Abstract
Magnetic imaging is a powerful tool for probing biological and physical systems. However, existing techniques either have poor spatial resolution compared to optical microscopy and are hence not generally applicable to imaging of sub-cellular structure (e.g., magnetic resonance imaging [MRI]1), or entail operating conditions that preclude application to living biological samples while providing sub-micron resolution (e.g., scanning superconducting quantum interference device [SQUID] microscopy2, electron holography3, and magnetic resonance force microscopy [MRFM]4). Here we demonstrate magnetic imaging of living cells (magnetotactic bacteria) under ambient laboratory conditions and with sub-cellular spatial resolution (400 nm), using an optically-detected magnetic field imaging array consisting of a nanoscale layer of nitrogen-vacancy (NV) colour centres implanted at the surface of a diamond chip. With the bacteria placed on the diamond surface, we optically probe the NV quantum spin states and rapidly reconstruct images of the vector components of the magnetic field created by chains of magnetic nanoparticles (magnetosomes) produced in the bacteria, and spatially correlate these magnetic field maps with optical images acquired in the same apparatus. Wide-field sCMOS acquisition allows parallel optical and magnetic imaging of multiple cells in a population with sub-micron resolution and >100 micron field-of-view. Scanning electron microscope (SEM) images of the bacteria confirm that the correlated optical and magnetic images can be used to locate and characterize the magnetosomes in each bacterium. The results provide a new capability for imaging bio-magnetic structures in living cells under ambient conditions with high spatial resolution, and will enable the mapping of a wide range of magnetic signals within cells and cellular networks5, 6.
Nitrogen vacancy (NV) colour centres in diamond enable nanoscale magnetic sensing and imaging under ambient conditions7, 8. As recently shown using a variety of methods 6, 9–10, NV centres within room-temperature diamond can be brought into few nm proximity of magnetic field sources of interest while maintaining long NV electronic spin coherence times (~ms), a large (~Bohr magneton) Zeeman shift of the NV spin states, and optical preparation and readout of the NV spin. Recent demonstrations of NV-diamond magnetometry include high-precision sensing and sub-micron imaging of externally applied and controlled magnetic fields 6, 9–11; detection of electron12 and nuclear13–15 spins; and imaging of a single electron spin within a neighbouring diamond crystal with ~10 nm resolution16. However, a key challenge for NV-diamond magnetometry is sub-micron imaging of spins and magnetic nanoparticles located outside the diamond crystal and within a target of interest. Here we present the first such demonstration of NV-diamond imaging of the magnetic field distribution produced by a living biological specimen.
Magnetotactic bacteria (MTB) are of considerable interest as a model system for the study of molecular mechanisms of biomineralization17, 18 and have often been employed for testing novel bio-magnetic imaging modalities3, 19–21. MTB form magnetosomes, membrane-bound organelles containing nanoparticles of magnetite (Fe3O4) or greigite (Fe3S4), that are arranged in chains with a net dipole moment, allowing them to orient and travel along geomagnetic field lines (magnetotaxis)17, 18. Magnetic nanoparticles (MN) produced in the magnetosomes are chemically pure, single-domain monocrystalline ferrimagnets, with species-specific morphologies and strikingly uniform size distributions17, 18. These features, combined with the ease of biofunctionalization and aqueous dispersion afforded by the magnetosome membrane22, make MN synthesis by MTB an attractive research area for various biomedical applications18, 22, including magnetic labelling, separation, and drug delivery, as well as local hyperthermic cancer treatment and MRI contrast enhancement. For the NV-diamond bio-magnetic imaging demonstrations presented here (see Fig. 1), we used Magnetospirillum magneticum AMB-1, an MTB strain that forms MN with cubo-octahedral morphology and an average diameter of ~50 nm. (Fig. 1c shows a TEM image exhibiting the characteristic morphology of AMB-1, including a chain of MN distributed over the length of the cell. Gaps between MN are common for AMB-123.)
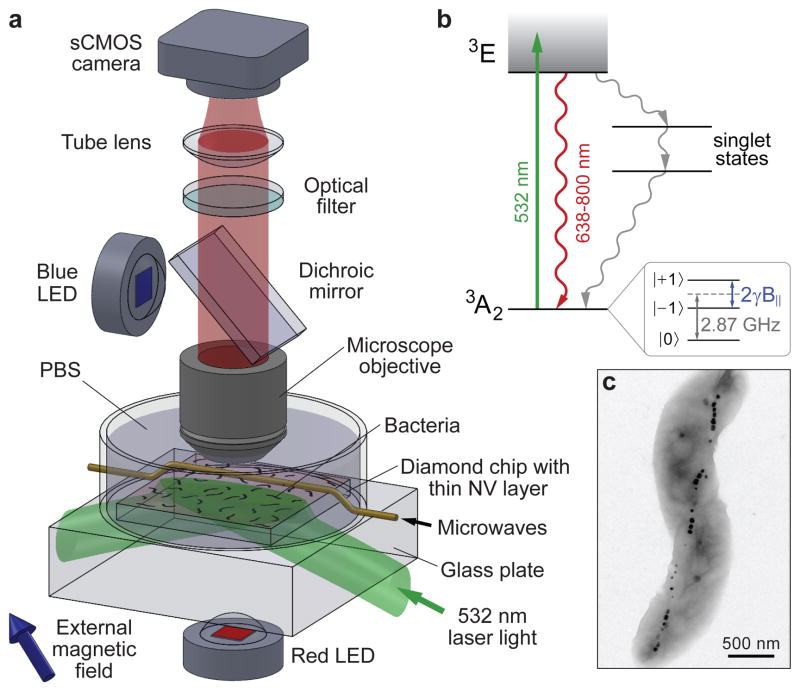
Figure 1. Wide-field magnetic imaging microscope.
