Summary
Mutations in the X-linked MECP2 cause Rett syndrome, a devastating neurological disorder typified by a period of apparently normal development followed by loss of cognitive and psychomotor skills. Data from rare male patients suggest symptom onset and severity can be influenced by the location of the mutation, with amino acids 270 and 273 marking the difference between neonatal encephalopathy and death, on the one hand, and survival with deficits on the other. We therefore generated two mouse models expressing either MeCP2-R270X or MeCP2-G273X. The mice developed phenotypes at strikingly different rates and showed differential ATRX nuclear localization within the nervous system, over time, coinciding with phenotypic progression. We discovered that MeCP2 contains three AT-hook-like domains over a stretch of 250 amino acids, like HMGA DNA-bending proteins; one conserved AT-hook is disrupted in MeCP2-R270X, lending further support to the notion that one of MeCP2’s key functions is to alter chromatin structure.
Introduction
Rett syndrome (RTT) is caused by mutations in the X-linked gene methyl-CpG-binding protein 2 (MECP2) (Amir et al., 1999). Girls with RTT achieve expected milestones for the first 6–18 months, only to experience a progressive loss of acquired linguistic, social, and motor skills. They also develop seizures, stereotypies, autonomic dysfunction, and spasticity—yet there is no evidence of neurodegeneration (Chahrour and Zoghbi, 2007). It is this pattern of normal development followed by functional impairment of the nervous system that is paradigmatic of RTT. The molecular mechanisms underlying this regression are unknown.
RTT-causing mutations have been identified throughout the entire length of MeCP2, which contains two functional domains: an N-terminal methyl-CpG binding domain (MBD) and a C-terminal transcriptional repression domain (TRD). Despite nearly two decades of investigation, the function of MeCP2 remains unclear. Its TRD can recruit an HDAC-containing complex via the Sin3A corepressor, suggesting it represses transcription (Nan et al., 1998). In vitro, MeCP2 associates with nucleosomal linker DNA, compacts nucleosomal arrays, and competes with histone H1 for chromatin binding (Ghosh et al., 2010; Nan et al., 1997; Nikitina et al., 2007a). More recent in vivo studies showed that MeCP2 loss results in greater neuronal H1 content, raising the possibility that MeCP2 serves as an alternative linker histone (Skene et al., 2010); the same work found that MeCP2 binds throughout the genome and might act to dampen overall transcriptional activity. Interestingly, MeCP2 loss-of-function or overexpression results in the inverse misregulation of thousands of genes in specific brain regions (Ben-Shachar et al., 2009; Chahrour et al., 2008; Samaco et al., 2012).
Given the clinical variability of RTT and the range of mutations, one way to clarify MeCP2 function is to correlate mutation with phenotype. Although this is difficult in females because of the confounding effect of X-chromosome inactivation, MECP2 mutations have been reported in about sixty boys (Villard, 2007) (http://mecp2.chw.edu.au/). Seventeen are non-mosaic, karyotypically normal males with truncating mutations. These boys can be grouped into two broad categories: (1) severe neonatal encephalopathy and death before 4 years of age or (2) survival for decades with either RTT-like phenotypes or neuropsychiatric deficits. Boys in category 1 tend to have early truncating mutations (e.g., G163fs, G252fs, G269fs, and R270fs) (Villard, 2007), while boys in category 2 tend to have late truncating mutations (e.g., G273fs, R294X, L386fs, Q406X, and E472fs) (Villard, 2007). Even though there is only one male reported to have the G273fs mutation (Ravn et al., 2003), he lived considerably longer than males with R270fs (Kankirawatana et al., 2006; Venancio et al., 2007). We postulated that the region, between amino acids R270 and G273, exerts a significant effect on the phenotype.
We therefore generated transgenic mice that express either MeCP2-R270X or MeCP2-G273X from the endogenous MECP2 locus. We characterized the mice over the course of disease, examined molecular phenotypes associated with MeCP2 dysfunction, and uncovered a key domain in MeCP2 critical for its role in chromatin organization.
Results
Generation of transgenic mice bearing the R270X and G273X alleles
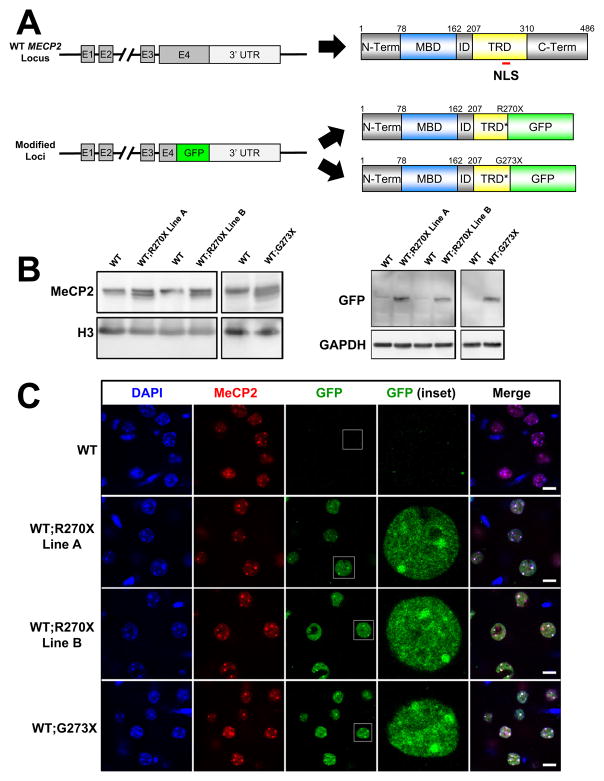
We modified a human PAC containing only the endogenous MECP2 locus with all known regulatory elements (Collins et al., 2004) to bear either a G273X or R270X mutation by recombineering (Figure 1A). We also inserted a C-terminal GFP tag within exon 4 to monitor protein level and localization in vivo. We injected the constructs into wild-type (WT) FVB embryos, generating four R270X and two G273X transgenic lines. We chose two R270X lines (termed “A” and “B”) and one G273X line that exhibited approximately 1x expression of the transgene product in brain compared to WT (Figures 1B and S1A). MeCP2 levels were stable across multiple generations in all lines (Figure S1A). Using immunofluorescence and confocal imaging we found the localization of the MeCP2-R270X and MeCP2-G273X proteins to mirror that of endogenous MeCP2 in the cortex, hippocampus, cerebellum, hypothalamus, and brainstem (Figures S1B–S1F). High-resolution images from the cortex showed that both MeCP2-R270X and MeCP2-G273X localized entirely to the nucleus and concentrated at heterochromatic foci, just like the endogenous protein (Figure 1C). In individual nuclei, the Overlap Coefficient and Manders’ Coefficient of MeCP2-R270X or MeCP2-G273X with endogenous MeCP2 were indistinguishable (k1xk2=0.9215, 0.9258, p=0.55 Student’s t-test; M2=0.6093, 0.6256, p=0.81 Student’s t-test; n=18 and n=25 nuclei, respectively). All three transgenic lines thus faithfully reproduce the distribution, abundance, and subnuclear localization of MeCP2 in the brain.
Figure 1.
Design and characterization of WT;R270X and WT;G273X transgenic mice
(A) Schematic of MECP2 locus the corresponding WT protein product (top). Diagrams are not to scale but positions along the primary sequence and location of the canonical NLS are indicated. Schematic indicating the final modified loci containing a GFP tag inserted in place of the codon for R270 or G273 and the corresponding mutant protein products (bottom). MBD=methyl-CpG binding domain and TRD=transcriptional repression domain. * indicates a truncated TRD. (B) Western blot analysis using whole brain lysates for each transgenic line and their WT littermates and an antibody against the N-terminus that recognizes WT and both mutant forms of MeCP2. Mutant MeCP2 fused with GFP migrates below MeCP2-WT. (C) Mutant MeCP2 localizes with MeCP2-WT in cortical tissue using double immunofluorescence. The C-terminus MeCP2 antibody is specific for MeCP2-WT. Scale bars represent 10 μm. See also Figure S1.
