Abstract
Purpose
A greater understanding of the biology of tumor recurrence should improve adjuvant treatment decision making. We conducted a validation study of the 12-gene recurrence score (RS), a quantitative assay integrating stromal response and cell cycle gene expression, in tumor specimens from patients enrolled onto Cancer and Leukemia Group B (CALGB) 9581.
Patients and Methods
CALGB 9581 randomly assigned 1,713 patients with stage II colon cancer to treatment with edrecolomab or observation and found no survival difference. The analysis reported here included all patients with available tissue and recurrence (n = 162) and a random (approximately 1:3) selection of nonrecurring patients. RS was assessed in 690 formalin-fixed paraffin-embedded tumor samples with quantitative reverse transcriptase polymerase chain reaction by using prespecified genes and a previously validated algorithm. Association of RS and recurrence was analyzed by weighted Cox proportional hazards regression.
Results
Continuous RS was significantly associated with risk of recurrence (P = .013) as was mismatch repair (MMR) gene deficiency (P = .044). In multivariate analyses, RS was the strongest predictor of recurrence (P = .004), independent of T stage, MMR, number of nodes examined, grade, and lymphovascular invasion. In T3 MMR-intact (MMR-I) patients, prespecified low and high RS groups had average 5-year recurrence risks of 13% (95% CI, 10% to 16%) and 21% (95% CI, 16% to 26%), respectively.
Conclusion
The 12-gene RS predicts recurrence in stage II colon cancer in CALGB 9581. This is consistent with the importance of stromal response and cell cycle gene expression in colon tumor recurrence. RS appears to be most discerning for patients with T3 MMR-I tumors, although markers such as grade and lymphovascular invasion did not add value in this subset of patients.
INTRODUCTION
The individualization of cancer care requires a deep understanding of tumor biology and the identification of subsets of tumors that offer targets for tumor-specific treatment. Colorectal cancer does not yet fit this model because the only clearly clinically applicable genomic information is KRAS status of advanced colorectal cancers, in which mutations predict lack of efficacy of epidermal growth factor receptor (EGFR) antibodies. This is in contrast to breast cancer, in which the status of estrogen receptor, progesterone receptor, human epidermal growth factor receptor 2 (HER2), and the 21-gene recurrence score (RS; among other factors) inform treatment decision making.1–5
No group of patients would reap more benefit from the identification of prognostic and predictive markers than those with stage II colon cancer. The need to balance the relatively low risk of disease recurrence with only modest benefit of adjuvant therapy in the face of toxicities and even treatment-related deaths challenges oncologists and patients alike.6 In practice, adjuvant therapy is generally offered to stage II patients believed to have higher recurrence risk based on the expectation that high-risk patients may derive larger absolute benefits with postoperative chemotherapy than patients at low risk of recurrence.7
Clinical factors thought to be associated with increased risk of recurrence in stage II colon cancer include clinical and pathologic tumor features such as T4 stage, bowel perforation or obstruction, inadequate nodal assessment (fewer than 12 lymph nodes examined), high tumor grade, and lymphovascular invasion (LVI).8 Any of these conventional features classifies a patient as high risk and may direct the recommendation toward adjuvant chemotherapy.7 However, the variability in the level of evidence supporting each of these factors and the lack of standardization in their assessment reduce the confidence that these features are informative.6,7,9 In reality, tumor grade, for example, is not always associated with increased recurrence risk in stage II disease,10–12 and grade and LVI are subjectively determined and often not reported.8,11
Due in part to this lack of clarity, there is an ongoing effort to identify genomic markers that could reliably predict recurrence risk and treatment benefit in stage II colon cancer. A major challenge, however, is the need for consistent results from well-powered, prospectively designed studies, and this conundrum partially explains the paucity of markers that have achieved the level of evidence to support clinical application.13 Deficiency of the MMR (mismatch repair genes) pathway is associated with lower recurrence risk in stage II colon cancer and may also predict a poorer outcome with fluorouracil-based adjuvant chemotherapy.14–16 The nearly universal finding of significantly lower recurrence risk in patients with MMR-deficient (MMR-D) tumors across multiple large, independent studies has led to its growing use as a factor arguing against adjuvant therapy in clinical practice for stage II patients.17,18
Multigene assays might more reliably give insight into tumor biology and the risk of recurrence than single-gene analysis, and this has motivated the development of several multigene panels. One such panel is the Oncotype DX Colon Cancer Recurrence Score (Genomic Health, Redwood City, CA). Developed by using tumor gene expression data from 1,851 patients with resected colon cancer in four independent trials,12 it has been validated as a predictor of recurrence risk in stage II patients with colon cancer from the QUASAR study.10 In that prospectively designed validation study, continuous RS was significantly associated with the risk of tumor recurrence, and in a prespecified multivariate analysis, RS predicted recurrence risk independently from MMR status, T stage, tumor grade, number of nodes examined, and LVI. The added value of the RS was most evident in patients with T3 MMR-intact (MMR-I) tumors, in which the classic prognostic factors demonstrated limited utility.10
The 12-gene RS is calculated from an analytically validated and standardized19 quantitative reverse transcriptase polymerase chain reaction assay that measures the expression of 12 genes (seven recurrence genes and five reference genes) in formalin-fixed paraffin-embedded (FFPE) primary colon tumor tissue. The applicability of the RS is reinforced by the identity of the recurrence genes and the measured changes in tumor biology produced by their dysregulation. Six of the seven genes are in two key biologic pathways: cell cycle control (MKI67, MYC, MYBL2) and stromal response (FAP, BGN, INHBA).12 The seventh recurrence gene (GADD45B) is a marker of genotoxic stress and may regulate activity of stromal response genes, including BGN.20 The association of cell cycle control and stromal response genes with colon tumor aggressiveness is supported in animal models as well as exploratory studies in human tumor samples.21–28
Cancer and Leukemia Group B (CALGB) 9581 was a randomized phase III clinical trial conducted by the CALGB, which found no effect of adjuvant edrecolomab (anti-EpCAM, monoclonal antibody 17-1A) compared with observation in patients with resected stage II colon cancer.29 Because of the absence of impact of therapy, the patients enrolled onto this trial provided a valuable resource for the study of pathologic and molecular determinants of outcome in a large cohort of stage II patients essentially treated with surgery alone with prospectively collected FFPE tumor tissue. CALGB 9581 excluded patients with highest risk, such as obstruction or perforation, and the overall risk of recurrence in the study was 14%. To further our understanding of the biologic pathways underlying recurrence risk in stage II colon cancer and to confirm and extend the findings from the QUASAR validation study, we conducted an independent, prospectively designed study in patient specimens from CALGB 9581 to test the 12-gene RS for association with risk of recurrence. Analyses were prespecified to determine the relationship of RS to recurrence in the setting of conventional clinical and pathologic risk factors.
