Abstract
Parkinson’s Disease (PD) is a devastating neurodegenerative disease that affects over one million patients in the US. Yet, no disease modifying drugs exist, only those that temporarily alleviate symptoms. Because of its poorly defined and highly complex disease etiology, it is essential to embrace unbiased and innovative approaches for identifying new chemical entities that target the underlying toxicities associated with PD. Traditional target-based drug discovery paradigm can suffer from a bias towards a small number of potential targets. Phenotypic screening of both genetic and pharmacological PD models offers an alternative approach to discover compounds that target the initiating causes and effectors of cellular toxicity. The relative paucity of reported phenotypic screens illustrates the intrinsic difficulty in establishing model systems that are both biologically meaningful and adaptable to high-throughput screening. Parallel advances in PD models and in vivo screening technologies will help create opportunities for identifying new therapeutic leads with unanticipated, breakthrough mechanisms of action.
Introduction
Parkinson’s Disease (PD) is the second most common neurodegenerative disease, affecting ~2% of the population over 65 years of age. Patients suffer from progressive loss of muscle control, including tremor, rigidity, bradykinesia, and postural instability. These movement deficits are largely caused by the selective degeneration of dopaminergic (DA) neurons in a brain region called the substantia nigra pars compacta. The histopathological hallmark of PD is the presence of large intracytoplasmic spherical structures (called Lewy Bodies, LBs) that contain the α-synuclein (α-syn) protein [1]. The misfolding and accumulation of a disease-specific protein that causes proteotoxic stress is a common theme among many neurodegenerative diseases, including Alzheimer’s and Huntington’s diseases. Although most PD arises sporadically, α-syn pathology is almost always detected. Importantly, a causal role for α-syn was strengthened with the identification of rare mutations in the α-syn gene (SNCA) [2]. Extra genomic copies of SNCA also cause familial PD, establishing that simply increasing α-syn dose can cause disease [3]. Intriguingly, aging, which is the most significant risk factor for PD, is accompanied by an increase in α-syn protein levels, possibly sensitizing cells to α-syn misfolding and toxicity [4,5].
The genetic complexity of PD is exceptionally rich as several other monogenic mutations cause related parkinsonisms. Some mutations, such as those in Parkin, PARK3, LRRK2, PLA2G6, and GBA (Gaucher’s locus), accumulate α-syn-positive Lewy Bodies. Others, including mutations in UCHL1, DJ-1, ATP13A2, PARK10, GIGYF2, PARK12, HTRA2, FBX07, PARK16, have less defined α-syn involvement [6]. The diverse functions of these disease-associated genes creates a complexity where different pathological mechanisms, including ubiquitin-proteasome system, oxidative stress, vesicle trafficking, and mitochondrial dysfunction, manifest in related parkinsonisms. Also, a number of environmental substances, including the mitochondrial toxins MPTP/MPP+ and rotenone, as well as manganese, can cause related parkinsonism’s.
Model organism studies of “PD” have provided key insights into some basic, underlying toxicities of PD. They do not, however, truly model “disease”, which integrates complex interactions between cellular pathology, neuronal networks, multiple systems, and patient heterogeneity. This distinction is critical in establishing dialogues between basic researchers and clinicians treating PD patients. Herein, we refer to “PD models” without making claims to the actual human disease, but rather to the underlying precipitating toxic events.
Model organisms used to study PD have provided key molecular insights into the underlying cellular pathology. The accumulation of α-syn has been linked to mitochondrial dysfunction, proteasome inhibition, oxidative stress, vesicle trafficking defects, lipid droplet accumulation, calcium dysregulation, α-syn aggregation, and cellular toxicity. These phenotypes are studied in several model systems, including immortalized cell lines, primary neuronal cultures, yeast, fruit flies, nematodes, and rodents [7,8]. Although no model organism faithfully recapitulates all cellular pathologies, the cross-validation of findings in multiple systems strengthens new connections. Indeed, the combination of diverse cellular pathologies with the aforementioned genetic complexity and environmental links necessitates innovative, unbiased phenotypic screening approaches in simple model organisms.
Despite the prevalence of PD and the substantial efforts in studying disease pathogenesis, no disease modifying agents exist. Current therapies largely manage symptoms through modulating neuronal activity, yet do not significantly modify disease progression [9]. Phenotypic screening provides an opportunity to both study toxic mechanisms and to provide new chemical entities that target the precipitating biology and may themselves modify disease. Herein, we will discuss the promise of phenotypic screens and early successes that can enrich the pipeline for potential therapeutics. We will not discuss the other significant hurdles in drug development, including compound optimization, the difficulties in testing drugs in a slowly progressing disease, or other approaches such as structure based design.
