Abstract
Fish use olfaction to sense a variety of nonvolatile chemical signals in water. However, the evolutionary importance of olfaction in species-rich cichlids is controversial. Here, we determined an almost complete sequence of the vomeronasal type 2 receptor-like (OlfC: putative amino acids receptor in teleosts) gene cluster using the bacterial artificial chromosome library of the Lake Victoria cichlid, Haplochromis chilotes. In the cluster region, we found 61 intact OlfC genes, which is the largest number of OlfC genes identified among the seven teleost fish investigated to date. Data mining of the Oreochromis niloticus (Nile tilapia) draft genome sequence, and genomic Southern hybridization analysis revealed that the ancestor of all modern cichlids had already developed almost the same OlfC gene repertoire, which was accomplished by lineage-specific gene expansions. Furthermore, comparison of receptor sequences showed that recently duplicated paralogs are more variable than orthologs of different species at particular sites that were predicted to be involved in amino acid selectivity. Thus, the increase of paralogs through gene expansion may lead to functional diversification in detection of amino acids. This study implies that cichlids have developed a potent capacity to detect a variety of amino acids (and their derivatives) through OlfCs, which may have contributed to the extraordinary diversity of their feeding habitats.
Keywords: gene duplication, olfaction, dN/dS
Introduction
Most fish rely on olfaction for social behaviors such as reproduction, kin recognition, and aggression as well as for feeding and migration (e.g., Laberge and Hara 2001). Because chemical signaling is the primary means by which fish communicate, many fish have developed a highly sophisticated olfactory system. However, the importance of olfaction in species-rich African cichlids remains to be elucidated. Given that highly advanced social behaviors are one aspect of the remarkable species diversity in cichlids, it is of great interest to know whether olfaction has contributed to such behaviors.
Each of the Great Eastern African Lakes—Tanganyika, Malawi, and Victoria—harbors several hundred endemic cichlid species that are ecologically and morphologically highly diverse (Fryer and Iies 1972; Turner et al. 2001; Kocher 2004; Turner 2007). Phylogenetic and geographical studies suggest that the cichlids of each lake have arisen independently from a small number of ancestral species followed by extensive diversification in a very short period (Kocher 2004). Therefore, biologists consider cichlids to be excellent model fish to understand the genetic mechanism of rapid radiation. Although vision has been traditionally thought to be the primary sense in cichlids (Seehausen and van Alphen 1998; Terai et al. 2002, 2006; Maan et al. 2004; Seehausen et al. 2008), several recent studies have proposed that olfaction may also substantially contribute to the social behaviors of haplochromines (Crapon de Caprona 1980; Plenderleith et al. 2005; Cole and Stacey 2006; Verzijden and ten Cate 2007) and tilapias (Barata et al. 2008; Miranda et al. 2005).
Vertebrates have four types of evolutionarily distinct multigene families of G-protein-coupled receptors (GPCRs) to detect chemicals in their environment; these receptors include OR, V1R, V2R, and TAAR (Shi and Zhang 2005; Nei et al. 2008; Grus and Zhang 2009). In mammals, vomeronasal type 2 receptors (V2Rs) are specifically expressed in the vomeronasal organ (Matsunami and Buck 1997) and are believed to encode pheromone receptors. Furthermore, V2Rs detect a peptide pheromone secreted from the lachrymal gland (Kimoto et al. 2005) and small peptides for the major histocompatibility complex (Loconto et al. 2003; Leinders-Zufall et al. 2004), and this system may constitute a fundamental mechanism for defining species individuality. These studies suggest that mammalian V2Rs are involved in social communication. In contrast, fish do not have a vomeronasal system. Accordingly, the receptors corresponding to V2Rs have recently been proposed to be named as OlfCs (olfactory receptors classified as type C GPCRs) (Alioto and Ngai 2006; Johnstone et al. 2009). The OlfCs are expressed in the olfactory epithelium of the nasal cavity. Several independent studies have shown that fish OlfCs instead detect amino acids and elicit feeding behavior. For example, one OlfC is expressed in microvillous sensory neurons and respond to amino acids but not to bile acids or sex pheromones (Speca et al. 1999); microvillous sensory neurons innervate the lateral chain glomeruli (Sato et al. 2005), which are activated by amino acids (Friedrich and Korsching 1997). Finally, genetic blockage of neural transmission in the olfactory sensory neurons innervating the lateral chain glomeruli completely abolishes the attractive response to a mixture of amino acids (Koide et al. 2009). Thus, although it is premature to rule out the possibility that fish OlfCs are involved in social interactions, most OlfCs are expected to detect amino acids and elicit feeding behavior.
The partial sequences of fish OlfC genes were first characterized in goldfish (Cao et al. 1998) and fugu (Naito et al. 1998). Fish OlfC genes were then extensively characterized from draft genome sequences of several fish (Alioto and Ngai 2006; Hashiguchi and Nishida 2006, 2009; Hashiguchi et al. 2008). In particular, Hashiguchi and Nishida (2006) characterized and compared the OlfC gene cluster regions among four fish species and found that lineage-specific gene gain and loss have contributed to highly variable gene repertoires. Furthermore, Johnstone et al. (2009) determined almost complete sequences of the OlfC gene clusters in the “non-model fish,” Atlantic salmon. The number of OlfC genes thus far identified varies from 11 in pufferfish to 54 in zebrafish (Alioto and Ngai 2006; Hashiguchi et al. 2008; Johnstone et al. 2009).
Given that the higher ability in the amino acid discrimination may provide the basis for the trophic diversity of the organisms, we focused on the OlfC receptor gene of the east African cichlids, which are often regarded as a textbook example of explosive trophic diversification. To investigate the OlfC gene cluster of cichlids, we determined an almost complete sequence of the OlfC gene cluster of a Lake Victoria cichlid, Haplochromis chilotes, by screening the H. chilotes bacterial artificial chromosome (BAC) library (Watanabe et al. 2003) and conducting shotgun sequencing. Investigation of the resultant high-quality sequence revealed that cichlids possess the largest number of intact OlfC genes (61 genes) among fishes and that this was achieved by lineage-specific gene expansion. Furthermore, the data mining of the Oreochromis niloticus (Nile tilapia) and the genomic Southern hybridization analyses revealed that that the common ancestor of all modern cichlids had already developed almost the same OlfC gene repertoire. Thus, the large number of OlfC genes in cichlids arose by gene duplication in the early stage of their evolution. In general, vision-oriented animals exhibit a reduced number of intact olfactory receptor genes because of the relative unimportance of olfaction in these animals (Nei et al. 2008). Conversely, having a larger number of intact genes may indicate the relative importance of olfaction among animals although there is an apparent exceptions in dogs (Young and Trask 2007; Young et al. 2010). Our detailed investigation also indicated that recently duplicated OlfC paralogs of one species are more variable than orthologs of different species at particular sites that were predicted to be involved in amino acid selectivity (Luu et al. 2004; Alioto and Ngai 2006). Thus, an increase in the number of paralogs through gene expansion may lead to functional diversification of amino acid detection. These two lines of data imply that cichlids have developed a keen ability to discriminate a variety of amino acids, which we speculate contributed to the observed extraordinary diversification of their feeding behaviors.