a, Home-built wide-field fluorescence microscope used for combined optical and magnetic imaging. Live magnetotactic bacteria (MTB) are placed in phosphate-buffered saline (PBS) on the surface of a diamond chip implanted with nitrogen vacancy (NV) centres. Vector magnetic field images are derived from optically detected magnetic resonance (ODMR)8–10 interrogation of NV centres excited by a totally-internally-reflected 532 nm laser beam, and spatially correlated with bright field optical images. b, Energy-level diagram of NV centre; see Methods for details. c, Typical transmission electron microscope (TEM) image of a Magnetospirillum magneticum AMB-1 bacterium. Magnetite nanoparticles appear as spots of high electron density.
We acquired correlated magnetic field and optical images of populations of MTB using the NV-diamond wide-field imager depicted schematically in Fig. 1a6. The system was operated in two distinct configurations, one optimized for rapid magnetic imaging of living cells in a liquid medium, and the other for high-precision measurements of stable magnetic field patterns produced by dry bacteria on the diamond surface. In both cases, magnetic imaging was carried out using a pure diamond chip doped with a 10 nm deep surface layer of NV centres. NV electronic spin states were optically polarized and interrogated with green illumination (λ = 532 nm), coherently manipulated using resonant microwave fields, and detected via spin-state-dependent fluorescence in the red (Fig. 1b). NV electronic spin resonance (ESR) frequencies are Zeeman-shifted in the presence of a local external magnetic field (such as from MN in an MTB), allowing NV-fluorescence-based magnetometry by optically-detected magnetic resonance (ODMR)8–10. Four independent ODMR measurements enabled determination of all vector components of the magnetic field within each imaging pixel (see Methods). For imaging of live samples, the green excitation beam was directed into the diamond chip at an angle greater than the critical angle for the diamond-water interface, resulting in total internal reflection of high-intensity green light within the diamond, while low-intensity red NV fluorescence passed freely to the objective and was imaged onto the sCMOS camera (Fig. 1a). Cells at the diamond surface were thereby decoupled from high optical intensity, allowing NV magnetic imaging times up to several minutes while maintaining cellular viability. For magnetic imaging of dry bacteria, the green excitation beam could be configured in the same manner as for live/wet samples, or be allowed to pass directly through the sample, normal to the diamond surface, with comparable optical and magnetic imaging results.
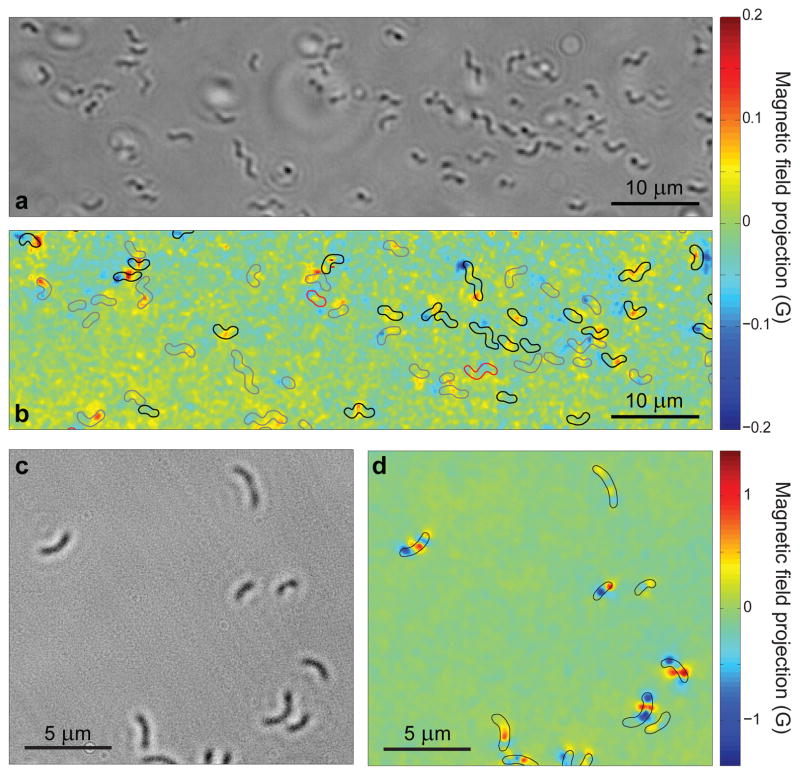
We obtained optical images of the magnetic field distributions produced by multiple cells on the diamond surface across a wide field of view (100 μm × 30 μm) and with high spatial resolution (~400 nm) using a sCMOS camera (Fig. 2). We concurrently acquired bright-field optical images using red (λ = 660 nm) LED illumination to enable correlation of cell positions and morphology with the observed magnetic field patterns. Immediately following magnetic imaging, the MTB were stained and imaged in fluorescence under blue (λ = 470 nm) LED excitation to perform a bacterial viability assay (see Methods), using a conservative viability threshold that excluded non-viable bacteria with 99% certainty (see Supplementary Methods). Under appropriate imaging conditions, the magnetic field patterns produced by the MTB could be measured within 4 minutes with minimal cellular radiation exposure, such that a significant fraction of the MTB remained alive after magnetic and bright-field imaging. For example, ~44% of the MTB in the field of view shown in Figs. 2a–b were found to be viable after magnetic and bright-field imaging, compared to 54% viability for cells directly from culture. Many of these living MTB produced magnetic field signals with large signal-to-noise ratios (SNR ~10). For high-precision characterization of the bacterial magnetic fields and comparison to electron microscope images, we also carried out a series of measurements using dried MTB samples on the diamond surface, imaged using a high-NA air objective (Fig 2c–d). Relaxing the requirement of maintaining cellular viability allowed for longer magnetic image averaging times, with concomitant reduction in photon shot-noise. Also, elimination of both the poly-L-lysine (PLL) adhesion layer (see Methods) and residual cellular Brownian motion in liquid brought the cells closer to the diamond substrate and improved their spatial stability, resulting in higher time-averaged magnetic fields at the layer of NV centres near the diamond surface. We thus expect that the dried cell technique may be the preferred approach for biological applications that do not require sustained imaging of magnetic fields produced by developing cells.