To generate mice expressing MeCP2-R270X or MeCP2-G273X in the absence of MeCP2-WT, we crossed transgenic male mice to Mecp2 +/− heterozygous females (Guy et al., 2001). The resulting male progeny were of four classes: (1) Mecp2 +/y (WT); (2) Mecp2 +/y; R270XTg or G273XTg (WT;R270X or WT;G273X); (3) Mecp2 −/y (knock-out or KO); and (4) Mecp2 −/y; R270XTg or G273XTg (R270X or G273X). All four classes were born at expected Mendelian ratios and appeared healthy at weaning; the fourth class serves as a model for male patients who express only the mutant form of MeCP2.
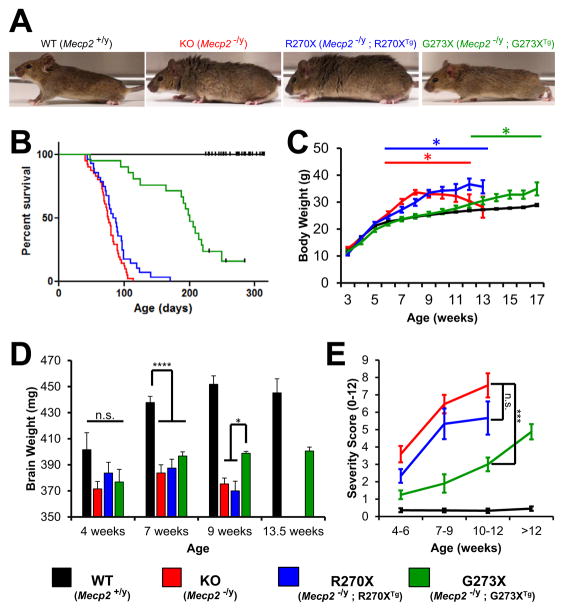
Both WT;R270X and WT;G273X mice appeared identical to their WT littermates (Figure S2A) and were indistinguishable from WT mice in a number of assays (Figures S2B–S2E). As previously reported (Chen et al., 2001; Guy et al., 2001), the KO mice began to develop phenotypes between 4 and 6 weeks of age and, by 8 weeks, were readily distinguished from their WT littermates (Figure 2A). The R270X mice looked very similar to the KO’s, but the G273X mice were leaner and displayed better habitus than either age-matched R270X mice or their KO littermates (Figure 2A).
Figure 2.
Phenotypic characterization of R270X and G273X mice
(A) Representative photographs of WT, KO, R270X and G273X mice at 8 weeks of age; KO and R270X mice have disheveled fur and are overweight. (B) Kaplan-Meier curves for 4 genotype classes. Censored animals are indicated with a black tick mark. n = 54, 41, 28, 21 for WT, KO, R270X, and G273X, respectively. (C) Average body weight plotted versus age. * p<0.05 compared to WT. Maximum n = 54, 41, 28, 21 for WT, KO, R270X, and G273X, respectively. (D) Average brain weights for 4 genotype classes shown at 4, 7, and 9 weeks. WT and G273X brains were also weighed at 13.5 weeks. **** p<0.0001 and * p<0.05. n = 6, 7, 8, 3 for WT and n = 6, 6, 6, 3 for G273X at 4, 7, 9, and 13.5 weeks, respectively. n = 8, 7, 8 for KO and n = 6, 6, 5 for R270X at 4, 7, and 9 weeks, respectively. (E) Average cumulative severity scores plotted against age. *** p<0.001, Mann-Whitney U Test. n = 25, 20, 15, 22 for WT and n = 12, 10, 10, 8 for G273X at 4–6, 7–9, 9–12, and >12 weeks, respectively. n = 17, 15, 11 for KO and n = 12, 6, 6 for R270X at 4–6, 7–9, and 9–12 weeks, respectively. All error bars show SEM. See also Figure S2.
G273X mice manifest disease later and survive longer than KO or R270X mice
KO and R270X mice died prematurely, with a median lifespan of 76 and 85 days, respectively (no significant difference, Gehan-Breslow-Wilcoxon test) (Figure 2B). The G273X mice lived significantly longer and had a median lifespan of 201 days (p<0.0001 compared with either KO or R270X mice, Gehan-Breslow-Wilcoxon test). MeCP2 loss of function has been shown to cause weight gain in both humans and mice, apparently through effects in the hypothalamus (Fyffe et al., 2008). KO animals became overweight compared to WT at postnatal week 6 and reached a maximum weight at 8 weeks of age (Figure 2C). R270X mice exhibited a similar but more gradual pattern during weeks 7 and 8. G273X mice did not begin to gain excessive weight until postnatal week 12 and thereafter slowly gained weight until 17 weeks, when their weight reached a level equal to the maximum KO weight (Figure 2C). R270X and G273X mice thus achieve a weight gain similar to KO animals, but the time courses are very different.
One feature often observed in boys with early truncating mutations is brain atrophy (Schüle et al., 2008). Males with later truncating mutations, however, are normocephalic or show brain growth deceleration leading to acquired microcephaly. We measured brain weights of our mice at 4, 7, and 9 weeks of age. WT mouse brains increase in weight until around 9 weeks of age, then stabilize. In contrast, the KO and R270X brain weights peak at 7 weeks and then decline. Interestingly, the brain weight of G273X mice also peaked at 7 weeks but did not diminish afterward, even at 13.5 weeks (Figure 2D). These data show a striking difference in the pattern of brain growth between R270X and G273X: even though the latter never achieves normal brain weight, the growth pattern is similar to WT.
We used a published severity scale (see Experimental Procedures) that measures tremor, gait abnormalities, and other motor features to evaluate a cohort of mice at the early (4–6 weeks old), middle (7–9 weeks old), and late (10–12 weeks old) symptomatic periods. The premature mortality of the KO and R270X lines precluded comparison at later ages. As expected, the KO mice had significantly higher severity scores than WT animals at all time points (Figure 2E). The average severity scores of both KO and R270X animals increased rapidly from the early to middle symptomatic period but more slowly as the mice entered the end stage of disease (Figure 2E). All features observed in KO mice were apparent in R270X mice and eventually G273X mice (e.g., hind-limb clasping) (Figure S2K). The severity scores of G273X mice were significantly higher than those of WT, but lower (and increasing more slowly) than either KO or R270X lines at all time points (Figure 2E). As G273X mice lived much longer than KO and R270X mice, we were able to analyze these mice after 12 weeks of age. The severity scores of older G273X mice continued to increase until the average reached a level indistinguishable from the middle symptomatic period of either KO or R270X mice. The KO, R270X, and G273X mice thus eventually developed the same phenotypes, but disease progression was significantly delayed in G273X animals.
In severity of symptoms, patterns of body and brain weight gain, and lifespan, then, the G273X mice show more moderate disease with later onset, while R270X mice exhibit the severe, early-onset disease course reminiscent of the more severely affected male patients. These results confirm our hypothesis that R270 and G273 mark a crucial region for MeCP2 function.
To exclude any potential effects of transgene insertion, we generated two additional lines of WT;G273X mice (lines “B” and “C”) and generated G273X mice from these independent lines (Figures S2F and S2G). Like the original G273X line, these mice lived significantly longer, had lower body weights, and had better severity scores relative to KO and R270X mice (Figures S2H–S2J). These data confirm that the loss of three additional amino acids in R270X mice renders them much sicker than G273X mice.
Both MeCP2-R270X and MeCP2-G273X exhibit genome-wide DNA binding
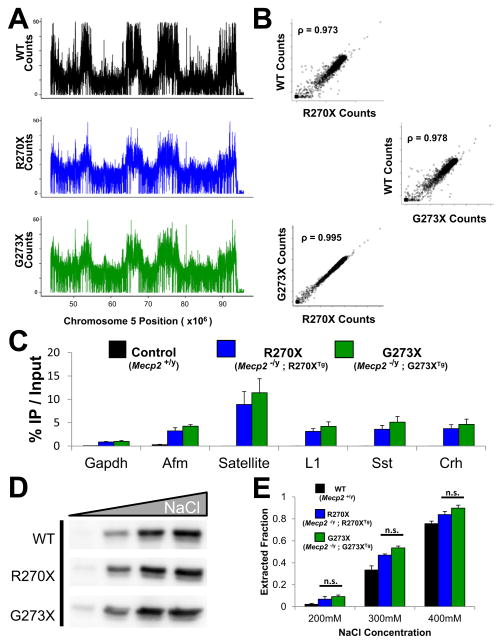
One possible explanation for the differences in phenotypes between the R270X and G273X mutations is differential DNA occupancy. To characterize the DNA binding profiles of these mutant proteins in vivo we performed chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-Seq) from mouse brain. We compared profiles of MeCP2-R270X and MeCP2-G273X to the distribution of MeCP2-WT (Figure 3A), which has been previously reported (Skene et al., 2010). All three profiles show striking similarities across the mouse genome (Figure 3B), including repetitive elements (Figure S3A). The binding pattern of both mutants was reminiscent of MeCP2-WT.