PATIENTS AND METHODS
Patients and Tissue Specimens
Patients with stage II colon cancer enrolled onto both arms of CALGB 9581 who had available tumor tissue were eligible. A cohort sampling study design was used to make the most efficient use of tissue samples in a population with relatively low risk of recurrence (14.6% at 5 years).30 All patients with recurrence and a random sample of patients without recurrence were identified for inclusion in the study (1:3 ratio). Sampling was stratified on treatment, and no matching was performed. CALGB statisticians identified the patient subsample, and the related archived specimens (FFPE colon tissue) were sent to Genomic Health by the CALGB Pathology Coordinating Office.
Pathology
Tumor grade and tumor type were centrally assessed according to the College of American Pathologists Consensus Statement, with mucinous carcinomas characterized as high-grade tumor.8 Data on pathologic T stage, number of nodes examined, tumor location, LVI, and evidence of bowel obstruction or perforation were obtained from the CALGB clinical database. MMR was assessed by immunohistochemistry for hMLH1 and hMSH211 by CALGB investigators, as previously described,31 with the modification that the threshold for scoring loss of expression was less than 5% tumor cells staining.
Gene Expression
RNA was extracted at Genomic Health from six 5-μm sections obtained from a single representative archived FFPE colon tumor tissue block for each patient. Nontumor elements were removed by manual microdissection before RNA extraction. Extracted RNA was quantified by RiboGreen (Invitrogen, Carlsbad, CA) and analyzed by reverse transcriptase polymerase chain reaction. The 12-gene RS was calculated by using the prespecified genes and algorithm previously validated in QUASAR.10 Prespecified percentile cut points (40th and 75th percentile) were used to define low, intermediate, and high RS groups. All centrally performed pathology and laboratory assay procedures were prespecified and conducted without knowledge of the clinical characteristics or outcomes.
Statistical Methods
The primary study end point was recurrence-free interval, defined as time from random assignment to documented first colon cancer recurrence or death as a result of colon cancer. Deaths resulting from other causes were censored. New primary colon cancers were ignored.
A weighted Cox proportional hazards model evaluated the association between RS and recurrence-free interval based on a Wald-type test statistic constructed by using a weighted partial pseudolikelihood estimate and robust variance estimate.30 A two-sided P value less than .04 was considered significant. An alternative score, not discussed in this article, was assessed for validation at a significance level of 0.01. Assessment of proportional hazards was performed by examining the relationship between scaled Schoenfeld residuals and time.32,33 Functional forms were examined by inclusion of splines in the Cox model. Patient characteristics were summarized by using weighted means and proportions. Analyses used SAS version 9.2 (SAS, Cary, NC), and all statistical analyses were conducted by CALGB statisticians in collaboration with Genomic Health.
RESULTS
Patient Characteristics
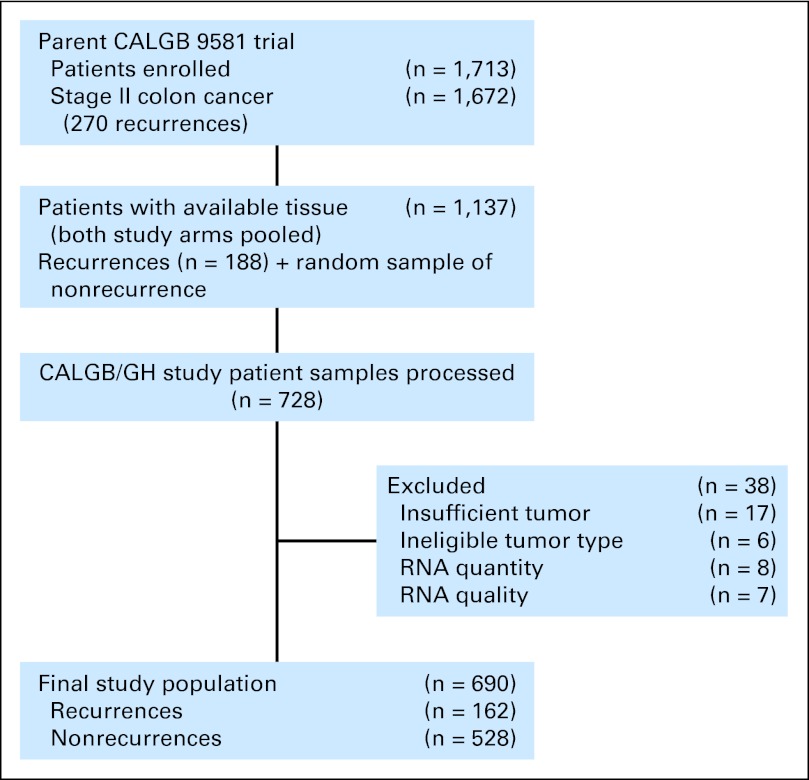
Tissue was collected for 1,137 (68%) of 1,672 patients with stage II colon cancer enrolled onto CALGB 9581. By using all recurrent patients with banked tissue and a random sample of nonrecurrent patients, 728 patient samples were processed. Following prespecified procedures for pathology and laboratory processing, 38 (5.2%) of 728 specimens were excluded, primarily for insufficient tumor tissue (Appendix Fig A1, online only). The final evaluable data set contained 690 patients, 162 of whom had recurrence.