Figure 2. Wide-field optical and magnetic images of magnetotactic bacteria.
a, Bright-field optical image of MTB adhered to the diamond surface while immersed in PBS. b, Image of magnetic field projection along the [111] crystallographic axis in the diamond for the same region as a, determined from NV ODMR. Superimposed outlines indicate MTB locations determined from a. Outline colours indicate results of the live-dead assay performed after measuring the magnetic field (black for living, red for dead, and grey for indeterminate). c, Bright-field image of dried MTB on the diamond chip. d, Image of magnetic field projection along [111] for the same region, with outlines indicating MTB locations determined from c.
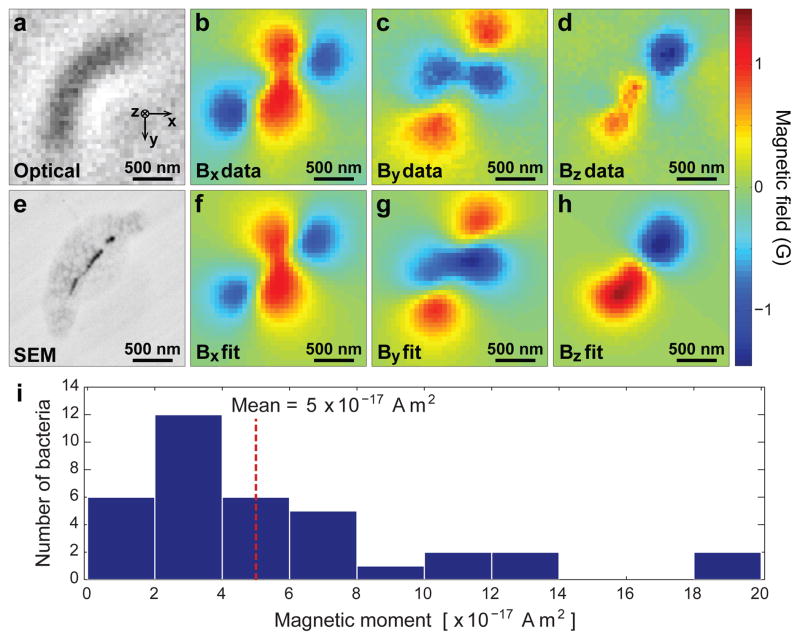
As shown in Figs. 2 – 4, the NV-diamond wide-field imager enables rapid, simultaneous measurement of biomagnetic particle distributions in many MTB, with magnetic field sensitivity and spatial resolution sufficient both to localize MN within individual MTB and to quantify the MTB magnetic moment from the magnetic field images. To verify these capabilities, we recorded scanning electron microscope (SEM) images of dried MTB in place on the surface of the diamond chip after the magnetic and bright-field imaging had been completed. Positions and relative sizes of the MN within each MTB were determined from the backscattered electron SEM images, and used to calculate the expected vector magnetic field pattern from the MTB (up to a normalization constant equivalent to the total magnetic moment of the particles - see Methods). The magnetic field patterns that we calculated (from SEM data) and measured (with the NV-diamond imager) were in excellent agreement (Figs. 3a–h), across a wide variety of MN distributions within the MTB (Fig. 4). We also determined the total magnetic moment of each MTB (e.g., [1.2 ± 0.1] × 10−16 A m2 for the MTB in Figs. 3a–h) by numerically fitting the modelled field distribution to the measured distribution, leaving the standoff distance and magnetic moment as free parameters. From such optical magnetic field measurements, we determined the distribution of magnetic moments from 36 randomly-sampled MTB on the diamond surface (Fig. 3i), with a mean value (0.5 × 10−16 A m2) that was consistent with previous estimates of the average moment per MTB for AMB-124, though our measurements showed that most AMB-1 cells had smaller moments. Note that most previously applied magnetic measurement techniques determine the average properties of large MTB populations24, 25 but are insensitive to variations among individuals within the population. In contrast, the ability of the NV-diamond wide-field magnetic imager to measure rapidly the magnetic properties of many individuals in an MTB population provides a robust tool to investigate the defects of various biomineralization mutants, making it possible to distinguish between defects that equally impact all cells in a population versus those that disproportionately disrupt magnetosome formation in a subset of cells. The AMB-1 bacteria studied here provided high signal-to-noise ratio magnetic imaging data, even though the typical magnetic moments of these bacteria are an order of magnitude smaller than many commonly studied MTB strains3, 21. This suggests that NV magnetic imaging will be applicable to a broad variety of MTB.
