Summary
NK-cell killing requires both the expression of activating receptor ligands and low MHC class I expression by target cells. Here we demonstrate that the expression of any of the murine ligands for the NK-cell activating receptor NKG2D results in a concomitant reduction in MHC class I expression. We show this both in tumor cell lines and in vivo. NK-cell lysis is enhanced by the decrease in MHC class I expression, suggesting the change is biologically relevant. These results demonstrate that NKG2D ligand expression on target cells not only allows for activating receptor recognition, but also actively reduces expression of the inhibitory ligand, MHC class I, leading to enhanced recognition and killing by NK cells.
Keywords: NK cells, NKG2D ligands, MHC class I
Introduction
NK cells play an important role in innate immunity against viral infection. These cells are among the first to respond to infection and are critical for controlling viral infection while adaptive immunity is generated [1]. NK-cell activation is determined by the balance of signals received through both activating and inhibitory receptors [2, 3]. Among the activating receptors constitutively expressed by both human and mouse NK cells is NKG2D [4]. Multiple ligands exist for NKG2D, all of which are structurally related to MHC class I molecules. These ligands are MICA/MICB and the RAET1 family members in the human, and RAE1α–ε, MULT-1 and the H60 family in the mouse (reviewed in [5]). NKG2D ligands are generally not expressed by healthy cells, but are often up-regulated under conditions of cellular stress, such as viral infection and cellular transformation [6].
The NK-cell inhibitory receptors include members of the killer immunoglobulin receptor (KIR) family in the human and the Ly49 family in the mouse [3]. The ligands for these inhibitory receptors are MHC class I molecules. Responses against healthy cells is inhibited due to the high endogenous expression of MHC class I proteins on all cells. Since MHC class I is the recognition element for CD8+ T cells, a common immune evasion strategy employed by viruses is the down-regulation of MHC class I expression. However, this decreased MHC class I expression dampens the signals transduced by NK-cell inhibitory receptors, enhancing NK-cell recognition [3]. If a ligand for an activating receptor, such as NKG2D, is coordinately induced, NK-cell killing and cytokine secretion occur.
Multiple studies demonstrate that engagement of NKG2D by its ligands leads to NK cell activation [4]. The presence of NKG2D ligands is thought to enhance activating signals, tipping the balance between activating versus inhibitory signals towards activation. In the current study, we found that expression of murine NKG2D ligands led to decreased MHC class I surface expression in multiple cell types both in vitro and in vivo. These results demonstrate that expression of NKG2D ligands by target cells can enhance NK-cell killing by two mechanisms: first, by generating an activating signal through the engagement of NKG2D, and second, by decreasing the inhibitory signal of MHC class I.
Results and Discussion
Expression of the NKG2D ligand RAE1ε results in decreased MHC class I surface expression in multiple cell lines
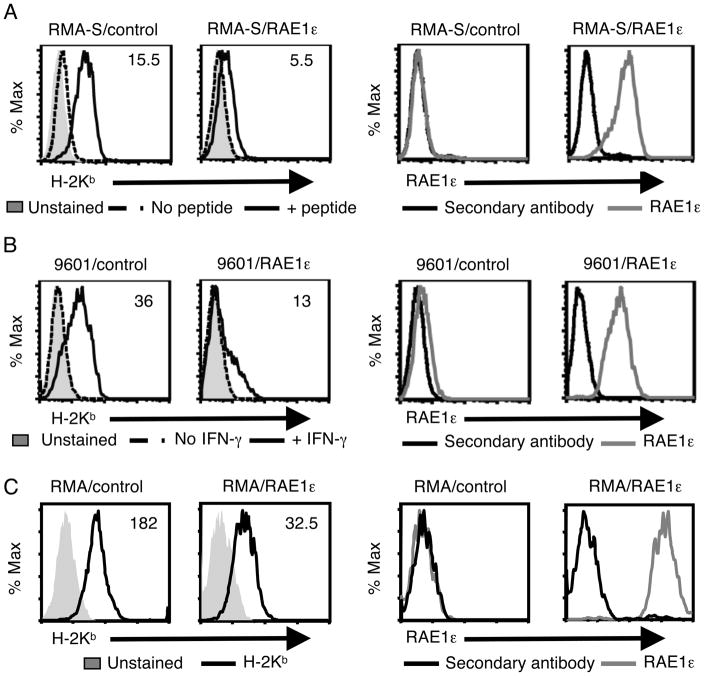