Target-based versus phenotypic drug screening
The two main high-throughput (HT) screening approaches for discovering potential new PD-modifying compounds are target-based and phenotype-based. Target-based approaches rely on experimentally validated (though sometimes poorly so) protein activities to screen in vitro. Phenotypic screens, in contrast, exploit unbiased cell-based assays to identify compounds that elicit a desired response, such as protecting cells from α-syn toxicity. These two fundamentally different approaches have advantages and disadvantages.
Target-based biochemical screening, currently the gold standard in drug discovery, is an effective approach with a well-defined pathology and highly validated target. Its biggest advantage is that the exact target and mechanism of action are known. However, many of our most commonly used drugs function through unknown mechanisms and target-based approaches do have a few key drawbacks. First, the success of target-centric approaches hinges on correctly predicting the link between the chosen target and disease pathogenesis such that modifying the target will modify disease. This bias may be based on false or incomplete information. Second, this biased approach may not actually target the best or most “druggable” protein to provide optimal rescue. Third, it is not always possible to accurately predict a compound’s activity in vivo (e.g., too high a potency may be toxic to cells). Finally, the small molecules obtained in vitro may have chemical liabilities that preclude efficacy in vivo, including entry into cells, solubility, metabolism, distribution, excretion, and off-target effects. These potential pitfalls can hinder validation of a compound within the context of a cellular or animal model.
Phenotypic screening against the underlying toxicities that cause PD has several advantages. First, without a preordained target, screens are fully unbiased and may identify compounds targeting completely unexpected proteins or pathways. Second, in vivo screening more likely identifies drug-like molecules that (1) can get into cells, (2) are not readily metabolized, and (3) are not overtly cytotoxic themselves at effective concentrations. Thus, the primary screen actually performs multiple functions by both identifying compounds and filtering out liabilities. The most significant barrier in phenotypic screens is determining a compound’s mechanism of action and protein target. However, the genetic tractability of model organisms offers new approaches for target discovery, though this is often quite difficult. Of course, a limiting factor for phenotypic screening is that a compound’s potential is only as valid as the model from which it was derived. It is interesting to note that, despite the heavy use of target-based drug screening, 37% of first-in-class new molecule entities (NMEs) from 1999 to 2008 were identified through phenotypic screens, while only 23% were identified from target-based approaches [10] (remaining were either modified natural substances or biologics). Second generation follower drugs were typically identified through directed target-centric methods (51% as compared to 18% for phenotypic screens).
Drug screening in cell culture models of PD-related toxicities
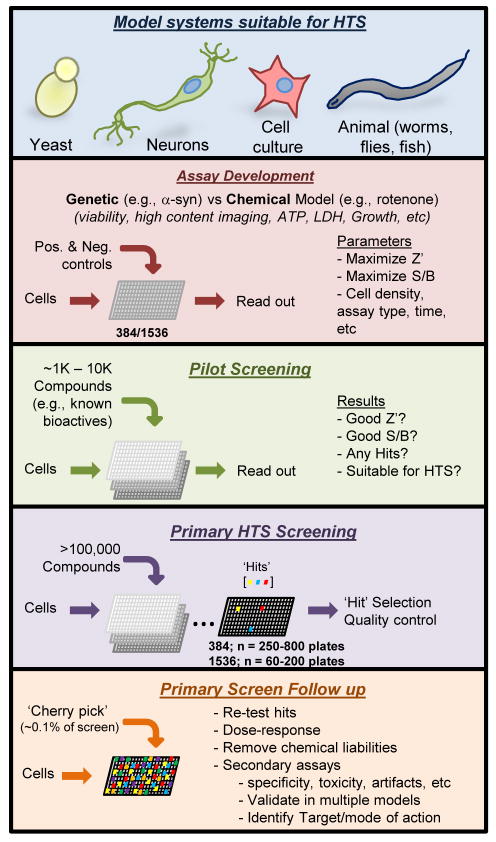
Academic screening centers provide an affordable and accessible entrée into HT phenotypic compound screening. However, few screens directed at ameliorating PD-relevant toxicities have been reported. This is likely because phenotypic screening requires robust, reproducible, and biologically meaningful assays. One must therefore examine the available model systems for their suitability to high-throughput drug screens (Figure 1). Both genetic and pharmacological approaches are used to model aspects of PD pathology. Overexpression of wild-type or mutant α-syn induces some cytotoxicity and recapitulates some key features of disease. Alternatively, treatment with mitochondrial toxins such as MPTP and rotenone mimics the mitochondrial dysfunction observed in PD [7]. Both genetic and pharmacological approaches provide meaningful insights and opportunities [7,8].
Figure 1. Phases of high-throughput phenotypic small molecule screens.
After choosing a model organism with a suitable phenotype for screening, the assay must be thoroughly vetted with robust, stereotypical behavior in high-throughput formats. A Z′, or measure of statistical effect, should be greater than 0.5, and a signal to background (S/B) should be maximized. Upon optimization, pilot screening of a small set of compound plates, often a known bioactives library, confirms acceptable signal to background ratios between positive and negative wells such that ‘hits’ can be reliably identified. Upon completion of a pilot screen, the primary HTS is carried out, typically surveying between 105 and 106 compounds. Statistically significant hits are then cherry-picked, validated, and interrogated in multiple secondary assays to focus on probes with the most desirable characteristics.