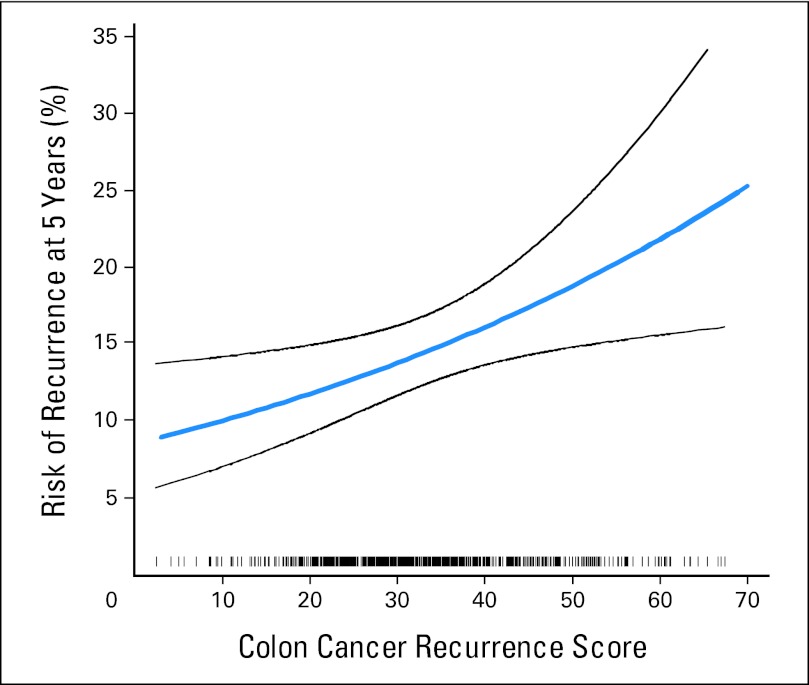
Fig 1.
Relationship between the continuous recurrence score and 5-year recurrence risk. Black lines represent 95% CIs. A rug plot depicting the distribution of recurrence score is included at the bottom of the figure.
Patients enrolled onto CALGB 9581 were generally older (35.5% were age ≥ 70 years) and had lower recurrence risk, as manifested by lower proportions of T4 patients (4.6%) and a higher proportion of MMR-D patients (21.4%) compared with other published series of patients with stage II colon cancer (Table 1).29 The distribution of demographic and baseline clinical characteristics for patients evaluated in this study was similar to that of CALGB 9581 patients who were not included, with the exception of T stage and number of nodes examined (both P <.05), although the differences were small (Table 1).
Table 1.
Demographics and Baseline Clinical Characteristics
| Characteristic | Patients With Stage II Colon Cancer |
|||||
|---|---|---|---|---|---|---|
| Included in Study (n = 690) |
Not Included in Study (n = 982) |
In CALGB 9581 (N = 1,672) |
||||
| No. | % (weighted) | No. | % (weighted) | No. | % | |
| Age, years | ||||||
| < 60 | 214 | 31.3 | 323 | 32.9 | 537 | 32.1 |
| 60-70 | 231 | 33.6 | 311 | 31.6 | 542 | 32.4 |
| ≥ 70 | 245 | 35.0 | 348 | 35.5 | 593 | 35.5 |
| Sex | ||||||
| Male | 360 | 51.9 | 513 | 52.4 | 873 | 52.2 |
| Race/ethnicity | ||||||
| White | 630 | 92.0 | 894 | 91.4 | 1,524 | 91.5 |
| Year of surgery | ||||||
| ≤ 1998 | 134 | 19.5 | 140 | 14.3 | 274 | 16.4 |
| 1999-2000 | 343 | 50.2 | 540 | 55.2 | 883 | 52.8 |
| ≥ 2001 | 213 | 30.3 | 302 | 30.5 | 515 | 30.8 |
| Treatment arm | ||||||
| Observation | 343 | 50.4 | 499 | 50.4 | 842 | 50.4 |
| Edrecolomab | 347 | 49.6 | 483 | 49.6 | 830 | 49.6 |
| T stage | ||||||
| T4 | 41 | 5.9 | 35 | 3.7 | 76 | 4.6 |
| < 12 Nodes examined | 327 | 47.1 | 427 | 44.2 | 754 | 45.1 |
| Lymphovascular invasion present | 78 | 10.9 | 112 | 11.5 | 190 | 11.4 |
| MMR deficient | 137 | 21.5 | 48 | 22.9 | 185 | 21.4 |
| Obstruction or perforation present | 11 | 1.7 | 19 | 2.1 | 30 | 1.8 |
| Tumor location (right side) | 360 | 52.8 | 492 | 50.1 | 852 | 51.0 |
| High-grade central tumor | 220 | 32.4 | N/A | N/A | N/A | N/A |
| Central mucinous histology present | 124 | 18.4 | N/A | N/A | N/A | N/A |
Abbreviations: CALGB, Cancer and Leukemia Group B; MMR, mismatch repair; N/A, not applicable.
Association of RS With Recurrence Risk
RS values ranged from 2 to 78, with a median score of 31.4 (interquartile range, 24.7 to 39.0) and a mean of 33 ± 11.5 standard deviation units. In the primary analysis, based on the Cox model, RS was significantly associated with recurrence risk, with a hazard ratio of 1.52 for a 25-unit increase in RS (95% CI, 1.09 to 2.12; P = .013; Fig 1). The proportional hazards assumption held (P = .42 for the test of nonzero slope of Shoenfeld residuals v time). There was no evidence of nonlinearity in the relationship between continuous RS and the log hazard of recurrence (P = .56 for the test of nonlinearity). In the overall study population, estimates of average 5-year recurrence risk from the Cox model with continuous RS for the low-, intermediate-, and high-percentile groups (defined by RS values of 29 [40th percentile] and 39 [75th percentile]) were 12% (95% CI, 10% to 15%), 15% (95% CI, 12% to 17%), and 18% (95% CI, 14% to 22%), respectively. Similar results were obtained by using RS cut points of 30 and 41 as defined in QUASAR10 (data not shown).
RS in the Context of Conventional Clinical/Pathologic Factors
Among conventional clinical/pathologic factors, MMR was the only covariate significantly (P < .05) associated with recurrence in univariate analyses, with MMR-D tumors associated with lower recurrence risk (Table 2). LVI and number of nodes examined (as a continuous variable) were of borderline significance, with effects in the expected directions. T4 stage was not a significant predictor of recurrence in this patient population, likely because of the exclusion of T4 tumors with invasion of adjacent organs or structures (pT4b) and the low overall frequency of T4 patients in CALGB 9581.
