Abstract
The mgtCBR operon from Salmonella enterica serovar Typhimurium specifies the virulence protein MgtC, the Mg2+ transporter MgtB and the regulatory peptide MgtR. The mgtCBR transcript includes a long leader region harbouring two short open reading frames (ORFs). Translation of these ORFs is anticipated to impact the formation of particular stem-loop structures and control transcription of the coding region by an attenuation-like mechanism. We previously reported that ORF mgtM enables Salmonella to promote transcription of the mgtC and mgtB coding regions when experiencing a rise in cytoplasmic ATP levels. We now show that the proline codon-rich ORF mgtP mediates an increase in transcription of the mgtC and mgtB coding regions under conditions predicted to decrease the levels of proline-charged tRNAPro. The high ATP and low proline signals act independently in an additive form. Replacing conserved mgtP proline codons with codons specifying other amino acids abolished the response to proline limitation but had no effect on the response to ATP. Substitution of conserved adenine nucleotides in mgtM abolished the response to ATP but had no effect in the response to proline limitation. This provides a singular example of a leader mRNA with tandem attenuators responding to different signals.
Introduction
Transcription attenuation is a bacterial regulatory mechanism that entails the formation of either of two alternative based-paired RNA structures in the leader region of a transcript: one promoting transcription termination and one advancing transcription elongation into the coding region. Which RNA structure forms is determined by growth conditions that favour (or hinder) the normal coupling of transcription of the leader region with translation of a short open reading frame (ORF) located within the leader (Landick et al., 1996; Henkin and Yanofsky, 2002; Merino and Yanofsky, 2005; Grundy and Henkin, 2006; Naville and Gautheret, 2009). Normally, a single attenuator responding to a specific signal controls expression of genes involved in nutrient biosynthesis. Here we describe an unusual leader mRNA with tandem attenuators, each responding to a different signal, which dictate genetic control of an operon involved in virulence and metal homeostasis.
The mgtCBR operon from Salmonella enterica serovar Typhimurium specifies the virulence protein MgtC, which is required for survival inside macrophages (Blanc-Potard and Groisman, 1997); the P-type ATPase MgtB, which transports Mg2+ from the periplasm to the cytoplasm (Snavely et al., 1991); and the peptide MgtR, which promotes the FtsH-mediated proteolysis of MgtC (Alix and Blanc-Potard, 2008). MgtR also promotes degradation of the Mg2+ transporter MgtA (Choi et al., 2012), which is specified somewhere else in the genome (Maguire, 2006). Unlike MgtC, MgtB and MgtR are not necessary to cause a lethal infection in BALB/c mice or to survive within the macrophage-like cell line J774.1 (Blanc-Potard and Groisman, 1997; Alix and Blanc-Potard, 2008).
Expression of the mgtCBR operon is regulated at multiple levels. Transcription initiation requires the PhoP/PhoQ two-component system (Soncini et al., 1996), which is activated when bacteria experience low Mg2+ (Garcia Vescovi et al., 1996), acidic pH (Prost et al., 2007) and/or certain antimicrobial peptides (Bader et al., 2005). Interestingly, PhoP also promotes transcription of AmgR, an anti-sense RNA for the mgtC portion of the polycistronic mgtCBR message (Lee and Groisman, 2010). Transcription elongation into the coding region is controlled by the 296 nt long mRNA leader, which responds to low cytosolic Mg2+ (Cromie et al., 2006; Spinelli et al., 2008) and to an increase in cytosolic ATP levels (Lee and Groisman, 2012) by advancing transcription of the mgtC and mgtB coding regions.
The response to cytosolic ATP requires a stretch of adenine nucleotides in a region of the mRNA leader located within a short ORF designated mgtM (Lee and Groisman, 2012). The deduced amino acid sequence of mgtM is not conserved in the mRNA leader of mgtC homologues. By contrast, the presence of adenine nucleotides and their location relative to regions with the potential to adopt particular stem-loop structures are conserved in the mgtCBR leader. The mRNA levels corresponding to the mgtC and mgtB coding regions increase dramatically when Salmonella is inside macrophages. This increase, and Salmonella’s ability to cause a lethal infection in mice, is dependent, in part, on the conserved adenine nucleotides in the mgtCBR leader mediating the response to ATP (Lee and Groisman, 2012).
The Salmonella mgtA gene specifies a protein that is 50% identical to MgtB (Maguire, 2006). Like mgtB, the mgtA gene is transcribed from a PhoP-dependent promoter (Garcia Vescovi et al., 1996) and harbours a Mg2+-responding mRNA leader (Cromie et al., 2006; Spinelli et al., 2008). Unlike mgtB, the mgtA coding region is not induced inside macrophages (Lee and Groisman, 2012), and this could be due to the absence of conserved adenine nucleotides in the mgtA mRNA leader (Lee and Groisman, 2012).
We now describe the identification of a short ORF rich in proline codons in the mgtCBR mRNA leader that enables Salmonella to stimulate transcription of the mgtC and mgtB coding regions under conditions that decrease the levels of proline-charged tRNAPro. This is reminiscent of the expression behaviour of the mgtA coding region, which is also regulated by changes in proline-charged tRNAPro sensed by an unrelated proline codon-rich ORF in the mgtA leader region (Park et al., 2010). The mgtCBR leader provides a singular example of a leader mRNA with tandem attenuators responding to distinct signals: ATP and proline.
Results
The mgtCBR leader mRNA harbours a translated proline codon-rich short ORF
We analysed the mgtCBR leader region seeking sequence elements that may suggest it can sense additional signals. We found a 17-codon long ORF that includes three consecutive proline codons and is preceded by a sequence resembling a Shine–Dalgarno sequence 9 nt from the putative start codon (Fig. 1A). This ORF, designated mgtP, is located in a region of the leader RNA that has the potential to adopt two alternative secondary structures: stem-loops C and D versus stem-loop E (Fig. 1A). Because the last four mgtP codons are part of the left arm of stem-loop C, translation of the complete mgtP is predicted to hinder formation of stem-loop C and to favour formation of stem-loop E. The presence of a proline codon-rich short ORF and its location relative to the stem-loop structures just discussed are conserved in the predicted leader mRNA regions of mgtC homologues (Fig. S1).
Figure 1.
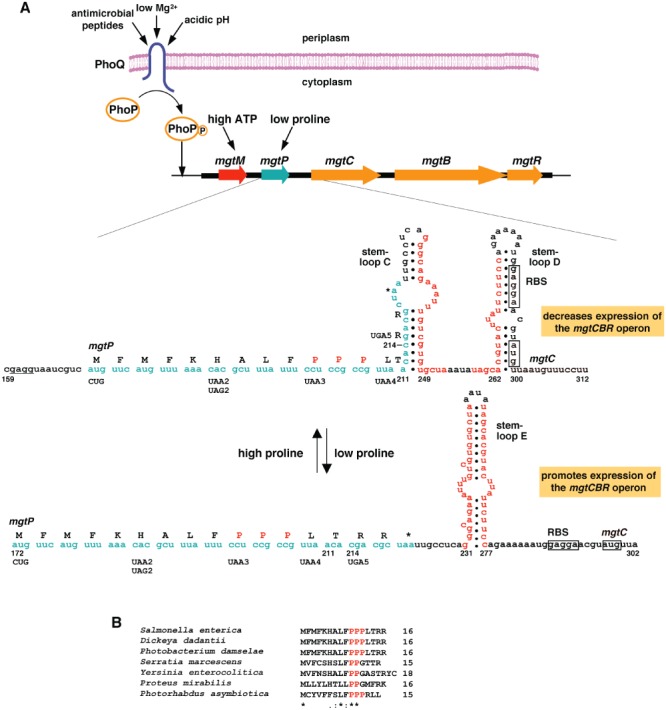
Regulation of the Salmonella mgtCBR virulence operon by the PhoP/PhoQ system and mgtCBR leader region. A. The sensor PhoQ responds to extracytoplasmic low Mg2+, acidic pH and antimicrobial peptides by promoting phosphorylation of the PhoP protein, which binds to the mgtCBR promoter resulting in transcription initiation. Transcription elongation into the coding region is controlled by the leader region, which can adopt alternative secondary structures depending on the coupling/uncoupling of transcription of the mgtCBR leader and translation of two short ORFs designated mgtM and mgtP. Which secondary structures form is determined by the cytoplasmic levels of ATP and proline. The alternative secondary structures potentially adopted by the segment of the mgtCBR leader that includes the mgtP ORF up to the second mgtC codon (i.e. stem-loops C and D versus stem-loop E) are shown with the predicted RBS for mgtP underlined. The mgtP sequences are indicated in cyan. The predicted RBS and mgtC start codon are boxed. Positions and sequences of stop codon mutations or nucleotide substitutions in the strains used in the experiments presented in Fig. 2 are indicated below the mgtP sequence. B. Alignment of the deduced amino acid sequences of mgtP in the mgtCBR leader regions from Salmonella enterica, Dikeya dadantii, Photobacterium damselae, Serratia marcescens, Yersinia enterocolitica, Proteus mirabilis, and Photorhadus asymbiotica. Sequences in red correspond to Pro codons. Asterisks correspond to positions conserved in all listed species.