To test the effect of NKG2D ligand expression on target cells, we first transduced the NK cell target cell line RMA-S (murine lymphoma) with a bicistronic vector containing the cDNA for RAE1ε and EGFP (RMA-S/RAE1ε) or EGFP alone (RMA-S/control) (Fig. 1A and Supporting Information Fig. 1A). Due to a deficiency in the TAP complex in RMA-S cells [7], a low level of the MHC class I molecule H-2Kb was detected on the surface of the RMA-S/control cells that was upregulated by culture with an exogenous H-2Kb-binding peptide. In contrast, the addition of peptide to RMA-S/RAE1ε cells, which did not alter RAE1ε expression (data not shown), resulted in little increase in MHC class I expression. Analysis of cell lines with variable RAE1ε expression demonstrated an inverse relationship between RAE1ε and MHC class I expression (Supporting Information Fig. 2). However, Thy-1 and ICAM-1 expression levels were similar on RMA-S/control and RMA-S/RAE1ε cells, suggesting RAE1ε expression did not lead to a generalized downregulation of membrane proteins (data not shown). Similar results were seen with the other MHC class I molecule expressed in these cells, H-2Db, as well as in RMA-S/RAE1ε cells generated by transfection rather than viral transduction (Supporting Information Fig. 3A and data not shown). These results demonstrate that RAE1ε expression in RMA-S cells inhibited MHC class I surface expression.
FIGURE 1. Ectopic expression of RAE1ε in tumor cell lines results in decreased MHC class I surface expression.
(A) The RMA-S/Control and RMA-S/RAE1ε cell lines were cultured with or without SIINFEKL peptide (1μM) for 16 hours, stained with a H-2Kb-specific (left) or RAE1ε-specific or secondary (right) antibody and analyzed by flow cytometry. (B) The 9601/control and 9601/RAE1ε cell lines were cultured with or without IFN-γ (1000U/ml) for 48 h, stained with a H-2Kb-specific (left) or RAE1ε-specific or secondary (right) antibody and analyzed by flow cytometry. (C) The RMA/control and RMA/RAE1ε cell lines stained with a H-2Kb-specific (left) or RAE1ε-specific or secondary (right) antibody and analyzed by flow cytometry. The mean fluorescence intensities (MFIs) of H-2Kb staining are shown. The data shown are representative of greater than ten experiments with 3 replicates in each experiment.
To determine whether MHC class I was similarly affected in another cell line, we tested the effect of RAE1ε expression in a fibrosarcoma cell line generated by methylcholanthrene treatment of C57BL/6 mice, 9601. This cell line has little MHC class I expression, but the expression can be up-regulated with IFN-γ treatment. 9601/control (EGFP) cells had a low surface expression of H-2Kb and H-2Db that was increased following 48 hours culture with IFN-γ (Fig. 1B, Supporting Information Fig. 1B, and Supporting Information Fig. 3B). In contrast, 9601/RAE1ε cells exhibited a much lower level of H2-Kb and H-2Db expression even when incubated with IFN-γ, which did not alter RAE1 expression (Fig. 1B, Supporting Information Fig. 1B, and Supporting Information Fig. 3B). These data demonstrate that, similarly to what was seen with RMA-S cells, RAE1ε expression led to decreased MHC class I surface expression on this fibrosarcoma cell line.
Next, to rule out the possibility that RAE1ε could only function to inhibit the induction of MHC class I surface expression on cells with normally low expression, we tested the effect of RAE1ε expression in a cell line with normal MHC class I surface expression by generating RMA/control (EGFP) and RMA/RAE1ε cell lines (Fig. 1C and Supporting Information Fig. 1C). Similar to what was seen in the RMA-S and 9601 cell lines, expression of RAE1ε in RMA cells resulted in decreased surface MHC class I expression.
