Abstract
The Drosophila oocyte has been established as a versatile system for investigating fundamental questions such as cytoskeletal function, cell organization, and organelle structure and function. The availability of various GFP-tagged proteins means that many cellular processes can be monitored in living cells over the course of minutes or hours, and using this technique, processes such as RNP transport, epithelial morphogenesis, and tissue remodeling have been described in great detail in Drosophila oocytes1,2.
The ability to perform video imaging combined with a rich repertoire of mutants allows an enormous variety of genes and processes to be examined in incredible detail. One such example is the process of ooplasmic streaming, which initiates at mid-oogenesis3,4. This vigorous movement of cytoplasmic vesicles is microtubule and kinesin-dependent5 and provides a useful system for investigating cytoskeleton function at these stages.
Here I present a protocol for time lapse imaging of living oocytes using virtually any confocal microscopy setup.
Keywords: Developmental Biology, Issue 73, Biochemistry, Genetics, Cellular Biology, Molecular Biology, Proteins, Anatomy, Physiology, Drosophila melanogaster, fruit fly, Cell Biology, Drosophila oocytes, oogenesis, oocytes, ovaries, GFP, Live Imaging, Time Lapse Video, imaging, confocal microscopy, dissection, animal model
Protocol
1. Preparing Flies for Dissection
Anesthetize healthy flies of the desired genotype (for example, expressing a GFP-labeled protein) that are less than one week old. Select 10-15 large females with rounded cream-colored abdomens.
Transfer the selected flies and a few (3-5) males to a vial containing new lightly yeasted food and allow the flies to fatten for two days at 25 °C. Refer to Weil et al.6 for details of Drosophila preparation. Fattening the flies ensures that a variety of stages, especially stages 8-12, will be available for imaging. Poorly fattened or unfattened flies usually contain only very early stages (1-6) and mature (stage 14) oocytes.
2. Preparation of Oocytes for Imaging Using an Upright Compound Microscope
Prepare a standard glass slide by affixing two coverslips to the slide, approximately 1 cm apart, using silicone grease. Only use enough grease to ensure a good seal between the coverslip and slide. Pressing down firmly on each the coverslip will spread the grease thinly and evenly. Add some halocarbon oil 27 (Sigma) to the juncture between the coverslips and slide. They will serve as spacers to prevent injuring isolated oocytes in subsequent steps. No. 1.5 coverslips work well, with an average thickness (0.16-0.18 mm) that matches that of late stage oocytes.
Place 2-3 drops of halocarbon oil 27 onto the surface of a 22 mm2 coverslip. To obtain the healthiest oocytes possible, an individual female is removed from the food vial using mouth pipetting and gently deposited into the halocarbon oil without first anesthetizing.
Using sharp dissecting forceps (such as Dumont #5), pin the fly against the coverslip and pull off the tip of the abdomen. Remove the ovaries by dragging them along the surface of the coverslip, under the oil. This will promote adhesion of the oocytes to the coverslip.
Gently tease apart the ovaries, looking for oocytes that are at least stage 10B of oogenesis. Oocytes at this stage can be identified by looking for egg chambers in which the oocyte occupies at least 50-60% of the total volume of the egg chamber. At this point in oogenesis, the follicle cells overlying the oocyte have become columnar, and surround the oocyte on all sides except for a small region where connections to the nurse cells remain. Use the forceps to remove individual oocytes from the ovariole sheath.
Proceed, using 1-3 females, until 5-8 undamaged oocytes have been isolated on the coverslip. Remove any unuseable tissue.
Carefully invert the coverslip containing the oocytes onto the pre-prepared slide so that the oocytes are positioned between the two spacers. Do not press down on the inverted coverslip as that will damage the oocytes. If necessary, add a small amount of halocarbon oil to the edges of the "sandwich" to ensure the space is filled. (Optional: Let slide rest for a several minutes to allow for settling of coverslip.) The slide is now ready for imaging.
3. Preparation of Oocytes for Imaging Using an Inverted Compound Microscope
Oocytes are dissected essentially as described in section 2, except that small Petri dishes with a glass bottom insert (MatTek, 35 mm) are used instead of coverslips. It is unnecessary to cover the specimen, and after dissection, tissue can be imaged directly in the dishes, but care must be taken to ensure the oocytes remain covered with oil and are not allowed to dry out.