We verified the formation of stem-loops C and D using in-line probing with a labelled RNA corresponding to nucleotides 196–385 [relative to the mgtC transcription start site (Lejona et al., 2003,2003; Zwir et al., 2012)]. The spontaneous RNA cleavage at regions not predicted to be part of stems differed in RNAs incubated in the presence of 1 mM versus 5 or 20 mM Mg2+. For instance, regions 1 and 4 were more accessible to cleavage at low than at high Mg2+ and the converse was true for regions 2, 3, 5 and 6 (Fig. S2). These data indicate that Mg2+ can modify the structure of this portion of the mgtCBR leader, which provides support for the genetic experiments suggesting that the mgtCBR leader functions as a Mg2+-sensing RNA (Cromie et al., 2006; Spinelli et al., 2008).
To examine whether mgtP is translated in vivo, we utilized our previously reported approach (Park et al., 2010) to determine the β-galactosidase activity produced by wild-type Salmonella harbouring a plasmid that expressed an mgtP–lacZ translational fusion and included the predicted Shine–Dalgarno sequence for mgtP. As a control, we used an isogenic derivative in which the truncated lacZ gene was placed after the mgtP stop codon. The former strain produced high levels of β-galactosidase whereas no activity was observed for the latter (Fig. S3). Taken together with our previous findings (Lee and Groisman, 2012), these results indicate that the mgtCBR leader mRNA includes two short ORFs, the translation of which is predicted to impinge the formation of particular stem-loop structures.
mgtP is part of a transcription attenuator
The possibility of the mgtCBR leader mRNA adopting alternative stem-loop structures (stem-loops C and D versus stem-loop E) in the region overlapping and adjacent to mgtP (Fig. 1A) suggested that mgtP might control transcription elongation into the mgtCBR coding region by a transcription attenuation mechanism similar to those governing expression of biosynthetic operons in enteric bacteria (Merino and Yanofsky, 2005). If this is the case, uncoupling transcription of the mgtCBR leader region and translation of mgtP might affect expression of the associated coding regions. To test this idea, we determined the β-galactosidase activity in a set of isogenic strains harbouring a lac transcriptional fusion in the chromosomal mgtC coding region and a wild-type or mutant mgtP.
A strain in which the mgtP start codon was changed from AUG to CUG produced seven times less β-galactosidase than the isogenic mgtP+ strain when grown in low Mg2+ to induce the PhoP/PhoQ system (Fig. 2A). Strains with stop codon mutations at the 6th or 10th positions of mgtP expressed similar low levels of β-galactosidase (Fig. 2A). We ascribe the low mgtC–lac expression of these mutants to a defect in mgtP translation as opposed to a structural change that ‘locked’ the mRNA leader in a low expression conformation. This is because a plasmid expressing the amber suppressor supF restored wild-type levels of mgtC–lac transcription to an mgtP mutant with an amber stop codon at the 6th position but not to one harbouring an ochre stop codon at that position (Fig. 2B). As expected, the supF-expressing plasmid had no effect on mgtC–lac transcription in a strain harbouring the wild-type mgtC leader (Fig. 2B).
Figure 2.
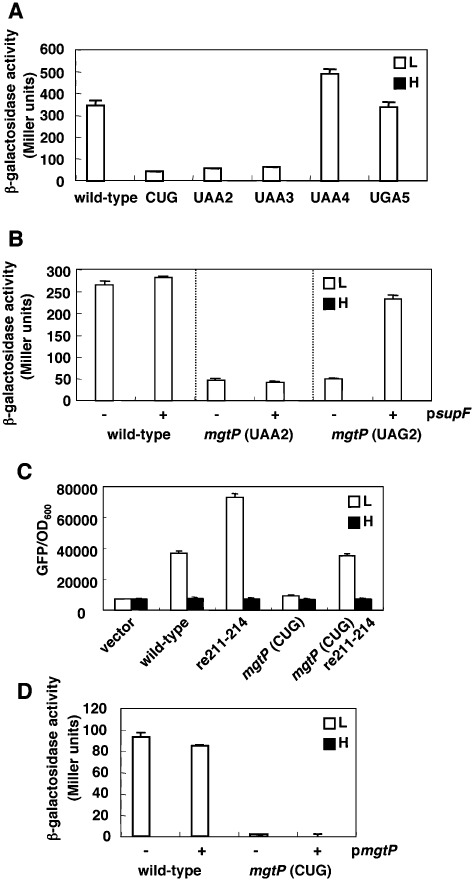
mgtCBR leader controls expression of the mgtCBR coding region by a transcription attenuation-like mechanism. A. β-Galactosidase activity (Miller units) produced by a Salmonella strain with a chromosomal mgtC–lac fusion (EG9527) and isogenic derivatives with mutation of the start codon (EG18799) or with stop codons at different positions (EG18801, EG19251, EG19272 and EG19253) in mgtP. Bacteria were grown in N-minimal media containing low (L; 10 µM) or high (H; 10 mM) Mg2+ for 4 h. Shown are the mean and SD from at least three independent experiments. B. β-Galactosidase activity (Miller units) produced by a Salmonella strain with a chromosomal mgtC-lac fusion (EG9527) harbouring either plasmid psupF or the empty vector pUH21-2lacIq or by isogenic derivatives with an amber stop codon (EG19840) or ochre stop codon (EG18801) at position 187–189 (i.e. mgtP’s 6th codon). Bacteria were grown as described above except in the presence of ampicillin (50 µg ml−1) and IPTG (0.2 mM). C. Fluorescence produced by wild-type Salmonella (14028s) harbouring a plasmid encoding a translational fusion to gfp and the wild-type mgtC leader, or derivatives with mutations that hinder stem-loop C formation (re211-214) and/or with mutation of the mgtP start codon. Bacteria were grown as described above except in the presence of ampicillin (50 µg ml−1). Shown are the mean and SD from at least three independent experiments. D. β-Galactosidase activity (Miller units) produced by a Salmonella strain with a chromosomal mgtC-lac fusion (EG9527) or an isogenic strain with mutation of the mgtP start codon (EG18799) harbouring either the plasmid vector or plasmid pmgtP. Bacteria were grown as described above except in the presence of ampicillin (50 µg ml−1) and IPTG (0.1 mM). Shown in (B) and (D) are the mean and SD from two independent experiments. For parts A, B and D, the activity was lower than the resolution of the figure following growth in high (H; 10 mM) Mg2+.
Derivatives with stop codon mutations at the 13th or 15th positions retained the expression behaviour of the strain with the wild-type leader (Fig. 2A). Given the space that a translating ribosome occupies on a transcript [12–15 nucleotides from the P site (Steitz, 1969)], mgtP translation beyond the 12th codon is anticipated to favour formation of stem-loop E. By contrast, when mgtP translation stops before the ribosome reaches the 13th mgtP codon, stem-loops C and D would form (Fig. 1), which reduces expression of the mgtCBR coding region.