Figure 3.
MeCP2-R270X and MeCP2-G273X bind DNA globally
(A) GFP antibodies were used to immunoprecipitate MeCP2-R270X and MeCP2-G273X from whole brain cross-linked chromatin of either R270X or G273X mice, respectively. Recovered DNA was subjected to deep sequencing. ChIP-Seq counts over a selected region of mouse chromosome 5 for MeCP2-R270X and MeCP2-G273X are compared to MeCP2-WT. Raw reads were binned per 100kb and the number of counts per bin are displayed over the indicated chromosome position. (B) Correlation plots between the number of ChIP-Seq reads per 100kb bin across the genome compared for WT vs. R270X (top), WT vs. G273X (middle) and R270X vs. G273X (bottom). ρ denotes Spearman’s Correlation. (C) Isolated DNA as in (A) was subjected to qPCR analysis using primers designed for the promoters of the indicated genes or repetitive elements. Satellite refers to mouse major satellite DNA, L1 refers to the mouse L1 retrotransposon. In addition to R270X and G273X mice ChIP-qPCR was also performed on WT mice that lack GFP as a control for antibody specificity and is plotted to the left of R270X for each gene or repetitive element. n = 3 for R270X and G273X. (D) Whole brain nuclei were purified from WT, R270X, or G273X mice and resuspended in buffers comprised of 200mM, 300mM, 400mM, or 1M NaCl. An antibody for the MeCP2 N-terminus shows the amount of MeCP2 extracted under each condition. n = 3. (E) The average extracted fractions from the experiment described in (D) are plotted for each genotype above the corresponding NaCl concentration. Data are normalized for the amount of MeCP2 extracted in 1M NaCl. n = 3 mice per genotype. Pooled data show mean +/− SEM. See also Figure S3.
We next assayed specific sites where MeCP2-WT has been reported to bind using ChIP followed by quantitative PCR (ChIP-qPCR) (Chahrour et al., 2008; McGill et al., 2006). We chose four gene promoters (Gapdh, Afm, Sst, and Crh) and two repetitive elements (major satellite DNA and the L1 retrotransposon). ChIP-qPCR revealed that MeCP2-R270X and MeCP2-G273X bound similarly to all of these regions (Figure 3C). Both mutants seemed to be enriched at major satellite DNA, consistent with MeCP2-WT (Lewis et al., 1992). These data suggest that, like the wild-type protein, both MeCP2-R270X and MeCP2-G273X bind widely throughout the genome.
We next assayed the ability of MeCP2-R270X and MeCP2-G273X to bind to chromatin in whole brain nuclei isolated from R270X and G273X mice. We extracted purified nuclei in buffers with increasing ionic strength (200mM, 300mM, 400mM, and 1M NaCl) and assayed the resulting supernatants for extracted MeCP2. Nuclei from WT animals were included for comparison. As expected, MeCP2-WT was increasingly extracted at higher salt concentrations (Figures 3D and 3E). Both MeCP2-R270X and MeCP2-G273X demonstrated similar profiles to MeCP2-WT (Figures 3D and 3E). We also assayed the levels and extractability of histone H1 in this assay and found no major difference in either the total levels (Figures S3B and S3C) or H1 extractability (Figure S3D). MeCP2-R270X and MeCP2-G273X thus show similar DNA-binding properties and localization within the mouse genome.
Both R270X and G273X disrupt the TRD of MeCP2
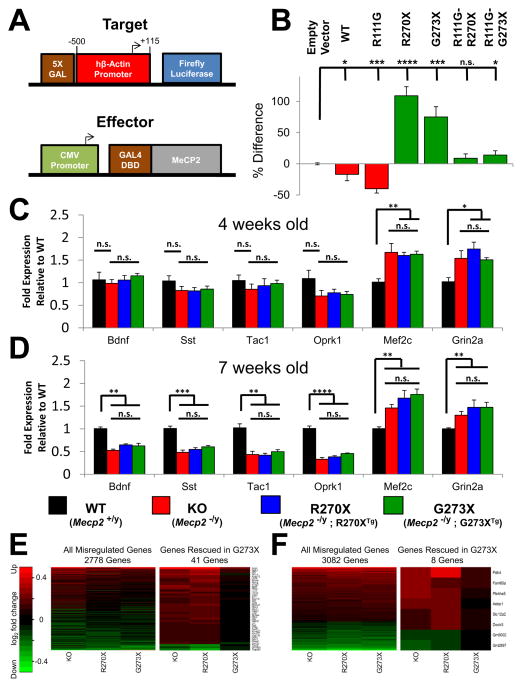
We next compared the effect of the R270X and G273X mutations on the functions of the TRD in a heterologous repression assay (Figure 4A). The initial report characterizing the MeCP2 TRD found that an intact MBD limits proper localization of the Gal4-MeCP2 fusion protein to the reporter construct (Nan et al., 1997), so we used both MeCP2-WT and a mutant form with a point mutation in the MBD, MeCP2-R111G, which abolishes methyl-CpG binding without disrupting MBD folding (Free et al., 2001). Both MeCP2-WT and MeCP2-R111G showed repressor activity (Figure 4B). Consistent with the idea that an intact MBD restricts the ability of the MeCP2 fusion product to localize to the Gal4 binding site, MeCP2-R111G showed stronger repressor activity.
Figure 4.
MeCP2-R270X and MeCP2-G273X have similar transcriptional regulatory function (A) Schematic of constructed transcriptional assay. The target reporter construct contains a 5X Gal DNA binding site immediately upstream of the human β-Actin promoter with endpoints relative to TSS indicated. The effector construct consists of the CMV promoter driving expression of WT or mutant MeCP2 fused at the N-terminus to the Gal4 DNA Binding Domain (Gal4 DBD). A control reporter lacking the 5X Gal site was cotransfected to monitor transfection efficiency. (B) Average luciferase activity of N2a lysates transfected with the target reporter construct and the indicated effector construct. Data are normalized to control reporter activity and the percent difference relative to empty effector is indicated. **** p<0.0001, *** p<0.001, * p<0.05, Student’s t-test. n = 3. (C and D) Hypothalamic RNA levels were quantified by qRT-PCR for 4 genes down-regulated in KO mice (Bdnf, Sst, Oprk1, and Tac1) and two up-regulated genes (Mef2c and Grin2a). Transcript levels were normalized to Gapdh and represented as the fold expression relative to WT. **** p<0.0001, *** p<0.001, ** p<0.01, * p<0.05
(C) Gene expression of 4-week old animals. n = 6, 7, 6, 6 for WT, KO, R270X, and G273X, respectively.(D) Gene expression of 7-week old animals. n = 7, 6, 6, 6 for WT, KO, R270X, and G273X, respectively. (E and F) Hippocampal RNA was quantified by microarray and the abundance of significantly altered genes is displayed as the log2(fold change) relative to WT. The intensity key indicates genes that are upregulated in red and genes that are downregulated in green. (E) (Left) All genes that were misregulated relative to WT in any mutant at 4 weeks of age. (Right) Genes that were misregulated in KO and R270X mice but rescued in G273X mice at 4 weeks of age. n = 4 animals per genotype. (F) (Left) All genes that were misregulated relative to WT in any mutant at 9 weeks of age. (Right) Genes that were misregulated in KO and R270X mice but rescued in G273X mice at 9 weeks of age. n = 4 animals per genotype.
Pooled data show mean +/− SEM. See also Figure S4.