4. Live Imaging
Bring an oocyte into focus using DIC or phase contrast optics and examine the oocyte for any signs of damage, such as leaking of cytoplasm, or bulging of follicle cells. Image only oocytes that appear healthy and undamaged.
Streaming can be monitored directly without the need for GFP labeling, since yolk vesicles in the cytoplasm are autofluorescent7. This signal can be captured in the FITC range of the scope but will likely require higher gain settings than those used for GFP-labeled proteins.
The best images of streaming are obtained from optical sections that are just below the surface of the oocyte that is resting against the glass coverslip/Petri glass. Here the oocyte is slightly flattened, making for a better plane of focus. Images can be captured at 200X - 1,000X. Using the arrangement described here, air objectives should be used, since the coverslip forms a barrier between the oocyte and objective. This is optimal because it greatly minimizes drift and movement of the sample.
Once a region of interest is in focus, turn off the brightfield optics and switch to confocal imaging. Using the fast scan/preview function of the software, adjust focus and gain as necessary.
Settings for capture will vary from microscope to microscope, but general parameters are as follows: Use an excitation wavelength of ~488 nm and an emission wavelength of ~ 515 nm. Set up Z-axis capturing so that the Z-axis remains constant, with a lag of 10 sec between frames. Capture a test sequence of 5-10 frames to ensure the oocyte is stationary and the plane of focus is appropriate.
A typical time lapse sequence will capture between 20 - 50 frames, and can be reviewed immediately after completion to assess video quality. The process can be repeated for other cells in the slide, or additional females can be dissected as needed.
5. Troubleshooting and Common Problems
Drift - When using an upright microscope, cells can sometimes drift due to spreading of oil. This can be minimized by using only enough oil to cover the area under the coverslip. If problems persist, use less silicone grease to affix spacers to the slide.
Note: Some confocal software packages allow for cell tracking, but in general it is better to abandon imaging of drifting oocytes and instead use another specimen.
No signs of streaming - If streaming is not evident, it is likely due to one of two causes; either the focal plane for image capture is not correct (usually too deep into the interior of the oocyte) or the oocyte is damaged. Time and practice can minimize both errors.
Representative Results
Imaging of GFP-labeled proteins will vary depending on how strongly the tagged protein is expressed. A mid-stage oocyte expressing reticulon-like 1 (Rtnl-1), exon tagged with GFP8,9 produces strong signal. Rtnl-1 appears to co-localize with the long microtubules present during ooplasmic streaming10 (Figure 1A). Rtnl-1 is a good marker for observing streaming, and is also an indicator of microtubule organization in late stage oocytes.
Ooplasmic streaming in a wildtype egg chamber is evident as noticeable movement of autofluorescent yolk vesicles when using a capture rate of one frame every 10 sec (Figure 1B). A maximum projection of five frames can be generated to mark the path of moving vesicles.
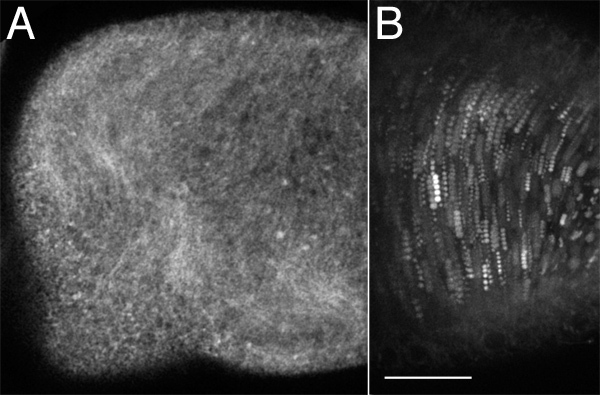 Figure 1. Live imaging of both autofluorescent and GFP-tagged structures.A) A single time-lapse image of a stage 11 oocyte expressing Rtnl1-GFP at 400X. Rtnl1-GFP fibers move with a wave-like motion in streaming oocytes. B) A five frame maximum projection of a stage 10B egg chamber in which the autofluorescent yolk vesicles were imaged every 10 sec for 50 sec total, at 400X magnification. Scale bar = 30 μm.
Figure 1. Live imaging of both autofluorescent and GFP-tagged structures.A) A single time-lapse image of a stage 11 oocyte expressing Rtnl1-GFP at 400X. Rtnl1-GFP fibers move with a wave-like motion in streaming oocytes. B) A five frame maximum projection of a stage 10B egg chamber in which the autofluorescent yolk vesicles were imaged every 10 sec for 50 sec total, at 400X magnification. Scale bar = 30 μm.