To further address the role of mgtP translation in mgtC expression, we measured fluorescence in wild-type Salmonella harbouring a plasmid in which the PhoP-dependent mgtC promoter, full-length mgtCBR leader and first two codon of mgtC gene were fused in frame to the third codon of the gfp gene. Fluorescence was fivefold higher following growth in low versus high Mg2+ (Fig. 2C). This is likely due to the PhoP-dependent promoter and Mg2+-responding mRNA leader because the expression levels were similarly low in an isogenic strain carrying the plasmid vector (Fig. 2C). A strain with a plasmid derivative in which the mgtP start codon AUG was substituted for CUG displayed decreased fluorescence (Fig. 2C), in agreement with the phenotype of a strain with the equivalent mutation in the chromosomal copy of mgtP (Fig. 2A). By contrast, a strain with a plasmid derivative substituted in nucleotides 211–214 of the mgtCBR leader exhibited higher fluorescence than the strain with the plasmid harbouring the wild-type mgtCBR leader (Fig. 2C). This mutation is expected to disrupt formation of stem-loop C, favour formation of stem-loop E, and release the ribosome binding site (RBS) and mgtC start codon sequestered in stem-loop D (Fig. 1). The substitution of the 211–214 region could overcome (partially) the decrease in expression resulting from mutation of the mgtP start codon (Fig. 2C).
Finally, a plasmid carrying the mgtP ORF failed to restore normal expression to a strain with a chromsomal mgtC–lac transcriptional fusion and a mutation of the mgtP start codon, behaving like the vector control (Fig. 2D); and it had no effect on an isogenic strain with a wild-type mgtC leader (Fig. 2D). This argues against the notion of mgtP exerting its regulatory effect by specifying a trans-acting peptide. Cumulatively, the results presented here indicate that mgtP acts as a cis-regulatory element, regulating the associated mgtCBR coding region by being part of a transcription attenuator.
Intracellular proline controls expression of the mgtC and mgtB coding regions dependent on conserved mgtP proline codons
We postulated that mgtP might confer regulation by intracellular proline levels because it includes three consecutive Pro codons, which is a disproportionately high frequency for a 17-codon long ORF (McCaldon and Argos, 1988), and also because the presence of consecutive Pro codons near the base of stem-loop C is conserved in the mgtCBR leader of other species (Figs 1 and S1). Furthermore, an unrelated short ORF harbouring four proline codons in the mgtA leader mRNA mediates expression of the mgtA coding region in response to changes in the levels of cytosolic proline (Park et al., 2010). Thus, conditions that decrease the levels of cytosolic proline available to charge tRNAPro may result in the ribosome stalling at the mgtP Pro codons thereby advancing a structure that alters transcription of the mgtCBR coding region.
We determined that the mRNA corresponding to the mgtC coding region was present at fivefold higher levels in a proline auxotroph grown in the absence of proline for 45 min than when it was grown in its presence (Fig. 3A). Proline limitation specifically induced mgtC expression in a manner dependent on the mgtP Pro codons because: first, it failed to promote an increase in the mRNA levels corresponding to the phoP coding region or the mgtCBR and mgtA leader regions (Fig. 3A). However, it did induce the mgtA coding region (Fig. 3A), which was used as a positive control (Park et al., 2010). Second, mgtC mRNA levels did not increase in a leucine auxotroph subjected to leucine limitation (Fig. 3B) despite the fact that mgtP includes two leucine codons (Fig. 1). And third, substitution of the three mgtP Pro codons by Gly codons prevented mgtC induction in response to proline limitation (Fig. 3A). As expected, the latter mgtP mutant retained wild-type expression of the phoP coding region and the mgtCBR and mgtA leader regions and still induced the mgtA coding region in response to proline limitation (Fig. 3A).
Figure 3.
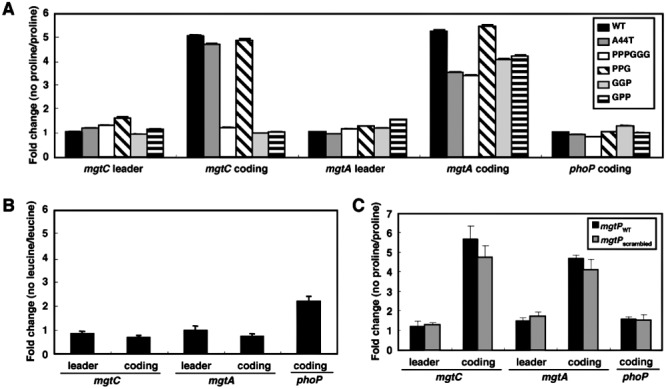
Proline limitation promotes transcription of the mgtCBR coding region in a manner dependent on conserved Pro codons in mgtP. A. Fold change in the mRNA levels of the leader regions of the mgtC and mgtA transcripts and the coding regions of the mgtC, mgtA, and phoP genes produced by a proline auxotroph harbouring either the wild-type mgtCBR leader (EL302), or derivatives where the A nucleotides at position 44–46 were substituted by Ts (A44T; EL339), or where the three mgtP Pro codons (PPPGGG; EL303), the Pro codons at the 12th (PPG; EL347), 10th and 11th (GGP; EL348) or 10th (GPP; EL349) positions were substituted by Gly codons. Bacteria were grown in N-minimal media with 500 µM Mg2+ in the presence of 1 mM proline for 1 h, and then grown for 45 min in media containing or lacking proline. Expression levels of target genes were normalized to that of 16S ribosomal RNA rrs gene. Fold change was calculated by dividing the mRNA levels of cells grown in the absence of proline by that of cells grown in the presence of proline. Shown are the mean and SD from three independent experiments. B. Fold change in the mRNA levels of the leader regions of mgtC and mgtA transcripts and the coding regions of the mgtC, mgtA, and phoP genes produced by a leucine auxotroph (EL337) under leucine limitation conditions analogous to that described above for proline limitation. Shown are the mean and SD from two independent experiments. C. Fold change in the mRNA levels of the genes listed above produced by a proline auxotroph with a wild-type mgtCBR leader (EG19886) sequence or with the mgtP sequence scrambled (EL379) following growth as described in (A). Shown are the mean and SD from two independent experiments.
Derivatives of the mgtCBR leader with substitutions of the mgtP Pro codons at the 10th position or the 10th and 11th positions failed to promote mgtC expression upon proline limitation (Fig. 3A). By contrast, a mutant with a substitution of the Pro codon at the 12th position exhibited a wild-type behaviour (Fig. 3A). This was expected given that mgtP harbours only two Pro codons in certain bacterial species (corresponding to the 10th and 11th positions; Fig. 1B). Furthermore, a strain in which the mgtP sequence was completely scrambled (except for the Pro codons at the 10th and 11th positions and the sequence required for formation of stem-loop C) still responded to proline limitation by enhancing the mRNA levels of the mgtC coding region (Fig. 3C). In sum, these experiments indicate that the mgtP Pro codons at the 10th and 11th positions are necessary, and possibly sufficient, for low cytosolic proline to induce transcription of the mgtC and mgtB coding regions.
Hyperosmotic stress promotes expression of the mgtC and mgtB coding regions
In addition to being a component of peptides and proteins, proline can function as an osmoprotectant (Csonka and Leisinger, 2007). This raised the possibility of hyperosmotic stress promoting transcription of the mgtCBR coding region by virtue of decreasing the amount of cytosolic proline available to charge tRNAPro. Indeed, when wild-type Salmonella experienced 0.3 M NaCl, the mRNA corresponding to the mgtC and mgtB coding regions increased fivefold (Fig. 4). This is similar to the induction of the mgtA coding region (Fig. 4), as we previously reported (Park et al., 2010). The increase in the mRNA levels of the mgtC and mgtB coding regions (but not that corresponding to the mgtA coding region) is dependent on the mgtP Pro codons (Fig. 4). By contrast, hyperosmotic stress had no effect on the transcript levels of the phoP coding region or the mgtCBR and mgtA leader regions (Fig. 4). As expected, the induction resulting from hyperosmotic stress was eliminated when proline was present in the media (Fig. 4). Thus, the Mg2+ transporter-specifying mgtA and mgtB genes rely on different Pro codon-rich short ORFs in their mRNA leaders to induce their respective coding regions under hyperosmotic stress.
Figure 4.