Materials and Methods
Fish and DNA Samples
The fish species used in this study are listed in supplementary table S1, Supplementary Material online. The cichlids were caught in the wild or purchased from a commercial source. Parts of fins or tissues from fresh-caught fishes were fixed in 100% ethanol and stored at 4 °C. DNA was extracted using the DNeasy Tissue kit (QIAGEN).
Polymerase Chain Reaction and Small-Scale Sequencing
The polymerase chain reaction (PCR) protocol consisted of 30 cycles with denaturation at 94 °C for 30 s, annealing at 55 °C for 45 s, and extension at 72 °C for 1 min. The PCR mixture contained 2.5 U Ex Taq polymerase (Takara), 1× Ex Taq buffer, 0.4 mM dNTPs, 0.1 μM of each primer, and 1 μl of template genomic DNA in a final volume of 50 μl. PCR products were confirmed by electrophoresis in a 3.0% agarose gel (Takara) and staining with ethidium bromide. The PCR products were then purified via precipitation with isopropanol. Purified PCR products were used for direct sequencing with 25 cycles of denaturation at 96 °C for 30 s, annealing at 50 °C for 15 s, and extension at 60 °C for 1 min. Reactions contained 1 μl BigDye ver. 3.1 terminator premix (Applied Biosystems), 1× sequencing buffer (Applied Biosystems), 1 μM sequence primer, and 2 μl purified PCR product in a final volume of 5 μl. Sequences were determined using an automated sequencer (Applied Biosystems, model 3100).
BAC Library Screening and Shotgun Sequencing
According to Hashiguchi and Nishida (2006) and Johnstone et al. (2009), the OlfC genes are clustered in one chromosomal region except for in zebrafish and Atlantic salmon genomes. Therefore, the OlfC genes of cichlid, which is phylogenetically close to medaka, were expected to cluster in one chromosomal region flanked by two genes encoding neprilysin and phospholipase C-η1. Thus, we sought to perform BAC walking to obtain the entire cluster region in cichlids. We first amplified a partial sequence of the transmembrane (TM) domain of the OlfC gene subfamily 4 using the genomic DNA of H. chilotes as a template. The PCR primer sequences were based on the corresponding regions of medaka and fugu. DNA fragments were then cloned into the pGEM-T Vector (Promega) and sequenced. On the basis of these sequences, we redesigned PCR primers that could specifically amplify cichlid OlfC subfamily 4 genes. We used this PCR primer set to screen the BAC library of H. chilotes and obtained clone 32K15. BAC clone DNA was extracted using the large construct kit (QIAGEN) and used for subsequent direct sequencing. The sequences of the SP6 and T7 ends of BACs were determined using M13 primers M4 and RV (Takara). The sequences of these BAC ends were used to further design primers to screen BAC clones overlapping 32K15 to extend the OlfC gene–containing genomic region (fig. 1). After several steps of BAC end walking, we checked for the presence of either neprilysin or phospholipase C-η1, which can be used as a landmark at the 5′- or 3′-end, respectively, of the OlfC cluster. The primers used for screening are summarized in the supplementary table S2, Supplementary Material online. The nucleotide sequences of the BAC clones were determined by the shotgun method using an automated sequencer (Applied Biosystems, model 3700). Sequences determined in this study are available at GenBank (accession numbers AB780549–AB780556).
Fig. 1.—
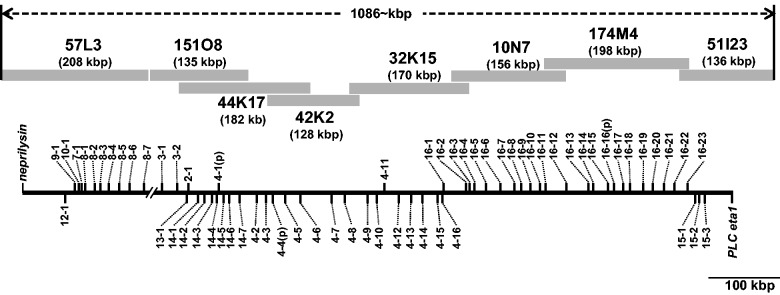
The OlfC gene cluster of the Lake Victoria cichlid Haplochromis chilotes. The cluster region spans more than 1 Mb covered by eight BAC clones, in which 61 intact OlfC genes are arranged. OlfC genes were categorized into 16 subfamilies according to Hashiguchi and Nishida (2006). Gray bars indicate the BACs that cover the region. The ID and the length of each BAC clone are indicated above each gray bar. The genes neprilysin and phospholipase C-η1 were used as landmarks at the 5′- and 3′-ends, respectively, of the cluster region. Genes in the sense orientation are shown above the black line, and genes in the antisense orientation are shown below the black line.
Data Mining
The identification of putative OlfC genes from the assembled sequence spanning more than 1 Mbp was performed according to Hashiguchi and Nishida (2006). Briefly, a TBLASTN search was conducted against the assembled sequence using the TM domains of all 16 subfamilies, which were classified in previous studies, as queries under the expect threshold E < 1e−10. Next, each TBLASTN hit region was extended in the 5′ and 3′ directions to perform a detailed prediction of OlfC coding sequences. Then, OlfC coding sequences were estimated for each sequence using the WISE2 program (Birney et al. 2004). The analysis identified 61 intact OlfC genes and three partial genes or pseudogenes. The deduced cDNA and amino acid sequences of the intact OlfC genes are provided in supplementary figures S1 and S2, Supplementary Material online, respectively. The data mining results for cichlid OlfC gene clusters, namely the position and orientation of each OlfC gene, are summarized in supplementary table S3, Supplementary Material online. We also explored the OlfC genes in the draft genome of O. niloticus (Nile tilapia), which was available in the Ensemble gene browser (http://www.ensembl.org/Oreochromis_niloticus/Info/Index/). The result of data mining is summarized in the supplementary figures S3 and S4 and table S4, Supplementary Material online.