Ubiquitous RAE1ε expression results in decreased MHC class I expression in vivo
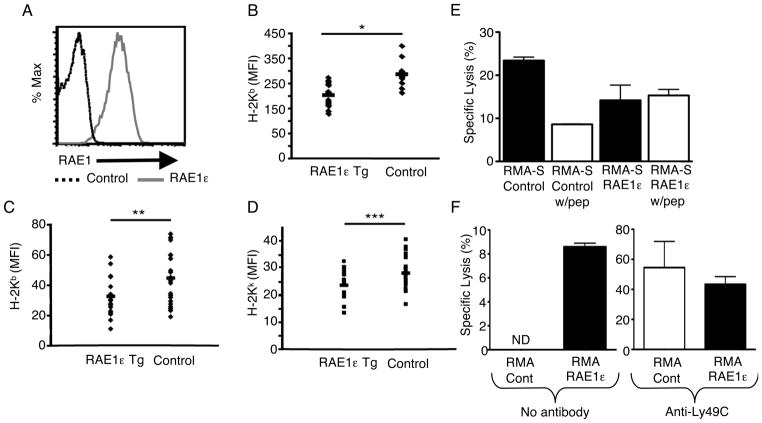
To determine whether RAE1ε expression had a similar effect on MHC class I expression in vivo, we generated mice that express RAE1ε ubiquitously on a C57BL/6 background. This was done by inserting a transgene containing the RAE1ε cDNA driven by the chicken β-actin promoter. RAE1ε is easily detectable in all PBMCs as well as cells in all tissues examined (Fig. 2A and data not shown). PBMCs from these mice expressed reduced H-2-Kb and H-2Db levels compared with non-transgenic mice from the same breeding colony (Fig. 2B and Supporting Information Fig. 3). This demonstrates that expression of RAE1ε can lead to the down-regulation of surface MHC class I expression on primary cells as well as tumor cell lines.
FIGURE 2. Transgenic expression of RAE1ε in vivo results in decreased MHC class I surface expression and increased NK-cell lysis.
(A) PBMCs from RAE1ε transgenic and non-transgenic (control) mice were stained with a RAE1-specific antibody and analyzed by flow cytometry. The data shown are representative of greater than twenty individual mice. (B) PBMCs from RAE1ε transgenic (n=15) and non-transgenic (n=11) mice were stained with a H-2Kb-specific antibody and analyzed by flow cytometry. (C and D) RAE1 transgenic mice were bred to B10.BR mice, and PBMCs from RAE1ε transgenic (n=18) and non-transgenic (n=21) mice were stained with H-2Kb and H-2Kk-specific antibodies and analyzed by flow cytometry. (E and F) NKG2D KO NK-cell lysis (E) against RMA-S/control and RMA-S/RAE1ε cells in the presence or absence of peptide (10μM), or (F) against RMA/control and RMA/RAE1ε, in the presence or absence of anti-Ly49C Fab fragments (200μg/ml), was measured with a standard chromium release assay at an E:T ratio of 30:1. The results are shown as mean + SD of 3 replicates and are representative of two independent experiments. *p=0.03 in a two-sided student’s t test. **p=0.02 in a two-sided student’s t test ***p=0.01 in a two-sided student’s t test. ND: Not detected.
To determine whether expression of MHC alleles of other haplotypes could be affected by RAE1 expression, we crossed the RAE1ε transgenic mice to B10.BR mice. In this way we generated RAE1ε transgenic mice bearing a mixed H-2b and H-2k haplotype. Expression of both H-2Kb and H-2Kk was decreased on PBMCs from these mice compared with non-transgenic littermates (Fig. 2C and D). These results suggest that RAE1ε expression can lead to decreased expression of MHC class I molecules of multiple haplotypes in vivo.
Decreased MHC class I expression induced by RAE1ε results in increased NK cell killing
Decreased MHC class I expression on target cells expressing RAE1ε is predicted to have functional consequences for NK cell killing of such targets. The decision for NK cell killing is made by the integration of signals received from both activating and inhibitory receptors. The murine inhibitory receptors include many members of the Ly49 family whose ligands are MHC class I molecules. RAE1 expression may decrease the inhibitory signal delivered by MHC class I in addition to delivering an activating signal via engagement with NKG2D. To test whether this was the case, we assayed the propensity of the RMA-S or RMA cell lines to be lysed by primary NK cells. Since we wanted to assess the contribution only of the decrease in MHC class I created by RAE1ε, and not the interference from its activating role, we used NK cells purified from NKG2D-deficient mice (NKG2D KO) [8].
