Abstract
Staphylococcus aureus is a leading human pathogen of both hospital and community-associated diseases worldwide. This organism causes a wealth of infections within the human host as a result of the vast arsenal of toxins encoded within its genome. Previous transcriptomic studies have shown that toxin production in S. aureus can be strongly impacted by the negative regulator CodY. CodY acts by directly, and indirectly (via Agr), repressing toxin production during times of plentiful nutrition. In this study we use iTRAQ based proteomics for the first time to study virulence determinant production in S. aureus, so as to correlate transcriptional observations with actual changes in protein synthesis. Using a codY mutant in the epidemic CA-MRSA clone USA300 we demonstrate that deletion of this transcription factor results in a major upregulation of toxin synthesis in both post-exponential and stationary growth. Specifically, we observe hyper-production of secreted proteases, leukocidins and hemolysins in both growth phases in the USA300 codY mutant. Our findings demonstrate the power of mass spectrometry-based quantitative proteomics for studying toxin production in S. aureus, and the importance of CodY to this central process in disease causation and infection.
Keywords: Staphylococcus aureus, regulation, toxins, virulence determinants, proteomics
The transcription factor CodY is well conserved within the low G+C Gram-positive bacteria, where it has been shown to be an important regulator of metabolism and virulence [1]. It functions by sensing intracellular levels of branched-chain amino acids (BCAAs) and GTP during growth, and responds by repressing genes involved in starvation behaviors in nutrient-rich conditions, such as amino acid transport, sporulation and competence [2–5]. As BCAA and GTP levels decline, CodY loses its affinity for DNA binding, bringing about de-repression of target genes [1]. This results in a physiological transition from growth and division to amino acid metabolism and stress tolerance. In this study we focus on the impact of CodY on toxin production in the major human pathogen Staphylococcus aureus. Previous reports have shown that, in addition to its role in nutritional regulation, CodY impacts toxin gene expression in S. aureus by repressing transcription of another regulator, agr. Agr is a quorum-sensing, two-component system that is maximally expressed during post-exponential growth, where it represses surface proteins, and induces transcription of toxins [6]. As such, CodY functions to couple toxin gene regulation in S. aureus to the nutritional status of the cell. Thus far, a variety of studies have documented the effects of CodY on S. aureus toxin gene transcription [7–9], but, to date, a study of how these changes affect toxin synthesis is lacking.
Accordingly, we have employed iTRAQ to assess the impact of CodY on toxin synthesis in the human clinical isolate, CA-MRSA USA300. A codY mutant was generated in strain USA300-HOU-MRSA [10] using techniques described previously [11]. Post-exponential phase (5h) or stationary phase (15h) cultures of wild-type and mutant strains (supplemental figure 1) were prepared in TSB as described previously [10,11]. Secretomes were harvested by centrifugation, sterilized by filtration, and concentrated using Millipore Centricon Plus-70 filter units with a 3 kDa cutoff. Proteins were precipitated overnight at 4° C using 10% trichloroacetic acid. Precipitates were collected by centrifugation and pellets washed 3 times with 100% ice-cold ethanol, before being air dried. Samples were resuspended in iTRAQ dissolution buffer at 100 μg/ml, in a volume of 20 μl. One μl of denaturant and 2 μl of reducing reagent were added to each sample, and incubated at 60° C for 1h. Next, 1 μl of cysteine blocking reagent was added and incubated for 10 minutes at room temperature. Trypsin was added in a ratio of 1:30, and samples digested for 12h–16h at 37° C. Labeling with iTRAQ reagents was completed according to the manufacturer’s instructions. Briefly, peptide samples were individually labeled with iTRAQ reagents (USA300-116, USA300-codY-117) for 1h at room temperature. After incubation, samples were combined and dried using a SpeedVac centrifuge, and resuspended in 1 ml of 0.1% formic acid in water. A C-18 Vydac column was used to desalt samples, with peptides eluted using 0.1% formic acid in acetonitrile. After peptide elution, iTRAQ-labeled samples were dried using a SpeedVac centrifuge, resuspended in 0.1% formic acid in water, and analyzed using a hybrid linear ion trap-Orbitrap instrument mass spectrometer (LTQ Orbitrap XL, Thermo) operated with Xcalibur (v2.0.7) data acquisition software. Five μl of protein digest was loaded onto a 75 μm i.d. × 2 cm ProteoPep II C18 trap (New Objective, Woburn, MA) and desalted online before injection onto a 75 μm i.d. × 10 cm Proteopep II C18 analytical column, where a linear gradient was carried out to 40% acetonitrile in 90 min at 250 nL/min, using a NanoUltra 2D-HPLC system (Eksigent, Dublin, CA). Data-dependent acquisition consisted of MS/MS analysis in the LTQ linear ion trap of the top 3 most intense precursor ions from initial high resolution (30,000 at m/z 400) survey scans, followed by high-energy collision dissociation (HCD) and Orbitrap fragment ion detection (7,500 at m/z 400) of the same 3 ions.