We then generated MeCP2-R270X and MeCP2-G273X both with and without the R111G mutation. Neither mutant showed repressor activity, either in the presence or the absence of the R111G mutation (Figure 4B). Curiously, we noted occasional transcriptional activation using Gal4-MeCP2 containing these mutated TRDs, a phenomenon previously observed with a truncated TRD (Nan et al., 1997). This assay indicates that both the R270X and G273X mutations disrupt the TRD, suggesting that both R270X and G273X mice express mutant proteins that are defective in transcriptional repression.
Transcriptional dysregulation is similar in R270X and G273X mice
We have identified a number of genes whose expression varies reliably with MeCP2 function in the hypothalamus (Chahrour et al., 2008). We therefore measured, in the hypothalami of R270X and G273X mice, the RNA levels of particular genes that are downregulated (Bdnf, Sst, Tak1, and Oprk1) or upregulated (Mef2c and Grin2) in KO mice. We focused on the expression of these genes at 4, 7, and 9 weeks of age so that we could compare these data with our phenotypic analyses (Figures 4C, 4D, and S4).
At 4 weeks of age, when KO mice appear largely asymptomatic, gene expression differences between WT and KO mice were not significant (Figure 4C); this is consistent with a previous report that found BDNF expression to be unchanged in presymptomatic mice and to decrease after symptom onset (Chang et al., 2006). Differences in gene expression between the mutant and WT mice began to appear at 7 weeks, but there were no significant differences between the KO, R270X, and G273X mice, either at these early time points (Figure 4D) or even by 9 weeks of age (Figure S4).
To determine if the differences in phenotypes are accompanied by differential gene expression changes in other tissues, we performed microarray analysis on RNA extracted from hippocampi of 4- and 9-week old WT, KO, R270X, and G273X mice. MeCP2 loss-of-function was associated with many subtle differences in gene expression (Table S1). Overall the transcriptional profiles of KO, R270X, and G273X mice appeared very similar at both ages (Figures 4E and 4F). The total number of genes misregulated in any mutant increased from 4 weeks of age (2778 genes) to 9 weeks of age (3082 genes). We searched for genes that were misregulated in KO and R270X mice but rescued in G273X mice. Interestingly, only 41 genes (38 upregulated and 3 downregulated) appeared similar to WT in G273X mice at 4 weeks of age (Figure 4E and Table S2), being reduced by 9 weeks of age to only 8 genes (6 upregulated and 2 downregulated) (Figure 4F and Table S2). Of the 41 genes that were rescued in G273X mice at 4 weeks, the majority (71%) were misregulated in G273X mice by 9 weeks. Thus the transcriptional profiles of KO, R270X, and G273X mice are similar but do reveal a delay in misregulation for a small subset of genes in G273X mice.
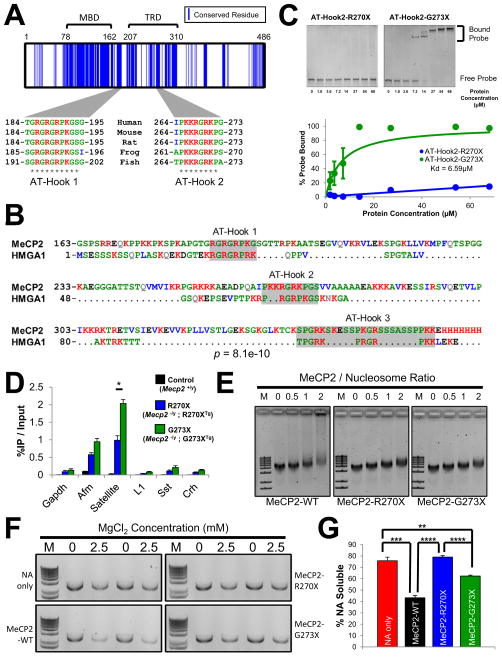
A conserved AT-hook domain is disrupted in MeCP2-R270X but not MeCP2-G273X
To understand the influence of the three amino acids positioned between R270 and G273, we examined the MeCP2 protein sequence surrounding this region. Sequence data indicate that MECP2 arose soon after the appearance of vertebrates, as homologs are detectable in jawless fish such as Petromyzon marinus but not in lower species (http://genome.ucsc.edu/). There is a relatively low level of conservation between the MeCP2 sequence from the single homologs found in zebrafish to humans (49% identity and 62% similarity for zebrafish vs. human) (Figure 5A). One major block of conservation overlaps with the MBD; the C-terminus, including the TRD, is poorly conserved and known to be highly disordered (Ghosh et al., 2010). Nevertheless, we found highly basic clusters of amino acids that are conserved from fish to humans within this region (Figure 5A). One of these conserved basic clusters (amino acids 185–194) is an AT-hook domain (AT-Hook 1) of unknown significance (Klose et al., 2005; Lewis et al., 1992). A second basic cluster (amino acids 265–272) contains another AT-hook (AT-Hook 2) that was uncharacterized although it had been annotated by InterPro (http://www.ebi.ac.uk/interpro/). Given the importance of AT-hooks to the non-histone, chromatin associated proteins of the high-mobility group AT-hook (HMGA) family, which contain two or three AT-hook domains connected by a flexible polypeptide linker (Reeves, 2010), we explored the possibility that MeCP2 and HMGA family members share a common ancestry.
Figure 5.
A conserved AT-hook domain between R270 and G273 enables MeCP2 to alter chromatin structure (A) Conserved MeCP2 residues shared by fish, frog, rat, mouse, and human. Sequences were aligned with ClustalW2 and blue lines indicate residues sharing identity. Two blocks of conserved amino acids corresponding to AT-Hook 1 and AT-Hook 2 are shown. * denotes identity within the aligned sequences. (B) PROMALS3D alignment of the human MeCP2 C-terminus (residues 163–486) and the human HMGA1 sequence. PairwiseStatSig method was used to compute significance (Ankit Agrawal, 2011). p=8.1e–10 after 200000 iterations. (C) (Top) A recombinant form of the MeCP2 AT-Hook 2 domain (amino acids 257–272) was used in an EMSA assay to test binding to a 64-mer double stranded DNA probe (66.7nM). The AT-Hook 2 domain truncated at R270 (R270X) failed to bind DNA. The full AT-Hook 2 domain ending at G273 (G273X) readily forms the indicated complexes with the probe DNA. (Bottom) For AT-Hook 2-G273X a hyperbolic model was fit to the data and the apparent Kd was calculated to be 6.59μM; quantification from 3 independent experiments.
(D) Native ChIP with anti-GFP on whole brain chromatin from R270X and G273X mice. Satellite refers to mouse major satellite DNA, L1 refers to the mouse L1 retrotransposon. WT mice that lack GFP were used as a control for antibody specificity. * p<0.05. n = 3.
(E) EMSA reaction with methylated nucleosomal arrays (NAs) and recombinant MeCP2. MeCP2-WT, MeCP2-R270X, and MeCP2-G273X were added at the indicated MeCP2/Nucleosome ratios. Each protein shifted the NAs at a similar concentration. MeCP2-WT induces the formation of higher order complexes, evident as a long trail above the shifted band. M stands for the 1Kb plus DNA marker. n = 6. (F) Recombinant MeCP2 was mixed with methylated NAs and after the addition of MgCl2 at the indicated concentration the oligomerized material was separated by centrifugation. The NAs that remain in the soluble fraction were run on an agarose gel and visualized by ethidium bromide staining. M stands for the 1Kb plus DNA marker. (G) Quantification of the experiment described in (F). **** p<0.0001, *** p<0.001, ** p<0.01, Student’s t-test. n = 4. Pooled data show mean +/− SEM. See also Figure S5.
Both of MeCP2’s AT-hooks align well with the corresponding AT-hook domains of HMGA1 (Figure 5B). There is an unexpectedly high degree of sequence identity between the third AT-hook of HMGA1 and a more C-terminal portion of MeCP2: this third AT-hook-like domain within MeCP2 appears to have been disrupted by the insertion of two serine-rich tracks. Further analysis showed that this domain exist in multiple extant fish species (e.g., Oryzias latipes and Takifugu rubripes) suggesting that it arose before fish and mammals diverged (http://genome.ucsc.edu/). To compare MeCP2 and HMGA1 using an unbiased method relative to the human proteome, we computed the percentage identity for MeCP2 versus all proteins of similar length to HMGA1 (Figure S5A and S5B). This analysis found that HMGA1 shared the greatest degree of identity among all similarly sized proteins in the human proteome. We found an analogous relationship when comparing HMGA1 to all human proteins of similar length to MeCP2 (Figures S5C and S5D). These data strongly suggest that MeCP2 shares a common ancestry with the HMGA family of proteins.