The NKG2D KO NK cells were able to lyse the RMA-S/control and RMA/RAE1ε cell line, but not the RMA/control cell line (Fig. 2E and F). The RMA-S lysis is due to the low level of MHC class I expression on RMA-S cells and expression of an unknown activating receptor [9]. Pulsing RMA-S cells with peptide resulted in increased MHC class I expression (Supporting Information Fig. 4), and consequently significantly decreased lysis of these cells. In contrast, pulsing RMA-S/RAE1ε with exogenous peptide did not result in increased MHC class I expression (Supporting Information Fig. 4), nor did it afford these cells protection from NK cell lysis (Fig. 2E). In addition, whereas RMA/control and RMA/RAE1ε cells were lysed equally in the presence of blocking Fab fragments specific for the H-2b-specific inhibitory receptor, Ly49C, only RMA/RAE1ε cells were lysed without this inhibition. These data suggest that the decrease in MHC class I expression mediated by RAE1ε expression in the RMA-S and RMA cells enhanced the ability of NK cells to lyse these target cells due to decreased inhibitory signaling in the NK cells.
Expression of other murine NKG2D ligands results in decreased MHC class I expression
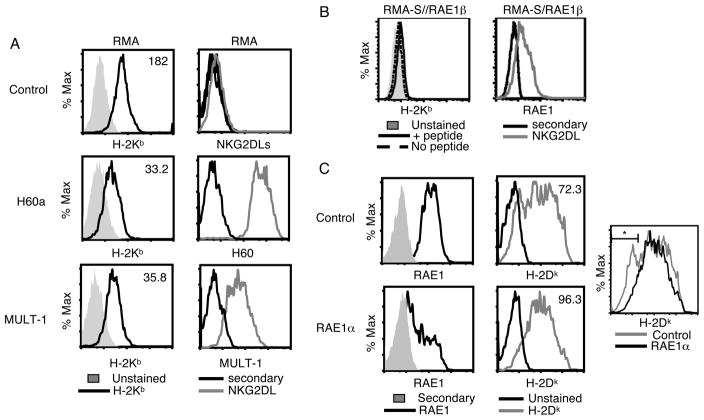
Having determined that expression of RAE1ε could decrease MHC class I expression in multiple cell types in vitro and in vivo, and could have the biological consequence of increasing NK cell lysis, we wondered whether expression of other NKG2D ligands could have a similar effect. To determine if this was the case, we determined the level of MHC class I expression on the surface of cells expressing MULT-1, H60a or RAE1β. (Fig. 3 and Supporting Information Fig. 1C). Expression of any of these NKG2D ligands resulted in decreased H-2Kb expression compared with control cells. These data suggest that the ability to decrease surface MHC class I expression may be a general property of all murine NKG2D ligands.
FIGURE 3. Multiple murine NKG2D ligands decrease MHC class I surface expression.
(A) RMA/control, RMA/H60, RMA/MULT-1 and (B) RMA-S/RAE1β (+/− 10μM peptide) cell lines were stained with a H-2Kb-specific antibody (left), H60, MULT1, and RAE1-specific antibodies, or secondary antibody alone (right). (C) RAW 264.7 cells expressing a RAE1α-specific shRNA or control shRNA were stained with a RAE-specific antibody or secondary antibody alone (left) or H-2Dk antibody (middle). An overly of the histograms of H-2Dk staining in the two cell lines is also shown (right). The MFIs of H-2Kb or H-2Dk staining are shown. *21.6% and 8.9% of cells expressing control shRNA or RAE1α-specific shRNA respectively, are present in this gate. These data are representative of two independent experiments with 3 replicates each.
Decrease in endogenous NKG2D ligand expression results in increased MHC class I expression
Finally, we wanted to determine whether endogenously expressed NKG2D ligands affected MHC class I expression similarly to ectopic NKG2D ligand expression. To do this we made use of RAW 264.7 cells, a macrophage cell line that endogenously expresses RAE1α. We transfected this cell line with a RAE1α-specific shRNA or control shRNA and compared RAE1 and MHC class I expression between the shRNA-expressing cells (Fig. 3C). Demonstrating efficient knockdown of RAE1, the cells expressing the RAE1α-specific shRNA expressed RAE1 at approximately 50% that expressed by the control transfected cells. Similar to the expression on non-transfected RAW cells (Supporting Information Fig. 5), there was broad MHC class I expression on RAW cells expressing the control shRNA. The expression of the RAE1α-specific shRNA increased MHC class I expression mainly of the cells with the lowest MHC class I expression, resulting in a 25% increase in MHC class I expression overall compared with the control cells. These results suggest that modulation of MHC class I expression is a function of endogenously expressed NKG2D ligands.