MS/MS spectra generated from data-dependent acquisition via the LTQ Orbitrap XL were extracted by extractMSN.exe (BioWorks v.3.3, Thermo), and searched against a concatenated subset database containing both normal and randomized sequences of Staphylococcus aureus USA300_FPR3757 obtained from Uniprot.org (downloaded 07-27-2010, 5214 entries) using the Mascot search algorithm (v2.2.2). Mascot was searched with a fragment ion mass tolerance of 0.60 Da, and a parent ion tolerance of 10 ppm, assuming full trypsin specificity with 1 possible missed cleavage. Oxidation of methionine, as well as iTRAQ labeling of peptide N-terminus, lysine and tyrosine, were included as variable modifications in Mascot. Fixed modifications included MMTS modification of cysteine. Mascot results were compiled in ProteoIQ (v2.1.11, Nusep) for final identification and relative quantitation. The Mascot database search score cutoff was adjusted within ProteoIQ to achieve a 1% false discovery rate at the protein level. Relative quantitation of iTRAQ reporter ions was performed by ProteoIQ followed by normalization to the dataset median (assuming a global median ratio of 1) using Microsoft Excel. Ratios are reported as 117 reporter ion intensity (USA300-codY)/116 reporter ion intensity (USA300), and considered significant if the ratio was at least ± 1 standard deviation away from the median. Quantitative proteomic analysis using iTRAQ was performed using three biological replicates. All Mascot identification data are deposited in the PRIDE proteomics identification database (http://www.ebi.ac.uk/pride) under accession numbers 17700–17705 (username: review49224, password: 2qQFem+7) [12]. The data were converted using PRIDE Converter [13].
Tables 1 and 2 show iTRAQ-based relative quantitation for proteins that are known or putative secreted virulence factors of S. aureus; however, all proteins and corresponding iTRAQ ratios are reported in Supplementary Tables 1 and 2. It should be noted that the complete datasets in the supplementary information contain a number of intracellular proteins within the secretomes of both strains. It has been our experience, and that of others [14], that sub-proteome contamination is common, and can be the result of a number of factors, including natural cell lysis during growth. Additionally, there is growing evidence to suggest that intracellular proteins are intentionally exported, and have novel functions outside the cell [14]. For the data presented herein, we analyzed only those proteins that were significantly altered between the two strains based on our filtering criteria, and are either known secreted proteins, or contained signal peptides, as determined by the SignalP algorithm.
Table 1.
Quantitative proteomic profiling of virulence determinant production in CA-MRSA USA300 and its codY mutant during exponential growth.
| UPREGULATED | ||||
|---|---|---|---|---|
| Accession Number | Gene | Protein Name | CodY Ratioa | Standard Deviationb |
| SAUSA300_0951 | sspA | V8 protease | 4.07 (16.8)* | 2.28 |
| SAUSA300_2572 | aur | Zinc metalloproteinase aureolysin | 3.82 (14.1)* | 2.71 |
| SAUSA300_1754 | splE | Serine protease | 2.86 (7.3) | 0.46 |
| SAUSA300_1382 | lukS-PV | Panton-Valentine leukocidin | 2.79 (6.9) | 1.73 |
| SAUSA300_1759 | Putative uncharacterized protein | 2.76 (6.8) | 2.06 | |
| SAUSA300_2366 | hlgC | Gamma-hemolysin component C | 2.75 (6.7) | 2.1 |
| SAUSA300_1757 | splB | Serine protease | 2.72 (6.6) | 2.29 |
| SAUSA300_1769 | lukE | Leukocidin | 2.34 (5.1) | 2.18 |
| SAUSA300_1381 | lukF-PV | Panton-Valentine leukocidin | 2.29 (4.9) | 2 |
| SAUSA300_0950 | sspB | Cysteine protease | 2.15 (4.4) | 2.13 |
| SAUSA300_0409 | Putative uncharacterized protein | 1.86 (3.6) | 0.58 | |
| SAUSA300_1768 | lukD | Leukocidin | 1.79 (3.5) | 1.89 |
| SAUSA300_2367 | hlgB | Gamma-hemolysin component B | 1.61 (3.0) | 1.91 |
| DOWNREGULATED | ||||
| SAUSA300_1067 | psmβ1 | Antibacterial protein | −2.23 (−4.8) | 1.02 |
| SAUSA300_1988 | hld | Delta-hemolysin | −2.31 (−5.0) | 1.48 |
ratio values are Log2 transformed; number in parenthesis represents fold change;
indicates ratio value that is ≥ 2 standard deviations away from median
standard deviation (SD) values are calculated based on ratio values obtained from all peptides across all replicates (n=3); SD values are Log2 transformed
Table 2.