Discussion
Drosophila oocytes are a versatile model system for addressing a variety of questions related to the function and organization of cells and subcellular components. A complete understanding of protein function requires the monitoring of both spatial and temporal aspects of protein behavior. Advances in both imaging systems and the genetic tools available to Drosophilists make it possible for even small labs to perform high quality imaging experiments in living oocytes. Here I outline a protocol to analyze the behavior of GFP-tagged or autofluorescent proteins and structures during mid- to late-oogenesis that can be used in conjunction with a variety of genetic backgrounds to probe protein function.
In this protocol I describe the process by which oocytes are prepared for imaging and the basic confocal settings that will enable acquisition of time lapse video of fluorescent proteins and structures. Additional details on oocyte preparation are described by Weil et al.6 A wide variety of fly strains expressing proteins that have been endogenously tagged via exon-trapping are available8, and infinitely more protein variants can be engineered and reintroduced to flies.
In working with living tissue, the health of the specimens is of paramount importance. Egg chambers from stage 10B can complete oogenesis in an essentially autonomous fashion11, making them ideal for short term imaging studies. Care must be taken at several key steps to get high quality data. During dissection it is important to manipulate ovaries and oocytes as little as possible, and to minimize the amount of time between dissection and the completion of imaging. Ideally, a small dissection station should be located in the same room that houses the confocal scope, and only a few egg chambers at a time should be imaged. The number recommended here (5-8) ensures that dissection time is minimized while maximizing the likelihood of obtaining good quality images. I recommend capturing a short series of images to assess image quality and confirm that capture settings are optimized, before capturing a longer time lapse sequence. Most software allows individual frames to be saved, and these images can then be analyzed in a variety of ways using ImageJ12 or other software.
Disclosures
I have nothing to disclose.
Acknowledgments
This work was supported by grant GM096076 from the NIH AREA program.
References
- Becalska AN, Gavis ER. Lighting up mRNA localization in Drosophila oogenesis. Dev. 2009;136:2493–2503. doi: 10.1242/dev.032391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Wang X, Montell DJ. Shining light on Drosophila oogenesis: Live imaging of egg development. Curr. Opin. Gen. Dev. 2011;21:612–619. doi: 10.1016/j.gde.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RC. Ovarian development Drosophila melanogaster. New York: Academic Press; 1970. [Google Scholar]
- Spradling AC. Developmental genetics of oogenesis. In: Bate M, Martinez-Arias A, editors. The Development of Drosophila melanogaster. Vol. 1. Cold Spring Harbor: Cold Spring Harbor Press; 1993. pp. 1–70. [Google Scholar]
- Serbus LR, Cha B-J, Theurkauf WE, Saxton WM. Dynein and the actin cytoskeleton control kinesin-driven cytoplasmic streaming in Drosophila oocytes. Dev. 2005;132:3743–3752. doi: 10.1242/dev.01956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil TT, Parton RM, Davis I. Preparing Individual Drosophila Egg Chambers for Live Imaging. J. Vis. Exp. 2012. p. e3679. [DOI] [PMC free article] [PubMed]
- Theurkauf WE. Premature microtubule-dependent cytoplasmic streaming in cappuccino and spire mutant oocytes. Science. 1998;265:2093–2096. doi: 10.1126/science.8091233. [DOI] [PubMed] [Google Scholar]
- Morin X, Daneman R, Zavortink M, Chia W. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. PNAS. 2001;98:15050–15055. doi: 10.1073/pnas.261408198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röper K. Rtnl1 is enriched in a specialized germline ER that associates with ribonucleoprotein granule components. J. Cell Sci. 2007;120:1081–1092. doi: 10.1242/jcs.03407. [DOI] [PubMed] [Google Scholar]
- Pokrywka NJ, Payne-Tobin A, Raley-Susman KM, Swartzman S. Microtubules, the ER and Exu: New associations revealed by analysis of mini spindles mutations. Mech. Dev. 2009;126:289–300. doi: 10.1016/j.mod.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petri WH, Mindrinos MN, Lombard MF, Margaritis LH. In vitro development of the Drosophila chorion.ih a chemically defined organ culture medium. Wilhelm Roux's Arch., Devl. Biol. 1979;186:351–362. doi: 10.1007/BF00848458. [DOI] [PubMed] [Google Scholar]
- ImageJ [Internet] 2013. Available from: http://rsbweb.nih.gov/ij/index.html.