Concluding Remarks
The data shown here demonstrate that expression of multiple murine NKG2D ligands can lead to decreased MHC class I surface expression. This was seen in multiple cell lines as well as in vivo. Additionally, our studies suggest that multiple MHC class I alleles and haplotypes can be affected.
It is currently unclear how a signal is transmitted by NKG2D ligands that results in decreased surface MHC class I levels. This is particularly true for the RAE1 proteins, which unlike MULT-1 and H60, do not contain a transmembrane sequence, but rather are attached to the plasma membrane by a GPI anchor. Studies with the RMA/control and RMA/RAE1ε cells demonstrated that mRNA and total protein levels are similar in these two cell lines (data not shown and Supporting Information Fig. 6). This suggests that NKG2D ligands do not affect total MHC class I protein expression, but instead control how much class I is presented at the cell surface. However, one possible explanation for the decreased MHC class I expression in our NKG2D ligand over-expression studies may be competition for chaperones required for the efficient folding and export of class I from the endoplasmic reticulum to the plasma membrane. We plan to address the mechanism of MHC class I downregulation further in future studies.
Previous studies demonstrate that IFN-γ, which has a well-known role in increasing MHC class I expression [10], can decrease NKG2D ligand expression in both mouse and human cell lines [11–13]. In addition, NKG2D can costimulate CD8+ T cell responses [14–19], a function that requires MHC class I expression. Our results suggest this costimulatory function may be necessary to overcome decreased MHC class I expression on cells when NKG2D ligands are present. Together, these studies suggest that there is a complex relationship between MHC class I and NKG2D ligand expression.
Before now the only described role for RAE1 proteins in NK cell biology was to provide an activation signal through engagement of NKG2D. Our data establish that this ligand family has dual functionality. In addition to providing an activating signal, these ligands also decrease an inhibitiory signal via down-regulation of MHC class I expression, leading to the generation of more potent NK cell targets.
Materials and Methods
Cell line generation
The 9601 fibrosarcoma cell line was generated previously by methylcholanthrene treatment of C57BL/6 mice [20]. RAE1, MULT1 or H60a cDNA [21] was introduced into RMA-S, 9601, or RMA cells by retroviral transduction using pMXIRES-enhanced GFP [22] courtesy of T. Kitamura (University of Tokyo, Tokyo, Japan), Plat-E packaging cells and Fugene (Roche). The control cell lines were generated similarly by transduction with the empty pMXIRES-EGFP vector. RMA-S/RAE1ε and RMA/control cells were separately generated by nucleofection (Lonza). EGFP-expressing cells were enriched using a FACSVantage SE cell sorter (BD Biosciences). In all experiments, cell lines with similar GFP expression were compared. The RMA-S/RAE1β cell line [23] was generously provided by M. Colonna (Washington University, St. Louis, MO). All cell lines were cultured in IMDM supplemented with 10% FCS and antibiotics.
Antibodies, cytokine and peptide
All MHC class I-specific antibodies, H-2Kb (clone AF6-88.5), H-2Kk (Clone 36-7-5), H-2Dk (Clone 15-5-5), were purchased from BD Biosciences. Antibodies specific for RAE1ε (Clone 205001), pan-RAE1 (Clone 18610), MULT-1 (Clone 237104) and H60 (Clone 205326) were purchased from R&D Systems. Anti-Rat IgG2a was purchased from Jackson Immunoresearch. All flow cytometry anaysis was done using a FACscan (BD Biosciences). rmIFN-γ was a gift from Genentech. The SIINFEKL peptide was generously provided by Dr. P. Allen (Washington University).
Mice
All mice were housed under specific pathogen-free conditions in the Washington University animal facilities in accordance with institutional guidelines. C57BL/6 and B10.BR mice were purchased from The Jackson Laboratory. To generate RAE1ε transgenic mice, RAE1ε cDNA was cloned into the PCCALL2 vector [23] (provided by A. Nagy, SLRI, Toronto, Canada). This construct contains a chicken β actin promoter, followed by a loxP-flanked bgeo fusion gene (lacZ and neomycin resistance), and RAE1ε inserted 3′ of the 3′ loxP site. Prior to Cre excision, the lacZ reporter and neomycin-resistance gene are expressed and RAE1ε is not. Following recombination of the loxP sites by Cre, however, the bgeo gene is removed allowing for Rae1ε expression. The PCCALL2/RAE1ε construct was transfected into C57BL/6/129 embryonic stem (ES) cells by electroporation and positive clones treated with Cre recombinase. The ES cells were then microinjected into blastocysts by the Washington University School of Medicine Department of Pathology and Immunology micro-injection core. Transgenic mice were then backcrossed to C57BL/6 for 5 generations. The generation of NKG2D-deficient mice was previously described [8].