Quantitative proteomic profiling of virulence determinant production in CA-MRSA USA300 and its codY mutant during stationary growth.
| UPREGULATED | ||||
|---|---|---|---|---|
| Accession Number | Gene | Protein Name | CodY Ratioa | Standard Deviationb |
| SAUSA300_1758 | splA | Serine protease | 4.06 (16.6)* | 0.96 |
| SAUSA300_2572 | aur | Zinc metalloproteinase aureolysin | 3.78 (13.7)* | 2 |
| SAUSA300_1754 | splE | Serine protease | 3.70 (13.0)* | 2.03 |
| SAUSA300_2366 | hlgC | Gamma-hemolysin component C | 3.54 (11.6)* | 2.24 |
| SAUSA300_1382 | lukS-PV | Panton-Valentine leukocidin | 3.50 (11.3)* | 1.94 |
| SAUSA300_0274 | Putative uncharacterized protein | 3.38 (10.4)* | 1.92 | |
| SAUSA300_1753 | splF | Serine protease | 3.12 (8.7)* | 1.46 |
| SAUSA300_1755 | splD | Serine protease | 3.12 (8.7)* | 1.8 |
| SAUSA300_1757 | splB | Serine protease | 3.08 (8.5)* | 2.01 |
| SAUSA300_1381 | lukF-PV | Panton-Valentine leukocidin | 3.04 (8.2)* | 1.61 |
| SAUSA300_0815 | ear | Ear protein | 3.04 (8.2)* | 2.21 |
| SAUSA300_0951 | sspA | V8 protease | 2.86 (7.3)* | 2.51 |
| SAUSA300_1759 | Putative uncharacterized protein | 2.76 (6.8)* | 2.28 | |
| SAUSA300_2365 | hlgA | Gamma-hemolysin component A | 2.38 (5.2)* | 2.42 |
| SAUSA300_0950 | sspB | Cysteine protease | 2.34 (5.0) | 2.11 |
| SAUSA300_1058 | hla | Alpha-hemolysin | 2.24 (4.7) | 1.47 |
| SAUSA300_0883 | Putative surface protein | 2.10 (4.3) | 1.93 | |
| SAUSA300_1890 | scpA | Staphopain A | 1.76 (3.4) | 1.75 |
| SAUSA300_2164 | Hypothetical protein | 1.58 (3.0) | 1.25 | |
| SAUSA300_1768 | lukD | Leukocidin | 1.44 (2.7) | 2.13 |
| SAUSA300_0955 | atl | Bifunctional autolysin | 1.38 (2.6) | 2.36 |
| SAUSA300_1988 | hld | Delta-hemolysin | 1.28 (2.4) | 1.89 |
ratio values are Log2 transformed; number in parenthesis represents fold change;
indicates ratio value that is ≥ 2 standard deviations away from median
standard deviation (SD) values are calculated based on ratio values obtained from all peptides across all replicates (n=3); SD values are Log2 transformed
Post-exponential secretomes of the codY mutant compared to the wild-type (Table 1) demonstrate a dramatic increase in several agr-regulated proteases, such as aureolysin (14.1-fold), Spl proteases (SplE = 7.3-fold, SplB = 6.6-fold), and Staphopain B (4.4-fold). Most pronounced, however, was V8 protease, which was produced at levels 16.8-fold higher in the mutant. Furthermore, synthesis of multiple leukocidins, including LukD (3.5-fold) and LukE (5.1-fold), along with the Panton-Valentine leukocidins, LukF-PV (4.9-fold) and LukS-PV (6.9-fold), were increased in the mutant during post-exponential growth. Additionally, subunits of the γ-hemolysin were found in greater abundance in the codY-null strain (HlgB = 3.0-fold, HlgC = 6.7-fold). Interestingly, during post-exponential growth in the mutant, we observed decreased levels of two phenol soluble modulins: δ-hemolysin and Psmβ1 (−4.8-fold and −5.0-fold, respectively). It is possible that this unexpected downregulation may result from overproduction of proteases in the codY mutant. Indeed, it has previously been suggested that the V8 protease may modulate self-derived toxin stability [15]; whilst more recently our group has shown that aureolysin degrades both Hla and the PSMs [16]. This observation might also explain why we did not observe significant alteration in α-hemolysin protein levels during post-exponential growth, despite our validation assays detecting elevated hla (and hld) transcription in the codY mutant (supplemental figure 2).