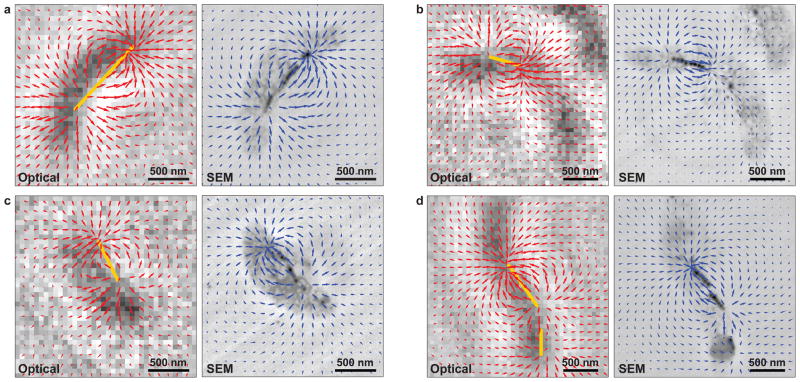
Figure 4. Localization of magnetosome chains using magnetic field measurements.
a, Vector plots of the measured (red arrows) and simulated (blue arrows) magnetic field projections in the x-y plane, for the same MTB as in Figs. 3a–h, superimposed on the optical and backscattered electron images, respectively. The estimated location of the magnetosome chain inside the MTB (yellow line), as determined from the divergence of the measured magnetic field, coincides well with the magnetosome positions found by SEM. b–d, The same information as presented in a, but for three different MTB. In panel d, two distinct magnetosome chains are identified.
Figure 3. Determining magnetic moments of individual bacteria from measured magnetic field distributions.
a, Bright-field image of an MTB. b–d, Measured magnetic field projections along the x, y, and z axes within the same field-of-view. e, Scanning electron microscope (SEM) image of the same bacterium. f–h, Simulated magnetic field projections along the x, y, and z axes, assuming that magnetic nanoparticle locations match those extracted from e. The total magnetic moment was determined from the best fit of the calculated field distribution to the measurement (see Methods for details). i, Magnetic moments of 36 randomly-sampled MTB, as determined from optical magnetic field images and modelled field distributions.
Furthermore, we were able to determine the positions of MN chains in individual MTB from the magnetic field distributions measured with the NV-diamond imager, even without the use of correlated SEM data, by noting that the MN chain endpoints occurred at locations of maximum field divergence (yellow bars in Fig. 4). Distinct groups of MN could be resolved if their separation was more than the 400 nm diffraction-limited resolution of our optical magnetometry measurements (e.g., Fig. 4d), and endpoints of single, well-isolated MN chains could be localized to within <100 nm (e.g., Fig. 4b). Using the MN chain positions and a simplified model for the MN field-source distribution, we estimated the total magnetic moment of an individual MTB from the magnetic field data alone (without correlated SEM measurements). The magnetic moments determined using this analysis procedure (e.g., 0.9 × 10−16 A m2 for the MTB in Figs. 3a–h and 4a, using the estimated MN chain position in Fig. 4a) agreed well with the values derived using the more detailed SEM-based models when the MN were arranged in long chains.
The NV-diamond wide-field imager provides powerful new capabilities that could shed light on unanswered questions regarding the development of MTB magnetic properties17, 18. Some existing methods can probe a single MTB’s internal magnetic structure3, 19, or measure the magnetic field20 or field gradient21 near a single MTB, but only NV magnetic imaging provides direct magnetic field measurements with sub-cellular resolution under ambient environmental conditions – opening the possibility for real-time imaging of MN formation and chain dynamics in single living MTB. Real-time magnetic measurements will enable observation of the transition of MN from superparamagnetic to permanent, single-magnetic-domain states as the MN grow18. The ability to locate MN chains from the magnetic images will make it possible to measure the movement of magnetosome chains across the cell-division cycle of individual MTB.
The measurements presented here are also directly applicable to studying MN formation in other organisms26, which is of interest for MRI contrast enhancement27, and has been linked with neurodegenerative disorders28 and proposed as a mechanism for magnetic navigation in higher organisms26, 29, 30. In particular, there is great current interest in identifying potential vertebrate magnetoreceptor cells30, which are believed to have a magnetic moment that is comparable to or larger than in MTB, suggesting that high-throughput NV-diamond magnetic imaging could be a valuable tool for localizing magnetic cells in a broad range of tissue samples. More generally, with further improvements in detector sensitivity and the use of spin-echo techniques for the detection of time-dependent fields6, 7, 11, NV-diamond magnetic imaging may be applied to a variety of biologically interesting systems including firing patterns in neuronal cultures5, 6, detection of free radicals generated by signalling or immune responses, and the localization of molecules tagged with specific spin labels.