A robust phenotype that maximizes the Z′ (i.e., statistical effect size that quantifies the suitability of an assay for use in a high-throughput screen) and signal to background (S/B) is paramount for accurate hit calling (Figure 1). Low Z′ or S/B values preclude confidently identifying hits. Though routine, stable immortalized cell lines conditionally expressing α-syn typically exhibit low S/B in HT formats (~15–30% ‘toxicity’). This limitation is in part because the same reduced apoptotic mechanisms that enable proliferation in culture also makes them relatively resistant to proteotoxic insults. However, assays that monitor cell health or death through mitochondrial reducing potential (e.g., MTT, MTS), ATP levels, membrane leakage (e.g., LDH, AK), or caspase activity are amenable to HT screens if the Z′ and S/B values are sufficiently high. Extensive optimization must be performed prior to screening. Less deranged cell types, such as primary rat neurons infected with α-syn-expressing lentivirus, may be more sensitive, but they also suffer from technical limitations in terms of required virus and cell amounts, as well as cost and labor constraints. Indeed, the lack of reported cell culture toxicity screens reflects the difficulty in their development and execution in HT formats (Table 1).
Table 1.
A summary of phenotypic small molecule screens for compounds that modulate phenotypes relevant to PD.
| Organism | Model | Screen Readout | Screen format | # of molecules | Year [Reference] |
|---|---|---|---|---|---|
| H4 Neuroglioma cells | α-syn 5′ UTR luciferase reporter | Specific activitation of α-syn-LUC expression | 384 well | 303,224 | 2011[20] |
| H4 Neuroglioma cells | α-syn 5′ UTR luciferase reporter | Specific inhibition of α-syn-LUC expression | 384 well | 303,224 | 2011[21] |
| SH-SY5Y Neuroblastoma cells | α-syn 5′ UTR luciferase reporter | Specific inhibition of α-syn-LUC expression | 96 well | 720 natural products | 2011[22] |
| S. cerevisiae | α-syn toxicity | Growth, OD600 | 384 well | ~115,000 | 2010[28] |
| SK-N-SH neuroblastoma cells | Rotenone | MTS assay | 96 well | 1120 | 2010[23] |
| S. cerevisiae | α-syn toxicity | Yeast colony growth | Selective agar plates | >5 × 106 | 2009[32] |
| SH-SY5Y | α-syn toxicity | MTT | 96 well | 99 | 2007[12] |
| C. elegans | 6-OHDA | # of DA neurons, microscopy | Liquid treatment, plate outgrowth | 11 | 2007[40] |
| S. cerevisiae | α-syn toxicity with Fe3+ and yca1 | Growth, OD600 | 96 well | 10,022 | 2006[30] |
| C. elegans | MPP+ | Mobility, microscopy | 96 well | 17 | 2004[41] |
| Screens against non-PD models that may target PD-relevant pathways or toxicities | |||||
| S. cerevisiae | TDP-43 toxicity | Growth, OD600 | 384 well | 200,000 | 2011[38] |
| S. cerevisiae | Activate human HSF1 | Growth, OD600 | 96 well | 10,440 | 2010[35] |
| S. cerevisiae | Modify Rapamycin | Growth, OD600 | 384 well | 50,729 | 2007[33] |
| PC12 cells | Polyglutamine levels | GFP quantitation | 96 well | 37,000 | 2006[16] |
Despite these caveats, α-syn toxicity has been assayed with viability or membrane permeabilization assays [11,12,13]. In addition to toxicity, α-syn oligomerization, which is thought to be a key feature of α-syn toxicity, can be monitored in cell-based assays. Expressing α-syn from separate constructs with a split GFP results in α-syn oligomerization and reconstitution of GFP, as well as modest toxicity [14,15]. In these systems, small molecule activity has been demonstrated for AGK2, a Sirt-1inhibitor [11] and B2, a compound that increases inclusion formation [16]. Hsp90 inhibitors also reduced α-syn oligomerization and cytotoxicity, consistent with previous experiments in flies and cells [13,14,17]. A large-scale screen, however, has not yet been reported against α-syn toxicity or oligomerization.
The toxicity elicited by α-syn is exquisitely dose-dependent [3,18], suggesting that reducing α-syn levels in vivo may be a potential therapeutic avenue. Three recent screens assayed for compounds that modulated translational regulation of α-syn through a unique 5′ UTR structure [19]. The first set of large scale screens looked for both activators and inhibitors of 5′ UTR-dependent expression (Figure 2A,B) [20,21]. An α-syn 5′ UTR luciferase construct was introduced into neuroglioma cells and modulating activity was interrogated with over 300K compounds. Indeed, two probes that activated (ML163) or inhibited (ML150) activity were identified and confirmed to modulate α-syn protein levels in cells [20,21]. A similar but smaller screen of 720 natural products identified the glycoside, Strophanthidine as a strong hit (Figure 2C) [22]. Together, these screens identified compounds that specifically down-regulate α-syn expression and may provide a direct means of ameliorating the apparent dose-dependent α-syn toxicity in PD [21,22]. Future experiments both in neurons and animal models will help determine if further pursuit of their therapeutic potential is warranted.