Cytolysis assay
NK cells were isolated from spleens of NKG2D-deficient mice using DX5 MicroBeads (Miltenyi Biotec). The cells were cultured in IMDM with 10% mouse serum and rhIL-2 (provided by Marina Cella, Washington University) for 5–7 days. The live cells harvested at the end of this culture were > 95% NK cells (NK1.1+ CD3−). Ly49C Fab fragments were generated from Ly49C-specific antibody (Bection Dickinson) using the Fab Preparation Kit (Thermo Scientific) following the manufacturer’s instructions. A standard chromium release assay was then performed and the supernatants read on a MicroBeta counter (PerkinElmer). The percent specific lysis was determined as: ((sample counts per minute – spontaneous counts per minute)/(maximum counts per minute – spontaneous counts per minute)) × 100.
Raw cell transfection
RAW 264.7 cells (ATCC) were transfected with pLKO.1-puro containing a RAE1α-specific or control shRNA (Sigma-Aldrich), along with pmaxGFP (Lonza) at a 1:10 ratio using an Amaxa Nucleofector (Lonza) following the manufacturer’s protocol. 48 hours later, the GFP+ cells were analyzed for RAE1 and H-2Dk expression by flow cytometry.
Supplementary Material
Acknowledgments
This work was supported by grants from the NIH (Diabetes Research and Training Center P60 DK020579 to MM and R37 AI057966 to AS) and the HHMI (AS).
Footnotes
Conflict of Interest
The authors declare no financial or commercial conflict of interest.
References
- 1.Biron CA, Nguyen KB, Pien GC, Cousens LP, Salazar-Mather TP. Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu Rev Immunol. 1999;17:189–220. doi: 10.1146/annurev.immunol.17.1.189. [DOI] [PubMed] [Google Scholar]
- 2.Pegram HJ, Andrews DM, Smyth MJ, Darcy PK, Kershaw MH. Activating and inhibitory receptors of natural killer cells. Immunol Cell Biol. 2011;89:216–224. doi: 10.1038/icb.2010.78. [DOI] [PubMed] [Google Scholar]
- 3.Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol. 2008;9:495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raulet DH. Roles of the NKG2D immunoreceptor and its ligands. Nat Rev Immunol. 2003;3:781–790. doi: 10.1038/nri1199. [DOI] [PubMed] [Google Scholar]
- 5.Champsaur M, Lanier LL. Effect of NKG2D ligand expression on host immune responses. Immunol Rev. 2010;235:267–285. doi: 10.1111/j.0105-2896.2010.00893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mistry AR, O’Callaghan CA. Regulation of ligands for the activating receptor NKG2D. Immunology. 2007;121:439–447. doi: 10.1111/j.1365-2567.2007.02652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Townsend A, Ohlen C, Foster L, Bastin J, Ljunggren HG, Karre K. A mutant cell in which association of class I heavy and light chains is induced by viral peptides. Cold Spring Harb Symp Quant Biol. 1989;54(Pt 1):299–308. doi: 10.1101/sqb.1989.054.01.038. [DOI] [PubMed] [Google Scholar]
- 8.Zafirova B, Mandaric S, Antulov R, Krmpotic A, Jonsson H, Yokoyama WM, Jonjic S, Polic B. Altered NK cell development and enhanced NK cell-mediated resistance to mouse cytomegalovirus in NKG2D-deficient mice. Immunity. 2009;31:270–282. doi: 10.1016/j.immuni.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takaki R, Watson SR, Lanier LL. DAP12: an adapter protein with dual functionality. Immunol Rev. 2006;214:118–129. doi: 10.1111/j.1600-065X.2006.00466.x. [DOI] [PubMed] [Google Scholar]
- 10.Farrar MA, Schreiber RD. The molecular cell biology of interferon-gamma and its receptor. Annu Rev Immunol. 1993;11:571–611. doi: 10.1146/annurev.iy.11.040193.003035. [DOI] [PubMed] [Google Scholar]