When exploring the effects of codY mutation on stationary phase toxin synthesis (Table 2), we observed additional alterations when compared to the wild-type. Specifically, the hyperactive agr phenotype of USA300 [17] is more pronounced in the absence of CodY during stationary phase, with significant overproduction of agr-regulated proteins. As with our post-exponential findings, the major class of proteins with increased synthesis was again secreted proteases. Of the ten known enzymes, 9 are increased in stationary phase cultures of the mutant, with only SplC not shown to be significantly altered. Specifically, Spl enzymes increased from 8.5-fold (SplB) to 16.6-fold (SplA). In addition, we observed upregulation of both cysteine proteases (SspB = 5.0-fold, ScpA = 3.4-fold), aureolysin (13.7-fold), and V8 protease (7.3-fold). These findings are again consistent with our transcriptional validation work, which reveals strong upregulation from major protease loci (aur, scp and ssp), throughout growth in the codY mutant (supplemental figure 2). Leading on from this, given that secreted proteases negatively impact S. aureus biofilm formation [10, 16], one would predict that a USA300 codY mutant would be impaired in this behavior. As such, we analyzed biofilm formation in the codY mutant, and found an approximately 3-fold reduction when compared to the parental and complemented strains (figure 1A).
Figure 1. Deletion of codY in S. aureus USA300 results in decreased biofilm formation and increased hemolytic activity.
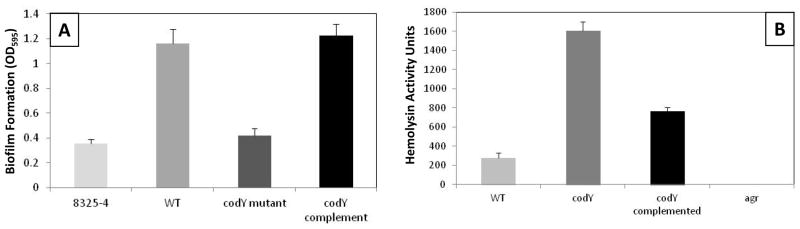
The USA300 codY mutant, and its parental and complemented strains, were assayed for biofilm formation (A) and hemolytic activity (B), as described previously [10, 13]. 8325-4 (A) and a USA300 agr mutant (B) were used as a negative controls. Hemolysis data is derived from 15h cultures. Data presented is from 3 biological replicates. Error bars are shown as ±SEM.
We also observed accumulation of cytolytic toxins in stationary phase cultures of the mutant, with both PVL components increased (LukS-PV = 11.3-fold, LukF-PV = 8.2-fold), as was LukD (2.7-fold). There was also a global increase in hemolysin production in the mutant, with α-hemolysin (4.7-fold), γ-hemolysin (HlgA = 5.2-fold, HlgC = 11.6-fold) and δ-hemolysin (2.4-fold) all displaying greater protein levels. This former finding is in strong agreement with our validation assays, which reveal a 5.8-fold increase in α-hemolysin activity in the codY mutant during this growth phase (figure 1B). This latter finding is of interest as it is in contrast to the post-exponential data, where we observed decreased δ-hemolysin. Whilst protease levels remain high during stationary phase, it is possible that they suffer a decrease in enzymatic activity as cultures age, explaining the increase in δ-hemolysin accumulation. Furthermore, δ-hemolysin is transcribed at very high levels in the run up to stationary phase (supplemental figure 2); therefore, despite proteolytic degradation, its abundance is still detectably increased.