Methods
NV Physics
The NV centre consists of a substitutional nitrogen atom adjacent to a vacancy in the diamond lattice (see Supplementary Fig. 1). The NV centre has a spin-triplet ground state with a 2.87 GHz zero-field splitting between the |0> and |±1> spin states (see Fig. 1b). Optical excitation of an NV centre primarily produces a spin-conserving excitation and decay process, resulting in the emission of a photon in the 640–800 nm wavelength band. However, the |±1> excited states also decay non-radiatively ~⅓ of the time to the |0> ground state via metastable singlet states. This leads to both optical polarization into the |0> ground state and state-dependent fluorescence rates that may be used to optically distinguish the |0> state from the |±1> states.
The magnetic field projection at an NV centre’s location can be measured by monitoring the fluorescence rate of the NV centre during continuous optical excitation, while varying the frequency of a continuous microwave drive8–10. When the applied microwave frequency is on resonance with either of the |0> ↔ |±1> state transitions, some of the NV state population is transferred from the |0> optically-pumped state to a mixed state, and consequently, the fluorescence rate decreases.
The NV centre’s zero-field splitting quantizes the spin states along the NV symmetry axis (indicated by a blue rod in Supplementary Fig. 1). Depending upon the relative positions of the nitrogen atom and vacancy, this symmetry axis can lie along one of four possible crystallographic directions within the diamond lattice (other possible crystallographic axes are indicated by yellow rods in Supplementary Fig. 1). In an external magnetic field, the |0> ↔ |±1> spin-flip transition frequencies shift by Δf = ±γB| (see Fig. 1c), where γ = 2.8 MHz/G is the gyromagnetic ratio of the NV electronic spin, and B| is the magnetic field projection along the NV symmetry axis.
Diamond Samples
Magnetic field sensing was carried out using high purity, single-crystal diamond chips. For imaging wet bacterial samples, we used an electronic-grade diamond (3 mm × 3 mm × 0.5 mm) grown using chemical vapour deposition (CVD) by Element Six Ltd. The diamond was implanted with 15N+ ions at 14 keV energy and annealed at 1200 °C to produce a 10 nm thick layer of NV centres 20 nm beneath the surface of the diamond (as estimated using the Stopping and Range of Ions in Matter [SRIM] software). The estimated NV surface density within the layer was 3×1011 NV/cm2. For imaging dry bacterial samples, we used a high-purity, single-crystal diamond chip (1.5 mm × 1.5 mm × 0.3 mm) manufactured by Sumitomo Electric Industries using the high-pressure, high-temperature (HPHT) method. This diamond was implanted with 15N2 + ions with 15 keV energy and then annealed at 800 °C to produce a 10 nm thick layer of NV centres 10 nm beneath the surface of the diamond (as estimated using SRIM), with an estimated surface density of 1×1012 NV/cm2.
Wide-field Magnetic Imaging Microscope
NV centres were optically excited with a 532 nm laser (Changchun New Industries Optoelectronics Tech. Co., MGL-H-532nm-800mW) switched on and off by an acousto-optic modulator (Isomet, M1133-aQ80L-1.5). A small fraction of the laser light was split off and directed onto a photodiode (Thorlabs, DET10A), and the resulting signal was sent to a servo-lock system (New Focus, LB1005 servo controller) to amplitude-stabilize the excitation beam using the same acousto-optic modulator. For imaging of bacterial samples in liquid, laser light was coupled into the diamond from below through a polished glass cube (constructed from two right-angle prisms, Thorlabs PS908), to which the diamond was affixed by optical adhesive (Norland, NBA107). The peak intensity of the totally-internally-reflected laser light at the interior surface of the diamond was measured in this case to be ~1 kW/cm2. We also note that for our angle of incidence at the diamond-water interface, θdw≈ 39°, the calculated attenuation length for the evanescent wave intensity is ddw = 58 nm. For imaging of dry samples, laser light could be configured in the same manner as for live/wet samples, or directed onto the bacteria from below, normal to the diamond surface. Dry sample data presented here was acquired using the latter method.
A 660 nm wavelength LED (Thorlabs, M660L3) was used to back-illuminate the sample for bright-field images. Excitation of fluorescence dyes used in the bacterial viability assays (see below) was carried out with a 470 nm LED (Thorlabs M470L2), directed onto the sample through the microscope objective. Optical fluorescence or transmitted red LED light was collected by the objective (Olympus, UIS2 LumFLN 60xW /1.1 NA for wet samples; Olympus, MPlan FL N 100x/0.90NA for dry samples), passed through a dichroic mirror (Thorlabs, DMLP505R for wet samples; Semrock, FF552-Di02-25×36 for dry samples) and an optical filter (Semrock, LP02-633RS-25 for NV fluorescence and transmitted red light; emission filters as described below for fluorescence from bacterial viability assay dyes), and imaged onto a digital camera (Andor, Neo sCMOS for wet samples; Starlight Xpress, SXVR-H9 CCD for dry samples). The output of a microwave synthesizer (SRS, SG384) was controlled by a switch (Mini-Circuits, ZASWA-2-50DR+), then amplified (Mini-Circuits, ZHL-16W-43-S+) and applied to the diamond with a wire. A permanent magnet was used to apply a uniform external magnetic field.