Phylogenetic Analysis
The sequences were edited by GENETYX-Windows version 5. ClustalX (Larkin et al. 2007) was used to align deduced amino acid sequences of OlfC genes from cichlids with those from zebrafish, Atlantic salmon, three-spined stickleback, green spotted pufferfish, fugu, and medaka. For nucleotide sequence comparison, CodonAlign 2.0 (http://www.sinauer.com/hall/2e/) was used to introduce gaps into OlfC coding sequences at positions corresponding to the gaps in the aligned protein sequences. To construct the OlfC tree for teleost fishes based on amino acid sequences, other family C GPCRs—CaSR and V2R2—were used as outgroups. MEGA 5.0 software (Tamura et al. 2011) was used for neighbor-joining tree construction and genetic distance calculation for aligned OlfC coding sequences. The PHYML program was used to construct the maximum-likelihood tree. For amino acid sequence comparison, we used WebLogo (Crooks et al. 2004) to visualize the functional residues of OlfC receptors. The site-specific dN/dS ratio within the coding sequence of the OlfC genes was performed with the Single Likelihood Ancestor Counting (SLAC) package, which implements the Suzuki–Gojobori method (Pond and Frost 2005).
Genomic Southern Hybridization
Genomic DNA (10 µg) of four different cichlid species was digested with EcoRI, HindIII, or PstI followed by 0.8% agarose gel electrophoresis. The DNAs were then transferred to GeneScreen Plus Charged Nylon Membrane (PerkinElmer USA) using the standard protocol (Sambrook et al. 1989). An approximately 600-bp fragment of the TM domain of OlfC subfamilies 4, 8, 14, and 16 was PCR amplified for each of the four different cichlids to prepare probes (supplementary table S2, Supplementary Material online). The PCR products were then cloned and sequenced to choose appropriate sequences for probes. The clones chosen for Southern hybridization were labeled with digoxigenin (DIG) using the PCR DIG probe synthesis kit (Roche). Hybridization was carried out in a solution containing 25% formamide, 7% SDS, 5× SSC, 0.1% N-lauroylsarcosine, 50 mM phosphate buffer (pH 7.0), and 2% blocking reagent (Roche) at 42 °C overnight, followed by washing with 0.1× SSC containing 0.1% SDS at 65 °C. Hybridized probes were detected using alkaline phosphatase-conjugated anti-DIG Fab fragment and CDP-Star (Roche), and bands were visualized using Kodak Image Station 2000R (Kodak).
Results and Discussion
Characterization of the OlfC Gene Cluster Region in H. chilotes
We characterized the entire OlfC gene cluster region of the Lake Victoria cichlid, H. chilotes, based on the BAC end-walking strategy (fig. 1). The primers used for BAC walking are summarized in the supplementary table S2, Supplementary Material online. Given that the OlfC gene clusters are flanked by neprilysin and phospholipase C-η1 in medaka, fugu, pufferfish, and stickleback (Hashiguchi and Nishida 2006), they can be used as landmarks at the 5′- and 3′-ends, respectively, of the clusters. Most of the BAC clones overlapped their 5′- and 3′-ends with 100% nucleotide sequence identity, which enabled us to connect BAC sequences. However, we could not find the BAC clone that connects 151O8 and 57L3. Because the 5′-end of BAC 151O8 was highly repetitive with the REX1 long interspersed element, it was difficult to design primers for BAC end walking. Accordingly, we used the neprilysin sequence to screen the OlfC gene cluster region from the opposite side and obtained the BAC clone 57L3, the 3′-tail of which was also highly repetitive with the REX1 sequence. Because REX1 sequences contain several recognition sites for Sac I, which was used to construct the BAC library, this genomic region could have been eliminated as short fragments during library construction. Accordingly, we treated this unconnected region as a gap (tentatively, we inserted a stretch of 300Ns). The OlfC gene cluster region was ultimately ascertained to span more than 1,000 kb that was covered by eight BAC clones (accession numbers AB780549–AB780556).
Next, we performed a TBLASTN search and GeneWise analyses to annotate the OlfC genes using the cDNA sequences of other teleost fishes. The arrangement and orientation of each OlfC gene in H. chilotes are indicated in figure 1 and the supplementary table S3, Supplementary Material online. Notably, the arrangement of OlfC genes of H. chilotes is similar to that of medaka, pufferfish (Hashiguchi and Nishida 2006), and stickleback (Hashiguchi and Nishida 2009), indicating that the data we obtained were reliable. Especially, the gene order around the gap-connected region, at which subfamily 3 follows subfamily 8 (fig. 1), is consistent with that of medaka. Furthermore, our analysis revealed that the gene arrangement of H. chilotes is consistent with that of the draft genome of O. niloticus (Nile tilapia, supplementary table S4, Supplementary Material online). Using PCR and sequencing, we further examined the presence of additional OlfC genes in the H. chilotes genome outside this cluster region, but none were found. These lines of evidence indicated that we identified almost all the OlfC genes of H. chilotes in a unique genomic region flanked by two of the landmark genes neprilysin and phospholipase C-η1.