Table 2.
Univariate Analysis: Relationship of Clinical and Pathologic Covariates With Risk of Recurrence
| Variable | HR | 95% CI | P |
|---|---|---|---|
| MMR (deficient v intact) | 0.62 | 0.39 to 0.99 | .044 |
| T stage (T4 v T3) | 1.19 | 0.60 to 2.37 | .62 |
| No. of nodes examined (< 12 v ≥ 12) | 1.17 | 0.85 to 1.62 | .34 |
| No. of nodes examined, continuous (per node) | 0.98 | 0.96 to 1.00 | .062 |
| Tumor grade (high v low) | 0.74 | 0.52 to 1.07 | .11 |
| Lymphovascular invasion (present v absent) | 1.56 | 0.98 to 2.50 | .062 |
| Mucinous histology (mucinous v not) | 0.73 | 0.46 to 1.16 | .18 |
| Tumor location (right-sided v other) | 0.79 | 0.57 to 1.10 | .16 |
| Age (≥ 70 v < 70 years) | 1.21 | 0.87 to 1.69 | .26 |
| Age, continuous (per year) | 1.01 | 1.00 to 1.03 | .15 |
| Sex (male v female) | 1.14 | 0.82 to 1.58 | .42 |
Abbreviations: HR, hazard ratio; MMR, mismatch repair.
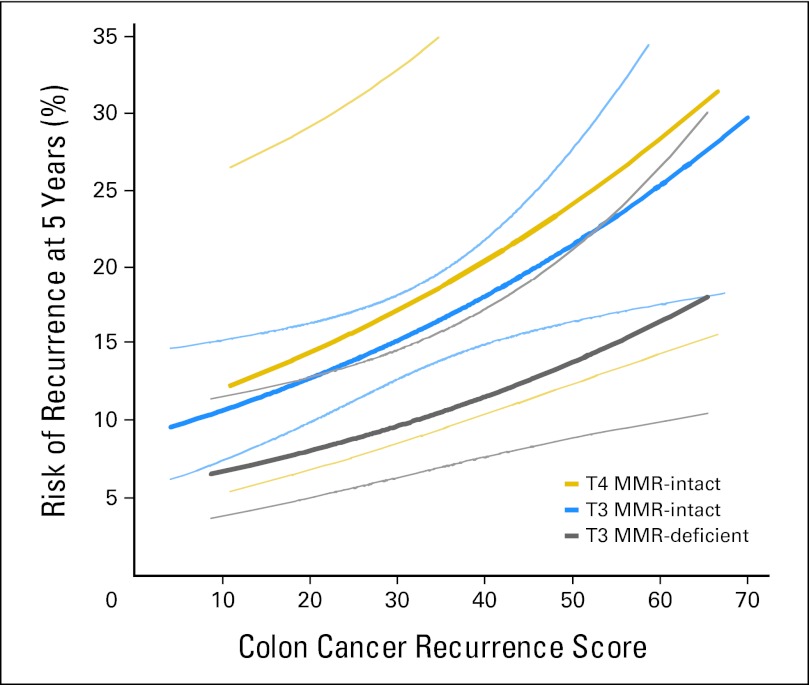
In multivariable analysis, RS was a significant predictor of recurrence risk (P = .004), providing independent value beyond MMR, T stage, number of nodes examined, grade, and LVI (Table 3). A wide range of RS values was observed within each level of the covariates, such as MMR, T stage, number of nodes examined, LVI, and tumor grade, indicating that RS revealed underlying biology not captured by the traditional clinical and pathologic factors (Fig 2). In addition, in a model examining the contribution of RS to prediction of recurrence risk in the context of MMR and T stage, the most consistent prognostic covariates in stage II colon cancer—RS (P = .007) and MMR (P = .02)—were significant predictors of recurrence (Appendix Table A1, online only; Fig 3). Among T3 MMR-I patients (74% of patients with MMR results), 44% were in the low RS and 22% were in the high RS groups. For T3 MMR-I patients, model-based estimates of average 5-year recurrence risk in the prespecified low, intermediate, and high RS groups were 13% (95% CI, 10% to 16%), 16% (95% CI, 13% to 19%), and 21% (95% CI, 16% to 26%), respectively.
Fig 2.
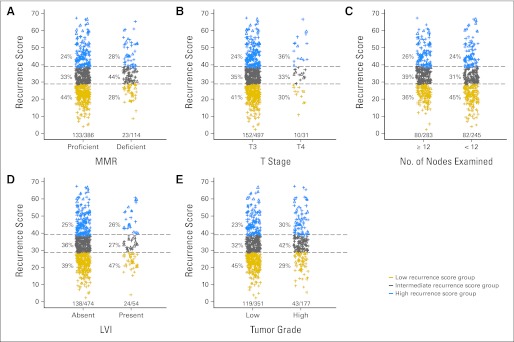
Distribution of the recurrence score by levels of covariates. Distribition of recurrence score values by (A) levels of mismatch repair (MMR) status, (B) T stage, (C) number of nodes examined, (D) lymphovascular invasion (LVI), and (D) tumor grade. Triangles represent recurrence scores for patients experiencing recurrence and plus signs represent recurrence scores from patients not experiencing recurrence. The numbers under each bar (X/Y) represent the number of recurrences/nonrecurrences for each level of the covariate.
Fig 3.
Relationship between the continuous recurrence score and 5-year recurrence risk in the context of T stage and mismatch repair (MMR), on the basis of the Cox model, including continuous recurrence score, MMR, T stage, and an interaction of T stage and MMR. Thin lines represent 95% CIs. The number of patients with T4 and MMR-deficient tumors was small (n = 7; no events), limiting reliable estimation of their recurrence risk. Thus, results for these patients, although they were included in the analysis, are not shown in the figure.
Table 3.