The conserved AT-Hook 2 domain terminates with G273, after which the sequence diverges among species (Figure 5A). Truncating the three amino acids between G273 and R270 would disrupt the central RGR motif of this AT-hook and could disrupt DNA binding. To test this possibility we purified a recombinant form of the AT-Hook 2 domain (amino acids 257–272) tagged with GST. Electrophoretic mobility shift assay (EMSA) showed that this domain binds double-stranded DNA with an apparent Kd = 6.59μM (Figure 5C). We then tested whether the AT-Hook 2 domain truncated at R270 could also bind DNA. As would be predicted based on the loss of the central RGR motif, this mutant form of AT-Hook 2 failed to bind DNA, even at very high concentrations (Figure S5E). This conserved feature of MeCP2 is therefore a DNA-binding domain that must retain amino acids 270–272 in order to function.
Although MeCP2-R270X contains a truncated AT-Hook 2 domain, we failed to uncover any differences in the occupancy of MeCP2-R270X and MeCP2-G273X using cross-linked ChIP-qPCR (see Figure 3C). We considered that native ChIP-qPCR might reveal subtle differences in DNA binding consistent with the loss of the AT-Hook 2 domain, as this assay requires that a protein stably interact with chromatin in the absence of cross-linking (Turner, 2001). Although native ChIP-qPCR binding of both mutant proteins was similar across the Gapdh, Afm, Sst, and Crh promoters and at the L1 retrotransposon, MeCP2-R270X exhibited half the binding of MeCP2-G273X within major satellite DNA (Figure 5D). Thus truncation of the AT-Hook 2 domain impairs the ability of MeCP2-R270X to stably interact with certain sequences in vivo.
Given the similarity of MeCP2 to HMGA1, we next sought to determine whether loss of the conserved AT-Hook 2 domain of MeCP2 has consequences for chromatin structure that might help explain the differences between R270X and G273X mice.
MeCP2-R270X and MeCP2-G273X differ in their activity towards nucleosomal arrays
Prior work has shown MeCP2 to form higher order structures with nucleosomal DNA in vitro; the RTT-causing R168X mutation impairs this ability (Georgel et al., 2003). We therefore sought to characterize the interactions of MeCP2-R270X and MeCP2-G273X with reconstituted nucleosomal arrays (NAs). Recombinant MeCP2-WT, MeCP2-R270X, and MeCP2-G273X were mixed with CpG methylated NAs and analyzed by EMSA. All three proteins were able to shift the methylated NAs at a similar concentration (Figure 5E). Consistent with previous reports MeCP2-WT formed higher order structures with the NAs evident as a broad smear above the shifted band (Figure 5E). MeCP2-R270X and MeCP2-G273X failed to exhibit this activity under identical conditions.
Adding magnesium to reconstituted NAs induces oligomerization and is thought to model the formation of compacted chromatin in vivo (Schwarz et al., 1996). NAs condensed in this manner become insoluble and can be separated under high-speed centrifugation. Interestingly, MeCP2 facilitates this process (Nikitina et al., 2007b). Using this assay, we evaluated the ability of MeCP2-R270X and MeCP2-G273X to oligomerize methylated NAs (Figure 5F). With the addition of 2.5mM MgCl2, MeCP2-WT greatly facilitated the oligomerization of NAs as evidenced by a reduction in the percent remaining soluble after centrifugation (Figures 5F and 5G). In contrast, MeCP2-R270X failed to facilitate NA oligomerization and was indistinguishable from NAs alone. MeCP2-G273X exhibited intermediate activity and facilitated NA oligomerization although not to the same extent as MeCP2-WT. Across a broad range of MgCl2 concentrations, MeCP2-R270X exhibited reduced activity for oligomerizing NAs whereas MeCP2-G273X showed activity more similar to MeCP2-WT (Figures S5F and S5G).
ATRX mislocalization distinguishes G273X mice
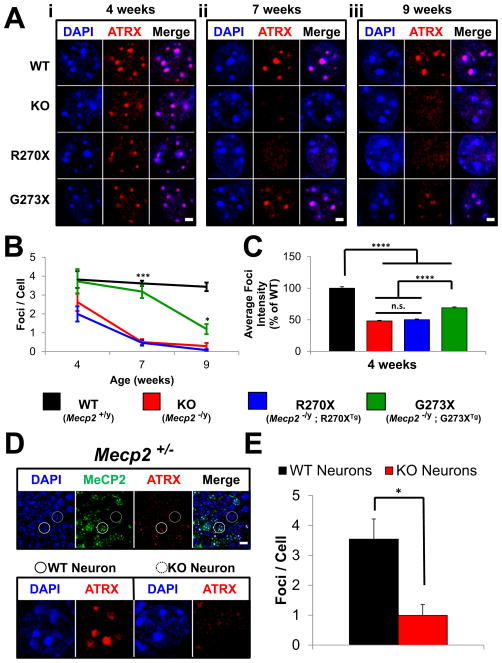
The loss of MeCP2 causes mislocalization of the chromatin remodeling protein alpha thalassemia/mental retardation syndrome X-linked (ATRX) in the hippocampus and cortex of symptomatic KO mice (Nan et al., 2007). We decided to investigate the nuclear localization of ATRX in the hippocampi of our mutant mice over the course of disease.
WT mice show ATRX foci colocalizing with pericentric heterochromatin (PCH) at 4, 7, and 9 weeks of age (Figures 6Ai–6Aiii and S6A). At 4 weeks of age, when mutant mice are largely asymptomatic, the average number of ATRX foci within PCH was similar among KO, R270X, G273X and WT mice, although these foci appeared less intense in the mutant mice (Figures 6Ai, 6B, and 6C). By 7 weeks of age the average number of ATRX foci in KO and R270X animals was significantly less than WT (p<0.0001) (Figures 6Aii and 6B). At 9 weeks of age only a few ATRX foci were detected in KO and R270X mice (Figure 6Aiii), consistent with the previous report on null mice (Nan et al., 2007).
Figure 6.
ATRX mislocalization in various Mecp2 mutants
(A) Immunofluorescence for ATRX in the hippocampus of WT, KO, R270X, and G273X mice. PCH appears as DAPI bright foci within neurons and colocalizes with ATRX staining in WT animals at all ages. (i) At 4 weeks of age ATRX localization is similar in WT and mutant mice. (ii) At 7 weeks of age KO and R270X mice show a noticeable reduction in the number of bright ATRX foci, whereas G273X mice remain similar to WT mice. (iii) At 9 weeks KO, R270X, G273X mice show a decrease in the number of bright ATRX foci relative to WT. Scale bars represent 2 μm. Images are of individual neuronal nuclei, for lower power images of more neurons see Figure S6A. (B) Quantification of the average number of ATRX foci detected per neuron in each genotype at 4, 7, and 9 weeks of age. *** p<0.001 and * p<0.05 for G273X compared to both KO and R270X. n = 6 high-power fields from 2 mice per genotype per age. (C) Quantification of the average foci intensities for each genotype at 4 weeks of age. **** p<0.0001. n = 651 WT foci, n = 513 KO foci, n = 344 R270X foci, and n = 735 G273X foci from 2 mice per genotype. (D) Double immunofluorescence for MeCP2 and ATRX in hippocampus of symptomatic Mecp2 +/− heterozygous female mice. (Top) MeCP2 expressing neurons (solid circle); neurons not expressing MeCP2 (dashed circle). ATRX foci are brighter in MeCP2 positive neurons compared to adjacent MeCP2 negative neurons. Scale bars represent 10 μm. n = 2 mice. (Bottom) Higher magnification image of the WT and KO neuron circled in the upper panel. (E) Quantification of the average number of ATRX foci detected per neuron in symptomatic Mecp2 +/− heterozygous female mice. n = 4 high-power fields from 2 mice. Pooled data show mean +/− SEM. See also Figure S6.