Figure 2. Structures of compounds derived from recent PD-relevant HT screens.
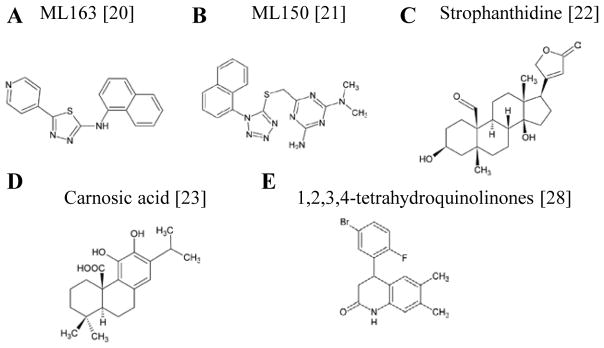
Structures of compounds identified in the last two years. (A,B) Compounds identified in a reporter-based cell culture screen modulate α-syn expression via its unique 5′ UTR structure. (C) A screen for natural products similarly identified Strophanthidine as a specific modulator of α-syn’s 5′ UTR translational regulation. (D) Carnosic acid, a compound identified by its ability to rescue cells from rotenone toxicity. (E) A series of tetrahydroquinolinones identified from a yeast model of α-syn toxicity were validated in multiple neuronal assays. Bracketed numbers indicate references.
Rotenone toxicity also provides a screenable phenotype whereby the toxin can be titrated to a desired intermediate level. Indeed, a small scale screen of rotenone toxicity in SH-SY5Y cells identified carnosic acid as a suppressor of toxicity (Figure 2D) [23]. Efficacy was confirmed using high-content imaging to assess apoptotic cells. Although only a secondary assay in this screen, the use of high-content imaging highlights its potential in high-throughput screens [24]. High-content imaging screening (HCI) utilizes visualization and quantitation of multiple cellular parameters by automated microscopy platforms and software to provide a rich data set for each compound. HCS, though more technically challenging than a live-dead assay, may be particularly useful in screens with mixed cell populations (e.g., primary neuronal cultures), where overt cellular toxicity is not readily evident, or where one non-cell autonomous effects are evident (e.g., glia and neurons). No PD screens have yet employed this approach.
Yeast as a screening platform for chemical suppressors of α-synuclein toxicity
A yeast model of α-syn toxicity is a robust HTS platform with potential for identifying therapeutic leads. In exploiting the dose-dependence of α-syn toxicity, yeast was engineered to express α-syn at different levels to cause clear dose-dependent α-syn foci formation (Figure 3A) and toxicity [18] (Figure 3B). Whereas nontoxic levels of α-syn localized to the plasma membrane, expressing toxic levels of α-syn caused the accumulation of α-syn foci and stalled vesicles [18,25]. Genetic overexpression screens of α-syn toxicity directly implicated disruption of vesicle trafficking, mitochondrial dysfunction, and involvement of manganese in α-syn toxicity, which were all confirmed in multiple neuronal models [26,27,28]. In one example, a yeast manganese pump, YPK9 (the homolog of human PARK9), was identified as a suppressor of α-syn toxicity, thereby creating a previously unappreciated link between manganese homeostasis, α-syn, and parkinsonism. These conserved mechanisms of toxicity in yeast highlight that this simple organism possesses the cellular functions required to model α-syn toxicity and identify new pathways and relationships through phenotypic screens. Therefore, drugs that alleviate α-syn toxicity in yeast may target disease-relevant proteins and pathways. Indeed, several major drugs have conserved targets in yeast, including statins, methotrexate, and omeprazole, highlighting evolutionary conservation among organisms (Figure 3C).
Figure 3. Yeast α-synuclein model as a small molecule screening platform.
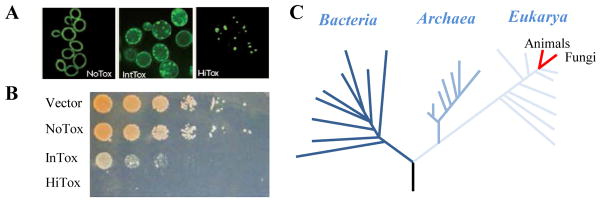
(A) Three yeast strains expressing increasing amounts of α-syn-GFP exhibit a dose-dependent increase in intracellular foci formation and loss of plasma membrane localization. (B) The same three yeast strains exhibit an increase in toxicity with increase α-syn dose. (C) Basic protein homeostasis machinery is conserved between yeast and man, allowing yeast-based screening to provide insights into human disease.