- 11.Bui JD, Carayannopoulos LN, Lanier LL, Yokoyama WM, Schreiber RD. IFN-dependent down-regulation of the NKG2D ligand H60 on tumors. J Immunol. 2006;176:905–913. doi: 10.4049/jimmunol.176.2.905. [DOI] [PubMed] [Google Scholar]
- 12.Schwinn N, Vokhminova D, Sucker A, Textor S, Striegel S, Moll I, Nausch N, et al. Interferon-gamma down-regulates NKG2D ligand expression and impairs the NKG2D-mediated cytolysis of MHC class I-deficient melanoma by natural killer cells. Int J Cancer. 2009;124:1594–1604. doi: 10.1002/ijc.24098. [DOI] [PubMed] [Google Scholar]
- 13.Yadav D, Ngolab J, Lim RS, Krishnamurthy S, Bui JD. Cutting edge: down-regulation of MHC class I-related chain A on tumor cells by IFN-gamma-induced microRNA. J Immunol. 2009;182:39–43. doi: 10.4049/jimmunol.182.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bauer S, Groh V, Wu J, Steinle A, Phillips JH, Lanier LL, Spies T. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 1999;285:727–729. doi: 10.1126/science.285.5428.727. [DOI] [PubMed] [Google Scholar]
- 15.Chalupny NJ, Sutherland CL, Lawrence WA, Rein-Weston A, Cosman D. ULBP4 is a novel ligand for human NKG2D. Biochem Biophys Res Commun. 2003;305:129–135. doi: 10.1016/s0006-291x(03)00714-9. [DOI] [PubMed] [Google Scholar]
- 16.Cosman D, Mullberg J, Sutherland CL, Chin W, Armitage R, Fanslow W, Kubin M, Chalupny NJ. ULBPs, novel MHC class I-related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity. 2001;14:123–133. doi: 10.1016/s1074-7613(01)00095-4. [DOI] [PubMed] [Google Scholar]
- 17.Diefenbach A, Jamieson AM, Liu SD, Shastri N, Raulet DH. Ligands for the murine NKG2D receptor: expression by tumor cells and activation of NK cells and macrophages. Nat Immunol. 2000;1:119–126. doi: 10.1038/77793. [DOI] [PubMed] [Google Scholar]
- 18.Markiewicz MA, Carayannopoulos LN, Naidenko OV, Matsui K, Burack WR, Wise EL, Fremont DH, et al. Costimulation through NKG2D enhances murine CD8+ CTL function: similarities and differences between NKG2D and CD28 costimulation. J Immunol. 2005;175:2825–2833. doi: 10.4049/jimmunol.175.5.2825. [DOI] [PubMed] [Google Scholar]
- 19.Sutherland CL, Chalupny NJ, Schooley K, VandenBos T, Kubin M, Cosman D. UL16-binding proteins, novel MHC class I-related proteins, bind to NKG2D and activate multiple signaling pathways in primary NK cells. J Immunol. 2002;168:671–679. doi: 10.4049/jimmunol.168.2.671. [DOI] [PubMed] [Google Scholar]
- 20.Kaplan DH, Shankaran V, Dighe AS, Stockert E, Aguet M, Old LJ, Schreiber RD. Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc Natl Acad Sci U S A. 1998;95:7556–7561. doi: 10.1073/pnas.95.13.7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carayannopoulos LN, Naidenko OV, Kinder J, Ho EL, Fremont DH, Yokoyama WM. Ligands for murine NKG2D display heterogeneous binding behavior. Eur J Immunol. 2002;32:597–605. doi: 10.1002/1521-4141(200203)32:3<597::aid-immu597>3.3.co;2-5. [DOI] [PubMed] [Google Scholar]
- 22.Onishi M, Kinoshita S, Morikawa Y, Shibuya A, Phillips J, Lanier LL, Gorman DM, et al. Applications of retrovirus-mediated expression cloning. Exp Hematol. 1996;24:324–329. [PubMed] [Google Scholar]
- 23.Gilfillan S, Ho EL, Cella M, Yokoyama WM, Colonna M. NKG2D recruits two distinct adapters to trigger NK cell activation and costimulation. Nat Immunol. 2002;3:1150–1155. doi: 10.1038/ni857. [DOI] [PubMed] [Google Scholar]
- 24.Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.