Multivariable Analysis: Relationship of Clinical and Pathologic Covariates With Risk of Recurrence
| Variable | HR | 95% CI | P |
|---|---|---|---|
| MMR (deficient v intact) | 0.70 | 0.42 to 1.17 | .17 |
| T stage (T4 v T3) | 0.93 | 0.44 to 1.97 | .85 |
| No. of nodes examined (< 12 v ≥ 12) | 1.14 | 0.81 to 1.60 | .46 |
| Tumor grade (high v low) | 0.78 | 0.51 to 1.18 | .24 |
| Lymphovascular invasion (present v absent) | 1.39 | 0.85 to 2.26 | .19 |
| RS, continuous (per 25 units) | 1.68 | 1.18 to 2.38 | .004 |
Abbreviations: HR, hazard ratio; MMR, mismatch repair; RS, recurrence score.
DISCUSSION
In this study, the 12-gene RS was significantly associated with risk of tumor recurrence in patients with stage II colon cancer and was prognostic for outcome independent of conventional clinical and pathologic features. Of the conventional risk parameters, only MMR status was also prognostic. The most common subset of patients was those with T3 MMR-I tumors, and the continuous RS was independently associated with recurrence risk in these patients. Consistency of these findings with findings from QUASAR convincingly confirms that RS can provide information regarding the likelihood of occult tumor metastasis and recurrence, which can help inform adjuvant therapy recommendations in practice.
From a biologic perspective, these results are consistent with the involvement of cell cycle control and stromal response as determinants of tumor recurrence in colon cancer. Unlike in breast cancer, studies of colon cancer show a weak correlation between MKI67 protein level and number of cells in S phase.25 Here, the association of cell cycle control gene expression with low risk of recurrence may reflect the importance of cell cycle regulation in response to DNA damage or misalignment of chromosomes during mitosis: tumors that possess at least some checkpoint control may have lower mutation rates and loss of heterozygosity, and therefore may have lower metastatic potential. In contrast, association of stromal response genes with higher recurrence risk is consistent with observations in multiple cancers, including breast, lung, and prostate cancer.24,26,34 The stromal response genes included in the RS—fibroblast activation protein (FAP), inhibin A (INHBA), and biglycan (BGN)—are part of a tightly coexpressed genomic program characterized by activated transforming growth factor beta (TGFβ) signaling. Activated tumor-stroma TGFβ signaling is known to produce a wound healing response, with upregulation of extracellular matrix proteins and an environment rich in growth, inflammatory, angiogenic, and invasion factors conducive to the growth of tumor cells.24,26
The possible role in recurrence of cancer stem cells (CSCs)35,36 provides further context for understanding the importance of cell cycle control and stromal response in colon cancer metastases. In the stage II setting, in which there is no macroscopic disease, the presumptive mechanism of recurrence includes the dissemination of CSCs that are capable of propagating into metastases at distant sites. The molecular predictors of recurrence risk, by logical extension, likely represent biologic pathways associated with these behaviors. It is thus reasonable to speculate that expression of cell cycle control and stromal response genes might relate to colon CSC behavior and to a tumor-stroma interaction in the dissemination of CSCs.
It is plausible that directing inhibitors toward elements of the pathways identified in the RS could lead to the development of novel adjuvant therapies for colon cancer. Given its limited expression in normal tissue, FAP is being targeted in early-phase clinical studies (eg, monoclonal antibody F19/sibrotuzumab).37–41 Activation of the TGFβ pathway upregulates COX-2 expression,42 and COX-2 inhibition can alter TGFβ pathway activity.43–45 Patients whose tumors have activated stromal response and TGFβ signaling may therefore have differential benefit from COX-2–targeted therapies. We expect that this and other clinical and translational studies will provide additional insights into the roles of cell cycle control and stromal response genes in colon tumor behavior.
The findings in this study also challenge us to re-examine the traditional approach to risk assessment and treatment planning in stage II colon cancer. Most of the conventional clinical and pathologic features in use suffer from lack of standardization, reproducibility, and prospective validation criteria which should apply to any marker, new or old, for clinical decision making.46 In this large study of well-defined stage II patients, many of the traditional markers, including tumor grade and LVI, proved to have little prognostic value. By contrast, the consistent results with the 12-gene RS across two large, prospectively designed validation studies (QUASAR and CALGB 9581) and multiple studies with consistent findings for MMR14–18 demonstrate that these molecular tools can complement and improve on existing approaches to risk assessment in clinical practice.
CALGB 9581 provides evidence of the importance of mandating the acquisition and storage of tumor tissue. Although the clinical intervention edrecolomab was ineffective, the prospective collection of biospecimens allowed us to test molecular markers in a population of patients with stage II colon cancer who had nearly 8 years of follow-up in a study that included a random assignment to observation. The cohort of patients was clearly a group with relatively low risk, because of exclusion of tumors with selected high-risk features (T4 with invasion of adjacent organs or structures, bowel perforation or obstruction, and positive margins). It should be noted that stage II patients who were ineligible for this study according to the exclusion criteria account for a small minority (15% to 20%) of patients with stage II disease. In addition, the age distribution (35% of patients age ≥ 70 years) and degree of nodal sampling make this population reasonably representative of the average patient likely to present with stage II colon cancer.
Given the low recurrence rate in CALGB 9581 (14%) compared with the benchmark of 20% for unselected stage II patients in the late 1990s, the finding that RS was the strongest predictor of recurrence risk in univariable and multivariable analyses was particularly notable. Although the degree of risk discrimination is modest when comparing low versus high RS groups, the greatest value of the RS is apparent when RS is applied as a continuous measure for individual patients (risk range, 9% to 26%), particularly for the subset of patients with T3 MMR-I and high RS disease (16% of the total population).
In QUASAR, patients across the range of RS derived similar relative risk reduction with adjuvant fluorouracil/leucovorin, meaning that patients with high RS disease would accrue larger absolute benefits with adjuvant chemotherapy compared with patients with low RS disease.10 Following on the results from QUASAR, the results presented here confirm the accuracy of the standardized, validated RS and its relevance for patients with T3 MMR-I tumors, in which further risk discrimination may be factored into the decision of whether or not to offer adjuvant chemotherapy. In addition, the genes comprising the RS highlight biologic pathways that may be most responsible for cancer recurrence and represent genes that should be interrogated further to identify promising targets in this new era of drug development. Hopefully, the results presented here will lead to both immediate and future improvements in treatment for patients with colorectal cancer.