Yeast is also exceptionally easy to genetically manipulate, grow, and monitor during HT screening where a simple optical density reading reflects the rescue of toxicity. This approach is also cost effective compared to expensive luciferase-based viability assays (though luciferase-based ATP assays are quite sensitive in yeast and are amenable to HTS). All that is required is that a compound rescues growth of a strain expressing a toxic protein. Importantly, yeast-based screens typically have robust Z′ factors of 0.5 to 0.8, which enables statistically significant scoring, and can be performed in only a couple of weeks. Compound activity, however, must be validated in mammalian neuronal systems. Upon validation, the power of yeast genetics can be used to study mechanisms of action [29].
Two screens targeting α-syn toxicity have been performed. First, a small scale screen identified a group of known protective flavonoids – quercetin and (−)-epigallocatechin gallate (EGCG) – that protected α-syn-expressing yeast in the presence of iron (~10,000 compounds) [30]. In a second larger scale screen, a class of structurally related 1,2,3,4-tetrahydroquinolinones rescued yeast from α-syn toxicity (~115,000 compounds; Figure 2E). These compounds reversed vesicle trafficking and mitochondrial defects [28]. Importantly, their confirmed efficacy in neuronal α-syn toxicity models argues for a highly conserved drug target. Interestingly, these compounds also rescued rotenone toxicity in neuronal cultures, supporting a deep-rooted activity linking rotenone and α-syn toxicities.
Yeast are also suitable to exploring novel classes of molecules, such as cyclic peptides (CPs) [31]. CP selections exploit the ability of yeast to express a plasmid-derived self-splicing intein that liberates a small peptide as a CP. These selections can assess the activity of over 107 CPs, roughly 10–100 times the size of a typical small molecule screen. The CP DNA sequence can be randomized in a plasmid-encoded library, the library transformed en masse into α-syn expressing yeast, and clones that express a protective CP construct selected and structurally examined through mutagenesis [32]. Again, CPs were validated in neuronal models of α-syn toxicity. Although not a quintessential ‘drug-like’ molecule, CPs may be useful to identify new drug targets and characterize the effects of modulating its target. It is likely that these CPs sample a different chemical space from traditional small molecules and thus may function in unique ways.
Small molecules from other screens with potential for suppressing PD-relevant toxicities
Screens not directly aimed at ameliorating PD-relevant toxicities may also identify small molecules that target potentially relevant proteins or pathways. For example, screens against basic protein homeostasis may provide protection in the context of PD-related toxicities. In one example, inducers of autophagy in yeast also functioned in mammalian cells to rescue polyglutamine toxicity and induce clearance of an A53T α-syn mutant protein [33,34]. In addition, a yeast screen for compounds that activate human HSF1 (master transcriptional regulator of the heat-shock response) and the heat-shock response identified compounds that protected cells from polyglutamine toxicity [35]. This compound may prove efficacious in α-syn toxicity models where heat-shock induction is known to ameliorate toxicity [36,37]. Screening of other models may also yield compounds protective of α-syn toxicity. For example, B2, a compound identified from screen for polyglutamine modulators, similarly rescued α-syn-expressing cells [16]. Finally, a recent screen of a TDP-43 yeast model identified multiple 8-hydroxyquinolines, some of which also protected the α-syn models [38]. Together, these screens highlight that compounds identified from alternative sources may ultimately provide potential benefit for the underlying toxicities associated with PD and that phenotypic approaches in model organisms can reveal these connections.
Untapped Models for Parkinson’s Disease drug screens
Surprisingly few in vivo screens have been performed when considering the prevalence and burden of PD. Proof of principle experiments have been reported for some models; however, their true potential has yet to be realized. For example, the soil nematode C. elegans, with its well-defined development, relative ease of growth and manipulation, and recapitulation of PD-related phenotypes, may also be amenable to HT drug screens [39]. Worms expressing α-syn recapitulate neuronal toxicity and have validated several drugs and genetic modifiers identified in yeast [26,28]. In pharmacological models, small scale screens have identified compounds protective against 6-OHDA and MPP+ toxicity [40,41]. However, large scale screens have yet to be realized. Technological advances, such as microfluidics, may help spur progress in establishing screening paradigms [42]. Zebrafish also offers an intact animal model that is amenable to drug treatments and screening [43,44]. While an α-syn zebrafish model has yet to be established, transgenic fish expressing Tau are amenable to drug treatments and likely screening [43]. At the very least, these genetically tractable animal models are strong secondary screening options for filtering primary screen hits from cell-based assays for efficacy in an intact animal.