The average number of ATRX foci observed within neurons of G273X mice at 7 weeks of age was indistinguishable from WT: more than six times that of either KO or R270X mice (p<0.0001 and p<0.001 for KO and R270X, respectively) (Figures 6Aii and 6B). By 9 weeks, significantly fewer foci were detectable in G273X mice than in WT (p<0.001) (Figures 6Aiii and 6B), but there were still more foci than in either KO or R270X mice (p<0.05 and p<0.01 for KO and R270X, respectively). ATRX targeting to PCH within the hippocampus is therefore disrupted in Mecp2 mutant lines, but much more gradually in G273X mice.
To understand the cause of ATRX loss from PCH we studied this phenomenon in other Mecp2 mutants. The brains of symptomatic Mecp2+/− heterozygous female mice (5–6 months old) present a mixture of MeCP2-positive and MeCP2-negative neurons, since Mecp2 is subject to X-chromosome inactivation. Within the hippocampus, only MeCP2-negative neurons showed loss of ATRX from PCH (Figure 6D and 6E). We found the same dichotomous pattern in the cortex (data not shown) and hippocampus of younger Mecp2+/− female mice (9 weeks old) (Figure S6B). The effects of MeCP2 loss on ATRX localization are therefore cell-autonomous. Interestingly, we found the opposite phenomenon within hippocampi of female MECP2-TG3 (TG3) mice, which overexpress MeCP2 from a transgene integrated on the X-chromosome (Collins et al., 2004). In TG3-positive neurons ATRX foci remained localized to PCH (Figures S6C and S6D), but these foci were often much brighter than in adjacent WT neurons (Figures S6C and S6E). Overexpression of MeCP2 thus leads to greater accumulation of ATRX within heterochromatin.
ATRX localization in mouse neurons was previously attributed to MeCP2 directly recruiting ATRX to PCH (Nan et al., 2007). The ATRX-interacting domain of MeCP2 overlaps with the MBD and remains intact in both MeCP2-R270X and MeCP2-G273X (amino acids 108–169). Consistent with the presence of this domain, both mutant proteins interact with ATRX to an equal extent as MeCP2-WT by coimmunoprecipitation (Figure S7A). To determine whether progressive loss of ATRX from PCH owes to an age-dependent mislocalization of these mutant forms of MeCP2 from these sites, we compared MeCP2-R270X and MeCP2-G273X localization at two time-points. Immunofluorescence staining detected both mutant forms of MeCP2 at PCH in the hippocampi of 4-week and 9-week old animals (Figure S7B). The staining pattern remained unchanged regardless of age. These truncated forms of MeCP2 appear to stably associate with PCH in vivo and ATRX loss from these sites is independent of MeCP2 presence.
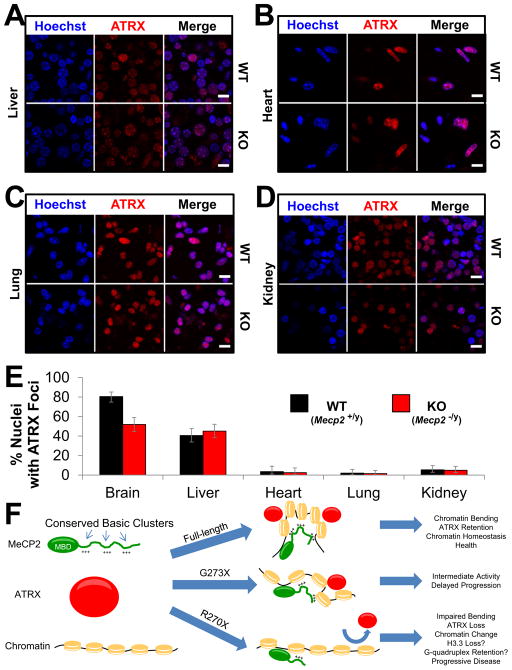
To rule out fixation artifacts as the cause of ATRX mislocalization, we purified fresh whole brain nuclei from both WT and KO mice at 9 weeks of age and stained them for ATRX. In WT brain nuclei ATRX appeared within dense foci colocalized with Hoechst bright PCH. In nuclei purified from KO brains, ATRX was depleted specifically from PCH (Figures S7C–S7E). We then used this assay to study the localization of ATRX in peripheral tissues. The number of nuclei with ATRX foci colocalizing with PCH were fewer in liver, heart, lung, and kidney than in the brain (Figures 7A–7E). There was no difference between WT and KO mice in the number of nuclei from peripheral tissues with ATRX foci (Figure 7E). The pathological processes leading to ATRX mislocalization are therefore brain-specific.
Figure 7.
ATRX mislocalization is specific to the brain
(A–D) Immunofluorescence for ATRX in fresh nuclei prepared from WT and KO mice at 9 weeks of age. Nuclei are counterstained with Hoechst from liver (A), heart (B), lung (C), and kidney (D). Scale bars represent 10 μm. n = 3 mice per genotype. (E) Quantification of the experiment described in (A–D and Figure S7C). The percentage of ATRX positive nuclei with foci localizing to PCH from WT and KO animals is plotted for each tissue. Error bars represent 95% confidence intervals. For brain n = 226 and 183; for liver n = 192 and 195; for heart n = 112 and 121; for lung n = 151 and 195; for kidney n = 184 and 204 nuclei from WT and KO mice respectively. (F) Proposed model for MeCP2 in chromatin homeostasis. MeCP2 contains highly conserved basic clusters (+++) within its disordered C-terminal region. MeCP2 first binds DNA through its MBD, but the presence of multiple DNA binding elements allow MeCP2 to alter chromatin conformation, leading to homeostasis as indicated by the recruitment of ATRX to PCH. When the C-terminus becomes truncated (e.g., G273X) this activity is reduced. Further truncation beyond the AT-Hook 2 domain (e.g., R270X) severely impairs the function of MeCP2, chromatin is no longer maintained in a physiological conformation, and ATRX is lost from PCH. See also Figure S7.
Discussion
Several models for MeCP2 function in the brain have been based on abnormalities observed in Mecp2 KO mice, but KO mice have not shed light on the postnatal regression so characteristic of RTT. In this study we generated two new mouse models of MeCP2 dysfunction that develop phenotypes at distinctly different rates, providing a parallel to the human male patients. We exploited these differences and the general features of G273X mice to gain insight into domains critical for MeCP2 function.
Many lines of evidence support a role for MeCP2 in transcriptional repression. Principal among these is the presence of a well-defined TRD mapped to amino acids 207–310 (Nan et al., 1997). When the TRD is shortened by even 10 amino acids from the C-terminus, terminating at amino acid 300, repression activity is completely lost (Nan et al., 1997). Neither MeCP2-R270X nor MeCP2-G273X displayed transcriptional repressor activity, consistent with the requirement for a fully intact TRD for these functions. Despite this shared loss, however, G273X mice live much longer and remain healthier for a significantly greater period of time than KO mice. MeCP2 clearly performs important functions besides classical transcriptional repression.
HMGA family members contain two or three AT-hooks connected by a flexible linker sequence (Reeves, 2010) and require multiple contacts with DNA in order to alter its conformation (Li et al., 2000). These features are highly reminiscent of the MeCP2 C-terminus. In contrast to the MBD, the C-terminus including the TRD region is highly disordered (Ghosh et al., 2010) and multiple non-overlapping fragments within the C-terminus are capable of binding DNA in vitro (Ghosh et al., 2010). One of these fragments (amino acids 261–330) contains the conserved AT-Hook 2 domain located between amino acids 265–272. In addition to binding naked DNA, this fragment can bind and compact nucleosomal arrays, a function that is enhanced by fusion with adjacent DNA-binding fragments of the C-terminus (Ghosh et al., 2010). Like MeCP2, HMGA proteins have a special relationship with histone H1. For example, HMGA proteins compete with H1 for binding sites (Catez et al., 2006) and overexpression of HMGA1 leads to reduced levels of certain H1 isoforms (Brocher et al., 2010). The homology of MeCP2 to HMGA1 could explain many of MeCP2’s effects on chromatin structure.