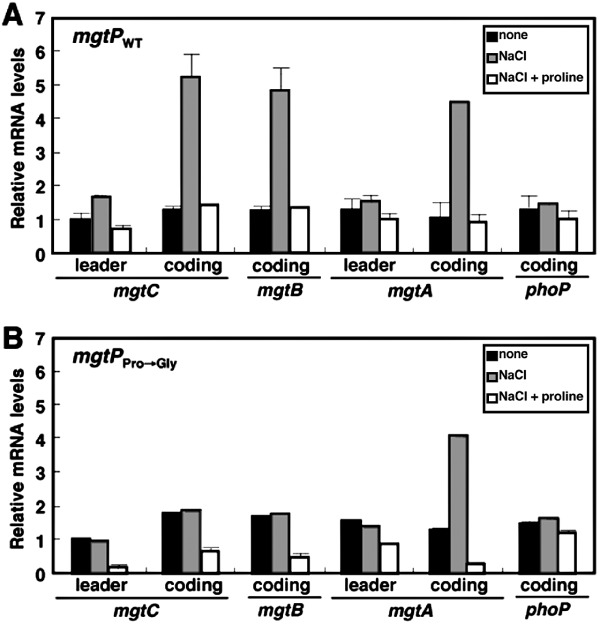
Hyperosmotic stress promotes transcription of the mgtCBR coding region. mRNA levels of the leader regions of the mgtC and mgtA transcripts and the coding regions of the mgtC, mgtA, and phoP genes in strains with the wild-type mgtCBR leader (EL296) or a derivative with the three mgtP Pro codons substituted by Gly codons (EL304). The RNA values were normalized relative to those corresponding to the rrs gene. Bacteria were grown for 1 h in modified N-minimal medium without casamino acids and containing 500 µM Mg2+, or in media that also had 0.3 M NaCl, or 0.3 M NaCl and 1 mM proline. Shown are the mean and SD from three independent experiments.
The proline and ATP signals act on the mgtCBR leader independently and additively
As discussed above, upstream of mgtP there is an additional short ORF, designated mgtM and including conserved adenine nucleotides, which enables an increase in intracellular ATP levels to promote transcription of the mgtC and mgtB coding regions (Lee and Groisman, 2012). We investigated whether the ability to respond to proline is independent from that mediating the response to ATP by exploring the response to these two signals in strains with mutations in the conserved regions of mgtM and mgtP. A chromosomal mutant with the adenine nucleotides at position 44–46 of the mgtCBR leader substituted for thymine nucleotides exhibited normal derepression of the mgtC coding region provoked by proline limitation (Fig. 3A) even though it no longer responded to an increase in ATP levels (Fig. 5A) (Lee and Groisman, 2012). Likewise, a chromosomal mutant in which the mgtP Pro codons were substituted by Gly codons displayed wild-type capacity to respond to changes in the adenine concentration in the media (Fig. 5A) despite lacking the ability to respond to proline (Fig. 3A).
Figure 5.
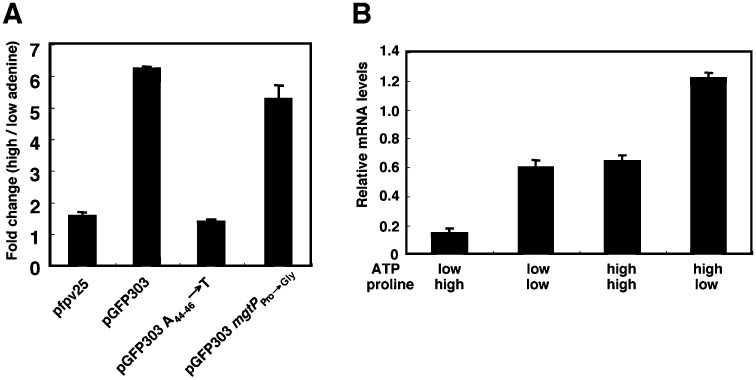
High ATP and low proline promote transcription of the mgtCBR coding region in an independent and additive manner. A. Fluorescence produced by an adenine auxotroph (EG9652) harbouring plasmid pGFP303 with the PhoP-dependent mgtCBR promoter and wild-type mgtCBR leader fused to a promoterless gfp gene, the plasmid vector pfpv25, or pGFP303 derivatives with conserved A nucleotides at position 44–46 substituted by Ts (pGFP303 A44–46→T) or with the three mgtP Pro codons substituted by Gly codons (pGFP303 mgtPPro→Gly). Bacteria were grown in N-minimal media with 10 µM Mg2+ in the presence of either 25 µM or 250 µM adenine. Fluorescence was monitored following growth for 6.5 h with shaking at 37°C under microaerophilic conditions in a Victor3 plate reader. Data correspond to a representative of four independent experiments. B. mRNA levels of the coding regions of the mgtC gene produced by a proline and adenine auxotroph (EL333) grown under different combinations of high and low levels proline and adenine. The RNA values were normalized relative to those corresponding to the rrs gene. Bacteria were grown in N-minimal media with 500 µM Mg2+ in the presence of 1 mM proline and 25 µM adenine for 1 h, and then grown for 1 h in media either containing or lacking proline, 25 µM or 250 µM adenine.
The effect of the low proline and high ATP signals is additive because the mRNA levels corresponding to the mgtC coding region were nearly two times higher in wild-type Salmonella experiencing both conditions than those subjected to either proline limitation or high levels of ATP alone (Fig. 5B). This provides a singular example of a leader mRNA that harbours tandem attenuators responding to different signals.
Discussion
We established that the mgtCBR leader mRNA is a complex sensing device that can respond to a variety of signals by altering the expression of a virulence protein and a Mg2+ transporter. On the one hand, it relies on the coupling of transcription of the mgtCBR leader mRNA and translation of two short ORFs located within the leader to alter the mRNA levels of the associated coding regions in response to the cytoplasmic levels of ATP and proline. On the other hand, the mgtCBR leader mRNA can respond to cytoplasmic Mg2+ (Cromie et al., 2006; Spinelli et al., 2008), perhaps acting as a Mg2+-sensing riboswitch like those preceding the coding regions of the Mg2+ transporter genes mgtA in Salmonella (Cromie et al., 2006) and mgtE in Bacillus subtilis (Dann et al., 2007).
Tandem attenuators in the mgtCBR leader mediate responses to two different signals
Classical attenuators regulate the expression of products that alter the levels of a metabolite affecting the coupling of transcription of the leader region and translation of an ORF located within the leader mRNA. For example, the transcript corresponding to the trp operon from Escherichia coli, which specifies enzymes responsible for tryptophan biosynthesis, includes a leader with a short ORF harbouring two adjacent Trp codons. This allows control of the genes specifying Trp biosynthetic enzymes by the levels of charged tRNATrp because when cytoplasmic tryptophan levels are low, ribosome stalling at the ORF Trp codons enables the leader RNA to adopt a conformation that favours transcription elongation into the trp operon coding region (Yanofsky, 2004). Likewise, the mRNA for the E. coli pyrBI operon, which encodes enzymes participating in the de novo synthesis of pyrimidine nucleotides, is preceded by a leader that includes a short ORF and an overlapping uridine-rich stretch. This allows intracellular UTP levels to alter the normal coupling of transcription and translation, and thus regulate expression of the pyrimidine biosynthetic genes (Turnbough and Switzer, 2008).
The mgtCBR leader mRNA is unusual both in harbouring two attenuators and in the nature of the signals that control these attenuators. We previously demonstrated that stop codon mutations in mgtM that resulted in translation of an ORF shorter than seven amino acids resulted in derepression of the mgtC coding region (Lee and Groisman, 2012). This was ascribed to formation of one stem-loop over the alternative stem-loop in the 5′ portion of the transcript (Lee and Groisman, 2012). It was also proposed that physiological conditions changing the intracellular ATP levels could affect the coupling/uncoupling between transcription of the mgtCBR leader and translation of mgtM. In other words, a ribosome-translating mgtM would be more likely to stay close to the RNA polymerase (RNAP) transcribing the mgtCBR leader when cytosolic ATP levels are low because RNAP may pause at the conserved adenine nucleotides hindering transcription elongation into the coding region. By contrast, mgtCBR transcription and mgtM translation could become uncoupled at high ATP levels, which would advance transcription into the coding region.