There is also considerable promise in using patient-derived iPSCs to study mechanisms of toxicity [45,46,47,48]. These cell lines generated from patients with sporadic or familial PD can be differentiated into mixed neuronal cultures enriched in DA neurons. Importantly, new targeted-recombination technologies enable the correction of disease alleles in PD patient cell lines such that an isogenic control line is available to compare potentially subtle phenotypes [45]. It is likely that phenotypes from patient-derived neurons will have subtle phenotypes. High-content screening (e.g., Cellomics) may help address these limitations by quantifying subtle cellular alterations rather than cell death. Although this approach is in its infancy, differentiated iPSCs may eventually provide a screenable platform of a disease-relevant cell type.
Conclusions
Our ever-aging population will continue to increase the number of patients suffering from age-related diseases, such as PD. Despite this trend, there are no disease-modifying agents on the market for most neurodegenerative diseases. Symptom management (e.g., with L-dopamine), and not disease modification, continues to be the focus of most therapeutics [9]. The limited number of published phenotypic high-throughput drug screens highlights the intrinsic difficulty in establishing robust, meaningful PD models amenable to screening. Yeast, despite its obvious differences from neuronal models, has actually provided potentially useful lead compounds against α-syn toxicity. Subsequent to discovery, there is a considerable bottleneck in generating the most effective probes as most academic labs lack the medicinal chemistry capabilities for carrying out detailed structure-activity relationships and compound optimization. This requires appropriate partnering – either through academic collaboration or biotech/pharma licensing – so that the most effective, bioavailable molecule can be pursued. Despite some of these limitations, the types of screens possible in academia are free from some the risk-benefit calculations made by the pharmaceutical industry [49]. Our desire to truly investigate the underlying biology in the absence of a need to develop a product, may, in the end, ultimately uncover new chemical entities that are both informative as biological probes and promising as therapeutic agents. Innovative approaches such as those described here hold promise for bridging the gap between discovery and therapeutic advancements. Although the distance between small molecule discoveries in vivo and an actual lead compound in a clinical trial is great, the grave impact on human patients requires that we make the effort to tackle this difficult task.
Acknowledgments
We thank members of the Lindquist lab for helpful comments on the manuscript. DFT was funded by a Ruth L. Kirschstein National Research Service Award Fellowship (NS614192). SL is an investigator of the Howard Hughes Medical Institute.
List of abbreviations
- PD
Parkinson’s Disease
- α-syn
α-synuclein
- HT
high throughput
- NME
new molecular entity
- S/B
signal to background ratio
- MPTP/MPP+
1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine
- MTT
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- HCI
high content imaging
- CP
cyclic peptide
- TDP-43
TAR DNA-binding Protein – 43 kDa
- 6-OHDA
6-hydroxydomapine
- iPSCs
induced pluripotent stem cells
- DA
dopaminergic
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, et al. Alpha-synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 2.Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 3.Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, et al. alpha-Synuclein locus triplication causes Parkinson’s disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 4.Li W, Lesuisse C, Xu Y, Troncoso JC, Price DL, et al. Stabilization of alpha-synuclein protein with aging and familial parkinson’s disease-linked A53T mutation. J Neurosci. 2004;24:7400–7409. doi: 10.1523/JNEUROSCI.1370-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu Y, Kordower JH. Age-associated increases of alpha-synuclein in monkeys and humans are associated with nigrostriatal dopamine depletion: Is this the target for Parkinson’s disease? Neurobiol Dis. 2007;25:134–149. doi: 10.1016/j.nbd.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 6.Martin I, Dawson VL, Dawson TM. Recent advances in the genetics of Parkinson’s disease. Annu Rev Genomics Hum Genet. 2011;12:301–325. doi: 10.1146/annurev-genom-082410-101440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hisahara S, Shimohama S. Toxin-induced and genetic animal models of Parkinson’s disease. Parkinsons Dis. 2010;2011:951709. doi: 10.4061/2011/951709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dawson TM, Ko HS, Dawson VL. Genetic animal models of Parkinson’s disease. Neuron. 2010;66:646–661. doi: 10.1016/j.neuron.2010.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meissner WG, Frasier M, Gasser T, Goetz CG, Lozano A, et al. Priorities in Parkinson’s disease research. Nat Rev Drug Discov. 2011;10:377–393. doi: 10.1038/nrd3430. [DOI] [PubMed] [Google Scholar]