ODMR Measurements
Magnetospirillum magneticum
AMB-1 cells were grown statically in 1.5 mL microcentrifuge tubes filled with 1.5 mL of growth medium (described in 31, but with 0.1 g/L of sodium thiosulfate). For measurements of wet samples, the diamond surface was prepared by placing a drop (~5 μL) of 0.01% poly-L-lysine solution (Sigma-Aldritch, P4707, mol. wt. 70–150 kDa) on its surface, which was then allowed to dry. The bath around the diamond (contained in a chamber consisting of a cut microcentrifuge tube glued to the glass mounting surface, volume ~200 μL) was filled with 50μL of bacterial solution, and topped up with phosphate-buffered saline (PBS). For dry measurements, a drop of bacterial solution was placed directly on the diamond above the NV layer, allowed to dry, rinsed with deionized water, and dried a second time. The sample was then placed in the imager with the active diamond surface facing the objective. A uniform 37 G external magnetic field was applied along a single NV axis to distinguish it from the other three NV axes. This magnetic field strength was an order of magnitude less than the coercive field typically required to flip the magnetic orientation of MTB3, 21, and we found that the magnetization of the MTB described here remained fixed as the external field was varied.
ODMR8 – 10 spectra were measured by imaging NV fluorescence from the whole field-of-view at different microwave frequency values. The typical total fluorescence collection time was 4 minutes for both wet and dry bacterial samples. For each pixel, Lorentzian fits were applied to the ODMR spectra and the magnetic field shifts along the NV axis were extracted. This procedure was repeated with the external field applied along each of the four NV axes, which in turn allowed the vector magnetic field in the NV layer to be determined for all three Cartesian directions across the field-of-view. For magnetic fields B1 to B4, corresponding to measurements along axes 1 to 4, respectively, the fields in the Cartesian coordinates were calculated from
Bacterial Viability Assay
Immediately after magnetic field imaging of wet samples, the viability of the bacteria was determined in place on the diamond surface using a standard fluorescence-based live-dead assay (Molecular Probes, BacLight kit L7007). A mixture of the fluorescent nucleic acid stains SYTO 9 (final concentration 5 μM) and propidium iodide (final concentration 30 μM) was added to the bath, and bright-field images were immediately collected to verify that the positions of the bacteria on the diamond surface were not perturbed. The sample was then incubated in the dark for 15 minutes, and fluorescence images were collected by exciting with a LED at 470 nm (Thorlabs M470L2). Green SYTO-9 fluorescence and red propidium iodide fluorescence were collected successively using appropriate emission filters (Thorlabs FELH0500 and Thorlabs FES0550 for green; Chroma HQ640/120 for red). Custom software was used to co-register the resulting fluorescence images and perform rolling-ball background subtraction, and a peak-finding algorithm was applied to determine the positions of the bacteria. The ratio of red to green fluorescence intensity, integrated over each cell, was calculated and compared to a live/dead calibration performed previously under the same conditions (see Supplementary Information for details). MTB with a fluorescence ratio less than 0.5 were taken to be alive, while those with a fluorescence ratio greater than 1.0 were assigned as dead. Bacteria with intermediate fluorescence ratios between 0.5 and 1.0 could not be assigned to either category with high certainty based upon assay calibration measurements, and were therefore labelled as indeterminate in experimental data.
Before collecting the data displayed in Fig. 2, we carried out a series of preliminary live-dead assays, including the calibrations described in the Supplementary Methods section. These assays revealed that, even after a full hour of exposure to f≈ 2.88 GHz microwave fields at the intensities used in our ODMR measurements, the fraction of bacteria remaining alive was essentially the same as that in unperturbed samples immediately after they were taken from culture. This suggests that any bacterial fatality during experiments was the result of residual evanescent coupling of laser light through the diamond surface. These observations were consistent with direct measurements of the bath temperature when microwave power was applied, which showed only a modest increase of 1–2 °C above room temperature.
Electron Microscopy
After magnetic field measurements were completed on dried samples, imaging was performed with a field emission SEM (Zeiss Sigma). The diamond substrate and intact bacteria were carbon-coated in a thermal evaporator (Edwards Auto 306) and mounted on silicon wafers using copper tape. The bacteria were imaged without dehydration or fixation. Images of magnetosomes were obtained using backscatter mode, at 30 kX magnification and with an accelerating voltage of 8 kV. The TEM image in Fig. 1d was recorded using the procedure outlined in 32.
Fitting the magnetic field of an MTB
Magnetic field patterns of the bacteria were fit with a constrained model using SEM measurements of the relative sizes and positions of the magnetosomes, with standoff distance from the diamond and magnetic moment scaling factors left as free parameters. First, a peak-finding algorithm was applied to locate magnetosomes in the image. Magnetosome chains were determined by assigning two adjacent magnetosomes to the same chain if their separation was less than 120 nm. For each chain, the orientation of the magnetic moment in the plane of the diamond surface was determined using a linear fit to the magnetosome positions. Gaussian curves were fit to the SEM images of each magnetosome along the direction perpendicular to the axis of the chain, and the fit amplitudes were used to assign relative magnetic moment densities along the chain. Each magnetosome in a chain was assumed to act as a point dipole with the same magnetic moment direction as its chain. (This approximation was motivated by the observation of highly aligned MN dipoles in previous work, e.g. Refs. 3, 19.) In some cases, individual magnetosomes were further than 120 nm from any chains; their dipole moment was estimated to be in the same direction as that of the nearest chain.