Comparative Analysis of OlfC Genes among Teleost Fishes
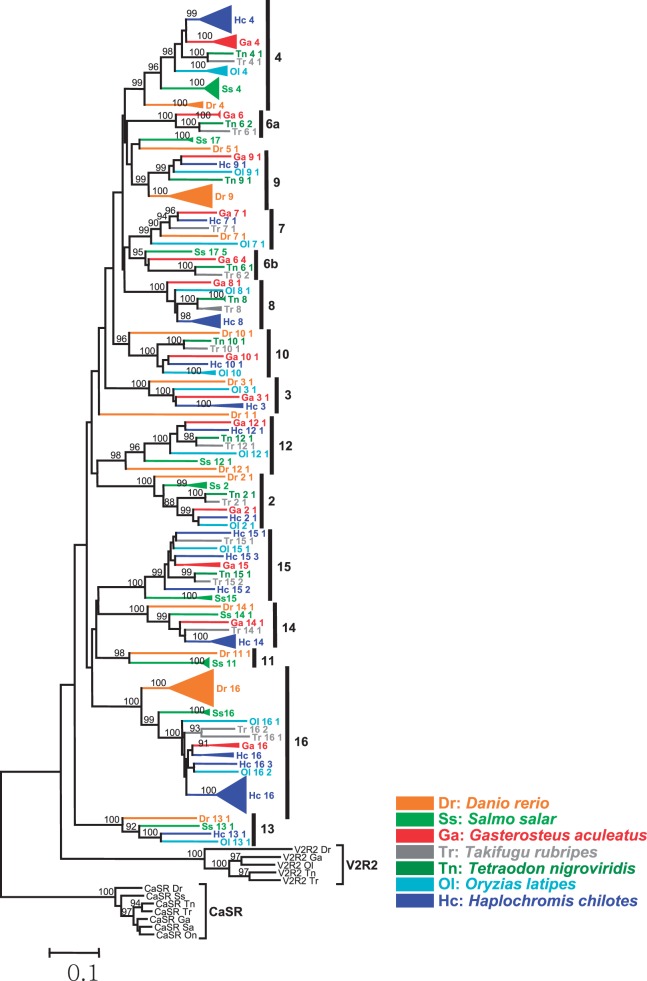
A neighbor-joining tree for teleost OlfC genes was constructed (fig. 2). The amino acid sequences of the putatively intact OlfC genes were included in this analysis. It is mostly consistent with previous studies (Hashiguchi and Nishida 2006; Johnstone et al. 2009) that the teleost OlfC genes were categorized into 16 or 17 subfamilies, which formed monophyletic groups at near-maximum bootstrap probabilities. However, subfamily 6 was divided into two groups, namely 6a and 6b (fig. 2). Johnstone et al. (2009) also indicated that subfamily 6 was not monophyletic in the tree. Indeed, Hashiguchi and Nishida (2006) indicated that the supporting bootstrap value for the monophyly of subfamily 6 was just 51. The inclusion of the additional data for Atlantic salmon and cichlids likely resulted in the splitting of subfamily 6. In the phylogenetic tree, gene expansions in the H. chilotes lineage were detected for subfamilies 4, 8, 14, and 16, which are indicated by blue triangles (fig. 2). The annotation of the OlfC genes of H. chilotes and the phylogenetic analysis were used to count the number of intact OlfC genes in this species. In the OlfC gene cluster of H. chilotes, 12 of 16 subfamilies were found. Members belonging to subfamilies 1, 5, 6, and 11 were not found in the H. chilotes OlfC cluster region. Although it is still possible that the OlfC genes belonging to these subfamilies are located in the other chromosomal region, the apparent absence of them in the draft genome of O. niloticus (Nile tilapia, see latter section) imply that they likely to be missing in the genome of H. chilotes. Furthermore, according to Hashiguchi and Nishida (2006) and Johnstone et al. (2009), Neoteleostei lineages, relatively recently diverged group of teleost, appear to have one unique OlfC cluster region. The maximum likelihood tree of teleost OlfC genes showed essentially the same topology as that of neighbor-joining tree (supplementary fig. S5A and B, Supplementary Material online).
Fig. 2.—
Neighbor-joining tree for OlfC genes of seven teleost fishes. The OlfC genes of zebrafish (Danio rerio [Dr]), Atlantic salmon (Salmo salar [Ss]), three-spined stickleback (Gasterosteus aculeatus [Ga]), fugu (Takihfugu rubripes [Tr]), green-spotted pufferfish (Tetraodon nigroviridis [Tn]), medaka (Oryzias latipes [Ol]), and Lake Victoria cichlid (Haplochromis chilotes [Hc]) are indicated by different colors. Triangles indicate the lineage-specific gene expansions. The vertical height of these triangles is proportional to the number of genes. All the nodes with bootstrap values higher than 85 in the neighbor-joining (NJ) tree were also supported by maximum likelihood (ML) tree.
Figure 3 compares the number and subfamily makeup of OlfC genes among seven teleost fishes investigated so far. Interestingly, the total number of intact OlfC genes was highest in H. chilotes—almost 6-fold more than in pufferfish. The extensive lineage-specific gene expansions in subfamilies 4 and 16 appear to have resulted in the large number of OlfC genes in H. chilotes. This large number was quite unexpected because traditionally cichlids have been thought to be guided primarily by vision (Fryer and Iies 1972) with respect to behaviors including predator avoidance, feeding, and social interactions, as illustrated by the recent demonstration of sensory-driven speciation (Seehausen et al. 2008). In general, vision-oriented animals are expected to be less dependent on olfaction, which leads to a decrease in the number of intact olfactory genes owing to pseudogenization. Conversely, the existence of greater numbers of olfactory genes in a particular genome may indicate the relative importance of olfaction in the species. Accordingly, given that most fish OlfC receptors are expected to detect amino acids and elicit feeding behaviors (Sato et al. 2005; Koide et al. 2009), the high OlfC gene copy number in H. chilotes among teleosts implies the importance of OlfC-mediated olfaction to feeding behavior in this species.
Fig. 3.—
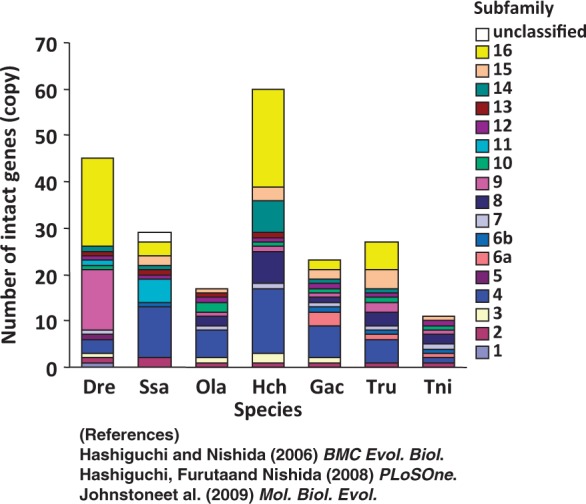
Comparison of OlfC gene copy number. The number of intact OlfC genes of seven teleost fishes is shown by a stacked-bar graph. Each of the subfamilies is indicated by a different color. Note that Haplochromis chilotes possesses the largest copy number among teleost fishes investigated so far. The copy number of each OlfC gene in species other than H. chilotes is according to Hashiguchi and Nishida (2006), Hashiguchi et al. (2008), and Johnstone et al. (2009). Abbreviation of species names is as follows: Dre (zebrafish), Ssa (Atlantic salmon), Gac (three-spined stickleback), Tru (fugu), Tni (green spotted pufferfish), Ola (medaka), and Hch (Lake Victoria cichlid H. chilotes).
The Timing of Lineage-Specific Gene Expansions in Cichlid Evolution
We were then prompted to estimate the number of OlfC genes in species other than H. chilotes to clarify the timing of lineage-specific expansions during the evolution of the family Cichlidae. First, we explored the OlfC genes in the draft genome of the O. niloticus (Nile tilapia), which was available in the Ensembl genome browser. Several phylogenetic analyses have suggested that the O. niloticus is a basal lineage of East African cichlids that diverged from the other cichlids approximately 10 Ma (Kocher 2004). Accordingly, investigation of this genome should elucidate the timing of lineage-specific OlfC gene expansions. Because short reads likely will result in sequence gaps in de novo assembly (see supplementary table S4, Supplementary Material online), it will not be possible to establish the OlfC cluster completely. Accordingly, it was difficult to perform detailed phylogenetic analyses, including the comparison of orthologous genes and tree construction. Although the quality of the sequences of this region is not high, we could estimate the number of putatively intact genes. In the genome of O. niloticus (Nile tilapia), the OlfC cluster region flanked by the two landmark genes is covered by scaffolds 162, 340, and 170 (see supplementary table S4, Supplementary Material online). Our assessment revealed a total of 58 putatively intact OlfC genes, which is similar to the number in H. chilotes, but 38 of them were truncated probably because of the assembling gaps. We found that the OlfC gene expansions occurred in same subfamilies in O. niloticus as in H. chilotes (subfamilies 4, 8, 14, and 16). Accordingly, the repertoire of OlfC genes of the common ancestor of East African cichlids was similar to that of extant Lake Victoria cichlids.