Our findings with regards to the high levels and activity of proteolytic enzymes raises an important technical point. Due to the high abundance of these enzymes in cultures at both time points, the standard deviation values for ratios determined may be elevated due to biological variability associated with proteolytic degradation. This result would be expected, giving variability at the peptide level within biological replicates, and producing alterations in sequence coverage obtained by mass spectrometric analysis. Additionally, iTRAQ-labeled peptides derived from the relatively complex secretomes used in this study were not fractionated prior to LC-MS/MS which may lead to iTRAQ ratio compression for certain peptides and therefore, underestimation and/or increased variability of relative changes determined at the protein level [18]. In spite of this, we suggest that such proteomic analyses are of significant value, and perhaps provide more insight than those focused on transcriptional changes. Specifically, changes in gene expression may not directly lead to alterations in protein abundance; resulting from the post-translational events described herein, and in other works [15, 16]. Even though these findings are from in vitro studies, one would predict that such events also occur in vivo, and are thus physiologically relevant and important.
We also observed an increase in the production of a number of uncharacterized secreted proteins in the codY mutant. Specifically, SAUSA300_0409 and SAUSA300_0274 were upregulated in post-exponential and stationary phases, respectively. Whilst no obvious domains or homology could be detected for these proteins, the same is not true for SAUSA300_1759, which was upregulated in the mutant during both growth phases at the same level (6.8-fold). SAUSA300_1759 has homology to the Ear protein, which was also found to be upregulated in the codY mutant (8.2-fold). The Ear protein currently has unknown function, but has been labeled a putative toxin in strain MW2, and is found on the υSa3 pathogenicity island [19, 20]. Other putative toxins were also found to have altered levels, with two Eap/Map domain proteins SAUSA300_0883 (4.3-fold) and SAUSA300_2164 (3.0-fold) both showing increases in the mutant. Whilst these two proteins have yet to be studied, they are part of a family of proteins in S. aureus that contain such domains. Eap/Map proteins are both surface associated and extracellular virulence determinants, with a variety of roles in pathogenesis, including adherence and immune-subversion [21, 22].
The present study is the first of its kind to use iTRAQ proteomic techniques to map toxin production in the major human pathogen Staphylococcus aureus. It is apparent from our studies that this approach can be an effective method for relative protein quantitation in this bacterium, as a way to corroborate transcriptional analyses. We show that a variety of known and novel toxins encoded within the genome of CA-MRSA USA300 are hyper-produced as a result of codY inactivation. As such, this information provides additional insight into the role of this important regulator, and an understanding of its contribution to disease causation.
Supplementary Material
Acknowledgments
This study was supported in part by grant 1R21AI090350-01 (LNS) from the National Institute of Allergies and Infectious Diseases. We gratefully thank Dr Chia Lee (UAMS) for kindly sharing the codY complementation construct.
Footnotes
Conflict of Interest
None.
References
- 1.Stenz L, Francois P, Whiteson K, Wolz C, et al. The CodY pleiotropic repressor controls virulence in Gram-positive pathogens. FEMS Immunol Med Microbiol. 2011 doi: 10.1111/j.1574-695X.2011.00812.x. [DOI] [PubMed] [Google Scholar]
- 2.Ferson AE, Wray LV, Jr, Fisher SH. Expression of the Bacillus subtilis gabP gene is regulated independently in response to nitrogen and amino acid availability. Mol Microbiol. 1996;22:693–701. doi: 10.1046/j.1365-2958.1996.d01-1720.x. [DOI] [PubMed] [Google Scholar]