Next, a non-linear fit routine using the Levenberg-Marquardt algorithm was performed to match simulated magnetic field images with those measured. The simulation first calculated the three components of the magnetic field on the diamond surface using the positions, directions, and relative magnetic strengths of each magnetosome. The ODMR signal for all NV axes was then calculated for each pixel, and these signals were convolved with a point-spread function (FWHM of 400 nm) to create simulated ODMR fluorescence data. As in the case of the measured data, images of Bx, By, and Bz were reconstructed on a pixel-by-pixel basis from the frequency shifts for the four NV axes extracted from Lorentzian fits. The algorithm was run independently to minimize x and y position offsets of the SEM images as well as the standoff distance from the diamond surface. Generally, Bx images were used for the fitting. Finally, the overall magnetic moment was calculated on a pixel-by-pixel basis for the best-fit geometry, and the optimal value was determined by least-squares fitting to the measured data. The best-fit magnetic moment did not depend strongly on the value of the best-fit standoff distance for typical distances of 100 nm – 200 nm, due to convolution of the NV fluorescence signal with the ~400 nm point spread function of the optical microscope. We note that this method cannot recover exact dipole orientations, particularly for isolated MN. Nevertheless, the overall magnetic moment is dominated by contributions from long chains, whose field patterns are well-described by this method.
Estimation of magnetosome chain locations and magnetic moments directly from magnetic field images
In cases where magnetic nanoparticles were organized into ordered chains that were well-approximated by finite solenoids, the chain positions and magnetic moments could be determined even without comparison to SEM data. Chain locations and orientations were estimated from the measured magnetic field divergence in the diamond plane (∂Bx/∂x + ∂By/∂y) by assigning chain endpoints to the local maxima and minima of the divergence. (The maximum precision of this estimate is given approximately by the diffraction-limited resolution of the ODMR measurement divided by the SNR of the calculated magnetic field divergence, which is approximately 40 nm.) The chain was then approximated as a continuous line of magnetic dipoles, which can be shown to have the same field as a magnetic source and sink separated by the chain length (i.e., a narrow finite solenoid). This provided a simple way to calculate Bz just below the chain. The magnetic moment could then be determined directly by spatially integrating the absolute value of Bz across the diamond surface. This integrated value is independent of standoff distance when the chain length is much larger than the standoff distance and the diameter of the field-of-view is much larger than the chain length. Moreover, it is independent of the point-spread function of the microscope objective.
Supplementary Material
Acknowledgments
We thank L. Qian for informative discussions about related experiments using a scanning SQUID microscope in K. A. Moler’s lab, J. W. Lichtman and R. Schalek for access to the SEM, S. G. Turney for access to the water-immersion objective and useful advice regarding cell immobilization, and P. R. Hemmer and H. Park for motivating technical discussions. A.K. was supported by a David and Lucille Packard Foundation Fellowship in Science and Engineering and the National Institute of Health (R01GM084122). This work was supported by the NSF and the DARPA QuASAR program.
Footnotes
Author contributions:
D.L. and R.L.W. conceived the study. K.A. developed modelling and fitting algorithms to interpret the data. D.L., K.A., D.R.G., S.J.D., and L.M.P. performed magnetic, optical, and SEM imaging experiments, and analysed data. L.R.-L. and A.K. provided bacteria cultures and TEM images. M.D.L., R.L.W., and A.Y conceived the application of the NV-diamond wide-field imager to biomagnetism. All authors discussed the results and participated in writing the manuscript.
Competing interests:
The authors declare that they have no competing financial interests.
References
- 1.Lee SC, et al. MR microscopy of micron scale structures. Magn Reson Imaging. 2009;27:828–833. doi: 10.1016/j.mri.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 2.Finkler A, et al. Self-Aligned Nanoscale SQUID on a Tip. Nano Lett. 2010;10 (3):1046–1049. doi: 10.1021/nl100009r. [DOI] [PubMed] [Google Scholar]
- 3.Dunin-Borkowski RE, et al. Magnetic Microstructure of Magnetotactic Bacteria by Electron Holography. Science. 1998;282:1868–1870. doi: 10.1126/science.282.5395.1868. [DOI] [PubMed] [Google Scholar]
- 4.Degen CL, Poggio M, Mamin HJ, Rettner CT, Rugar D. Nanoscale magnetic resonance imaging. Proc Natl Acad Sci USA. 2009;106:1313–1317. doi: 10.1073/pnas.0812068106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall LT, et al. Monitoring ion-channel function in real time through quantum decoherence. Proc Natl Acad Sci USA. 2010;107 (44):18777–18782. doi: 10.1073/pnas.1002562107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pham LM, et al. Magnetic field imaging with nitrogen-vacancy ensembles. New J Phys. 2011;13:045021. [Google Scholar]