- 10.Swinney DC, Anthony J. How were new medicines discovered? Nat Rev Drug Discov. 2011;10:507–519. doi: 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- 11.Outeiro TF, Kontopoulos E, Altmann SM, Kufareva I, Strathearn KE, et al. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson’s disease. Science. 2007;317:516–519. doi: 10.1126/science.1143780. [DOI] [PubMed] [Google Scholar]
- 12.Zhao DL, Zou LB, Zhou LF, Zhu P, Zhu HB. A cell-based model of alpha-synucleinopathy for screening compounds with therapeutic potential of Parkinson’s disease. Acta Pharmacol Sin. 2007;28:616–626. doi: 10.1111/j.1745-7254.2007.00539.x. [DOI] [PubMed] [Google Scholar]
- 13.McLean PJ, Klucken J, Shin Y, Hyman BT. Geldanamycin induces Hsp70 and prevents alpha-synuclein aggregation and toxicity in vitro. Biochem Biophys Res Commun. 2004;321:665–669. doi: 10.1016/j.bbrc.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 14.Putcha P, Danzer KM, Kranich LR, Scott A, Silinski M, et al. Brain-permeable small-molecule inhibitors of Hsp90 prevent alpha-synuclein oligomer formation and rescue alpha-synuclein-induced toxicity. J Pharmacol Exp Ther. 2010;332:849–857. doi: 10.1124/jpet.109.158436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Outeiro TF, Putcha P, Tetzlaff JE, Spoelgen R, Koker M, et al. Formation of toxic oligomeric alpha-synuclein species in living cells. PLoS One. 2008;3:e1867. doi: 10.1371/journal.pone.0001867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bodner RA, Outeiro TF, Altmann S, Maxwell MM, Cho SH, et al. Pharmacological promotion of inclusion formation: a therapeutic approach for Huntington’s and Parkinson’s diseases. Proc Natl Acad Sci U S A. 2006;103:4246–4251. doi: 10.1073/pnas.0511256103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Auluck PK, Bonini NM. Pharmacological prevention of Parkinson disease in Drosophila. Nat Med. 2002;8:1185–1186. doi: 10.1038/nm1102-1185. [DOI] [PubMed] [Google Scholar]
- 18.Outeiro TF, Lindquist S. Yeast cells provide insight into alpha-synuclein biology and pathobiology. Science. 2003;302:1772–1775. doi: 10.1126/science.1090439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedlich AL, Tanzi RE, Rogers JT. The 5′-untranslated region of Parkinson’s disease alpha-synuclein messengerRNA contains a predicted iron responsive element. Mol Psychiatry. 2007;12:222–223. doi: 10.1038/sj.mp.4001937. [DOI] [PubMed] [Google Scholar]
- 20.Ross NT, Metkar SR, Le H, Burbank J, Cahill C, et al. Identification of a small molecule that selectively activates alpha-synuclein translational expression. 2010. [PubMed] [Google Scholar]
- 21.Ross NT, Metkar SR, Le H, Burbank J, Cahill C, et al. Identification of a small molecule that selectively inhibits alpha-synuclein translational expression. 2010. [PubMed] [Google Scholar]
- 22.Rogers JT, Mikkilineni S, Cantuti-Castelvetri I, Smith DH, Huang X, et al. The alpha-synuclein 5′ untranslated region targeted translation blockers: anti-alpha synuclein efficacy of cardiac glycosides and Posiphen. J Neural Transm. 2011;118:493–507. doi: 10.1007/s00702-010-0513-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoon IS, Au Q, Barber JR, Ng SC, Zhang B. Development of a high-throughput screening assay for cytoprotective agents in rotenone-induced cell death. Anal Biochem. 2010;407:205–210. doi: 10.1016/j.ab.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 24.Abraham VC, Taylor DL, Haskins JR. High content screening applied to large-scale cell biology. Trends Biotechnol. 2004;22:15–22. doi: 10.1016/j.tibtech.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 25.Gitler AD, Bevis BJ, Shorter J, Strathearn KE, Hamamichi S, et al. The Parkinson’s disease protein alpha-synuclein disrupts cellular Rab homeostasis. Proc Natl Acad Sci U S A. 2008;105:145–150. doi: 10.1073/pnas.0710685105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooper AA, Gitler AD, Cashikar A, Haynes CM, Hill KJ, et al. Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson’s models. Science. 2006;313:324–328. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gitler AD, Chesi A, Geddie ML, Strathearn KE, Hamamichi S, et al. Alpha-synuclein is part of a diverse and highly conserved interaction network that includes PARK9 and manganese toxicity. Nat Genet. 2009;41:308–315. doi: 10.1038/ng.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Su LJ, Auluck PK, Outeiro TF, Yeger-Lotem E, Kritzer JA, et al. Compounds from an unbiased chemical screen reverse both ER-to-Golgi trafficking defects and mitochondrial dysfunction in Parkinson’s disease models. Dis Model Mech. 2010;3:194–208. doi: 10.1242/dmm.004267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoon S, Smith AM, Wallace IM, Suresh S, Miranda M, et al. An integrated platform of genomic assays reveals small-molecule bioactivities. Nat Chem Biol. 2008;4:498–506. doi: 10.1038/nchembio.100. [DOI] [PubMed] [Google Scholar]