That mutation of the mgtP start codon mutant decreased expression of the mgtC coding region (Figs 1 and 2A) might be due to a genetic situation favouring formation of stem-loops C and D. This would also apply to the reduced expression displayed by strains with stop codon mutations at the 6th and 10th codons of mgtP (Fig. 2A). By contrast, stop codon mutations at 13th or 15th positions retained wild-type mgtC–lac expression. This is because the ribosome occupies 12–15 nt from P site (Steitz, 1969) and a ribosome translating beyond the 13th position would cover the left side of stem-loop C and favour formation of stem-loop E (Figs 1 and 2A). When bacteria experience physiological conditions that decrease the levels of cytosolic proline available to charge tRNAPro, the ribosome would stall at the mgtP Pro codons thereby favouring formation of stem-loop E, which furthers transcription elongation into the mgtCBR coding region. Because formation of stem-loop D would sequester the RBS and start codon of mgtC (Fig. 1A), it is possible that mgtP translation affects translation of mgtC in addition to the transcriptional effects discussed above.
The vast majority of transcription attenuators described to date respond to a signal by affecting the formation of an intrinsic transcription terminator [i.e. a GC-rich stem-loop structure followed by a string of uridine nucleotides (Peters et al., 2011)]. However, the stem-loop structures identified in the mgtCBR leader mRNA (Lee and Groisman, 2012) (Fig. 1) do not resemble intrinsic transcription terminators. This is reminiscent of the mgtA leader, which also lacks an intrinsic transcription terminator. Given that the mgtA leader governs transcription elongation into the coding region by affecting access of the termination factor Rho (Hollands et al., 2012), this raises the possibility of a Rho-dependent terminator(s) controlling transcription elongation beyond the mgtCBR leader as well.
We determined that the ATP- and the proline-sensing attenuators present in the mgtCBR leader act independently and that the effect of the two inducing conditions is additive (Fig. 5). In other words, high ATP stimulates transcription of the mgtC and mgtB coding regions even when there is no proline limitation, and low proline stimulates mgtC and mgtB expression even if the ATP levels are not high. This bears interesting similarities and differences with the mRNA leader of the B. clausii metE gene, which harbours tandem riboswitches that monitor two different signals (i.e. S-adenosylmethionine and coenzyme B12) (Sudarsan et al., 2006). Like in the tandem attenuators in the mgtCBR leader, the tandem riboswitches in the metE leader work independently and the effect of the two signals is greater than that of each signal alone. Unlike the mgtCBR leader, the metE leader harbours two sites anticipated to function as intrinsic transcription terminators and the signals are sensed directly by the mRNA leader.
Mg2+, hyperosmotic stress and ATP control transcription of the mgtCBR operon
Transcription initiation of the Mg2+ transporter loci mgtA and mgtCBR is controlled by the PhoP/PhoQ two-component system (Soncini et al., 1996), which is activated in low periplasmic Mg2+ (Garcia Vescovi et al., 1996). In addition, the mgtA and mgtCBR transcripts include long leader sequences that respond to low cytoplasmic Mg2+ by stimulating transcription of their respective coding regions (Cromie et al., 2006; Spinelli et al., 2008). This may allow Salmonella to differentially regulate expression of gene products that mediate cytoplasmic Mg2+ homeostasis (i.e. Mg2+ uptake systems) from those involved in extracytoplasmic Mg2+ homeostasis (i.e. proteins that modify Mg2+ binding sites in the bacterial cell surface).
We propose that hyperosmotic stress promotes expression of the mgtB and mgtA genes because both of them encode P-type ATPases (Snavely et al., 1991) that can transport Mg2+ even when Salmonella experiences a decrease in membrane potential (Csonka, 1989), a condition that compromises the activity of the constitutively expressed Mg2+ transporter CorA (Smith and Maguire, 1998). We previously reported that up-regulation of the mgtA coding region by hyperosmotic stress is mediated by a short ORF harbouring four Pro codons located in the mgtA leader region (Park et al., 2010). That the mgtB gene is also up-regulated by hyperosmotic stress (Fig. 4) and that it harbours an unrelated ORF with three consecutive Pro codons (Fig. 1) mediating the response to low proline (Fig. 3A) provides further support to the notion that Salmonella, and likely other enteric bacteria, utilize short ORFs rich in Pro codons to control expression of related Mg2+ transporters. Moreover, it argues against the proposal that the role of the Pro codon-rich ORF in the mgtA leader region is to mediate the response to low levels of cytoplasmic Mg2+ (Zhao et al., 2011).
The proline levels present at any given time in Salmonella result from the combined activities of proline biosynthetic enzymes and proline uptake systems (Csonka, 1989). The hyperosmotic stress induction of the mgtC and mgtB genes was detected in a proline prototroph with functional proline transporters during growth in the absence of casamino acids. Given that the total proline content does not change when Salmonella experiences hyperosmotic stress (Csonka, 1988), if proline is used for osmoprotection, then less proline would be available to charge tRNAPro with proline. This could then give rise to a situation where hyperosmotic stress results in ribosome stalling at the mgtP proline codons thereby resulting in transcription of the mgtCB coding region.
The similarities in expression behaviour discussed above for the mgtA and mgtB genes suggest that the corresponding Mg2+ transporters might be operating at the same time. However, this does not appear to be the case because: First, the PhoP-activated mgtA and mgtCBR promoters have different architectures (Zwir et al., 2012). Second, the mgtC and mgtB coding regions are induced under mild acidic pH conditions and inside macrophages whereas mgtA’s is not (Lee and Groisman, 2012). And third, the MgtR peptide specified in the mgtCBR operon has been shown to bind to the MgtA protein promoting its degradation (Choi et al., 2012). Given that the mgtA gene can be transcribed by the Rob protein independently of the PhoP/PhoQ system (Barchiesi et al., 2008), it appears that expression of the MgtA protein versus the MgtB protein is favour under different circumstances. This could reflect the 50% differences in amino acid identity between these transporters, which likely accounts for dissimilar substrate specificity (Maguire, 2006).
Experimental procedures
Bacterial strains, plasmids, oligodeoxynucleotides and growth conditions
Bacterial strains and plasmids used in this study are listed in Table 1. All S. enterica serovar Typhimurium strains are derived from the wild-type strain 14028s (Fields et al., 1986) and were constructed by phage P22-mediated transductions as described (Davis et al., 1980). All DNA oligonucletides are listed in Table S1. Bacteria were grown at 37°C in Luria–Bertani broth (LB), N-minimal media (Snavely et al., 1991) supplemented with 0.1% casamino acids, 38 mM glycerol and the indicated concentrations of MgCl2. To examine the effect of hyperosmotic stress on gene expression, we used a modified N-minimal medium containing 0.2% glucose instead of 38 mM glycerol. Escherichia coli DH5α was used as the host for preparation of plasmid DNA. Ampicillin was used at 50 µg ml−1, chloramphenicol was used at 20 µg ml−1, tetracycline at 10 µg ml−1 and fusaric acid at 12 µg ml−1.
Table 1.
Bacterial strains and plasmids used in this study.
