;5:321–332. doi: 10.1517/17425250902787616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Talal AH, Ribeiro RM, Powers KA, Grace M, Cullen C, Hussain M, Markatou M, Perelson AS. Pharmacodynamics of PEG-IFN alpha differentiate HIV/HCV coinfected sustained virological responders from nonresponders. Hepatology. 2006;43:943–953. doi: 10.1002/hep.21136. [DOI] [PubMed] [Google Scholar]
- 23.Dahari H, Affonso de Araujo ES, Haagmans BL, Layden TJ, Cotler SJ, Barone AA, Neumann AU. Pharmacodynamics of PEG-IFN-a-2a in HIV/HCV co-infected patients: Implications for treatment outcomes. J Hepatol. 2010;53:460–467. doi: 10.1016/j.jhep.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guedj J, Rong L, Dahari H, Perelson AS. A perspective on modelling hepatitis C virus infection. Journal of viral hepatitis. 2010;17:825–833. doi: 10.1111/j.1365-2893.2010.01348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dahari H, Shudo E, Cotler SJ, Layden TJ, Perelson AS. Modelling hepatitis C virus kinetics: the relationship between the infected cell loss rate and the final slope of viral decay. Antiviral therapy. 2009;14:459–464. [PMC free article] [PubMed] [Google Scholar]
- 26.Dahari H, Ribeiro RM, Perelson AS. Triphasic decline of hepatitis C virus RNA during antiviral therapy. Hepatology. 2007;46:16–21. doi: 10.1002/hep.21657. [DOI] [PubMed] [Google Scholar]
- 27.Herrmann E, Lee JH, Marinos G, Modi M, Zeuzem S. Effect of ribavirin on hepatitis C viral kinetics in patients treated with pegylated interferon. Hepatology. 2003;37:1351–1358. doi: 10.1053/jhep.2003.50218. [DOI] [PubMed] [Google Scholar]
- 28.Adiwijaya BS, Hare B, Caron PR, Randle JC, Neumann AU, Reesink HW, Zeuzem S, Herrmann E. Rapid decrease of wild-type hepatitis C virus on telaprevir treatment. Antivir Ther. 2009;14:591–595. [PubMed] [Google Scholar]
- 29.Guedj J, Perelson AS. Second-phase hepatitis C virus RNA decline during telaprevir-based therapy increases with drug effectiveness: implications for treatment duration. Hepatology. 2011;53:1801–1808. doi: 10.1002/hep.24272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adiwijaya BS, Herrmann E, Hare B, Kieffer T, Lin C, Kwong AD, Garg V, Randle JCR, Sarrazin C, Zeuzem S, Caron PR. A multi-variant, viral dynamic model of genotype 1 HCV to assess the in vivo evolution of protease-inhibitor resistant variants. PLoS Comput Biol. 2010:6. doi: 10.1371/journal.pcbi.1000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adiwijaya BS, Kieffer TL, Henshaw J, Eisenhauer K, Kimko H, Alam JJ, Kauffman RS, Garg V. A viral dynamic model for treatment regimens with direct acting anitvirals for chronic hepatitis C infection. PLoS Comput Biol. 2012;8:e1002339. doi: 10.1371/journal.pcbi.1002339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feld JJ, Lutchman GA, Heller T, Hara K, Pfeiffer JK, Leff RD, Meek C, Rivera M, Ko M, Koh C. Ribavirin improves early responses to peginterferon through improved interferon signaling. Gastroenterology. 2010;139:154–162. e154. doi: 10.1053/j.gastro.2010.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guedj J, Neumann AU. Understanding hepatitis C viral dynamics with direct-acting antiviral agents due to the interplay between intracellular replication and cellular infection dynamics. J Theor Biol. 2010;267:330–340. doi: 10.1016/j.jtbi.2010.08.036. [DOI] [PubMed] [Google Scholar]