- 30.Griffioen G, Duhamel H, Van Damme N, Pellens K, Zabrocki P, et al. A yeast-based model of alpha-synucleinopathy identifies compounds with therapeutic potential. Biochim Biophys Acta. 2006;1762:312–318. doi: 10.1016/j.bbadis.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 31.Scott CP, Abel-Santos E, Wall M, Wahnon DC, Benkovic SJ. Production of cyclic peptides and proteins in vivo. Proc Natl Acad Sci U S A. 1999;96:13638–13643. doi: 10.1073/pnas.96.24.13638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kritzer JA, Hamamichi S, McCaffery JM, Santagata S, Naumann TA, et al. Rapid selection of cyclic peptides that reduce alpha-synuclein toxicity in yeast and animal models. Nat Chem Biol. 2009;5:655–663. doi: 10.1038/nchembio.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarkar S, Perlstein EO, Imarisio S, Pineau S, Cordenier A, et al. Small molecules enhance autophagy and reduce toxicity in Huntington’s disease models. Nat Chem Biol. 2007;3:331–338. doi: 10.1038/nchembio883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang J, Zhu H, Haggarty SJ, Spring DR, Hwang H, et al. Finding new components of the target of rapamycin (TOR) signaling network through chemical genetics and proteome chips. Proc Natl Acad Sci U S A. 2004;101:16594–16599. doi: 10.1073/pnas.0407117101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neef DW, Turski ML, Thiele DJ. Modulation of heat shock transcription factor 1 as a therapeutic target for small molecule intervention in neurodegenerative disease. PLoS Biol. 2010;8:e1000291. doi: 10.1371/journal.pbio.1000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Auluck PK, Chan HY, Trojanowski JQ, Lee VM, Bonini NM. Chaperone suppression of alpha-synuclein toxicity in a Drosophila model for Parkinson’s disease. Science. 2002;295:865–868. doi: 10.1126/science.1067389. [DOI] [PubMed] [Google Scholar]
- 37.Yeger-Lotem E, Riva L, Su LJ, Gitler AD, Cashikar AG, et al. Bridging high-throughput genetic and transcriptional data reveals cellular responses to alpha-synuclein toxicity. Nat Genet. 2009;41:316–323. doi: 10.1038/ng.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tardiff DF, Tucci ML, Caldwell KA, Caldwell GA, Lindquist S. Different 8-Hydroxyquinolines Protect Models of TDP-43 Protein, alpha-Synuclein, and Polyglutamine Proteotoxicity through Distinct Mechanisms. J Biol Chem. 2012;287:4107–4120. doi: 10.1074/jbc.M111.308668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harrington AJ, Hamamichi S, Caldwell GA, Caldwell KA. C. elegans as a model organism to investigate molecular pathways involved with Parkinson’s disease. Dev Dyn. 2010;239:1282–1295. doi: 10.1002/dvdy.22231. [DOI] [PubMed] [Google Scholar]
- 40.Marvanova M, Nichols CD. Identification of neuroprotective compounds of caenorhabditis elegans dopaminergic neurons against 6-OHDA. J Mol Neurosci. 2007;31:127–137. doi: 10.1385/jmn/31:02:127. [DOI] [PubMed] [Google Scholar]
- 41.Braungart E, Gerlach M, Riederer P, Baumeister R, Hoener MC. Caenorhabditis elegans MPP+ model of Parkinson’s disease for high-throughput drug screenings. Neurodegener Dis. 2004;1:175–183. doi: 10.1159/000080983. [DOI] [PubMed] [Google Scholar]
- 42.Samara C, Rohde CB, Gilleland CL, Norton S, Haggarty SJ, et al. Large-scale in vivo femtosecond laser neurosurgery screen reveals small-molecule enhancer of regeneration. Proc Natl Acad Sci U S A. 2010;107:18342–18347. doi: 10.1073/pnas.1005372107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paquet D, Bhat R, Sydow A, Mandelkow EM, Berg S, et al. A zebrafish model of tauopathy allows in vivo imaging of neuronal cell death and drug evaluation. J Clin Invest. 2009;119:1382–1395. doi: 10.1172/JCI37537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bowman TV, Zon LI. Swimming into the future of drug discovery: in vivo chemical screens in zebrafish. ACS Chem Biol. 2010;5:159–161. doi: 10.1021/cb100029t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soldner F, Laganiere J, Cheng AW, Hockemeyer D, Gao Q, et al. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell. 2011;146:318–331. doi: 10.1016/j.cell.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, et al. Parkinson’s disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nguyen HN, Byers B, Cord B, Shcheglovitov A, Byrne J, et al. LRRK2 mutant iPSC-derived DA neurons demonstrate increased susceptibility to oxidative stress. Cell Stem Cell. 2011;8:267–280. doi: 10.1016/j.stem.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Devine MJ, Ryten M, Vodicka P, Thomson AJ, Burdon T, et al. Parkinson’s disease induced pluripotent stem cells with triplication of the alpha-synuclein locus. Nat Commun. 2011;2:440. doi: 10.1038/ncomms1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Munos BH, Chin WW. How to revive breakthrough innovation in the pharmaceutical industry. Sci Transl Med. 2011;3:89cm16. doi: 10.1126/scitranslmed.3002273. [DOI] [PubMed] [Google Scholar]