The most likely explanation for the functional differences between MeCP2-R270X and MeCP2-G273X is the loss-of-function of the AT-Hook 2 domain. We propose a model (Figure 7F) wherein MeCP2 binds with high affinity to methyl-CpG sites located throughout the genome and at concentrated domains of heterochromatin using its MBD. Once there, MeCP2 manipulates the nearby chromatin structure in a manner similar to other AT-hook-containing proteins, e.g., by altering local DNA conformation or adjusting nucleosome positioning. Additional residues C-terminal to G273 might perform separate roles or enhance the activity of the N-terminal amino acids such that without the full C-terminus MeCP2 function is reduced. Loss of the three amino acids between G273 and R270 largely abolishes the already impaired ability of MeCP2 to maintain chromatin order, hastening the loss of ATRX from PCH. The presence of disorganized chromatin might well prevent neurons from responding appropriately to stimuli important for normal synaptic function. Ultimately, impaired synaptic function causes the clinical symptoms observed in RTT. At the opposite end of the spectrum, overabundance of MeCP2 leads to a progressive restructuring of chromatin that would ordinarily be maintained by a balance between MeCP2 and other factors. The excessive accumulation of chromatin modifiers such as ATRX to domains such as PCH might induce hyper-responsiveness to stimuli and excessive synaptic function. This would lead to progressive but less severe symptoms, as observed in mice and humans that overexpress MeCP2.
The impaired binding of ATRX to PCH has itself been associated with mutations that cause the Alpha-thalassemia X-linked intellectual disability (ATRX) syndrome (Iwase et al., 2011). In Mecp2 mutant mice it will be important to investigate the consequences of ATRX loss from PCH. Knock-down of ATRX leads to genomic instability in cultured cells and is associated with impairments in metaphase chromatin condensation (Ritchie et al., 2008). ATRX interacts with the H3.3 chaperone DAXX (Drané et al., 2010) and loss of ATRX leads to depletion of H3.3 from telomeric DNA (Goldberg et al., 2010). In addition, ATRX binds to non-B form DNA structures (i.e., G-quadruplex) that are associated with repetitive sequences (Law et al., 2010). ATRX is highly related to the SNF2 family members Rad54 and ARIP4, and it has been suggested that ATRX may resolve non-B forms of DNA to regular forms (Law et al., 2010).
ATRX syndrome shows a high degree of overlap with RTT (and other MECP2 related disorders) (Gibbons and Higgs, 2000). Cognitive deficits are one common feature of both diseases. Microcephaly is another. Males are more sensitive to identical mutations than females, because of XCI. One distinction between RTT patients and those affected by ATRX syndrome is that the latter never show a period of normal development. Like MeCP2, ATRX is widely expressed in many tissues, and both proteins seem important for the proper functioning of mature neurons. If progressive ATRX dysfunction occurs downstream of MeCP2 loss only in neurons, this observation could account for the absence of hematopoetic, craniofacial, and urological abnormalities in RTT. The clinical course of males with MECP2 mutations is often more severe than male patients with disruptions in ATRX (premature lethality is not common in ATRX syndrome), but this could reflect the spectrum of mutations in ATRX that cause disease (many of which are hypomorphic alleles) (Gibbons and Higgs, 2000). Indeed, male Atrx mutant mice die in utero (Garrick et al., 2006). Given the severity of phenotypes in male patients with MECP2 mutations, it is possible that other proteins in addition to ATRX are also disrupted in neurons lacking MeCP2. The underlying processes that lead to improper ATRX targeting may also affect these other, as-yet unidentified proteins, with consequences for chromatin maintenance.
Experimental Procedures
Animals and phenotypic analysis
Generation, breeding and characterization of phenotypic severity scores is detailed in supplemental material. All mouse studies were approved by the Institutional Animal Care and Use Committee for Baylor College of Medicine. Mice were evaluated for six phenotypic categories as previously described (Guy et al., 2007).
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 5 software, Microsoft Excel, or R statistical software with the exception of the ChIP-Seq analysis and the alignments of MeCP2 and HMGA1 (see Extended Experimental Procedures). One-way ANOVA followed by Bonferroni-Holm post-hoc analysis were used to test for significance unless otherwise specified.
PAC Recombineering
Modification of the human PAC PAC671D9 to insert the Enhanced Green Fluorescent Protein (GFP) tag along with a poly-serine linker in MECP2 in place of the codons for amino acids R270 and G273 was performed as previously described (Chao et al., 2010).
Gene Expression Analysis
RNA extraction, processing, and RT-qPCR analysis were performed as previously described (Chao et al., 2010). Hippocampal RNA was polyA selected and measured using GeneChip Mouse Gene 1.0 ST Arrays (Affymetrix) by the Genomic and RNA Profiling Core at Baylor College of Medicine.
Supplementary Material
Highlights.
A 3 amino-acid shift in mutation location within MeCP2 transforms clinical course
Patients with severest disease lose 3 amino acids that encode an annotated AT-hook
MeCP2 has 3 AT-hook-like domains in a 250 amino acid stretch homologous to HMGA1
AT-hook2 domain in MeCP2 is important for chromatin maintenance and ATRX localization
Acknowledgments
We thank Christopher Woodcock for the pUC19-601×12 vector, Chris McGraw, Hsiao-Tuan Chao, Laura Lombardi, and Jeffrey Neul for critical reading of this manuscript, and Gabriele Schuster for pronuclear injections. This work was supported by the Howard Hughes Medical Institute, NINDS grant HD053862 (HYZ), the cores of the Baylor College of Medicine IDDRC (HD024064), and NINDS F30NS066527 (SAB).
Footnotes
Accession Numbers
ChIP-Seq and microarray data are deposited under GEO records GSE36536 and GSE42987, respectively.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Ankit Agrawal XH. Pairwise Statistical Significance of Local Sequence Alignment Using Sequence-Specific and Position-Specific Substitution Matrices. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 2011;8:194–205. doi: 10.1109/TCBB.2009.69. [DOI] [PubMed] [Google Scholar]
- Ben-Shachar S, Chahrour M, Thaller C, Shaw CA, Zoghbi HY. Mouse models of MeCP2 disorders share gene expression changes in the cerebellum and hypothalamus. Human Molecular Genetics. 2009;18:2431–2442. doi: 10.1093/hmg/ddp181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocher J, Vogel B, Hock R. HMGA1 down-regulation is crucial for chromatin composition and a gene expression profile permitting myogenic differentiation. BMC Cell Biology. 2010;11:64. doi: 10.1186/1471-2121-11-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catez F, Ueda T, Bustin M. Determinants of histone H1 mobility and chromatin binding in living cells. Nat Struct Mol Biol. 2006;13:305–310. doi: 10.1038/nsmb1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong STC, Qin J, Zoghbi HY. MeCP2, a Key Contributor to Neurological Disease, Activates and Represses Transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour M, Zoghbi HY. The Story of Rett Syndrome: From Clinic to Neurobiology. Neuron. 2007;56:422–437. doi: 10.1016/j.neuron.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Chang Q, Khare G, Dani V, Nelson S, Jaenisch R. The Disease Progression of Mecp2 Mutant Mice Is Affected by the Level of BDNF Expression. Neuron. 2006;49:341–348. doi: 10.1016/j.neuron.2005.12.027. [DOI] [PubMed] [Google Scholar]