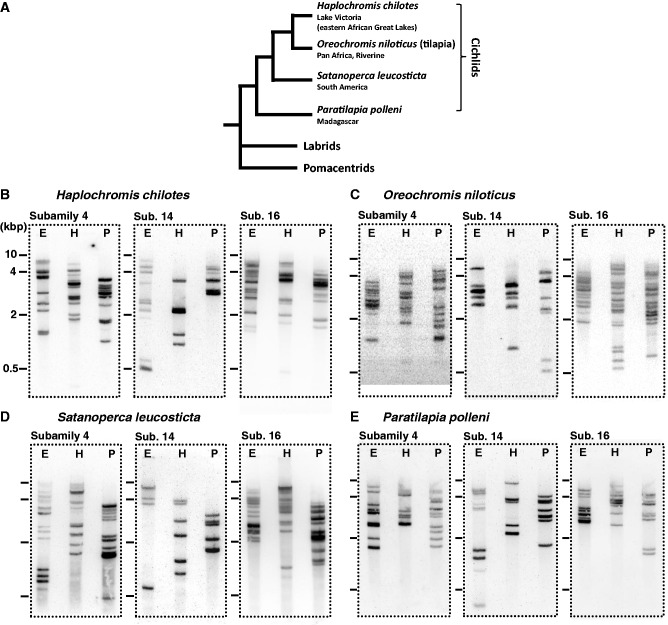
To further elucidate the timing of OlfC gene expansions, we examined additional representative cichlids by genomic Southern hybridization to include a broad range of taxa from South America (Satanoperca leucosticta) and Madagascar (Paratilapia polleni) (supplementary table S1, Supplementary Material online). The phylogenetic relationship of these cichlids is shown in figure 4A. The genomes of H. chilotes and O. niloticus were used as standards, for which the OlfC gene number was estimated from the genome data. Previous studies indicated that the African and South American cichlids form sister groups, and the Malagasy cichlids are the most basal group in the family Cichlidae (Streelman et al. 1998; Azuma et al. 2008).
Fig. 4.—
Genomic Southern hybridization results for cichlids. (A) The phylogenetic relationships of the cichlids used in the genomic Southern hybridization analysis. The tree was drawn according to Streelman et al. (1998). (B–E) Each OlfC gene copy number was estimated for three subfamilies in which gene expansion occurred in H. chilotes. Four cichlid species, which are distantly related in the phylogenetic tree, were chosen for this analysis. Note that the number of hybridizing bands for the most basal group of cichlids, Paratilapia polleni, appears to be almost the same as that for Lake Victoria cichlids. The E, H, and P above the lanes indicate the restriction enzymes used for the digestion of genomic DNA (EcoRI, HindIII, and PstI, respectively).
Our preliminary PCR and sequencing analyses suggested that only one or two genes exist in each of subfamilies 2, 3, 7, 9, 10, 12, and 15 in the three cichlid species, indicating that gene expansion did not occur in these subfamilies (data not shown). For further analysis, we focused on subfamilies 4, 8, 14, and 16, in which lineage-specific gene expansions were detected in H. chilotes and O. niloticus. In particular, we examined whether lineage-specific gene expansions have occurred in these subfamilies in three species of cichlids. Figure 4 shows the genomic Southern hybridization analyses using the PCR fragments of the TM region of each subfamily as probes (the primers used for the PCRs were summarized in the supplementary table S2, Supplementary Material online). The number of hybridizing bands detected in subfamilies 4, 14, and 16 of H. chilotes was consistent with the results of the BAC sequencing, demonstrating the reliability of this genomic Southern hybridization experiment. However, an unexpectedly large number of hybridizing bands was observed in subfamily 8 (supplementary fig. S6, Supplementary Material online). This result may have been caused by cross-hybridization of the probe with members of the other OlfC subfamilies such as subfamilies 3, 7, 9, and 10, which are phylogenetically close to subfamily 8. Because the nucleotide sequence similarity among members of subfamily 8 was relatively low compared with that of subfamilies 4, 14, and 16, it was quite difficult to design probes that would specifically detect subfamily 8. Therefore, we used the genomic Southern hybridization data to estimate the gene numbers only of subfamilies 4, 14, and 16. Overall, the number of hybridizing bands in O. niloticus, S. leucosticta, and P. polleni appears similar to that of H. chilotes (fig. 4), except for the slightly larger number of bands in subfamily 4 of S. leucosticte and the slightly smaller number of bands in subfamily 16 of P. polleni. Regarding subfamily 8, we found almost the same number of genes in the draft genome of O. niloticus (Nile tilapia, supplementary table S4, Supplementary Material online) as was found in H. chilotes, indicating that the gene expansion event of subfamily 8 has already occurred at least before the radiation of African cichlids. These results indicate that most of the lineage-specific expansions of OlfC genes preceded the splitting of African, South American, and Malagasy cichlids (more than 100 MYA, Azuma et al. 2008) that was due to the breakup of the Gondwana supercontinent. In addition, we examined the presence or absence of the orthologous OlfC genes among east African cichlids using PCRs, which were designed to distinguish each OlfC gene found in the genome of H. chilotes (supplementary fig. S7, Supplementary Material online). As a result, we can detect the PCR bands in most of the other cichlids, implying that the ancestor of modern cichlids possess mostly the similar OlfC gene repertoire observed in H. chilotes. Although the detection of the PCR bands does not directly indicate the presence of “intact” gene in the genome, the PCR data are consistent with the genomic Southern hybridization analysis. Accordingly, the ancestral founder group(s) of the extant cichlids already possessed almost the same repertoire of OlfC genes as is observed in present-day cichlids.