Glossary Terms
- COX-2:
A prostaglandin endoperoxide synthase, COX enzymes are responsible for the production of prostaglandins, intracellular messengers found at high levels at inflammation sites. Of COX-1 and COX-2, the latter has received much attention due to drug development that has targeted COX-2 for selectively down regulating inflammatory processes.
- Cox Proportional hazards:
The Cox proportional hazards regression model is a statistical model for regression analysis of censored survival data. It examines the relationship of censored survival distribution to one or more covariates. It produces a baseline survival curve, covariate coefficient estimates with their standard errors, risk ratios, 95% CIs, and significance levels.
- MMR (mismatch repair genes):
Mismatch repair genes recognize and correct errors in DNA replication leading to single base-pair mismatches or insertions/deletions in small repetitive tracts of DNA known as microsatellites.
- Recurrence-free interval:
Time from randomization to documented first recurrence or death due to original cancer.
- Stromal response genes:
Genes within the surrounding tissues that control the reaction of the supporting stromal cells to the presence of cancer cells.
Appendix
The following institutions participated in this study: Cancer Centers of the Carolinas, Greenville, SC: Jeffrey K. Giguere, MD, supported by CA29165; Christiana Care Health Services Community Clinical Oncology Program (CCOP), Wilmington, DE: Stephen Grubbs, MD, supported by CA45418; Dana-Farber Cancer Institute, Boston, MA: Harold J. Burstein, MD, PhD, supported by CA32291; Dartmouth Medical School-Norris Cotton Cancer Center, Lebanon, NH: Konstantin Dragnev, MD, supported by CA04326; Duke University Medical Center, Durham, NC: Jeffrey Crawford, MD, supported by CA47577; Georgetown University Medical Center, Washington, DC: Minetta C. Liu, MD, supported by CA77597; Monter Cancer Center of North Shore-Long Island Jewish Health Systems, Lake Success, NY: Daniel Budman, MD, supported by CA35279; Massachusetts General Hospital, Boston, MA: Jeffrey W. Clark, MD, supported by CA32291; Memorial Sloan-Kettering Cancer Center, New York, NY: Clifford A. Hudis, MD, supported by CA77651; Mount Sinai School of Medicine, New York, NY: Lewis R. Silverman, MD, supported by CA04457; Nevada Cancer Research Foundation CCOP, Las Vegas, NV: John A. Ellerton, MD, supported by CA35421; Northern Indiana Cancer Research Consortium CCOP, South Bend, IN: Rafat Ansari, MD, supported by CA86726; Rhode Island Hospital, Providence, RI: William Sikov, MD, supported by CA08025; Roswell Park Cancer Institute, Buffalo, NY: Ellis Levine, MD, supported by CA59518; Southeast Cancer Control Consortium CCOP, Goldsboro, NC: James N. Atkins, MD, supported by CA45808; State University of New York Upstate Medical University, Syracuse, NY: Stephen L. Graziano, MD, supported by CA21060; The Ohio State University Medical Center, Columbus, OH: Clara D. Bloomfield, MD, supported by CA77658; University of California at San Diego, San Diego, CA: Barbara A. Parker, MD, supported by CA11789; University of California at San Francisco, San Francisco, CA: Charles J. Ryan, MD, supported by CA60138; University of Chicago, Chicago, IL: Hedy L. Kindler, MD, supported by CA41287; University of Illinois Minority-Based CCOP, Chicago, IL: David J. Peace, MD, supported by CA74811; University of Iowa, Iowa City, IA: Daniel A. Vaena, MD, supported by CA47642; University of Maryland Greenebaum Cancer Center, Baltimore, MD: Martin Edelman, MD, supported by CA31983; University of Massachusetts Medical School, Worcester, MA: William V. Walsh, MD, supported by CA37135; University of Minnesota, Minneapolis, MN: Bruce A. Peterson, MD, supported by CA16450; University of Missouri/Ellis Fischel Cancer Center, Columbia, MO: Karl E. Freter, MD, supported by CA12046; University of Nebraska Medical Center, Omaha, NE: Apar Ganti, MD, supported by CA77298; University of North Carolina at Chapel Hill, Chapel Hill, NC: Thomas C. Shea, MD, supported by CA47559; University of Tennessee Memphis, Memphis, TN: Harvey B. Niell, MD, supported by CA47555; University of Vermont, Burlington, VT: Steven M. Grunberg, MD, supported by CA77406; Wake Forest University School of Medicine, Winston-Salem, NC: David D. Hurd, MD, supported by CA03927; Walter Reed Army Medical Center, Washington, DC: Brendan M. Weiss, MD, supported by CA26806; Washington University School of Medicine, St. Louis, MO: Nancy Bartlett, MD, supported by CA77440; Weill Medical College of Cornell University, New York, NY: John Leonard, MD, supported by CA07968.
Fig A1.
Study flow diagram. CALGB, Cancer and Leukemia Group B; GH, Genomic Health.
Table A1.
Multivariable Analysis: Relationship of RS, T Stage, and MMR With Risk of Recurrence
| Variable | HR | 95% CI | P |
|---|---|---|---|
| MMR (deficient v intact) | 0.58 | 0.37 to 0.93 | .023 |
| T4 v T3 | 1.00 | 0.48 to 2.11 | .995 |
| RS, continuous (per 25 units) | 1.62 | 1.14 to 2.29 | .007 |
Abbreviations: HR, hazard ratio; MMR, mismatch repair; RS, recurrence score.
Footnotes
Supported in part by Grants No. CA31946 from the National Cancer Institute to the Alliance for Clinical Trials in Oncology for CALGB 9581 (M.M.B.) and No. CA33601 to the Alliance Statistics and Data Center.
Terms in blue are defined in the glossary, found at the end of this article and online at www.jco.org.
Presented in part at the 47th Annual Meeting of the American Society of Clinical Oncology, Chicago, IL June 3-7, 2011.
The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute.