| Strain or plasmid | Description | Reference or source | ||
|---|---|---|---|---|
| S. enterica serovar Typhimurium | ||||
| 14028s | Wild type | Fields et al. (1989) | ||
| TT206 | LT2 leu-1151::Tn10 | John R. Roth | ||
| EG9527 | mgtCB9232::MudJ | Blanc-Potard and Groisman (1997) | ||
| EG9652 | purB877:: Tn10 | Blanc-Potard and Groisman (1997) | ||
| EG18715 | mgtCB leader::tetRA/pKD46 | Lee and Groisman (2012) | ||
| EG18798 | mgtCBR leader::tetRA mgtC-lac/pKD46 | Lee and Groisman (2012) | ||
| EG18799 | mgtP (CUG) mgtC-lac | This work | ||
| EG18801 | mgtP (UAA2) mgtC-lac | This work | ||
| EG19251 | mgtP (UAA3) mgtC-lac | This work | ||
| EG19253 | mgtP (UGA5) mgtC-lac | This work | ||
| EG19272 | mgtP (UAA4) mgtC-lac | This work | ||
| EG19840 | mgtP (UAG2) mgtC-lac | This work | ||
| EG19886 | proB1657::Tn10 | Park et al. (2010) | ||
| EL296 | yicL::CmR | This work | ||
| EL302 | yicL::CmR, proB1657::Tn10 | This work | ||
| EL303 | yicL::CmR, mgtP (Pro10,11,12 → Gly), proB1657::Tn10 | This work | ||
| EL304 | yicL::CmR, mgtP (Pro10,11,12 → Gly) | This work | ||
| EL333 | proB::CmR, purB877::Tn10 | This work | ||
| EL337 | yicL::CmR, leuB1151::Tn10 | This work | ||
| EL339 | yicL::CmR, mgtM (A44-46 → T), proB1657::Tn10 | This work | ||
| EL347 | yicL::CmR, mgtP (Pro12 → Gly), proB1657::Tn10 | This work | ||
| EL348 | yicL::CmR, mgtP (Pro10,11 → Gly), proB1657::Tn10 | This work | ||
| EL349 | yicL::CmR, mgtP (Pro10 → Gly), proB1657::Tn10 | This work | ||
| EL350 | proB::CmR | This work | ||
| EL379 | mgtPscrambled, proB1657::Tn10 | This work | ||
| Plasmid | ||||
| pACYC-′lacZ | repp15A CmR′lacZ | Park et al. (2010) | ||
| pCP20 | reppSC101ts ApRCmRFLP+λcI857+ | Datsenko and Wanner (2000) | ||
| pKD3 | repRR6KApR FRT CmR FRT | Datsenko and Wanner (2000) | ||
| pKD46 | reppSC101ts ApR paraBADγβ exo | Datsenko and Wanner (2000) | ||
| pmgtP-′lacZ | repp15A CmR plac1-6 mgtP-′lacZ | This work | ||
| pmgtP stop-′lacZ | repp15A CmRplac1-6 mgtP stop-′lacZ | This work | ||
| pUHE21-2lacIq | reppMBI ApRlacIq | Soncini et al. (1995) | ||
| pmgtP | pUHE21- mgtP | This work | ||
| psupF | pUHE21-supF | Park et al. (2010) | ||
| pfpv25 | pMB1ori, ApR, promoterless gfp | Valdivia and Falkow (1996) | ||
| pGFP303 | pfpv25 pmgtC-mgtC leader 303-gfp | Lee and Groisman (2012) | ||
| pGFP303 A44-46 → T | pfpv25 pmgtC-mgtC leader 303 (A44-46 → T)-gfp | Lee and Groisman (2012) | ||
| pGFP303 mgtPpro→gly | pfpv25 pmgtC-mgtC leader 303 (mgtPpro→gly)-gfp | This work | ||
| ptGFP | ColE1 ori ApR′gfp | This work | ||
| ptGFP303 | pmgtC-mgtC leader 303-′gfp | This work | ||
| ptGFP303 re211-214 | pmgtC-mgtC leader 303 (re211-214)-′gfp | This work | ||
| ptGFP303 mgtP (CUG) | pmgtC-mgtC leader 303 (mgtP(CUG))-′gfp | This work | ||
| ptGFP303 mgtP (CUG) re211-214 | pmgtC-mgtC leader 303 (mgtP(CUG) re211-214)-′gfp | This work | ||
Construction of plasmids harbouring a lacZ translational fusion to mgtP
Polymerase chain reaction (PCR) fragments corresponding to nucleotides 148–219 of the mgtCBR leader were amplified with primer 9804, which includes the sequence corresponding to the plac1–6 promoter, and either primer 9805 or 9806 (creating a stop codon) using 14028s genomic DNA as a template. The resulting PCR products were digested with SmaI and XbaI and cloned into plasmid pACYC-′lacZ digested with the same enzymes. The sequence of the resulting constructs was verified by DNA sequencing.
Construction of a plasmid harbouring the mgtP ORF
Plasmid pmgtP was constructed as follows: a PCR fragment corresponding to the mgtP ORF generated by PCR with primers 8587 and 8589 using 14028s genomic DNA as a template, was digested with HindIII and BamHI and cloned into pUHE 21-2lacIq digested with the same enzymes. The sequence of the resulting constructs was verified by DNA sequencing.
Construction of plasmids harbouring fusions to a promoterless gfp gene
pGFP303, a gfp plasmid with the PhoP-dependent mgtCBR promoter and the wild-type mgtC leader, and its derivative, the A44–46→T substitutions in the mgtC leader were constructed as described (Lee and Groisman, 2012). Derivatives pGFP303 with nucleotide substitutions in the mgtC leader were constructed by cloning PCR fragments generated by two rounds of PCR reactions. For the mgtPPro→Gly substitution in the mgtC leader, a first PCR fragment was generated with primers 1746 and 8826, and a second fragment was generated with primers 8827 and 8117 and 14028s genomic DNA as a template. A third PCR was performed with primers 1746 and 8117 using the two PCR-generated DNA fragments as templates. The resulting PCR product was digested with EcoRI and XbaI and cloned into plasmid pfpv25 digested with the same enzymes. The sequence of the resulting construct was verified by DNA sequencing.
ptGFP, a gfp plasmid for translational fusion, was constructed as follows: a PCR fragment corresponding to the gfp gene starting from the third codon generated by PCR with primers 10109 and 10110 using pfpv25 plasmid as a template, was digested with BamHI and HindIII and cloned back into pfpv25 digested with the same enzymes, creating a gfp plasmid lacking its own RBS and the first two codons of the gfp gene (′gfp).
ptGFP303, a plasmid with the PhoP-dependent mgtCBR promoter, the wild-type mgtCBR leader and first two codons of mgtC fused in frame to the gfp gene was constructed as follows: a PCR fragment generated with primers 1746 and 10111 using 14028s genomic DNA as a template and digested with EcoRI and BamHI was cloned into plasmid ptGFP digested with the same enzymes.
Derivatives of ptGFP303 with nucleotide substitutions in the mgtCBR leader region were constructed by cloning PCR fragments generated by two rounds of PCR reactions. For the substitution in the position at 211–214 in the mgtC leader to hinder formation of stem-loop C, a first PCR fragment was generated with primers 1746 and 10113, and a second fragment was generated with primers 10112 and 10111 using 14028s genomic DNA as a template. A third PCR reaction was performed with primers 1746 and 10111 using the two PCR-generated DNA fragments as templates. The resulting PCR product was cloned into ptGFP using the same restriction enzymes used for construction of ptGFP303. All other substitutions were generated in a similar way using the following primer pairs: mgtP (CUG) (1746/8344 and 8347/8117) using 14028s genomic DNA as a template and mgtP (CUG) re211-214 (1746/10113 and 10112/10111) using ptGFP mgtP (CUG) plasmid as a template. DNA sequencing was used to verify the nucleotide sequences of all constructs.
Construction of a strain with a chromosomal deletion of the proB gene
A Salmonella strain deleted for the proB gene was generated by the one-step gene inactivation method (Datsenko and Wanner, 2000). A CmR cassette was PCR amplified from plasmid pKD3 using primers 11729 and 11730 and the resulting PCR product was integrated into the 14028s chromosome to generate EL350 (proB::CmR). A P22 phage lysate grown in strain EL350 was used to transduce EG9652 (purB877::Tn10) Salmonella selecting for chloramphenicol resistance to generate EL333 (proB::CmRpurB877::Tn10).
Construction of strains with chromosomal mutations in the mgtCBR leader region
Two different methods were used to generate strains with chromosomal mutations in the mgtCBR leader. For strain EL379, we used the fusaric acid method as described (Lee and Groisman, 2010). DNA fragments carrying 10 out of 16 sense codons substitution in the mgtP were prepared by a two-step PCR reaction. For the first PCR reaction, we used two primer pairs 8118/11963 and 11962/7308, and 14028s genomic DNA as a template. For the second PCR reaction, we mixed two PCR products from the first PCR reaction as templates and amplified a DNA fragment using primers 8118 and 7308. The resulting PCR products were purified and integrated into the EG18715 chromosome and selected against TetR with media containing fusaric acid to generate EL379, a TetS AmpS chromosomal mutant. The presence of the expected substitution was verified by sequencing.