The Evolutionary Consequence of Lineage-Specific OlfC Gene Expansion
We sought to determine the contribution of lineage-specific expansion of OlfC genes to the evolution of teleost fishes. The observed marked differences in OlfC gene copy number between fish species probably led to significant differences in their abilities to detect amino acids. We here raise two alternative possibilities that explain the effect of the OlfC gene expansion to the cichlid olfaction: 1) it might increase the sensitivity to a particular amino acid and 2) it might increase the ability to distinguish broader amino acids and/or its derivatives. Regarding the first possibility, most olfactory receptor genes are expressed in olfactory sensory neurons in a mutually exclusive and monoallelic manner, and their expression is believed to be stochastic. Therefore, if the OlfCs encoded by paralogs of the same subfamily bind to the same amino acid, a greater number of paralogs may indicate a greater absolute number of olfactory sensory neurons that can bind a specific amino acid. This is expected to directly lead to increased sensitivity to that specific amino acid. Regarding the second possibility, it is more reasonable that paralogs that emerged by gene duplication bind to different amino acids and their derivatives or even to small peptides. This scenario is well accepted in evolutionary theory of gene duplication and functional diversification. Namely, it is likely that the cichlids that underwent lineage-specific gene expansion improved their capability to detect broader range of amino acids and their derivatives, and that this may provide the basis for trophic diversification.
To examine the above two possibilities, we need to investigate whether the paralogs emerged by gene duplications bind to the same amino acid or not. Accordingly, we conducted evolutionary analyses based on the ratio of nonsynonymous and synonymous divergence (dN/dS), which can be an indicator of selective pressure acting on a protein-coding gene. Figure 5A shows the plots of dN vs. dS between paralogs of the same subfamilies. We investigated nine subfamilies in which lineage-specific gene expansions were observed in teleost fishes. Given that synonymous substitutions are evolutionarily neutral, the dS values (x axis) are expected to be proportional to the divergence time for each paralog comparison. The data points for each subfamily tended to cluster, implying that the lineage-specific gene expansions occurred episodically in each subfamily. For example, the gene duplications of zebrafish appear to have occurred earlier in subfamily 16 than in the other subfamilies. In contrast, episodic gene expansion in subfamily 4 of Atlantic salmon occurred very recently. Importantly, the values of dN/dS were higher among comparisons of paralogs that diverged relatively recently. After a duplication event, dN/dS values progressively decrease over time. These data indicated that purifying selection acting on the OlfC genes subsided after the gene duplication that led to the accumulation of amino acid substitutions between paralogs. Subsequently, purifying selection may have again become strict to avoid loss of OlfC receptor function owing to an excess of amino acid changes. These observations imply that the effect of gene duplication in OlfC may have led to functional differentiation. Thus, our analysis favors the second possibility.
Fig. 5.—
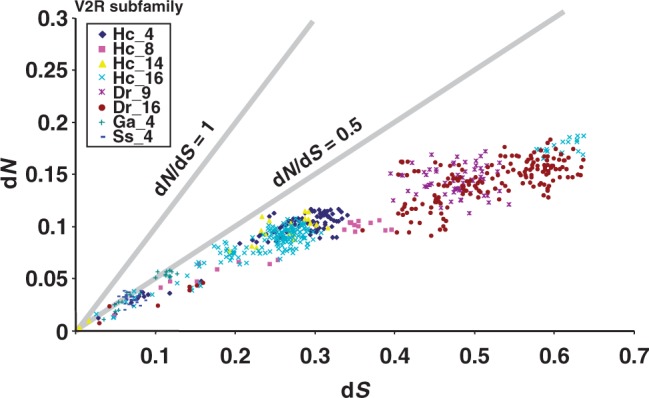
Synonymous and nonsynonymous substitutions after lineage-specific gene expansion. Plot of dN vs. dS. The gray lines indicate dN/dS at 1 or 0.5.
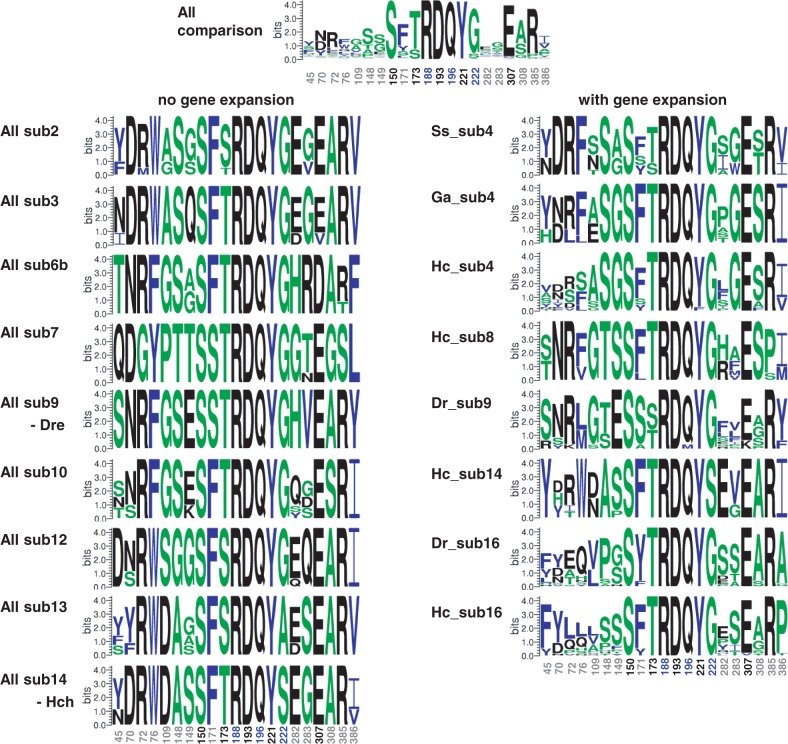
To further examine whether lineage-specific gene expansion contributed to functional diversification, we constructed sequence logos to compare the degree of sequence conservation among OlfC subfamilies with (fig. 6, right) or without (fig. 6, left) lineage-specific gene expansions. The amino acid residues used for the analysis were 5 proximal binding residues (black), 13 distal binding residues (gray), and 3 structural residues (blue) (Luu et al. 2004; Alioto and Ngai 2006). The proximal residues are predicted to be essential for the direct “binding,” whereas the distal binding residues are predicted to determine “selectivity.” Although the structural residues do not directly contact with amino acids, they are predicted to be involved in structural interaction. Interestingly, the logos reveal apparent differences in the degree of sequence conservation at residues responsible for “selectivity” of amino acid binding. Namely, for the sequence logos without gene expansion (fig. 6, left), the sequences were mostly conserved regardless of their predicted functions. For the logos with gene expansion (fig. 6, right), however, the sequences were variable at positions responsible for amino acid “selectivity” but were highly conserved at those positions responsible for essential functions of OlfC receptors.
Fig. 6.—
Sequence logos of predicted binding-pocket residues in teleost OlfCs. Sequence logos were constructed for two comparisons: The OlfC genes among subfamilies with (right) or without (left) lineage-specific expansions. The logos show the difference in the degree of sequence conservation at residues responsible for selectivity of amino acid binding (the positions of these residues are indicated in gray). The number of positions of residues predicted to be responsible for amino acid binding and structure maintenance are indicated in black and blue, respectively. The roles for each OlfC residue were predicted according to Alioto and Ngai (2006).