- Chao HT, Chen H, Samaco RC, Xue M, Chahrour M, Yoo J, Neul JL, Gong S, Lu HC, Heintz N, et al. Dysfunction in GABA signalling mediates autism-like stereotypies and Rett syndrome phenotypes. Nature. 2010;468:263–269. doi: 10.1038/nature09582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RZ, Akbarian S, Tudor M, Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet. 2001;27:327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- Collins AL, Levenson JM, Vilaythong AP, Richman R, Armstrong DL, Noebels JL, David Sweatt J, Zoghbi HY. Mild overexpression of MeCP2 causes a progressive neurological disorder in mice. Human Molecular Genetics. 2004;13:2679–2689. doi: 10.1093/hmg/ddh282. [DOI] [PubMed] [Google Scholar]
- Drané P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes & Development. 2010;24:1253–1265. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Free A, Wakefield RID, Smith BO, Dryden DTF, Barlow PN, Bird AP. DNA Recognition by the Methyl-CpG Binding Domain of MeCP2. Journal of Biological Chemistry. 2001;276:3353–3360. doi: 10.1074/jbc.M007224200. [DOI] [PubMed] [Google Scholar]
- Fyffe SL, Neul JL, Samaco RC, Chao HT, Ben-Shachar S, Moretti P, McGill BE, Goulding EH, Sullivan E, Tecott LH, et al. Deletion of Mecp2 in Sim1-Expressing Neurons Reveals a Critical Role for MeCP2 in Feeding Behavior, Aggression, and the Response to Stress. Neuron. 2008;59:947–958. doi: 10.1016/j.neuron.2008.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrick D, Sharpe JA, Arkell R, Dobbie L, Smith AJH, Wood WG, Higgs DR, Gibbons RJ. Loss of Atrx Affects Trophoblast Development and the Pattern of X-Inactivation in Extraembryonic Tissues. PLoS Genet. 2006;2:e58. doi: 10.1371/journal.pgen.0020058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgel PT, Horowitz-Scherer RA, Adkins N, Woodcock CL, Wade PA, Hansen JC. Chromatin Compaction by Human MeCP2: ASSEMBLY OF NOVEL SECONDARY CHROMATIN STRUCTURES IN THE ABSENCE OF DNA METHYLATION. Journal of Biological Chemistry. 2003;278:32181–32188. doi: 10.1074/jbc.M305308200. [DOI] [PubMed] [Google Scholar]
- Ghosh RP, Nikitina T, Horowitz-Scherer RA, Gierasch LM, Uversky VN, Hite K, Hansen JC, Woodcock CL. Unique Physical Properties and Interactions of the Domains of Methylated DNA Binding Protein 2. Biochemistry. 2010;49:4395–4410. doi: 10.1021/bi9019753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons RJ, Higgs DR. Molecular–clinical spectrum of the ATR-X syndrome. American Journal of Medical Genetics. 2000;97:204–212. doi: 10.1002/1096-8628(200023)97:3<204::AID-AJMG1038>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, et al. Distinct Factors Control Histone Variant H3.3 Localization at Specific Genomic Regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J, Gan J, Selfridge J, Cobb S, Bird A. Reversal of Neurological Defects in a Mouse Model of Rett Syndrome. Science. 2007;315:1143–1147. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27:322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- Iwase S, Xiang B, Ghosh S, Ren T, Lewis PW, Cochrane JC, Allis CD, Picketts DJ, Patel DJ, Li H, et al. ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome. Nat Struct Mol Biol. 2011;18:769–776. doi: 10.1038/nsmb.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankirawatana P, Leonard H, Ellaway C, Scurlock J, Mansour A, Makris CM, Dure LS, Friez M, Lane J, Kiraly-Borri C, et al. Early progressive encephalopathy in boys and MECP2 mutations. Neurology. 2006;67:164–166. doi: 10.1212/01.wnl.0000223318.28938.45. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Sarraf SA, Schmiedeberg L, McDermott SM, Stancheva I, Bird AP. DNA Binding Selectivity of MeCP2 Due to a Requirement for A/T Sequences Adjacent to Methyl-CpG. Molecular Cell. 2005;19:667–678. doi: 10.1016/j.molcel.2005.07.021. [DOI] [PubMed] [Google Scholar]
- Law MJ, Lower KM, Voon HPJ, Hughes JR, Garrick D, Viprakasit V, Mitson M, De Gobbi M, Marra M, Morris A, et al. ATR-X Syndrome Protein Targets Tandem Repeats and Influences Allele-Specific Expression in a Size-Dependent Manner. Cell. 2010;143:367–378. doi: 10.1016/j.cell.2010.09.023. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to Methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- Li L, Yoder K, Hansen MST, Olvera J, Miller MD, Bushman FD. Retroviral cDNA Integration: Stimulation by HMG I Family Proteins. Journal of Virology. 2000;74:10965–10974. doi: 10.1128/jvi.74.23.10965-10974.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill BE, Bundle SF, Yaylaoglu MB, Carson JP, Thaller C, Zoghbi HY. Enhanced anxiety and stress-induced corticosterone release are associated with increased Crh expression in a mouse model of Rett syndrome. Proceedings of the National Academy of Sciences. 2006;103:18267–18272. doi: 10.1073/pnas.0608702103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Campoy FJ, Bird A. MeCP2 Is a Transcriptional Repressor with Abundant Binding Sites in Genomic Chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- Nan X, Hou J, Maclean A, Nasir J, Lafuente MJ, Shu X, Kriaucionis S, Bird A. Interaction between chromatin proteins MECP2 and ATRX is disrupted by mutations that cause inherited mental retardation. Proceedings of the National Academy of Sciences. 2007;104:2709–2714. doi: 10.1073/pnas.0608056104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Nikitina T, Ghosh RP, Horowitz-Scherer RA, Hansen JC, Grigoryev SA, Woodcock CL. MeCP2-Chromatin Interactions Include the Formation of Chromatosome-like Structures and Are Altered in Mutations Causing Rett Syndrome. Journal of Biological Chemistry. 2007a;282:28237–28245. doi: 10.1074/jbc.M704304200. [DOI] [PubMed] [Google Scholar]
- Nikitina T, Shi X, Ghosh RP, Horowitz-Scherer RA, Hansen JC, Woodcock CL. Multiple Modes of Interaction between the Methylated DNA Binding Protein MeCP2 and Chromatin. Molecular and Cellular Biology. 2007b;27:864–877. doi: 10.1128/MCB.01593-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravn K, Nielsen JB, Uldall P, Hansen FJ, Schwartz M. No correlation between phenotype and genotype in boys with a truncating MECP2 mutation. Journal of Medical Genetics. 2003;40:e5. doi: 10.1136/jmg.40.1.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R. Nuclear functions of the HMG proteins. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 2010;1799:3–14. doi: 10.1016/j.bbagrm.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie K, Seah C, Moulin J, Isaac C, Dick F, Bérubé NG. Loss of ATRX leads to chromosome cohesion and congression defects. The Journal of Cell Biology. 2008;180:315–324. doi: 10.1083/jcb.200706083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaco RC, Mandel-Brehm C, McGraw CM, Shaw CA, McGill BE, Zoghbi HY. Crh and Oprm1 mediate anxiety-related behavior and social approach in a mouse model of MECP2 duplication syndrome. Nat Genet. 2012;44:206–211. doi: 10.1038/ng.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüle B, Armstrong DD, Vogel H, Oviedo A, Francke U. Severe congenital encephalopathy caused by MECP2 null mutations in males: central hypoxia and reduced neuronal dendritic structure. Clinical Genetics. 2008;74:116–126. doi: 10.1111/j.1399-0004.2008.01005.x. [DOI] [PubMed] [Google Scholar]
- Schwarz PM, Felthauser A, Fletcher TM, Hansen JC. Reversible Oligonucleosome Self-Association:ý Dependence on Divalent Cations and Core Histone Tail Domains†. Biochemistry. 1996;35:4009–4015. doi: 10.1021/bi9525684. [DOI] [PubMed] [Google Scholar]
- Skene PJ, Illingworth RS, Webb S, Kerr ARW, James KD, Turner DJ, Andrews R, Bird AP. Neuronal MeCP2 Is Expressed at Near Histone-Octamer Levels and Globally Alters the Chromatin State. Molecular Cell. 2010;37:457–468. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B. ChIP with Native Chromatin: Advantages and Problems Relative to Methods Using Cross-Linked Material. Paris: Institut national de la santé et de la recherche médicale (INSERM); 2001. [PubMed] [Google Scholar]
- Venancio M, Santos M, Pereira SA, Maciel P, Saraiva JM. An explanation for another familial case of Rett syndrome: maternal germline mosaicism. Eur J Hum Genet. 2007;15:902–904. doi: 10.1038/sj.ejhg.5201835. [DOI] [PubMed] [Google Scholar]
- Villard L. MECP2 mutations in males. Journal of Medical Genetics. 2007;44:417–423. doi: 10.1136/jmg.2007.049452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.