- 34.Lin K, Kwong AD, Lin C. Combination of a hepatitis C virus NS3-NS4A protease inhibitor and alpha interferon synergistically inhibits viral RNA replication and facilitates viral RNA clearance in replicon cells. Antimicrob Agents Chemother. 2004;48:4784. doi: 10.1128/AAC.48.12.4784-4792.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dahari H, Sainz B, Jr, Perelson AS, Uprichard SL. Modeling subgenomic hepatitis C virus RNA kinetics during treatment with alpha interferon. Journal of virology. 2009;83:6383–6390. doi: 10.1128/JVI.02612-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jacobson IM, McHutchison JG, Dusheiko G, Di Bisceglie AM, Reddy KR, Bzowej NH, Marcellin P, Muir AJ, Ferenci P, Flisiak R, George J, Rizzetto M, Shouval D, Sola R, Terg RA, Yoshida EM, Adda N, Bengtsson L, Sankoh AJ, Kieffer TL, George S, Kauffman RS, Zeuzem S, Team AS. Telaprevir for previously untreated chronic hepatitis C virus infection. N Eng J Med. 2011;364:2405–2416. doi: 10.1056/NEJMoa1012912. [DOI] [PubMed] [Google Scholar]
- 37.Domingo E. Biological significance of viral quasispecies. Viral Hepatitis Rev. 1996;2:247–261. [Google Scholar]
- 38.Cuevas JM, Gonzalez-Candelas F, Moya A, Sanjuan R. Effect of ribavirin on the mutation rate and spectrum of hepatitis C virus In vivo. Journal of virology. 2009;83:5760–5764. doi: 10.1128/JVI.00201-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rong L, Dahari H, Ribeiro RM, Perelson AS. Rapid emergence of protease inhibitor resistance in hepatitis C virus. Sci Transl Med. 2010:2. doi: 10.1126/scitranslmed.3000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kieffer T, Zhou Y, Zhang E, Marcial M, Byrn R, Pfeiffer T, Miller J, Tigges A, Bartels D, Kwong A, Ferenci P, Dusheiko G, Zeuzem S, Pawlotsky J-M. Evaluation of viral variants during a Phase 2 study (PROVE2) of telaprevir with peginterferon alfa-2a and ribavirin in treatment-naive HCV genotype I-infected patients. Hepatology. 2007;46:862A–862A. [Google Scholar]
- 41.Blight KJ, Kolykhalov AA, Rice CM. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290:1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- 42.Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 43.Lohmann V, Hoffmann S, Herian U, Penin F, Bartenschlager R. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. Journal of virology. 2003;77:3007–3019. doi: 10.1128/JVI.77.5.3007-3019.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Appel N, Zayas M, Miller S, Krijnse-Locker J, Schaller T, Friebe P, Kallis S, Engel U, Bartenschlager R. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 2008;4:e1000035. doi: 10.1371/journal.ppat.1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hughes M, Griffin S, Harris M. Domain III of NS5A contributes to both RNA replication and assembly of hepatitis C virus particles. J Gen Virol. 2009;90:1329–1334. doi: 10.1099/vir.0.009332-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Masaki T, Suzuki R, Murakami K, Aizaki H, Ishii K, Murayama A, Date T, Matsuura Y, Miyamura T, Wakita T, Suzuki T. Interaction of hepatitis C virus nonstructural protein 5A with core protein is critical for the production of infectious virus particles. Journal of virology. 2008;82:7964–7976. doi: 10.1128/JVI.00826-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tellinghuisen TL, Foss KL, Treadaway J. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 2008;4:e1000032. doi: 10.1371/journal.ppat.1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao M, Nettles RE, Belema M, Snyder LB, Nguyen VN, Fridell RA, Serrano-Wu MH, Langley DR, Sun J-H, O’Boyle DR, Lemm JA, Wang C, Knipe JO, Chien C, Colonno RJ, Grasela DM, Meanwell NA, Hamann LG. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature. 2010;465:96–U108. doi: 10.1038/nature08960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guedj J, Dahari H, Rong L, Nettles RE, Cotler SJ, Layden TJ, Perelson AS. Modeling shows that the NS5A inhibitor daclatasvir has two modes of action and yields a new estimate of the hepatitis C virus half-life. 2011 doi: 10.1073/pnas.1203110110. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guedj J, Dahari H, Shudo E, Smith P, Perelson AS. Hepatitis C viral kinetics with the nucleoside polymerase inhibitor mericitabine (RG7128) Hepatology. doi: 10.1002/hep.24788. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lawitz E, Rodriguez-Torres M, Denning J, Cornpropst M, Clemons D, McNair L, Berrey M, Symonds W. Once daily dual nucleotide combination of PSI-938 and PSI-7977 provides 94% HCV-RNA< LOD at day 14: first purine/pyrimidine clinical combination data (the NUCLEAR study) Journal of hepatology. 2011:S54. [Google Scholar]
- 52.Mihm U, Welker M, Teuber G, Wedemeyer H, Berg T, Breske A, Zeuzem S, Sarrazin C, Herrmann E. HCV viral kinetics during ribavirin monotherapy: results of a randomized partially double bling placebo controlled clinical trial. Journal of hepatology. 2011;54:S185–S185. [Google Scholar]
- 53.Ferenci P, Scherzer TM, Kerschner H, Rutter K, Beinhardt S, Hofer H, Schoniger-Hekele M, Holzmann H, Steindl-Munda P. Silibinin is a potent antiviral agent in patients with chronic hepatitis C not responding to pegylated interferon/ribavirin therapy. Gastroenterology. 2008;135:1561–1567. doi: 10.1053/j.gastro.2008.07.072. [DOI] [PubMed] [Google Scholar]
- 54.Guedj H, Guedj J, Negro F, Lagging M, Westin J, Bochud P-Y, Bibert S, Neumann AU. The impact of fibrosis and steatosis on early viral kinetics in HCV genotype 1 infected patients treated with PEG-IFN-alfa-2a and ribavirin. J Viral Hepatology. doi: 10.1111/j.1365-2893.2011.01569.x. (In press) [DOI] [PubMed] [Google Scholar]
- 55.Ahmed-Belkacem A, Ahnou N, Barbotte L, Wychowski C, Pallier C, Brillet R, Pohl R-T, Pawlotsky J-M. Silibinin and related compounds are direct inhibitors of hepatitis C virus RNA-dependent RNA polymerase. Gastroenterol. 2010;138:1112–1122. doi: 10.1053/j.gastro.2009.11.053. [DOI] [PubMed] [Google Scholar]
- 56.Wagoner J, Morishima C, Graf TN, Oberlies NH, Teissier E, Pecheur E-I, Tavis JE, Polyak SJ. Differential in vitro effects of intravenous versus oral formulations of silibinin on the HCV life cycle and inflammation. Plos One. 2011:6. doi: 10.1371/journal.pone.0016464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wagoner J, Negash A, Kane OJ, Martinez LE, Nahmias Y, Bourne N, Owen DM, Grove J, Brimacombe C, McKeating JA, Pecheur E-I, Graf TN, Oberlies NH, Lohmann V, Cao F, Tavis JE, Polyak SJ. Multiple effects of silymarin on the hepatitis C virus lifecycle. Hepatol. 2010;51:1912–1921. doi: 10.1002/hep.23587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Polyak SJ, Morishima C, Shuhart MC, Wang CC, Liu Y, Lee DYW. Inhibition of T-cell inflammatory cytokines, hepatocyte NF-kappa B signaling, and HCV infection by standardized silymarin. Gastroenterol. 2007;132:1925–1936. doi: 10.1053/j.gastro.2007.02.038. [DOI] [PubMed] [Google Scholar]
- 59.Herrmann E, Zeuzem S, Sarrazin C, Hinrichsen H, Benhamou Y, Manns MP, Reiser M, Reesink H, Calleja JL, Forns X, Steinmann GG, Nehmiz G. Viral kinetics in patients with chronic hepatitis C treated with the serine protease inhibitor BILN 2061. Antiviral therapy. 2006;11:371–376. [PubMed] [Google Scholar]
- 60.Reesink HW, Fanning GC, Abou Farha K, Weegink C, Van Vliet A, van ‘t Klooster G, Lenz O, Aharchi F, Marien K, Van Remoortere P, de Kock H, Broeckaert F, Meyvisch P, Van Beirendonck E, Simmen K, Verloes R. Rapid HCV-RNA decline with once daily TMC435: A phase I study in healthy volunteers and hepatitis C patients. Gastroenterol. 2010;138:913–921. doi: 10.1053/j.gastro.2009.10.033. [DOI] [PubMed] [Google Scholar]