To examine the above possibility in more detail, we counted the number of amino acid differences in positions responsible for “selectivity” among each subfamily and compared them between subfamilies with and without gene expansion. The Student’s t test indicated that the number of amino acid differences was significantly larger in subfamilies with gene expansions than in those without gene expansions (P < 0.01). Furthermore, given that the lineage-specific gene expansions occurred after the splitting of each fish lineage, the paralogs that emerged by gene expansions in a given lineage are expected to be more closely related than orthologs of different lineages. However, we revealed that recently duplicated paralogs of a given lineage are more variable than those among orthologs of different lineages at the particular sites that were predicted to be involved in amino acid “selectivity.” Thus, the contrasting mode of sequence variability found in the OlfC genes with or without lineage-specific gene expansion suggests that the increase in the number of OlfC genes may have led to diversification of the binding to various amino acids and their derivatives. Hence, the highly diversified OlfC repertoire in cichlids found in this study suggests that this group has developed a greater capability to discriminate a variety of amino acids (and derivatives), which probably contributed to the observed diversity of their feeding habitats. Furthermore, we examined the operation of natural selection acting on OlfCs with gene expansion. Supplementary figure S8, Supplementary Material online, is the schematic representation of the SLAC analysis (Pond and Frost 2005) showing the sites under positive and negative selection in OlfCs. Apparently, the negative selection was dominant in any of the subfamilies that we investigated. We did not find the signature of the operation of positive selection in the residues responsible for the “ligand selectivity” except for only one site in subfamily 9 of zebrafish. We interpreted the results of SLAC analysis that the sequence variation in the residues for ligand selectivity was caused by the relaxation of purifying selection or random genetic drift rather than positive selection.
Although several lines of evidence suggest that OlfC-mediated olfaction is involved in feeding behavior (Speca et al. 1999; Sato et al. 2005; Koide et al. 2009), it is also possible that OlfCs are involved in social communication. Namely, Yambe et al. (2006) showed that an amino acid derivative, l-kynurenine, secreted in the female urine acts as the male-attracting pheromone in masu salmon, suggesting that OlfC-mediated amino acid detection might also be involved in social interactions. Furthermore, Hashiguchi and Nishida (2006) and Johnstone et al. (2009) suggested that the small peptides cleaved by neprilysin during ovulation may bind OlfC receptors. Because such peptides are possibly used to convey social information regarding reproductive status and/or individuality, it is also possible that OlfC-mediated olfaction is involved in social behaviors. Thus, the unexpected diversity of the OlfC repertoire is worthy of further study to elucidate perhaps unrecognized behaviors in cichlids elicited by OlfC-mediated olfaction.
In conclusion, the high-quality sequence data we generated from the OlfC gene cluster region of the Lake Victoria cichlids were of great utility. Namely, we extensively explored the single-nucleotide polymorphisms by mapping the huge data set of short reads of closely related species obtained by next-generation sequencing techniques. Such study is of primary importance to reveal the direct association between the phenotype (i.e., feeding habitat) and genotype that enables us to understand the genetic mechanism of explosive radiation in cichlids.
Supplementary Material
Supplementary figures S1–S8 and tables S1–S4 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by a grant from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (21227002) to N.O., a JSPS Asia-Africa Science Platform Program grant to N.O., and a Grant-in-Aid for Scientific Research on Innovative Areas to N.O.
Literature Cited
- Alioto TS, Ngai J. The repertoire of olfactory C family G protein-coupled receptors in zebrafish: candidate chemosensory receptors for amino acids. BMC Genomics. 2006;7:309. doi: 10.1186/1471-2164-7-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma Y, Kumazawa Y, Miya M, Mabuchi K, Nishida M. Mitogenomic evaluation of the historical biogeography of cichlids toward reliable dating of teleostean divergences. BMC Evol Biol. 2008;8:215. doi: 10.1186/1471-2148-8-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barata EN, et al. A sterol-like odorant in the urine of Mozambique tilapia males likely signals social dominance to females. J Chem Ecol. 2008;34:438–449. doi: 10.1007/s10886-008-9458-7. [DOI] [PubMed] [Google Scholar]
- Birney E, Clamp M, Durbin R. GeneWise and Genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Oh BC, Stryer L. Cloning and localization of two multigene receptor families in goldfish olfactory epithelium. Proc Natl Acad Sci U S A. 1998;95:11987–11992. doi: 10.1073/pnas.95.20.11987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole TB, Stacey NE. Olfactory responses to steroids in an African mouth-brooding cichlid, Haplochromis burtoni (Gunther) J Fish Biol. 2006;68:661–680. [Google Scholar]
- Crapon de Caprona MD. Olfactory communication in a cichlid fish, Haplochromis burtoni. Z Tierpsychol. 1980;52:113–134. doi: 10.1111/j.1439-0310.1980.tb00706.x. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich RW, Korsching SI. Combinatorial and chemotopic odorant coding in the zebrafish olfactory bulb visualized by optical imaging. Neuron. 1997;18:737–752. doi: 10.1016/s0896-6273(00)80314-1. [DOI] [PubMed] [Google Scholar]
- Fryer G, Iies TD. The cichlid fishes of the great lakes of Africa. Edinburgh (United Kingdom): Oliver and Boyd; 1972. [Google Scholar]
- Grus WE, Zhang J. Origin of the genetic components of the vomeronasal system in the common ancestor of all extant vertebrates. Mol Biol Evol. 2009;26:407–419. doi: 10.1093/molbev/msn262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Lethiec F, Duroux P, Gascuel O. PHYML online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005;33:W557–W559. doi: 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiguchi Y, Furuta Y, Nishida M. Evolutionary patterns and selective pressures of odorant/pheromone receptor gene families in teleost fishes. PLoS One. 2008;3:e4083. doi: 10.1371/journal.pone.0004083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiguchi Y, Nishida M. Evolution and origin of vomeronasal-type odorant receptor gene repertoire in fishes. BMC Evol Biol. 2006;6:76. doi: 10.1186/1471-2148-6-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiguchi Y, Nishida M. Screening the V2R-type putative odorant receptor gene repertoire in bitterling Tanakia lanceolata. Gene. 2009;441:74–79. doi: 10.1016/j.gene.2008.07.022. [DOI] [PubMed] [Google Scholar]