To create mutations with the start codon or stop codons at different positions in mgtP, DNA fragments carrying the mutation at the start codon or stop codons in the mgtP were prepared as follows: we used primer pairs 8118/8344 and 8347/7308 (for CUG), 8118/8348 and 8349/7308 (for UAA2), 8118/8699 and 8698/7308 (for UAA3), 8118/8809 and 8808/7308 (for UAA4), 8118/8704 and 8703/7308 (for UGA5) or 8118/9853 and 9852/7308 (for UAG2) and 14028s genomic DNA as a template in the first PCR reaction. For the second PCR reaction, we mixed the two PCR products from the first PCR reaction as templates and amplified the DNA fragment with the expected substitutions using primers 8118 and 7308. The resulting PCR products were purified and integrated into the EG18798 chromosome and selected against TetR in media containing fusaric acid to generate strains EG18799, EG18801, EG19251, EG19272, EG19253 and EG19840, which were TetS AmpS.
All other chromosomal mutants with substitutions in the mgtC leader were constructed by a multiple step PCR process. Strain EL296 was constructed by inserting a CmR cassette in the yicL gene, which is 278 nt upstream from mgtC transcription start site. The CmR cassette was amplified from plasmid pKD3 using primers 4801 and 4802 and the resulting PCR products were integrated into the 14028s chromosome to generate EL296 (yicL::CmR). Then, we prepared DNA fragments containing a CmR cassette and the proper nucleotide substitutions in the mgtC leader using two primer pairs and EL296 genomic DNA as a template: 10077/8826 and 8827/7308 for the mgtPPro10,11,12→Gly substitution; 10077/11732 and 11731/7308 for the mgtPPro12→Gly substitution; 10077/11734 and 11733/7308 for the mgtPPro10,11→Gly substitution; 10077/11736 and 11735/7308 for the mgtPPro10→Gly substitution; and 10077/11727 and 11726/7308 for the A44–46→T substitution in the mgtCBR leader. The two resulting DNA fragments from the first PCR reactions were mixed and used as PCR templates to amplify DNA fragments containing CmR cassette and the proper nucleotide substitution using primers 10077 and 7308. The resulting DNA fragments were purified and integrated into the 14028s chromosome by the one-step inactivation method (Datsenko and Wanner, 2000) and mutants were selected for resistance to chloramphenicol. The presence of the expected substitution was verified by DNA sequencing.
Effect of exogenous adenine on gene expression
Experiment was carried out using the adenine auxotrophic strain EG9652 harbouring a plasmid harbouring the mgtC–gfp fusion (or the plasmid vector) as described (Lee and Groisman, 2012).
Effect of proline limitation on gene expression
The proline limitation experiment was performed as described (Park et al., 2010) with the following modifications: proline auxotrophic strains with a wild-type or mutant mgtC leader were grown overnight in N-minimal medium containing 10 mM Mg2+, and 1 mM proline. 1/100 dilution of the overnight culture was used to inoculate 20 ml of the same medium and grown for 3 h. Cells were then washed and transferred to 20 ml of N-minimal medium containing 500 µM Mg2+ and 1 mM proline and grown for 1 h. The cells were harvested and washed with N-minimal medium containing 500 µM Mg2+ without proline and resuspended in a small volume of the same media. Then, the resuspended cells were split into two cultures in 10 ml of N-minimal medium containing a mixture of 19 amino acids (all essential amino acids except proline) and 500 µM Mg2+ with or without 1 mM proline and growth continued for 45 min. Bacteria were stabilized using RNAprotect Bacteria Reagent (Qiagen) and RNA was isolated for further analysis.
Effect of leucine limitation on gene expression
Leucine limitation was performed as described above except that we used a leucine auxotroph and a 19 amino amino acid mixture (all essential amino acids except leucine).
Effect of hyperosmotic stress on gene expression
Experiment was performed as described (Park et al., 2010).
Effect of proline and/or adenine on gene expression
Proline and adenine auxotrophic strains were grown overnight in N-minimal medium containing 10 mM Mg2+, 1 mM proline and 250 µM adenine. 1/50 dilution of the overnight culture was used to inoculate 40 ml of the same medium and grown for 3 h. Cells were then washed and transferred to 40 ml of N-minimal medium containing 500 µM Mg2+, 1 mM proline and 25 µM adenine and grown for 1 h. The cells were harvested and washed with N-minimal medium containing 500 µM Mg2+ and 25 µM adenine without proline and resuspended in a small volume of the same media. Then, the resuspended cells were split into four cultures in 10 ml of N-minimal medium containing a mixture of 19 amino acids (all essential amino acids except proline) and 500 µM Mg2+ with or without 1 mM proline in the presence of 25 µM or 250 µM adenine and growth continued for 1 h. Bacteria were stabilized using RNAprotect Bacteria Reagent (Qiagen) and RNA was isolated for further analysis.
Quantitative real-time PCR
Total RNA was isolated using RNeasy Kit (Qiagen) according to the manufacturer’s instructions. The purified RNA was quantified using a Nanodrop machine (NanoDrop Technologies). cDNA was synthesized using High Capacity RNA-to-cDNA Master Mix (Applied Biosystems). The mRNA levels of the mgtC, mgtB, mgtA, phoP and rrs genes were measured by quantification of cDNA using SYBR Green PCR Master Mix (Applied Biosystems, Foster City) and appropriate primers (mgtC leader: 6962/6963, mgtC coding: 7530/7531, mgtB coding: 7763/7764, mgtA leader: 7225/7226, mgtA coding: 4308/4309, and phoP coding: 4489/4490) and monitored using a Fast ABI7500 machine (Applied Biosystems, Foster City). Data were normalized to the levels of 16S ribosomal RNA amplified with primers 6970 and 6971.
β-Galactosidase assays
Cells were grown overnight in N-minimal media and washed once in N-minimal media before resuspending them in N-minimal media with different MgCl2 concentrations for 4 h at 37°C with shaking. The activity was determined as described (Miller, 1972). Data correspond to two or more independent experiments conducted in duplicate.
In-line probing
Experiments were carried out as described (Regulski and Breaker, 2008) with the following modifications: the mgtC leader RNA was synthesized in vitro with T7 RiboMAX Large Scale RNA production system (Promega) from the DNA template amplified from wild-type 14028s and primers 10336 and 6140 for the mgtC leader 196–385. To probe the structures at different Mg2+ concentrations, 1 pmol of 5′-end-labelled mgtC leader RNA was incubated in buffer [100 mM KCl, 50 mM Tris (pH 8.0)] with 1, 5 or 20 mM Mg2+ for 40 h at room temperature. Reactions were quenched with urea gel loading buffer II (Ambion) and analysed on a 10% denaturing polyacrylamide gel.
Acknowledgments
This research was supported, in part, by Grant AI49561 from the National Institutes of Health to E.A.G., who is an investigator of the Howard Hughes Medical Institute.