Author's disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) and/or an author's immediate family member(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships markedwith a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors
Employment or Leadership Position: Margarita Lopatin, Genomic Health (C); Mark Lee, Genomic Health (C); Kim Clark-Langone, Genomic Health (C); Carl Millward, Genomic Health (C); Steven Shak, Genomic Health (C) Consultant or Advisory Role: Richard L. Schilsky, Foundation Medicine (C) Stock Ownership: Margarita Lopatin, Genomic Health; Mark Lee, Genomic Health; Kim Clark-Langone, Genomic Health; Carl Millward, Genomic Health; Steven Shak, Genomic Health; Richard L. Schilsky, Foundation Medicine, Universal Oncology Honoraria: None Research Funding: Alan P. Venook, Genomic Health Expert Testimony: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: Alan P. Venook, Donna Niedzwiecki, Margarita Lopatin, Mark Lee, Steven Shak, Monica M. Bertagnolli
Provision of study materials or patients: Paula N. Friedman
Collection and assembly of data: Alan P. Venook, Donna Niedzwiecki, Kim Clark-Langone, Carl Millward, Richard M. Goldberg, Najjia N. Mahmoud, Robert S. Warren, Richard L. Schilsky, Monica M. Bertagnolli
Data analysis and interpretation: Alan P. Venook, Donna Niedzwiecki, Margarita Lopatin, Xing Ye, Mark Lee, Paula N. Friedman, Wendy Frankel, Steven Shak, Monica M. Bertagnolli
Manuscript writing: All authors
Final approval of manuscript: All authors
References
- 1.Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 2.Early Breast Cancer Trialists' Cooperative Group (EBCTCG) Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: An overview of the randomised trials. Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 3.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 4.Paik S, Tang G, Shak S, et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol. 2006;24:3726–3734. doi: 10.1200/JCO.2005.04.7985. [DOI] [PubMed] [Google Scholar]
- 5.Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 6.Vicuna B, Benson AB., 3rd Adjuvant therapy for stage II colon cancer: Prognostic and predictive markers. J Natl Compr Canc Netw. 2007;5:927–936. doi: 10.6004/jnccn.2007.0080. [DOI] [PubMed] [Google Scholar]
- 7.National Comprehensive Cancer Network. NCCN Clinical Practice Guidelines for Oncology: Colon Cancer v1.2013. [DOI] [PubMed] [Google Scholar]
- 8.Compton CC, Fielding LP, Burgart LJ, et al. Prognostic factors in colorectal cancer: College of American Pathologists Consensus Statement 1999. Arch Pathol Lab Med. 2000;124:979–994. doi: 10.5858/2000-124-0979-PFICC. [DOI] [PubMed] [Google Scholar]
- 9.Kelley RK, Venook AP. Prognostic and predictive markers in stage II colon cancer: Is there a role for gene expression profiling? Clin Colorectal Cancer. 2011;10:73–80. doi: 10.1016/j.clcc.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gray RG, Quirke P, Handley K, et al. Validation study of a quantitative multigene reverse transcriptase-polymerase chain reaction assay for assessment of recurrence risk in patients with stage II colon cancer. J Clin Oncol. 2011;29:4611–4619. doi: 10.1200/JCO.2010.32.8732. [DOI] [PubMed] [Google Scholar]
- 11.Harris EI, Lewin DN, Wang HL, et al. Lymphovascular invasion in colorectal cancer: An interobserver variability study. Am J Surg Pathol. 2008;32:1816–1821. doi: 10.1097/PAS.0b013e3181816083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Connell MJ, Lavery I, Yothers G, et al. Relationship between tumor gene expression and recurrence in four independent studies of patients with stage II/III colon cancer treated with surgery alone or surgery plus adjuvant fluorouracil plus leucovorin. J Clin Oncol. 2010;28:3937–3944. doi: 10.1200/JCO.2010.28.9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelley RK, Van Bebber SL, Phillips KA, et al. Personalized medicine and oncology practice guidelines: A case study of contemporary biomarkers in colorectal cancer. J Natl Compr Canc Netw. 2011;9:13–25. doi: 10.6004/jnccn.2011.0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hutchins G, Southward K, Handley K, et al. Value of mismatch repair, KRAS, and BRAF mutations in predicting recurrence and benefits from chemotherapy in colorectal cancer. J Clin Oncol. 2011;29:1261–1270. doi: 10.1200/JCO.2010.30.1366. [DOI] [PubMed] [Google Scholar]
- 15.Ribic CM, Sargent DJ, Moore MJ, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–257. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sargent DJ, Marsoni S, Monges G, et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol. 2010;28:3219–3226. doi: 10.1200/JCO.2009.27.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de la Chapelle A, Hampel H. Clinical relevance of microsatellite instability in colorectal cancer. J Clin Oncol. 2010;28:3380–3387. doi: 10.1200/JCO.2009.27.0652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sinicrope FA, Foster NR, Thibodeau SN, et al. DNA mismatch repair status and colon cancer recurrence and survival in clinical trials of 5-fluorouracil-based adjuvant therapy. J Natl Cancer Inst. 2011;103:863–875. doi: 10.1093/jnci/djr153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark-Langone KM, Sangli C, Krishnakumar J, et al. Translating tumor biology into personalized treatment planning: Analytical performance characteristics of the Oncotype DX Colon Cancer Assay. BMC Cancer. 2010;10:691. doi: 10.1186/1471-2407-10-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ungefroren H, Groth S, Ruhnke M, et al. Transforming growth factor-beta (TGF-beta) type I receptor/ALK5-dependent activation of the GADD45beta gene mediates the induction of biglycan expression by TGF-beta. J Biol Chem. 2005;280:2644–2652. doi: 10.1074/jbc.M411925200. [DOI] [PubMed] [Google Scholar]
- 21.Allegra CJ, Paik S, Colangelo LH, et al. Prognostic value of thymidylate synthase, Ki-67, and p53 in patients with Dukes' B and C colon cancer: A National Cancer Institute-National Surgical Adjuvant Breast and Bowel Project collaborative study. J Clin Oncol. 2003;21:241–250. doi: 10.1200/JCO.2003.05.044. [DOI] [PubMed] [Google Scholar]