- Johnstone KA, et al. Genomic organization and evolution of the vomeronasal type 2 receptor-like (OlfC) gene clusters in Atlantic salmon, Salmo salar. Mol Biol Evol. 2009;26:1117–1125. doi: 10.1093/molbev/msp027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimoto H, Haga S, Sato K, Touhara K. Sex-specific peptides from exocrine glands stimulate mouse vomeronasal sensory neurons. Nature. 2005;437:898–901. doi: 10.1038/nature04033. [DOI] [PubMed] [Google Scholar]
- Kocher TD. Adaptive evolution and explosive speciation: the cichlid fish model. Nat Rev Genet. 2004;5:288–298. doi: 10.1038/nrg1316. [DOI] [PubMed] [Google Scholar]
- Koide T, et al. Olfactory neural circuitry for attraction to amino acids revealed by transposon-mediated gene trap approach in zebrafish. Proc Natl Acad Sci U S A. 2009;106:9884–9889. doi: 10.1073/pnas.0900470106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Laberge F, Hara TJ. Neurobiology of fish olfaction: a review. Brain Res Rev. 2001;36:46–59. doi: 10.1016/s0165-0173(01)00064-9. [DOI] [PubMed] [Google Scholar]
- Leinders-Zufall T, et al. MHC class I peptides as chemosensory signals in the vomeronasal organ. Science. 2004;306:1033–1037. doi: 10.1126/science.1102818. [DOI] [PubMed] [Google Scholar]
- Loconto J, et al. Functional expression of murine V2R pheromone receptors involves selective association with the M10 and M1 families of MHC class Ib molecules. Cell. 2003;112:607–618. doi: 10.1016/s0092-8674(03)00153-3. [DOI] [PubMed] [Google Scholar]
- Luu P, Acher F, Bertrand HO, Fan J, Ngai J. Molecular determinants of ligand selectivity in a vertebrate odorant receptor. J Neurosci. 2004;24:10128–10137. doi: 10.1523/JNEUROSCI.3117-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maan ME, et al. Intraspecific sexual selection on a speciation trait, male coloration, in the Lake Victoria cichlid Pundamilia nyererei. Proc Biol Sci. 2004;271:2445–2452. doi: 10.1098/rspb.2004.2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunami H, Buck LB. A multigene family encoding a diverse array of putative pheromone receptors in mammals. Cell. 1997;90:775–784. doi: 10.1016/s0092-8674(00)80537-1. [DOI] [PubMed] [Google Scholar]
- Miranda A, Almeida OG, Hubbard PC, Barata EN, Canário AV. Olfactory discrimination of female reproductive status by male tilapia (Oreochromis mossambicus) J Exp Biol. 2005;208:2037–2043. doi: 10.1242/jeb.01584. [DOI] [PubMed] [Google Scholar]
- Naito T, et al. Putative pheromone receptors related to the Ca2+-sensing receptor in Fugu. Proc Natl Acad Sci U S A. 1998;95:5178–5181. doi: 10.1073/pnas.95.9.5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Niimura Y, Nozawa M. The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat Rev Genet. 2008;9:951–963. doi: 10.1038/nrg2480. [DOI] [PubMed] [Google Scholar]
- Plenderleith M, van Oosterhout C, Robinson RL, Turner GF. Female preference for conspecific males based on olfactory cues in a Lake Malawi cichlid fish. Biol Lett. 2005;1:411–414. doi: 10.1098/rsbl.2005.0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond SL, Frost SD. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 2005;21:2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
- Sato Y, Miyasaka N, Yoshihara Y. Mutually exclusive glomerular innervation by two distinct types of olfactory sensory neurons revealed in transgenic zebrafish. J Neurosci. 2005;25:4889–4897. doi: 10.1523/JNEUROSCI.0679-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Seehausen O, et al. Speciation through sensory drive in cichlid fish. Nature. 2008;455:620–626. doi: 10.1038/nature07285. [DOI] [PubMed] [Google Scholar]
- Seehausen O, van Alphen JJM. The effect of male coloration on female mate choice in closely related Lake Victoria cichlids (Haplochromis nyererei complex) Behav Ecol Sociobiol. 1998;42:1–8. [Google Scholar]
- Shi P, Zhang J. Comparative genomic analysis identifies an evolutionary shift of vomeronasal receptor gene repertoires in the vertebrate transition from water to land. Genome Res. 2007;17:166–174. doi: 10.1101/gr.6040007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speca DJ, et al. Functional identification of a goldfish odorant receptor. Neuron. 1999;23:487–498. doi: 10.1016/s0896-6273(00)80802-8. [DOI] [PubMed] [Google Scholar]
- Streelman JT, Zardoya R, Meyer A, Karl SA. Multilocus phylogeny of cichlid fishes (Pisces: Perciformes): evolutionary comparison of microsatellite and single-copy nuclear loci. Mol Biol Evol. 1998;15:798–808. doi: 10.1093/oxfordjournals.molbev.a025985. [DOI] [PubMed] [Google Scholar]
- Tamura K, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28: 2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terai Y, et al. Divergent selection on opsins drives incipient speciation in Lake Victoria cichlids. PLoS Biol. 2006;4:e433. doi: 10.1371/journal.pbio.0040433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terai Y, Mayer WE, Klein J, Tichy H, Okada N. The effect of selection on a long wavelength-sensitive (LWS) opsin gene of Lake Victoria cichlid fishes. Proc Natl Acad Sci U S A. 2002;99:15501–15506. doi: 10.1073/pnas.232561099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner GF. Adaptive radiation of cichlid fish. Curr Biol. 2007;17:R827–R831. doi: 10.1016/j.cub.2007.07.026. [DOI] [PubMed] [Google Scholar]
- Turner GF, Seehausen O, Knight ME, Allender CJ, Robinson RL. How many species of cichlid fishes are there in African lakes? Mol Ecol. 2001;10:793–806. doi: 10.1046/j.1365-294x.2001.01200.x. [DOI] [PubMed] [Google Scholar]
- Verzijden MN, ten Cate C. Early learning influences species assortative mating preferences in Lake Victoria cichlid fish. Biol Lett. 2007;3:134–136. doi: 10.1098/rsbl.2006.0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Kobayashi N, Fujiyama A, Okada N. Construction of a BAC library for Haplochromis chilotes, a cichlid fish from Lake Victoria. Genes Genet Syst. 2003;78:103–105. doi: 10.1266/ggs.78.103. [DOI] [PubMed] [Google Scholar]
- Yambe H, et al. L-Kynurenine, an amino acid identified as a sex pheromone in the urine of ovulated female masu salmon. Proc Natl Acad Sci U S A. 2006;103:15370–15374. doi: 10.1073/pnas.0604340103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JM, Massa HF, Hsu L, Trask BJ. Extreme variability among mammalian V1R gene families. Genome Res. 2010;20:10–18. doi: 10.1101/gr.098913.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JM, Trask BJ. V2R gene families degenerated in primates, dog and cow, but expanded in opossum. Trends Genet. 2007;23:212–215. doi: 10.1016/j.tig.2007.03.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.