References
- Alix E, Blanc-Potard AB. Peptide-assisted degradation of the Salmonella MgtC virulence factor. EMBO J. 2008;27:546–557. doi: 10.1038/sj.emboj.7601983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, et al. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell. 2005;122:461–472. doi: 10.1016/j.cell.2005.05.030. [DOI] [PubMed] [Google Scholar]
- Barchiesi J, Castelli ME, Soncini FC, Vescovi EG. mgtA Expression is induced by rob overexpression and mediates a Salmonella enterica resistance phenotype. J Bacteriol. 2008;190:4951–4958. doi: 10.1128/JB.00195-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc-Potard AB, Groisman EA. The Salmonella selC locus contains a pathogenicity island mediating intramacrophage survival. EMBO J. 1997;16:5376–5385. doi: 10.1093/emboj/16.17.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi E, Lee KY, Shin D. The MgtR regulatory peptide negatively controls expression of the MgtA Mg2+ transporter in Salmonella enterica serovar Typhimurium. Biochem Biophys Res Commun. 2012;417:318–323. doi: 10.1016/j.bbrc.2011.11.107. [DOI] [PubMed] [Google Scholar]
- Cromie MJ, Shi Y, Latifi T, Groisman EA. An RNA sensor for intracellular Mg(2+) Cell. 2006;125:71–84. doi: 10.1016/j.cell.2006.01.043. [DOI] [PubMed] [Google Scholar]
- Csonka LN. Regulation of cytoplasmic proline levels in Salmonella typhimurium: effect of osmotic stress on synthesis, degradation, and cellular retention of proline. J Bacteriol. 1988;170:2374–2378. doi: 10.1128/jb.170.5.2374-2378.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csonka LN. Physiological and genetic responses of bacteria to osmotic stress. Microbiol Rev. 1989;53:121–147. doi: 10.1128/mr.53.1.121-147.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csonka LN, Leisinger T. Biosynthesis of proline. In: Cohen G, editor. Escherichia Coli and Salmonella: Cellular and Molecular Biology. Washington, DC: ASM; 2007. doi: 10.1128/ecosal 3.6.1.4. [Google Scholar]
- Dann CE, 3rd, Wakeman CA, Sieling CL, Baker SC, Irnov I, Winkler WC. Structure and mechanism of a metal-sensing regulatory RNA. Cell. 2007;130:878–892. doi: 10.1016/j.cell.2007.06.051. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RW, Bolstein D, Roth JR. Advanced Bacterial Genetics. Cold Spring Harbor: Cold Spring Harbor Lab; 1980. [Google Scholar]
- Fields PI, Swanson RV, Haidaris CG, Heffron F. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci USA. 1986;83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields PI, Groisman EA, Heffron F. A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science. 1989;243:1059–1062. doi: 10.1126/science.2646710. [DOI] [PubMed] [Google Scholar]
- Garcia Vescovi E, Soncini FC, Groisman EA. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell. 1996;84:165–174. doi: 10.1016/s0092-8674(00)81003-x. [DOI] [PubMed] [Google Scholar]
- Grundy FJ, Henkin TM. From ribosome to riboswitch: control of gene expression in bacteria by RNA structural rearrangements. Crit Rev Biochem Mol Biol. 2006;41:329–338. doi: 10.1080/10409230600914294. [DOI] [PubMed] [Google Scholar]
- Henkin TM, Yanofsky C. Regulation by transcription attenuation in bacteria: how RNA provides instructions for transcription termination/antitermination decisions. Bioessays. 2002;24:700–707. doi: 10.1002/bies.10125. [DOI] [PubMed] [Google Scholar]
- Hollands K, Proshkin S, Sklyarova S, Epshtein V, Mironov A, Nudler E, Groisman EA. Riboswitch control of Rho-dependent transcription termination. Proc Natl Acad Sci USA. 2012;109:5376–5381. doi: 10.1073/pnas.1112211109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landick R, Turnbough C, Yanofsky C. Transcription attenuation. In: Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, et al., editors. Escherichia coli and Salmonella Cellular and Molecular Biology. Washington, DC: ASM Press; 1996. pp. 1263–1286. [Google Scholar]
- Lee EJ, Groisman EA. An antisense RNA that governs the expression kinetics of a multifunctional virulence gene. Mol Microbiol. 2010;76:1020–1033. doi: 10.1111/j.1365-2958.2010.07161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EJ, Groisman EA. Control of a Salmonella virulence locus by an ATP-sensing leader messenger RNA. Nature. 2012;486:271–275. doi: 10.1038/nature11090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejona S, Aguirre A, Cabeza ML, Garcia Vescovi E, Soncini FC. Molecular characterization of the Mg2+-responsive PhoP-PhoQ regulon in Salmonella enterica. J Bacteriol. 2003;185:6287–6294. doi: 10.1128/JB.185.21.6287-6294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaldon P, Argos P. Oligopeptide biases in protein sequences and their use in predicting protein coding regions in nucleotide sequences. Proteins. 1988;4:99–122. doi: 10.1002/prot.340040204. [DOI] [PubMed] [Google Scholar]
- Maguire ME. Magnesium transporters: properties, regulation and structure. Front Biosci. 2006;11:3149–3163. doi: 10.2741/2039. [DOI] [PubMed] [Google Scholar]
- Merino E, Yanofsky C. Transcription attenuation: a highly conserved regulatory strategy used by bacteria. Trends Genet. 2005;21:260–264. doi: 10.1016/j.tig.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor: Cold Spring Harbor Lab; 1972. [Google Scholar]
- Naville M, Gautheret D. Transcription attenuation in bacteria: theme and variations. Brief Funct Genomic Proteomic. 2009;8:482–492. doi: 10.1093/bfgp/elp025. [DOI] [PubMed] [Google Scholar]
- Park SY, Cromie MJ, Lee EJ, Groisman EA. A bacterial mRNA leader that employs different mechanisms to sense disparate intracellular signals. Cell. 2010;142:737–748. doi: 10.1016/j.cell.2010.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Vangeloff AD, Landick R. Bacterial transcription terminators: the RNA 3′-end chronicles. J Mol Biol. 2011;412:793–813. doi: 10.1016/j.jmb.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prost LR, Daley ME, Sage VL, Bader MW, Moual HL, Klevit RE, Miller SI. Activation of the bacterial sensor kinase PhoQ by acidic pH. Mol Cell. 2007;26:165–174. doi: 10.1016/j.molcel.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Regulski EE, Breaker RR. In-line probing analysis of riboswitches. Methods Mol Biol. 2008;419:53–67. doi: 10.1007/978-1-59745-033-1_4. [DOI] [PubMed] [Google Scholar]
- Smith RL, Maguire ME. Microbial magnesium transport: unusual transporters searching for identity. Mol Microbiol. 1998;28:217–226. doi: 10.1046/j.1365-2958.1998.00810.x. [DOI] [PubMed] [Google Scholar]
- Snavely MD, Miller CG, Maguire ME. The mgtB Mg2+ transport locus of Salmonella typhimurium encodes a P-type ATPase. J Biol Chem. 1991;266:815–823. [PubMed] [Google Scholar]
- Soncini FC, Vescovi EG, Groisman EA. Transcriptional autoregulation of the Salmonella typhimurium phoPQ operon. J Bacteriol. 1995;177:4364–4371. doi: 10.1128/jb.177.15.4364-4371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soncini FC, Garcia Vescovi E, Solomon F, Groisman EA. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J Bacteriol. 1996;178:5092–5099. doi: 10.1128/jb.178.17.5092-5099.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli SV, Pontel LB, Garcia Vescovi E, Soncini FC. Regulation of magnesium homeostasis in Salmonella: Mg(2+) targets the mgtA transcript for degradation by RNase E. FEMS Microbiol Lett. 2008;280:226–234. doi: 10.1111/j.1574-6968.2008.01065.x. [DOI] [PubMed] [Google Scholar]
- Steitz JA. Polypeptide chain initiation: nucleotide sequences of the three ribosomal binding sites in bacteriophage R17 RNA. Nature. 1969;224:957–964. doi: 10.1038/224957a0. [DOI] [PubMed] [Google Scholar]
- Sudarsan N, Hammond MC, Block KF, Welz R, Barrick JE, Roth A, Breaker RR. Tandem riboswitch architectures exhibit complex gene control functions. Science. 2006;314:300–304. doi: 10.1126/science.1130716. [DOI] [PubMed] [Google Scholar]
- Turnbough CL, Jr, Switzer RL. Regulation of pyrimidine biosynthetic gene expression in bacteria: repression without repressors. Microbiol Mol Biol Rev. 2008;72:266–300. doi: 10.1128/MMBR.00001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia RH, Falkow S. Bacterial genetics by flow cytometry: rapid isolation of Salmonella typhimurium acid-inducible promoters by differential fluorescence induction. Mol Microbiol. 1996;22:367–378. doi: 10.1046/j.1365-2958.1996.00120.x. [DOI] [PubMed] [Google Scholar]
- Yanofsky C. The different roles of tryptophan transfer RNA in regulating trp operon expression in E. coli versus B. subtilis. Trends Genet. 2004;20:367–374. doi: 10.1016/j.tig.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Zhao G, Kong W, Weatherspoon-Griffin N, Clark-Curtiss J, Shi Y. Mg2+ facilitates leader peptide translation to induce riboswitch-mediated transcription termination. EMBO J. 2011;30:1485–1496. doi: 10.1038/emboj.2011.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwir I, Latifi T, Perez JC, Huang H, Groisman EA. The promoter architectural landscape of the Salmonella PhoP regulon. Mol Microbiol. 2012;84:463–485. doi: 10.1111/j.1365-2958.2012.08036.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