- 7.Maze JR, et al. Nanoscale magnetic sensing with an individual electronic spin in diamond. Nature. 2008;455:644–647. doi: 10.1038/nature07279. [DOI] [PubMed] [Google Scholar]
- 8.Balasubramanian G, et al. Nanoscale imaging magnetometry with diamond spins under ambient conditions. Nature. 2008;455:648–651. doi: 10.1038/nature07278. [DOI] [PubMed] [Google Scholar]
- 9.Maletinsky P, et al. A robust scanning diamond sensor for nanoscale imaging with single nitrogen-vacancy centres. Nature Nanotech. 2012;7:320–324. doi: 10.1038/nnano.2012.50. [DOI] [PubMed] [Google Scholar]
- 10.Steinert S, et al. High sensitivity magnetic imaging using an array of spins in diamond. Rev Sci Instrum. 2010;81:043705. doi: 10.1063/1.3385689. [DOI] [PubMed] [Google Scholar]
- 11.Le Sage D, et al. Efficient photon detection from color centers in a diamond optical waveguide. Phys Rev B. 2012;85:121202(R). [Google Scholar]
- 12.Hanson R, Mendoza FM, Epstein RJ, Awschalom DD. Polarization and readout of coupled single spins in diamond. Phys Rev Lett. 2006;97 (8):087601. doi: 10.1103/PhysRevLett.97.087601. [DOI] [PubMed] [Google Scholar]
- 13.Childress L, et al. Coherent Dynamics of Coupled Electron and Nuclear Spin Qubits in Diamond. Science. 2006;314:281–285. doi: 10.1126/science.1131871. [DOI] [PubMed] [Google Scholar]
- 14.Mamin HJ, et al. Nanoscale Nuclear Magnetic Resonance with a Nitrogen-Vacancy Spin Sensor. Science. 2013;339:557–560. doi: 10.1126/science.1231540. [DOI] [PubMed] [Google Scholar]
- 15.Staudacher T, et al. Nuclear Magnetic Resonance Spectroscopy on a (5-Nanometer)3 Sample Volume. Science. 2013;339:561–563. doi: 10.1126/science.1231675. [DOI] [PubMed] [Google Scholar]
- 16.Grinolds MS, et al. Nanoscale magnetic imaging of a single electron spin under ambient conditions. Nature Physics. doi: 10.1038/NPHYS2543. [DOI] [Google Scholar]
- 17.Komeili A. Molecular mechanisms of compartmentalization and biomineralization in magnetotactic bacteria. FEMS Microbiol Rev. 2012;36:232–255. doi: 10.1111/j.1574-6976.2011.00315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faivre D, Schüler D. Magnetotactic Bacteria and Magnetosomes. Chem Rev. 2008;108:4875–4898. doi: 10.1021/cr078258w. [DOI] [PubMed] [Google Scholar]
- 19.Lam KP, et al. Characterizing magnetism of individual magnetosomes by X-ray magnetic circular dichroism in a scanning transmission X-ray microscope. Chem Geol. 2010;270:110–116. [Google Scholar]
- 20.Qian L, et al. Magnetic Characterization of Individual Magnetotactic Bacteria. presented at APS March Meeting; 2011; Dallas, Texas. 2011. (unpublished) [Google Scholar]
- 21.Proksch RB, et al. Magnetic force microscopy of the submicron magnetic assembly in a magnetotactic bacterium. Appl Phys Lett. 1995;66:2582. [Google Scholar]
- 22.Matsunaga T, Suzuki T, Tanaka M, Arakaki A. Molecular analysis of magnetotactic bacteria and development of functional bacterial magnetic particles for nano-biotechnology. Trends Biotechnol. 2007;25 (4):182–188. doi: 10.1016/j.tibtech.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 23.Draper O, et al. MamK, a bacterial actin, forms dynamic filaments in vivo that are regulated by the acidic proteins MamJ and LimJ. Mol Microbiol. 2011;82 (2):342–354. doi: 10.1111/j.1365-2958.2011.07815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krichevsky A, et al. Trapping motile magnetotactic bacteria with a magnetic recording head. J Appl Phys. 2007;101:014701. [Google Scholar]
- 25.Moskowitz BM, Frankel RB, Bazylinski DA. Rock magnetic criteria for the detection of biogenic magnetite. Earth Planet Sci Lett. 1993;120:283–300. [Google Scholar]
- 26.Pósfai M, Dunin-Borkowski RE. Magnetic Nanocrystals in Organisms. Elements. 2009;5:235–240. [Google Scholar]
- 27.Zurkiya O, Chan AWS, Hu X. MagA Is Sufficient for Producing Magnetic Nanoparticles in Mammalian Cells, Making it an MRI Reporter. Magn Reson Med. 2008;59:1225–1231. doi: 10.1002/mrm.21606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dobson J. Magnetic iron compounds in neurological disorders. Ann N Y Acad Sci. 2004;1012:183–192. doi: 10.1196/annals.1306.016. [DOI] [PubMed] [Google Scholar]
- 29.Mora CV, Davison M, Wild JM, Walker MM. Magnetoreception and its trigeminal mediation in the homing pigeon. Nature. 2004;432:508–511. doi: 10.1038/nature03077. [DOI] [PubMed] [Google Scholar]
- 30.Eder SHK, et al. Magnetic characterization of isolated candidate vertebrate magnetoreceptor cells. Proc Natl Acad Sci USA. 2012;109 (30):12022–12027. doi: 10.1073/pnas.1205653109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Komeili A, Vali H, Beveridge TJ, Newman DK. Magnetosome vesicles are present before magnetite formation, and MamA is required for their activation. Proc Natl Acad Sci USA. 2004;101 (11):3839–3844. doi: 10.1073/pnas.0400391101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murat D, Quinlan A, Vali H, Komeili A. Comprehensive genetic dissection of the magnetosome gene island reveals the step-wise assembly of a prokaryotic organelle. Proc Natl Acad Sci USA. 2010;107 (12):5593–5598. doi: 10.1073/pnas.0914439107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.