- 22.Anjomshoaa A, Lin YH, Black MA, et al. Reduced expression of a gene proliferation signature is associated with enhanced malignancy in colon cancer. Br J Cancer. 2008;99:966–973. doi: 10.1038/sj.bjc.6604560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhowmick NA, Neilson EG, Moses HL. Stromal fibroblasts in cancer initiation and progression. Nature. 2004;432:332–337. doi: 10.1038/nature03096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang HY, Nuyten DS, Sneddon JB, et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc Natl Acad Sci U S A. 2005;102:3738–3743. doi: 10.1073/pnas.0409462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garrity MM, Burgart LJ, Mahoney MR, et al. Prognostic value of proliferation, apoptosis, defective DNA mismatch repair, and p53 overexpression in patients with resected Dukes' B2 or C colon cancer: A North Central Cancer Treatment Group Study. J Clin Oncol. 2004;22:1572–1582. doi: 10.1200/JCO.2004.10.042. [DOI] [PubMed] [Google Scholar]
- 26.Kim H, Watkinson J, Varadan V, et al. Multi-cancer computational analysis reveals invasion-associated variant of desmoplastic reaction involving INHBA, THBS2 and COL11A1. BMC Med Genomics. 2010;3:51. doi: 10.1186/1755-8794-3-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mesker WE, Junggeburt JM, Szuhai K, et al. The carcinoma-stromal ratio of colon carcinoma is an independent factor for survival compared to lymph node status and tumor stage. Cell Oncol. 2007;29:387–398. doi: 10.1155/2007/175276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salminen E, Palmu S, Vahlberg T, et al. Increased proliferation activity measured by immunoreactive Ki67 is associated with survival improvement in rectal/recto sigmoid cancer. World J Gastroenterol. 2005;11:3245–3249. doi: 10.3748/wjg.v11.i21.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niedzwiecki D, Bertagnolli MM, Warren RS. Documenting the natural history of patients with resected stage II adenocarcinoma of the colon after random assignment to adjuvant treatment with edrecolomab or observation: Results of CALGB 9581. J Clin Oncol. 2011;29:3146–3152. doi: 10.1200/JCO.2010.32.5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gray RJ. Weighted analyses for cohort sampling designs. Lifetime Data Anal. 2009;15:24–40. doi: 10.1007/s10985-008-9095-z. [DOI] [PubMed] [Google Scholar]
- 31.Bertagnolli MM, Niedzwiecki D, Compton CC, et al. Microsatellite instability predicts improved response to adjuvant therapy with irinotecan, fluorouracil, and leucovorin in stage III colon cancer: Cancer and Leukemia Group B Protocol 89803. J Clin Oncol. 2009;27:1814–1821. doi: 10.1200/JCO.2008.18.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grambsch PM, Therneau TM. Proportional hazards tests and diagnostics based on weighted residuals. Biometrika. 1994;8:515–526. [Google Scholar]
- 33.Therneau TM, Grambsch PM. New York, NY: Springer; 2001. Modeling Survival Data: Extending the Cox Model. [Google Scholar]
- 34.Verona EV, Elkahloun AG, Yang J, et al. Transforming growth factor-beta signaling in prostate stromal cells supports prostate carcinoma growth by up-regulating stromal genes related to tissue remodeling. Cancer Res. 2007;67:5737–5746. doi: 10.1158/0008-5472.CAN-07-0444. [DOI] [PubMed] [Google Scholar]
- 35.Boman BM, Fields JZ, Cavanaugh KL, et al. How dysregulated colonic crypt dynamics cause stem cell overpopulation and initiate colon cancer. Cancer Res. 2008;68:3304–3313. doi: 10.1158/0008-5472.CAN-07-2061. [DOI] [PubMed] [Google Scholar]
- 36.Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng JD, Dunbrack RL, Jr, Valianou M, et al. Promotion of tumor growth by murine fibroblast activation protein, a serine protease, in an animal model. Cancer Res. 2002;62:4767–4772. [PubMed] [Google Scholar]
- 38.Hofheinz RD, al-Batran SE, Hartmann F, et al. Stromal antigen targeting by a humanised monoclonal antibody: An early phase II trial of sibrotuzumab in patients with metastatic colorectal cancer. Onkologie. 2003;26:44–48. doi: 10.1159/000069863. [DOI] [PubMed] [Google Scholar]
- 39.Loeffler M, Krüger JA, Niethammer AG, et al. Targeting tumor-associated fibroblasts improves cancer chemotherapy by increasing intratumoral drug uptake. J Clin Invest. 2006;116:1955–1962. doi: 10.1172/JCI26532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scott AM, Wiseman G, Welt S, et al. A phase I dose-escalation study of sibrotuzumab in patients with advanced or metastatic fibroblast activation protein-positive cancer. Clin Cancer Res. 2003;9:1639–1647. [PubMed] [Google Scholar]
- 41.Welt S, Divgi CR, Scott AM, et al. Antibody targeting in metastatic colon cancer: A phase I study of monoclonal antibody F19 against a cell-surface protein of reactive tumor stromal fibroblasts. J Clin Oncol. 1994;12:1193–1203. doi: 10.1200/JCO.1994.12.6.1193. [DOI] [PubMed] [Google Scholar]
- 42.Neil JR, Johnson KM, Nemenoff RA, et al. Cox-2 inactivates Smad signaling and enhances EMT stimulated by TGF-beta through a PGE2-dependent mechanisms. Carcinogenesis. 2008;29:2227–2235. doi: 10.1093/carcin/bgn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berger DH, O'Mahony CA, Sheng H, et al. Intestinal transformation results in transforming growth factor-beta-dependent alteration in tumor cell-cell matrix interactions. Surgery. 2003;133:568–579. doi: 10.1067/msy.2003.125. [DOI] [PubMed] [Google Scholar]
- 44.Carothers AM, Davids JS, Damas BC, et al. Persistent cyclooxygenase-2 inhibition downregulates NF-{kappa}B, resulting in chronic intestinal inflammation in the min/+ mouse model of colon tumorigenesis. Cancer Res. 2010;70:4433–4442. doi: 10.1158/0008-5472.CAN-09-4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sagiv E, Sheffer M, Kazanov D, et al. Gene expression following exposure to celecoxib in humans: Pathways of inflammation and carcinogenesis are activated in tumors but not normal tissues. Digestion. 2011;84:169–184. doi: 10.1159/000322688. [DOI] [PubMed] [Google Scholar]
- 46.Simon RM, Paik S, Hayes DF. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J Natl Cancer Inst. 2009;101:1446–1452. doi: 10.1093/jnci/djp335. [DOI] [PMC free article] [PubMed] [Google Scholar]