- 3.Fisher SH, Rohrer K, Ferson AE. Role of CodY in regulation of the Bacillus subtilis hut operon. J Bacteriol. 1996;178:3779–3784. doi: 10.1128/jb.178.13.3779-3784.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fujita M, Losick R. Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev. 2005;19:2236–2244. doi: 10.1101/gad.1335705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mirel DB, Estacio WF, Mathieu M, Olmsted E, et al. Environmental regulation of Bacillus subtilis sigma(D)-dependent gene expression. J Bacteriol. 2000;182:3055–3062. doi: 10.1128/jb.182.11.3055-3062.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Novick RP. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol. 2003;48:1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- 7.Majerczyk CD, Sadykov MR, Luong TT, Lee C, et al. Staphylococcus aureus CodY negatively regulates virulence gene expression. J Bacteriol. 2008;190:2257–2265. doi: 10.1128/JB.01545-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pohl K, Francois P, Stenz L, Schlink F, et al. CodY in Staphylococcus aureus: a regulatory link between metabolism and virulence gene expression. J Bacteriol. 2009;191:2953–2963. doi: 10.1128/JB.01492-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Somerville GA, Proctor RA. At the crossroads of bacterial metabolism and virulence factor synthesis in staphylococci. Microbiol Mol Biol Rev. 2009;73:233–248. doi: 10.1128/MMBR.00005-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kolar SL, Nagarajan V, Oszmiana A, Rivera FE, et al. NsaRS is a Cell-Envelope-Stress Sensing Two-Component System of Staphylococcus aureus. Microbiology. 2011 doi: 10.1099/mic.0.049692-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaw LN, Lindholm C, Prajsnar TK, Miller HK, et al. Identification and characterization of σS, a novel component of the Staphylococcus aureus stress and virulence responses. PLoS One. 2008;3:e3844. doi: 10.1371/journal.pone.0003844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martens L, Hermjakob H, Jones P, Adamski M, et al. PRIDE: the proteomics identifications database. Proteomics. 2005;5:3537–3545. doi: 10.1002/pmic.200401303. [DOI] [PubMed] [Google Scholar]
- 13.Barsnes H, Vizcaino JA, Reisinger F, Eidhammer I, Martens L. Submitting proteomics data to PRIDE using PRIDE Converter. Methods Mol Biol. 2011;694:237–253. doi: 10.1007/978-1-60761-977-2_16. [DOI] [PubMed] [Google Scholar]
- 14.Dreisbach A, Hempel K, Buist G, Hecker M, et al. Profiling the surfacome of Staphylococcus aureus. Proteomics. 2010;10:3082–3096. doi: 10.1002/pmic.201000062. [DOI] [PubMed] [Google Scholar]
- 15.Lindsay JA, Foster SJ. Interactive regulatory pathways control virulence determinant production and stability in response to environmental conditions in Staphylococcus aureus. Mol Gen Genet. 1999;262:323–331. doi: 10.1007/s004380051090. [DOI] [PubMed] [Google Scholar]
- 16.Zielinska AK, Beenken KE, Joo HS, Mrak LN, et al. Defining the strain-dependent impact of the staphylococcal accessory regulator (sarA) on the alpha toxin phenotype of Staphylococcus aureus. J Bacteriol. 2011 doi: 10.1128/JB.01517-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montgomery CP, Boyle-Vavra S, Adem PV, Lee JC, et al. Comparison of virulence in community-associated methicillin-resistant Staphylococcus aureus pulsotypes USA300 and USA400 in a rat model of pneumonia. J Infect Dis. 2008;198:561–570. doi: 10.1086/590157. [DOI] [PubMed] [Google Scholar]
- 18.Ow SY, Salim M, Noirel J, Evans C, Rehman I, Wright PC. iTRAQ underestimation in simple and complex mixtures: “the good the bad the ugly”. J Proteome Res. 2009;8:5347–5355. doi: 10.1021/pr900634c. [DOI] [PubMed] [Google Scholar]
- 19.Shukla SK, Karow ME, Brady JM, Stemper ME, et al. Virulence genes and genotypic associations in nasal carriage, community-associated methicillin-susceptible and methicillin-resistant USA400 Staphylococcus aureus isolates. J Clin Microbiol. 2010;48:3582–3592. doi: 10.1128/JCM.00657-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baba T, Bae T, Schneewind O, Takeuchi F, Hiramatsu K. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J Bacteriol. 2008;190:300–310. doi: 10.1128/JB.01000-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jonsson K, McDevitt D, McGavin MH, Patti JM, Hook M. Staphylococcus aureus expresses a major histocompatibility complex class II analog. J Biol Chem. 1995;270:21457–21460. doi: 10.1074/jbc.270.37.21457. [DOI] [PubMed] [Google Scholar]
- 22.Hussain M, Haggar A, Heilmann C, Peters G, et al. Insertional inactivation of Eap in Staphylococcus aureus strain Newman confers reduced staphylococcal binding to fibroblasts. Infect Immun. 2002;70:2933–2940. doi: 10.1128/IAI.70.6.2933-2940.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


